Abstract
ATP sulfurylase and 5′-adenylylsulfate (APS) reductase catalyze two reactions in the sulfate assimilation pathway. Cell fractionation of Arabidopsis leaves revealed that ATP sulfurylase isoenzymes exist in the chloroplast and the cytosol, whereas APS reductase is localized exclusively in chloroplasts. During development of Arabidopsis plants the total activity of ATP sulfurylase and APS reductase declines by 3-fold in leaves. The decline in APS reductase can be attributed to a reduction of enzyme during aging of individual leaves, the highest activity occurring in the youngest leaves and the lowest in fully expanded leaves. By contrast, total ATP sulfurylase activity declines proportionally in all the leaves. The distinct behavior of ATP sulfurylase can be attributed to reciprocal expression of the chloroplast and cytosolic isoenzymes. The chloroplast form, representing the more abundant isoenzyme, declines in parallel with APS reductase during aging; however, the cytosolic form increases over the same period. In total, the results suggest that cytosolic ATP sulfurylase plays a specialized function that is probably unrelated to sulfate reduction. A plausible function could be in generating APS for sulfation reactions.
The sulfate assimilation pathways serve for the synthesis of sulfur compounds necessary for growth and development. Sulfur is assimilated in either the oxidized or the reduced form (Leustek and Saito, 1999). When incorporated in the oxidized form it is used for esterification of a variety of compounds including polysaccharides such as carrageenans and agaroids (McCandless and Craigie, 1979), sulfated flavonoids (Varin et al., 1997), glucosinolates, and many others (Leustek et al., 2000). Alternatively, sulfate is reduced to sulfide and is then incorporated as the thiol group of Cys. Cys is incorporated into proteins, coenzymes, and glutathione and serves as the thiol donor for Met and a multitude of other compounds. Most assimilated sulfur is in the reduced form. Sulfolipids are a special class of compound in which partly reduced sulfur, in the form of sulfite, is incorporated (Mulichak et al., 1999).
Inorganic sulfate is chemically stable and therefore must be activated prior to further metabolism. The activation reaction is catalyzed by ATP sulfurylase (ATP:sulfate adenylyl transferase, EC 2.7.7.4) forming 5′-adenylylsulfate (APS), a mixed anhydride of phosphate and sulfate. APS can be further phosphorylated by APS kinase (EC 2.7.1.25), forming 3′-phospho-5′-adenylylsulfate (PAPS). PAPS is the sulfuryl donor for esterification of metabolites catalyzed by sulfotransferases (Varin et al., 1997). In contrast, APS is used directly as the substrate for the reduction pathway. Reduced glutathione-dependent APS reductase (EC 1.8.99.-) transfers two electrons to form sulfite (Suter et al., 2000). Then ferredoxin-dependent sulfite reductase completes the reduction to sulfide. Cys is formed when sulfide reacts with O-acetyl Ser (OAS) mediated by OAS thiol-lyase (OASTL).
A few preliminary studies document that the activity of the reductive sulfate assimilation pathway varies in plants during development. The highest activities of specific enzymes in this pathway occur in the youngest plant leaves, and the activity declines as leaves mature (Adams and Rinne, 1969; Schmutz and Brunold, 1982; von Arb and Brunold, 1985; von Arb and Brunold, 1986). Similarly, ATP sulfurylase activity is highest in the elongation zone of roots (Cacco et al., 1977). Little is known about the temporal and spatial distribution of the sulfotransferases responsible for sulfate ester synthesis.
Some but not all sulfur assimilation enzymes exist as isoenzymes localized in multiple subcellular compartments. Plastids contain all the enzymes necessary for reductive assimilation of sulfate (Schiff, 1983; Schmidt, 1986). However, all known sulfotransferases that carry out sulfate esterification are cytosolic enzymes (Varin et al., 1997), indicating the need for activated sulfate in the cytosol. A cytosolic isoenzyme of ATP sulfurylase has been reported in some plant species (Lunn et al., 1990; Renosto et al., 1993). Two catalytically distinct forms of ATP sulfurylase have been purified from Euglena gracilis, one of which exists in mitochondria (Li et al., 1991). APS reductase, formerly termed APS sulfotransferase (Suter et al., 2000), was found to be exclusively plastid-localized (Fankhauser and Brunold, 1978; Brunold and Suter, 1989; Rüegsegger and Brunold, 1993). In Euglena an isoform is also localized in mitochondria (Saidha et al., 1988). One form of APS kinase may be localized in plastids (Lee and Leustek, 1998), but there are three genes in Arabidopsis that may encode proteins associated with a membrane system, plasma membrane, endoplasmic reticulum, or mitochondria (Leustek and Saito 1999). Cytosolic and mitochondrial forms of OASTL (Fankhauser and Brunold, 1979; Schmidt, 1986; Lunn et al., 1990) and Ser acetyltransferase (Smith, 1972) have been reported. Since the primary site for reductive sulfate assimilation is the chloroplast, the function of cytosolic and mitochondrial isoenzymes is not certain.
Since the localization of sulfur assimilation has been studied in only a few species it is by no means certain whether the information is generally applicable to all flowering plants. The problem is particularly acute with respect to Arabidopsis, which is the primary source of cloned sulfur assimilation genes (Leustek and Saito, 1999) but from which the subcellular localization of the corresponding enzymes has not been studied experimentally. The gap in understanding has resulted in confusion relating to ATP sulfurylase. Genes encoding four different isoenzymes have been cloned from Arabidopsis (Leustek et al., 1994; Murillo and Leustek, 1995; Hatzfeld et al., 2000), yet all encode enzymes with a characteristic plastid transit peptide. By contrast, two genes are present in potato that can clearly be distinguished as encoding plastid and cytosolic ATP sulfurylases (Klonus et al., 1994). The results suggest that Arabidopsis either does not contain a cytosolic form of ATP sulfurylase or that the gene encoding the cytosolic isoform has not yet been identified. The present study was undertaken to resolve this question. The results of subcellular fractionation indicate that both cytosolic and chloroplast forms of ATP sulfurylase exist in Arabidopsis leaves, whereas APS reductase is exclusively localized within chloroplasts. The localization results confirm that Arabidopsis is similar to other flowering plants. The study also revealed that the plastid form of ATP sulfurylase and APS reductase decline with leaf age. However, the cytosolic form of ATP sulfurylase increases with leaf age. The finding suggests that chloroplast and cytosolic forms of ATP sulfurylase may have specialized functions.
