Abstract
Background
Vectors of mosquito-borne diseases in Sri Lanka, except for malaria, belong to the subfamily Culicinae, which includes nearly 84% of the mosquito fauna of the country. Hence, accurate and precise species identification of culicine mosquitoes is a crucial factor in implementing effective vector control strategies. During the present study, a combined effort using morphology and DNA barcoding was made to characterize mosquitoes of the subfamily Culicinae for the first time from nine districts of Sri Lanka. Cytochrome c oxidase subunit 1 (cox1) gene from the mitochondrial genome and the internal transcribed spacer 2 (ITS2) region from the nuclear ribosomal DNA were used for molecular characterization.
Results
According to morphological identification, the field collected adult mosquitoes belonged to 5 genera and 14 species, i.e. Aedes aegypti, Ae. albopictus, Ae. pallidostriatus, Aedes sp. 1, Armigeres sp. 1, Culex bitaeniorhynchus, Cx. fuscocephala, Cx. gelidus, Cx. pseudovishnui, Cx. quinquefasciatus, Cx. tritaeniorhynchus, Cx. whitmorei, Mansonia uniformis and Mimomyia chamberlaini. Molecular analyses of 62 cox1 and 36 ITS2 sequences were exclusively comparable with the morphological identifications of all the species except for Ae. pallidostriatus and Aedes sp. 1. Although the species identification of Armigeres sp. 1 specimens using morphological features was not possible during this study, DNA barcodes of the specimens matched 100% with the publicly available Ar. subalbatus sequences, giving their species status. Analysis of all the cox1 sequences (14 clades supported by strong bootstrap value in the Neighbor-Joining tree and interspecific distances of > 3%) showed the presence of 14 different species. This is the first available DNA sequence in the GenBank records for morphologically identified Ae. pallidostriatus. Aedes sp. 1 could not be identified morphologically or by publicly available sequences. Aedes aegypti, Ae. albopictus and all Culex species reported during the current study are vectors of human diseases. All these vector species showed comparatively high diversity.
Conclusions
The current study reflects the significance of integrated systematic approach and use of cox1 and ITS genetic markers in mosquito taxonomy. Results of DNA barcoding were comparable with morphological identifications and, more importantly, DNA barcoding could accurately identify the species in the instances where the traditional morphological identification failed due to indistinguishable characters of damaged specimens and the presence of subspecies.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2810-z) contains supplementary material, which is available to authorized users.
Keywords: DNA barcoding, Culicinae, Mosquitoes, Aedes, Culex, Sri Lanka, cox1, ITS2
Background
Correct species recognition of mosquito vectors is a vital component in implementing effective vector control strategies. Morphology based taxonomy and molecular characterization are the two major approaches used in species identification. Taxonomic keys used in morphological identification of mosquitoes mainly depend on the external features of adult females and fourth-instar larvae where the specimens must be handled and stored cautiously without damaging the external features, which is not practically possible all the time. Moreover, this approach needs expertise and is time consuming. Also, it does not identify the factors such as genetic variations and phenotypic plasticity that can have an impact on species level identification [1]. Alternatively, DNA barcoding or molecular characterization has become increasingly popular as an efficient method of species identification since it produces results with high precision even from a part of the specimen, within a short period of time [1]. Molecular taxonomy enables researchers to identify mosquitoes to species or subspecies level, understand genetic diversity and make predictions on evolution and phylogenetic relationships. Among all the barcoded insect groups, mosquitoes are the most intensely barcoded [2], probably because of their importance as vectors of many life threatening human diseases.
A region of the cytochrome c oxidase subunit 1 (cox1) gene located in the mitochondrial genome and a region in the nuclear ribosomal DNA internal transcribed spacer 2 (ITS2) have been the most frequently used genetic markers in DNA barcoding of mosquitoes. cox1 has been used as the only molecular marker in characterizing 37 Canadian, 62 Indian, 122 Chinese, 32 Pakistani and 45 Singaporean mosquito species [3–7]. A study conducted in India has used the same marker in investigating the molecular evolution of mosquito vectors of medical and veterinary importance [8]. The ITS2 region has been used in distinguishing closely related mosquito species belonging to various genera of the subfamily Culicianae, such as Culex [9] and Aedes [10]. Another barcode region used in mosquito barcoding studies is cox2 of the mitochondrial genome [11, 12]. D3 expansion segment (~1000 bp) a coding region located in the large subunit of the nuclear ribosomal DNA is also used as a molecular marker in mosquito barcoding studies since it shows a higher inter-/intraspecific variations.
Molecular markers, as a group, will provide more reliable information on the genetic variation between and within a species. Sri Lankan anophelines that included major and potential vectors of malaria have been studied successfully using both cox1 and ITS2 markers [13]. A study has used both these marker regions in barcoding the vector mosquitoes Ae. aegypti, Ae. albopictus, Culex tritaeniorhynchus, Cx. vishnui and Cx. pseudovishnui [14]. A combination of cox1, ITS2 and D3 has been used in another study to recognize sibling species of several mosquito species complexes [15]. However, researchers state that characterization of mosquitoes through integrated approaches using both morphological and molecular identification is the best approach for species identification [5].
