Abstract
The Enhancing Neuroimaging Genetics through Meta-analysis (ENIGMA) Consortium is a global team science effort, now including over 800 scientists spread across 340 institutions in 35 countries, with the shared goal of understanding disease and genetic influences on the brain. This “crowdsourcing” approach to team neuroscience has unprecedented power for advancing our understanding of both typical and atypical human brain development.
Background and History
The ENIGMA Consortium (http://enigma.usc.edu/) was founded in 2009 with the goal of identifying genetic influences on brain structure. Named after the codebreaking effort led by Alan Turing and colleagues in World War II, the ENIGMA effort brings together scientists from around the world with an interest in “cracking” the complex codes that determine genotype-phenotype relationships in humans. Despite consistent evidence for the high heritability (~80%) of human brain anatomy, specific genes that determine brain structure had been hard to identify due to underpowered studies. As large-scale brain imaging studies are costly to conduct, ENIGMA took a novel, team-science approach to the problem by combining existing genomic and imaging data from around the globe. Initial efforts focused on identifying common variants that reliably influence normal brain structural variation. In studies of over 30,000 individuals, these efforts led to the discovery of the first significant, genome-wide variants associated with human hippocampal structure and other subcortical brain volumes (Hibar et al., 2017a; Stein et al., 2012). ENIGMA has since become a global alliance of over 800 scientists spread across 340 institutions in 35 countries, collectively analyzing brain imaging, clinical data, and other phenotypic and genomic data (Figure 1).
Figure 1. ENIGMA Worldwide Map.
Since 2009, ENIGMA has grown to become a global network of over 800 neuroscientists working together—spanning over 300 institutions across 35 countries.
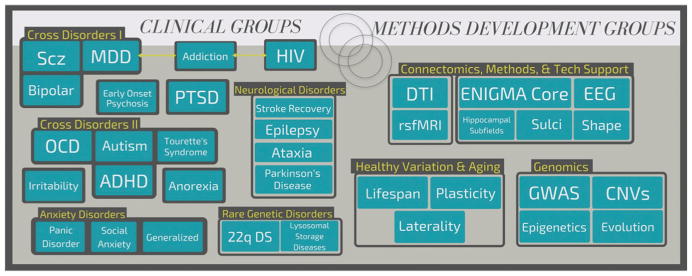
Building on ENIGMA’s initial successes in genetics, clinical working groups formed to study specific brain diseases, pooling data worldwide to study patterns of brain abnormalities in over 20 major psychiatric, neurodevelopmental, and neurogenetic disorders (Figure 2). Using sample sizes 10 to 30 times greater than a typical brain imaging study, ENIGMA recently published the world’s largest structural magnetic resonance imaging (sMRI) studies of major depressive disorder (MDD; n = 10,105; Schmaal et al., 2016), schizophrenia (n = 4,568; van Erp et al., 2016), bipolar disorder (n = 4,304; Hibar et al., 2017b), obsessive compulsive disorder (OCD; n = 3,589; Boedhoe et al., 2017), and attention-deficit/hyperactivity disorder (ADHD; n = 3,242; Hoogman et al., 2017), combining MRI data from >20,000 people. Large samples are critical to the study of these complex neuropsychiatric disorders, as they are highly heterogeneous at both the phenomenological and biological levels. Three of these studies focused on adolescent and adult-onset disorders—MDD, bipolar disorder, and schizophrenia—revealing consistent patterns for these diseases worldwide. Hippocampal volume deficits were common to all three disorders, whereas volumetric deficits in frontal regions were greater in bipolar disorder than in MDD and still greater in schizophrenia. ADHD and OCD—typically disorders with a developmental onset—showed distinct anatomical patterns, with previously unsuspected deficits in the amygdala (ADHD) and thalamus (OCD). These efforts leave no question that these illnesses—despite an absence of any objective laboratory diagnostic test—are all, in fact, disorders of the brain, and point to both common and unique effects of these disorders on brain structure. The initial studies led to many additional lines of inquiry; ENIGMA working groups began over 100 secondary projects to relate brain differences to risk factors, treatment response, and long-term outcomes. Cross-disorder comparisons have begun, seeking features that may prove useful for differential diagnosis (see Cross-Disorder groups, Figure 2).
Figure 2.