RESULTS
Regulation of ATP Sulfurylase and APS-Reductase in Arabidopsis Leaves during Plant Development
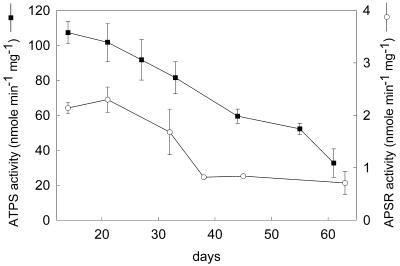
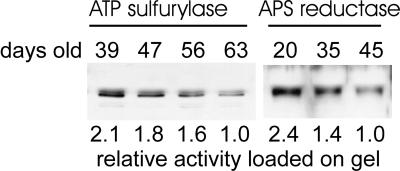
The activity of ATP sulfurylase and APS reductase in leaves was analyzed as plants age to identify the optimal developmental stage for subcellular fractionation experiments. The activity of both sulfur assimilation enzymes in entire shoots of plants at various stages of development was maximal in the youngest plants and declined progressively during development (Fig. 1). ATP sulfurylase activity showed a linear, 3-fold decline between 14 and 61 d after germination. APS reductase also declined during development but not at a steady rate. Its activity was stable between 14 and 21 d, then it fell approximately 3-fold between 21 and 38 d, and the activity stabilized between 39 and 63 d. Thus, both ATP sulfurylase and APS reductase declined approximately 3-fold during development, but the decline in APS reductase was much more rapid and occurred at an earlier time than did ATP sulfurylase. The decline in activity of both enzymes correlated with a decline in the steady-state level of ATP sulfurylase and APS reductase protein as measured by immunoblotting (Fig. 2). The ATP sulfurylase antibody reacts specifically with a doublet of approximately 50 kD and 51.5 kD. The ratio between the two bands varied between samples, but the 50-kD protein was always the most prominent. The APS reductase antibody reacted with a single protein of approximately 50.5 kD.
Figure 1.
ATP sulfurylase (ATPS) and APS reductase (APSR) activity in leaves of Arabidopsis during growth. Each value represents the mean ± sd of six independent measurements.
Figure 2.
Immunoblot analysis of ATP sulfurylase and APS reductase in leaves of Arabidopsis during growth. Representative samples from the experiments shown in Figure 1 were analyzed by immunoblotting. For the ATP sulfurylase blot, 5 μg of total protein was loaded, whereas 15 μg was loaded for APS reductase. The relative enzyme activity loaded onto the gel is indicated below the blot with the lowest activity normalized to a value of 1. For ATP sulfurylase the absolute activity corresponding to the relative value of 1 was 32 nmol min−1 mg−1, whereas for APS reductase it was 0.45 nmol min−1 mg−1. The relative activity values correspond closely with the signal intensity determined by densitometry.
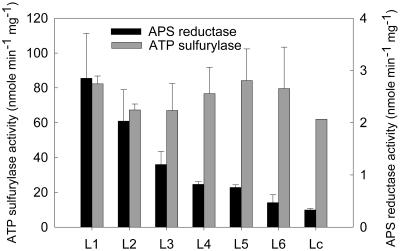
To determine whether the decline in both enzymes is limited to specific leaves or occurs in all leaves ATP sulfurylase and APS reductase were measured in leaves of various ages from 42-d plants. The results in Figure 3 show that ATP sulfurylase activity was approximately the same in the leaves at different stages of development. By contrast, APS reductase was highest in the youngest leaves, and its activity fell sharply in the successively more expanded leaves and fell even further with increasing age. The results from immunoblot analysis correlated with the enzyme activity measurements in that the level of ATP sulfurylase was similar in leaves of various ages, whereas the level of APS reductase protein fell with increasing leaf age (data not shown). Thus, the level of ATP sulfurylase correlates more closely with the age of the entire plant than with the developmental stage of individual leaves. By contrast, APS reductase correlates closely with developmental leaf stage rather than plant age. Taken together with the data shown in Figures 1 and 2 the results indicate that the decline in total APS reductase likely occurs as the mass of the plant shifts progressively to more mature leaves.
Figure 3.
ATP sulfurylase and APS reductase activity in leaves of different developmental stages from 42-d-old Arabidopsis plants. Each value represents the mean ± sd of six independent measurements.
Subcellular Localization of Sulfur Assimilation Enzymes in Shoots of Arabidopsis
In initial subcellular fractionation experiments, 21- and 47-d Arabidopsis plants were selected for analysis. The localization of ATP sulfurylase and APS reductase was studied in the leaves of these plants by differential fractionation of protoplasts. The distribution of marker enzymes in the fractions enriched in chloroplasts, mitochondria, or cytosol revealed that chloroplast fractions were contaminated at a relatively low level with the cytosolic marker but were significantly contaminated with the mitochondrial and peroxisomal enzymes (Table I). The mitochondrial fractions were not significantly contaminated with other markers, but the yield was very low. The cytosolic fractions showed relatively low contamination with the chloroplast and mitochondrial markers but were significantly contaminated with the peroxisomal marker. In these same samples APS reductase was predominantly found in the chloroplast fractions from plants of both ages. Negligible activity was associated with the mitochondrial fraction, and the low level of activity associated with the cytosolic fraction was not greater than the level of contamination with the chloroplast or peroxisome markers. The result suggests that APS reductase is very likely exclusively localized in the chloroplasts.
Table I.
