Abstract
Studies of the cellular autophagy pathway have exploded over the past twenty years. Now appreciated as a constitutive degradative mechanism that promotes cellular homeostasis, autophagy is also required for a variety of developmental processes, cellular stress responses, and immune pathways. Autophagy certainly acts as both an anti-viral and pro-viral pathway, and the roles of autophagy depend on the virus, the cell type, and the cellular environment. The goal of this review is to summarize, in brief, what we know so far about the relationship between autophagy and viruses, particularly for those who are not familiar with the field. With a massive amount of relevant published data, it is simply not possible to be comprehensive, or to provide a complete “parade of viruses”, and apologies are offered to researchers whose work is not described herein. Rather, this review is organized around general themes regarding the relationship between autophagy and animal viruses.
Keywords: Autophagy, virus, amphisome, endosome, Beclin 1, IFN, SNARE
Observation of autophagosomes during infection
In 1965, George Palade's group published electron microscopy images of poliovirus-infected HeLa cells. (Dales et al., 1965) Late in infection, they identified vesicles with two lipid bilayers. Between the bilayers was an electron-light lumen, and the region within the inner bilayer appeared to be cytosol-like. Dales and Palade observed apparent virions near, and often within, these vesicles. The authors suggested these resembled “autolytic vesicles,” which “appear to represent a secondary response to infection.” Similar vesicles, termed “compound membrane vesicles,” were later identified in images of Coxsackievirus-infected mouse pancreata, indicating that these structures were present during infection of multiple picornaviruses. (Burch and Harb, 1979; Harb and Burch, 1975)
These unique, double-membraned, “autolytic vesicles” are now better known as autophagosomes, the hallmark organelles of the cellular autophagy pathway. (Carlsson and Simonsen, 2015) Autophagy is a process of cellular homeostasis and stress response, in which cytosolic cargo is engulfed by forming autophagosomes and degraded when these vesicles fuse with lysosomes. (Figure 1) Autophagy is a constitutive pathway, but the amount of degradation varies by cell type, availability of nutrients, environment, or other cellular stresses, including infection. (Galluzzi et al., 2014) Autophagy itself provides a survival mechanism during cell starvation, and plays crucial roles in development, post-natal survival, and the immune response. (Choi et al., 2013) Examination of the immune response led to early studies genetically linking the autophagic pathway and the cellular response to virus infection.
Figure 1. Viral regulation of the autophagic pathway.
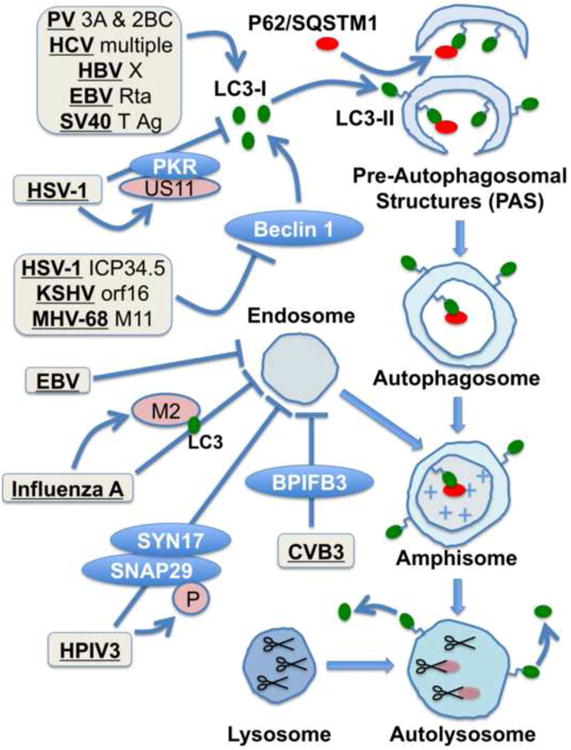
Autophagy is initiated when a signal is sent, through the Beclin 1 complex and other signaling complexes, to convert the cellular LC3 protein from its non-lipidated LC3-I form to phosphotidylamine-conjugated LC3-II. The newly lipidated LC3-II localizes to cup-shaped Pre-Autophagosomal Structures (PAS), and is required for the PAS to self-fuse into double-membraned autophagosomes with cytosolic contents. Autophagosomes fuse with endosomes, which provide vacuolar ATPases and promote acidification of the newly formed vesicle, termed an amphisome. Acidic amphisomes fuse with lysosomes, delivering proteases for degradation of internal contents. LC3-II is recycled to LC3-I, so it is difficult to monitor flux through the pathway using LC3 lipidation. The autophagic cargo adapter p62/SQSTM1 directly interacts with LC3, which directs its localization to PAS. LC3-I and LC3-II can be distinguished by western blot, but the recycling pathway makes it difficult to use LC3-II levels to monitor autophagy. However, lower steady state levels of p62 indicate higher levels of active autophagic degradation. Many of the known virus-encoded activators of autophagic signaling, and in some cases active autophagy, are listed, although the mechanisms of activation are poorly understood. Many other viruses induce autophagic signaling, but the specific proteins which provide the signal are unknown. Several viruses are known to inhibit either autophagic signaling, or autophagic maturation, as shown. HSV-1 US11 protein inhibits autophagic signaling through inhibition of PKR. HSV-1 ICP34.5, KSHV orf16, and MHV-68 M11 inhibit autophagosome formation by binding to the Beclin 1 autophagy signaling molecule. Influenza A, CVB3, HPIV3, and EBV induce autophagosomes, but block autophagosome maturation and autophagic degradation. Influenza M2 protein binds to LC3 to inhibit amphisome formation. The HPIV3-encoded P protein binds to SNAP29, inhibiting endosome-autophagsome fusion. In the case of CVB3, the virus requires host BPIFB3 to inhibit vesicle maturation.