Roadmap of ENIGMA working groups
In addition to the disease-focused working groups described above, dedicated working groups focus on large-scale testing of imaging and genomics methods. One group relates epigenetic processes such as methylation to brain aging; others develop novel methods to harmonize and compare other types of brain imaging data and signals, such as diffusion MRI, resting state functional MRI (rs-fMRI), and electroencephalography (EEG). In collaboration with other consortia, including the Psychiatric Genomics Consortium (PGC), the Alzheimer’s Disease Neuroimaging Initiative (ADNI), Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE), IMAGEN, and others, ENIGMA’s genomic screens number over 30,000 MRI scans. Initial screens identified over 20 genetic loci associated with variation in brain volumes at a genome-wide, significant level and also designed a road-map to identify genetic overlap between brain measures and risk for psychiatric disorders (Franke et al., 2016). Newer methods, such as partitioning-based heritability analysis, show that multiple brain phenotypes are influenced by genetic variants that overlap with those associated with risk for schizophrenia (Lee et al., 2017). In Parkinson’s disease (PD), a genetic overlap analysis implicated MAPT, a microtubule-associated protein gene, in both PD risk and overall intracranial volume, suggesting that some variants may affect brain size in childhood and also boost the risk of PD later in life (Adams et al., 2016). Further elucidation of genes and pathways shared between brain structure, neuropsychiatric disorders, and/or cognitive traits may suggest new treatment targets related to a common underlying neurobiology.
Precision Medicine
Ultimately, one major goal of the ENIGMA consortium is to make predictions regarding individual prognosis. Detailed information on genetic and environmental contributions to brain structure over time can inform individual prediction.
ENIGMA’s Lifespan group has compiled charts of regional brain volumes and how they change throughout life, with percentiles based on over 10,000 people aged 3 to 90. Results to date indicate a developmental sequence in which the hippocampus achieves peak volume later in childhood than components of the basal ganglia; after age 60, the hippocampus tends to lose volume rapidly, while other structures only gradually decline. ENIGMA’s Laterality group found unsuspected right-left hemisphere differences that vary by sex in these brain metrics. A large, genome-wide screen is now underway to detect common genetic loci that influence individual rates of brain growth and aging (ENIGMA-Plasticity). Multiple working groups are now incorporating machine learning methods in the service of individual prediction of outcome. One such effort is the analysis of brain markers in MDD to determine which combination of brain measures best predicts diagnosis and future outcomes. Intriguingly, both MDD and OCD initiatives have identified specific structural abnormalities that differ between the adolescent and adult-onset forms of illness; these findings suggest that some disorders may shift in their effects throughout life, with some brain markers declining and others tending to normalize with development or treatment.
Lessons Learned: Are Brain Phenotypes More Efficient Targets for Genetic Analysis Than Disease Diagnosis?
Before such large datasets were amassed, many researchers expected that intermediate traits such as brain structure would have a simpler genetic architecture than neuropsychiatric disorders and thus might not require such large samples to identify genome-wide, significant effects. In the largest study to date of human hippocampal structure (n = 33,000), almost 20% of the variance in hippocampal volume was explained (in the aggregate) by genotyped common genetic variation (Hibar et al., 2017a). However, each of these variants individually explained only about 1% of the variance in brain metrics; in other words, effect sizes appeared comparable to those observed in other genome-wide association studies of complex traits. Notably, recent empirical modeling studies have nevertheless estimated the polygenicity of schizophrenia to be greater than that of putamen volume, a basal ganglia structure implicated in the disorder; thus, the sample size needed to fully explain chip heritability of schizophrenia is an order of magnitude greater than that required for putamen volume (Holland et al., 2016). Functional ENIGMA groups amassing data on rs-fMRI and EEG may discover functional connectivity phenotypes with different genetic architectures; parallel work in image-wide, genome-wide search and machine learning (Yang et al., 2010) show promise in detecting complex genotype-phenotype relationships.
CNV Effects on Brain Development
ENIGMA now encompasses a large body of scientific knowledge regarding genotype-phenotype relationships. While the past decade has seen vast advances in our understanding of the contribution of common genetic variation to neuropsy-chiatric disease, parallel evidence has emerged indicating that other types of genetic variation—deletions and duplications of DNA segments—occur more frequently in disorders such as autism and schizophrenia. Effects of these rare variants may be more challenging to identify, but their individual effects may greatly exceed those typical for common genetic variants.
Three of ENIGMA’s international disease working groups—22q-ENIGMA, ENIGMA-CNV, and a new group on lysosomal storage diseases—focus on the effects of highly penetrant copy number variants (CNVs) on brain development. The ENIGMA-CNV working group looks at CNVs in large-scale datasets and identifies microdeletions or microduplications with possible influence on brain imaging phenotypes. The next step is to more precisely define breakpoint regions in CNV carriers in order to conduct more precise gene-brain-behavior mapping. A recurrent CNV at the 22q11.2 locus is of particular of interest, as it represents one of the greatest known genetic risk factors for psychosis. Parallel work on rare syndromes such as Gaucher’s disease, an inherited lysosomal storage disorder, is using the ENIGMA model to aggregate data from major medical centers where enough patients are scanned to build up the first picture of the disease.