Distribution of ATP sulfurylase and APS reductase in subcellular fractions from Arabidopsis leaf protoplasts isolated from 21- and 47-d-old plants
| Fraction | Percent of Total Enzyme Activity
|
|||||
|---|---|---|---|---|---|---|
| Marker enzyme
|
Experimental enzyme
|
|||||
| PFP (cytosol) | NADP-GAPDH (chloroplast) | Cyt c oxidase (mitochondria) | HPR (peroxisome) | ATP sulfurylase | APS reductase | |
| 21-d-old plants | ||||||
| Chloroplasts | 8 ± 2 | 93 ± 4 | 90 ± 5 | 72 ± 15 | 86 ± 7 | 96 ± 1 |
| Mitochondria | <1 | <1 | 10 ± 5 | 1 ± 1 | 2 ± 2 | <1 |
| Cytosol | 92 ± 2 | 7 ± 4 | <1 | 27 ± 16 | 12 ± 4 | 4 ± 1 |
| Recovery | 119 ± 30 | 94 ± 12 | 103 ± 30 | 105 ± 26 | 102 ± 26 | 81 ± 8 |
| 47-d-old plants | ||||||
| Chloroplasts | 7 ± 3 | 95 ± 3 | 93 ± 3 | 82 ± 7 | 67 ± 3 | 95 ± 2 |
| Mitochondria | 1 ± 1 | <1 | 7 ± 3 | 1 ± 1 | <1 | <1 |
| Cytosol | 93 ± 6 | 5 ± 3 | <1 | 17 ± 6 | 33 ± 3 | 4 ± 3 |
| Recovery | 108 ± 30 | 104 ± 16 | 97 ± 20 | 104 ± 16 | 87 ± 23 | 85 ± 14 |
Each value is presented as a percentage of the total enzyme activity recovered in the three fractions, chloroplast, mitochondria, and cytosol. Also presented is the percentage of the enzyme recovered compared with an unfractionated protoplast lysate. Each value represents the mean ± sd of three independent fractionations. The enzyme activity value for each fractionation represents the mean of two measurements. The known localization of marker enzymes is indicated in parentheses. The marker enzymes are: HPR, hydroxypyruvate reductase; PFP, pyrophosphate: Fru-6-P 1-phosphotransferase; NADP-GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Cyt c oxidase, cytochrome c oxidase.
ATP sulfurylase was primarily distributed in the chloroplast fraction. But there was significant activity in the cytosolic fraction above the level of contamination with the chloroplast marker. There was negligible ATP sulfurylase activity in the mitochondrial fraction. This result indicates that ATP sulfurylase is localized in both the chloroplast and the cytosol. Moreover, the level of ATP sulfurylase in the cytosol fraction was approximately 3-fold greater in the 47-d plants than in 21-d plants.
Due to the high level of mitochondrial contamination in the chloroplast fraction Percoll density gradient centrifugation was used to isolate chloroplasts of greater purity. The additional step significantly reduced the contamination with mitochondria (Table II), yet the level of ATP sulfurylase and APS reductase was unaffected, indicating that the sulfur assimilation enzymes are not localized in mitochondria, a possibility that could not be ruled out by the experiment shown in Table I.
Table II.
ATP sulfurylase and APS reductase activity in chloroplasts free of mitochondrial contamination
| Chloroplast Sample | Specific Enzyme Activity
|
|||
|---|---|---|---|---|
| NADP-GAPDH | Cyt c oxidase | ATP sulfurylase | APS reductase | |
| nmole · min−1 · mg protein−1 | ||||
| Crude chloroplasts | 498 | 28 | 33 | 2.1 |
| Percoll-purified chloroplasts | 812 | 0 | 32 | 1.8 |
Chloroplasts isolated from 47-d-old plants were prepared by low-speed centrifugation of lysed protoplasts (crude chloroplasts) or were subjected to further purification by Percoll density gradient centrifugation. The values for crude chloroplasts represent the mean of three independent measurements and the Percoll-purified chloroplasts of two independent measurements. The marker enzymes are as described in Table I.
The subchloroplast localization of ATP sulfurylase and APS reductase was studied, and the results are shown in Table III. After osmotic lysis and centrifugation of Percoll-purified chloroplasts both enzymes fractionated with the soluble chloroplast components. The results indicate that APS reductase and the chloroplast form of ATP sulfurylase are probably localized in the stroma. The chloroplast marker, NADP-GAPDH, fractionated to both stromal and thylakoid fractions, just as it does in pea chloroplasts (Anderson et al., 1996).
Table III.
Distribution of ATP sulfurylase and APS reductase activity in subchloroplast fractions
| Sample | Specific Enzyme Activity
|
|||||
|---|---|---|---|---|---|---|
| PFP | NADP-GAPDH | Cyt c oxidase | HPR | ATP sulfurylase | APS reductase | |
| nmole · min−1 · mg protein−1 | ||||||
| Leaf extract | 2.4 | 418 | 11.2 | 460 | 47 | 1.6 |
| Stromal fraction | 0 | 840 | 0.0 | 46 | 32 | 1.8 |
| Thylakoid fraction | 0 | 135 | 0.0 | 32 | 0 | 0.02 |
Crude chloroplasts isolated from 47-d-old plants were fractionated by osmotic lysis and the activity of marker enzymes, and ATP sulfurylase and APS reductase were measured and compared with the activity in a leaf extract. Each value represents the mean of two independent measurements. The marker enzymes are as described in Table I.
The localization of cytosolic ATP sulfurylase was studied to determine whether it is truly cytosolic or associated with a particulate microsomal fraction. After subjecting the cytosolic preparation to a centrifugation force of 140,000g for 1 h, sufficient to pellet microsomes, ATP sulfurylase activity was found to be localized entirely in the soluble fraction (Table IV).
Table IV.
ATP sulfurylase in high-speed supernatant and pellet fractions from isolated cystosol
| Fraction | Volume | Protein Concentration | Protein | SA | Total Activity | Recovery of Total Cytosol |
|---|---|---|---|---|---|---|
| mL | mg/mL | mg | nmole min−1 mg−1 | nmol min−1 | % | |
| Total cytosol | 1.50 | 0.6 | 0.90 | 45 | 41 | 100 |
| S140 | 1.49 | 0.6 | 0.89 | 56 | 50 | 122 |
| P140 | 0.08 | 0.2 | 0.02 | 0 | 0 | 0 |
The cytosolic fraction isolated from 47-d-old plants was subjected to ultracentrifugation and 140,000g. The activity of cytosolic ATP sulfurylase in the soluble and particulate fractions was measured. Each value represents the mean of two independent measurements. SA, Specific activity.
Immunoblot analysis was carried out as an independent method for assessing the localization of the sulfur assimilation enzymes. The results were consistent with the enzyme activity measurements in that ATP sulfurylase APS reductase were detected only in the fractions containing activity (data not shown).