Autophagy as an anti-viral response
The autophagy field came to the spotlight with identification of mutant strains of Saccharomyces cerevisae which are sensitive to starvation. (Harding et al., 1995; Tsukada and Ohsumi, 1993) The comprehensive set of genes identified in these studies moved autophagy from a largely descriptive field to one with the tools to dissect the genetics and molecular biology of the pathway. While the primary sequence of autophagy genes rarely suggested a cellular function, mammalian homologues began to be identified. In one case, a binding partner of the anti-apoptotic regulatory protein Bcl-2, Beclin 1, was identified as a homologue of the gene now known as yeast ATG6. (Liang et al., 1999; 1998) Beclin 1, when expressed from a recombinant Sindbis Virus, reduced virus load and protected mice from fatal encephalitis.
Subsequent work extended the idea of Beclin 1 as a main regulator of an anti-viral response. Several viruses encode a Beclin 1-binding protein, such as a Herpes Simplex Virus 1 protein, ICP34.5, and viruses lacking this domain are less pathogenic in a mouse encephalitis model. (Orvedahl et al., 2007) ICP34.5 binding to Beclin 1 inhibits the formation of autophagosomes in neurons, suggesting that the virus has evolved to actively inhibit autophagy. Other viral proteins inhibiting through Beclin 1 binding include Bcl-2 homologs, such as the KSHV orf16 protein and the MHV-68 M11 protein. (Ku et al., 2008; Su et al., 2014) In addition to ICP34.5′s Beclin 1 binding, HSV also encodes the US11 protein, which inhibits autophagy through direct interaction with the PKR kinase, indicating that HSV encodes at least two proteins capable of inhibiting autophagy, which may speak to the importance of autophagy as an antiviral against this particular virus. (Lussignol et al., 2013)
The hypothesis that a cellular process dedicated to degrading cytosolic contents would also engulf and destroy pathogens has proven to be true for several pathogens, and the process has been termed “xenophagy.” (Paulus and Xavier, 2015) One effector of xenophagy, the STimulator of INterferon Genes (STING), a transmembrane protein, senses dsDNA viruses and targets them for autophagic degradation. (Reviewed in (Barber, 2014)) Movement of STING itself within a cell is dependent on components of the autophagy machinery. (Ishikawa et al., 2009) STING also induces type I IFN, which indicates a role for autophagy in cell-to-cell immune signaling. Degradation of viral antigens by xenophagy can feed into the MHC Class II presentation pathway as well, indicating that xenophagy can play a role in both clearing a cell of pathogens and prolonged presentation of peptides from those pathogens. (Paludan et al., 2005)
Autophagy and the immune response
The anti-viral activity of autophagy in systemic immunity depends on the overall environment of the host. Toll-like receptor-dependent autophagy is required for an antiviral response against Rift Valley fever virus, Vesicular Stomatitis Virus, and others. (Moy et al., 2014) The machinery of autophagy is required for a successful IFNγ response against murine norovirus (MNV), although this effect does not involve autophagic degradation. (Hwang et al., 2012) Anti-viral autophagic states can be induced through cell to cell signaling as well. Evidence suggests that placental trophoblasts protect the fetus from viruses by secreting exosomes, which contain miRNAs from the chromosome 19 cluster that are capable of inducing autophagy in neighboring cells, conferring anti-viral resistance. (Delorme-Axford et al., 2013) In this case, autophagy is anti-viral even for viruses which have been shown to be resistant to the anti-viral effects of autophagy, suggesting there may be something cell-type specific about nature of the relationship of viruses and the autophagic machinery.
