Summary
The Cancer Genome Atlas (TCGA) has catalyzed systematic characterization of diverse genomic alterations underlying human cancers. At this historic junction marking the completion of genomic characterization of over 11,000 tumors from 33 cancer types, we present our current understanding of the molecular processes governing oncogenesis. We illustrate our insights into cancer through synthesis of the findings of the TCGA PanCancer Atlas project on three facets of oncogenesis: 1) somatic driver mutations, germline pathogenic variants, and their interactions in the tumor; 2) the influence of the tumor genome and epigenome on transcriptome and proteome; and 3) the relationship between tumor and the microenvironment, including implications for drugs targeting driver events and immunotherapies. These results will anchor future characterization of rare and common tumor types, primary and relapsed tumors, and cancers across ancestry groups and will guide the deployment of clinical genomic sequencing.
Keywords: Oncogenic process, TCGA, Omics, Cancer, Cancer Genomics
ITI
A synthesized view on oncogenic processes based on PanCancer Atlas analyses highlights the complex impact of genome alterations on the signaling and multi-omic profiles of human cancers, as well as their influence on tumor microenvironment.
Introduction
In the nearly half century of the “War on Cancer,” prevention and treatment have progressed significantly, but many forms of the disease remain incurable. The advent of large-scale DNA sequencing ushered in new possibilities. Beginning with coding regions (Sjöblom et al., 2006), sequencing has sparked a revolution in cancer research. Genomic studies have identified numerous cancer driver genes (Kandoth et al., 2013; Lawrence et al., 2014) and germline variants that increase disease susceptibility (Lu et al., 2015). We increasingly understand the molecular determinants of oncogenesis, including tumor suppressor inactivation and pathway alteration. Significant progress has been made in identifying driver mutations (Porta-Pardo et al., 2017), assessing their druggability (Niu et al., 2016), disease subtyping (Waddell et al., 2015), prognosis (Cancer Genome Atlas Research et al., 2015), and residual disease detection (Martinez-Lopez et al., 2014).
Gene and protein expression are also key aspects. Studies have reported new fusions (Klijn et al., 2015), alternatively spliced transcripts (Oltean and Bates, 2014), expression-based stratification (Stricker et al., 2017), and implications of viral infection (Cao et al., 2016). Proteomic studies have made progress on sub-typing (Lawrence et al., 2015), biomarker identification (Sogawa et al., 2016), and drug sensitivity and resistance (Ji et al., 2017). Advancements have also been made in immune response (Bieging et al., 2014), infiltrate-based subtyping (Akbani et al., 2015), associations of PD-1/PD-L1 with prognosis (Danilova et al., 2016), interactions between immune reprogramming and angiogenesis (Tian et al., 2017), and immune cytolytic activity (Rooney et al., 2015). Each area shows enormous promise.
The era of the first large genome sequences was called the “end of the beginning” of genomics. It seems fitting to call the conclusion of The Cancer Genome Atlas (TCGA) the end of the beginning of cancer genomics. TCGA has systematized large-scale genomics-based cancer research, with its projects and data on 11,000 tumors from 33 cancer types having led to enormous advancements. The TCGA PanCancer Atlas project has a special focus on the oncogenic processes governing cancer development and progression, with its ten analysis working groups (AWGs) presenting their findings. Together we synthesized findings from consensus somatic mutation calling, fusion detection, splicing events, aneuploidy, image analysis, and the immune system in oncogenesis (Figure 1). Here, we concentrate on three themes: 1) interactions between somatic drivers and germline pathogenic variants; 2) links across genomic substrates i.e., methylome, transcriptome, and proteome; and 3) tumor microenvironment and implications for targeted and immune therapies. We begin each section with an overview from AWG results and follow with additional analyses addressing questions not explored in individual AWG papers. The results of the PanCancer Atlas project will provide a foundation for subsequent phases of deeper, broader, and more sophisticated work that holds great promise for personalized cancer care.
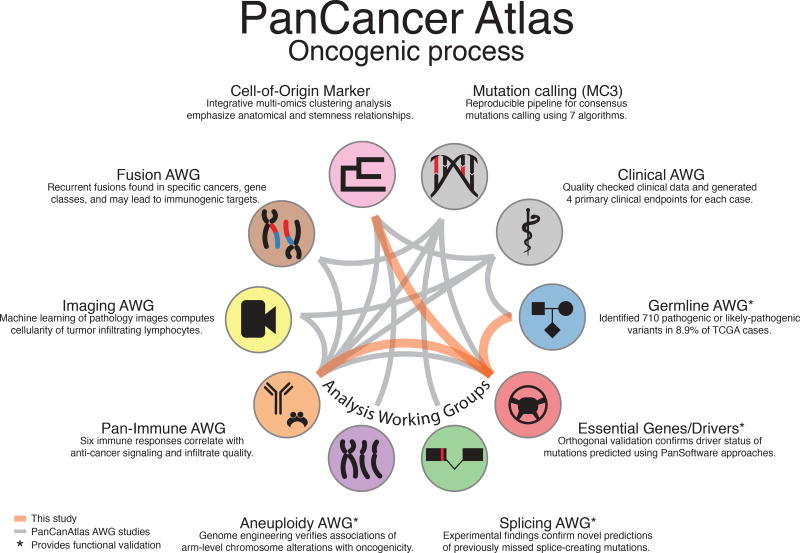
Figure 1. Overview of the PanCancer Atlas oncogenic process group.
PanCan Atlas studies use data from multiple working groups, with relationships shown by gray edges between associated studies. New connections described in this study are shown as orange edges.
Results
Insights into germline and somatic alterations
Previous TCGA studies often concentrated on focal copy number alterations rather than chromosomal-level aneuploidy. The PanCancer Atlas Aneuploidy AWG systematically quantified aneuploidy (Reference Aneuploidy), correlated its degree with genomic features, such as TP53 status, mutational load, and level of lymphocytic infiltrate, and provided experimental evidence confirming some predictions.
Gene fusions, which can drive overexpression or create fusion proteins, are another important class of drivers. The Fusion AWG systematically characterized fusions (Reference Fusions), finding that they are recurrent and disease defining in some neoplasms (e.g. SS18/SSX1 or SSX2 fusion in synovial sarcoma). In others, fusion drivers are present in small subsets of tumors (ALK or ROS1 fusions in lung adenocarcinoma). The accompanying mutational events and how they differ among cancers provide functional insights (Reference Fusions).
Two other AWGs systematically characterized germline and somatic variants across 33 cancer types (Table S1, Reference Germline and MC3). They generated and analyzed 1.5 billion germline (Reference Germline) and ~3.6 million somatic calls (Reference MC3), making TCGA PanCancer Atlas the largest resource for investigating joint variant contributions to cancer. The germline group highlighted the two-hit hypothesis through loss of heterozygosity (LOH) and compound heterozygosity, rare copy number events, and additional evidence supporting variant pathogenicity. The somatic dataset anchored a comprehensive analysis using 26 bioinformatic tools, identifying 299 driver genes and over 3,200 oncogenic mutations (Reference Driver). Similarly, the PanCancer Atlas Germline group identified >800 pathogenic or likely pathogenic germline variants in 99 predisposition genes affecting ~8% of all cases (Reference Germline).
Properties of oncogenic germline and somatic variants
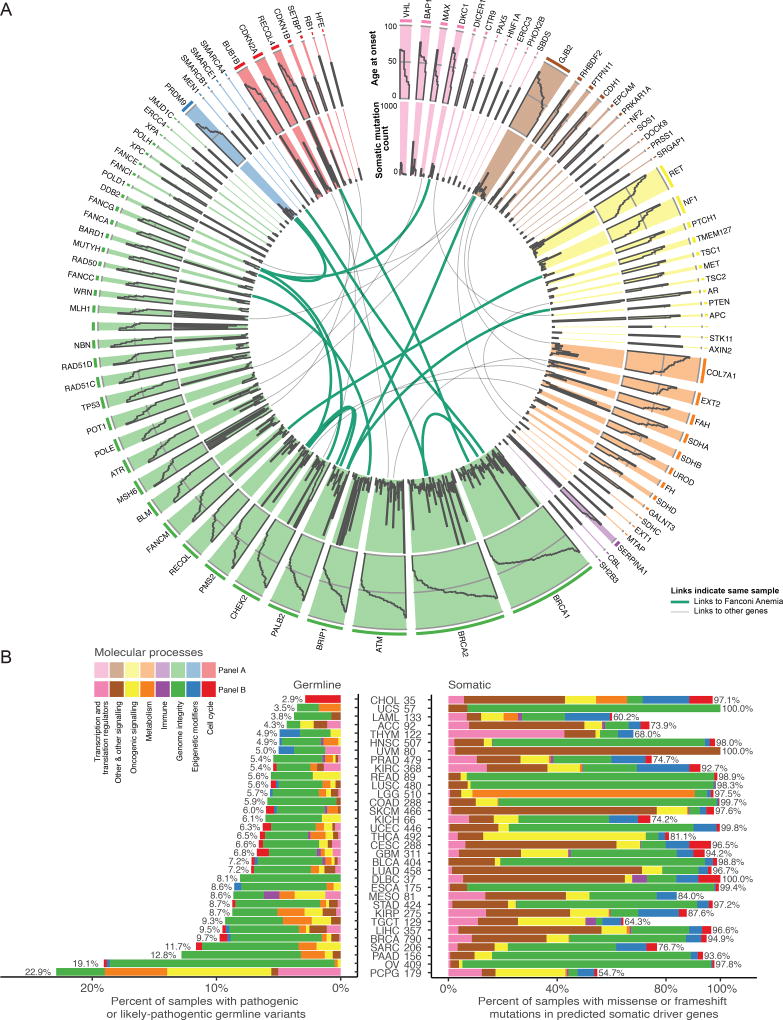
Here, we used the 299 driver and 99 predisposition genes to study interactions of germline and somatic events in 9,389 samples (STAR Methods, Table S1). Many predisposition genes play roles in genome integrity (Green bars, Figure 2A, Table S2). Alterations in these genes represent a higher fraction of germline variants (63%, 490/769) versus somatic drivers (14%, 8850/75825, p-value=7e-151 Fisher’s Exact Test), highlighting the role of genome integrity in cancer predisposition. The remaining somatic alterations are largely from genes involved in cell cycle, epigenetic modifiers, metabolism, oncogenic signaling, and transcriptional/translational regulation. We surveyed the frequency of cases showing disruptions of genome integrity in individual cancer types. Of the 8 molecular process categories examined (STAR Methods), genome integrity dominates both germline and somatic alterations in OV due to BRCA1 or BRCA2 predisposition variants and a high fraction of TP53 mutations. Other cancers are further skewed with respect to percent of cases carrying mutations involved in genome integrity i.e., 4% of samples in LUSC have germline compared to 89% somatic (Figure 2B, Table S3).
Figure 2. Sequence level evaluation of samples with pathogenic germline mutations.
A Circos plot for each predisposition cancer gene. Width of each slice is proportional to germline variant frequency. The outermost tier shows age at onset, while middle indicates total number of somatic mutations for each sample. Links designate one sample that has multiple pathogenic or likely pathogenic germline mutations and are green if one of the genes is from the Fanconi anemia pathway. B shows somatic and germline driver genes grouped into 8 molecular process categories. On the x-axis, germline and somatic proportions are plotted using number of samples as the denominator. Cancers are sorted by increasing germline contribution.
DNA damage response (DDR) pathway
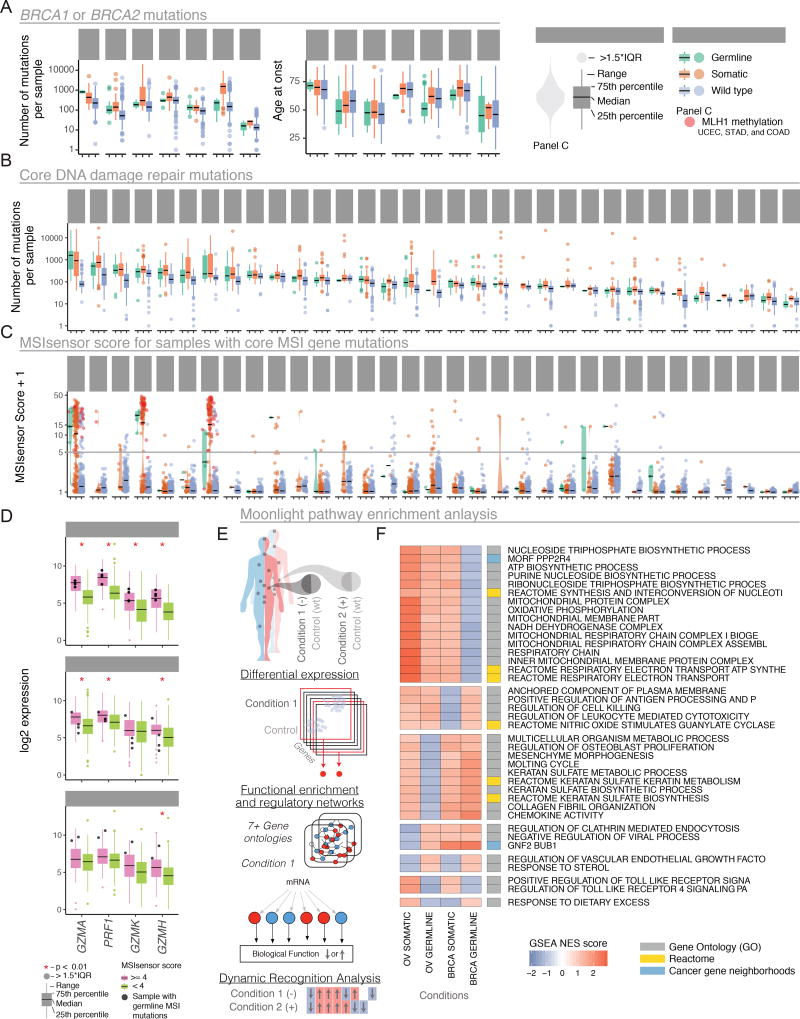
Most predisposition genes affecting genome integrity (23 of 36, 64%) belong to the Core DDR genes (Reference DDR AWG, Table S2). Several show high germline variant counts, including BRCA1, BRCA2, CHEK2, ATM, BRIP1, PALB2, and PMS2. When considering germline and somatic mutations jointly, the most frequently mutated genes are BRCA1 and BRCA2, together having 854 (571 samples) somatic and 153 (152 samples) germline mutations. We grouped samples with germline mutations, somatic, or no/low-impact mutations in these two genes by cancer type to establish associations between age of onset and somatic mutation load. Patients with germline BRCA1/2 mutations develop cancer at younger ages compared to wild type samples in OV, LUSC, and BRCA (FDR 9.12e-6, 9.23e-3 and 1.15e-2 respectively, t-test). Mean age of diagnosis in patients with germline mutations is 54.4+/−13.0 years (standard deviation), compared to 62.3+/−13.4 years when the mutation is somatic across the pan-cancer cohort (P-value = 2.07e-10, 95% CI = (−10.27, −5.57), Figure 3A, Table S4). As expected, germline or somatic variants associate with higher mutation load across cancer types (Figure 3B), being observed in OV samples with germline BRCA1/2 mutations (FDR 3e-3 t-test) and BLCA, STAD somatic (FDR 5.6e-3, 9.2e-6, t-test).
