Abstract
BRASSINOSTEROID-INSENSITIVE 1 (BRI1) encodes a putative Leucine-rich repeat receptor kinase in Arabidopsis that has been shown by genetic and molecular analysis to be a critical component of brassinosteroid signal transduction. In this study we examined some of the biochemical properties of the BRI1 kinase domain (BRI1-KD) in vitro, which might be important predictors of in vivo function. Recombinant BRI1-KD autophosphorylated on serine (Ser) and threonine (Thr) residues with p-Ser predominating. Matrix-assisted laser desorption/ionization mass spectrometry identified a minimum of 12 sites of autophosphorylation in the cytoplasmic domain of BRI1, including five in the juxtamembrane region (N-terminal to the catalytic KD), five in the KD (one each in sub-domains I and VIa and three in sub-domain VIII), and two in the carboxy terminal region. Five of the sites were uniquely identified (Ser-838, Thr-842, Thr-846, Ser-858, and Thr-872), whereas seven were localized on short peptides but remain ambiguous due to multiple Ser and/or Thr residues within these peptides. The inability of an active BRI1-KD to transphosphorylate an inactive mutant KD suggests that the mechanism of autophosphorylation is intramolecular. It is interesting that recombinant BRI1-KD was also found to phosphorylate certain synthetic peptides in vitro. To identify possible structural elements required for substrate recognition by BRI1-KD, a series of synthetic peptides were evaluated, indicating that optimum phosphorylation of the peptide required R or K residues at P − 3, P − 4, and P + 5 (relative to the phosphorylated Ser at P = 0).
Brassinosteroids (BRs) are polyhydroxylated sterol derivatives that occur at nanogram per kilogram fresh weight levels in pollen and immature vegetative tissues throughout the plant kingdom (Fujioka, 1999). The role of animal steroids in regulating growth and development is well known and BRs, in addition to sharing structural similarity with animal steroid hormones, also play many of the corresponding functional roles in plant development. Like their animal steroid counterparts, BRs have been shown to regulate gene expression, stimulate cell division and differentiation, and modulate reproductive biology (for review, see Clouse and Sasse, 1998). BRs also mediate growth responses unique to plants, including promotion of cell elongation in the presence of a complex cell wall and coordinating multiple developmental responses to darkness and light. The physiological, biochemical, and genetic characterization of several BR biosynthetic mutants has resulted in the widespread acceptance of BRs as a new class of plant hormone that is as essential in controlling normal plant development as auxins and gibberellins (Clouse and Feldmann, 1999).
In an attempt to identify components of the BR signal transduction pathway, we screened for BR insensitivity based on the ability of mutant Arabidopsis seedlings to elongate roots in the presence of BR concentrations inhibitory to wild-type root elongation (Clouse et al., 1993). This analysis uncovered a mutant brassinosteroid-insensitive1 (bri1) that conferred pleiotropic phenotypic effects, including severely dwarfed stature, reduced apical dominance, delayed flowering and senescence, male sterility, and nearly complete insensitivity to BRs in a variety of assays (Clouse et al., 1996). The severity of the phenotype strongly suggested that the BRI1 protein played a critical role in BR signal perception or transduction; this suggestion was further supported when the cloning and sequencing of BRI1 by Li and Chory (1997) revealed that this gene encoded a putative Leu-rich repeat receptor kinase. BRI1 exhibits homology with plant and animal receptor kinases and Leu-rich repeat proteins in three major domains including the putative extracellular ligand-binding domain, the single membrane-spanning domain, and the cytoplasmic kinase domain (KD). Moreover, sequence analysis of several mutant alleles confirmed that the putative ligand-binding domain and the KD are essential for in vivo function (Li and Chory, 1997; Noguchi et al., 1999). Based on mutant phenotype and sequence homology, it is obvious that BRI1 is a critical component of the BR signal transduction pathway. However, BRI1's role as the BR receptor has not been confirmed by direct binding studies nor have substrates for the KD been reported in the literature.
Receptor kinases have been thoroughly studied in animal systems and play a proven role in many signal transduction pathways. For example, binding of vertebrate epidermal growth factor to its cognate receptor kinase results in receptor dimerization and autophosphorylation on Tyr residues in the KD (Heldin, 1995). The activated kinase phosphorylates an intracellular transcription factor, Stat3, which is then translocated to the nucleus where it transcriptionally activates specific epidermal growth factor responsive genes (Park et al., 1996). Ligand-dependent dimerization followed by autophosphorylation is believed to be a conserved mechanism in plants, although no complete analysis of any plant receptor kinase, including conclusive identification of extracellular ligand- and dimerization-dependent autophosphorylation sites, has yet been accomplished. Based on biochemical and genetic evidence, the CLAVATA3 peptide has recently been proposed to serve as the ligand for the Arabidopsis CLAVATA1 receptor-like kinase (Fletcher et al., 1999; Trotochaud et al., 1999) and a putative ligand for the S locus receptor kinase has been proposed (Schopfer et al., 1999), but both of these hypotheses await final confirmation. However, the KDs of numerous plant receptor-like kinases have been expressed as recombinant proteins in Escherichia coli and they do indeed behave as functional kinases in vitro (Braun and Walker, 1996; Schulze-Muth et al., 1996; Wang et al., 1996; Braun et al., 1997; Williams et al., 1997; Muschietti et al., 1998; Stone et al., 1998; Coello et al., 1999; van der Knaap et al., 1999). Moreover, the recombinant KDs have been used as molecular probes for interaction cloning (Stone et al., 1994; Braun et al., 1997) and yeast two-hybrid screens (Bower et al., 1996; Gu et al., 1998) have been used to identify intracellular substrates for plant receptor-like kinases. Phosphoamino acid analyses reveal that plant receptor-like kinases autophosphorylate on Ser and Thr residues (as opposed to Tyr in most animal receptor kinases), but a thorough analysis of specific autophosphorylation sites using biophysical techniques such as matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) has not yet been reported.
The identification of the bri1 mutant and subsequent cloning of the BRI1 gene were significant advances in our understanding of BR action and signal transduction mechanisms, and suggests that a receptor kinase plays an important role in the response of cells to BR. Several critical questions need to be addressed to further understand how BRI1 functions in BR-signaling pathways: (a) Does BRI1 bind BR directly or indirectly in the presence of a sterol binding peptide? (b) Does BRI1 undergo homo- or heterodimerization? (c) What are the specific amino acids that undergo autophosphorylation in the BRI1-KD? (d) What are the intracellular substrates for the BRI1-KD, how are these substrates involved in BR signal transduction, and are they uniquely modified by BRI1 or are they shared in other signal transduction pathways? In this paper we report the initial biochemical characterization of recombinant BRI1-KD, including MALDI-MS determination of 12 sites of autophosphorylation (five of which were uniquely identified), some enzymatic properties of the recombinant kinase, and an analysis of putative sequence requirements for substrate recognition, determined by assaying the recombinant kinase with a range of synthetic peptides.
RESULTS
Recombinant BRI1 Autophosphorylates on Ser and Thr Residues
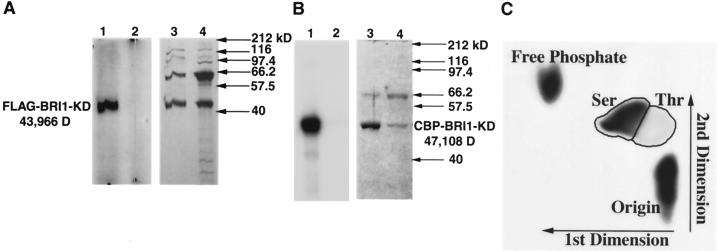
Expression of recombinant KDs of plant receptor-like kinases in E. coli generally leads to constitutive kinase activity in the absence of ligand (Braun and Walker, 1996). We also found that recombinant BRI1-KD possessed Ser/Thr kinase activity. Affinity-purified FLAG-BRI1-KD protein showed strong autophosphorylation when incubated with [γ-32P]ATP, whereas a mutant construct, FLAG-BRI1-K911E, had no kinase activity (Fig. 1A). CBP-BRI1-KD also showed similar autophosphorylation, whereas the mutant kinase CBP-BRI1-K911E failed to autophosphorylate, as expected (Fig. 1B). Phosphoamino acid analysis of autophosphorylated CBP-BRI1-KD using TLE showed heavy phosphorylation of Ser residues, with weaker phosphorylation of Thr (Fig. 1C).
Figure 1.