Pathogen subversion of autophagy
The idea of xenophagy derived from early work showing that some bacterial pathogens, including group A Streptococcus, Mycobacterium tuberculosis, and others have increased intracellular survival when autophagy is suppressed. (Reviewed in (J. Huang and Brumell, 2014)) However, studies of Legionella pneumophila first revealed that some bacteria can manipulate, avoid, and sometimes subvert the autophagy pathway. Legionella can be enveloped in autophagosomes, but the bacterium actively inhibits autophagosome formation, while also restricting the maturation of any autophagosomes that do engulf the bacterium, allowing bacterial replication in the autophagosome. (Amer and Swanson, 2005) Many other bacteria have evolved mechanisms to subvert autophagy to the benefit of the pathogen. (J. Huang and Brumell, 2014) Thus, when autophagosomes are observed in response to an intracellular invader, the first question must be whether it is a successful host response or whether the vesicles are specifically triggered by the pathogen.
Viral proteins that trigger the autophagy pathway
In the case of picornaviruses, autophagosomes are specifically triggered by two viral proteins. Co-expression of the poliovirus proteins 2BC and 3A induces double-membraned vesicle formation, similar at the EM level to those seen during infection. 2BC alone induces LC3 modification but not DMVs, while 3A causes ER swelling but no vesicles or LC3 modification. (Suhy et al., 2000; Taylor and Kirkegaard, 2007) The requirement for dual expression makes it unlikely that immune recognition of either foreign protein leads to an autophagic response. It has since been shown that several viruses express proteins that specifically induce autophagic signaling, including Hepatitis B Virus X protein, Epstein Barr Virus Rta transcription factor, SV40 T antigen, and three Hepatitis C Virus proteins: NS4B, NS5A, and. NS5B. (Guévin et al., 2010; Hung et al., 2014; Liu et al., 2014; Mao et al., 2011; Shrivastava et al., 2012; Su et al., 2011; Tang et al., 2009; Zhang et al., 2014) Multiple RNA viruses, including Hepatitis C Virus and HIV, target the cellular protein immunity-associated GTPase family M (IRGM) to induce autophagic signaling, if not degradative autophagy. (Grégoire et al., 2012) These data indicate a common evolutionary selection across divergent viruses for viral signaling to the autophagic pathway.
The vesicles induced by PV infection, or viral protein expression, were studied using fractionation of infected cells were found to contain markers from many organelles, which is a hallmark of autophagosomes. (Suhy et al., 2000) Later, the identification of cellular LC3 as a specific marker of autophagosomes allowed confirmation of an autophagic origin for PV-induced vesicles. (Jackson et al., 2005) As the autophagic origin of the vesicles became clear, it also became clear that autophagy benefits polio and many other viruses. An increase in cellular autophagy increases virus production, and autophagic inhibition decreases viral load. (Jackson et al., 2005) This is true for other picornaviruses, and a large number of viruses, particularly positive-strand RNA viruses, have been shown to benefit from some aspect of the autophagic pathway. (S.-C. Huang et al., 2009; Klein and Jackson, 2011; O'Donnell et al., 2011; Yoon et al., 2008) For poliovirus, the DMVs can act as a substrate for RNA replication, although they play other important roles, discussed below. For Hepatitis C Virus (HCV), autophagy appears be required for initiation of, but not maintenance of, viral replication. (Dreux et al., 2009) For dengue virus (DENV), viral entry, translation, and replication have all been linked to the autophagic pathway. (Panyasrivanit et al., 2009) Autophagy of lipids (lipophagy) also provides metabolic energy for DENV replication. (Heaton and Randall, 2010)
Induction of autophagic signaling vs. bona fide autophagy
The finding that genuine degradative autophagy benefits DENV demonstrated that active degradation, not just proliferation of membranes, might be important for promoting the replication of some viruses. This is also true for HCV, which induces degradative autophagy, one benefit of which is the inhibition of RIG-I-mediated IFN-γ activation. (Ke and Chen, 2011) DENV itself may also benefit from degradation of immune signals through autophagy. (Ke and Chen, 2011) The benefits of autophagic degradation during infection are the topic of a recent review. (Richards and Jackson, 2013) In several cases, it has been shown that closely related viruses have very different effects on autophagic degradation vs. signaling. Coxsackievirus, for example, induces autophagic signaling and the generation of autophagosomes, but there is little to no degradation of autophagosome contents during infection. (Kemball et al., 2010; Wong et al., 2008) It has been reported that in some circumstances the best-studied cargo adapter for autophagic degradation, p62, is cleaved by a viral protease, although other studies report that p62 remains intact. (Shi et al., 2013) This may be a cell-type specificity issue. However, studies agree that autophagic degradation is inhibited during CVB3 infection. In contrast, poliovirus induces complete autophagy, including lysosome-dependent degradation of p62. (Richards and Jackson, 2012) One cellular target used by CVB3 to inhibit degradation, the bactericidal/permeability-increasing protein (BPI) fold-containing family B, member 3 (BPIFB3), was recently identified in an siRNA screen for factors. (Coyne et al., 2011) Silencing of BPIFB3 enhances both autophagic degradation and replication of CVB3, but does not affect PV replication. (Figure 1) (Delorme-Axford et al., 2014) While this indicates there are specific targets for inhibiting degradation, it paradoxically indicates that the virus is limiting its own replication, relative to other picornaviruses, by inhibiting autophagic degradation.