Challenges in Global Neuroscience
The challenges we faced in making ENIGMA a worldwide effort encompass both ethical and computational challenges regarding data sharing as well as complex issues regarding variations in science policy across countries. For a variety of reasons, many investigators may be unable to share raw imaging data; thus, ENIGMA has developed open-source methods for each contributing site to analyze their data in exactly the same way and provide summary statistics for meta-analysis (Box 1). Each group thus has full control over their own data. With some projects, this proved to be an intermediate step before moving to a “mega-analysis” approach that aggregates de-identified, individual-level data. With mega-analysis, the effects of individual risk scores (e.g., polygenic risk score for schizophrenia) are easier to model. Machine learning models such as “deep learning” that seek diagnostic or prognostic features in images are also easier to implement on individual-level data.
Box 1. Summary of the ENIGMA Consortium.
GOALS OF THE ENIGMA NETWORK
To create a network of like-minded individuals interested in pushing forward the field of imaging and genetics
To ensure promising findings are replicated via member collaborations
To share ideas, algorithms, data, and information on research studies and methods
To facilitate training, including workshops and conferences on key methods and emerging directions in imaging and genetics
GOVERNANCE
ENIGMA began in 2009 as a partnership focused on genome-wide association studies of the brain and grew to include groups from over 340 institutions in 35 countries. Working groups on schizophrenia, 22q11.2 deletion syndrome, bipolar disorder, depression, ADHD, OCD, and other disorders function as their own international consortia and are led by two to six leaders in those respective fields. Analysis methods for imaging and genomics are harmonized across groups, allowing cross-disorder projects. Participating members can propose and lead their own projects, which are supported by ENIGMA’s technical working groups.
FUNDING
The “ENIGMA Center for Worldwide Medicine, Imaging, and Genomics” (PI: Thompson) received 11 million dollars in funding in 2014 from NIH Big Data to Knowledge (BD2K) U54 EB020403, supporting nine of the ENIGMA working groups. Personnel and infrastructure used in ENIGMA have been supported by federal and private agencies from 35 countries; funders and major contributors are acknowledged in full in Hibar et al. (2017b).
METHODS DEVELOPMENT
Protocols for analyzing anatomical MRI, diffusion MRI, and genome-wide genetic data are available at http://enigma.usc.edu.
ENIGMA was recently described as “crowdsourcing meets neuroscience” (Mohammadi, 2015). Global collaboration, sharing of open source code and analysis tools, and multidisciplinary exchange of ideas are central to our approach. “Team science” projects like ENIGMA facilitate the engagement of scientists from low- and middle-income countries who may not have the resources to conduct large-scale investigations independently. Furthermore, greater opportunities for training are extended to more junior researchers who are able to take advantage of the network and its resources.
Multiple factors can influence reproducibility of imaging genomics findings, including differences in cohort inclusion criteria, demographics, severity and duration of illness, unmodeled risk factors, and variations in scanning protocols and analysis methods. ENIGMA maintains a strong commitment to reproducibility of scientific findings; its broad surveys of data from populations with different environments, experiences, and genetic backgrounds eases replication and confidence in the applicability of the results. ENIGMA has used standardized protocols for imaging and genomics, showing strong consistency in patterns of findings across Asia, the Americas, Europe, and the Middle East, as well as Russia, Australasia, and Africa. This international coordination has been facilitated by ENIGMA training workshops in Russia and Asia and by the fact that working groups are coordinated by junior scientists across many continents.
Future methods developments for ENIGMA are seeking deeper understanding of gene and disease effects on the brain. Anatomical MRI and diffusion image analyses are well-harmonized across diverse scanning protocols, but ENIGMA is beginning to test how reproducible and heritable functional MRI measures are across sites. Protocols are being designed to extract functional measures that are sensitive to disease biology but consistent and simple enough to apply on a large scale across many centers. Statistical methods also continue to evolve: gene-brain associations assessing a single genetic variant and a single brain measure at a time are being extended to newer “deep learning methods” that seek latent patterns in brain images and networks. One intriguing merger of methods is the “connectome-wide genome-wide” search—a quest to scan the genome for markers that predict the connectivity between brain regions and even their higher-order organization, growth, and decline.