Sri Lanka is a tropical country with high mosquito diversity supported by both natural and man-made factors for breeding and survival of mosquitoes. A total of 141 mosquito species belonging to 17 genera and two subfamilies have been reported from the country [16–18]. Among these 17 genera, Anopheles (23 species) belongs to the subfamily Anophelinae and the rest, i.e. Aedeomyia (1 species), Aedes (48 species), Armigeres (4 species), Coquillettidia (1 species), Culex (37 species), Ficalbia (1 species), Heizmannia (1 species), Hodgesia (2 species), Malaya (1 species), Mansonia (3 species), Mimomyia (4 species), Orthopodomyia (2 species), Topomyia (2 species), Toxorhynchites (2 species), Tripteroides (3 species) and Uranotaenia (7 species), belong to the subfamily Culicinae [16–18]. Culicinae is the largest group in Sri Lanka, represented by 118 species (~84% of the total mosquito species in the country). Mosquito-borne diseases have adversely affected the economy and the public health of the country. Nearly 150,000 dengue cases and 300 deaths by dengue have been recorded for the first eight months of 2017 [19]. Dengue and chikungunya are transmitted by Aedes aegypti and Ae. albopictus [20, 21]. Culex tritaeniorhynchus and Cx. gelidus transmit Japanese encephalitis [22], whereas Cx. quinquefasciatus transmits filariasis [23]. Anopheles culicifacies and An. subpictus act as the primary and secondary vectors, respectively, of malaria in Sri Lanka [24, 25]. Previously we reported molecular characterization of 16 anopheline species from Sri Lanka [13]. Here were report characterization of culicinae mosquitoes collected from nine administrative districts of Sri Lanka, using both morphology and molecular based taxonomy. Two commonly used genetic markers, cox1and ITS2, were used in molecular characterization.
Methods
Study sites
Mosquitoes were collected from nine administrative districts located in the three main climatic zones in Sri Lanka, i.e. Kandy, Matale and Nuwara-Eliya in wet zone (annual rainfall > 2500 mm rainfall); Badulla and Kurunegala in the intermediate zone (annual rainfall 1750–2500 mm rainfall) and Ampara, Batticaloa, Jaffna and Mannar in dry zone (annual rainfall < 1750 mm rainfall) Ampara, Batticaloa and Jaffna were coastal areas covered with little vegetation, few paddy fields and few human settlements. Kurunegala had little vegetation but a river running along the collection site. A dense vegetation cover was observed in the vicinity of Badulla and Matale study sites. Kandy site had a moderate vegetation cover and was surrounded by a significant human settlement. Mannar site was largely surrounded by paddy fields with few human settlements (Fig. 1).
Fig. 1.
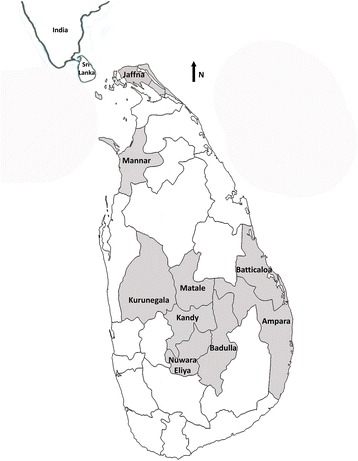
Map of Sri Lanka showing the nine administrative districts from which the mosquitoes were collected for the study
Mosquito collection
Adult mosquitoes were collected using cattle baited trap-huts, backpack aspirators, light traps and BG Sentinel traps. Dried specimens (a minimum of 10 individual mosquitoes) were used in morphological and molecular identifications. Mosquitoes were morphologically identified to generic and species level using standard taxonomic keys [26–29]. Voucher specimens of each species were pin mounted for a reference collection at the Invertebrate Systematic Diversity Facility (ISDF) at the Department of Zoology, Faculty of Science, University of Peradeniya, Peradeniya, Sri Lanka (UP/ZOO/BT011 - KU015). The reminder were dried and stored for molecular characterization.
DNA extraction, polymerase chain reaction (PCR) and sequencing
Genomic DNA was extracted from head and thoracic regions of each individual using following the method described by Livak [30]. Cytochrome c oxidase subunit 1 (cox1) gene was amplified using forward primer C1-J-1718 (5'-GGA GGA TTT GGA AAT TGA TTA GTT CC-3') and reverse primer C1-N-2191 (5'-CCC GGT AAA ATT AAA ATA TAA ACT TC-3') [31]. ITS2 region was amplified using the forward primer 5'-ATC ACT CGG CTC ATG GAT CG-3' and the reverse primer 5'-ATG CTT AAA TTT AGG GGG TAG TC-3' [32]. Each amplification was performed in 15 μl that included 1 μl of DNA template, 1.5 μl of 10× KAPA buffer A (Cape Town, South Africa), 0.12 μl of KAPA Taq, 0.12 μl of 2.5 mM dNTP mix, 0.75 μl of 50 mM MgCl2, 0.51 μl of each primer (10 mmol) and 10.49 μl of ddH2O. PCR parameters were 95 °C for 5 min and 35 cycles of 94 °C for 30 s, 51 °C (for cox1) / 55 °C (for ITS2) for 40 s and 72 °C for 45 s, followed by a final extension step of 72 °C for 10 min. PCR products were run in 1.5% agarose gel stained with ethidium bromide and visualized in a gel imaging system.
PCR products showing positive clear bands were purified using QIAquick® PCR Purification kits (Hilden, Germany) according to the manufacturers’ protocol. A maximum of three PCR positive samples of each species were sequenced bi-directionally at the Department of Molecular Biology and Biotechnology, Faculty of Science, University of Peradeniya.