Two related herpesviruses also have different relationships with autophagic degradation. As with HSV, the herpesvirus Varicella Zoster Virus (VZV) induces an autophagic response during infection. (Carpenter et al., 2011; Takahashi et al., 2009) However, the outcome of this response is quite different. HSV encodes two factors, ICP34.5 and US11, to inhibit autophagic signaling, but VZV does not encode homologues for either of these proteins, and VZV infection induces degradative autophagy. (Buckingham et al., 2014) The role of degradative autophagy in VZV replication is unclear, but the radically different relationship of two herpesviruses to autophagy suggests that VZV has, at a minimum, evolved a strategy to avoid the negative effects of autophagic degradation. Whether these herpesviruses encode a specific inducer of autophagy, or, as has been suggested, autophagy is a general anti-herpesvirus response, is not yet known.
Some paramyxoviruses have been shown to induce autophagy and usurp the pathway to promote virus production. (Manuse et al., 2010) One paramyxovirus, Human Parainflueza Virus 3 (HPIV3), specifically inhibits autophagosome maturation during infection. The virus encodes a protein, P, which is central to viral RNA replication. During normal autophagosome maturation, the intermediate SNARE protein SNAP29 mediates the interaction of the autophagosomal SNARE Syntaxin17 (SYN17) with the endosome/lysosome SNARE VAMP8. (Ding et al., 2014) HPIV3 P protein binds to SNAP29, disrupting the SNARE interactions and preventing autophagosome maturation. (Figure 1) P protein expression is sufficient to inhibit autophagosome maturation, indicating that SNAP29 inhibition is a powerful mechanism for blocking autophagy.
Autophagic Exit Without Lysis (AWOL)
During the initial demonstration that poliovirus subverts autophagy to promote its own growth, the Kirkegaard group observed that autophagy levels affect pre-lytic release of virus more than intracellular virus yields. (Jackson et al., 2005) The significance of this was unclear at the time; however, the phenomenon was named Autophagic exit With Out Lysis, or AWOL. Later, studies of Pichia Pastoris and Dictyostelium discoideum showed that autophagy promotes a non-canonical secretory pathway. (Duran et al., 2010; Manjithaya et al., 2010) This pathway is ordinarily minor, but can be detected with sensitive bioassays, such as the sporulation response of P. Pastoris and D. discoideum to concentrated conditioned medium, or detection of individual plaque forming units of poliovirus by HeLa cell plaque assay. The discovery of an autophagic secretory pathway opened the possibility that picornaviruses might be utilizing the autophagic secretion pathway for cell exit. However, there was a problem of topology - how would a non-enveloped virus escape three lipid bilayers (two in the autophagosome, and a third from the cellular membrane) to exit without a membrane? An explanation would not appear for another few years. First, researchers focused on another question - is there any evidence that viruses which subvert autophagy spend time within autophagosomes as part of a successful infection strategy?
The role of autophagy in virion maturation and egress
Two studies, one of the enveloped Dengue Virus, and one of the supposedly non-enveloped poliovirus, demonstrated a direct role for autophagy in the maturation of the virus particle into an infectious virus. In the case of Dengue, the cleavage of the viral structural protein prM into pr and M by cellular Furin is required for generation of infectious virus. If autophagy is inhibited, release of the pr peptide from the virion is reduced. (Mateo et al., 2013) For poliovirus, the cleavage of the viral structural protein VP0 into VP2 and VP4, which occurs after the genome-containing virion has been assembled, is required to to generate infectious virus. This event is inhibited if acidification of cellular vesicles is blocked, and autophagosomes are known to become acidic. (Richards et al., 2014) However, autophagosomes also provide substrates for viral RNA replication, so there are clearly multiple roles for autophagy in picornavirus production. (Belov et al., 2012; Richards et al., 2014; Richards and Jackson, 2012) Any hypothesis proposing autophagic release of virus still had to overcome the aforementioned topology problem, however.