The Future of ENIGMA
ENIGMA’s worldwide collaboration has brought together over 800 scientists to answer questions about genetic and environmental factors that affect the brain. Each disease working group is an autonomous consortium with its own projects and research goals (Figure 2), but common genomic and image analysis protocols across groups facilitate cross-disorder efforts. Current cross-disorder projects boost power to discover how risk factors affect the brain; these efforts are moving from simple anatomical MRI and diffusion tensor imaging into more complex functional phenotypes, which are more challenging to harmonize but may offer a deeper evaluation of brain dysfunction worldwide. In parallel, advances in statistical modeling will further our understanding of the genetic architecture of complex brain and disease phenotypes.
Acknowledgments
We thank Agnes McMahon for assistance in figure preparation and all ENIGMA participants worldwide for all their contributions. This work was supported in part by a consortium grant (U54 EB 020403) from the NIH institutes contributing to the Big Data to Knowledge (BD2K) Initiative. Names, affiliations, funding, and financial support of all ENIGMA Consortium members are acknowledged in individual ENIGMA publications available at http://enigma.usc.edu.
Biography
Paul Thompson is Professor of Neurology and Director of the Imaging Genetics Center at the University of Southern California, and he has served as PI for the ENIGMA Consortium since 2009. Carrie Bearden is Professor of Psychiatry and Biobehavioral Sciences and Professor of Psychology at UCLA; she joined the team as the leader of the 22q-ENIGMA disease working group. Both authors are passionate about the potential of global neuroscience for advancing our understanding of human brain development, both typical and atypical.
References
- Adams HH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Rentería ME, Trompet S, Arias-Vasquez A, Seshadri S, Desrivières S, et al. Nat Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedhoe PS, Schmaal L, Abe Y, Ameis SH, Arnold PD, Batistuzzo MC, Benedetti F, Beucke JC, Bollettini I, Bose A, et al. Am J Psychiatry. 2017;174:60–69. doi: 10.1176/appi.ajp.2016.16020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke B, Stein JL, Ripke S, Anttila V, Hibar DP, van Hulzen KJ, Arias-Vasquez A, Smoller JW, Nichols TE, Neale MC, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2; Enigma Consortium. Nat Neurosci. 2016;19:420–431. [Google Scholar]
- Hibar DP, Adams HH, Jahanshad N, Chauhan G, Stein JL, Hofer E, Renteria ME, Bis JC, Arias-Vasquez A, Ikram MK, et al. Nat Commun. 2017a;8:13624. doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Westlye LT, Trung Doan N, Jahanshad N, Cheung JW, Ching CRK, Versace A, Bilderbeck AC, Uhlmann A, Mwangi B, et al. Mol Psychiatry. 2017b http://dx.doi.org/10.1038/mp.2017.73.
- Holland D, Wang Y, Thompson WK, Schork A, Chen CH, Lo MT, Witoelar A, Werge T, O’Donovan M, Andreassen OA, Dale AM Schizophrenia Working Group of the Psychiatric Genomics Consortium; Enhancing Neuro Imaging Genetics through Meta Analysis Consortium. Front Genet. 2016;7:15. doi: 10.3389/fgene.2016.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogman M, Bralten J, Hibar DP, Mennes M, Zwiers MP, Schweren LS, van Hulzen KJ, Medland SE, Shumskaya E, Jahanshad N, et al. Lancet Psychiatry. 2017;4:310–319. doi: 10.1016/S2215-0366(17)30049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, Baker JT, Holmes AJ, Jahanshad N, Ge T, Jung JY, Cruz Y, Manoach DS, Hibar DP, Faskowitz J, et al. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.42. Published online February 21, 2017 http://dx.doi.org/10.1038/mp.2017.42. [DOI] [PubMed]
- Mohammadi D. Lancet Neurol. 2015;14:462–463. doi: 10.1016/S1474-4422(15)00005-8. [DOI] [PubMed] [Google Scholar]
- Schmaal L, Hibar DP, Sämann PG, Hall GB, Baune BT, Jahanshad N, Cheung JW, van Erp TG, Bos D, Ikram MA, et al. Mol Psychiatry. 2016 Published online May 3, 2016 http://dx.doi.org/10.1038/mp.2016.60.
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann Ø, et al. Alzheimer’s Disease Neuroimaging Initiative; EPIGEN Consortium; IMAGEN Consortium; Saguenay Youth Study Group; Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium; Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium. Nat Genet. 2012;44:552–561. [Google Scholar]
- van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, Agartz I, Westlye LT, Haukvik UK, Dale AM, et al. Mol Psychiatry. 2016;21:585. doi: 10.1038/mp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu J, Sui J, Pearlson G, Calhoun VD. Front Hum Neurosci. 2010;4:192. doi: 10.3389/fnhum.2010.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]