Autophosphorylation and phosphoamino acid analysis of recombinant BRI1-KD. A, Affinity-purified FLAG-BRI1-KD (lane 1) or the mutant FLAG-BRI1-K911E (lane 2) was incubated with 20 μCi [γ-32P]ATP in kinase buffer for 1 h at ambient temperature, followed by PAGE and visualization of incorporated isotopes with a phosphor imager. Lanes 3 and 4 represent the silver-stained gel corresponding to lanes 1 and 2. To confirm the identity of the labeled protein in lane 1, an SDS-PAGE gel was transferred to a polyvinylidene difluoride (PVDF) membrane, and the labeled band was digested with trypsin and subjected to MALDI-MS. Molecular mass was calculated from the predicted amino acid sequence of the recombinant protein. B, A similar analysis to that described above using calmodulin (CaM)-binding peptide (CBP)-BRI1-KD (lanes 1 and 3) and CBP-BRI1-K911E (lanes 2 and 4). The gel was stained with Coomassie Blue rather than silver. C, CBP-BRI1-KD was autophosphorylated and transferred to a PVDF membrane as described above. The membrane was digested with HCl and subjected to phosphoamino acid analysis by thin-layer electrophoresis (TLE) with p-Ser, p-Thr, and p-Tyr standards. CBP-BRI1-KD autophosphorylated primarily on Ser residues with a minor Thr component and no detectable phospho-Tyr residues.
Identification of Autophosphorylation Sites by MALDI-MS
To more precisely identify specific p-Ser or p-Thr residues in CBP-BRI1-KD, MALDI-MS was performed on radioactive HPLC fractions of a tryptic digest of affinity-purified, [γ-32P]ATP-autophosphorylated CBP-BRI1-KD. MALDI-MS is a powerful technique for precise Mr determination of a complex mixture of peptides. When diammonium citrate is included in the MALDI matrix, the peak intensity of phosphorylated peptides is increased relative to unphosphorylated peptides (Asara and Allison, 1999). Moreover, when the amino acid sequence of the protein is known, the mass of each potential tryptic peptide can be calculated and the phosphorylation state of a specific peptide can then be assigned from mass-to-charge ratio (m/z) values in MALDI-MS spectra. The addition of each phosphate group (plus HPO3) causes an increase in molecular mass of 80 D compared to the unphosphorylated peptide. Post-source decay (PSD) mass spectra of individual peptides can then be used to confirm phosphorylation. Annan and Carr (1996) have shown that during PSD, phosphate groups are lost before skeletal breakdown of the peptide and that phosphorylated Ser and Thr residues eliminate predominately H3PO4 (98 D) with a relatively minor loss of HPO3 (80 D).
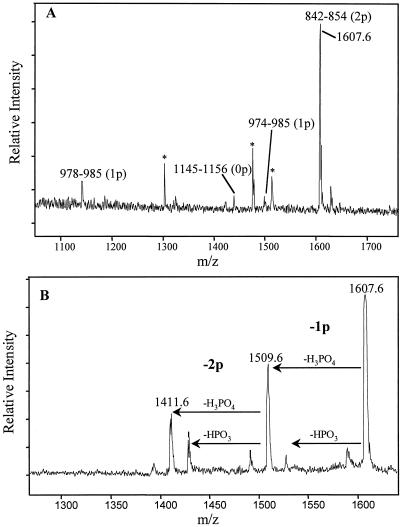
Figure 2A shows a representative MALDI mass spectrum from one HPLC fraction of the tryptic BRI1-KD digest. A prominent peak at m/z 1,607.6 is predicted to represent the tryptic peptide 842-TANNTNWKLTGVK-854 of the BRI1-KD with the addition of two phosphates. Figure 2B shows the MALDI-PSD spectrum of this peptide, confirming the presence of two phosphates by the sequential loss of 98 D for each phosphate group. An exhaustive MALDI-MS/MALDI-PSD analysis of all HPLC fractions generated the data presented in Table I. Subsequent digests were performed on specific fractions, followed by further MALDI-MS, to eliminate some of the ambiguities in the exact location of phosphate groups within a tryptic peptide. For example, peptide 842 to 854 (containing an uncleaved tryptic site) was subjected to a second digestion with trypsin that produced peptides 842 to 849 and 850 to 854, and MALDI-MS analysis of the digest confirmed that only Thr-842 and Thr-846 were phosphorylated. Tryptic peptide 1,033 to 1,062 was cleaved under acidic conditions with cyanogen bromide which resulted in peptides 1,033 to 1,037 and 1,038 to 1,062. Further MALDI-MS analysis of the cleavage products showed that there were three sites in peptide 1,038 to 1,062 and none in 1,033 to 1,037. Tryptic peptide 870 to 899 was cleaved with AspN protease to yield peptides 870 to 874, 875 to 885, 886 to 895, and 896 to 899. MALDI-MS revealed that peptides 870 to 874 and 886 to 895 each contained one phosphorylation site. Tryptic peptide 1,157 to 1,171 was also cleaved with AspN protease to yield peptide 1,165 to 1,171, eliminating S-1163 as a possible phosphorylation site. In all, at least 12 sites of in vitro autophosphorylation in the BRI-KD were identified, five uniquely and seven with some remaining ambiguity about the specific phosphorylated residue(s) within particular peptide fragments.
Figure 2.
Determination of autophosphorylation sites by MALDI-MS. A, A portion of the MALDI mass spectrum from one HPLC fraction of the tryptic digest of BRI1-KD. The numbers identify the proteolysis products and p represents a phosphate group. The peak at m/z 1,607.6 represents peptide 842 to 854, which contains two phosphate groups. The peaks marked with an asterisk result from non-specific cleavage products. B, A portion of the MALDI-PSD spectrum of peptide 842 to 854. The presence of two phosphate groups is confirmed by the sequential loss of 98 D H3PO4 and 80 D HPO3 for each phosphate group. 2,5- Dihydroxybenzoic acid with diammonium citrate was used as the matrix in both spectra.
Table I.
Phosphorylation sites of BRI1 kinase domain determined by MALDI-MS
| Sequence | [M+H]+ | Sites of Phosphorylation |
|---|---|---|
| CBP vector tag | ||
| 8-NFIAVSAANR-17 | 1,143.2 | S-13 |
| 21-ISSSGALLVPR-31 | 1,180.3 | S-23 |
| BRI1 kinase domain | ||
| 825-EAELEMYAEGHGNSGDR-841a | 1,942.0 | S-838 |
| 842-TANNTNWKLTGVK-854 | 1,607.6 | T-842, T-846 |
| 855-EALSINLAAFEKPLR-869 | 1,753.0 | S-858 |
| 870-KLTFA-874 | 659.3 | T-872 |
| 886-DSLIGSGGFG-895 | 989.2 | One site: S-887 or S-891 |
| 978-LNWSTR-983 | 856.9 | One site: S-981 or T-982 |
| 1,038-DTHLSVSTLAGTPGYVPPEYYQSFR-1,062 | 2,786.0 | Three sites: T-1039, S-1042, S-1044, T-1045, T-1049, S-1060 |
| 1,165-DSQSTIR-1,171 | 886.3 | One site: S-1166, S-1168, T-1169 |
| 1,172-SIEDGGFSTIEMVDMSIK-1,189 | 2,040.2 | One site: S-1172, S-1179, T-1180, S-1187 |
Amino acid numbering corresponds to Li and Chory (1997) and GenBank accession no. AAC49810.
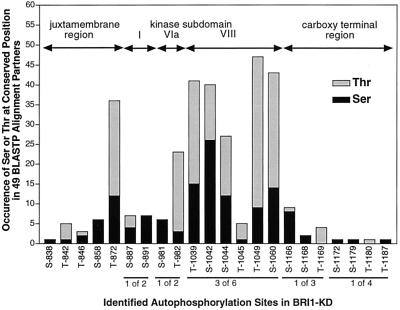
To determine if the Ser and Thr residues autophosphorylated in vitro in BRI1-KD are conserved at a corresponding position in related plant Ser/Thr kinases, we performed a BLASTP analysis (Altschul et al., 1990) of amino acids 815 to 1196 of BRI1 against the non-redundant GenBank database. The 49 sequences most closely related to BRI1-KD (ranging from 69% identical/83% similar to 41% identical/59% similar) were selected for further analysis. Figure 3 shows the number of times a Ser or Thr residue occurs in these 49 sequences at specific locations within the alignments corresponding to phosphorylated (or potentially phosphorylated, if ambiguity remains) sites in the BRI1-KD. The region where the greatest occurrence of Ser or Thr residues occurred at positions corresponding to potentially phosphorylated sites in the BRI1-KD was in the peptide 1,038-DTHLSVSTLAGTPGYVPPEYYQSFR-1,062, which lies in the highly conserved activation loop of domain VIII (Lease et al., 1998). Outside of domain VIII, the only other strongly conserved site for Ser or Thr in related kinases was the position equivalent to T-872 in BRI1-KD.
Figure 3.
Occurrence of Ser or Thr in related kinases at sites corresponding to autophosphorylation in BRI1-KD. BLASTP analysis was performed with amino acids 815 through 1196 of BRI1, comprising the entire KD. The top 49 matches were examined for the number of times a Ser or Thr occurred at a position in the alignment partner equivalent to a confirmed or possible autophosphorylation site in BRI1, as determined by MALDI-MS.