Regulation of ATP Sulfurylase Isoforms during Development
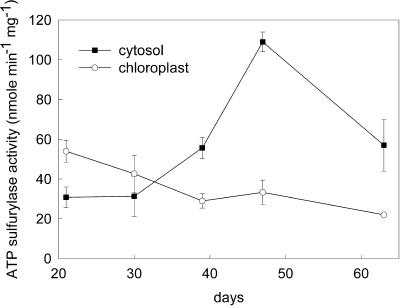
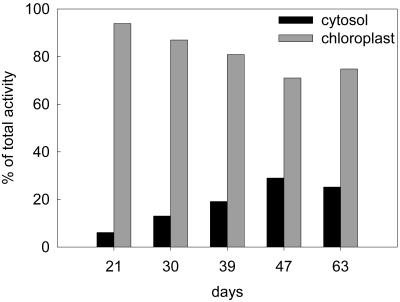
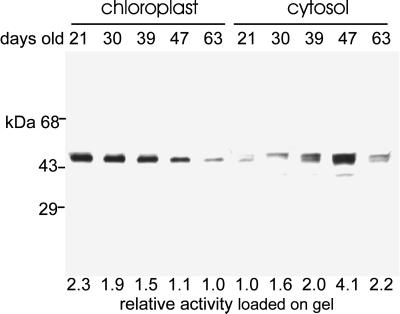
The results of cell fractionation showed that the cytosolic form of ATP sulfurylase is approximately 3 times as abundant in 47-d plants as in 21-d plants, whereas the chloroplast form decreases in relation to the age of the plant (Table I), suggesting that the isoforms are reciprocally regulated. The observation was explored further by performing subcellular fractionation on plants from a range of ages. Over the period from 21 to 63 d, the ATP sulfurylase specific activity in the cytosolic fraction increased 3.5-fold between 21 and 47 d, and then declined to 1.7 times the activity of 21-d plants (Fig. 4). Over the same period the specific activity in the chloroplast fraction showed a linear, 2-fold decline. Correcting for the level of cross contamination, the values are presented in Figure 5 as a percentage of the total ATP sulfurylase activity. At 21 d, 6% of the total ATP sulfurylase fractionated with the cytosol, whereas 94% fractionated with chloroplasts. The total ATP sulfurylase in the cytosol increased with age reaching 29% of the total activity at 47 d and then declined to 25% after 63 d. During the same period the enzyme fractionating with chloroplasts declined to 75% of the total activity. Over the entire growth period APS reductase activity remained restricted to the chloroplast fraction and showed the same decline in specific enzyme activity as measured in crude leaf extracts (Fig. 1) and the plastid form of ATP sulfurylase (data not shown). Immunoblot analysis showed that chloroplast ATP sulfurylase declines with plant age, whereas the cytosolic enzyme accumulates and reaches a peak at 47 d (Fig. 6), similar to the results obtained through activity measurement. The chloroplast enzyme appears to migrate as a single approximately 50-kD band, whereas the antibody reacts with a doublet of 50 and 51.5 kD in the cytosolic fractions. The intensity of the two bands in the cytosolic fraction changes coordinately during development.
Figure 4.
ATP sulfurylase activity in chloroplast and cytosolic fractions of Arabidopsis leaves during growth. Each value represents the mean ± sd from three independent fractionations for the first four developmental stages and estimated sd from two independent fractionations for 63-d-old plants. Two activity measurements were carried out for each fractionation.
Figure 5.
Distribution of ATP sulfurylase in chloroplast and cytosolic fractions of Arabidopsis leaves during growth. The data from Figure 4 were corrected for cross contamination as described in Table I. The data are presented here as a percentage of total activity in protoplasts.
Figure 6.
Immunoblot analysis of ATP sulfurylase in chloroplast and cytosolic fractions of Arabidopsis leaves during growth. Representative samples from the experiments shown in Figure 4 were analyzed by immunoblotting. Five micrograms of total protein was analyzed for each sample. The relative enzyme activity loaded onto the gel is indicated below the blot with the lowest activity normalized to a value of 1. For ATP sulfurylase the absolute activity corresponding to the relative value of 1 was 23 nmol min−1 mg−1 for the chloroplast sample and 30 nmol min−1 mg−1 for the cytosolic sample. The relative activity values correspond closely with the signal intensity determined by densitometry.
RNA-blot analysis was performed on leaves of plants of various ages to determine whether the increase in cytosolic ATP sulfurylase can be accounted for by changes in the steady-state level of mRNA for any of the ATP sulfurylase genes. However, none of the ATP sulfurylase mRNA species showed an increase with age (data not shown).
DISCUSSION
This study was initiated to examine the localization of ATP sulfurylase and APS reductase in Arabidopsis. Gene cloning had been inconclusive on the question of whether Arabidopsis contains a cytoplasmic ATP sulfurylase, suggesting that either Arabidopsis is an atypical flowering plant or the gene for cytosolic ATP sulfurylase had not yet been identified in this species. Second, APS reductase localization had only been studied in a single plant species and only in roots (Brunold and Suter, 1989). However, the existence of a cytosolic isoform of ATP sulfurylase, OASTL, and Ser acetyltransferase in a range of other plant species raised the possibility that a complete reductive assimilatory pathway might exist in the cytosol of flowering plants and Arabidopsis specifically.
The results from subcellular fractionation clearly show the existence of chloroplast and cytosolic isoforms of ATP sulfurylase and that APS reductase is exclusively localized in chloroplasts, confirming earlier studies and affirming that Arabidopsis is indeed a typical flowering plant. Moreover the results indicate either that a fifth ATP sulfurylase gene exists in Arabidopsis that has so-far eluded discovery or that one of the known genes produces the cytosolic isoenzyme. Several lines of evidence suggest that a fifth ATP sulfurylase gene is unlikely. The Arabidopsis genome sequencing effort is 85% complete and more than approximately 46,000 Arabidopsis expressed sequence tags have been sequenced, yet only the known ATP sulfurylase genes are represented in these databases (Hatzfeld et al., 2000). Also, only four ATP sulfurylase genes can be detected by cross hybridization on genomic blots (Murillo and Leustek, 1995), so if a fifth gene exists its nucleotide sequence must be highly divergent from the known genes. However, the ability of an antibody raised against one of the isoenzymes (APS3) to react with chloroplast and cytosolic isoenzymes (this study) suggests that Arabidopsis ATP sulfurylases are highly conserved. Thus, the idea that one of the known ATP sulfurylase genes encodes the cytosolic enzyme is a more likely possibility. But it remains to be experimentally determined. Based on the position of possible translational initiation codons within the coding sequences of the APS genes Hatzfeld et al. (2000) speculate that it could be APS2, however, there is as yet no direct evidence for this hypothesis.