Figure 3. Evaluation of BRCA1/BRCA2 DDR, and MSI genes using somatic and germline variation.
A Samples with BRCA1 or BRCA2 mutations are grouped by cancer type and stratified by somatic, germline, or wild-type status. Box-plots highlight mutations per-sample (left) and age at onset (right). Outlier samples are plotted as points. B Box-plots for samples having mutations in DNA damage response genes grouped by cancer. C Violin plots of MSIsensor scores with samples grouped based on mutation status of MSI genes. Samples with MLH1 promoter methylations status are shown in red. D Gene expression differences for cytokine activators for three cancer types. Black dots are samples with predisposition germline mutation in MSI genes. Red stars highlight significant differences between groups. E Moonlight workflow shows how samples were stratified based on germline vs. wild type (condition 1) and somatic vs. wild type (condition 2) and integrated across pathways with genes that are labeled as differentially expressed. These were then compared using dynamic recognition analysis to identify patterns. F Normalized scores from gene set enrichment analysis for germline and somatic mutations in BRCA1 and/or BRCA2 only, as conditions of OV and BRCA cancer types. Only the first 50 characters of each pathway are shown (additional information in Supplemental Figure 1).
Germline/somatic associated microsatellite instability (MSI) phenotypes
Many samples (250 out of 1464) with non-synonymous somatic mutations in DNA mismatch repair (MMR) genes have high MSI status (MSIsensor score > 4, Figure 3C, Table S5) (Niu et al., 2013). Samples with germline pathogenic variants in MMR genes (18 out of 60) also have high MSI status. Notably, 16 of these 18 samples have both predisposition germline variants and somatic mutations in MMR genes (Table S2), representing a population with potentially higher neoantigen load and response to checkpoint blockade therapy. Indeed, samples with MSIsensor scores >4 had higher expression of immune response marker genes (GZMA, PRF1, GZMK, and GZMH) in the three cancer types with enough MSI high samples: COADREAD, STAD, and UCEC (two-sample Kolmogorov-Smirnov p < 0.01, Figure 3D). This highlights the influence of mutations and MMR genes and the MSI phenotype in the immune response against tumors. Finally, using Moonlight we found several pathways that are differentially expressed depending on whether the mutations affecting BRCA1 and/or BRCA2 are somatic or germline (Figure 3E, 3F). For example, BRCA samples with somatic mutations in BRCA1/2 downregulate genes involved in antigen processing and leukocyte cytotoxicity, whereas BRCA samples with germline BRCA1/2 mutations downregulate genes involved in mitochondrial respiratory chain complex and metabolic pathways. The impact of BRCA1/2 mutations may depend on both their somatic or germline status and the tissue of origin.
Somatic-somatic interactions
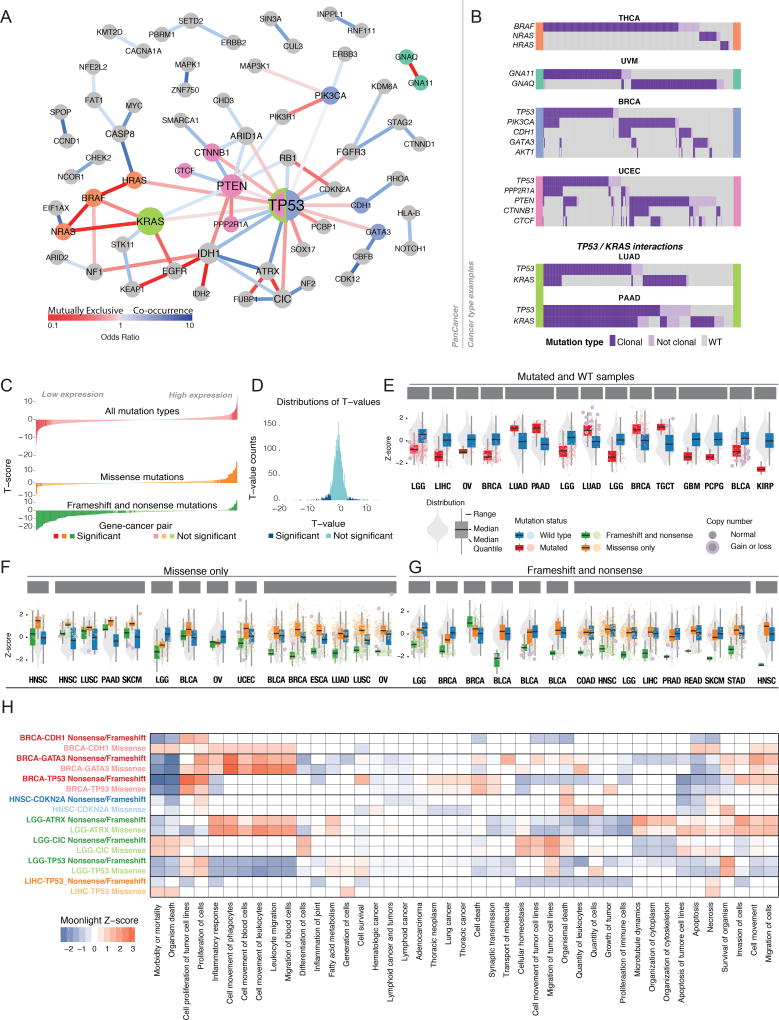
Interactions among somatic driver genes, ranging from sequential dynamics to interactions of pathway and synthetic lethality, hold potential for therapeutic exploitation. We used the MC3 somatic mutation (Reference MC3) dataset and the driver gene list (Reference Driver) to identify pairs of drivers that are mutually exclusive or tend to co-occur (STAR Methods). We found an extensive network of interactions (Cochran Mantel test FDR < 0.1, Figure 4A, Table S6). TP53 is the prime hub, co-occurring with IDH1, ATRX, PPP2R1A, RB1, and CDKN2A and mutually exclusive of PIK3CA, HRAS, CTNNB1, ARID1A, and FGFR3. As expected, driver genes and mutations that act via certain pathways/mechanisms show strong exclusivity, a primary example being BRAF and HRAS/NRAS/KRAS, all of which affect the Ras signaling pathway. Other examples are pairs of homologous genes, such as IDH1/IDH2 and GNAQ/GNA11, and interacting genes, such as PIK3CA and PIK3R1. These patterns held across virtually all 33 tumor types, indicating discovery of a key oncogenic relationship. We also observed exclusivity in specific tissues (Figure 4B), for example BRAF, NRAS, and HRAS in THCA and GNAQ and GNA11 in uveal melanoma.
Figure 4. Interactions between somatic driver events.
A Mutual exclusivity and cooccurrence of driver events. Nodes sized according to degree and edges colored according to odds ratio of pairs of drivers: red for mutually exclusive (OR < 1) and blue for co-occurrence (OR > 1). B Tissue-specific interactions of driver events. Waterfall plots show whether each patient has clonal (dark purple), sub-clonal (light purple), or no driver mutation (gray). Each plot is flanked with a color corresponding to genes in panel A. C Landscape of cis-expression changes shown for three mutation types, with FDR < 0.1 considered significant. D Distribution of T-values for gene expression analyses. ECis-effects of mutations in expression of driver genes. Gray violin plot depicts expression in all samples of driver gene in the tissue marked below each plot. Red boxes show expression of samples with any mutations in that gene blue boxes show expression for samples with no mutation in that gene. Each dot represents a sample and is red if there is a copy number alteration of the gene. F Same information as in E, but separating samples according to frameshift and nonsense (green) versus missense mutations (orange). Selected genes show the top-15 t-values when comparing between the missense and no-mutation groups (FDR < 0.1). G Same as in F, but genes selected by top-15 t-values between nonsense/frameshift and no-mutations groups. H Moonlight scores for groups of mutations in driver genes in specific cancer types (y-axis) and genes annotated with several GO terms (x-axis). Boxes colored red or blue if Moonlight Z-score is positive (overexpression of the biological function) or negative (downregulation), respectively. See also supplemental figure 2.
At a larger scale, some cancer types require cooperation between gene networks. For example, in UCEC, there are two mutually exclusive networks, the first consisting of TP53 and PPP2R1A (and occasionally PTEN) and the second CTNNB1, PTEN, and CTCF. This is consistent with previous descriptions of UCEC subtypes, with TP53-driven endometrial tumors having a copy-number high phenotype and PTEN-driven endometrial tumors being copy-number low or hypermutated (either via MSI and/or POLE). Finally, we observed cancer-specific somatic-somatic interactions. For instance, TP53 and KRAS are mutually exclusive in COAD, READ, and LUAD (Table S6), but significantly co-occur in PAAD (Table S6). These observations highlight the importance of investigating both at the pan-cancer level and by tissue of origin (Park and Lehner, 2015).
Insights into interactions at -omics levels
The tumor genome and transcriptome interact at multiple levels. For example, 1–2% of genome mutations have detectable effects on splicing, with potential to alter the transcriptome and biochemical pathways (Wang and Cooper, 2007). Locally, cis-mutations can disrupt or activate splicing factor binding sites or splice sites. The Splicing AWG analyzed 8,656 TCGA tumors, finding that 1,964 mostly missense and synonymous mutations create novel splice junctions (Table S1) (Reference splicing). They also produce neo-antigens, often accompanied by an elevated immune response. Mutations in splice-governing genes result in large-scale abnormal splicing, providing potential biomarkers and therapeutic targets (Dvinge et al., 2016) and acting as proto-oncogenes or tumor suppressors (Yoshida et al., 2011). The Spliceosome Pathway AWG surveyed 33 tumor types for somatic mutations of over 400 splicing factor genes, identifying 119 genes with likely driver mutations (Reference Spliceosome Pathways AWG). They confirmed aberrant splicing of frequently mutated genes, suggesting that splicing deregulation in cancer is broader than previously reported.
Integrating profiles from individual molecular platforms can provide insights into the molecular state of tumors and identify samples with shared regulation (sample clusters) across multiple assays. A recent analysis (Reference Cell of origin) performed clustering of individual platforms and subsequent clustering of cluster assignments (COCA) (Hoadley et al., 2014) on clusters derived from aneuploidy levels (10 clusters; 10,522 samples), mRNA (25 clusters with at least 40 samples; 10,165 samples), miRNA (15 clusters; 10,170 samples), DNA methylation (25; 10,814), and RPPA (10; 7,858). They also performed integrative molecular subtyping with the iCluster method (Shen et al., 2009) in a joint analysis of aneuploidy, DNA methylation, mRNA, and miRNA levels across 9,759 tumor samples, identifying 28 iClusters. Consistent with previous multiplatform analyses (Hoadley et al., 2014), samples cluster primarily by tissue of origin.
Cis- and trans- effects of driver mutations and mutation types
We analyzed the impact of somatic mutations in the cis-expression of driver genes. We grouped samples for each gene according to whether they contained frameshift or nonsense mutations (group I), missense (group II), or no mutations (group III). This analysis shows clear up-regulation of cancer driver genes affected by missense mutations and down-regulation of those affected by nonsense or frameshift mutations (Figures 4C, 4D, Table S7), consistent with previous findings (Hu et al., 2017, Alvarez et al., 2016). We observed reduced expression for tumor suppressors, such as ATRX, BRCA1, NF1, and RB1 and elevated expression of oncogenes, like EGFR and KIT (FDR < 0.1, Figure 4E). We highlight the top 15 genes showing significant expression differences between at least two of the three groups in at least one cancer type (Figures 4F, 4G, S2). In most cases, the frameshift/nonsense group had significantly lower mRNA than the others, consistent with the hypothesis that they induce nonsense-mediated decay (NMD) (Lindeboom et al., 2016). The exception is GATA3 in breast cancer, where samples with frameshift or nonsense mutations have higher mRNA levels (FDR = 4.54e-18 Welch’s test, Figure 4G), likely because GATA3 frameshift mutations can have gain-of-function, oncogenic effect (Mair et al., 2016). In cases such as CASP8, samples with missense mutations also overexpress the driver gene (FDR < 0.1, Figure 4G).
We used Moonlight to identify gene programs that are differentially expressed in each of the two mutated groups when compared against non-mutated samples (Figure 4H, Methods). Remarkably, several genes seem to affect different transcriptional programs, depending on the type of mutation affecting them. Following on the GATA3 mutations in BRCA, samples with frameshift/nonsense mutations associate with downregulated genes related to microtubule dynamics or organization of cytoskeleton, an effect not seen in those with missense mutations. Similar effects also happen with CDH1 in BRCA: samples with nonsense and frameshift mutations associate with upregulated genes involved in leukocyte migration, but not in samples with missense CDH1 mutations. The tissue of origin seems to also influence the transcriptional effects. For example, LGG samples with any kind of TP53 mutations associate with downregulated expression of leukocyte migration genes, but the expression of these genes remains unaltered in LIHC or BRCA samples with TP53 mutations (Figure 4H). Overall, associations of driver mutations and the transcriptome of the cancer cell seem to be affected by both the original cell type and the type of driver gene mutation.