Putative BRI1 Kinase Substrate Recognition Sequences Determined by Synthetic Peptides
MALDI-MS also uncovered the surprising result that the N-terminal tag of CBP-BRI1-KD was phosphorylated when recombinant CBP-BRI1-KD was incubated with [γ-32P]ATP in vitro (Table I). Although two tryptic peptides of the CBP tag were radioactive, phosphor imager analysis showed that the peptide 21-ISSSGALLVPR-31 was more strongly labeled. To test the ability of BRI1-KD to phosphorylate synthetic peptides, we recloned BRI1-KD into the FLAG vector to eliminate the potentially confusing effects of phosphorylation of the CBP tag. The FLAG N-terminal tag is only 11 amino acids long and contains no Ser, Thr, or Tyr residues. Affinity-purified FLAG-BRI1-KD showed very strong autophosphorylation within the BRI-kinase sequence (Fig. 1A), so we then proceeded to examine a variety of synthetic peptides for in vitro phosphorylation by the recombinant kinase.
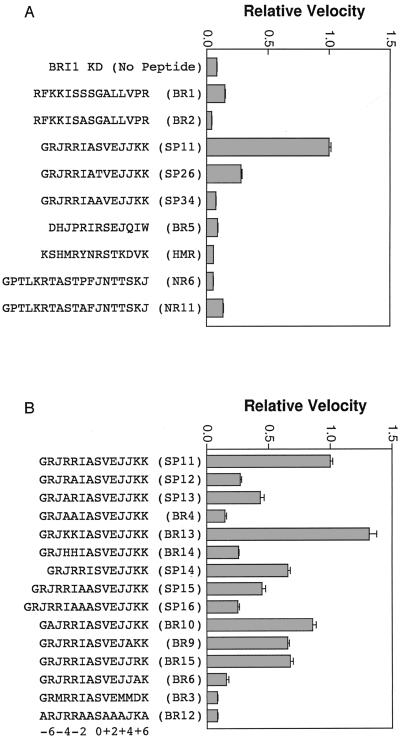
The synthetic peptide BR1 (RFKKISSSGALLVPR), based on the CBP tag sequence, was produced and found to be phosphorylated in vitro by recombinant FLAG-BRI1-KD (Fig. 4A). When the middle Ser of the peptide was replaced by Ala (BR2, Fig. 4A), the peptide was no longer phosphorylated and autophosphorylation of CBP-BRI1-KD was slightly inhibited, strongly suggesting that Ser-23 of CBP-BRI1-KD is the residue autophosphorylated in the CBP tag of the recombinant protein.
Figure 4.
Phosphorylation of synthetic peptides in vitro by FLAG-BRI1-KD. A typical 40-μL reaction mixture contained 1 μg of affinity-purified FLAG-BRI1-KD, 0.1 mm [γ-32P]ATP (500 cpm/pmol), and 100 μg/mL synthetic peptide in kinase buffer. Reactions were incubated for 20 min at ambient temperature and incorporation of 32P into the synthetic peptide was quantitated by binding to P81 phosphocellulose paper, followed by liquid scintillation spectrometry. Peptide BR1 corresponds to residues surrounding the phosphorylated Ser in the CBP vector tag of CBP-BRI1-KD. SP11, NR6, and hydroxymethyl glutaryl-coenzyme A reductase (HMR) are based on sequences surrounding the regulatory phosphorylation sites of spinach SPS, spinach nitrate reductase (NR), and Arabidopsis HMR, respectively. All other peptides are sequence variants of BR1, SP11 or NR6. J indicates nor-Leu, a nonoxidizing functional equivalent of Met. All velocities are normalized to SP11 = 1.0. Error bars are se, n = 3. A and B represent two separate experiments.
It is interesting that the BR1 peptide has basic residues at P − 3, P − 4, P − 6, and a hydrophobic residue at P − 5, relative to the phosphorylated Ser (P = 0), that is similar to the arrangement of amino acids in the regulatory phosphorylation sites of Suc-P synthase (SPS), nitrate reductase (NR), and HMG-CoA reductase (HMR) (Toroser and Huber, 1998). Figure 4A shows that FLAG-BRI1-KD did not phosphorylate synthetic peptides corresponding to either the regulatory phosphorylation sites of NR or HMR, but had strong activity on a peptide (SP11) based on the regulatory phosphorylation site (Ser-158) of spinach SPS (McMichael et al., 1995).
Using variants of SP11, we found that there were strict structural requirements for phosphorylation of synthetic peptides by FLAG-BRI1-KD in vitro (Fig. 4): (a) Replacing the Ser of SP11 with a Thr (SP26) reduced peptide phosphorylation by 79%, showing that the BRI1-KD has a preference for Ser over Thr, as also observed with phosphoamino acid analysis of autophosphorylation (Fig. 1C). (b) Substituting the Ser of SP11 with Ala (SP34) verifies Ser phosphorylation of the peptide by BRI1-KD, without inhibition of BRI-KD autophosphorylation, suggesting that the autophosphorylation recognition sites in BRI-KD may be substantially different than those found in the synthetic peptide. (c) Altering the spacing of the basic residues relative to the Ser (SP14–SP16) dramatically reduced phosphorylation of the peptide by BRI1-KD. Replacing the basic residues at P − 3 and/or P − 4 relative to the Ser with Ala (SP12, SP13, and BR4) also negatively affected phosphorylation, with the loss of both Arg residues resulting in an 87% drop in activity. Replacement of Arg at P − 3 (26% of SP11 phosphorylation) was more significant than at P − 4 (42% of SPll phosphorylation). (d) Exchanging the Arg residues of SP11 at P − 3 and P − 4 with Lys (BR13) enhanced kinase activity by about 32% whereas replacement with His residues (BR14) reduced activity by 75%. (e) Replacing the Arg residue at P − 6 with Ala (BR10) resulted in only a 15% decrease in activity, suggesting that a basic residue at this position is not as critical as at P − 3 and P − 4. (f) Replacing the basic residue at P + 5 relative to the Ser with Ala (BR6) reduced phosphorylation by 86%, whereas substitution of a negatively charged Asp residue at P + 5 (BR3) resulted in even greater loss of activity. Substitution of Arg for Lys at P + 5 (BR15) yielded only 67% of the activity of SP11, roughly equivalent to the preference for Lys over Arg observed at P − 3 and P − 4. (g) Replacing the hydrophobic residue at P + 4 of SP11 with Ala (BR9) resulted in a 35% decrease in peptide phosphorylation, suggesting that this position also has some role in substrate recognition. (h) NR6, which had positively charged groups at P − 3 and P − 4 and a hydrophobic residue at P − 5 but did not have a positive group at P − 6 or P + 5, was a very poor substrate, confirming the importance of a positive group at P + 5 in a different sequence context. NR6 also contained a Pro at P + 2 that would be expected to result in a bend in the peptide. Replacement of the Pro with Ala (NR11) improved phosphorylation slightly, but the new peptide remained a poor substrate. HMR retained the positively charged groups at P + 5, P − 4, P − 6, and the hydrophobic residue at P − 5. However, HMR had no positive charge at P − 3 and was not a substrate for BRI1-KD, again pointing to the critical requirement for a positive group at P − 3. Results nearly identical to those presented in Figure 4 were obtained when CBP-BRI1-KD was used in place of FLAG-BRI1-KD (data not shown).
Because the original SP11 peptide was based on spinach SPS, it was of interest to determine whether the corresponding Arabidopsis sequence would also be phosphorylated. It is not currently known whether Arabidopsis SPS is regulated by phosphorylation as it is in spinach. However, a recent search of the databases revealed a putative Arabidopsis SPS (65% similarity to spinach SPS) that has a Ser at position 180 with basic amino acids at P − 3 and P − 6 and a hydrophobic amino acid at P − 5, similar to the spinach Ser-158 motif (although the Arabidopsis SPS contains a Pro rather than a positively charged group at P − 4). We synthesized a peptide corresponding to the exact sequence of the Arabidopsis SPS in this region (BR5) but found that it was a poor substrate for in vitro phosphorylation by FLAG-BRI1-KD (Fig. 4A), which is not surprising considering the lack of a positively charged residue at P + 5. Although it is possible that Arabidopsis has other members of the SPS family (yet to be sequenced) that contain the appropriate recognition sequence, it is our current belief that the clear preference of BRI1-KD for in vitro phosphorylation of a peptide based on the spinach SPS regulatory phosphorylation site is simply fortuitous and is unlikely to reflect a direct interaction with SPS and BRI-KD in vivo. Even if SPS is not a true substrate of BRI1 in planta, our synthetic peptide analysis has identified some essential features required for substrate phosphorylation in vitro, which may give us a clue to the target sequences in intracellular substrates of BRI1 that are phosphorylated in vivo. Based on the peptide results, it is clear that R or K residues at P − 3, P − 4, and P + 5 are essential with a hydrophobic group at P + 4 and a positive group at P − 6 contributing to the highest observed activity.