DNA sequence analysis
The trace files/chromatograms of cox1 and ITS2 sequences were manually edited using BioEdit software. Sequences of low quality were excluded during data analysis. Consensus sequences were aligned using Clustal W in BioEdit software. Once the alignment was completed, species identification was confirmed by comparison to publicly available sequence data in GenBank using Basic Logical Alignment Search Tool (BLAST) [https://blast.ncbi.nlm.nih.gov/Blast.cgi] and the Barcode of Life Database (BOLD) interface [www.boldsystems. org]. The number of parsimony informative sites, number of variable sites, number of haplotypes, haplotype diversity and GC content were analyzed using the DNA Sequences Polymorphism software (DnaSP, version 5.1.10). MEGA version 6.0 was used to calculate intraspecific and interspecific pairwise sequence divergence using the Kimura-2 parameter distance model [33]. Neighbor-Joining (NJ) phylogenetic trees of cox1 and ITS2 sequences were constructed in MEGA 6.0 using Kumura-2 Parameter distances. Branch support of NJ trees were assessed by bootstrapping with 1000 replicates. Codon positions included 1st + 2nd + 3rd + noncoding regions. All the haplotype sequences of cox1 and ITS2 were deposited in the GenBank database (see below).
Results
According to morphological identification, the mosquitoes collected belonged to 14 species of 5 genera Aedes, Armigeres, Culex, Mansonia and Mimomyia (Table 1). A total of 62 cox1 sequences obtained from all 14 species and 36 ITS2 sequences obtained from only 10 species were used in genetic diversity and phylogenetic analysis. The fragment sizes of cox1 was 428 bp and that of ITS2 sequences ranged from 335 bp (Ae. aegypti) to 403 bp (Ae. albopictus) (Table 1). ITS2 sequences generated for Cx. bitaeniorhynchus, Cx. fuscocephala, Cx. gelidus and Cx. whitmorei were not considered for analysis as they were not of good quality. Fifty cox1 and 29 ITS2 haplotypes were obtained (Table 1).
Table 1.
The intraspecific distances and haplotype diversity of cox1 sequences. Intraspecific distances were calculated using Kimura 2-Parameter distance algorithm
| Species | n | h | GenBank accession numbers | Mean distance | Pairwise distance range | Haplotype diversity |
|---|---|---|---|---|---|---|
| Aedes aegypti (Linnaeus, 1762) | 4 | 3 | KY352243–KY352244, KY352256 | 0.010 ± 0.004 | 0.000–0.019 | 0.833 ± 0.049 |
| Aedes albopictus (Skuse, 1894) | 6 | 3 | KY352245–KY352247 | 0.002 ± 0.001 | 0.000–0.005 | 0.600 ± 0.046 |
| Aedes pallidostriatus (Theobald, 1907) | 6 | 4 | KY352248–KY352251 | 0.009 ± 0.003 | 0.000–0.017 | 0.800 ± 0.030 |
| Aedes sp. 1 | 4 | 4 | KY352252–KY352255 | 0.003 ± 0.002 | 0.002–0.005 | 1.000 ± 0.031 |
| Armigeres subalbatus (Coquillett, 1898) | 9 | 7 | KY352257–KY352263 | 0.004 ± 0.002 | 0.000–0.007 | 0.944 ± 0.005 |
| Culex bitaeniorhynchus (Giles, 1901) | 1 | 1 | KY040659 | n/c | – | n/c |
| Culex fuscocephala (Theobald, 1907) | 2 | 2 | KY040660–KY040661 | 0.014 ± 0.006 | – | 1.000 ± 0.250 |
| Culex gelidus (Theobald, 1901) | 2 | 1 | KY053491 | n/c | – | n/c |
| Culex pseudovishnui (Colless, 1957) | 4 | 4 | KY040662–KY040665 | 0.009 ± 0.004 | 0.005–0.012 | 1.000 ± 0.016 |
| Culex quinquefasciatus (Say, 1823) | 4 | 4 | KY040666–KY040669 | 0.002 ± 0.002 | 0.000–0.005 | 1.000 ± 0.050 |
| Culex tritaeniorhynchus (Giles, 1901) | 5 | 5 | KY040670–KY040674 | 0.007 ± 0.002 | 0.002–0.012 | 1.000 ± 0.031 |
| Culex whitmorei (Giles, 1904) | 1 | 1 | KY040675 | n/c | – | n/c |
| Mansonia uniformis (Theobald, 1901) | 11 | 9 | KY352264–KY352272 | 0.008 ± 0.003 | 0.000–0.017 | 0.945 ± 0.004 |
| Mimomyia chamberlaini (Ludlow, 1904) | 3 | 2 | KY352273–KY352274 | 0.002 ± 0.002 | 0.000–0.002 | 0.667 ± 0.099 |
Abbreviations: n total number of cox1 sequences, h number of haplotypes
Species identifications were performed using cox1 and ITS2 sequences compared to already available sequences in GenBank and BOLD system (Additional file 1: Table S1). Sequences obtained for Aedes aegypti, Aedes albopictus, all 7 Culex species, Mansonia uniformis and Mimomyia chamberlaini were 100% corresponding with their morphological species identifications. cox1 and ITS2 sequences generated for morphologically identified Armigeres sp. 1 specimens showed 99% similarity to the publicly available Armigeres subalbatus sequences, confirming its distinct species status.
Sequence data for cox1 and ITS2 markers from Aedes pallidostriatus were not available in the GenBank or BOLD systems to compare the sequence data generated for the morphologically identified Aedes pallidostriatus. The closest available sequences were from Aedes ochraceus (92% cox1 sequence similarity and 90% ITS2 similarity). Aedes sp. 1 could not be identified using taxonomic keys due to damages in identification features in the samples collected. According to molecular data, Ae. vexans was the closest species to Aedes sp. 1 specimens, with a cox1 similarity of 94% and ITS2 similarity of 82%. Hence, the species level identification of Aedes sp. 1 could not be confirmed using the available morphological and molecular data.