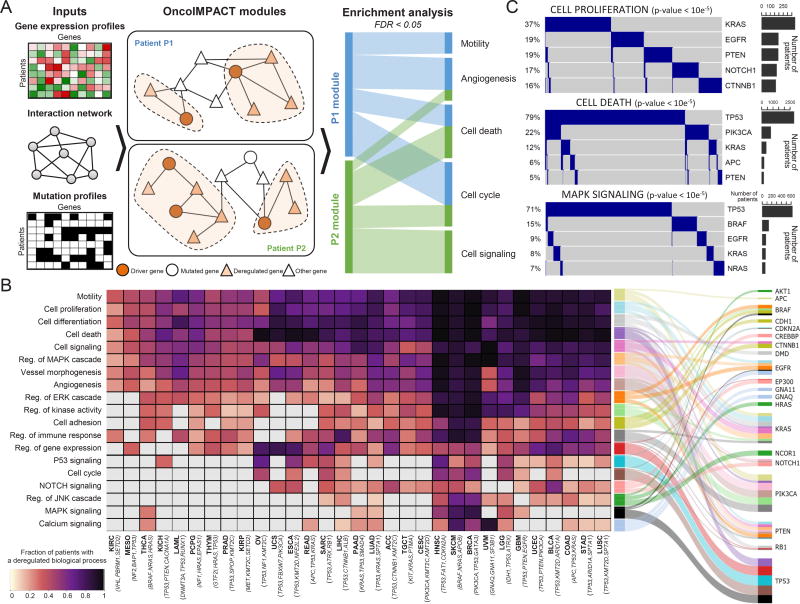
Impacts of genome mutations on transcriptomic activities
Driver mutations often affect the expression of interacting genes and genes in the same pathway. We investigated this phenomenon by integrating protein interaction, transcriptomic, and mutation information using OncoIMPACT (Figure 5A). To reveal key deregulated oncogenic processes occurring in each cancer type, we calculated the fraction of patients for which an oncogenic process was associated with a driver mutation (Figure 5B). With few exceptions (e.g. KIRC), general tumorigenic processes, such as cell proliferation, death, signaling, and motility, are frequently deregulated across cancer types. These processes are mostly deregulated by TP53, PTEN, KRAS, and PIK3CA. Processes were more frequently deregulated in some cancers (e.g. HNSC, SKCM, and BRCA). We also observed associations between oncogenic process and cancer types, e.g. calcium signaling pathway deregulation and Uveal Melanoma (UVM), with frequent activating mutations in GNA11 and GNAQ that are upstream members of the Calcium signaling pathway (Moore et al., 2016) and frequent deregulation of the Notch signaling pathway in bladder urothelial carcinoma (BLCA) due to inactivating driver mutations in this pathway (Rampias et al., 2014).
Figure 5. Relationships between oncogenic processes and driver genes.
A Identifying processes deregulated by driver gene modules using OncoIMPACT. Pathways associated with each module were identified using enrichment analysis (Methods). B Relationships among oncogenic processes, cancer types, and driver genes. (Left) Heatmap shows fraction of samples with deregulated processes associated with sample-specific driver mutations. The three most frequently mutated driver genes are shown with each cancer type. (Right) Graph of associations between processes and top three genes predicted to be responsible for their deregulation. Grey cells represent non-significant fraction of patients (binomial test, p-value Bonferroni corrected > 0.05). Edge widths represent relative fraction of samples with deregulated processes associated to each driver gene. C Oncoprint of mutational profile of the 5 most mutated genes associated with deregulation of 3 biological processes. (Left) Different samples harbor driver genes in a mutually exclusive manner, suggesting many samples have only one process driver gene. (Right) Number of samples having driver gene mutated. P-values are computed using R-exclusivity test (Methods).
We also observed known pairs of significantly mutually exclusive mutated genes such as TP53 and PIK3CA (Kandoth et al., 2013) and KRAS and BRAF (Loes et al., 2016) in cell death and MAPK signaling processes (Figure 5C, permutation test, p-value < 10−5), suggesting that a single driver suffices to perturb these processes and that mutations in multiple drivers are functionally interchangeable in certain contexts. In heterogeneous tumors, this functional redundancy might serve as an important source of drug resistance and metastatic clones.
Interactions between different molecular layers
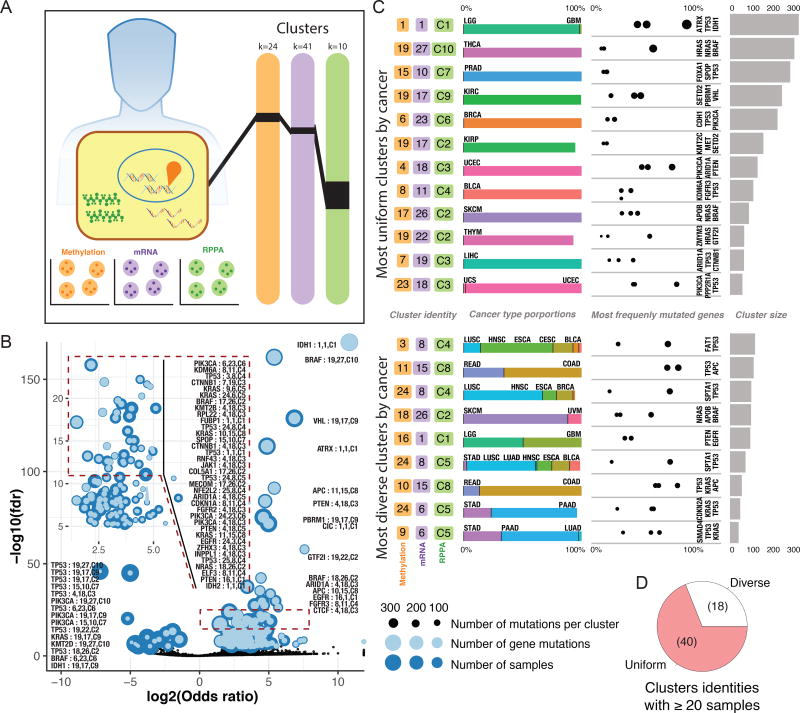
Having established the connections between driver events and the transcriptome, we investigated the relationship between driver genes and the methylomic, transcriptomic, and proteomic profiles of tumors (Figure 6A). We used the clustering data from the Cell of origin AWG (Reference Cell of origin) to search for cluster combinations enriched in driver events (Figure 6B), identifying 40 genes associated with multiplatform clusters; TP53, KRAS, and PIK3CA mutations were enriched in 10 or more multiplatform clusters, and ARID1A, BRAF, CTNNB1, KMT2D, PTEN, and APC mutations were significantly enriched in 4 or more clusters (Tables S8 and S9).
Figure 6. Complexities of multi dimensional molecular evaluation.
A Clustering analysis was performed using 3 substrates: methylation, mRNA, and RPPA. Samples divided into 24 methylation clusters, 41 mRNA, and 10 RPPA clusters. Links show each tumor was given a unique cluster combination identifier. B Gene enrichment analysis for each cluster assignment is displayed as a volcano plot. Dashed square is enlarged in an inset. Overlapping dots show number of samples in the cluster assignment (dark blue) and the number of samples with a given mutation superimposed (light blue), jointly indicating the mutated proportion in that cluster. C The 21 most gene enriched cluster identities, with breakdown by tissue type proportion and most frequently mutated gene from that cluster identity. Sample size for each identity appears in bar plot. D The 58 cluster identities having ≥20 samples. Pie chart illustrates fraction of uniform clusters, where 90% of samples within a cluster are from a single cancer type.
Interestingly, we found similar multiplatform clusters that differ in their associated genes. One notable case is comprised of LGG and GBM samples, which are predominantly covered by mRNA cluster 1 and RPPA cluster C1, but which differ markedly in their methylome profiles. IDH1-driven LGGs are in methylation cluster 1, where 330 of the 351 samples carried IDH1 mutations, while EGFR-driven LGG and GBM are in methylation cluster 16 (Figure 6C). Another example is that APC and KRAS-driven COAD/READ tumors are strongly enriched in mRNA cluster 15 and RPPA cluster C8, but separate in methylation clusters 10 and 11. Similar circumstances are observed for PIK3CA-driven BRCA tumors, which are enriched in mRNA and proteome clusters 23 and C6, respectively, but which can belong to methylation clusters 24 or 6.
Notably, we also found instances where specific driver genes differentiate among cluster combinations. For example, UCEC samples belong mostly to multiplatform clusters 4/18/C3 and 23/18/C3, which again differ only in methylation profile. The first multi-cluster is enriched in ARID1A, PTEN, CTNNB1, and PIK3CA mutations and has fewer TP53 mutations. The second cluster is conversely dominated by TP53 and PPP2R1A mutations, indicating that differences in driver prevalences can be reflected in the methylation profile. While multiplatform clusters are largely driven by tissue of origin (Figure 6D), they may also be affected by the mutations that drive tumor growth.
Insights into interactions in the tumor microenvironment
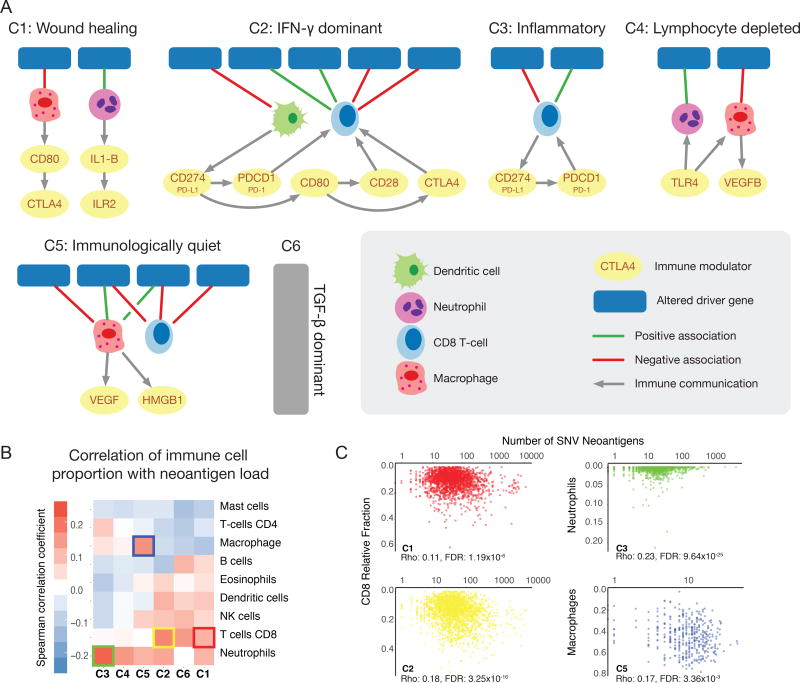
A third frontier involves interactions between cancer cells and the tumor microenvironment (TME), comprising stromal cells and the immune infiltrate. Results from the Immune Response Working Group (IRWG) (PanImmune reference) indicate that the TME can be characterized as belonging to one of six immune subtypes, namely Wound Healing (C1), IFN-γ Dominant (C2), Inflammatory (C3), Lymphocyte Depleted (C4), Immunologically Quiet (C5), and TGF-β Dominant (C6) (Tables S8 and S10).
While immune signatures can infer levels of lymphocytic infiltrates in tumors, they provide no information on spatial distribution of the lymphocytes. The IRWG exploited high resolution imaging of hematoxylin and eosin (H&E) to estimate tumor associated lymphocytes across all samples from 13 of the 33 TCGA tumor types (Reference Imaging). These data revealed relationships between degree of lymphocytic infiltrates measured by gene expression and feature extraction from imaging data using neural network algorithms. Further correlations were made with cancer molecular subtypes, oncogenic events and outcome, highlighting the power of the underutilized image resources of the TCGA.
Impact of driver mutations on the immune communication network
Here, we further study the relationship between specific driver events, composition of the immune infiltrate, and the signaling network among different cell types within distinct immune subtypes. The networks identified for each immune subtype (STAR Methods) might be relevant to identifying synergistic interventions between targeted drugs and immuno-therapies.
BRAF-driven tumors have a higher proportion of CD8 T-cells than NRAS-driven tumors (ANOVA p < 2×10−5 in both cases) (Figure 7A, Table S11) in the C3 immune subtype. Elevated CD8 T-cell proportion, considered an important effector of checkpoint inhibition (Ji et al., 2012), correlates with better outcomes. We also identified a signaling loop involving CD8 T-cells, CD274 (PD-L1), and PCDC1 (PD-1) (Methods) in C3, where targeting BRAF and PD-L1 might have synergistic effects. The analysis also reveals an interesting network within the C5 subtype. Samples having mutations in ATRX or TP53 have higher presence of macrophages and lower of CD8 (ANOVA p < 2×10−8 in both cases). Interestingly, these macrophages secrete HMGB1, which promotes proliferation and metastasis in glioma (Bassi et al., 2008), a prominent cancer type in C5.
Figure 7. Statistical associations and predicted interactions within the tumor microenvironment.
A Networks of driver gene events in distinct cancer immune subtypes C1-C6 shown in each subpanel. Lines between events and immune cells are green if correlation between immune cell in samples with the driver event is positive and red if negative. Lines between cell types, ligands, and receptors denote interaction pairs known to occur in other contexts and for which there are concordant values across multiple tumor samples in the subtype. B Heatmap shows Spearman correlation between number of predicted neoantigens in each sample of each immune subtype and proportion of different types of immune cells. Colored outline boxes are detailed in the next panel. C In subtypes C1 and C2, proportion of CD8 T cells increases with burden of predicted neoantigens (left two plots). Correlation between number of neoantigens and Neutrophils in samples of C3 subtype (top right) and between number of neoantigens and fraction of macrophages in the TME in samples with C5 immune response (bottom right).
Driver mutations in KRAS/NRAS/HRAS and BRAF V600 are among the most frequently predicted neoantigens in cancer (Reference PanImmune) and could thus, as presented peptides, be directly steering immune response. Additionally, driver gene mutations may impact the transcriptional regulation that guides immune response. For example, IDH1-driven gliomas associate with lower levels of STAT1, which can decrease levels of immune infiltrate by ultimately decreasing the secretion of CXCL10, a critical chemokine for T-cell trafficking in brain (Kohanbash et al., 2017). Also models of transcriptional networks (Reference PanImmune) implicate Ras family members and other driver genes in transcriptional control of genes affecting TME composition.
Mutation burden and immune fraction
Another way in which somatic mutations interact with the immune system is through neo-antigens presented on Class I or II major histocompatibility complex (MHC) proteins, which can activate immune cells. This has been studied by various PanCancer Atlas groups, describing splice-creating mutations and fusion events creating immunogenic neoantigens (Reference Splice, Reference Fusion) and neoantigens based on the derived HLA type and their predicted binding affinity (Reference PanImmune).
Using neoantigen predictions and immune infiltrate composition, we investigated associations between numbers of presented neoantigens and relative proportion of immune cells comprising immune subtypes (Table S12). These associations differ by immune subtype (Figure 7B). C2 has the greatest overall immune activity. Here, the CD8 T-cell fraction increases with neoantigen load (FDR < 1e-15, Figure 7C), suggesting that CD8 T-cells may respond to neoantigen burden. CD4 T-cell fraction and neutrophil fraction increase in relation to neoantigen burden in C3, perhaps reflective of the overall balanced immune response and good prognosis of C3 tumors (FDR < 1e-25, Figure 7C). Macrophages have greater infiltration with neoantigen burden in C5, which contains many gliomas and for which TAMs (tumor-associated macrophages) support tumor growth (FDR < 5e-3, Figure 7C).