The localization of APS reductase suggests that sulfate reduction occurs exclusively in chloroplasts. However, the finding raises the question of the function of cytosolic isoforms of the other sulfur assimilation enzymes. Either APS and sulfide are transported into and out of chloroplasts or the cytosolic forms of sulfur assimilation enzymes serve a function other than for Cys synthesis. The localization results reported here are in agreement with results from Arabidopsis gene cloning (Gutierrez-Marcos et al., 1996; Setya et al., 1996; Chen and Leustek, 1998). Three APS reductase genes have been characterized from this species and all encode proteins with plastid transit peptides. A remaining challenge for discovery is whether the three APS reductases are redundant or whether they have specific functions.
The finding that ATP sulfurylase and APS reductase are most active in the youngest leaves is consistent with previous studies (Schmutz and Brunold, 1982). The result suggests that reductive sulfate assimilation leading to Cys synthesis occurs predominantly in the youngest leaves and the finding supports what is known about the physiological changes that accompany sulfate deficiency. One of the first symptoms is chlorosis of the youngest leaves, by contrast to nitrogen deficiency that initially causes chlorosis of the oldest leaves (Clarkson et al., 1993). It has been proposed that when reduced sulfur is fixed into organic compounds it becomes relatively immobile, unlike nitrogen, which is rapidly remobilized to the growing points of the plant.
A potentially important observation that emerged from the subcellular fractionation experiments is that the cytosolic and chloroplast isoforms of ATP sulfurylase are differentially regulated during development. The activity of the chloroplast form declines in parallel with APS reductase as plants age. By contrast the cytosolic isoform increases during development. The result explains why the decline in ATP sulfurylase lags behind that of APS reductase when measured in whole shoots (Fig. 1) and why total ATP sulfurylase activity appears not to decline with increasing leaf age (Fig. 3). Chloroplasts are known to be the primary site for sulfate reduction and Cys synthesis. Chloroplasts contain all the enzymes necessary for synthesis of Cys from sulfate and are capable of doing so autonomously under in vitro conditions (Trebst and Schmidt, 1969; Schürmann and Brunold, 1980). By contrast, the cytoplasm does not contain all the enzymes necessary for sulfate reduction. Thus, if cytosolic ATP sulfurylase were to participate in Cys synthesis a mechanism would be necessary for transport of APS into chloroplasts. However, since most of the total ATP sulfurylase is plastid localized (Lunn et al., 1990; Renosto et al., 1993; this study) and ATP sulfurylase activity is present in excess over that required for normal rates of sulfate assimilation (Lee, 1999) it is difficult to imagine the conditions under which cytosolic ATP sulfurylase might contribute significantly toward sulfate reduction or why the level of this enzyme increases just at the time when APS reductase is declining. More likely is the idea that cytosolic ATP sulfurylase has a specialized function.
If it is not involved in sulfate reduction and Cys synthesis what possible function could cytosolic ATP sulfurylase serve? One possibility is in providing activated sulfate for sulfate ester biosynthesis. A number of enzymes involved in formation of sulfated compounds have been characterized from plants. All of the known sulfotransferases from plants and animals contain signature sequences thought to be involved in PAPS binding (Varin et al., 1997). Indeed all are strictly dependent on PAPS as a sulfuryl donor. Without exception the enzymes have been demonstrated to be localized in the cytosol or are predicted to be cytosolic based on the absence of organellar transit peptides deduced from the cloned gene sequences. A gap in the understanding of sulfation is that the localization of APS kinase has not yet been studied. However, there are three APS kinase genes in Arabidopsis (Leustek and Saito, 1999). The product from one of them is able to enter isolated intact chloroplasts in vitro (Lee and Leustek, 1998), but its localization is uncertain, and the others could be localized in the cytosol based on analysis with the PSORT program (http://psort.nibb.ac.jp/) for prediction of protein localization.
The number of different sulfated metabolites in Arabidopsis is uncertain, however a major group of related sulfated compounds are the glucosinolates. These are amino acid derived thioglucosides produced by Arabidopsis and other members of the Brassicaceae. When the integrity of cells is disrupted glucosinolates are hydrolyzed to toxic isothiocyanates. Thus, they are believed to play a defensive role against insects and pathogens (Chew, 1988). In Arabidopsis the Trp-oxidizing enzyme, thought to be involved in synthesis of indole glucosinolate, is induced with increasing plant age (Ludwig-Müller et al., 1999). In Brassica napus glucosinolate content of leaves peaks as the leaves reach the fully expanded stage (Porter et al., 1991). Although the temporal production of glucosinolates in Arabidopsis has not been specifically studied it is interesting to note that the peak of cytosolic ATP sulfurylase activity is approximately the time when the bulk of leaf tissue of the plants has reached the fully expanded stage. Thus, it seems possible that the induction of cytosolic ATP sulfurylase may correlate with the initiation of maximal glucosinolate biosynthesis. Further experimentation will be necessary to explore this hypothesis.
In another study, an ATP sulfurylase cDNA was identified, corresponding to an mRNA that accumulates in B. napus during leaf senescence (Buchanan-Wollaston and Ainsworth, 1997). In the present study none of the four ATP sulfurylase genes of Arabidopsis were observed to increase as plants aged, although senescing plants were not analyzed.
MATERIALS AND METHODS
General Methods
Protein concentration was measured using the Bradford protein assay reagent (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin (BSA) as a standard. Chlorophyll was measured by the method of Arnon (1949). SDS-PAGE and immunoblotting was carried out as described in Harlow and Lane (1988).
Plant Material
The Columbia ecotype of Arabidopsis was grown in Promix (Premier Horticulture, Ltd., Rivére-du-Loup, Quebec) at 20°C in a growth chamber with a 10-h photoperiod at a light intensity of approximately 90 μE m−2 s−1, 24-h diurnal cycle. The growth conditions were chosen to delay bolting and extend the vegetative growth phase. For the purpose of this study it was of interest maximize the yield of vegetative material and to avoid the phase change associated with bolting.