Phylogenetic analysis
The NJ tree constructed using the cox1 sequences formed 14 strongly supported clades (bootstrap value of 100%) 100% compatible with morphological identification (Figs. 2 and 3). All 7 species of genus Culex clustered together with Mi. chamberlaini specimens to form a single major clade, highlighting their close relationship. These two genera also share many common morphological characters. Haplotypes of Aedes aegypti and Ae. albopictus formed a separate clade while Ae. pallidostriatus and Aedes spp. 1 grouped into another clade with Ae. ochraceus and Ae. vexans sequences.
Fig. 2.
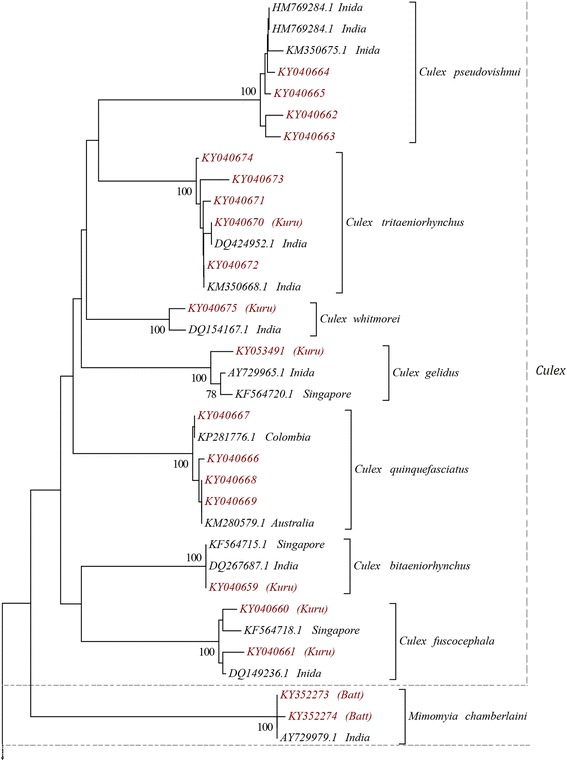
NJ phylogenetic tree (based on Kimura 2-parameter genetic distance model) based on cox1 sequences of all the 50 haplotypes of 14 species belonging to the subfamily Culicinae, collected from Sri Lanka during the study (red labels) and the sequences retrieved from the GenBank database (back labels). Only nodal support > 70% is shown
Fig. 3.
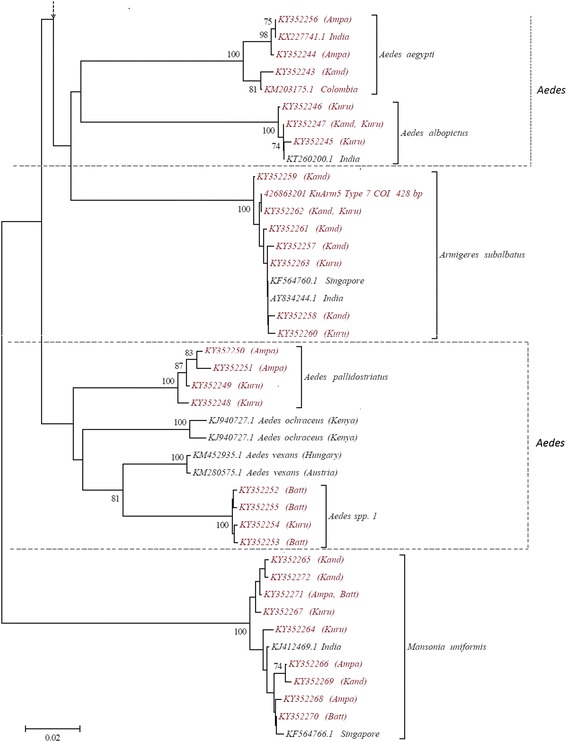
NJ phylogenetic tree based on Kimura 2-parameter genetic distance model for cox1 sequences of all the 50 haplotypes of 14 species belonging to the subfamily Culicinae, collected from Sri Lanka during the study (red labels) and the sequences retrieved from the GenBank database (back labels); continuation of Fig.2. Only nodal support > 70% is shown
Figure 4 shows the NJ tree constructed using the ITS2 sequences generated for the 10 mosquito species (except Culex bitaeniorhynchus, Culex fuscocephala, Culex gelidus and Culex whitmorei). All 10 species formed 10 strongly supported clades, each representing individual species. Further, 5 genera formed 5 major clades.
Fig. 4.
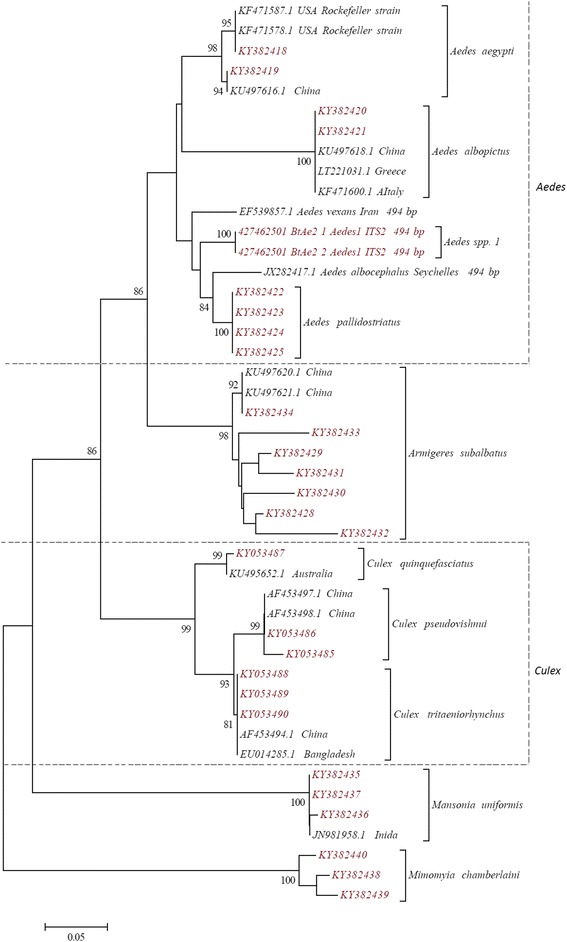
NJ phylogenetic tree based on Kimura 2-parameter genetic distance model for ITS2 sequences of all the 29 haplotypes of 10 species belonging to the subfamily Culicinae, collected from Sri Lanka during the study (red labels) and the sequences retrieved from the GenBank (back labels). Only nodal support > 70% is shown
DNA polymorphism analysis of cox1 sequences
Cox1 sequences were AT rich and the G+C content was 0.333. The number of variable bases was 145, accounting for 33.88% of the total number of sites. Among these variable sites, 142 were parsimony informative sites and only 3 represented singleton mutations. These nucleotide variations were heavily skewed to the third codon position which had 112 variable sites and 1 singleton mutation. First codon position had 28 variable sites and 2 singletons. The remaining 5 variable sites were at the second codon position without any singleton mutations.