Discussion
This study summarizes and expands the findings of the TCGA PanCancer Atlas project investigating oncogenic processes. The germline genome has far-ranging, pathway-dependent influences on the somatic landscape, often promoting somatic mutations. Interactions between driver genes and the transcriptome are context-dependent, as is the impact of driver mutations in both cis- and trans-. Some oncogenic processes that tend to be deregulated in few cancer types, such as cell adhesion, are more related to specific genes rather than to prominent drivers. Findings also suggest that networks involving driver mutations, cell types, and cytokines might be used as blueprints for combining two or more immunomodulatory therapies (Tian et al., 2017) in selected tumors.
In summary, this work illuminates the complex milieu of oncogenic processes by integrating an enormous corpus of data obtained over the course of TCGA into organized themes. In effect, biomedical science is now graduating from studying the tumor in isolation to assessing it within its larger environmental context. The findings described here suggest drastic changes in clinical practice and drug development. For example, molecular treatments will increasingly be developed with “multi-omics”. This strategy is being used to create small molecule inhibitors for druggable mutations (Drilon et al., 2017), mutation signatures (Davies et al., 2017), and gene expression (Li et al., 2017), immunotherapeutic agents (Le et al., 2017), and vaccines (Ott et al., 2017). Bioinformatic systems will help efficiently design optimized treatment plans lurking within large combinatorial spaces with respect to dosage, efficacy, side-effects, etc.
As we look to the future, there are many questions. For example, we are only beginning to realize that oncogenic mutations, such as BRAF V600E, frequently occur in healthy people (Martincorena et al., 2015). Could some somatic mutations be tolerated in normal development? If so, how does this impact our understanding of oncogenic mutations? TCGA data come mostly from primary tumors, yet patients usually succumb to metastases: can we find the alterations that drive this process? The next leaps to be taken by the Cancer Moonshot Initiative and Human Tumor Atlas Network (HTAN) will involve pre-cancer, primary, and metastatic tumors associated with treatment sensitivity or resistance and will advance the multidimensional mapping of human cancers over time for informing future cancer research and clinical decision-making.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Li Ding (lding@wustl.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
For this research we used data collected by The Cancer Genome Atlas. Under the direction of the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), TCGA collected both tumor and non-tumor biospecimens from more than 10,000 human samples with informed consent under that authorization of local Institutional Review Boards (https://cancergenome.nih.gov/abouttcga/policies/informedconsent). These steps ensured that patients were exposed to no unnecessary risks and that the resulting research is legal, ethical, and well designed. Mutation and clinical data (including age and sex) used for this manscript are deposited by the GDC (https://gdc.cancer.gov/about-data/publications).
METHOD DETAILS
Germline variant calling
TCGA sequence information was obtained from the database of Genotypes and Phenotypes (dbGaP). Data from paired tumor and germline samples were independently aligned to human reference GRCh37-lite using BWA (Li and Durbin, 2009) v0.5.9 and de-duplicated using Picard 1.29. GenomeVIP (Mashl et al., 2017) was used to orchestrate germline calling using the following tools. Germline single nucleotide variants (SNVs) were identified using Varscan (Koboldt et al., 2012) version 2.3.8 (default parameters, except where –min-var-freq 0.10, --p-value 0.10, --min-coverage 3, --strand-filter 1) operating on an mpileup stream produced by samtools (Li et al., 2009) version 1.2 (default parameters, except where -q 1 -Q 13) and GATK (McKenna et al., 2010) version 3.5 using the haplotype caller in single-sample mode with duplicate or unmapped reads removed and calls with quality threshold of 10 retained. Germline indels were identified using Varscan and GATK, both as configured as above, along with Pindel (Ye et al., 2016) version 0.2.5b8. We specified an insert size of 500 whenever this information was not present in the BAM header. Variants were limited to coding regions of full length transcripts obtained from Ensembl release 70 plus two additional base pairs flanking each exon that cover splice donor/acceptor sites. The union of GATK and VarScan SNVs was processed through our in-house false-positive filter (Kanchi et al., 2014). We included indels called by at least two out of the three callers (GATK, Varscan, Pindel) and high-confidence, Pindel-unique calls (at least 30× coverage and 20% VAF). The combined indels set was again processed through our false-positive filter (default parameters, except where --min-homopolymer 10 --min-var-freq 0.2 --min-var-count=6). The entire process is described in more detail in (Reference germline). For germline and somatic variant comparision we restricted our data to the overlap of samples with at least one mutations in the MC3 MAF after restricting variants as outlied below. This overlap removed one gene from the germline predisposition list (CYLD).
Somatic variant calling
A publicly available MAF file (syn7824274, GDC LINK) was compiled by the TCGA MC3 Working Group and annotated with filter flags to highlight potential artifacts and discrepancies (Reference MC3). A host of possible artifacts were flagged, including strand-bias, contamination, Oxo-guanine artifacts, and low normal read depth. If a mutation escaped flagging and was called by 2 or more variant calling tools, it was labeled a ‘PASS’. We restricted analysis to PASS calls, except for samples from OV and LAML, which were early entrants in TCGA that were whole genome amplified (WGA). Of the 412 OV and 141 LAML samples in our data set, 347 (84%) and 141 (100%), respectively, had artificial variants induced by WGA. In order to maintain sample sizes and uniformity in mutation calling, we did not filter mutations containing only ‘wga’ filter tags from these two cancer types. Seven bioinformatic tools were applied, five for Single Nucleotide Variants (SNV) and three for short Insertion Deletion (INDEL) events, with Varscan 2 providing both types of analysis. This list is comprised of MuTect (Cibulskis et al., 2013), VarScan2 (Koboldt et al., 2012), Indelocator (Chapman et al., 2011), Pindel (Ye et al., 2016), SomaticSniper (Larson et al., 2012), RADIA (Radenbaugh et al., 2014), and MuSE (Fan et al., 2016). The final call set was filtered to identify cohort level artifacts and was subject to extensive variant, subject, and cohort level QC. In total, 22,485,627 putative variants were identified and 2,907,335 high confidence mutations were retained after filtering.
Association testing between biological processes and germline and/or somatic BRCA1/2 mutations
Additionally, Moonlight (Colaprico et al., 2018) analysis was considered to incorporate multiple molecular levels to identify differentially expressed genes in the context of biological pathways (Figure 3 and Figure S1). For this analysis samples with germline predisposition variants in the BRCA1 and/or BRCA2 were considered for OV and BRCA. Similarly if a sample harbored somatic missense, frameshift, nonsense, splice site, or in-frame in BRCA1 or BRCA2, that sample was aggregated into the somatic group. If a sample had both germline and somatic mutations, it was not considered for this comparison. A full table of GSEA results is publically available at https://github.com/ibsquare/MoonlightOP “Moonlight_GSEA_NES_results_Rebut_v3”.
Germline and somatic gene assignment to pathway analysis
Assignment of genes to specific pathways was performed to provide a landscape of frequently mutated biological processes across 33 cancer types. Primarily genes were classified into 24 unique categories comprised of which combined the drivers and essentiality working group classification supplemented by Kegg pathway designations provided by Moonlight. These pathways included: apoptosis, cell cycle, chromatin SWI/SNF complex, chromatin histone modifiers, chromatin other, epigenetics DNA modifiers, genome integrity, histone modification, immune signaling, MAPK signaling, metabolism, NFKB signaling, NOTCH signaling, other, other signaling, PI3K signaling, protein homeostasis/ubiquitination, RNA abundance, RTK signaling, splicing, TGFB signaling, TOR signaling, Transcription factor, and Wnt/B-catenin signaling. This was then further reduced to the 8 molecular processes shown on Figure 2.
In order to calculate the prominent molecular process in each tumor type, a single process was assigned to each sample. This was calculated as follows. If a sample did not carry a predisposing germline variant or missense/frameshift mutation in a driver gene then it was merely added to the denominator of that cancer type. Otherwise, if a sample carried a mutations in a germline and/or somatic driver gene, each driver mutation was compared to the ranked order molecular processes based on the cancer type as a whole. For example, if the top molecular processes, by frequency, for LGG were ranked metabolism, genome integrity, and oncogenic signalling, and a sample only carried mutations in both a metabolic gene and a genome integrity gene, then that sample would be classified for the highest rank of that particular cancer.
Detection of gene programs differentially expressed in samples with indels or nonsense mutations (FSN) and missense mutations (MIS)
Cancer Genome Atlas (TCGA) cohort were available in Genomic Data Commons (GDC) Data Portal and were used in this study in September 2017. We focused on these 16 cancer types because the top 15 cases of cancer-gene combinations for two groups (30 combinations in total) from the frameshift / missense from the significant cis-expression associations RNA-seq raw counts of 7668 cases as legacy archive, and using the reference of hg19 were downloaded, normalized and filtered using the R/Bioconductor package TCGAbiolinks version 2.5.9 (Colaprico et al., 2016) using GDCprepare for tumor types (level 3, and platform "IlluminaHiSeq_RNASeqV2") using data.type as “Gene expression quantification” and file.type as "results". This allowed us to extract the raw signal for expression of a gene for each case following the TCGA pipeline used to create Level 3 expression data from RNA Sequence data that uses MapSplice (Wang et al., 2010) to do the alignment and RSEM to perform the quantificiation (Li et al., 2010). Integrative analysis using mutation, clinical and gene expression were performed following our recent TCGA’s workflow (Silva et al., 2016).
For this study we used TCGAbiolinks version 2.7.6 and MoonlightR Version 1.2.0 in October 2017 with the following parameters: (i) for Differential Phenotype Analysis (DPA) we filtered out differentially expressed genes with fdr.cut = 0.01 and logFC.cut = 1, (ii) for Functional Enrichment Analysis (FEA) we considered significantly enriched BPs by each signature of DEGs with a Fisher Test FDR less than 0.01, (iii) for Gene regulatory network (GRN) the pairwise mutual information was computed using entropy estimates from k-nearest (k=3) neighbor distances filtering out non-significant interactions using a permutation test (nboot=100, nGenesPerm = 1000), (iv) Upstream Regulator Analysis (URA) was performed considering the output of previous steps with nCores = 64. Hierarchical cluster analysis using a complete linkage method to finds similar cluster of BPs was applied to generate the heatmap (Figure 4H) sorted by each cancer type. A full list of Moonlight significance scores are pubically available at https://github.com/ibsquare/MoonlightOP (“Moonlight_FrameShift_Missense_SupplementalData”)
We used Moonlight (Colaprico et al., 2018) to find pathways and biological processes that show differences in the expression levels of their genes based on the presence and type of mutations in driver genes. We had three groups: WT, missense and frameshift/nonsense. Samples with both types of mutations, missense and frameshift/nonsense were excluded from this analysis.
Identification of biological processes associated with cancer driver genes
OncoIMPACT (Bertrand et al., 2015) integrates genomic and transcriptomic profiles using a gene interaction network model to discern patient-specific drivers based on their “phenotypic” effect. We used this tool to predict patient-specific modules of deregulated genes associated with mutational driver genes. Modules are constructed by: 1) identifying phenotype genes defined as significantly deregulated genes associated with a driver mutation (deregulated in ≥5% of patients, permutation test, FDR < 0.1) for a particular cancer type, 2) aggregating patient specific modules by linking driver genes to the phenotypes genes using the protein interaction network. For each cancer type, deregulated genes of a patient were identified by calculating the log2 fold-change between the patient gene expression value and the cancer type median gene expression value. After obtaining the gene modules predicted by OncoIMPACT based on patients’ transcriptomic and mutational profiles (SNV, indels and CNA), we selected, for each patient, the largest module containing at least one driver gene from the PanCancer Atlas oncogenic process working group cancer driver genes list. Genes affected by a focal amplification/deletion were filtered out from the modules, as their change in expression may be associated with the copy number change. Biological processes associated with each module were identified by using enrichment analysis on MSigDB’s GO_BP and KEGG_PATHWAY gene lists (Fisher exact test, FDR < 0.05). Patient-specific predictions were then combined at the cancer type level to obtain the fraction of patients for which an oncogenic process was associated with a driver mutation. To control for Type 1 errors introduced by the FDR threshold (0.05 of the predictions are expected to be false positive), we performed a binomial test for each fraction reported (expected frequency 0.05) and filtered out any fraction with a Bonferroni corrected p-values > 0.05. The total number of samples used in this analysis was 6,224 (samples from DLBC and CHOL were excluded due to their small module sizes).
Additionally, we tested if the five most frequently mutated driver genes were significantly mutually exclusive in each oncogenic process using the R-exclusivity test (Leiserson et al., 2016). For each oncogenic process, we constructed a mutation matrix where rows are driver genes and columns are samples. We then counted the number of samples harboring mutually exclusive driver mutations and performed a permutation test by maintaining frequencies of all five driver genes. The reported p-value is based on the number of permuted matrices (100,000) showing higher numbers of samples harboring mutually exclusive driver mutations. The full table of results from this anlayis can be located at https://github.com/CSB5/OncoIMPACT/blob/development/TCGA_PAN_CAN_ANALYSIS/gene_list_driver.csv.
Integration for cell of origin clusters with mutations
Sample and cluster information was provided in the private communication with the cell-of-origin group for 3 additional molecular levels, methylation, mRNA, and reverse phase protein array (RPPA). These sets had varying samples sizes based on data quality and availability (Table S8). These 3 level identifiers were concatenated to create a new cluster identifcaiton number that was utilized for down stream analysis and investigation. From the data provided we identifed 166 samples with one a single sample in the classifier. Samples is missense, indel, or splice site mutations (considered drivers for this analysis) in any of the 299 genes identifed by the PanCancer Atlas drivers group were merged in by sample and a gene enrichement analysis was performed comparing clusters sizes (by sample) to the number of samples with a driver mutation. FDR ≤ 0.05 was considered significant. We also determined what fraction of the cluster ids originate from a single tissue of origin. To address this, we implented a simple heuristic to estmate cluster homogeneity. We define cluster homogeneity as those clusters with ≥20 samples that have ≥90% of the samples from a single cancer type (Figure 6D). 58/414 cluster have 20 or more samples, of which, 69% are homogeneous (40/58), however there are a number of clusters that capture more universal molecular patterns are shared across cancer types.
The cell-to-cell communication network
A network of documented ligand-receptor, cell-receptor, and cell-ligand pairs was retrieved from the FANTOM5 resource at (http://fantom.gsc.riken.jp/5/suppl/Ramilowski et al 2015/). Because CIBERSORT cell types are more granular than immune cells in FANTOM5, CIBERSORT abundance estimates were aggregated by summing to yield estimates for FANTOM5 immune cell abundances, as defined above. This network was augmented with additional known interactions of immumodulators, and only ligand-receptor edges that contained at least one cell or one immune modulator were retained, yielding a ‘scaffold’ of possible interactions.
