Abstract
Phytophthora cactorum is a homothallic oomycete pathogen, which has a wide host range and high capability to adapt to host defense compounds and fungicides. Here we report the 121.5 Mb genome assembly of the P. cactorum using the third-generation single-molecule real-time (SMRT) sequencing technology. It is the second largest genome sequenced so far in the Phytophthora genera, which contains 27,981 protein-coding genes. Comparison with other Phytophthora genomes showed that P. cactorum had a closer relationship with P. parasitica, P. infestans and P. capsici. P. cactorum has similar gene families in the secondary metabolism and pathogenicity-related effector proteins compared with other oomycete species, but specific gene families associated with detoxification enzymes and carbohydrate-active enzymes (CAZymes) underwent expansion in P. cactorum. P. cactorum had a higher utilization and detoxification ability against ginsenosides–a group of defense compounds from Panax notoginseng–compared with the narrow host pathogen P. sojae. The elevated expression levels of detoxification enzymes and hydrolase activity-associated genes after exposure to ginsenosides further supported that the high detoxification and utilization ability of P. cactorum play a crucial role in the rapid adaptability of the pathogen to host plant defense compounds and fungicides.
Introduction
Phytophthora cactorum (Lebert & Cohn) J. Schröt is a devastating homothallic oomycete pathogen1,2, which produces both sporangia (asexual reproduction) and oospores (sexual reproduction) in its life cycle in the field1. Oospores can remain dormant in soil for many years, making them difficult to manage1. In addition, sexual reproduction may allow this pathogen to maintain high adaptability to its host and environment. P. cactorum has a broad range of hosts, which includes over 200 species of trees, ornamentals, and fruit crops3. Other Phytophthora species, such as P. sojae and P. ramorum, have a narrow range of hosts1,4,5. Wide host range of pathogens or insects may be partly due to a high detoxification ability against the defense compounds from their hosts6,7. P. cactorum is an important pathogen of Panax species. It causes leaf blight, stem canker, and root rot in these plants1,8,9. Panax species can synthesize a high level of ginsenosides, a group of defense compounds that account for more than 6% of dry biomass in the plants10. Previous studies found that ginsenosides could inhibit the growth of leaf pathogen of Alternaria panax and nonpathogenic Trichoderma spp, whereas the growth of root pathogens (P. cactorum, Fusarium solani, Fusarium oxysporum, and Cylindrocarpon destructans) could not be inhibited by ginsenosides at the physiological concentration in roots of P. notoginseng or P. quinquefolius11,12. Our experimental data demonstrated that wide host range species–P. cactorum, P. capsici, and P. parasitica–had higher utilization or detoxification ability against ginsenosides compared with narrow host pathogen P. sojae (Supplementary Figs 1 and 2).
It has been reported that detoxification pathways used by organisms against plant defense compounds are co-opted for pesticide tolerance6. Most oomycete fungicides, including dimethomorph, flumorph, pyraclostrobine, kresoxim-methyl, fluopicolide, cymoxanil, and metalaxyl-M, have been widely used for the control of Phytophthora infection. Our fungicide sensitivity test found that the wide host range pathogens showed a high tolerance ability against these fungicides compared with narrow host range species (Supplementary Fig. 3). Previous study also showed that P. cactorum had a stronger ability to obtain fungicide tolerance when being cultured in increasing concentrations of fungicide13. Proteomic analysis revealed that many proteins involved in the detoxification metabolic pathway are responsible for the tolerance of P. cactorum to fungicides14. Over the past decades, it is apparent that P. cactorum has gradually developed tolerance to many fungicides in the field9,13,15–17. The above described data implied that P. cactorum had a high ability to detoxify plant defense compounds or fungicides. Thus, P. cactorum can provide a good system to understand the genetic and molecular bases of how Phytophthora species adapt to the defense compounds of their hosts and the fungicides in the environment.
The genome of P. cactorum is highly heterozygous, and it is difficult to de novo assembly using the next-generation sequencing technology. Here we report the 121.5 Mb genome assembly of the P. cactorum using the third-generation single-molecule real-time (SMRT) sequencing technology to generate super long reads to facilitate the genome assembly process. The genome of P. cactorum is the second largest genome sequenced in the Phytophthora genus so far. Comparative analyses of Phytophthora genomes showed extensive expansion of genes encoding detoxification enzymes and carbohydrate-active enzymes (CAZymes). These data provide important references to investigate the adaptation process in P. cactorum to plant defense compounds and fungicides.
Results and Discussion
Genome sequencing, assembly and characterization of Phytophthora cactorum genome
Based on the 5.2 Gb PacBio single-molecule sequencing data, the 121.5 Mb reference genome was assembled using the PBcR pipeline18. This process resulted in 5,449 scaffolds with an N50 of 30.67 Kb. The lengths of 97.3% scaffolds were greater than 5 Kb (Table 1). The genome of P. cactorum is the second largest among the sequenced Phytophthora species, only smaller than P. infestans (~240 Mb)19, but larger than P. lateralis (~44 Mb)20, P. capsici (~64 Mb)21, P. ramorum (~65 Mb)22, P. fragariae (~73.68 Mb)23, P. parasitica (~64.5 Mb)24, and P. sojae (~95 Mb)22. Based on the protists dataset in BUSCO25, the genome captured 170 (79.1%) complete BUSCOs (Benchmarking Universal Single-Copy Orthologs). There were 36 (16.7%) missing BUSCOs (Table 1; Supplementary Table 1). The P. cactorum genome showed a highly syntenic relationship with the genomes of P. infestans, P. sojae, and P. capsici (Supplementary Fig. 4).
Table 1.
Summary of genome assembly and annotation for the P. cactorum genome.
| Assembly | |
| Assembled genome size (bp) | 121,526,021 |
| Genome-sequencing depth (×) | 42.8 |
| No. of contigs | 5,449 |
| N50 of contigs (bp) | 30,670 |
| Longest contig (bp) | 1,025,155 |
| GC content of the genome (%) | 52.15 |
| Completeness evaluation | |
| CEGMA | 95.16% |
| BUSCO | 79.1%* |
| Annotation | |
| Percentage of repeat sequences (%) | 46.69 |
| Repeat sequence length (bp) | 56,743,788 |
| No. of predicted protein-coding genes | 27,981 |
| Percentage of average gene length (bp) | 1,692.53 |
| Average exon length (bp) | 363.33 |
| Average exon per gene | 3.45 |
| Total intron length (bp) | 12,218,887 |
| tRNAs | 6731 |
| rRNAs | 376 |
| snRNAs | 376 |
| miRNAs | 2 |
| Family number | 11,674 |
| Genes in families | 19,783 |
*Based on protists_ensembl database.