The picornavirus field was fundamentally changed by the findings of the Lemon group in 2013, when they showed that Hepatitis A Virus (HAV), a non-lytic picornavirus, is not a purely non-enveloped virus as had been previously thought, but can be released in vesicular packets. For HAV this process is not dependent on autophagy, but by an endosomal-sorting complex required for transport (ESCRT)-dependent release pathway. (Feng et al., 2013) Quickly the field has come to accept that picornaviruses are not entirely non-enveloped, and that membrane-bound packets of virus may even be the primary form of the virus, particularly for cell-to-cell transmission within a host. (Feng and Lemon, 2014) This solved the topology problem, as fusion of the double-membranes autophagosome with the plasma membrane could release a single-membraned vesicle. A subsequent study of Coxsackievirus B3 showed that virus is released in extracellular microvesicles (EMVs) containing the lipidated form of the autophagy marker LC3, indicating that these EMVs derive from the autophagy pathway. (Robinson et al., 2014) For both poliovirus and the negative stranded, conventionally enveloped morbilliviruses, non-lytic spread from cell to cell depends on cellular autophagy machinery. (Bird et al., 2014; Delpeut et al., 2012)
Autophagic membranes in classical viral envelope formation
The large DNA tumor virus Hepatitis B Virus (HBV) is associated with hepatocellular carcinomas. The X protein of HBV binds to, and activates, both phosphatidylinositol-3-kinase class 3 (PI3KC3) and death-associated protein kinase (DAPK), initiating autophagic signaling. (Sir et al., 2010; Zhang et al., 2014) However, autophagic degradation is inhibited. (Liu et al., 2014) The autophagic membranes generated during infection are used for viral envelopment. (Li et al., 2011) Recently, studies revealed a role for autophagy in HBV pathogenesis. Autophagy promotes degradation of cellular microRNA-224, whose overexpression is associated with hepatocellular carcinoma. (Lan et al., 2014a) Autophagy specifically has been shown to suppress HBV-associated tumor formation by degrading microRNA-224. (Lan et al., 2014b)
Recent studies of Epstein-Barr virus (EBV) indicate that the virus also subverts the autophagy pathway to physically generate viral membranes. EBV, which is a large enveloped DNA virus, takes from the cellular membrane pool to build a viral membrane. In the past several months, it was shown that the Rta transcription factor of EBV promotes expression of autophagy genes. (Hung et al., 2014) However, while autophagosomes are induced during EBV infection, they do not fuse with lysosomes, and therefore, degradative autophagy is inhibited. (Granato et al., 2014) Autophagy promotes viral packaging and assembly, and LC3 is found in viral particles, indicating that EBV subversion of autophagic machinery generates an envelope of the virion. (Nowag et al., 2014)
A role for viroporins
The Matrix 2 Protein (M2) of influenza virus is required for virion morphogenesis. (Nayak et al., 2009) M2 is sufficient to inhibit the fusion of autophagosomes with lysosomes. (Gannagé et al., 2009) M2 also contains an LC3-interacting domain, which is required for influenza virus' subversion of autophagy. (Beale et al., 2014) Presumably, then, M2 inhibits autophagosome-lysosome fusion through its activity as a viroporin, the same activity required for disassembly during viral entry. However, the LC3-interacting motif of M2 is in the cytoplasmic tail domain, not the viroporin domain. Rotavirus NSP4 induces release of calcium from the ER lumen, triggering autophagic signaling, an effect dependent upon a calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) and 5′ adenosine monophosphate-activated protein kinase (AMPK). (Crawford et al., 2012) It should be noted, however, that rotavirus does not appear to induce formation of classical autophagosome structures, so the autophagy machinery may play a role in rotavirus replication independent of double-membraned vesicle formation. (Arnoldi et al., 2014) Poliovirus 3A and 2BC have also been reported to be viroporins, although co-expression of both is required to induce autophagosomes. (Suhy et al., 2000)
The complex relationship between HIV and autophagy
In this review, some viruses have been discussed for which autophagy is an anti-viral response. Other viruses have been discussed for which autophagy promotes viral replication. For perhaps the most-studied virus of the past several years, Human Immunodeficiency Virus (HIV), both pro- and anti- viral roles for autophagy have been proposed. Doing justice to the literature regarding HIV and autophagy would double the length of this article. Separate reviews on this topic have appeared, and the interested reader is directed to an excellent recent article for a much more detailed analysis. (Dinkins et al., 2014) However, a brief survey of the HIV-autophagy literature is informative with respect to the very different relationships viruses and autophagy have depending on the cellular context.