From the scaffold of possible interactions, interactions were identified that could be playing a role within the TME in each subtype as follows. Cellular fractions were binned into tertiles (low, medium, high), as were gene expression values for ligands and receptors, yielding ternary values for all ‘nodes’ in the network. The binning was performed over all TCGA samples. In subsequent processing, nodes and edges were treated uniformly in processing, without regard to type (cell, ligand, receptor). From the scaffold, interactions predicted to take place in the TME were identified first by a criterion for the nodes to be included (‘present’ in the network), then by a criterion for inclusion of edges. For nodes, if at least 66% of samples within a subtype map to mid or high value bins, the node is entered into the subtype-network. An edge present in the scaffold network between any two nodes is then evaluated for inclusion. A contingency table is populated for the ternary values of the two nodes, over all samples in the subtype, and a concordance vs discordance ratio (“concordance score”) is calculated for the edge in terms of the values of ((high,high)+(low,low))/((low,high)+(high,low)). Edges were retained with concordance score > 2.9, set based on evaluation of quantile distributions (Table S11). Additional details in (Reference Pan-Immune).
QUANTIFICATION AND STATISTICAL ANALYSIS
Comparison of clinical and mutational impact of somatic and germline BRCA1 and BRCA2 variants
We grouped samples according to whether they had BRCA1 and/or BRCA2 germline, somatic or no mutations. We then compared the number of somatic mutations (Reference MC3) in each group using a Wilcoxon test. We also used the clinical data (https://www.synapse.org/#!Synapse:syn4983466.1) to compare the age at onset of each group using also a Wilcoxon test. Samples with both, germline and somatic BRCA1/2 mutations were included in both categories. These results are reported in Table S4 and distiguishable with the column header AnalysisGrouping (Figure 3A).
Comparison of clinical and mutational impact of somatic and germline DDR pathway alterations
We grouped samples according to whether they had germline, somatic or no mutations in the core DDR pathway (Figure 3B). This pathway consists of 80 genes according to genes from the Pathways DDR AWG (Table S2). The number of mutations was compared using Wilcoxon test. Samples with both, germline and somatic in DDR genes mutations were included in both categories. These results are reported in Table S4 and distiguishable with the column header AnalysisGrouping.
Comparison of clinical and mutational impact of somatic and germline MSI pathway alterations
We grouped the samples as in Figure 3C, but using the MSI pathway definition instead, which consists of 33 genes (Table S2). We used MSIsensor (Niu et al., 2013) to determine the MSI score of each sample and compared the scores in each group using a Wilcoxon test (Table S3). In addition to stratifiying our analysis by mutation status in MSI and germline predisposition genes, promoter methylation status for MLH1 was appended to UCEC, COAD, and STAD and was obtained from MIRMRR (Foltz et al., 2017).
Correlation between MSI scores and expression of immune-related genes
We grouped samples according to whether they had high or low MSI scores (MSIsensor score ≥4 and MSIsensor score < 4 respectively). Then we compared the log2 expression of immune-related genes (GZMA, PRF1, GZMK and GZMH) in both groups using both student’s t-test and a two sample Kolmogorov–Smirnov test (KS-test). We limited our analysis to those cancer types because there were sufficient number of MSIhigh samples: UCEC, STAD and COADREAD. We used the KS-test significance of p-value < 0.01 for (Figure 2D). All groups indicated as significnat also showed significance using the t-test except when comparing GZMH abundence in UCEC (t-test p-value= 0.49; KS-test pvalue = 0.003).
Mutation mutual exclusivity and co-occurrence analysis
We performed a mutually exclusivity/co-occurring mutation analysis of samples between all official pairs (258/299) of consensus driver genes from (Reference Driver), which included splice site mutations, but excluded non-coding and silent mutations. The analysis was run at the gene level. We used a two-sided exact Mantel-Haenszel test (mantelhaen.test R function) to identify significant patterns for each individual cancer type and for the PanCancer set as a whole, with multiple test correction of FDR < 0.1. The covariate stratum for this test used mutation burden and the identity of the cancer type for the PanCancer analysis. Mutation burden was dichotomized at a 500 mutations threshold based on an even split of the minimum hypermutated sample threshold (1,000 mutations per sample). This was intended to control for spurious co-occurrence inferences induced by samples with very high mutation burden. Odds ratios of greater or less than one indicate tendencies toward co-occurrence and mutual exclusivity, respectively. Note that in the tissue-specific analyses, this amounts to the tables being 2×2×2 (Gene1 / Gene2 / Mutation burden) whereas in the Pancan analysis they are 2×2×66 (Gene1 / Gene2 /Tissue + mutation burden). We corrected for multiple hypotheses using the Benjamini-Hochberg FDR method, reporting all gene pairs having a FDR < 0.1.
Association testing between different types of mutations and biological processes
We conducted this analysis on the extended consensus driver list of 299 genes, grouping the associated samples for each cancer type into three categories; (i) samples having only frameshift indels or nonsense mutations (FSN), (ii) those having only missense mutations (MIS), and (iii) those having no mutations (WT). Samples with both types of mutations, missense and frameshift/nonsense, were not included in this analysis. For each combination of cancer type and gene, we compiled subsets of samples for these three categories. Any cancer-gene combination not having at least five samples in each of the three categories was excluded for lack of power.
RNA-Seq gene expression data were obtained for each sample category for the above cancer-gene combinations. All RSEM value sets were transformed into normal distributions with Box-Cox transformations, after which Z-Scores were calculated. For a given cancer type, gene, and respective subsets of samples (distinguished by mutation category), Welch’s t-Test was performed to assess the significance of the difference of expression distributions between the test subset and the subset of wild type samples from the same cancer type and gene. Here, the t-statistic is
where, Xi, Si, and Ni are the respective sample mean, standard deviation, and tally of the ith distribution. Welch’s test is especially appropriate, since we do not always find equal variances or sample numbers between the distributions. The t-scores and degrees of freedom generated by the t-test were used to perform a two-tailed significance test against the t-distributions. The distribution of t-scores and their corresponding significance status is depicted in Figure 4. The results from this analysis are reported in Table S7, and seperated by Mutated (any non-silent mutation) and “Frame_Shift_And_Nonsense” or “Missense_Only” under the column header “AnalysisGrouping”. These two groups (“Mutated” and “Frame_Shift_And_Nonsense”/ “Missense_Only”) were tested independent of each other. Additionally, we have included results by expanding our analysis to all non-silent mutations and show the top results in Figure S2.
Correlation between driver events and immune cell types
We focused our analysis on the set of 299 driver genes and >3200 driver mutations from (Reference Driver). We considered that a sample had a driver event if it carried a frameshift or truncating mutation, or a missense mutation detected by at least 2 different signals of oncogenicity (Reference Driver). In order to reduce the issues related to multiple-testing we analyzed only driver events present in 10 or more samples. We considered both individual driver mutations and entire driver genes that met these criteria.
Then, for each of the six immune subtypes (Reference PanImmune) we checked for a correlation between the presence of the driver event and the quantity of different immune cells in the tumor microenvironment. The quantification of immune cells is described in “Immune Fraction Estimates” below. Then, we used domainXplorer to identify driver events that correlate with the presence of different immune cell types (Porta-Pardo and Godzik, 2016). Briefly, domainXplorer uses a linear correlation model that accounts for different variables that might bias the results, such as the tissue of origin or the number of mutations in the tumor sample. The model is:
where CF is the cell fraction of each sample, T is the tissue of origin for each sample, N the total number of mutations in the sample and D is a binary variable showing whether the sample has a certain driver event or not. To correct for multiple testing, the Benjamini-Hochberg method was applied to p-values of the D factor from the ANOVA test of each driver event (Table S11).
DATA AND SOFTWARE AVAILABILITY
Germline predisposition variant list
The list of germline variants was obtained from (Reference Germline). While the details on how to obtain the final 1,461 germline variants are explained in detail in the manuscript, in brief the group first selected for cancer-relevant pathogenic variants, based on whether they were found in the curated cancer variant database or in the curated cancer predisposition gene list, and their associated ClinVar trait. This resulted in 1,678 variants for manual review using the Integrative Genomics Viewer (IGV). For candidate germline variants having the same genomic change as somatic mutations, we further filtered for the germline variants that may have originated from contaminated adjacent normal samples by eliminating variants called from adjacent normal, the VAF in normal < 30%, and co-localizing with any known somatic mutation.
Driver gene list
The list of driver genes was obtained from (Reference Driver genes). The details about how this list was created are further detailed in that manuscript, but in brief, the Driver AWG combined the predictions of 8 different tools comprising algorithms based on mutation frequency (MuSiC2(Dees et al., 2012) and MutSig2CV(Lawrence et al., 2014)), features (20/20 (Tokheim et al., 2016), CompositeDriver(in preparation) and OncodriveFML(Mularoni et al., 2016)), clustering (OncodriveCLUST(Tamborero et al., 2013)), and externally defined regions (e-Driver(Porta-Pardo and Godzik, 2014) and ActiveDriver(Reimand and Bader, 2013))
The preliminary total of 2,101 potential driver genes was identified by taking the union of genes predicted by the eight driver-gene discovery tools. They refined this list by calculating, for each gene predicted in each cancer type, a consensus score that compensated for outlier results and correlation among tools. The consensus score was defined as a weighted sum of the number of tools that predicted the gene to be a driver in each cancer type (see Gene Discovery Weighting Strategy). They required a minimum of two tools to agree, where both could not be outliers (score≥1.5).
To maximize the coverage of the analysis and ensure the accuracy of the final list, they reviewed previous findings in 31 individual cancer types and PanCancer-12 from TCGA. For cancer types not yet having a TCGA publication, they consulted with the relevant analysis working groups (LIHC, TGCT, UVM, SARC, PAAD, and THYM). They included in the final consensus list all those genes that were previously described as drivers by experts in the cancer-specific analysis of TCGA datasets and that were also identified by at least one of the eight algorithms, even if they did not meet the consensus score threshold (≥1.5). Then, to limit false positives in the expanded list, they applied linear discriminant analysis, removing 45 genes from the consensus we detected as likely false positives.
Finally, given the limitations of a systematic approach, they additionally manually rescued 41 genes based on supportive evidence from the following sources: hypermutator phenotype related genes (since they excluded hypermutated samples in our systematic discovery), established cancer genes from LAML because of low quality variant calling originating from tumor contamination of the normal samples, genes supported by omic network tools: OncoIMPACT (Bertrand et al., 2015) and DriverNet (Bashashati et al., 2012). Addition of genes to the final list was subjected to expert manual curation.
Cell of origin transcript data
The PanCancer Atlas Cell Origin manuscript provided us with cluster data for 3 additional substrates: methylation, mRNA, and RPPA (Table S9). This overview supports notion that cancers should be classified by their molecular characteristics and can effectly identify molecular subgroup patterns. Methylation data used unsupervised clustering of 10,814 tumors using Ward’s method to cluster the distance matrix computed with the Jaccard index. This resulted in 25 number of clusters. Unsupervised consensus clustering using Consensus Cluster Plus (Wilkerson and Hayes, 2010) was performed on RSEM (mRNA normalized expression) for 10,165 smamples and 15,363 genes and resulted in 43 clusters. And finally, reverse phase protein arrays (RPPA) was also clustered using Pearson’s correlation coefficient as the distance metric and Ward’s method as the linkage function, which resulted in 10 clusters.
Expression and copy number data
Gene expression and copy number information for each sample were retrieved from the Genomic Data Commons unless indicated otherwise in specific sections of STAR Methods
Cancer Immune Subtypes
To characterize the commonality and diversity of intratumoral immune states, we scored 160 published immune expression signatures on all available TCGA PanCancerAtlas tumor samples and performed cluster analysis to identify similarity modules of multiple immune signature sets. The 160 immune expression signatures were selected based on extensive literature search, utilizing diverse resources considered to be reliable and comprehensive based on expert opinions of immuno-oncologists. 83 signatures were derived in the context of immune response studies in cancer and the remaining 77 are of general validity for immunity. TCGA RNA-seq values from the PanCancer Atlas normalized gene expression matrix were scored for each of the 160 identified gene expression signatures using single-sample gene set enrichment (ssGSEA) analysis, using the R package GSVA. Clusters of similar signature scores were identified by weighted gene correlation network analysis (WGCNA)(Langfelder and Horvath, 2008). Based on the WGCNA analysis, five immuno-oncology-related immune expression signatures: activation of macrophages/monocytes (Beck et al., 2009), overall lymphocyte infiltration (dominated by T and B cells) (Calabrò et al., 2009), TGF-β response (Teschendorff et al., 2010), IFN-γ response (Wolf et al., 2014), and wound healing (Chang et al., 2004)), robustly reproduced co-clustering of the immune signature sets, and were selected to perform cluster analysis of all cancer types, with the exception of hematologic neoplasias (acute myeloid leukemia, LAML; diffuse large B-cell lymphoma, DLBC; and thymoma, THYM). Clustering of tumor samples scored on these five signatures was performed using model based clustering, using the mclust R package (Scrucca et al., 2016), with the number of clusters, K, determined by maximization of Bayesian Information Criterion (BIC). Maximal BIC was found with a six cluster solution, and the six resulting clusters C1-C6 (with 2416, 2591, 2397, 1157, 385 and 180 cases, respectively) were characterized by a distinct distribution of scores over the five representative signatures, and effectively categorized each TCGA sample as belonging to one of six cancer “immune subtypes”, namely Wound Healing (C1), IFN-γ Dominant (C2), Inflammatory (C3), Lymphocyte Depleted (C4), Immunologically Quiet (C5), or TGF-β Dominant (C6). Additional details in (Reference PanImmune, Table S11, Table S12).
FANTOM5 network
A network of documented ligand-receptor, cell-receptor, and cell-ligand pairs was retrieved from the FANTOM5 resource at (http://fantom.gsc.riken.jp/5/suppl/Ramilowski et al 2015/).