Biochemical Characterization of BRI1 Kinase Activity in Vitro
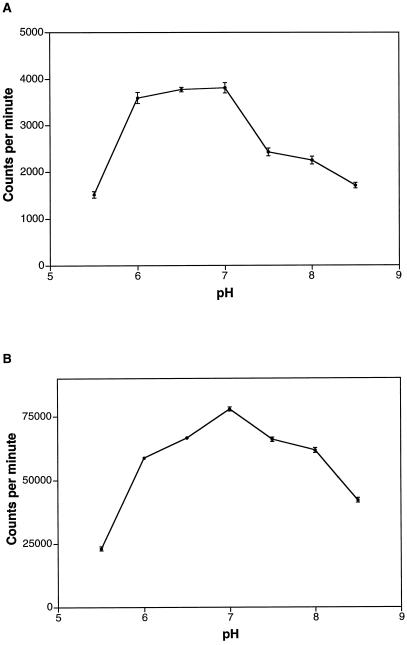
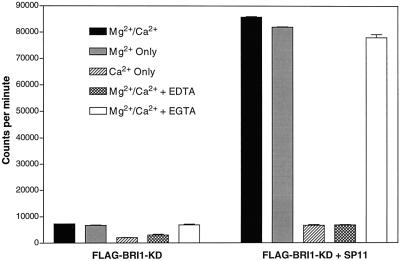
Recombinant FLAG-BRI1-KD and the synthetic peptide SP11 were used to further evaluate some biochemical properties of BRI1 kinase activity. To verify that the dramatic increase in bound radioactivity that occurred when SP11 was added to the reaction was a result of peptide phosphorylation, and not an allosteric activation of autophosphorylation by the peptide, we compared autophosphorylation of BRI1-KD in the presence and absence of SP11 (Fig. 5). The peptide clearly had no significant effect (either positive or negative) on BRI1-KD autophosphorylation and thus the increase in bound radioactivity truly was due to phosphorylation of SP11 by BRI1-KD, and phosphorylation of the synthetic peptide did not occur at the expense of autophosphorylation. Both FLAG-BRI1-KD autophosphorylation (Fig. 6A) and phosphorylation of SP11 (Fig. 6B) had pH optima near 7.0, and both activities required Mg2+ but not Ca2+ (Fig. 7).
Figure 5.
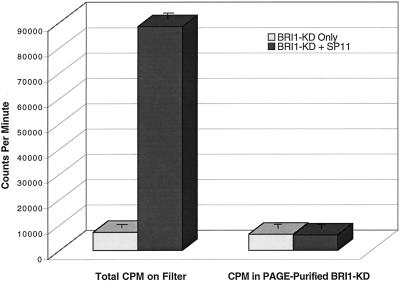
The synthetic peptide SP11 is a true substrate of BRI1-KD. To show that SP11 was in fact phosphorylated by BRI1-KD rather than stimulating BRI1-KD autophosphorylation, parallel replicate reactions were performed with BRI1-KD only, or BRI1-KD plus SP11, using conditions described in Figure 4. Reaction pairs were spotted on P81 paper, washed, and counted by liquid scintillation spectrometry to determine total radioactivity incorporated, or were separated by 10% (w/v) SDS-PAGE and the BRI1-KD band was excised and counted. SP11 had no detectable affect on incorporation of 32P into BRI1-KD, showing that increased filter-bound radioactivity in BRI1-KD + SP11 was due to phosphorylation of the peptide by BRI1-KD. Error bars are se, n = 3
Figure 6.
Dependence of BRI1-KD kinase activity in vitro on pH. Affinity-purified FLAG-BRI1-KD was assayed as described in Figure 4 except that the pH was varied from 5.5 to 8.5. A, Autophosphorylation (no synthetic peptide present). B, Phosphorylation of SP11. Error bars are se, n = 3
Figure 7.
Effect of Mg2+ and Ca2+ on BRI1-KD in vitro kinase activity. Affinity-purified FLAG-BRI1-KD was assayed as described in Figure 4 in buffer containing 10 mm Mg2Cl2 plus 0.2 mm CaCl2; 10 mm Mg2Cl2 only; 0.2 mm CaCl2 only; 10 mm Mg2Cl2 plus 0.2 mm CaCl2 with 6.0 mm EDTA; and 10 mm Mg2Cl2 plus 0.2 mm CaCl2 with 1.0 mm EGTA. Both FLAG-BRI1-KD autophosphorylation and phosphorylation of SP11 peptide were dependent on Mg2+ but not CaCl2. Error bars are se, n = 3.
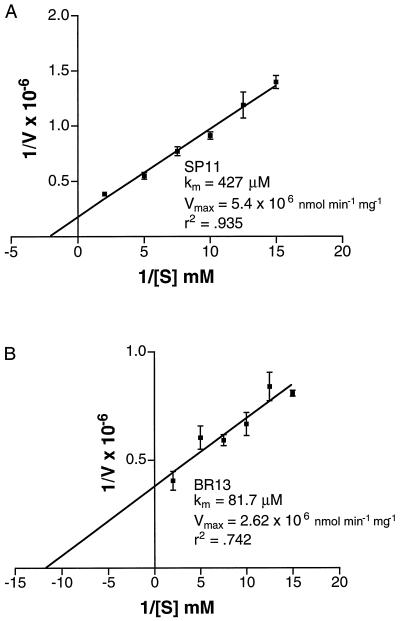
To determine if the two most active peptides, SP11 and BR13, were reasonable substrates compared with other known peptide substrates of plant kinases in vitro, we determined Km and Vmax for both peptides using double-reciprocal plots with six substrate concentrations (Fig. 8). The kinase clearly had a greater affinity for BR13 over SP11 (Km of 81.7 versus 427 μm, respectively) and the Vmax to Km ratio for BR13 was 2.5-fold higher than for SP11, suggesting that BR13 was a preferred substrate over SP11. Although BR13 and SP11 are not based on known physiological substrates for BRI1-KD, their Km values are within the range of those found for soluble plant kinases with peptide substrates that do reflect true physiological substrates. For example, cauliflower HMR kinase had a Km of 95 μm for the SAMS peptide based on conserved residues of known physiological substrates (Weekes et al., 1993), whereas a synthetic peptide modeled on the N-terminal phosphorylation domain of phosphoenolpyruvate carboxylase had an apparent Km of 540 μm for maize phosphoenolpyruvate carboxylase kinase (Li et al., 1997).
Figure 8.
Peptide substrate kinetics of BRI1-KD. Lineweaver-Burk double reciprocal plots for SP11 (A) and BR13 (B) were constructed using the indicated substrate concentrations, 1 μg of FLAG-BRI1-KD and 0.1 mm [γ-32P]ATP (500 cpm/pmol), in kinase buffer. Reactions were incubated for 20 min at ambient temperature and processed as described in Figure 4. Linear regression lines, Km and Vmax were determined using Prism 2.0 graphics software (San Diego).
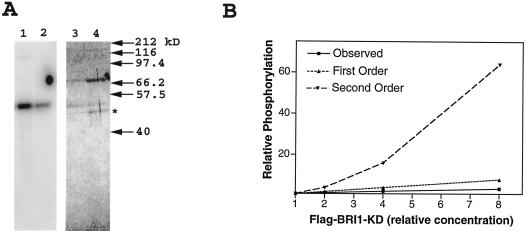
Examination of the sequence context of the confirmed and putative autophosphorylation sites in BRI1-KD revealed that the positive residues found at P − 3, P − 4, and P + 5 in the most active peptide substrates were not conserved in the autophosphorylation sites. One possible explanation for this difference is that autophosphorylation and substrate phosphorylation might proceed using different mechanisms with different sequence recognition motifs. The phosphorylation of a peptide substrate is, by definition, intermolecular. However, autophosphorylation might proceed by either intermolecular or intramolecular mechanisms. The availability of mutant and wild-type KDs with N-terminal tags of different molecular mass allowed us to distinguish between these two possibilities. Figure 9A shows that an active CBP-BRI1-KD failed to transphosphorylate an inactive FLAG-BRI1-K911E, suggesting an intramolecular rather than an intermolecular reaction mechanism. Moreover, when relative phosphorylation rate was plotted against increasing enzyme concentration (Fig. 9B), the kinetics more closely approximated first-order reaction kinetics (a linear increase in rate with increasing enzyme) than second order (rate increases with the square of enzyme concentration). Thus kinetic analysis also supports an intramolecular reaction.
Figure 9.
BRI1-KD autophosphorylation reaction mechanism. A, CBP-BRI1-KD (lane 1) or CBP-BRI1-KD + FLAG-BRI1-K911E (lane 2) was incubated with [γ-32P]ATP as described in Figure 1, separated by SDS-PAGE, and the incorporated isotope was visualized with a phosphor imager. Lanes 3 and 4 represent the Coomassie Blue-stained gel corresponding to lanes 1 and 2. The band marked with an asterisk corresponds to the FLAG-BRI1-K911E protein. Lack of a labeled band of the same size in lane 2 shows that CBP-BRI1-KD cannot transphosphorylate the mutant kinase, suggesting an intramolecular autophosphorylation mechanism. B, A plot of relative phosphorylation rate versus FLAG-BRI1-KD concentration. Actual amounts of FLAG-BRI1-KD varied from 0.5 μg to 4.0 μg per 40-μL reaction. Reaction conditions were as described in Figure 4. The approximation of first-order kinetics again suggests an intramolecular autophosphorylation mechanism.