All plant material was harvested within 1 h into the light period. Plants were harvested at different ages ranging up to 63 d after germination. In experiments where plant age was the variable the entire shoot was harvested and analyzed as a homogenous sample. When leaf age was the variable, six 42-d-old plants were selected at random and the leaves of different ages were harvested and analyzed separately. Starting with the youngest, every three leaves forming a rosette were pooled together into six batches designated L1 (youngest) to L6 (oldest). The cotyledons formed a separate pool designated Lc. The L1, L2, and L3 pools consisted of leaves expanded to approximately 10%, approximately 30%, and approximately 90% of fully expanded leaves, respectively. L4 and L5 leaf pools were fully expanded and bright green. The L6 pool showed some yellowing on the margins. Lc were yellow but succulent at the time of harvest.
Subcellular and Subchloroplast Fractionation
Protoplasts were prepared as described by Robinson (1987) and Lunn et al. (1990) with optimizations. The sample plants were stored in the dark overnight at 20°C. Approximately 15 g (fresh weight) of freshly harvested shoots, or for plants 40 d and older, the whole shoot of four to seven plants (approximately 15 g fresh weight), were combined in a 19-cm diameter Petri dish with 100 mL of buffer containing 0.5 m sorbitol, 1 mm CaCl2, 0.05% (w/v) BSA, and 20 mm MES [2-(N-morpholino)ethanesulfonic acid]-NaOH (pH 5.5). The material was cut into fine strips with a sharp razor blade and the buffer replaced with 0.5 m sorbitol, 1 mm CaCl2, 0.5% (w/v) BSA, 1% (w/v) cellulase (Worthington Biochemical, Lakewood, NJ), and 0.3% (w/v) macerozyme R-10 (ICN Pharmaceuticals, Costa Mesa, CA). The material was incubated at 27°C for 4 h with illumination from two 15-W fluorescent bulbs positioned 15 cm above the Petri dish. All subsequent procedures were carried out at 4°C. The protoplasts were released from the digested plant tissue by gentle agitation for 10 min and then filtered through a nylon mesh with 100-μm pores. The protoplasts were collected by centrifugation at 100g for 5 min using a swinging bucket rotor and then gently resuspended in a total volume of 30 mL of 0.5 m Suc, 1 mm CaCl2, 5 mm MES-NaOH, pH 6.0. The protoplast suspension was distributed among six 15-mL glass centrifuge tubes, overlaid first with 2 mL of 0.4 m Suc, 0.1 m sorbitol, 1 mm CaCl2, 5 mm MES-NaOH, pH 6.0, and then with 2 mL of 0.5 m sorbitol, 1 mm CaCL2, 5 mm MES-NaOH, pH 6.0. The gradients were centrifuged in two steps at 100g for 10 min and then at 300g for 5 min in a swinging bucket rotor. The material at the interface of the Suc/sorbitol and sorbitol layers was collected and examined under a light microscope to ensure that the protoplast preparation was free of cell material and chloroplasts. After determining the chlorophyll concentration the protoplasts were centrifuged at 100g for 5 min, and the pellet was carefully resuspended in 0.5 m sorbitol, 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH, pH 7.0 at a chlorophyll concentration of 0.2 mg/mL. Protoplast lysis was achieved using a 2.5-cc syringe fitted with nylon mesh held in place over the open end of the barrel with a rubber O-ring. The protoplasts were slowly drawn into the syringe and then slowly ejected through the mesh. The suspension was examined under a light microscope after each passage to determine the minimum number of passages necessary to achieve complete breakage. Usually, two passages through the 10-μm mesh or three passages through the 20-μm mesh, respectively, were sufficient to break most of the protoplasts. The 10-μm mesh was used for plants up to 30 d of age and a 20-μm mesh was used for older plants.
Cell fractions were recovered by centrifuging the lysate at 500g for 90 s using a swinging bucket rotor. The supernatant was collected and centrifuged again at 500g for 2 min to recover remaining chloroplasts. The chloroplast pellets were resuspended in 0.5 m sorbitol, 50 mm HEPES-NaOH, pH 7.0 and pooled. The supernatant was decanted and centrifuged at 5,000g for 10 min. This pellet comprised the mitochondrial fraction and the supernatant comprised the cytosolic fraction. The pelleted mitochondria were resuspended in the same buffer as the chloroplasts. All three fractions and the purified protoplasts were aliquoted into small volumes and stored at −70°C.
For some experiments chloroplasts were further purified by centrifugation on Percoll step gradients as described by Gruissem et al. (1986). Protoplasts from 15 g (fresh weight) of leaf material were ruptured and the resulting chloroplast pellet was suspended in 3 mL of resuspension buffer containing 0.33 m sorbitol, 2 mm EDTA, 2 mm MgCl2, 2 mm MnCl, and 50 mm HEPES-KOH, pH 8.0. The chloroplast suspension was layered onto an 11-mL 40%/80% (v/v) Percoll step gradient and centrifuged at 7,000g for 20 min at 4°C, using a swinging bucket rotor. The band of intact chloroplasts was removed with a pipette, diluted with 4 volumes of resuspension buffer, and then centrifuged at 8,000g for 1 min at 4°C in a swinging bucket rotor.
Chloroplast fractionation was performed by resuspension of the pellet in lysis buffer containing 5 mm dithiothreitol (DTT), 1 mm EDTA, 10 mm Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 8.0, at a ratio of 5 volumes of buffer per 1 volume of packed chloroplasts (approximately 0.2 mg of chlorophyll per mL). The suspension was kept on ice for 10 min, vigorously vortexed, and then centrifuged at 14,000g for 5 min. The pelleted thylakoid membranes were resuspended in lysis buffer to a volume equal to that of the supernatant.
The cytoplasmic fraction was further divided into a microsomal component and a high-speed supernatant by centrifuging at 140,000g for 1 h.
Leaf Protein Extract Preparation and Enzymatic Assays
Leaf extracts were prepared in 100 mm Tris-HCl, pH 8.0, or in this buffer supplemented with 2 mm DTT for assay of pyrophophate:Fru-6-P-1-phosphotransferase, or supplemented with 100 mm Na2SO4 for assay of APS reductase. Homogenates were centrifuged at 14,000g for 10 min, and the supernatant was collected and centrifuged further for 5 min. The supernatant comprised the crude extract and was used immediately for enzyme assays. All procedures were carried out at 4°C.