The combination of de novo prediction and homology-based comparisons resulted in the identification of 56.7 Mb repetitive elements in the P. cactorum genome (Table 1; Supplementary Table 2), accounting for about 46.7% of the assembled genomes. 45.3% of the repeats in the P. cactorum genome were transposable elements (TEs), of which 20.3% were long terminal repeats (LTR) (Supplementary Table 3). The P. cactorum draft genome has more repeat sequences than P. capsici (19%), P. sojae (39%), and P. ramorum (28%), but less than P. infestans (74%).
We predicted 27,981 protein-coding genes in the assembled genome following a combination of homology and ab initio methods (Table 1). The average coding length was 1692.53 bp, and the average exon per gene was 3.45. P. cactorum had a noticeable expansion of gene content compared to P. capsici (19,805), P. infestans (17,797), P. sojae (16,988), and P. ramorum (14,451). The gene density in P. cactorum (241/Mb) was less than P. capsici (268/Mb), but was higher than other Phytophthora species (74/Mb in P. infestans, 179/Mb in P. sojae, and 222/Mb in P. ramorum). Gene structure-based evaluation was performed to confirm the annotation of protein-coding genes (Supplementary Figs S5 and 6b,c). The analysis of local gene density in P. cactorum showed that most genes with intergenic regions were 400 bp to 15 kb long. The main distribution of flanking distances is wider in P. cactorum but not the other three sequenced genomes (Supplementary Fig. 6a). In addition, 63% of the predicted genes (17,566) showed expression levels (FPKM > 0.05) with the alignment of ~2.2 Gb RNA-seq data26 to the our genome (Supplementary Table 4). In total, 25,225, 11,533, 10,480 and 13,287 of the predicted genes were assigned with a functional annotation in the NR, Swiss-Prot, KEGG, and InterProScan databases, respectively (Supplementary Table 5).
An overview of annotated ncRNA is shown in Supplementary Table 6. A total of 6,731, 5,947, 143, and 218 tRNAs were identified in P. cactorum, P. infestans, P. sojae, and P. ramorum, respectively. The numbers of Leu-tRNA, Glu-tRNA, and Pro-tRNA in P. cactorum were most abundant. 376 rRNAs and 376 snRNAs were predicted in P. cactorum. Two mature miRNAs and four potential target genes of these miRNAs were identified (Table 1; Supplementary Table 7). These four target genes encoded a nuclear pore complex protein, a poly(A) polymerase, an acid/auxin permease, and an unknown protein, respectively.
Comparative genomics and evolution of Phytophthora species
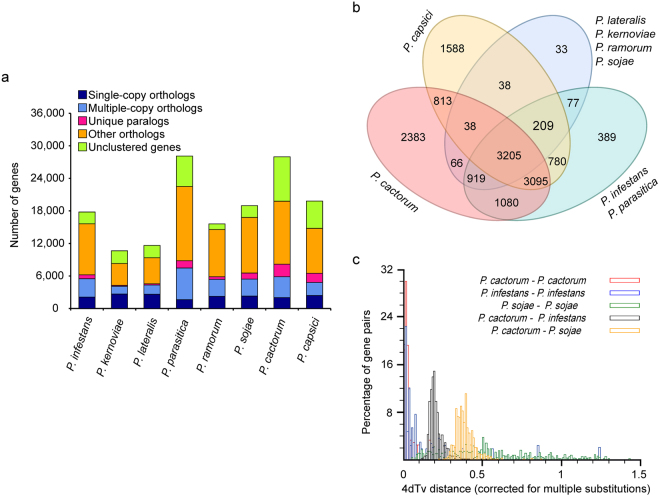
Gene family clustering analysis of eight Phytophthora species identified 11,674 gene families with a total of 19,783 genes in P. cactorum (Table 1; Fig. 1a). The numbers of single-copy orthologs in eight Phytophthora species were comparable. P. cactorum had 8,198 unclustered genes and 893 unique gene families (2,310 unique paralogs) (Supplementary Table 8). Among the genes unique to P. cactorum, the majority were enriched in defense response, cell cycle, interaction between organisms, peptidyl-amino acid modification, regulation of cell cycle, and TOR signaling pathway in the biological process (Supplementary Table 9). The Venn diagram showed that the eight Phytophthora species shared a common core set of 3,205 gene families (Fig. 1b). The number of P. cactorum-specific gene families was 2,383 (Fig. 1b).
Figure 1.
Evolutionary analysis of the P. cactorum genome assembly. (a) The orthologous gene distribution in eight sequenced Phytophthora species. (b) Venn diagram showing the number of unique and shared gene families among eight Phytophthora species. (c) Whole-genome duplications in P. cactorum, P. infestans and P. sojae as revealed by the distribution of 4DTv distance (four-fold degenerate third-codon transversion) between orthologous genes.
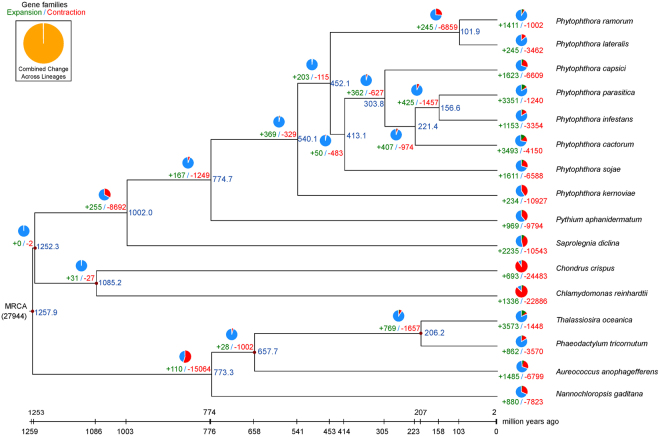
To systematically study the evolutionary dynamics of Stramenopile species, species phylogeny was performed utilizing 5,103 single-copy orthologous genes among 16 species, which included red algae (Chondrus crispus) and green algae (Chlamydomonas reinhardtii) (Fig. 2). The phylogenetic analysis revealed that red algae and green algae were grouped into one branch. The pathogenic oomycetes (such as Phytophthora, Pythium, and Saprolegnia) were separated from the nonpathogenic stramenochromes (such as Thalassiosira, Aureococcus, and Nannochloropsis). P. cactorum was more closely related to wide host range species (P. parasitica, P. infestans, and P. capsici) than to other sequenced Phytophthora species. Furthermore, the phylogenetic analysis mostly resembles the known topology of the tree of Stramenopile27,28. The exact topology of the eight Phytophthora species based on genomic data is highly consistent with the phylogenetic relationships of Phytophthroa species studies by Blair et al.29 and Runge et al.30 using multi-locus analysis. With the sequencing of more oomycete species genomes, the true phylogeny between Pythiaceae and Peronosporaceae in Peronosporales will gradually become clear. The estimated divergence time between P. parasitica, P. infestans and P. cactorum was 221.4 (138.6–342.4) million years ago (MYA) (Fig. 2). This most comprehensive and robust study of Phytophthora relationships to date based on genomic data will provide a phylogenetic framework for interpreting the evolutionary events of the genus.