DISCUSSION
BRI1 Encodes an Active Ser/Thr Kinase
Based on sequence similarity to known Ser/Thr kinases, including the presence of all 12 kinase subdomains with appropriate invariant amino acids (Li and Chory, 1997), it was expected that the BRI1-KD would be a functional kinase that autophosphorylates on Ser and/or Thr residues. To test this hypothesis, we expressed BRI-KD as a fusion protein with a small (4 kD) CBP to generate CBP-BRI1-KD. The KDs of over a dozen plant receptor-like kinases from eight different species have been successfully expressed as active recombinant kinases in E. coli, and it was therefore not surprising that affinity-purified CBP-BRI1-KD showed strong autophosphorylation when incubated with [γ-32P]ATP.
To rigorously verify that the major band seen in the autoradiograph of the autophosphorylation assay (Fig. 1B) corresponded to CBP-BRI1-KD, we excised this band, digested it with trypsin, and subjected the tryptic peptides to MALDI-MS analysis. The MALDI mass spectra were consistent with the expected molecular mass of the tryptic peptides calculated from the predicted amino acid sequence of CBP-BRI1-KD, indicating that the autophosphorylated band resulted from the recombinant protein and not an E. coli contaminant that copurified with CBP-BRI1-KD. Furthermore, a mutant kinase in which the essential Lys at position 911 in sub-domain II was replaced with Glu did not exhibit autophosphorylation, confirming that E. coli contaminants were not responsible for the phosphorylation observed when CBP-BRI1-KD was incubated with [γ-32P]ATP in vitro. The identity of the FLAG-BRI1-KD was also verified by MALDI-MS and similar results were obtained with mutant analysis (Fig. 1A). Moreover, CBP-BRI1-KD and FLAG-BRI1-KD showed the same sequence specificity for phosphorylation of synthetic peptides, with strong kinase activity for a peptide analog of the regulatory phosphorylation site of SPS, but not for NR, Suc synthase, or HMR. It is extremely unlikely that any E. coli kinase contaminant could be purified by both CBP- and FLAG-affinity chromatography and have the same kinetics on synthetic peptides. Thus all of the auto- and peptide-phosphorylation activities observed can be assumed to reflect BRI-KD activity.
Phosphoamino acid analysis of autophosphorylated CBP-BRI1-KD showed that, at least in vitro, BRI1-KD is a functional Ser/Thr kinase with a preference for Ser residues. The majority of the incorporated phosphates in CBP-BRI1-KD occurred in the area corresponding to the p-Ser standard, with a minor amount in p-Thr and none in p-Tyr. Thus BRI1 resembles CLV1 (Williams et al., 1997; Stone et al., 1998), RLK5 (Horn and Walker, 1994), Ath.lecRK1 (Herve et al., 1996), and SRK (Stein and Nasrallah, 1993) in its preference for Ser over Thr. However, other plant receptor-like kinases have either an equal propensity for autophosphorylation on Ser and Thr, i.e. RKF1 (Takahashi et al., 1998), or have a preference for Thr over Ser, including CrRLK1 (Schulze-Muth et al., 1996), TMK1 (Chang et al., 1992), SRK (Goring and Rothstein, 1992), KIK1 (Braun et al., 1997), RLK4 (Coello et al., 1999), and OsTMK (van der Knaap et al., 1999). A petunia receptor-like kinase has been described that apparently is a dual-function kinase that phosphorylates both Ser and Tyr residues (Mu et al., 1994). Pairwise alignment of BRI1-KD with other receptor-like kinases did not reveal any obvious motif that would predict whether the kinases autophosphorylated primarily on Ser or Thr. For example, based on BLASTP alignment scores and sequence identity, CrRLK1 (Schulze-Muth et al., 1996) and TMK1 (Chang et al., 1992), which autophosphorylate predominantly on Thr, were as closely related to BRI1 as RLK5 (Horn and Walker, 1994) and CLV1 (Stone et al., 1998), both of which have a preference for Ser. Nor was the type of extracellular domain any predictor of Ser versus Thr autophosphorylation, with both Leu-rich repeat and S locus-type glycoprotein domains occurring in receptor-like kinases that preferred either Ser or Thr.
BRI1-KD Autophosphorylates on Multiple Ser and Thr Residues in Vitro
Previous studies using two-dimensional TLE/thin-layer chromatography to analyze tryptic digests of recombinant KDs have suggested that several plant receptor-like kinases autophosphorylate on multiple sites, but individual residues cannot be identified by this method (Horn and Walker, 1994; Stone et al., 1998; Coello et al., 1999). To our knowledge, only one previous study has identified a specific site of autophosphorylation in a plant receptor-like kinase. Using formic acid digests and in vitro mutagenesis, it was shown that Thr-720 was required for autophosphorylation of CRLK1, a novel receptor-like kinase from Catharanthus roseus (Schulze-Muth et al., 1996). We used MALDI-MS analysis to show that FLAG-BRI1-KD autophosphorylates on at least 12 residues within the cytoplasmic domain. Exhaustive digestion with a variety of reagents uniquely identified five of these sites, whereas seven of the sites have been located within short peptides, but with some ambiguity remaining due to the number of Ser and Thr residues within the peptides. To resolve this ambiguity, sited-directed mutagenesis is currently being performed in which each of the Ser or Thr residues in question is successively replaced by Ala to assess their roles in autophosphorylation.
The cytoplasmic domain of receptor kinases can be subdivided into a juxtamembrane region, the kinase catalytic domain (with 12 conserved sub-domains), and a carboxy-terminal region. Autophosphorylation in all three of these regions is common (Johnson et al., 1996). Based on alignments of conserved amino acids in many kinases (Hanks and Quinn, 1991), the catalytic domain of the BRI1 kinase likely extends from Phe-883 through Phe-1155. Amino acids 815 through 882 then represent the juxtamembrane region and 1,156 through 1,196 comprise the carboxy-terminal segment. MALDI-MS analysis of BRI1-KD showed that at least five Ser or Thr residues were autophosphorylated in the juxtamembrane region and at least two sites were autophosphorylated in the carboxy-terminal region. The rather large number of autophosphorylation sites in these areas, if reflected in vivo, might indicate multiple, interacting cytoplasmic partners for BRI1, each with a specific phosphorylated target sequence within the juxtamembrane or the carboxy-terminal region of BRI1.
Plant receptor-like kinases function in many diverse physiological processes including growth and development, embryogenesis, fertilization, abscission, disease resistance, and response to light (Lease et al., 1998). Diversity of ligands and extracellular ligand-binding domains are expected to account for much of this functional diversity, but mechanisms for signal transduction pathway-specific cytoplasmic components to bind to specific KDs of receptor kinases are also required. Autophosphorylation of juxtamembrane and carboxy-terminal regions might be one way to achieve this specificity. BLASTP analysis of amino acids 815 through 882 and 1,156 through 1,196 of BRI1 revealed no sequence identity with any other kinases in the database. Moreover, when the 49 most closely related kinases to BRI1 (Fig. 3) were examined for conserved Ser or Thr residues in positions corresponding to autophosphorylated residues in the juxtamembrane and carboxy terminal regions of BRI1, only those corresponding to Thr-872 showed any significant number of conserved Ser or Thr residues.
The remaining five autophosphorylation sites we identified in BRI1-KD occur within the kinase catalytic domain (one in sub-domain I, one in sub-domain VIa, and three in sub-domain VIII). The latter three sites are of particular interest given that the activation of many protein kinases occurs by autophosphorylation of one to three residues within the activation loop of sub-domain VIII (Johnson et al., 1996). The activation loop, also referred to as the “activation segment,” begins with the invariant Asp in sub-domain VII (Asp-1,027 in BRI1) and terminates with the invariant Glu in sub-domain VIII (Glu-1,056 in BRI1). All kinases have an invariant Asp in sub-domain VIb (Asp-1,009 in BRI1) that is required for catalytic activity, and those kinases that require autophosphorylation of the activation loop for kinase activity also have an Arg immediately upstream of this Asp residue. The phosphorylated amino acid of the activation loop is thought to interact with the Arg to allow substrate access to the catalytic Asp (Johnson et al., 1996). BRI1 and many other plant receptor-like kinases also contain an Arg at this position. Genetic analysis also points to the functional importance of this region because numerous mutations in plant receptor-like kinases fall within the activation loop (Lease et al., 1998), including bri1-104 and bri1-115 (Li and Chory, 1997).
MALDI-MS analysis clearly showed that there were three sites of phosphorylation within the tryptic peptide 1038 to 1062, which overlaps with the majority of the activation loop, but we could not uniquely identify specific sites within the activation loop because six Ser or Thr residues occurred within this peptide (Table I). Further biochemical analysis and in vitro mutagenesis will be required to identify the specific Ser and/or Thr residues phosphorylated in this important region. However, it is clear that BRI1 autophosphorylates at least two residues within the activation loop (three if Ser-1,060 is not a site) and thus BRI1 might share the common requirement with many animal kinases for autophosphorylation of the activation loop for kinase activity. Autophosphorylation within this region is also likely to occur in other plant receptor-like kinases because inspection of sequences in the 49 plant kinases most closely related to BRI1 showed that over 50% of these related kinases had a Ser or Thr residue in positions corresponding to Thr-1,039, Ser-1,042, Ser-1,044, and Thr-1,049 in the activation loop of BRI1 (Fig. 3).