Aedes sp. 1, Cx. fuscocephala, Cx. pseudovishnui, Cx. quinquefasciatus and Cx. tritaeniorhynchus had a haplotype diversity of 1.000, followed by Ma. uniformis. Aedes albopictus had the lowest haplotype diversity (0.600 ± 0.046) (Table 1).
As shown in Table 2, the mean intraspecific K2P distances for all the species were less than 2%. The maximum distance was seen among the haplotypes of Cx. fuscocephala which was 1.4% while Ae. albopictus, Cx. quinquefasciatus and Mi. chamberlaini reported the lowest mean intraspecific distance of 0.2%. The interspecific distances ranged from 6.8% between Cx. whitmorei and Cx. tritaeniorhynchus to 21.6% between Ma. uniformis and Ae. albopictus (Table 2).
Table 2.
Interspecific (below the diagonal) and mean intraspecific distances (along the diagonal) for cox1 sequences. Distances were calculated using Kimura 2-parameter distance algorithm
| Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ae. aegy | 0.010 | |||||||||||||
| 2 | Ae. albo | 0.147 | 0.002 | ||||||||||||
| 3 | Ae. pall | 0.151 | 0.144 | 0.009 | |||||||||||
| 4 | Aedes 1 | 0.140 | 0.143 | 0.095 | 0.003 | ||||||||||
| 5 | Ar. suba | 0.145 | 0.150 | 0.138 | 0.133 | 0.004 | |||||||||
| 6 | Cx. bita | 0.156 | 0.148 | 0.133 | 0.146 | 0.153 | 0.000 | ||||||||
| 7 | Cx. fusc | 0.164 | 0.160 | 0.142 | 0.137 | 0.147 | 0.084 | 0.014 | |||||||
| 8 | Cx. geli | 0.150 | 0.142 | 0.146 | 0.153 | 0.152 | 0.077 | 0.107 | 0.000 | ||||||
| 9 | Cx. pseu | 0.184 | 0.163 | 0.132 | 0.142 | 0.144 | 0.114 | 0.101 | 0.102 | 0.009 | |||||
| 10 | Cx. quin | 0.129 | 0.138 | 0.116 | 0.138 | 0.144 | 0.086 | 0.086 | 0.087 | 0.109 | 0.002 | ||||
| 11 | Cx. trit | 0.140 | 0.145 | 0.117 | 0.146 | 0.135 | 0.090 | 0.101 | 0.086 | 0.086 | 0.077 | 0.007 | |||
| 12 | Cx. whit | 0.153 | 0.134 | 0.129 | 0.128 | 0.144 | 0.079 | 0.092 | 0.071 | 0.086 | 0.069 | 0.068 | 0.000 | ||
| 13 | Ma. unif | 0.183 | 0.216 | 0.164 | 0.162 | 0.197 | 0.198 | 0.205 | 0.181 | 0.196 | 0.181 | 0.175 | 0.185 | 0.008 | – |
| 13 | Mi. cham | 0.141 | 0.168 | 0.145 | 0.162 | 0.178 | 0.112 | 0.131 | 0.140 | 0.150 | 0.133 | 0.126 | 0.134 | 0.190 | 0.002 |
Abbreviations: Ae. aegy Aedes aegypti, Ae. albo, Ae. albopictus, Ae. pall Ae. pallidostriatus, Aedes 1 Aedes sp. 1, Ar. suba Armigeres subalbatus, Cx. bita Culex bitaeniorhynchus, Cx. fusc Cx. fuscocephala, Cx. geli Cx. gelidus, Cx. pseu Cx. pseudovishnui, Cx. quin Cx. quinquefasciatus, Cx. trit Cx. tritaeniorhynchus, Cx. whit Cx. whitmorei, Ma. unif Mansonia uniformis, Mi. cham Mimomyia chamberlaini
Discussion
The present study provides both morphological and molecular characterization of a collection of mosquitoes belonging to subfamily Culicinae in Sri Lanka for the first time. Using the traditional morphology-based taxonomy, 14 mosquito species belonging to the genera Aedes, Armigeres, Culex, Mansonia and Mimomyia were identified. Three Aedes species were identified to the species level: Ae. aegypti, Ae. albopictus and Ae. pallidostriatus. The species referred to as Aedes sp. 1 and Armigeres sp. 1 could not be identified into its species level due to physical damages.