Figure 2.
Phylogenetic relationship, the divergence time and gene contract and expand among oomycetes (Phytophthora sp, Pythium aphanidermatum and Saprolegnia diclina), stramenochromes [diatoms (Thalassiosira oceanica, Phaeodactylum tricornutu), Aureococcus (Pelagomonadales) and Nannochloropsis (Eustigmatales)], red algae (Chondrus crispus) and green algae (Chlamydomonas reinhardtii). The estimated divergence time was 221.4 (138.6–342.4) million years ago (MYA) between P. parasitica, P. infestans and P. cactorum. The number of contracting and expanding gene families among 16 species is shown in the pie chart, the estimates of divergence time are indicated at each node. The red dot on branches means divergence time has been adjusted by fossil evidence.
The expansion of gene families was frequently reported to directly or indirectly involve in pathogenicity in fungal pathogens19,22,28,31–33. 3,493 and 4,150 gene families of P. cactorum had undergone expansion and contraction, respectively (Fig. 2). The number of expanded gene families in P. cactorum was the largest among Phytophthora species, whereas the number of contracted gene families was relatively small. The expanded genes in P. cactorum were enriched in membrane, cytoskeleton, transport, carbohydrate metabolism, nucleotide binding, transporter activity, transferase activity, and hydrolase activity (p < 0.01, Supplementary Table 10). The contracted genes in P. cactorum were enriched in cellular catabolism, DNA metabolism, chromosome, nucleic acid binding, and nucleotide binding (p < 0.01, Supplementary Table 11). The expansion of gene families is likely the result of the transposons33. The expansion of gene families in P. cactorum may be due to the large number of transposable elements in genome. However, more analysis should be performed to elucidate the relationship of transposons and gene family expansion.
Whole-genome duplication (WGD) followed by gene loss has been found in most eudicots and is regarded as the major evolutionary force that gives rise to gene neofunctionalization in both plants and animals34. It has been speculated that the increased genome size and gene content of Phytophthora species may be due to WGD or their divergent repertoires of transposable elements19,35,36. WGD analysis of three Phytophthora species indicated that both P. cactorum and P. infestans have experienced a recent WGD event (Fig. 1c). This WGD event helps clarify some of the evolutionary history of Phytophthora species. The time of divergence between P. cactorum and P. infestans was later than that between P. cactorum and P. sojae, which was consistent with the phylogenetic tree (Fig. 2). And some evidence indicated the WGD in P. cactorum was followed by gene loss (Supplementary Fig. 7).
Positive selection was proposed to contribute to fitness. About 428 to 634 P. cactorum genes were determined as positive-selected genes comparing with other Phytophthora species (p < 0.05; Supplementary Table 12 sheet 1–7). GO enrichments analysis demonstrated that most of these genes in P. cactorum were involved in cellular macromolecule metabolism, hydrolase activity, and transferase activity (Supplementary Table 13).
P. cactorum, P. parasitica, and P. capsici have a wide spectrum of hosts, and P. sojae has a narrow spectrum of hosts. It has been reported that oomycete pathogens have a large and diverse repertoire of expanded gene families19,22,28,37. Most of the expanded gene families were reported to be directly or indirectly involved in pathogenicity, such as glycoside hydrolases28,38 or secreted proteins characterized by the presence of either the RXLR or the LXLFLAK (Crinkler) motifs19,39–41. Comparative genomic analysis demonstrated that the numbers of pathogenicity-associated genes, including proteinase inhibitor, protein toxins, secretomes, secondary metabolites biosynthesis, were similar to or smaller than the numbers reported for P. capsici, P. sojae, and P. parasitica. The numbers of genes involved in CAZymes and detoxification metabolism were increased in the genome of P. cactorum (Table 2). Thus, the divergence of wide host range species and narrow host range species may be partly associated with the expansion and extraction of genes involved in detoxification enzymes and CAZymes.
Table 2.
Potential infection-related genes in the P. cactorum, P. sojae, P. capsici and P. parasitica.
| Gene product | P. cactorum | P. capsici* | P. parasitica* | P. sojae |
|---|---|---|---|---|
| Proteases, all | 87 | 40 | 64 | 186 |
| Serine proteases | 47 | 18 | 40 | 119 |
| Cysteine proteases | 40 | 22 | 24 | 67 |
| Carbohydrate-active enzymes (CAZys) | 901 | 628 | 839 | 786 |
| Glycosyl hydrolases (GHs) | 374 | 261 | 312 | 125 (314) |
| Glycosyl transferases (GTs) | 190 | 130 | 220 | (155) |
| Polysaccharide lyases (PLs) | 73 | 54 | 44 | (58) |
| Auxiliary Activities (AAs) | 50 | 43 | 50 | 48 |
| Carbohydrate-binding modules (CBMs) | 103 | 54 | 102 | 92 |
| Carbohydrate esterases (CEs) | 111 | 86 | 111 | 119 |
| Pectinases | 68 | 55 | 54 | 62 |
| Pectin esterases | 24 | 7 | 16 | 19 |
| Pectate lyases | 44 | 48 | 38 | 43 |
| Cutinases | 7 | 6 | 6 | 16 |
| Chitinases | 3 | 2 | 4 | 5 |
| Lipases | 10 | 12 | 15 | 171 |
| Phospholipases | 55 | 29 | 44 | >50 |
| Protease inhibitors | 17 | 25 | 30 | 19 |
| Kazal | 14 | 23 | 28 | 15 |
| Cystatin | 3 | 2 | 2 | 4 |
| Protein toxins | 41 | 45 | 51 | 48 |
| NPP family | 37 | 39 | 49 | 29 |
| PcF family | 4 | 6 | 2 | 19 |
| Secondary metabolite biosynthesis | 4 | 3 | 13 | 4 |
| Nonribosomal peptide synthetases | 3 | 2 | 9 | 4 |
| Polyketide synthases | 1 | 1 | 4 | 0 |
| Effectors | 174 | 156 | 294 | 218 |
| Elicitins | 39 | 48 | 54 | 57 |
| Avh (RXLR) family | 135 | 108 | 240 | 350 (120) |
| Crn family (Crinklers) | 16 | 25 | 13 | 40 (41) |
| Detoxification metabolism | 896 | 695 | 794 | 585 |
| ABC transporters (ABC) | 60 | 40 | 48 | 134 (42) |
| Major facilitator superfamily (MFS) | 239 | 217 | 242 | 228 |
| Cytochrome P450’s (CYPs) | 46 | 36 | 40 | 30 (33) |
| Alcohol dehydrogenase (ADH) | 101 | 58 | 71 | 52 |
| Short-chain dehydrogenase/reductase (SDR) | 84 | 68 | 79 | 67 |
| Peroxidase (POD) | 56 | 31 | 35 | 34 |
| Glutathione S-transferases (GSTs) | 45 | 32 | 37 | 41 |
| Methyltransferase (MTR) | 265 | 213 | 242 | 163 |
* and () indicated the data obtained according our method.