The following enzymes were measured as markers of subcellular compartments: peroxisomes, hydroxypyruvate reductase (EC 1.1.1.81) (Tolbert et al., 1970); cytoplasm, pyrophosphate:Fru- 6-P-1-phosphotransferase (EC 2.7.1.90) (Weiner et al., 1987); chloroplasts, glyceraldehyde-3-phosphate dehydrogenase (phosphorylating), (EC 1.2.1.13) (Lunn et al., 1990); and mitochondria, cytochrome c oxidase (EC 1.9.3.1) (Storrie and Madden, 1990). ATP sulfurylase was measured using the ATP synthesis assay (Renosto et al., 1991; as modified by Murillo and Leustek, 1995). All of the preceding enzymes were assayed spectrophotometrically at 340 nm in reactions coupled to reduction of NAD+ or oxidation of NAD(P)H. The assays were conducted as described in the references with the exception that 2 mm DTT was added to the pyrophosphate:Fru- 6-P-1-phospho-transferase reaction buffer and the reaction was started with Fru-6-P. APS reductase (EC 1.8.99.-) was measured as described by Setya et al. (1996). The radioactive substrate [35S]APS was prepared from [35S]PAPS (Dupont NEN, Inc., Boston) by treatment with P1 nuclease (N-8630, Sigma, St. Louis). [35S]APS was used at a specific activity of approximately 500 Bq· nmole−1.
Immunoblotting
Immobilon-P-membrane was used for immunoblotting and immune complexes were detected with the Renaissance kit (DuPont NEN, Inc.). Rabbit antibodies against recombinant APS3 ATP sulfurylase (Murillo and Leustek, 1995) or against APS reductase (Gao et al., 2000) were used for analysis. The antibodies were used at a dilution of 1:5,000.
ACKNOWLEDGMENT
We thank Julie-Ann Bick for help and advice.
Footnotes
This work was supported by the National Science Foundation (grant nos. IBN–9601146 and IBN–9817594) and by the Studienstiftung des deutschen Volkes (to C.R.). The work was carried out in part as a Diplomarbeit Thesis from the Carl von Ossietzky Universität Oldenburg, Germany.
LITERATURE CITED
- Adams CA, Rinne RW. Influence of age and sulfur metabolism on ATP sulfurylase activity in the soybean and a survey of selected species. Plant Physiol. 1969;44:1241–1246. doi: 10.1104/pp.44.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LE, Gibbons JT, Wang X. Distribution of 10 enzymes of carbon metabolism in pea (Pisum sativum) chloroplasts. Int J Plant Sci. 1996;157:525–538. [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunold C, Suter M. Localization of enzymes of assimilatory sulfate reduction in pea roots. Planta. 1989;179:228–234. doi: 10.1007/BF00393693. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Ainsworth C. Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridization. Plant Mol Biol. 1997;33:821–834. doi: 10.1023/a:1005774212410. [DOI] [PubMed] [Google Scholar]
- Cacco G, Saccomani M, Ferrari G. Development of sulfate uptake capacity and ATP sulfurylase activity during root elongation in maize. Plant Physiol. 1977;60:582–584. doi: 10.1104/pp.60.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Leustek T. Three genomic clones from Arabidopsis encoding 5′-adenylylsulfate reductase (accession nos. AF016282, AF016283, and AF016284) (PGR–030) Plant Physiol. 1998;116:869. [Google Scholar]
- Chew FS. Biological effects of glucosinolates. In: Cutler HG, editor. Biologically Active Natural Products: Potential Use in Agriculture. Washington, DC: American Chemical Society; 1988. pp. 155–181. [Google Scholar]
- Clarkson DT, Hawkesford M, Davidian J-C. Membrane and long distance transport of sulfate. In: De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, editors. Sulfur Nutrition and Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 3–20. [Google Scholar]
- Fankhauser H, Brunold C. Localization of adenosine 5′-phosphosulfate sulfotransferase in spinach leaves. Planta. 1978;143:285–289. doi: 10.1007/BF00392000. [DOI] [PubMed] [Google Scholar]
- Fankhauser H, Brunold C. Localization of O-acetyl-serine sulfhydrylase in Spinacea oleracea L. Plant Sci Lett. 1979;14:185–192. [Google Scholar]
- Gao Y, Schofield O, Leustek T. Characterization of sulfate assimilation in marine algae focusing on the enzyme 5′-adenylylsulfate (APS) reductase. Plant Physiol. 2000;123:1087–1096. doi: 10.1104/pp.123.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W, Greenberg BM, Zurawski G, Hallick RB. Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 1986;118:253–270. doi: 10.1016/0076-6879(86)18077-3. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL. Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA. 1996;93:13377–13382. doi: 10.1073/pnas.93.23.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies, a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hatzfeld Y, Lee S, Lee M, Leustek T, Saito K. Functional characterization of a gene encoding a fourth ATP sulfurylase isoform from Arabidopsis thaliana. Gene. 2000;248:51–58. doi: 10.1016/s0378-1119(00)00132-3. [DOI] [PubMed] [Google Scholar]
- Klonus D, Höfgen R, Willmitzer L, Riesmeier JW. Isolation and characterization of two cDNA clones encoding ATP-sulfurylase from potato by complementation of a yeast mutant. Plant J. 1994;6:105–112. doi: 10.1046/j.1365-313x.1994.6010105.x. [DOI] [PubMed] [Google Scholar]
- Lee S. Molecular analysis of sulfate assimilation in higher plants: effect of cysteine, sulfur and nitrogen nutrients, heavy metal stress, and genomic DNA cloning. PhD thesis. 1999. Rutgers, The State University of New Jersey, New Brunswick. [Google Scholar]