Seven Culex species, i.e. Cx. bitaeniorhynchus, Cx. fuscocephala, Cx. gelidus, Cx. pseudovishnui, Cx. quinquefasciatus, Cx. tritaeniorhynchus and Cx. whitmorei, one Mansonia species, Ma. uniformis, and one Mimomyia species, Mi. chamberlaini, were recognized with the aid of taxonomic keys. DNA barcoding conducted with cox1 and ITS2 confirmed the identity of these species and the sequence similarity with the publicly available sequences in the GenBank and BOLD system was 99–100%, except for Ae. pallidostriatus and Aedes sp. 1. There were no available sequences for Ae. pallidostriatus and ours was the first GenBank record of this species. Although Ae. vexans showed the closest sequence similarity to Aedes sp. 1, its species identity could not be finalized using both molecular data and morphological identification. Although species status of Armigeres samples was not identified morphologically, sequence data clearly support its identification as Ar. subalbatus.
The NJ phylogenetic trees constructed using both cox1 and ITS2 sequences, and genetic distance analysis, further supported the species identity and displayed the phylogenetic relationships between the species. Each individual species was represented by well supported clades (> 98% bootstrap support) confirming the morphological identification of 14 species. Interspecific distance of more than 3% between the cox1 sequences is considered as the threshold in differentiating species [1, 34] and this has been applied in many mosquito phylogenetic studies [4–6, 13]. According to the genetic distance estimates of cox1 sequences, the intraspecific distances of all the species identified during the present study was less than 3% (ranged between 0.2–1.4 %) while the interspecific distances were above 3% (6.8–21.6%). Mosquito barcoding studies have previously reported divergence ranges similar to the present study [4–6, 13]. Ten of the mosquitoes with ITS2 sequences (except Cx. bitaeniorhynchus, Cx. fuscocephala, Cx. gelidus and Cx. whitmorei) were represented by more than one ITS2 haplotype. Previous studies, had also reported ITS2 haplotype variations within a single species [35–37]. However, a study on anopheline mosquitoes from Sri Lanka reported the presence of species-specific ITS2 haplotypes only [13].
Aedes pallidostriatus and Aedes sp. 1 showed highest sequence similarity to Ae. ochraceus and Ae. vexans, respectively. Based on the morphological features, Ae. aegypti and Ae. albopictus belong to subgenus Stegomyia while Ae. pallidostriatus, Ae. ochraceus and Ae. vexans belong to subgenus Aedimorphus. In the phylogenetic tree based on cox1 sequences, the two members of the subgenus Stegomyia formed a single clade while Aedes sp. 1 grouped with the members of the subgenus Aedimorphus. Therefore, the molecular data strongly suggest that Aedes sp. 1 is a member of Aedimorphus.
Culex species recognition is mainly based on morphology of adult females and fourth-instar larvae. However, absence and overlapping of morphological characters have often been identified as factors that lead to misidentification of Culex species [38]. Hence, molecular characterization is important in accurate and precise identification of them. The present study provides a basis for Culex species identification in Sri Lanka using molecular approach. The mean cox1 intraspecific distance ranged between 0.2–1.4% and the interspecific distance ranged between 7.0–11.2% for Culex species. Similar interspecific distances have been reported for Culex species from eastern Amazonian Ecuador [39] and India [8]. The intraspecific distances evaluated for 22 Culex species in Argentina and Brazil varied between 0.09–3.00% [38] and that for 13 Culex species in Turkey between 0–0.8% [40].
Accurate and precise identification of mosquito vectors and determination of their genetic diversity is important especially in determining the vectorial capacities and planning vector control strategies. Among the mosquitoes studied, Ae. aegypti and Ae. albopictus are the primary and secondary vectors, respectively, of dengue and chikungunya, Cx. tritaeniorhynchus is the major vector of Japanese encephalitis (JE) and Cx. quinquefasciatus major vector of filariasis in Sri Lanka. Also, JE virus has been isolated from wild-caught Cx. pseudovishnui, Cx. gelidus, Cx. fuscocephala and Cx. whitmorei from Sri Lanka [22], and these too were barcoded during the current study. According to our results, almost all the species tested showed high genetic diversity which, in turn, demands a greater attention since uniform control measures might not work in the same manner for all the populations of a single species. The present study highlights the importance of molecular characterization in species recognition of mosquitoes which can be successfully incorporated to future development and implementation of vector control strategies.
Conclusions
The study showed that molecular characterization can be successfully employed for species identification of Culicinae mosquitoes. Results of DNA barcoding, using a combination of the genetic markers cox1 and ITS2, were comparable with morphological identifications and more importantly, DNA barcoding could accurately identify the species in the instances where the traditional morphological identification failed due to damaged specimens and indistinguishable characters. The cox1 and ITS2 sequences generated and submitted to the GenBank database could be used as reference sequences in future mosquito taxonomic studies. High genetic diversities observed in vectors of mosquito-borne diseases such as dengue, chickungunya and Japanese encephalitis should be taken into account in planning future vector control programmes in the country. Implementation of vector control programmes must be planned cautiously as uniform control measures may not be equally effective for genetically different populations.
Additional file
Table S1. Morphologically identified mosquito species, cox1 (fragment size of 428 bp) and ITS2 sequences (fragment sizes are separately listed for each species) generated from them, and the GenBank accession numbers for relevant submissions. Details of the closest publicly available sequences are also presented for comparison. (DOCX 19 kb)
Acknowledgements
Dr Devika Perera (Regional Malaria officer, Anti-Malaria Campaign, Kurunegala, Sri Lanka) and the Entomological team of Anti-Malaria Campaign, Kurunegala, Sri Lanka are greatly acknowledged for the assistance rendered during field collections.
Funding
This study was funded by the International Research Center (InRC), University of Peradeniya, Peradeniya, Sri Lanka (Grant No. InRC/RG/13/21).