Immune cellular fraction estimates
The relative fraction of 22 immune cell types within the leukocyte compartment were estimated by applying CIBERSORT (Newman et al., 2015) to TCGA RNASeq data (Table S12). CIBERSORT (cell-type identification by estimating relative subsets of RNA transcripts) uses a set of 22 immune cell reference profiles to derive a base (signature) matrix which can be applied to mixed samples to determine relative proportions of immune cells. As several key immune genes used in the signatures are absent from TCGA GAF (Generic Annotation File) Version 3.0, we applied CIBERSORT to a re-quantification of the TCGA data using Kallisto and the Gencode GTF, which includes the missing genes. A version of the entire TCGA RNA-seq data normalized to Gencode with Kallisto was computed on the ISB Cancer Genomics Cloud by Steve Piccolo’s group at BYU (https://osf.io/gqrz9/wiki/home/) (Tatlow and Piccolo, 2016). In this study, the 22 CIBERSORT values were aggregated into 9 overall cell types as follows
Mast.cells=Mast.cells.resting + Mast.cells.activated,
Dendritic.cells=Dendritic.cells.resting + Dendritic.cells.activated,
Macrophage=Macrophages.M0 + Macrophages.M1 + Macrophages.M2,
NK.cells=NK.cells.resting+NK.cells.activated,
B.cells=B.cells.naive + B.cells.memory,
T.cells.CD4=T.cells.CD4.naive+T.cells.CD4.memory.resting+T.cells.CD4.memory.activated
Neutrophils=Neutrophils,
Eosinophils=Eosinophils,
T.cells.CD8=T.cells.CD8
Additional details in (Reference PanImmune), where this particular combination is referred to as “Aggregate 2”.
HLA typing and Predicting mutant peptide-MHC binding (neoantigens [pMHCs]) from SNVs
HLA class I typing of samples (raw RNA-Seq from 8872 samples and aligned reads from 715 samples) was performed on the Seven Bridges Cancer Genomics Cloud using a Common Workflow Language (CWL) description of the OptiType tool (version 1.2) (Szolek et al., 2014). The aligned RNA-Seq samples were first converted to raw sequences using a CWL description of the Picard SamtoFastq tool (version 1.140). The reads from each raw RNA-Seq sample were first aligned to the HLA class I database using a CWL description of the yara aligner (version 0.9.9) (Siragusa et al., 2013) with its error rate parameter set to 3%. Next, the CWL description of OptiType was used to compute the HLA class I types for the sample. Potential neoantigenic peptides were identified using NetMHCpan v3.0 (Nielsen and Andreatta, 2016), based on HLA types. For each sample, all pairs of MHC and minimal mutant peptide were input into NetMHCpan v3.0 using default settings. NetMHCpan will automatically extract all 8–11mer peptides from a minimal peptide sequence and predict binding for each peptide-MHC pair. After computation, the results were parsed to only retain peptides which included the mutated position. Peptides containing amino acid mutations were identified as potential antigens on the basis of a predicted binding to autologous MHC (IC50 < 500 nM) and detectable gene expression meeting an empirically determined threshold of 1.6 transcripts-per-million (TPM). This threshold was selected in order to divide the bimodal distribution in the expression data. Additional details in (Reference PanImmune)
CIBERSORT
CIBERSORT (cell-type identification by estimating relative subsets of RNA transcripts, Newman et. al., 2015) uses a set of 22 immune cell reference profiles to derive a base (signature) matrix which can be applied to mixed samples to determine relative proportions of immune cells. It can be accessed at https://cibersort.stanford.edu
MOONLIGHT
Moonlight (Colaprico et al., 2018) is a new methodology available as R bioconductor package, (https://bioconductor.org/packages/release/bioc/html/MoonlightR.html, DOI: 10.18129/B9.bioc.MoonlightR) that does not only identify driver genes playing a dual role (e.g. tumor suppressor genes (TSGs) in one cancer type and oncogenes (OCGs) in another), but also helps in elucidating the biological processes underlying their specific roles.
For this study we used MoonlightR Version 1.2.0 in July 2017 with the following parameters: (i) for DPA we filtered out differentially expressed genes with fdr.cut = 0.01 and logFC.cut = 1, (ii) for FEA we considered significantly enriched BPs by each signature of DEGs with a Fisher Test FDR less than 0.01, (iii) for GRN the pairwise mutual information was computed using entropy estimates from k-nearest (k=3) neighbor distances filtering out non-significant interactions using a permutation test (nboot=100, nGenesPerm = 1000), (iv) URA was performed considering the output of previous steps with nCores = 64, (v) Firstly we retrieved a list of validated OCGs and TSGs from the Catalogue of somatic mutations in cancer (COSMIC). The list consists of 84 OCGs, 55 TSGs, 17 dual role genes and 439 genes without validated role. Secondly PRA was performed considering the URA output as input for the random forest learning approach together with the list of known OCGs and TSGs (COSMIC) used to construct the training set and using a permutation test with nrand = 1000 for obtaining p-values filtered by FDR = 0.01.
domainXplorer
This pipeline identifies events that show statistically significant correlations with the presence of immune cells in the tumor microenvironment (Porta-Pardo and Godzik, 2016). It accounts for several potentially confounding factors, such as the presence of neo-antigens. It can be accessed at https://github.com/eduardporta/domainXplorer.git
OncoIMPACT
Integrates genomic and transcriptomic profiles using a gene interaction network model to discern patient-specific drivers based on their “phenotypic” effect. It can be accessed at https://github.com/CSB5/OncoIMPACT.git.
ABSOLUTE
We used ABSOLUTE (Carter et al., 2012) calls to infer whether each mutation was clonal or sub-clonal. ABSOLUTE optimizes/solves a mixture model for the observed allelic fraction for each mutation (i.e. the mutated reads could have arisen from 1 copy, 2 copies, 3 copies, etc. or from a subclonal population). We defined ‘clonal’ as all mutations that were predicted only as clonal by ABSOLUTE (n = 910,138 out of a total 1,451,623 mutations, 62%). It can be accessed at http://software.broadinstitute.org/cancer/software/genepattern/modules/docs/ABSOLUTE
Supplementary Material
Highlights.
An overview of PanCancer Atlas analyses on oncogenic molecular processes
Germline genome affects somatic genomic landscape in a pathway-dependent fashion
Genome mutations have impacts on expression, signaling, and multi-omic profiles
Mutation burdens and drivers influence immune cell composition in microenvironment
Significance.
At this historic juncture of the completion of The Cancer Genome Atlas project, the PanCancer Atlas consortium carried out a broad set of analyses on more than 11,000 tumor samples spanning 33 cancer types. Here we present an overview and additional new results of the PanCancer Atlas oncogenic process analyses: somatic driver events vs. germline pathogenic variants, influence of the tumor DNA alterations on the transcriptome and proteome, and multi-faceted interactions with immune cells infiltrating the tumor microenvironment. These analyses of this remarkable data set have important ramifications for both basic cancer research and clinical intervention within the cancer development process.
Acknowledgments
We thank patients who contributed to this study and the NCI Office of Cancer Genomics and acknowledge NIH grants U54 HG003273, U54 HG003067, U54 HG003079, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845,U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025, U24 CA211006 and P30 CA016672.
Appendix
Amy Blum, Samantha J. Caesar-Johnson, John A. Demchok, Ina Felau, Melpomeni Kasapi, Martin L. Ferguson, Carolyn M. Hutter, Heidi J. Sofia, Roy Tarnuzzer, Peggy Wang, Zhining Wang, Liming Yang, Jean C. Zenklusen, Jiashan (Julia) Zhang, Sudha Chudamani, Jia Liu, Laxmi Lolla, Rashi Naresh, Todd Pihl, Qiang Sun, Yunhu Wan, Ye Wu, Juok Cho, Timothy DeFreitas, Scott Frazer, Nils Gehlenborg, Gad Getz, David I. Heiman, Jaegil Kim, Michael S. Lawrence, Pei Lin, Sam Meier, Michael S. Noble, Gordon Saksena, Doug Voet, Hailei Zhang, Brady Bernard, Nyasha Chambwe, Varsha Dhankani, Theo Knijnenburg, Roger Kramer, Kalle Leinonen, Yuexin Liu, Michael Miller, Sheila Reynolds, Ilya Shmulevich, Vesteinn Thorsson, Wei Zhang, Rehan Akbani, Bradley M. Broom, Apurva M. Hegde, Zhenlin Ju, Rupa S. Kanchi, Anil Korkut, Jun Li, Han Liang, Shiyun Ling, Wenbin Liu, Yiling Lu, Gordon B. Mills, Kwok-Shing Ng, Arvind Rao, Michael Ryan, Jing Wang, John N. Weinstein, Jiexin Zhang, Adam Abeshouse, Joshua Armenia, Debyani Chakravarty, Walid K. Chatila, Ino de Bruijn, Jianjiong Gao, Benjamin E. Gross, Zachary J. Heins, Ritika Kundra, Konnor La, Marc Ladanyi, Augustin Luna, Moriah G. Nissan, Angelica Ochoa, Sarah M. Phillips, Ed Reznik, Francisco Sanchez-Vega, Chris Sander, Nikolaus Schultz, Robert Sheridan, S. Onur Sumer, Yichao Sun, Barry S. Taylor, Jioajiao Wang, Hongxin Zhang, Pavana Anur, Myron Peto, Paul Spellman, Christopher Benz, Joshua M. Stuart, Christopher K. Wong, Christina Yau, D. Neil Hayes, Joel S. Parker, Matthew D. Wilkerson, Adrian Ally, Miruna Balasundaram, Reanne Bowlby, Denise Brooks, Rebecca Carlsen, Eric Chuah, Noreen Dhalla, Robert Holt, Steven J.M. Jones, Katayoon Kasaian, Darlene Lee, Yussanne Ma, Marco A. Marra, Michael Mayo, Richard A. Moore, Andrew J. Mungall, Karen Mungall, A. Gordon Robertson, Sara Sadeghi, Jacqueline E. Schein, Payal Sipahimalani, Angela Tam, Nina Thiessen, Kane Tse, Tina Wong, Ashton C. Berger, Rameen Beroukhim, Andrew D. Cherniack, Carrie Cibulskis, Stacey B. Gabriel, Galen F. Gao, Gavin Ha, Matthew Meyerson, Steven E. Schumacher, Juliann Shih, Melanie H. Kucherlapati, Raju S. Kucherlapati, Stephen Baylin, Leslie Cope, Ludmila Danilova, Moiz S. Bootwalla, Phillip H. Lai, Dennis T. Maglinte, David J. Van Den Berg, Daniel J. Weisenberger, J. Todd Auman, Saianand Balu, Tom Bodenheimer, Cheng Fan, Katherine A. Hoadley, Alan P. Hoyle, Stuart R. Jefferys, Corbin D. Jones, Shaowu Meng, Piotr A. Mieczkowski, Lisle E. Mose, Amy H. Perou, Charles M. Perou, Jeffrey Roach, Yan Shi, Janae V. Simons, Tara Skelly, Matthew G. Soloway, Donghui Tan, Umadevi Veluvolu, Huihui Fan, Toshinori Hinoue, Peter W. Laird, Hui Shen, Wanding Zhou, Michelle Bellair, Kyle Chang, Kyle Covington, Chad J. Creighton, Huyen Dinh, HarshaVardhan Doddapaneni, Lawrence A. Donehower, Jennifer Drummond, Richard A. Gibbs, Robert Glenn, Walker Hale, Yi Han, Jianhong Hu, Viktoriya Korchina, Sandra Lee, Lora Lewis, Wei Li, Xiuping Liu, Margaret Morgan, Donna Morton, Donna Muzny, Jireh Santibanez, Margi Sheth, Eve Shinbrot, Linghua Wang, Min Wang, David A. Wheeler, Liu Xi, Fengmei Zhao, Julian Hess, Elizabeth L. Appelbaum, Matthew Bailey, Matthew G. Cordes, Li Ding, Catrina C. Fronick, Lucinda A. Fulton, Robert S. Fulton, Cyriac Kandoth, Elaine R. Mardis, Michael D. McLellan, Christopher A. Miller, Heather K. Schmidt, Richard K. Wilson, Daniel Crain, Erin Curley, Johanna Gardner, Kevin Lau, David Mallery, Scott Morris, Joseph Paulauskis, Robert Penny, Candace Shelton, Troy Shelton, Mark Sherman, Eric Thompson, Peggy Yena, Jay Bowen, Julie M. Gastier-Foster, Mark Gerken, Kristen M. Leraas, Tara M. Lichtenberg, Nilsa C. Ramirez, Lisa Wise, Erik Zmuda, Niall Corcoran, Tony Costello, Christopher Hovens, Andre L. Carvalho, Ana C. de Carvalho, José H. Fregnani, Adhemar Longatto-Filho, Rui M. Reis, Cristovam Scapulatempo-Neto, Henrique C.S. Silveira, Daniel O. Vidal, Andrew Burnette, Jennifer Eschbacher, Beth Hermes, Ardene Noss, Rosy Singh, Matthew L. Anderson, Patricia D. Castro, Michael Ittmann, David Huntsman, Bernard Kohl, Xuan Le, Richard Thorp, Chris Andry, Elizabeth R. Duffy, Vladimir Lyadov, Oxana Paklina, Galiya Setdikova, Alexey Shabunin, Mikhail Tavobilov, Christopher McPherson, Ronald Warnick, Ross Berkowitz, Daniel Cramer, Colleen Feltmate, Neil Horowitz, Adam Kibel, Michael Muto, Chandrajit P. Raut, Andrei Malykh, Jill S. Barnholtz-Sloan, Wendi Barrett, Karen Devine, Jordonna Fulop, Quinn T. Ostrom, Kristen Shimmel, Yingli