Features of the Putative Substrate Phosphorylation Site
In plant kinase research, the use of synthetic peptides has been limited to soluble kinases, particularly those involved in the phosphorylation of regulatory enzymes involved in metabolic pathways. Several soluble kinases have been partially purified from spinach leaf and cauliflower inflorescences that are able to inactivate SPS, NR, and HMR in vitro by phosphorylation on specific Ser residues (McMichael et al., 1993; Dale et al., 1995a; Douglas et al., 1997). Further analysis showed that some of these kinases were in fact homologs of the yeast SNF1 kinase and mammalian AMP-activated protein kinase, both of which limit the activity of specific metabolic pathways during stress by phosphorylation of regulatory enzymes such as HMR (Ball et al., 1995; Dale et al., 1995b; Sugden et al., 1999). Sequence alignments around the known regulatory phosphorylation sites of spinach SPS (Ser-158), spinach NR (Ser-543), and Arabidopsis HMR (Ser-577) from numerous species revealed several conserved residues (McMichael et al., 1993; Dale et al., 1995a; Douglas et al., 1997). It was then found that short synthetic peptides based on the sequences surrounding the phosphorylation sites of SPS, NR, and HMR were good substrates for the SNF1-related plant kinases in vitro (McMichael et al., 1995; Toroser and Huber, 1998). Extensive analysis with synthetic peptide analogs in which specific residues were substituted in turn by Ala showed that the conserved residues identified by sequence alignment were required for in vitro phosphorylation by the SNF1-related kinases (Weekes et al., 1993; Dale et al., 1995b; McMichael et al., 1995; Sugden et al., 1999). A consensus motif was generated which included the requirement for a positively charged residue at P − 3 and/or P − 4, a hydrophobic residue at P − 5, and in some cases, a hydrophobic residue at P + 4. SPS also required a positively charged residue at P − 6, whereas HMR had an invariant His at this position.
The finding that CBP-BRI1-KD has a prominent autophosphorylation site in the CBP tag with the same configuration of basic and hydrophobic amino acids at P − 3 through P − 6 as the configuration found for target sequences of SNF1-related kinases prompted us to test the ability of recombinant BRI-KD to phosphorylate a variety of synthetic peptide analogs of this motif in vitro. Initial experiments showed that FLAG-BRI1-KD had the highest in vitro activity on SP11, a modified form of the sequence surrounding Ser-158 of spinach SPS. SP11 differed from the native spinach sequence by the substitution of nor-Leu for Met, Ala for the non-phosphorylated Ser-157, and the truncation of the native sequence after P + 4 with the addition of two Lys residues to increase binding to the P81 paper (McMichael et al., 1995). Thus a positive charge was introduced at P + 5 that does not occur in the native sequence. Kinetic assays showed that spinach SPS kinase (PKIII, an SNF1-related kinase) had similar affinities for the native peptide and SP11 (McMichael et al., 1995). Using analogs of SP11, we found that the positioning of residues at both the N and C termini of the phosphorylated Ser was critical for optimal activity of BRI1-KD. Positive residues at P − 3, P − 4, and P + 5 were essential with a preference for Lys over Arg. A moderate reduction of activity was observed after substitution of the hydrophobic group at P + 4, with a lesser effect seen when the positive group at P − 6 was replaced with Ala. We have not yet tested the effect of replacing the hydrophobic group at P − 5.
Based on the data discussed above, we hypothesized that the putative consensus sequence X-[RK]-[LMVIFY]-[RK]-[RK]-X(2)-S-X(3)-[LMVIFY]-[RK]-X was required for peptide substrate recognition by BRI1-KD. However, when we synthesized a peptide (BR12) in which the residues marked as X in the putative consensus sequence were replaced with Ala, the peptide was a poor substrate (Fig. 4B). Thus at least one of the replaced residues is essential for recognition and we are currently synthesizing a new set of peptides in which residues at P − 2, P − 5, P − 7, P + 1 through P + 3, and P + 7 are substituted individually to more accurately define the BRI1-KD in vitro recognition sequence. Using the preliminary sequence [RK]-[RK]-X(2)-S-X(3)-[LMVIFY]-[RK] (corresponding to the most important residues identified to date) to search the Arabidopsis non-redundant protein database resulted in 350 hits. A variety of interesting proteins were found, some of which had obvious connections to signal transduction pathways. With continued refinement of the putative consensus sequence, a number of these will be eliminated and those remaining proteins with the putative BRI1-KD substrate recognition sequence might provide valuable molecular tools for further analysis. For example, peptides based on the conserved sequences could be used for in vitro assays with BRI1-KD. Those peptides that are efficiently phosphorylated would then be subjected to whole protein interaction analysis with recombinant BRI1-KD and any showing positive interaction in vitro could be further tested for in vivo interaction.
We also note that the identified autophosphorylation sites of BRI1-KD are not in the context of the proposed substrate recognition motif, suggesting that the mechanisms of substrate phosphorylation and autophosphorylation by BRI1-KD are likely to be different. Two methods have been employed to study the mechanism of autophosphorylation in plant receptor kinases: incubation of active and mutant KDs with different molecular mass vector tags to allow differentiation by SDS-PAGE and examination of reaction order with respect to enzyme concentration. By using one or the other of these methods, it was shown that RLK4 (Coello et al., 1999), RLK5 (Horn and Walker, 1994), and OsTMK (van der Knaap et al., 1999) followed intermolecular autophosphorylation, whereas CrRLK1 exhibited intramolecular autophosphorylation (Schulze-Muth et al., 1996). We employed both methods to show that BRI1-KD, like CrRLK1, followed an intramolecular autophosphorylation mechanism.
CONCLUSIONS
In summary, we have applied MALDI-MS and synthetic peptide substrate studies to plant receptor-like kinase analysis, and we have identified several novelbiochemical features of BRI1 in vitro. A critical step in the biochemical characterization of any receptor kinase is to determine the specific amino acid residues that are autophosphorylated. Ligand-dependent autophosphorylation of these residues leads to activation of the cytoplasmic domain, including competence to bind intracellular signal transduction partners and further phosphorylation of downstream components. In vivo characterization of specific autophosphorylation sites is ultimately necessary for a complete molecular understanding of receptor kinase function, but initial studies in vitro with recombinant KDs can yield a great deal of information and serve as a guide for designing in vivo studies.
The identification of protein substrates that are regulated by interaction with the cytoplasmic domain of receptor kinases is also critical in understanding signal transduction mechanisms. A variety of molecular genetic and biochemical approaches have been employed with plant receptor-like kinases in an attempt to identify putative in vivo binding partners, including yeast two-hybrid analysis (Bower et al., 1996; Gu et al., 1998), interaction cloning (Stone et al., 1994; Braun et al., 1997), and immunoprecipitation and purification of receptor-protein complexes (Trotochaud et al., 1999). In addition to these methods, synthetic peptides have been extensively used in animal Tyr receptor kinase research to understand binding motifs and KD substrate recognition consensus sequences (for review, see Kuriyan and Cowburn, 1997). It has been generally assumed in these studies that recombinant KD and synthetic peptide interactions in vitro will provide useful tools in predicting recognition sequences of in vivo substrates (Himpel et al., 2000). Our identification of critical residues for substrate recognition for BRI1-KD should soon lead to a consensus sequence that will likely provide an additional tool for identification of true in vivo substrates of this important plant-signaling molecule. At the very least, the conserved peptide motif provides a biochemical reagent for further mechanistic and kinetic characterization of the BRI1-KD.
MATERIALS AND METHODS
Cloning of BRI1 into Expression Vectors and Purification of Recombinant Protein
Ligation-independent cloning (LIC) was performed with PCR-amplified BRI1-KD and the Escherichia coli expression vector pCAL-n-EK (Stratagene, La Jolla, CA) which resulted in a CBP N-terminal fusion with BRI1-KD. This 40-amino-acid N-terminal addition (MKRRWKKNFIAVSAANRFKKISSSGALLVPRGSGSGDDDDK) allows efficient purification on a CaM-affinity resin in the presence of calcium. The Arabidopsis expressed sequence tag clone ATTS 4702, containing the BRI1-KD, was obtained from the Arabidopsis Stock Center (Ohio State University, Columbus) and amplified with the sense primer (5′-gacgacgacaagagagagatgaggaagagacgg-3′) and the antisense primer (5′-ggaacaagacccgtttggctctgtttctaactctc-3′). The underlined sequences represent vector-compatible LIC overhangs for cloning, whereas the gene-specific primer sequences resulted in amplification of DNA encoding amino acids 815 (first amino acid after the membrane spanning domain) through 1,196 (carboxy-terminal amino acid) of BRI1 (Li and Chory, 1997). The 50-μL PCR reaction contained buffer [20 mm Tris-HCl, pH 8.8, 10 mm KCl, 10 mm (NH4)2SO4, 2 mm MgSO4; 0.1% (v/v) Triton X-100; and 0.1 mg/mL bovine serum albumin], 200 μm dNTPs, 100 ng of ATTS 4702, 50 pmol of each primer, and 5 units of cloned pfu DNA polymerase (Stratagene). After pre-incubation at 94°C for 3 min, 20 cycles (of 94°C, 30 s; 50°C, 30 s; 72°C, 2.5 min) were performed followed by an extension of 10 min at 72°C. PCR products were gel purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) and single-stranded LIC overhangs were generated by treating the purified DNA with pfu DNA polymerase in the presence of dATP. Cloning into the pCAL-n-EK vector, transformation into E. coli BL21(DE2) pLysS cells, overexpression of recombinant protein, and purification of CBP-BRI1-KD by CaM-affinity chromatography were all performed exactly as described in the Affinity LIC Cloning and Protein Purification Kit Manual (Stratagene).