For successful infection, phytopathogenic microorganisms have the ability to adapt to the plant defense system through detoxification or direct utilization of plant defense compounds42,43. In this study, we found that P. cactorum made good use of ginsenosides as the sole carbon source to growth (Supplementary Fig. 1). Four tested Phytophthora species showed similar abilities to utilize glucose, xylan, pectin, cellulose, and gum guar. However, P. cactorum, P. capsici, and P. parasitica showed higher ability to utilize ginsenosides than P. sojae (Supplementary Fig. 2). Ginsenosides can be hydrolyzed by microbial CAZY enzymes44. Based on genomic analysis, we predicted 901 genes that putatively encode CAZY enzymes in P. cactorum. This number was larger than those of other three sequenced Phytophthora species (Table 2). Especially, the members of GHs, GT, and PL families were expanded in P. cactorum.
Microbes have evolved the ability to detoxify xenobiotics through enzymes and transporters45–47. We identified the ATP-binding cassette (ABC) transporter families and major facilitator superfamily (MFS), as well as the cytochrome P450 (CYPs), peroxidase (POD), glutathione S-transferases (GSTs), methyltransferase (MTR), and dehydrogenase in the P. cactorum genome (Table 2). Our previous proteomic analysis also showed that P. cactorum employed detoxification enzymes to tolerate a series of fungicides13. These data imply that the expansion of detoxification metabolism genes enable P. cactorum with higher detoxification ability against host defense compounds or commercial fungicides. However, more genetic studies should be performed to elucidate the function and relationship of these key genes.
Transcriptome
Transcriptomes from four Phytophthora species (P. cactorum, P. capsici, P. parasitica, and P. sojae) exposed to ginsenosides for 24 hours contained a total of 267 (Supplementary Table 14), 408 (Supplementary Table 15), 18 (Supplementary Table 16), and 28 (Supplementary Table 17) differentially expressed genes (DEGs), respectively (Supplementary Fig. 8). A set of the up-regulated DEGs in P. cactorum and P. capsici were involved in the detoxification metabolism and glycosyl hydrolase, highlighting the potential roles of these genes in adaptation to ginsenosides. Furthermore, time-course transcriptome analyses respectively identified 179, 355, 45, 270, 341, and 127 DEGs in P. cactorum after exposure to ginsenosides for 1 h, 3 h, 6 h, 12 h, 24 h, and 48 h (Supplementary Fig. 9). The GO classifications showed that nine GO terms were unique for up-regulated genes, which included drug binding and transporter, transcription related factors, and enzyme activator GO terms (Supplementary Fig. 10). Further analyses showed that a series of glycoside hydrolase and transferase genes as well as detoxification-related genes were induced after the exposure to ginsenosides (Fig. 3; Supplementary Tables 18 and 19). Previous study demonstrated that ginsenosides could be hydrolyzed by microbial glycosyl hydrolases to release glycosyl as nutrient for microbes44. Although the function of ginsenosides induced detoxification-related genes in P. cactorum should be proven by further genetic studies, these genes were frequently reported in chemoresistance45–47. Thus, P. cactorum could not only detoxify ginsenosides through detoxification-related enzymes but also utilize them as nutrient for growth through glycosyl hydrolases. It may be an important strategy for P. cactorum to infect P. notoginseng.
Figure 3.
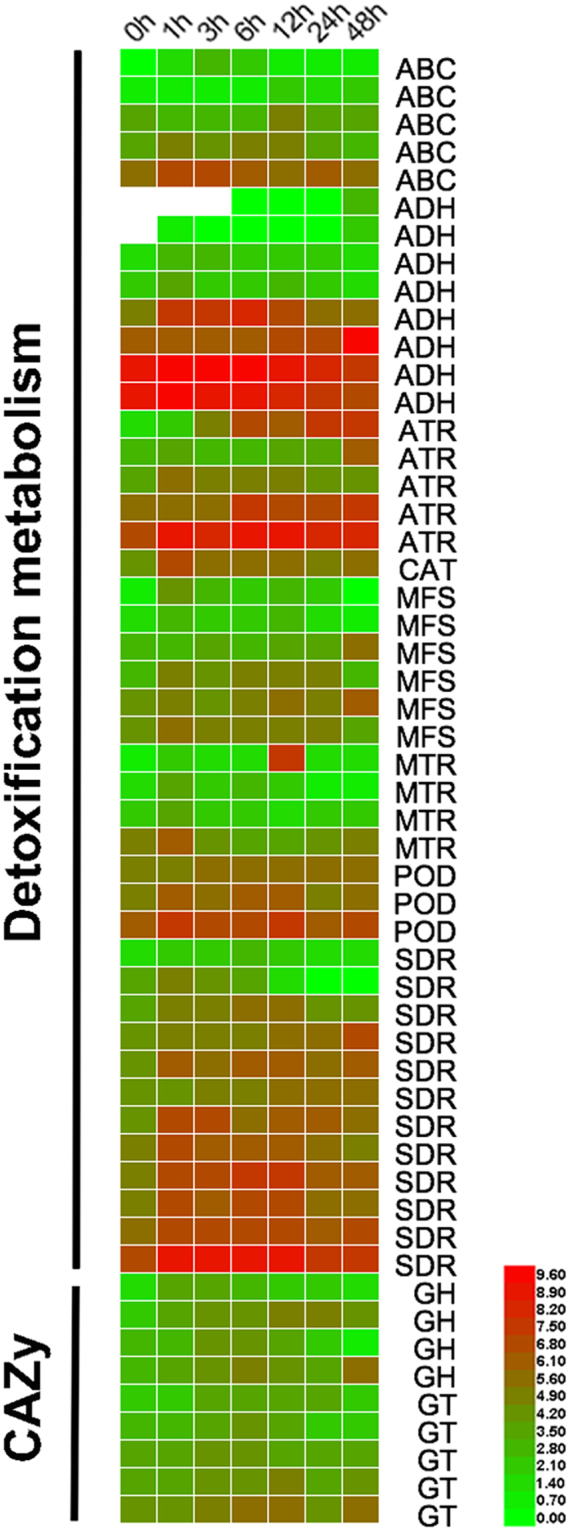
Heat-map depicting the changes of the up-regulated expressed genes involved in detoxification metabolism and CAZymes after exposure to ginsenosides in time-course treatments of P. cactorum. ABC, ABC transporter; MFS, major facilitator superfamily; ADH, alcohol dehydrogenase; SDR, short-chain dehydrogenase/reductase; MTR, methyltransferase; ATR, acyltransferase; CAT, catalases; POD, peroxidase; GH, glycoside hydrolase; GT, glycosly transferase. Detailed descriptions of these metabolites are shown in Supporting Information Table S18.