Availability of data and materials
All data generated during this study are included in this published article. The datasets generated during the current study are available in the GenBank database.
Abbreviations
- BOLD
Barcode of Life Database
- cox1
Cytochrome c oxidase subunit 1
- ITS2
Internal transcribed spacer 2 region
- K2P
Kumura-2 Parameter
- NJ
Neighbor-Joining
Authors’ contributions
SHPPK and SNS conceptualized and designed the work. TCW did mosquito collections, morphological identifications and molecular laboratory work. All authors contributed in analyzing the data and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2810-z) contains supplementary material, which is available to authorized users.
Contributor Information
Thilini Chathurika Weeraratne, Email: thiliniw@pdn.ac.lk.
Sinnathamby Noble Surendran, Email: surendransn@gmail.com.
S. H. P. Parakrama Karunaratne, Email: shppk@pdn.ac.lk
References
- 1.Hebert PDN, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc R Soc Lond Biol. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ondrejicka DA, Locke SA, Morey K, Borisenko AV, Hanner RH. Status and prospects of DNA barcoding in medically important parasites and vectors. Trends Parasitol. 2014;30:582–591. doi: 10.1016/j.pt.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Cywinska A, Hunter FF, Hebert PDN. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20:413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- 4.Kumar NP, Rajavel AR, Natarajan R, Jambuligam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) J Med Entomol. 2007;44:1–7. doi: 10.1093/jmedent/41.5.01. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, et al. Identifying the main mosquito species in China based on DNA barcoding. PLoS One. 2012;7:e47051. doi: 10.1371/journal.pone.0047051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashfaq M, Hebert PDN, Mirza JH, Khan AM, Zafar Y, Mirza MS. Analyzing mosquito (Diptera:Culicidae) diversity in Pakistan by DNA barcoding. PLoS One. 2014;9:e97268. doi: 10.1371/journal.pone.0097268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan A, Chiang L, Hapuarachchi HC, Tan C, Pang S, Lee R, et al. DNA barcoding: complementing morphological identification of mosquito species in Singapore. Parasit Vectors. 2014;7:569. doi: 10.1186/s13071-014-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murugan K, Vadivalagan C, Karthika P, Panneerselvam C, Paulpandi M, Subramaniam J, et al. DNA barcoding and molecular evolution of mosquito vectors of medical and veterinary importance. Parasitol Res. 2016;115:107–121. doi: 10.1007/s00436-015-4726-2. [DOI] [PubMed] [Google Scholar]
- 9.Toma T, Miyagi I, Crabtree MB, Miller BR. Identification of Culex vishnui subgroup (Diptera: Culicidae) mosquitoes from the Ryukyu Archipelago, Japan: development of a species-diagnostic polymerase chain reaction assay based on sequence variation in ribosomal DNA spacers. J Med Entomol. 2000;37:554–558. doi: 10.1603/0022-2585-37.4.554. [DOI] [PubMed] [Google Scholar]
- 10.Beebe NW, Whelan PI, Van den Hurk AF, Ritchie SA, Corcoran S, Cooper RD. A polymerase chain reaction based diagnostic to identify larvae and eggs of container mosquito species from the Australian region. J Med Entomol. 2007;44:376–380. doi: 10.1093/jmedent/44.2.376. [DOI] [PubMed] [Google Scholar]
- 11.Cook S, Diallo M, Sall AA, Cooper A, Holmes EC. Mitochondrial markers for molecular identification of Aedes mosquitoes (Diptera: Culicidae) involved in transmission of arboviral disease in West Africa. J Med Entomol. 2005;42:19–28. doi: 10.1093/jmedent/42.1.19. [DOI] [PubMed] [Google Scholar]
- 12.Sharpe RG, Harbach RE, Butlin RK. Molecular variation and phylogeny of members of the Minimus group of Anopheles subgenus Cellia (Diptera: Culicidae) Syst Entomol. 2000;25:263–272. doi: 10.1046/j.1365-3113.2000.00118.x. [DOI] [Google Scholar]
- 13.Weeraratne TC, Surendran SN, Reimer LJ, Wondji CS, Perera MDB, Walton C, et al. Molecular characterization of Anopheline (Diptera:Culicidae) mosquitoes from eight geographical locations of Sri Lanka. Malar J. 2017;16:234. doi: 10.1186/s12936-017-1876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhananjeyan KJ, Paramasivan R, Tewari SC, Rajendran R, Thenmozhi V, Leo SV, et al. Molecular identification of mosquito vectors using genomic DNA isolated from eggshells, larval and pupal exuvium. Trop Biomed. 2010;27:47–53. [PubMed] [Google Scholar]
- 15.Garros C, Harbach RE, Manguin S. Systematics and biogeographical implications of the phylogenetic relationships between members of the Funestus and Minimus Groups of Anopheles (Diptera: Culicidae) J Med Entomol. 2005;42:7–18. doi: 10.1093/jmedent/42.1.7. [DOI] [PubMed] [Google Scholar]
- 16.Gunathilaka N, Fernando T, Hapugoda M, Abeyewickreme W, Wickremasinghe R. Revised morphological identification key to the larval anopheline (Diptera: Culicidae) of Sri Lanka. Asian Pac J Trop Biomed. 2014;4:222–227. doi: 10.12980/APJTB.4.2014C941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amerasingh FP, Munasingha NB, Kulasekera VL. Some new records of mosquitoes occurring in Sri Lanka. Mosq Syst. 1987;19:162–166. [Google Scholar]
- 18.Jayasekara N, Chelliah RV. An annotated checklist of mosquitoes of Sri Lanka (Diptera: Culicidae). UNESCO: Man and the Biosphere National Committee for Sri Lanka publication No. 8. Colombo: National Science Council of Sri Lanka; 1981. [Google Scholar]
- 19.Epidemiology Unit, Sri Lanka. Dengue update. http://www.dengue.health.gov.lk/index.php/updates. Accessed 26 Sep 2017.