A second E. coli expression construct was generated in the pFLAG-MAC vector (Sigma, St. Louis). Expressed sequence tag clone ATTS 4702 was amplified with the sense primer (5′-gtcagcaagcttagagagatgaggaagagacgg-3′) and the antisense primer (5′-gtcagcggtaccttggctctgtttctaactctc-3′) using PCR conditions described above except that 25 cycles (of 94°C, 45 s; 64°C, 45 s; and 72°C, 2.5 min) were used. The PCR reaction was ethanol precipitated, resuspended in the appropriate buffer, and digested with HindIII followed by a second ethanol precipitation and digestion with KpnI. The digested PCR product was gel purified as described above and ligated with HindIII/KpnI digested, gel-purified pFLAG-MAC vector to yield FLAG-BRI-KD, consisting of an 11-amino acid N-terminal tag (DYKDDDDKVKL) followed by amino acids 815 through 1,196 of BRI1. Overexpression of FLAG-BRI-KD in BL21(DE2) pLysS cells and purification on an anti-FLAG M2 affinity gel were conducted as described in the manufacturer's instructions (Sigma). Both CBP-BRI1-KD and FLAG-BRI-KD were sequenced at the 5′- and 3′-vector/insert junctions to verify the constructs.
To generate the mutant kinases FLAG-BRI1-K911E and CBP-BRI1-K911E, the invariant Lys at position 911 in sub-domain II was substituted with Glu, which is predicted to eliminate kinase activity (Hanks et al., 1988). In vitro mutagenesis was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) with the primers 5′-ggaagcgcggtggctatcgagaaactgattc-3′ and 5′-gaatcagtttctcgatagccaccgcgcttcc-3′. Recombinant proteins were purified as described above and constructs were verified by DNA sequence analysis.
Autophosphorylation Assay and Phosphoamino Acid Analysis
For autophosphorylation assays, 1 to 5 μg of affinity-purified CBP-BRI1-KD or FLAG-BRI1-KD was incubated at ambient temperature for 1 h in a final volume of 40 μL with 20 μCi of [γ-32P]ATP (3,000 Ci/mmol; New England Nuclear, Boston) in kinase buffer (50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-KOH, pH 7.9, 10 mm MnCl2, 1.0 mm dithiothreitol, and 0.2 mm unlabeled ATP). Reactions were terminated by adding 40 μL of 2× Laemmli loading buffer (Laemmli, 1970), followed by 10% (w/v) SDS-PAGE and autoradiography. Phosphoamino acid analysis was performed with p-Ser, p-Thr, and p-Tyr standards on a model HTLE 7,000 TLE system (C.B.S. Scientific, Del Mar, CA). CBP-BRI1-KD was labeled with [γ-32P]ATP as described above, separated by 10% (w/v) SDS-PAGE and electrophoretically transferred to PVDF membranes. After autoradiography, the labeled band was excised, rewetted with methanol, blocked in 1 mL of 2% (w/v) bovine serum albumin (in 25 mm Tris-HCl, pH 7.5) and rinsed five times with 1 mL of distilled water. The treated membrane fraction was then incubated for 1 h at 110°C in 200 μL of 5.7 n HCl and taken to dryness in a vacuum microcentrifuge. Five microliters of first-dimension TLE buffer (1:10:89, formic acid:pyridine:water; pH 1.9) containing 20 mg/mL of phosphoamino acid standards (Sigma) was then added and the mixture was applied as a single spot to Kodak (Rochester, NY) cellulose thin-layer chromatography sheets. First-dimension electrophoresis was performed for 20 min at 1,500 V (pH 1.9), whereas the second dimension was performed for 16 min at 1,300 V (pH 3.5).
MALDI-MS
CBP-BRI1-KD or FLAG-BRI1-KD was affinity purified, incubated with [γ-32P]ATP, electrophoresed, and transferred to a PVDF membrane as described above. The PVDF membrane segment containing the autophosphorylated KD was rinsed with methanol and cut into 1-mm2 pieces. The membrane was then digested at 37°C for 24 h with 1.5 μg of porcine trypsin (Promega, Madison, WI) in 10% (v/v) acetonitrile (Aldrich, Milwaukee, WI), 1% (v/v) n-octyl-β-d-gluco- pyranoside (Sigma), and 100 mm Tris-HCl (Serva, New York). The solution containing the membrane pieces was then vortexed, sonicated for 5 min, and centrifuged for 3 min. The buffer containing the digestion products was then transferred to a microcentrifuge tube. Two washings of the remaining membrane pieces were carried out with 0.1% (v/v) trifluoroacetic acid (Spectrum, Gardena, CA), followed by transfer of the wash solution to the tube containing the digestion products. The digest was then separated on a Michrom BioResources (Auburn, CA) Ultrafast Microprotein Analyzer HPLC system equipped with a 1-mm i.d. C18 column. Solvent A contained 95% (v/v) water, 5% (v/v) ACN, and 0.1% (v/v) TFA; whereas solvent B contained 10% (v/v) water, 90% (v/v) ACN, and 0.1% (v/v) TFA. The gradient was as follows: 5% (v/v) B to 65% (v/v) B from 0 to 50 min; 65% (v/v) B to 95% (v/v) B from 50 to 51 min; 95% (v/v) B from 51 to 53 min; 95% (v/v) B to 5% (v/v) B from 53 to 55 min; and 5% (v/v) B from 55 to 63 min. Eluent was monitored at 214 nm. Fractions were collected by hand and 1 μL of each fraction was checked for 32P incorporation using a phosphor imager. The radioactive fractions were then analyzed by MALDI-MS to determine the sites of phosphorylation as previously described (Asara and Allison, 1999).
MALDI-MS spectra were recorded on a PerSeptive Biosystems (Framingham, MA) Voyager Elite delayed extraction time-of-flight reflectron mass spectrometer equipped with a nitrogen laser (337 nm, 3-ns pulse). Spectra were acquired in the linear mode with an accelerating voltage of 21 kV (128 laser shots average) to obtain a spectrum. PSD experiments were performed in the reflectron mode with an accelerating voltage of 21 kV, a grid voltage of 73.0%, and a guide wire voltage of 0.15% of the accelerating voltage. The timed ion selector was set for the precursor's m/z value and three spectra obtained with mirror ratios of 1.00, 0.96, and 0.67 were stitched together by the data system to obtain the PSD spectra (128 laser shots average). The samples were prepared by mixing 1 μL of analyte solution with 1 μL of saturated 2,5-dihydroxybenzoic acid (Sigma) containing 25 mm of diammonium citrate (J.T. Baker, Phillipsburg, NJ).
Peptide Synthesis and Assay Conditions
SP11 was designed around the regulatory phosphorylation site (Ser-158) of spinach SPS (McMichael et al., 1993). BR5 was based on sequences in Arabidopsis SPS homologous to the Ser-158 region of spinach SPS. NR6 and HMR were designed based on the known regulatory phosphorylation sites of plant NR and HMR, respectively (Toroser and Huber, 1998). BR1 corresponded to a portion of the CBP encoded by the pCAL-n-EK vector. All other peptides were structural variations of those listed above. Peptides were either synthesized on a Synergy 432A peptide synthesizer (Perkin-Elmer, Norwalk, CT) and purified by reverse-phase HPLC, or were synthesized by Research Genetics (Huntsville, AL). All peptides were greater than 90% pure. In most peptides, nor-Leu (J) was used as a nonoxidizing replacement for Met, which previous experiments had shown to have no effect on kinase enzyme kinetics (McMichael et al., 1995; Toroser and Huber, 1998).
For synthetic peptide assays, 40-μL reactions typically contained 0.1 mg/mL synthetic peptide, 1.0 μg FLAG-BRI1-KD, and 0.1 mm [γ-32P]ATP (500 cpm/pmol) in a buffer consisting of 50 mm MOPS [3-(N-morpholino)propanesulfonic acid], pH 7.5; 10 mm MgCl2, and 0.2 mm CaCl2. Following a 20-min incubation at ambient temperature, 20 μL of the reaction was spotted on a 2- × 2-cm piece of P81 phosphocellulose paper. The paper was washed three times in excess 75 mm H3PO4 (5 min per wash) and 32P incorporation into the peptide plus BRI1-KD was determined by liquid scintillation counting of the washed squares. Each experimental point was determined in triplicate, and the results are presented as means ± se.