Transcription factors regulate gene expression and protein kinases regulate cellular activities by phosphorylating target proteins in response to internal or external signals. We identified a total of 566 transcription factors and 536 protein kinases in the P. cactorum. The numbers were smaller than those found in P. parasitica (689 transcription factors, 577 protein kinases), but larger than those found in P. capsici (458, 406), P. sojae (431, 423), P. infestans (381, 413), P. ramorum (367, 399), and P. kernoviae (252, 231) (Supplementary Tables 20 and 21). The C2H2, MYB-related and SET transcription factors were comparatively abundant in P. cactorum, as well as the group CAMK and TKL protein kinases. P. cactorum has an expanded RLK Pelle group proteins of 22 members. There were also a large number of unclassified kinases, suggesting novel functions performed by the P. cactorum. After exposure to ginsenosides, a set of transcription factors and protein kinases-related genes were significantly up- or down- regulated (Supplementary Tables 18 and 19), which may facilitate the adaptation of P. cactorum to defense compounds.
Conclusions
In summary, we sequenced the P. cactorum using the third-generation single-molecule real-time (SMRT) sequencing technology and revealed the relationship between P. cactorum and other sequenced Phytophthora species. Comparative genomics analyses identified the expansion of gene families associated with the detoxification and carbohydrate-active enzymes (CAZymes) against plant defense compounds. These genes may enable P. cactorum with a high ability to tolerate or utilize plant defense compounds and commercial fungicides. This may partly explain the pathogenicity of P. cactorum in a broad range of hosts. Together, our genomic analyses provide insights into the adaptive mechanisms of P. cactorum to plant defense compounds and fungicides, which will facilitate future studies on pathogenesis and disease management.
Methods
DNA isolation, sequencing and assembly
P. cactorum was isolated from an infected P. notoginseng in Yunnan. The mycelia were harvested after the strains grown in 150 mL of carrot liquid medium in a 500-mL shaker culture flask on a shaker for three days at room temperature, respectively. Then genomic DNA used for sequencing was extracted from mycelia using Omega Fungal DNA Kit according to the manufacturer’s instructions. Briefly, fresh fungal tissue was disrupted and then lysed in a specially formulated buffer containing detergent. Contaminants were further removed after DNA precipitation using isopropanol. Binding conditions were then adjusted and the sample was applied to a spin-colum. Trace contaminants such as residual polysaccharides were removed with two rapid wash steps, and pure DNA was eluted using low ionic strength buffer. In total of 50 mg DNA were used to construct the PacBio sequencing libraries.
Then sequencing was performed to produce raw reads. Totally, 4.84 Gb filtered subreads were obtained for P. cactorum from raw data. Though the error rates of single-molecule reads are high, but we yielded a considerable amount of long reads (43×), which required for self-correction and for subsequent de novo assembly, to assemble the genome. Automatic assembly was performed using PBcR pipeline of Celera Assembler version 8.3rc118. Syntenies was yielded through aligning the scaffolds of P. cactorum to that genome of P. infestans, P. sojae and P. capsici using NUCmer in MUMmer 3.2348, respectively.
Repeats annotation
First, we searched for tandem repeats across the genome using the program Tandem Repeat Finder (TRF)49. The transposable elements (TEs) in the genome were identified by a combination of homology-based and de novo approaches. For homolog based prediction, known repeats were identified using RepeatMasker50 and RepeatProteinMask50 against Repbase51 (Repbase Release 16.10; http://www.girinst.org/repbase/index.html). Repeat Masker was applied for DNA-level identification using a custom library. At the protein level, RepeatProteinMask was used to perform an RMBLAST search against the TE protein database. For de novo prediction, RepeatModeler (http://repeatmasker.org/) and LTR FINDER52 were used to identify de novo evolved repeats inferred from the assembled genome.
Gene prediction and functional annotation
We used the MAKER253 pipeline to predict protein-coding genes in the genome. First, the genome was repeat masked using the result of repeat annotation. Then the masked genome was fed to the MAKER2 pipeline with the ab initio gene predictors being GeneMark-ES, FGENESH, Augustus and SNAP. And for the evidence-driven gene prediction, cDNA of P. infestans and proteins of six related species from Ensembl (release-28, P. infestans, P. kernoviae, P. lateralis, P. parasitica, P. ramorum, and P. sojae) were fed to the pipeline. Gene functions were assigned according to the best match alignment using BLASTp against NR, Swiss-Prot and KEGG databases. InterProScan functional analysis and Gene Ontology IDs were obtained using InterProScan54.
Non-coding gene annotation
Software tRNAscan-SE55 is specified for Eukaryotic tRNA and was deployed for tRNA annotation. We used homologous method to identify rRNA. The rRNA sequence data downloaded from Rfam56 database was used as a reference. INFERNAL57 was used to identify snRNA.
Gene family cluster
To identify different sets of gene clusters, protein-coding genes sequences of 16 species were downloaded from Esembl and JGI (http://genome.jgi.doe.gov) and used to locate gene clusters. After pairwise aligning was conducted using BLASTp with an E-value cutoff of 10−5, OrthoMCL package58 was performed to identify the gene family clusters using the BLASTp output with default parameters, final paralogous and orthologous genes were defined using MCL software in OrthoMCL.