Wolinsky, Andrew E. Sloan, Agostino De Rose, Felice Giuliante, Marc Goodman, Beth Y. Karlan, Curt H. Hagedorn, John Eckman, Jodi Harr, Jerome Myers, Kelinda Tucker, Leigh Anne Zach, Brenda Deyarmin, Hai Hu, Leonid Kvecher, Caroline Larson, Richard J. Mural, Stella Somiari, Ales Vicha, Tomas Zelinka, Joseph Bennett, Mary Iacocca, Brenda Rabeno, Patricia Swanson, Mathieu Latour, Louis Lacombe, Bernard Têtu, Alain Bergeron, Mary McGraw, Susan M. Staugaitis, John Chabot, Hanina Hibshoosh, Antonia Sepulveda, Tao Su, Timothy Wang, Olga Potapova, Olga Voronina, Laurence Desjardins, Odette Mariani, Sergio Roman-Roman, Xavier Sastre, Marc-Henri Stern, Feixiong Cheng, Sabina Signoretti, Andrew Berchuck, Darell Bigner, Eric Lipp, Jeffrey Marks, Shannon McCall, Roger McLendon, Angeles Secord, Alexis Sharp, Madhusmita Behera, Daniel J. Brat, Amy Chen, Keith Delman, Seth Force, Fadlo Khuri, Kelly Magliocca, Shishir Maithel, Jeffrey J. Olson, Taofeek Owonikoko, Alan Pickens, Suresh Ramalingam, Dong M. Shin, Gabriel Sica, Erwin G. Van Meir, Hongzheng Zhang, Wil Eijckenboom, Ad Gillis, Esther Korpershoek, Leendert Looijenga, Wolter Oosterhuis, Hans Stoop, Kim E. van Kessel, Ellen C. Zwarthoff, Chiara Calatozzolo, Lucia Cuppini, Stefania Cuzzubbo, Francesco DiMeco, Gaetano Finocchiaro, Luca Mattei, Alessandro Perin, Bianca Pollo, Chu Chen, John Houck, Pawadee Lohavanichbutr, Arndt Hartmann, Christine Stoehr, Robert Stoehr, Helge Taubert, Sven Wach, Bernd Wullich, Witold Kycler, Dawid Murawa, Maciej Wiznerowicz, Ki Chung, W. Jeffrey Edenfield, Julie Martin, Eric Baudin, Glenn Bubley, Raphael Bueno, Assunta De Rienzo, William G. Richards, Steven Kalkanis, Tom Mikkelsen, Houtan Noushmehr, Lisa Scarpace, Nicolas Girard, Marta Aymerich, Elias Campo, Eva Giné, Armando López Guillermo, Nguyen Van Bang, Phan Thi Hanh, Bui Duc Phu, Yufang Tang, Howard Colman, Kimberley Evason, Peter R. Dottino, John A. Martignetti, Hani Gabra, Hartmut Juhl, Teniola Akeredolu, Serghei Stepa, Dave Hoon, Keunsoo Ahn, Koo Jeong Kang, Felix Beuschlein, Anne Breggia, Michael Birrer, Debra Bell, Mitesh Borad, Alan H. Bryce, Erik Castle, Vishal Chandan, John Cheville, John A. Copland, Michael Farnell, Thomas Flotte, Nasra Giama, Thai Ho, Michael Kendrick, Jean-Pierre Kocher, Karla Kopp, Catherine Moser, David Nagorney, Daniel O’Brien, Brian Patrick O’Neill, Tushar Patel, Gloria Petersen, Florencia Que, Michael Rivera, Lewis Roberts, Robert Smallridge, Thomas Smyrk, Melissa Stanton, R. Houston Thompson, Michael Torbenson, Ju Dong Yang, Lizhi Zhang, Fadi Brimo, Jaffer A. Ajani, Ana Maria Angulo Gonzalez, Carmen Behrens, Jolanta Bondaruk, Russell Broaddus, Bogdan Czerniak, Bita Esmaeli, Junya Fujimoto, Jeffrey Gershenwald, Charles Guo, Alexander J. Lazar, Christopher Logothetis, Funda Meric-Bernstam, Cesar Moran, Lois Ramondetta, David Rice, Anil Sood, Pheroze Tamboli, Timothy Thompson, Patricia Troncoso, Anne Tsao, Ignacio Wistuba, Candace Carter, Lauren Haydu, Peter Hersey, Valerie Jakrot, Hojabr Kakavand, Richard Kefford, Kenneth Lee, Georgina Long, Graham Mann, Michael Quinn, Robyn Saw, Richard Scolyer, Kerwin Shannon, Andrew Spillane, Jonathan Stretch, Maria Synott, John Thompson, James Wilmott, Hikmat Al-Ahmadie, Timothy A. Chan, Ronald Ghossein, Anuradha Gopalan, Douglas A. Levine, Victor Reuter, Samuel Singer, Bhuvanesh Singh, Nguyen Viet Tien, Thomas Broudy, Cyrus Mirsaidi, Praveen Nair, Paul Drwiega, Judy Miller, Jennifer Smith, Howard Zaren, Joong-Won Park, Nguyen Phi Hung, Electron Kebebew, W. Marston Linehan, Adam R. Metwalli, Karel Pacak, Peter A. Pinto, Mark Schiffman, Laura S. Schmidt, Cathy D. Vocke, Nicolas Wentzensen, Robert Worrell, Hannah Yang, Marc Moncrieff, Chandra Goparaju, Jonathan Melamed, Harvey Pass, Natalia Botnariuc, Irina Caraman, Mircea Cernat, Inga Chemencedji, Adrian Clipca, Serghei Doruc, Ghenadie Gorincioi, Sergiu Mura, Maria Pirtac, Irina Stancul, Diana Tcaciuc, Monique Albert, Iakovina Alexopoulou, Angel Arnaout, John Bartlett, Jay Engel, Sebastien Gilbert, Jeremy Parfitt, Harman Sekhon, George Thomas, Doris M. Rassl, Robert C. Rintoul, Carlo Bifulco, Raina Tamakawa, Walter Urba, Nicholas Hayward, Henri Timmers, Anna Antenucci, Francesco Facciolo, Gianluca Grazi, Mirella Marino, Roberta Merola, Ronald de Krijger, Anne-Paule Gimenez-Roqueplo, Alain Piché, Simone Chevalier, Ginette McKercher, Kivanc Birsoy, Gene Barnett, Cathy Brewer, Carol Farver, Theresa Naska, Nathan A. Pennell, Daniel Raymond, Cathy Schilero, Kathy Smolenski, Felicia Williams, Carl Morrison, Jeffrey A. Borgia, Michael J. Liptay, Mark Pool, Christopher W. Seder, Kerstin Junker, Larsson Omberg, Mikhail Dinkin, George Manikhas, Domenico Alvaro, Maria Consiglia Bragazzi, Vincenzo Cardinale, Guido Carpino, Eugenio Gaudio, David Chesla, Sandra Cottingham, Michael Dubina, Fedor Moiseenko, Renumathy Dhanasekaran, Karl-Friedrich Becker, Klaus-Peter Janssen, Julia Slotta-Huspenina, Mohamed H. Abdel-Rahman, Dina Aziz, Sue Bell, Colleen M. Cebulla, Amy Davis, Rebecca Duell, J. Bradley Elder, Joe Hilty, Bahavna Kumar, James Lang, Norman L. Lehman, Randy Mandt, Phuong Nguyen, Robert Pilarski, Karan Rai, Lynn Schoenfield, Kelly Senecal, Paul Wakely, Paul Hansen, Ronald Lechan, James Powers, Arthur Tischler, William E. Grizzle, Katherine C. Sexton, Alison Kastl, Joel Henderson, Sima Porten, Jens Waldmann, Martin Fassnacht, Sylvia L. Asa, Dirk Schadendorf, Marta Couce, Markus Graefen, Hartwig Huland, Guido Sauter, Thorsten Schlomm, Ronald Simon, Pierre Tennstedt, Oluwole Olabode, Mark Nelson, Oliver Bathe, Peter R. Carroll, June M. Chan, Philip Disaia, Pat Glenn, Robin K Kelley, Charles N. Landen, Joanna Phillips, Michael Prados, Jeff Simko, Jeffry Simko, Karen Smith-McCune, Scott VandenBerg, Kevin Roggin, Ashley Fehrenbach, Ady Kendler, Suzanne Sifri, Ruth Steele, Antonio Jimeno, Francis Carey, Ian Forgie, Massimo Mannelli, Michael Carney, Brenda Hernandez, Benito Campos, Christel Herold-Mende, Christin Jungk, Andreas Unterberg, Andreas von Deimling, Aaron Bossler, Joseph Galbraith, Laura Jacobus, Michael Knudson, Tina Knutson, Deqin Ma, Mohammed Milhem, Rita Sigmund, Andrew K Godwin, Rashna Madan, Howard G. Rosenthal, Clement Adebamowo, Sally N. Adebamowo, Alex Boussioutas, David Beer, Thomas Giordano, Anne-Marie Mes-Masson, Fred Saad, Therese Bocklage, Lisa Landrum, Robert Mannel, Kathleen Moore, Katherine Moxley, Russel Postier, Joan Walker, Rosemary Zuna, Michael Feldman, Federico Valdivieso, Rajiv Dhir, James Luketich, Edna M. Mora Pinero, Mario Quintero-Aguilo, Carlos Gilberto Carlotti, Jr., Jose Sebastião Dos Santos, Rafael Kemp, Ajith Sankarankuty, Daniela Tirapelli, James Catto, Kathy Agnew, Elizabeth Swisher, Jenette Creaney, Bruce Robinson, Carl Simon Shelley, Eryn M. Godwin, Sara Kendall, Cassaundra Shipman, Carol Bradford, Thomas Carey, Andrea Haddad, Jeffey Moyer, Lisa Peterson, Mark Prince, Laura Rozek, Gregory Wolf, Rayleen Bowman, Kwun M. Fong, Ian Yang, Robert Korst, W. Kimryn Rathmell, J. Leigh Fantacone-Campbell, Jeffrey A. Hooke, Albert J. Kovatich, Craig D. Shriver, John DiPersio, Bettina Drake, Ramaswamy Govindan, Sharon Heath, Timothy Ley, Brian Van Tine, Peter Westervelt, Mark A. Rubin, Jung Il Lee, Natália D. Aredes, Armaz Mariamidze, Anant Agrawal, Jaeil Ahn, Jordan Aissiou, Dimitris Anastassiou, Jesper B. Andersen, Jurandyr M. Andrade, Marco Antoniotti, Jon C. Aster, Donald Ayer, Matthew H. Bailey, Rohan Bareja, Adam J. Bass, Azfar Basunia, Oliver F. Bathe, Rebecca Batiste, Oliver Bear Don't Walk, Davide Bedognetti, Gloria Bertoli, Denis Bertrand, Bhavneet Bhinder, Gianluca Bontempi, Dante Bortone, Donald P. Bottaro, Paul Boutros, Kevin Brennan, Chaya Brodie, Scott Brown, Susan Bullman, Silvia Buonamici, Tomasz Burzykowski, Lauren Averett Byers, Fernando Camargo, Joshua D. Campbell, Francisco J. Candido dos Reis, Shaolong Cao, Maria Cardenas, Helio HA. Carrara, Isabella Castiglioni, Anavaleria Castro, Claudia Cava, Michele Ceccarelli, Shengjie Chai, Kridsadakorn Chaichoompu, Matthew T. Chang, Han Chen, Haoran Chen, Hu Chen, Jian Chen, Jianhong Chen, Ken Chen, Ting-Wen Chen, Zhong Chen, Zhongyuan Chen, Hui Cheng, Hua-Sheng Chiu, Cai Chunhui, Giovanni Ciriello, Cristian Coarfa, Antonio Colaprico, Lee Cooper, Daniel Cui Zhou, Aedin C. Culhane, Christina Curtis, Patrycja Czerwinska, Aditya Deshpande, Lixia Diao, Michael Dill, Di Du, Charles G. Eberhart, James A. Eddy, Robert N. Eisenman, Mohammed Elanbari, Olivier Elemento, Kyle Ellrott, Manel Esteller, Farshad Farshidfar, Bin Feng, Camila Ferreira de Souza, Esla R. Flores, Steven Foltz, Mitchell T. Frederick, Qingsong Gao, Carl M. Gay, Zhongqi Ge, Andrew J. Gentles, Olivier Gevaert, David L. Gibbs, Adam Godzik, Abel Gonzalez-Perez, Marc T. Goodman, Dmitry A. Gordenin, Carla Grandori, Alex Graudenzi, Casey Greene, Justin Guinney, Margaret L. Gulley, Preethi H Gunaratne, A. Ari Hakimi, Peter Hammerman, Leng Han, Holger Heyn, Le Hou, Donglei Hu, Kuan-lin Huang, Joerg Huelsken, Scott Huntsman, Peter Hurlin, Matthias Hüser, Antonio Iavarone, Marcin Imielinski, Mirazul Islam, Jacek Jassem, Peilin Jia, Cigall Kadoch, Andre Kahles, Benny Kaipparettu, Bozena Kaminska, Havish Kantheti, Rachel Karchin, Mostafa Karimi, Ekta Khurana, Pora Kim, Leszek J. Klimczak, Jia Yu Koh, Alexander Krasnitz, Nicole Kuderer, Tahsin Kurc, David J. Kwiatkowski, Teresa Laguna, Martin Lang, Anna Lasorella, Thuc D. Le, Adrian V. Lee, Ju-Seog Lee, Steve Lefever, Kjong Lehmann, Jake Leighton, Chunyan Li, Lei Li, Shulin Li, David Liu, Eric Minwei Liu, Jianfang Liu, Rongjie Liu, Yang Liu, William J.R. Longabaugh, Nuria Lopez-Bigas, Li Ma, Wencai Ma, Karen MacKenzie, Andrzej Mackiewicz, Dejan Maglic, Raunaq Malhotra, Tathiane M. Malta, Calena Marchand, R. Jay Mashl, Sylwia Mazurek, Pieter Mestdagh, Chase Miller, Marco Mina, Lopa Mishra, Younes Mokrab, Raymond Monnat, Jr., Nate Moore, Nathanael Moore, Loris Mularoni, Niranjan Nagarajan, Aaron M. Newman, Vu Nguyen, Michael L. Nickerson, Akinyemi I. Ojesina, Catharina Olsen, Sandra Orsulic, Tai-Hsien Ou Yang, James Palacino, Yinghong Pan, Elena Papaleo, Sagar Patil, Chandra Sekhar Pedamallu, Shouyong Peng, Xinxin Peng, Arjun Pennathur, Curtis R. Pickering, Christopher L. Plaisier, Laila Poisson, Eduard Porta-Pardo, Marcos Prunello, John L. Pulice, Charles Rabkin, Janet S. Rader, Kimal Rajapakshe, Aruna Ramachandran, Shuyun Rao, Xiayu Rao, Benjamin J. Raphael, Gunnar Rätsch, Brendan Reardon, Christopher J. Ricketts, Jason Roszik, Carlota Rubio-Perez, Ryan Russell, Anil Rustgi, Russell Ryan, Mohamad Saad, Thais Sabedot, Joel Saltz, Dimitris Samaras, Franz X. Schaub, Barbara G. Schneider, Adam Scott, Michael Seiler, Sara Selitsky, Sohini Sengupta, Jose A. Seoane, Jonathan S. Serody, Reid Shaw, Yang Shen, Tiago Silva, Pankaj Singh, I.K Ashok Sivakumar, Christof Smith, Artem Sokolov, Junyan Song, Pavel Sumazin, Yutong Sun, Chayaporn Suphavilai, Najeeb Syed, David Tamborero, Alison M. Taylor, Teng Teng, Daniel G. Tiezzi, Collin Tokheim, Nora Toussaint, Mihir Trivedi, Kenneth T. Tsai, Aaron D. Tward, Eliezer Van Allen, John S. Van Arnam, Kristel Van Steen, Carter Van Waes, Christopher P. Vellano, Benjamin Vincent, Nam S. Vo, Vonn Walter, Chen Wang, Fang Wang, Jiayin Wang, Sophia Wang, Wenyi Wang, Yue Wang, Yumeng Wang, Zehua Wang, Zeya Wang, Zixing Wang, Gregory Way, Amila Weerasinghe, Michael Wells, Michael C. Wendl, Cecilia Williams, Joseph Willis, Denise Wolf, Karen Wong, Yonghong Xiao, Lu Xinghua, Bo Yang, Da Yang, Liuqing Yang, Kai Ye, Hiroyuki Yoshida, Lihua Yu, Sobia Zaidi, Huiwen Zhang, Min Zhang, Xiaoyang Zhang, Tianhao Zhao, Wei Zhao, Zhongming Zhao, Tian Zheng, Jane Zhou, Zhicheng Zhou, Hongtu Zhu, Ping Zhu, Michael T. Zimmermann, Elad Ziv, and Patrick A. Zweidler-McKay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
LD, GG, and DW conceived the project. LD supervised the project. MCW, AJL, EPP, MHB, SS, KH, VT, AC, DB, RJ, FC, LY, and LD drafted the manuscript. JS, GBM, CH, JCZ, DW, GG, and LD provided scientific input. MHB, MAW, and EPP produced figures. Analysis was performed by MHB, KH, AC, CO, I,C-C, JK, EPP, CT, AW, DB, CS, NN, RJ, FC, LY, KAH, RA, VT, DLG, IS, BV, and AJL. All authors approved submission.