ACKNOWLEDGMENTS
We thank John Allison for many useful discussions on MALDI-MS and Dikran Toroser for advice on peptide assays and TLE.
Footnotes
This work was supported by the National Science Foundation (Integrative Plant Biology Program), the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (Plant Growth and Development), and the North Carolina Agricultural Research Service.
LITERATURE CITED
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Annan RS, Carr SA. Phosphopeptide analysis by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal Chem. 1996;68:3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- Asara J, Allison J. Enhanced detection of phosphopeptides in matrix-assisted laser desorption/ionization mass spectrometry using ammonium salts. J Am Soc Mass Spectrom. 1999;10:35–44. doi: 10.1016/S1044-0305(98)00129-9. [DOI] [PubMed] [Google Scholar]
- Ball KL, Barker J, Halford NG, Hardie DG. Immunological evidence that HMG-CoA reductase kinase-A is the cauliflower homologue of the RKIN1 subfamily of plant protein kinases. FEBS Lett. 1995;377:189–192. doi: 10.1016/0014-5793(95)01343-1. [DOI] [PubMed] [Google Scholar]
- Bower M, Matias D, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein S, Goring D. Two members of the thioredoxin-h family interact with the kinase domain of a BrassicaS locus receptor kinase. Plant Cell. 1996;8:1641–1650. doi: 10.1105/tpc.8.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D, Stone J, Walker J. Interaction of the maize and Arabidopsiskinase interaction domains with a subset of receptor-like protein kinases: implications for transmembrane signaling in plants. Plant J. 1997;12:83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- Braun DM, Walker JC. Plant transmembrane receptors: new pieces in the signaling puzzle. Trends Biochem Sci. 1996;21:70–73. [PubMed] [Google Scholar]
- Chang C, Schaller G, Patterson S, Kwok S, Meyerowitz E, Bleecker A. The TMK1 gene from Arabidopsiscodes for a protein with structural and biochemical characteristics of a receptor protein kinase. Plant Cell. 1992;4:1263–1271. doi: 10.1105/tpc.4.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S, Feldmann K. Molecular genetics of brassinosteroid action. In: Sakurai A, Yokota T, Clouse S, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer; 1999. pp. 163–190. [Google Scholar]
- Clouse S, Sasse J. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Hall AF, Langford M, McMorris TC, Baker ME. Physiological and molecular effects of brassinosteroids on Arabidopsis thaliana. J Plant Growth Regul. 1993;12:61–66. [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thalianaexhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P, Sassen A, Haywood V, Davis K, Walker J. Biochemical characterization and expression of RLK4, a receptor-like kinase from Arabidopsis thaliana. Plant Sci. 1999;142:83–91. [Google Scholar]
- Dale S, Arro M, Becerra B, Morrice NG, Boronat A, Hardie DG, Ferrer A. Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem. 1995a;233:506–513. doi: 10.1111/j.1432-1033.1995.506_2.x. [DOI] [PubMed] [Google Scholar]
- Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995b;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- Douglas P, Pigaglio E, Ferrer A, Halford N, MacKintosh C. Three spinach leaf nitrate reductase-3-hydroxy-3-methylglutaryl-CoA reductase kinases that are regulated by reversible phosphorylation and/or Ca2+ions. Biochem J. 1997;325:101–109. doi: 10.1042/bj3250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J, Brand U, Running M, Simon R, Meyerowitz E. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Fujioka S. Natural occurrence of brassinosteroids in the plant kingdom. In: Sakurai A, Yokota T, Clouse S, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer; 1999. pp. 21–45. [Google Scholar]
- Goring D, Rothstein S. The S-locus receptor kinase gene in a self-incompatible Brassica napusline encodes a functional serine/threonine kinase. Plant Cell. 1992;4:1273–1281. doi: 10.1105/tpc.4.10.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Heldin C. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–224. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- Herve C, Dabos P, Galaud J, Rouge P, Lescure B. Characterization of an Arabidopsis thalianagene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. J Mol Biol. 1996;258:778–788. doi: 10.1006/jmbi.1996.0286. [DOI] [PubMed] [Google Scholar]
- Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- Horn MA, Walker JC. Biochemical properties of the autophosphorylation of RLK5, a receptor-like protein kinase from Arabidopsis thaliana. Biochim Biophys Acta. 1994;1208:65–74. doi: 10.1016/0167-4838(94)90160-0. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Kuriyan J, Cowburn D. Molecular peptide recognition domains in eukaryotic signaling. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lease K, Ingham E, Walker J. Challenges in understanding RLK function. Curr Opin Plant Biol. 1998;1:388–392. doi: 10.1016/s1369-5266(98)80261-6. [DOI] [PubMed] [Google Scholar]
- Li B, Pacquit V, Jiao J, Duff S, Maralihalli G, Sarath G, Condon S, Vidal J, Chollet R. Structural requirements for phosphorylation of C4-leaf phosphoenolpyruvate carboxylase by its highly regulated protein serine kinase: a comparative study with synthetic peptide substrates and mutant target proteins. Aust J Plant Physiol. 1997;24:443–449. [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinsteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- McMichael RW, Klein R, Salvucci M, Huber S. Identification of the major regulatory phosphorylation site in sucrose-phosphate synthase. Arch Biochem Biophys. 1993;307:248–252. doi: 10.1006/abbi.1993.1586. [DOI] [PubMed] [Google Scholar]
- McMichael RW, Jr, Kochansky J, Klein RR, Huber SC. Characterization of the substrate specificity of sucrose-phosphate synthase protein kinase. Arch Biochem Biophys. 1995;321:71–75. doi: 10.1006/abbi.1995.1369. [DOI] [PubMed] [Google Scholar]
- Mu J-H, Lee H-S, Kao T-H. Characterization of a pollen-expressed receptor-like kinase gene of Petunia inflataand the activity of its encoded kinase. Plant Cell. 1994;6:709–721. doi: 10.1105/tpc.6.5.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti J, Eyal Y, McCormick S. Pollen tube localization implies a role in pollen-pistil interactions for the tomato receptor-like protein kinases LePRK1 and LePRK2. Plant Cell. 1998;10:319–330. doi: 10.1105/tpc.10.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann K, Tax F. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park O, Schaefer T, Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc Natl Acad Sci USA. 1996;93:13704–13708. doi: 10.1073/pnas.93.24.13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer C, Nasrallah M, Nasrallah J. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- Schulze-Muth P, Irmler S, Schroder G, Schroder J. Novel type of receptor-like protein kinase from a higher plant (Catharanthus roseus) J Biol Chem. 1996;271:26684–26689. doi: 10.1074/jbc.271.43.26684. [DOI] [PubMed] [Google Scholar]
- Stein J, Nasrallah J. A plant receptor-like gene, the S-locus receptor kinase of Brassica oleraceaL., encodes a functional serine/threonine kinase. Plant Physiol. 1993;101:1103–1106. doi: 10.1104/pp.101.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, Collinge M, Smith R, Horn M, Walker J. Interaction of a protein phosphatase with an Arabidopsisserine-threonine receptor kinase. Science. 1994;266:793–795. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- Stone J, Trotochaud A, Walker J, Clark S. Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 1998;117:1217–1225. doi: 10.1104/pp.117.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Mu JH, Gasch A, Chua NH. Identification by PCR of receptor-like protein kinases from Arabidopsis flowers. Plant Mol Biol. 1998;37:587–596. doi: 10.1023/a:1005924817190. [DOI] [PubMed] [Google Scholar]
- Toroser D, Huber S. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase kinase and sucrose-phosphate synthase kinase activities in cauliflower florets: Ca2+dependence and substrate specificities. Arch Biochem Biophys. 1998;355:291–300. doi: 10.1006/abbi.1998.0740. [DOI] [PubMed] [Google Scholar]
- Trotochaud A, Hao T, Wu G, Yang Z, Clark S. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;11:393–405. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Song WY, Ruan DL, Sauter M, Ronald PC, Kende H. Expression of a gibberellin-induced leucine-rich repeat receptor-like protein kinase in deepwater rice and its interaction with kinase- associated protein phosphatase. Plant Physiol. 1999;120:559–570. doi: 10.1104/pp.120.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zafian P, Choudhary M, Lawton M. The PR5K receptor protein kinase from Arabidopsis thalianais structurally related to a family of plant defense proteins. Proc Natl Acad Sci USA. 1996;93:2598–2602. doi: 10.1073/pnas.93.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes J, Ball KL, Caudwell FB, Hardie DG. Specificity determinants for the AMP-activated protein kinase and its plant homologue analyzed using synthetic peptides. FEBS Lett. 1993;334:335–339. doi: 10.1016/0014-5793(93)80706-z. [DOI] [PubMed] [Google Scholar]
- Williams R, Wilson J, Meyerowitz E. A possible role for kinase-associated protein phosphatase in the ArabidopsisCLAVATA1 signaling pathway. Proc Natl Acad Sci USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]