Phylogenetic tree construction
The single-copy orthologous genes defined by OrthoMCL58 were formed, then multiple single-copy genes was aligned using MUSCLE59, and the aligned sequences were extracted to feed to MrBayes (http://mrbayes.sourceforge.net) to inferred the species phylogeny using a maximum likelihood (ML) approach. To estimate the divergence time of each species, the information about the already known divergence time data between these species from http://www.timetree.org/ were collected. The topology of the ML tree was fed to MCMCTREE in paml version 4.460 for constructing a divergence time tree and calculated the divergence time. Based on the calculated phylogeny and the divergence time, CAFÉ61 (Computational Analysis of Gene Family Evolution, version 2.1), a tool based on the stochastic birth and death model for the statistical analysis of the evolution of gene family size, was applied to identify gene families that had undergone expansion and/or contraction. The GO enrichment was done with Ontologizer 2.062 by using one-sided Fisher’s exact test, the Parent-Child-Union method, with a p-value cut-off of 0.01. All genes with GO annotation were used as reference, and the genes undergone expansion or contraction was used as study set.
Detection of positively selected genes
To detect genes under positive selection, BLASTn was performed to align the coding sequence (CDS) libraries of P. infestans, P. lateralis, P. capsici, P. ramorum, P. kernoviae, P. parasitica and P. sojae against the P. cactorum CDS library, respectively, in order to find the gene pairs with the best alignments. The resulting orthologous gene pairs were aligned again using lastz with the default parameters as a preparation for KaKs_Calculator 1.263, which finally yielded a dataset of each gene pair’s Ka/Ks ratio, and the Ka/Ks ratio >1 was determined a positively selected gene (significance, p-value < 0.05). The GO enrichment was done with Ontologizer 2.062 by using one-sided Fisher’s exact test, the Parent-Child-Union method, with a p-value cut-off of 0.05. All genes of P. cactorum were used as reference, and of all positively selected genes in P. cactorum were used as study set.
Characterization of protein families
Transcription factors and protein kinases were identified using iTAK v1.5 (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi). Carbohydrate-active enzymes (CAZymes) were identified by scanning using HMMER 3.064 against the Hidden Markov Model (HMMs) corresponding to the Pfam65 CAZyme family and subfamily (download from http://csbl.bmb.uga.edu/dbCAN/)66. Secondary metabolism genes were annotated based on their genomic context and domain content using an automatic web-based software SMURF (www.jcvi.org/smurf/)67.
The gene families of potential infection-related genes were scanned using HMMER with HMMs against the Pfam families (E-value cutoff of 0.01, PF00067 for cytochrome P450, PF05630 for NPP1 family). The candidates of NPP1 family were further identified to confirm the existence of a signal peptide in N-terminal. Transporters were identified by scanning for the PFAM domains representing both two ABC transporters domains (PF00005 and PF00664) and assisted with manual inspection. Statistics of other proteins in Table 2 were based on the annotation of InterProScan database.
The families of CRN effectors in P. cactorum, P. sojae, P. capsici and P. parasitica dataset were initially predicted based on the BLASTp comparisons (E-value cut-off of 10−5) against the collection of CRN effectors of Phytophthora species and NCBI databases, and confirmed the existence of a signal peptide in N-terminal.
RXLR effector prediction
A reference method68 to identify sequences containing a signal peptide and the predicted cleavage site must be within first 40 amino acids in N-terminal using SignalP4.069, the RXLR motif was extended to incorporate the presence of an [ED][ED][KR] motif down-stream and within 40 amino acids of the RXLR motif. The RXLR position must be downstream of the signal peptide cleavage site, and the RXLR motif and [ED][ED][KR] motif must be within the first 100 amino acids downstream of the signal peptide cleavage site.
The sensitivity test of Phytophthora species to plant defense compounds and fungicides
The cultures of P. cactorum, P. capsici, P. parasitica, and P. sojae, growing on carrot agar medium (CA) plates were transferred onto new CA plates amended with crude ginsensides at concentrations of 0, 0.10, 0.50, 1.0, 5.0, and 10.0 g L−1 or fungicides at the following concentrations: fluopicolide, dimethomorph and flumorph, at 0, 0.1, 0.5, 1.0, and 2.0 mg L−1; pyraclostrobine and kresoxim-methyl, at 0, 0.1, 0.5, 1.0, 2.0, and 5.0 mg L−1; cymoxanil, at 0, 10, 20, 40, and 80 mg L−1; metalaxyl-M, at 0, 0.1, 1.0, 10.0, and 50.0 mg L−1. Fungicide or ginsenosides was dissolved in methanol (OmniSolv, HPLC grade) to prepare stock solutions. To prepare agar plates supplemented with serial dilutions of fungicides or ginsenosides, the stock solutions were added into CA medium (200 g boiled carrot and 15 g agar in a total volume of 1 L of distilled water) when CA medium was cooled to 50 °C. The final concentration of methanol in any tested media was limited to 0.1% (vol/vol). The experiment was performed three times with four replicates and incubated for 4 days in the dark at 25 °C. The diameters of the colonies were measured perpendicularly. The ginsenosides was extracted from three-year-cultivated P. notoginseng roots with MeOH:H2O (80:20) and identified by HPLC-MS as described previously70.
Growth profiles of Phytophthora species on single carbon source
Citrus pectin, glucose, xylan, cellulose, gum guar and crude ginsenosides (all from Sigma) were used as single carbon source separately in agar medium, to evaluate the growth of different Phytophthora species. Inocula of P. cactorum and other Phytophthora species (P. capsici, P. parasitica, and P. sojae) were placed on these media and incubated at 25 °C for four days. These tests were repeated three times, and the results were analysed to evaluate mycelium growth ability on different single carbon source.
RNA-seq
P. cactorum and other Phytophthora species (P. capsici, P. parasitica, and P. sojae) were grown from mycelial inocula at 27 °C (150 mL of carrot liquid medium in a 500-mL shaker culture flask) shaking at 115 rpm. 10 fresh plug (5 mm in diameter) was taken from the growing edge of a CA culture and transferred into 150 mL of medium in each shock culture flask, and the mixture was incubated in an orbital shaker (ZHWY-111B, Shanghai ZHICHENG Analytical Instruments Manufacturing Co., Ltd.) at 150 rpm at 25 °C. After 24 h pre-incubation, ginsenosides stock solution was added to the medium to a final concentration of 1.0 mg L−1. For the control culture, only methanol was added. Mycelia were collected at 24 h after exposure to ginsenosides. For P. cactorum, mycelia were collected at 1, 3, 6, 12, 24, and 48 h after treatment with ginsenosides. And then, the mycelial mat was separated from the medium by filtration, quickly washed three times with deionized water, then immediately frozen in liquid nitrogen and lyophilized. Each treatment had two independent replicates.