HIV infection has been shown to both induce (Denizot et al., 2008; Espert et al., 2006; Meng et al., 2013; Wang et al., 2012) and inhibit (Van Grol et al., 2010; Zhou and Spector, 2008) autophagy. The disparities in the data are almost certainly due to differences between cell types. (Borel et al., 2012; Killian, 2012) In a broad sense, the induction of autophagy has been shown to be both a positive and negative influence on HIV infection. In some cases, autophagy has been shown to inhibit viral replication and spread. In long-term nonprogressor patients, enhanced levels of autophagic markers are found, suggesting a role for autophagy in keeping the virus at bay. (Sagnier et al., 2014) In CD4+ T lymphocytes, autophagy selectively degrades HIV Tat protein, restricting viral replication. (Nardacci et al., 2014) However, it has also been shown that knockdown of autophagy factors in T-cell lines can inhibit HIV replication. (Eekels et al., 2012) Certainly there is a complex relationship between HIV and autophagy, and the nature of the interactions between virus and pathway depend on the cell and the circumstances. This may be true for most viruses, which are often studied in easy-to-grow cell lines. The role of autophagy may be very different in cells with direct relevance to in vivo infection.
Summary
As a constitutive, cytosolic, degradative process, autophagy can directly degrade intracellular pathogens. However, studies have revealed that there are myriad other ways in which the autophagic machinery, and autophagic degradation, can either inhibit or actively promote virus production. Our understanding of the multi-layered relationship between viruses and autophagy is sure to expand in the next several years to reveal host-cell interactions and mechanisms for viral avoidance and subversion of the pathway that are impossible to even consider today. As a pathway for which approved therapeutics already exist, autophagy will no doubt provide a rich source of targets for anti-viral treatments in the coming years.
Autophagy acts as both an anti-viral and pro-viral pathway.
The roles of autophagy depend on the virus, the cell type, and the cellular environment.
What we know so far about the relationship between autophagy and viruses is summarized.
Viral proteins known to induce or inhibit the autophagy pathway are highlighted.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amer AO, Swanson MS. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol. 2005;7:765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoldi F, De Lorenzo G, Mano M, Schraner EM, Wild P, Eichwald C, Burrone OR. Rotavirus Increases Levels of Lipidated LC3 Supporting Accumulation of Infectious Progeny Virus without Inducing Autophagosome Formation. PLoS ONE. 2014;9:e95197. doi: 10.1371/journal.pone.0095197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe. 2014;15:239–247. doi: 10.1016/j.chom.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird SW, Maynard ND, Covert MW, Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci USA. 2014;111:13081–13086. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel S, Espert L, Biard-Piechaczyk M. Macroautophagy Regulation during HIV-1 Infection of CD4+ T Cells and Macrophages. Front Immunol. 2012;3:97. doi: 10.3389/fimmu.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham EM, Carpenter JE, Jackson W, Zerboni L, Arvin AM, Grose C. Autophagic flux without a block differentiates varicella-zoster virus infection from herpes simplex virus infection. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1417878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch GE, Harb JM. Electron microscopic studies of viral pancreatitis in coxsackie B4 virus infected mice. Exp Mol Pathol. 1979;31:23–35. doi: 10.1016/0014-4800(79)90004-2. [DOI] [PubMed] [Google Scholar]
- Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. J Cell Sci. 2015 doi: 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- Carpenter JE, Jackson W, Benetti L, Grose C. Autophagosome formation during varicella-zoster virus infection following endoplasmic reticulum stress and the unfolded protein response. J Virol. 2011;85:9414–9424. doi: 10.1128/JVI.00281-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AMK, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Coyne CB, Bozym R, Morosky SA, Hanna SL, Mukherjee A, Tudor M, Kim KS, Cherry S. Comparative RNAi screening reveals host factors involved in enterovirus infection of polarized endothelial monolayers. Cell Host Microbe. 2011;9:70–82. doi: 10.1016/j.chom.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Hyser JM, Utama B, Estes MK. Autophagy hijacked through viroporin-activated calcium/calmodulin-dependent kinase kinase-β signaling is required for rotavirus replication. Proc Natl Acad Sci USA. 2012;109:E3405–13. doi: 10.1073/pnas.1216539109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S, Eggers HJ, Tamm I, Palade GE. Electron Mcroscopic Study of the Formation of Poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE, Sadovsky Y, Coyne CB. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci USA. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E, Morosky S, Bomberger J, Stolz DB, Jackson WT, Coyne CB. BPIFB3 regulates autophagy and coxsackievirus B replication through a noncanonical pathway independent of the core initiation machinery. MBio. 2014;5:e02147. doi: 10.1128/mBio.02147-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpeut S, Rudd PA, Labonté P, von Messling V. Membrane fusion-mediated autophagy induction enhances morbillivirus cell-to-cell spread. J Virol. 2012;86:8527–8535. doi: 10.1128/JVI.00807-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot M, Varbanov M, Espert L, Robert-Hebmann V, Sagnier S, Garcia E, Curriu M, Mamoun R, Blanco J, Biard-Piechaczyk M. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy. 2008;4:998–1008. doi: 10.4161/auto.6880. [DOI] [PubMed] [Google Scholar]
- Ding B, Zhang G, Yang X, Zhang S, Chen L, Yan Q, Xu M, Banerjee AK, Chen M. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe. 2014;15:564–577. doi: 10.1016/j.chom.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Dinkins C, Pilli M, Kehrl JH. Roles of autophagy in HIV infection. Immunol Cell Biol. 2014 doi: 10.1038/icb.2014.88. [DOI] [PubMed] [Google Scholar]
- Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eekels JJ, Sagnier S, Geerts D, Jeeninga RE, Biard-Piechaczyk M, Berkhout B. Inhibition of HIV-1 replication with stable RNAi-mediated knockdown of autophagy factors. Virol J. 2012;9:69. doi: 10.1186/1743-422X-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Lemon SM. Peek-a-boo: membrane hijacking and the pathogenesis of viral hepatitis. Trends Microbiol. 2014;22:59–64. doi: 10.1016/j.tim.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic Control of Autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, García-Sastre A, Münz C. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, Santarelli R, Farina A, Gonnella R, Lotti LV, Faggioni A, Cirone M. Epstein-barr virus blocks the autophagic flux and appropriates the autophagic machinery to enhance viral replication. J Virol. 2014;88:12715–12726. doi: 10.1128/JVI.02199-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégoire IP, Rabourdin-Combe C, Faure M. Autophagy and RNA virus interactomes reveal IRGM as a common target. Autophagy. 2012;8:1136–1137. doi: 10.4161/auto.20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guévin C, Manna D, Bélanger C, Konan KV, Mak P, Labonté P. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010;405:1–7. doi: 10.1016/j.virol.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb JM, Burch GE. Spherical aggregates of coxsackie B4 virus particles in mouse pancreas. Beitr Pathol. 1975;156:122–127. doi: 10.1016/s0005-8165(75)80145-4. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Chang CL, Wang PS, Tsai Y, Liu HS. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J Med Virol. 2009;81:1241–1252. doi: 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CH, Chen LW, Wang WH, Chang PJ, Chiu YF, Hung CC, Lin YJ, Liou JY, Tsai WJ, Hung CL, Liu ST. Regulation of Autophagic Activation by Rta of Epstein-Barr Virus via the Extracellular Signal-Regulated Kinase Pathway. J Virol. 2014;88:12133–12145. doi: 10.1128/JVI.02033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, Green KY, López-Otín C, Xavier RJ, Thackray LB, Virgin HW. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke PY, Chen SSL. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol. 2010;84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian MS. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res Ther. 2012;9:16. doi: 10.1186/1742-6405-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KA, Jackson WT. Human rhinovirus 2 induces the autophagic pathway and replicates more efficiently in autophagic cells. J Virol. 2011;85:9651–9654. doi: 10.1128/JVI.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy-preferential degradation of MIR224 participates in hepatocellular carcinoma tumorigenesis. Autophagy. 2014a;10 doi: 10.4161/auto.29959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014b;59:505–517. doi: 10.1002/hep.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319–6333. doi: 10.1128/JVI.02627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, Liu W. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10:416–430. doi: 10.4161/auto.27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussignol M, Queval C, Bernet-Camard MF, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J Virol. 2013;87:859–871. doi: 10.1128/JVI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuse MJ, Briggs CM, Parks GD. Replication-independent activation of human plasmacytoid dendritic cells by the paramyxovirus SV5 Requires TLR7 and autophagy pathways. Virology. 2010;405:383–389. doi: 10.1016/j.virol.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Da L, Tang H, Yang J, Lei Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun. 2011;415:68–74. doi: 10.1016/j.bbrc.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Mateo R, Nagamine CM, Spagnolo J, Méndez E, Rahe M, Gale M, Yuan J, Kirkegaard K. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol. 2013;87:1312–1321. doi: 10.1128/JVI.02177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Zhang Z, Xu K, Qi G. HIV-1 gp120 induces autophagy in cardiomyocytes via the NMDA receptor. Int J Cardiol. 2013;167:2517–2523. doi: 10.1016/j.ijcard.2012.06.067. [DOI] [PubMed] [Google Scholar]