TCGA PanCanAtlas Declaration of Interest:
Michael Seiler, Peter G. Smith, Ping Zhu, Silvia Buonamici, and Lihua Yu are employees of H3 Biomedicine, Inc. Parts of this work are the subject of a patent application: WO2017040526 titled “Splice variants associated with neomorphic sf3b1 mutants.” Shouyoung Peng, Anant A. Agrawal, James Palacino, and Teng Teng are employees of H3 Biomedicine, Inc. Andrew D. Cherniack, Ashton C. Berger, and Galen F. Gao receive research support from Bayer Pharmaceuticals. Gordon B. Mills serves on the External Scientific Review Board of Astrazeneca. Anil Sood is on the Scientific Advisory Board for Kiyatec and is a shareholder in BioPath. Jonathan S. Serody receives funding from Merck, Inc. Kyle R. Covington is an employee of Castle Biosciences, Inc. Preethi H. Gunaratne is founder, CSO, and shareholder of NextmiRNA Therapeutics. Christina Yau is a part-time employee/consultant at NantOmics. Franz X. Schaub is an employee and shareholder of SEngine Precision Medicine, Inc. Carla Grandori is an employee, founder, and shareholder of SEngine Precision Medicine, Inc. Robert N. Eisenman is a member of the Scientific Advisory Boards and shareholder of Shenogen Pharma and Kronos Bio. Daniel J. Weisenberger is a consultant for Zymo Research Corporation. Joshua M. Stuart is the founder of Five3 Genomics and shareholder of NantOmics. Marc T. Goodman receives research support from Merck, Inc. Andrew J. Gentles is a consultant for Cibermed. Charles M. Perou is an equity stock holder, consultant, and Board of Directors member of BioClassifier and GeneCentric Diagnostics and is also listed as an inventor on patent applications on the Breast PAM50 and Lung Cancer Subtyping assays. Matthew Meyerson receives research support from Bayer Pharmaceuticals; is an equity holder in, consultant for, and Scientific Advisory Board chair for OrigiMed; and is an inventor of a patent for EGFR mutation diagnosis in lung cancer, licensed to LabCorp. Eduard Porta-Pardo is an inventor of a patent for domainXplorer. Han Liang is a shareholder and scientific advisor of Precision Scientific and Eagle Nebula. Da Yang is an inventor on a pending patent application describing the use of antisense oligonucleotides against specific lncRNA sequence as diagnostic and therapeutic tools. Yonghong Xiao was an employee and shareholder of TESARO, Inc. Bin Feng is an employee and shareholder of TESARO, Inc. Carter Van Waes received research funding for the study of IAP inhibitor ASTX660 through a Cooperative Agreement between NIDCD, NIH, and Astex Pharmaceuticals. Raunaq Malhotra is an employee and shareholder of Seven Bridges, Inc. Peter W. Laird serves on the Scientific Advisory Board for AnchorDx. Joel Tepper is a consultant at EMD Serono. Kenneth Wang serves on the Advisory Board for Boston Scientific, Microtech, and Olympus. Andrea Califano is a founder, shareholder, and advisory board member of DarwinHealth, Inc. and a shareholder and advisory board member of Tempus, Inc. Toni K. Choueiri serves as needed on advisory boards for Bristol-Myers Squibb, Merck, and Roche. Lawrence Kwong receives research support from Array BioPharma. Sharon E. Plon is a member of the Scientific Advisory Board for Baylor Genetics Laboratory. Beth Y. Karlan serves on the Advisory Board of Invitae.
References
- Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, Arachchi H, Arora A, Auman JT, Ayala B. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, Califano A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashashati A, Haffari G, Ding J, Ha G, Lui K, Rosner J, Huntsman DG, Caldas C, Aparicio SA, Shah SP. DriverNet: uncovering the impact of somatic driver mutations on transcriptional networks in cancer. Genome biology. 2012;13:R124. doi: 10.1186/gb-2012-13-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R, Giussani P, Anelli V, Colleoni T, Pedrazzi M, Patrone M, Viani P, Sparatore B, Melloni E, Riboni L. HMGB1 as an autocrine stimulus in human T98G glioblastoma cells: role in cell growth and migration. J Neurooncol. 2008;87:23–33. doi: 10.1007/s11060-007-9488-y. [DOI] [PubMed] [Google Scholar]
- Bertrand D, Chng KR, Sherbaf FG, Kiesel A, Chia BK, Sia YY, Huang SK, Hoon DS, Liu ET, Hillmer A. Patient-specific driver gene prediction and risk assessment through integrated network analysis of cancer omics profiles. Nucleic acids research. 2015;43:e44–e44. doi: 10.1093/nar/gku1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Wendl MC, Wyczalkowski MA, Wylie K, Ye K, Jayasinghe R, Xie M, Wu S, Niu B, Grubb R., III Divergent viral presentation among human tumors and adjacent normal tissues. Scientific reports. 2016;6:28294. doi: 10.1038/srep28294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Olsen C, Cava C, Terkelsen T, Silva TC, Olsen A, Cantini L, Bertoli G, Zinovyev A, Barillot E, et al. Moonlight: a tool for biological interpretation and driver genes discovery. bioRxiv 2018 [Google Scholar]
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic acids research. 2015;44:e71–e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova L, Wang H, Sunshine J, Kaunitz GJ, Cottrell TR, Xu H, Esandrio J, Anders RA, Cope L, Pardoll DM. Association of PD-1/PD-L axis expression with cytolytic activity, mutational load, and prognosis in melanoma and other solid tumors. PNAS. 2016;113:E7769–E7777. doi: 10.1073/pnas.1607836113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, Ramakrishna M, Martin S, Boyault S, Sieuwerts AM, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, Mooney TB, Callaway MB, Dooling D, Mardis ER. MuSiC: identifying mutational significance in cancer genomes. Genome research. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, Bauer TM, Farago AF, Wheler JJ, Liu SV, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7:400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, McLellan M, Sofia HJ, Hutter C, Getz G, et al. Automating Somatic Mutation calling for Ten Thousand Tumor Exomes. in review [Google Scholar]
- Fan Y, Xi L, Hughes DS, Zhang J, Zhang J, Futreal PA, Wheeler DA, Wang W. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol. 2016;17:178. doi: 10.1186/s13059-016-1029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz SM, Liang W-W, Xie M, Ding L. MIRMMR: binary classification of microsatellite instability using methylation and mutations. Bioinformatics. 2017;33:3799–3801. doi: 10.1093/bioinformatics/btx507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MD, Niu B, McLellan MD, Uzunangelov V. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yau C, Ahmed AA. A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay. Nat Commun. 2017;8:15943. doi: 10.1038/ncomms15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Wei S, Hou J, Zhang C, Xue P, Wang J, Chen X, Guo X, Yang F. Integrated proteomic and N-glycoproteomic analyses of doxorubicin sensitive and resistant ovarian cancer cells reveal glycoprotein alteration in protein abundance and glycosylation. Oncotarget. 2017;8:13413–13427. doi: 10.18632/oncotarget.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, Degenhardt J, Mayba O, Gnad F, Liu J, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanbash G, Carrera DA, Shrivastav S, Ahn BJ, Jahan N, Mazor T, Chheda ZS, Downey KM, Watchmaker PB, Beppler C, et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest. 2017;127:1425–1437. doi: 10.1172/JCI90644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RT, Perez EM, Hernández D, Miller CP, Haas KM, Irie HY, Lee S-I, Blau CA, Villén J. The proteomic landscape of triple-negative breast cancer. Cell reports. 2015;11:630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Reyna MA, Raphael BJ. A weighted exact test for mutually exclusive mutations in cancer. Bioinformatics. 2016;32:i736–i745. doi: 10.1093/bioinformatics/btw462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Karanika S, Yang G, Wang J, Park S, Broom BM, Manyam GC, Wu W, Luo Y, Basourakos S, et al. Androgen receptor inhibitor-induced "BRCAness" and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci Signal. 2017;10 doi: 10.1126/scisignal.aam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loes IM, Immervoll H, Sorbye H, Angelsen JH, Horn A, Knappskog S, Lonning PE. Impact of KRAS, BRAF, PIK3CA, TP53 status and intraindividual mutation heterogeneity on outcome after liver resection for colorectal cancer metastases. Int J Cancer. 2016;139:647–656. doi: 10.1002/ijc.30089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xie M, Wendl MC, Wang J, McLellan MD, Leiserson MD, Huang K-l, Wyczalkowski MA, Jayasinghe R, Banerjee T. Patterns and functional implications of rare germline variants across 12 cancer types. Nat com. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair B, Konopka T, Kerzendorfer C, Sleiman K, Salic S, Serra V, Muellner MK, Theodorou V, Nijman SM. Gain-and loss-of-function mutations in the breast cancer gene GATA3 result in differential drug sensitivity. PLoS genetics. 2016;12:e1006279. doi: 10.1371/journal.pgen.1006279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez J, Lahuerta JJ, Pepin F, Gonzalez M, Barrio S, Ayala R, Puig N, Montalban MA, Paiva B, Weng L, et al. Prognostic value of deep sequencing method for minimal residual disease detection in multiple myeloma. Blood. 2014;123:3073–3079. doi: 10.1182/blood-2014-01-550020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashl RJ, Scott AD, Huang KL, Wyczalkowski MA, Yoon CJ, Niu B, DeNardo E, Yellapantula VD, Handsaker RE, Chen K, et al. GenomeVIP: a cloud platform for genomic variant discovery and interpretation. Genome Res. 2017;27:1450–1459. doi: 10.1101/gr.211656.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Ceraudo E, Sher JJ, Guan Y, Shoushtari AN, Chang MT, Zhang JQ, Walczak EG, Kazmi MA, Taylor BS, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome biol. 2016;17:128. doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Scott AD, Sengupta S, Bailey MH, Batra P, Ning J, Wyczalkowski MA, Liang WW, Zhang Q, McLellan MD, et al. Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat Genet. 2016;48:827–837. doi: 10.1038/ng.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, Wendl MC, Ding L. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2013;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltean S, Bates D. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Lehner B. Cancer type-dependent genetic interactions between cancer driver alterations indicate plasticity of epistasis across cell types. Molecular systems biology. 2015;11:824. doi: 10.15252/msb.20156102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-Pardo E, Godzik A. e-Driver: a novel method to identify protein regions driving cancer. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu499. btu499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-Pardo E, Godzik A. Mutation drivers of immunological responses to cancer. Cancer immunology research. 2016;4:789–798. doi: 10.1158/2326-6066.CIR-15-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-Pardo E, Kamburov A, Tamborero D, Pons T, Grases D, Valencia A, Lopez-Bigas N, Getz G, Godzik A. Comparison of algorithms for the detection of cancer drivers at subgene resolution. Nat meth. 2017 doi: 10.1038/nmeth.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radenbaugh AJ, Ma S, Ewing A, Stuart JM, Collisson EA, Zhu J, Haussler D. RADIA: RNA and DNA integrated analysis for somatic mutation detection. PLoS One. 2014;9:e111516. doi: 10.1371/journal.pone.0111516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampias T, Vgenopoulou P, Avgeris M, Polyzos A, Stravodimos K, Valavanis C, Scorilas A, Klinakis A. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014;20:1199–1205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- Reimand J, Bader GD. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Molecular systems biology. 2013;9:637. doi: 10.1038/msb.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Olshen AB, Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–2912. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva TC, Colaprico A, Olsen C, D'Angelo F, Bontempi G, Ceccarelli M, Noushmehr H. TCGA Workflow: Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Research. 2016;5 doi: 10.12688/f1000research.8923.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Sogawa K, Takano S, Iida F, Satoh M, Tsuchida S, Kawashima Y, Yoshitomi H, Sanda A, Kodera Y, Takizawa H, et al. Identification of a novel serum biomarker for pancreatic cancer, C4b–binding protein alpha-chain (C4BPA) by quantitative proteomic analysis using tandem mass tags. Br J Cancer. 2016;115:949–956. doi: 10.1038/bjc.2016.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker TP, Brown CD, Bandlamudi C, McNerney M, Kittler R, Montoya V, Peterson A, Grossman R, White KP. Robust stratification of breast cancer subtypes using differential patterns of transcript isoform expression. PLoS Genet. 2017;13:e1006589. doi: 10.1371/journal.pgen.1006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- Tian L, Goldstein A, Wang H, Ching Lo H, Sun Kim I, Welte T, Sheng K, Dobrolecki LE, Zhang X, Putluri N, et al. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature. 2017;544:250–254. doi: 10.1038/nature21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokheim CJ, Papadopoulos N, Kinzler KW, Vogelstein B, Karchin R. Evaluating the evaluation of cancer driver genes. PNAS. 2016 doi: 10.1073/pnas.1616440113. 201616440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-S, Cooper Ta. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat rev Genetics. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wolf DM, Lenburg ME, Yau C, Boudreau A, van 't Veer LJ. Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLoS One. 2014;9:e88309. doi: 10.1371/journal.pone.0088309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Wang J, Jayasinghe R, Lameijer EW, McMichael JF, Ning J, McLellan MD, Xie M, Cao S, Yellapantula V, et al. Systematic discovery of complex insertions and deletions in human cancers. Nat Med. 2016;22:97–104. doi: 10.1038/nm.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.