3 μg of total RNA per sample was used as input material for the RNA sample preparation. Beads with oligo (dT) were used to isolate poly(A) mRNA from total RNA. RNA sequencing libraries were constructed from these mRNA using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, USA). Briefly, the Elution 2-Frag-Prime (94 °C for 8 minutes, 4 °C hold) was used to elute, fragment and prime the mRNA with Elute, Prime, Fragment Mix (Illumina). First strand cDNA synthesis was performed with First Strand Master Mix and SuperScript II mix (ratio: 1 µl SuperScript II/7 µl First Strand Master Mix) (Invitrogen). The second strand was synthesized with Second Strand Master Mix (Illumina) and Ampure XP beads (Illumina) were used to separate the double-stranded (ds) cDNA from the 2nd strand reaction mix. After end repair and the addition of a 3′-dA overhang, the cDNA was ligated to Illumina PE adapter oligo mix (Illumina), and size-selected for 400 ± 10% bp fragments by gel purification. After 15 cycles of PCR amplification, the paired-end libraries were sequenced using the paired-end sequencing module (150 bp at each end) of the Illumina HiSeq 4000 platform.
These raw reads were processed through Trimmomatic (Version 0.32)71 to remove reads containing adapter, reads containing poly-N and low quality reads from the raw data and yielded clean data for downstream analyses. The corresponding trimmed clean reads were aligned to the related reference genome (P. parasitica and P. sojae were downloaded from Ensembl, and the P. capsici genome was downloaded from JGI database) employing TopHat272 software with default settings. Calculation of gene expression level and identification of differentially expressed genes (DEGs) between the time-course (for P. cactorum) or treatments (for P. parasitica, P. sojae and P. capsici) and control groups were conducted using Cufflinks v2.2.173. Fragments per kilobase of exon per million fragments mapped (FPKM) were used to normalize RNA-seq fragment counts and estimate the relative abundance of each gene. The Cuffdiff package in Cufflinks was used to perform pairwise comparisons of the expressions of each gene between treatments and control in the four species and to report DEGs and transcripts. The DEGs were decided based on a p-value < 0.05 and at least a 2-fold change between the two FPKM values.
Data availability
All raw genome sequence data have been deposited in the Short Read Archive (SRA) at NCBI under accession number SRR3386345 (PRJNA318145). Raw RNA-seq data have been deposited in the SRA under accession number SRP111895.
Electronic supplementary material
Acknowledgements
We acknowledge grant support from the Major Programme for Bio-medicine in Yunnan Province (2016ZF001), the National Key Research and Development Program of China (SQ2017YFC170021-02), the programme for outstanding talent in Yunnan Agricultural University (2015JY02) and the Natural Science Foundation of China (31660605). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
S.Z., X.H. and Y.D. conceived the study and initiated the genome project. Y.D., X.L. and X.H. designed major scientific objectives. S.D., H.H. and W.C. performed genome assembly and functional annotation. S.D., Y.D. and M.Y. conducted gene family analysis and comparative genomics. S.D., W.W., C.G., H.H. and Y.L. performed transcriptomes sequencing, assembly, and analyses. M.Y., X.M., T.Y., C.G. participated in sample collection and sensitivity test. S.Z., S.D., W.C., X.L., X.H., Y.D. participated in discussions and provided suggestions for manuscript improvement. S.Z., M.Y., S.D. and W.C. wrote the paper with input from all authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Min Yang, Shengchang Duan and Xinyue Mei contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24939-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiahong He, Email: hexiahong@ynau.edu.cn.
Yang Dong, Email: loyalyang@163.com.
Shusheng Zhu, Email: shushengzhu79@126.com.
References
- 1.Darmono TW, Owen ML, Parke JL. Isolation and pathogenicity of Phytophthora cactorum from forest and ginseng garden soils in Wisconsin. Plant Dis. 1991;75:610. doi: 10.1094/PD-75-0610. [DOI] [Google Scholar]
- 2.Judelson HS, Blanco FA. The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 2005;3:47–58. doi: 10.1038/nrmicro1064. [DOI] [PubMed] [Google Scholar]
- 3.Hantula J, et al. Pathogenicity, morphology and genetic variation of Phytophthora cactorum from strawberry, apple, rhododendron, and silver birch. Mycol. Res. 2000;104:1062–1068. doi: 10.1017/S0953756200002999. [DOI] [Google Scholar]
- 4.Tyler BM. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007;8:1–8. doi: 10.1111/j.1364-3703.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 5.Rizzo DM, Garbelotto M, Hansen EM. Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 2005;43:309–335. doi: 10.1146/annurev.phyto.42.040803.140418. [DOI] [PubMed] [Google Scholar]
- 6.You, M. et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genetics45 (2013). [DOI] [PubMed]
- 7.Pedras MS, Ahiahonu PW. Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi. Phytochemistry. 2005;66:391–411. doi: 10.1016/j.phytochem.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Liu YZ, Yang JZ, Liu YL, Chen YJ. A study on biological characters of Phytophthora cactorum on Panax notoginseng. Southwest Chin. J Agric. Sci. 2008;21:671–674. [Google Scholar]
- 9.Hill SN, Hausbeck MK. Virulence and fungicide sensitivity of Phytophthora cactorum isolated from American ginseng gardens in Wisconsin and Michigan. Plant Dis. 2008;92:1183–1189. doi: 10.1094/PDIS-92-8-1183. [DOI] [PubMed] [Google Scholar]
- 10.Zhou JM, et al. Study on chemical composition of root exudates of Panax notogineseng. Special Wild Economic Animal & Plant Res. 2009;3:37–39. [Google Scholar]
- 11.Nicol RW, Traquair JA, Bernards MA. Ginsenosides as host resistance factors in American ginseng (Panax quinquefolius) Can. J. Bot. 2002;80:557–562. doi: 10.1139/b02-034. [DOI] [Google Scholar]
- 12.Yang M, et al. Sensitivity of the pathogens of Panax notoginseng to ginsenosides. Plant Protection. 2014;40:71–86. [Google Scholar]
- 13.Mei XY, et al. Proteomic analysis on zoxamide-induced sensitivity changes in Phytophthora cactorum. Pesticide Biochem. and Physiol. 2015;123:9–18. doi: 10.1016/j.pestbp.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Mei XY, et al. Proteomic analysis of zoxamide-induced changes in Phytophthora cactorum. Pesticide Biochem. and Physiol. 2014;113:31–39. doi: 10.1016/j.pestbp.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Jeffers SN, Schnabel G, Smith JP. First report of resistance to mefenoxam in Phytophthora cactorum in the United States and elsewhere. Plant Dis. 2004;88:576–576. doi: 10.1094/PDIS.2004.88.5.576A. [DOI] [PubMed] [Google Scholar]
- 16.Thomidis T, Michailidis Z. Preliminary evaluation of nine fungicides for control of Phytophthora cactorum and P. citrophthora associated with crown rot in peach trees. Phytoparasitica. 2002;30:52–60. doi: 10.1007/BF02983970. [DOI] [Google Scholar]
- 17.Rebollaralviter A, Madden LV, Ellis MA. Pre- and post-infection activity of azoxystrobin, pyraclostrobin, mefenoxam, and phosphite against leather rot of strawberry, caused by Phytophthora cactorum. Plant Dis. 2007;91:559–564. doi: 10.1094/PDIS-91-5-0559. [DOI] [PubMed] [Google Scholar]
- 18.Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 19.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 20.Quinn L, et al. Genome-wide sequencing of Phytophthora lateralis reveals genetic variation among isolates from Lawson cypress (Chamaecyparis lawsoniana) in Northern Ireland. FEMS Microbiol. Lett. 2013;344:179–185. doi: 10.1111/1574-6968.12179. [DOI] [PubMed] [Google Scholar]
- 21.Lamour KH, et al. Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol. PlantMicrobe Int. 2012;25:1350–1360. doi: 10.1094/MPMI-02-12-0028-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 23.Gao R, et al. Genome Sequence of Phytophthora fragariae var. fragariae, a quarantine plant-pathogenic fungus. Genome Announcements. 2014;3:25–30. doi: 10.1128/genomeA.00034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phytophthora parasitica Assembly Dev initiative, B. I. b. o. https://olive.broadinstitute.org/ projects/phytophthora_parasitica. (2017).