- Moy RH, Gold B, Molleston JM, Schad V, Yanger K, Salzano MV, Yagi Y, Fitzgerald KA, Stanger BZ, Soldan SS, Cherry S. Antiviral autophagy restrictsRift Valley fever virus infection and is conserved from flies to mammals. Immunity. 2014;40:51–65. doi: 10.1016/j.immuni.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardacci R, Amendola A, Ciccosanti F, Corazzari M, Esposito V, Vlassi C, Taibi C, Fimia GM, Del Nonno F, Ippolito G, D'Offizi G, Piacentini M. Autophagy plays an important role in the containment of HIV-1 in nonprogressor-infected patients. Autophagy. 2014;10:1167–1178. doi: 10.4161/auto.28678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowag H, Guhl B, Thriene K, Romao S, Ziegler U, Dengjel J, Münz C. Macroautophagy Proteins Assist Epstein Barr Virus Production and Get Incorporated Into the Virus Particles. EBioMedicine. 2014;1:116–125. doi: 10.1016/j.ebiom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell V, Pacheco JM, LaRocco M, Burrage T, Jackson W, Rodriguez LL, Borca MV, Baxt B. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology. 2011;410:142–150. doi: 10.1016/j.virol.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Linking dengue virus entry and translation/replication through amphisomes. Autophagy. 2009;5:434–435. doi: 10.4161/auto.5.3.7925. [DOI] [PubMed] [Google Scholar]
- Paulus GLC, Xavier RJ. Autophagy and checkpoints for intracellular pathogen defense. Curr Opin Gastroenterol. 2015;31:14–23. doi: 10.1097/MOG.0000000000000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jackson WT. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog. 2012;8:e1003046. doi: 10.1371/journal.ppat.1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jackson WT. How positive-strand RNA viruses benefit from autophagosome maturation. J Virol. 2013;87:9966–9972. doi: 10.1128/JVI.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Soares-Martins JAP, Riddell GT, Jackson WT. Generation of unique poliovirus RNA replication organelles. MBio. 2014;5 doi: 10.1128/mBio.00833-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, Williams W, Kha N, Cruz C, Hancock BM, Nguyen DP, Sayen MR, Hilton BJ, Doran KS, Segall AM, Wolkowicz R, Cornell CT, Whitton JL, Gottlieb RA, Feuer R. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014;10:e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagnier S, Daussy CF, Borel S, Robert-Hebmann V, Faure M, Blanchet FP, Beaumelle B, Biard-Piechaczyk M, Espert L. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol. 2014 doi: 10.1128/JVI.02174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wong J, Piesik P, Fung G, Zhang J, Jagdeo J, Li X, Jan E, Luo H. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy. 2013;9:1591–1603. doi: 10.4161/auto.26059. [DOI] [PubMed] [Google Scholar]
- Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol. 2012;86:8705–8712. doi: 10.1128/JVI.00616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D, Ann DK, Ou JHJ. Autophagy by hepatitis B virus and for hepatitis B virus. Autophagy. 2010;6:548–549. doi: 10.4161/auto.6.4.11669. [DOI] [PubMed] [Google Scholar]
- Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S. Targeting γ-herpesvirus 68 Bcl-2-mediated down-regulation of autophagy. J Biol Chem. 2014;289:8029–8040. doi: 10.1074/jbc.M113.515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MMC. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol. 2011;85:10561–10571. doi: 10.1128/JVI.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy DA, Giddings TH, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi MN, Jackson W, Laird DT, Culp TD, Grose C, Haynes JI, Benetti L. Varicella-zoster virus infection induces autophagy in both cultured cells and human skin vesicles. J Virol. 2009;83:5466–5476. doi: 10.1128/JVI.02670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Van Grol J, Subauste C, Andrade RM, Fujinaga K, Nelson J, Subauste CS. HIV-1 inhibits autophagy in bystander macrophage/monocytic cells through Src-Akt and STAT3. PLoS ONE. 2010;5:e11733. doi: 10.1371/journal.pone.0011733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gao Y, Tan J, Devadas K, Ragupathy V, Takeda K, Zhao J, Hewlett I. HIV-1 and HIV-2 infections induce autophagy in Jurkat and CD4+ T cells. Cell Signal. 2012;24:1414–1419. doi: 10.1016/j.cellsig.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kweon HS, Lee JY, Kim DH. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol. 2008;82:11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HT, Chen GG, Hu BG, Zhang ZY, Yun JP, He ML, Lai PBS. Hepatitis B virus x protein induces autophagy via activating death-associated protein kinase. J Viral Hepat. 2014;21:642–649. doi: 10.1111/jvh.12191. [DOI] [PubMed] [Google Scholar]
- Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]