- Lee S, Leustek T. APS kinase from Arabidopisis thaliana, genomic organization, expression, and kinetic analysis of the recombinant enzyme. Biochem Biophys Res Commun. 1998;247:171–175. doi: 10.1006/bbrc.1998.8751. [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick J-A, Davies JP. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:141–166. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- Leustek T, Murillo M, Cervantes M. Cloning of a cDNA encoding ATP sulfurylase from Arabidopsis thaliana by functional expression in Saccharomyces cerevisiae. Plant Physiol. 1994;105:897–902. doi: 10.1104/pp.105.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Saito K. Sulfate transport and assimilation in plants. Plant Physiol. 1999;120:637–643. doi: 10.1104/pp.120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Saidha T, Schiff JA. Purification and properties of two forms of ATP sulfurylase from Euglena. Biochim Biophys Acta. 1991;1078:68–76. doi: 10.1016/0167-4838(91)90094-g. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Pieper K, Ruppel M, Cohen JD, Epstein E, Kiddle G, Bennett R. Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana (L.) Heynh. glucosinolate mutants and the development of clubroot disease. Planta. 1999;208:409–419. doi: 10.1007/s004250050576. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Droux M, Martin J, Douce R. Localization of ATP sulfurylase and O-acetylserine(thiol) lyase in spinach leaves. Plant Physiol. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandless EL, Craigie JS. Sulfated polysaccharides in red and brown algae. Annu Rev Plant Physiol. 1979;30:41–53. [Google Scholar]
- Mulichak AM, Theisen MJ, Essigmann B, Benning C, Garavito RM. Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose. Proc Natl Acad Sci USA. 1999;96:13097–13102. doi: 10.1073/pnas.96.23.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo M, Leustek T. Adenosine-5′-triphosphate-sulfurylase from Arabidopsis thaliana and Escherichia coli are functionally equivalent but structurally and kinetically divergent: nucleotide sequence of two adenosine-5′-triphosphate-sulfurylase cDNAs from Arabidopsis thaliana and analysis of a recombinant enzyme. Arch Biochem Biophys. 1995;323:195–204. doi: 10.1006/abbi.1995.0026. [DOI] [PubMed] [Google Scholar]
- Porter AJR, Morton AM, Kiddle G, Doughty KJ, Wallsgrove RM. Variation in the glucosinolate content of oilseed rape (Brassica napus L.) leaves: I. Effect of leaf age and position. Ann Appl Biol. 1991;118:461–467. [Google Scholar]
- Renosto F, Martin RL, Borrell JL, Nelson DC, Segel IH. ATP sulfurylase from trophosome tissue of Riftia pachyptila (hydrothermal vent tube worm) Arch Biochem Biophys. 1991;290:66–78. doi: 10.1016/0003-9861(91)90592-7. [DOI] [PubMed] [Google Scholar]
- Renosto F, Patel HC, Martin RL, Thomassian C, Zimmerman G, Segel IH. ATP sulfurylase from higher plants: kinetic and structural characterization of the chloroplast and cytosol enzymes from spinach leaf. Arch Biochem Biophys. 1993;307:272–285. doi: 10.1006/abbi.1993.1590. [DOI] [PubMed] [Google Scholar]
- Robinson SP. Separation of chloroplasts and cytosol from protoplasts. Methods Enzymol. 1987;148:188–195. [Google Scholar]
- Rüegsegger A, Brunold C. Localization of γ-gluta-mylcysteine synthetase and glutathione synthetase activity in maize seedlings. Plant Physiol. 1993;101:561–566. doi: 10.1104/pp.101.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidha T, Na SQ, Li JY, Schiff JA. A sulfate metabolizing center in Euglena mitochondria. Biochem J. 1988;253:533–539. doi: 10.1042/bj2530533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff JA. Reduction and other metabolic reactions of sulfate. In: Läuchli A, Bieleski RL, editors. Encyclopedia of Plant Physiology. 15A. New York: Springer; 1983. pp. 401–421. [Google Scholar]
- Schmidt A. Regulation of sulfur metabolism in plants. Prog Bot. 1986;48:133–150. [Google Scholar]
- Schmutz D, Brunold C. Regulation of sulfate assimilation in plants: XIII. Assimilation of sulfate reduction during ontogenesis of primary leaves of Phaseolus vulgaris L. Plant Physiol. 1982;70:524–527. doi: 10.1104/pp.70.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann P, Brunold C. Formation of cysteine from adenosine 5′-phosphosulfate (APS) in extracts from spinach chloroplasts. Z Pflanzenphysiol. 1980;100:257–268. [Google Scholar]
- Setya A, Murillo M, Leustek T. Sulfate reduction in higher plants: molecular evidence for a novel 5′-adenylylphosphosulfate (APS) reductase. Proc Natl Acad Sci USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IK. Studies of l-cysteine biosynthetic enzymes in Phaseolus vulgaris L. Plant Physiol. 1972;50:477–479. doi: 10.1104/pp.50.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B, Madden EA. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- Suter M, von Ballmoos P, Kopriva S, den Camp RO, Schaller J, Kuhlemeier C, Schurmann P, Brunold C. Adenosine 5′-phosphosulfate sulfotransferase and adenosine 5′-phosphosulfate reductase are identical enzymes. J Biol Chem. 2000;275:930–936. doi: 10.1074/jbc.275.2.930. [DOI] [PubMed] [Google Scholar]
- Tolbert NE, Yamazaki RK, Oeser A. Localization and properties of hydroxypyruvate and glyoxylate reductases. J Biol Chem. 1970;245:5129–5136. [PubMed] [Google Scholar]
- Trebst A, Schmidt A. Photosynthetic sulfate and sulfite reduction by chloroplasts. Prog Photosynth Res. 1969;3:1510–1516. [Google Scholar]
- Varin L, Marsolais F, Richard M, Rouleau M. Biochemistry and molecular biology of plant sulfotransferases. FASEB J. 1997;11:517–525. doi: 10.1096/fasebj.11.7.9212075. [DOI] [PubMed] [Google Scholar]
- von Arb C, Brunold C. Ferredoxin-sulfite reductase and ferredoxin-nitrite reductase activities in leaves of Pisum sativum: changes during ontogeny and in vitro regulation by sulfide. Physiol Plant. 1985;64:290–294. [Google Scholar]
- von Arb C, Brunold C. Enzymes of assimilatory sulfate reduction in leaves of Pisum sativum: changes during ontogeny and in vivo regulation by H2S and cysteine. Physiol Plant. 1986;67:81–86. [Google Scholar]
- Weiner H, Stitt M, Heldt HW. Subcellular compartmentation of pyrophosphate and alkaline pyrophosphatase in leaves. Biochim Biophys Acta. 1987;893:13–21. [Google Scholar]