- 25.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics (Oxford, England) 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 26.Chen XR, et al. Transcriptomic analysis of the phytopathogenic oomycete Phytophthora cactorum provides insights into infection-related effectors. BMC Genomics. 2014;15:980. doi: 10.1186/1471-2164-15-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang RH, et al. Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLOS Genet. 2013;9:e1003272. doi: 10.1371/journal.pgen.1003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidl MF, V den Ackerveken G, Govers F, Snel B. Reconstruction of oomycete genome evolution identifies differences in evolutionary trajectories leading to present-day large gene families. Genome Biol. Evol. 2012;4:199–211. doi: 10.1093/gbe/evs003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blair JE, Coffey MD, Park SY, Geiser DM, Kang S. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet. Biol. 2008;45:266–277. doi: 10.1016/j.fgb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Runge F, et al. The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphyly in Phytophthora. IMA Fungus. 2011;2:163–171. doi: 10.5598/imafungus.2011.02.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 32.Morales-Cruz A, et al. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC genomics. 2015;16:469. doi: 10.1186/s12864-015-1624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartze, V. U. et al. Gene expansion shapes genome architecture in the human pathogen Lichtheimia corymbifera: an evolutionary genomics analysis in the ancient terrestrial mucorales (Mucoromycotina). PLoS genetics10 (2014). [DOI] [PMC free article] [PubMed]
- 34.Huang S, et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013;4:2640. doi: 10.1038/ncomms3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang RH, et al. Elicitin genes in Phytophthora infestans are clustered and interspersed with various transposon-like elements. Mol. Genet. Genomics. 2005;273:20–32. doi: 10.1007/s00438-005-1114-0. [DOI] [PubMed] [Google Scholar]
- 36.Martens C, de Peer V. Y. The hidden duplication past of the plant pathogen Phytophthora and its consequences for infection. BMC Genomics. 2010;11:353. doi: 10.1186/1471-2164-11-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baxter L, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ospina-Giraldo MD, Griffith JG, Laird EW, Mingora C. The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genomics. 2010;11:525. doi: 10.1186/1471-2164-11-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 40.Dou DL, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang RH, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl Acad. Sci. USA. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duffy B, Schouten A, Raaijmakers JM. Pathogen self-defense: mechanisms to counteract microbial antagonism. Annu. Rev. Phytopathol. 2003;41:501–538. doi: 10.1146/annurev.phyto.41.052002.095606. [DOI] [PubMed] [Google Scholar]
- 43.Coleman JJ, White GJ, Rodriguez-Carres M, Vanetten HD. An ABC transporter and a cytochrome P450 of Nectria haematococca MPVI are virulence factors on pea and are the major tolerance mechanisms to the phytoalexin pisatin. Mol. Plant Microbe Int. 2011;24:368–376. doi: 10.1094/MPMI-09-10-0198. [DOI] [PubMed] [Google Scholar]
- 44.Wang RF, et al. Enzymatic transformation of vina-ginsenoside R(7) to rare notoginsenoside ST-4 using a new recombinant glycoside hydrolase from Herpetosiphon aurantiacus. Appl. Microbiol. Biot. 2015;99:3433–3442. doi: 10.1007/s00253-015-6446-z. [DOI] [PubMed] [Google Scholar]
- 45.Coleman J, Blake-Kalff M, Davies E. Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci. 1997;2:144–151. doi: 10.1016/S1360-1385(97)01019-4. [DOI] [Google Scholar]
- 46.Windsor B, Roux SJ, Lloyd A. Multiherbicide tolerance conferred by AtPgp1 and apyrase overexpression in Arabidopsis thaliana. Nat. Biotechnol. 2003;21:428–433. doi: 10.1038/nbt809. [DOI] [PubMed] [Google Scholar]
- 47.Campbell EJ, et al. Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 2003;133:1272–1284. doi: 10.1104/pp.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delcher, A. L., Salzberg, S. L. & Phillippy, A. M. Using MUMmer to identify similar regions in large sequence sets in Current protocols in bioinformatics (ed. Baxevanis, A. D.) Chapter 10, Unit 10.13 (2003). [DOI] [PubMed]
- 49.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences in Current protocols in bioinformatics (ed. Baxevanis, A. D.) Chapter 4, Unit 4.10 (2009). [DOI] [PubMed]
- 51.Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenetic Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 52.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:265–268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zdobnov EM, Apweiler R. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 55.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:226–232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 61.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 62.Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0–a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Z, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics, Proteomics and Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin Y, et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khaldi N, et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Win J, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 70.Yang M, et al. Autotoxic ginsenosides in the rhizosphere contribute to the replant failure of Panax notoginseng. PLOS One. 2015;10:e0118555. doi: 10.1371/journal.pone.0118555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw genome sequence data have been deposited in the Short Read Archive (SRA) at NCBI under accession number SRR3386345 (PRJNA318145). Raw RNA-seq data have been deposited in the SRA under accession number SRP111895.