- 20.Karunaratne SHPP, Weeraratne TC, Perera MDB, Surendran SN. Insecticide resistance and, efficacy of space spraying and larviciding in the control of dengue vectors Aedes aegypti and Aedes albopictus in Sri Lanka. Pest Biochem Physiol. 2013;107:98–105. doi: 10.1016/j.pestbp.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Weeraratne TC, Perera MDB, Mansoor MACM, Karunaratne SHPP. Prevalence and breeding habitats of the dengue vectors Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in the semi-urban areas of two different climatic zones in Sri Lanka. Int J Trop Insect Sci. 2013;33:216–226. doi: 10.1017/S174275841300026X. [DOI] [Google Scholar]
- 22.Peiris JSM, Amerasinghe PH, Amerasinghe FP, Charlesh CH, Perera LP, Arunagiri CK, et al. Viruses isolated from mosquitoes collected in Sri Lanka. Am J Trop Med Hyg. 1994;51:154–161. doi: 10.4269/ajtmh.1994.51.154. [DOI] [PubMed] [Google Scholar]
- 23.Wondji CS, De Silva WAPP, Hemingway J, Ranson H, Karunaratne SHPP. Characterization of knockdown resistance in DDT-and pyrethroid-resistant Culex quinquefasciatus populations from Sri Lanka. Trop Med Int Health. 2008;13:548–555. doi: 10.1111/j.1365-3156.2008.02033.x. [DOI] [PubMed] [Google Scholar]
- 24.Kelly-Hope LA, Yapabandara AMGM, Wickramasinghe MB, Perera MDB, Karunaratne SHPP, Fernando WP, et al. Spatio-temporal distribution of insecticide resistance in Anopheles culicifacies and Anopheles subpictus in Sri Lanka. Trans R Soc Trop Med Hyg. 2005;99:751–761. doi: 10.1016/j.trstmh.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Karunaratne SHPP, Hawkes NJ, Perera MDB, Ranson H, Hemingway J. Mutated sodium channel genes and elevated monooxygenases are found in pyrethroid resistant populations of Sri Lankan malaria vectors. Pest Biochem Physiol. 2007;88:108–113. doi: 10.1016/j.pestbp.2006.10.001. [DOI] [Google Scholar]
- 26.Amerasinghe FP. Illustrated keys to the genera of mosquitoes (Diptera: Culicidae) in Sri Lanka. J Natl Sci Found. 1995;23:183–221. [Google Scholar]
- 27.Amerasinghe FP. Keys for the identification of the adults of Genus Culex (Diptera: Culicidae) in Sri Lanka. J Natl Sci Found. 1995;23:221–239. [Google Scholar]
- 28.Barraud PJ. The fauna of British India, including Ceylon and Burma. Diptera. Vol V. Family Culicidae. Tribes Megarhinini and Culicini. London: Taylor and Francis; 1934. [Google Scholar]
- 29.Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Coleman RE, Richardson JH. Illustrated keys to the mosquitoes of Thailand. VI. Tribe Aedini. Trop Med Public Health. 2010;41:1–225. [PubMed] [Google Scholar]
- 30.Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon C, Frati F, Beckenbach A, Crepsi B, Liu H, Flook K. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- 32.Paskewitz SM, Collins FH. Use of the polymerase chain reaction to identify mosquito species of the Anopheles gambiae complex. Med Vet Entomol. 1990;4:367–373. doi: 10.1111/j.1365-2915.1990.tb00453.x. [DOI] [PubMed] [Google Scholar]
- 33.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 34.Hebert PDN, Ratnasingham S, de Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond Biol. 2003;270:96–99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manni M, Gomulski LM, Aketarawong N, Tait G, Scolari F, Somboon P, et al. Molecular markers for analyses of intraspecific genetic diversity in the Asian tiger mosquito, Aedes albopictus. Parasit Vectors. 2015;58:188. [DOI] [PMC free article] [PubMed]
- 36.Vinogradova EB, Shaikevich EV. Morphometric, physiological and molecular characteristics of underground populations of the urban mosquito Culex pipiens Linnaeus f. molestus Forskål (Diptera: Culicidae) from several areas of Russia. J Euro Mosq Control Assoc. 2007;22:17–24. [Google Scholar]
- 37.Miller BR, Crabtree MB, Savage HM. Phylogeny of fourteen Culex mosquito species, including the Culex pipiens complex, inferred from the internal transcribed spacers of ribosomal DNA. Insect Mol Biol. 1996;5:93–107. doi: 10.1111/j.1365-2583.1996.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 38.Laurito M, de Oliveira TMP, Almirón WR, Sallum MAM. COI barcode versus morphological identification of Culex (Culex) (Diptera: Culicidae) species: a case study using samples from Argentina and Brazil. Mem Inst Oswaldo Cruz. 2013;108:110–122. doi: 10.1590/0074-0276130457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linton Y, Pecor JE, Porter CH, Mitchell LB, Garzón-Moreno A, Foley DH, et al. Mosquitoes of eastern Amazonian Ecuador: biodiversity, bionomics and barcodes. Mem Inst Oswaldo Cruz. 2013;108:100–109. doi: 10.1590/0074-0276130440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guany F, Alten B, Simsek F, Aldemir A, Linton Y. Barcoding Turkish Culex mosquitoes to facilitate arbovirus vector incrimination studies reveals hidden diversity and new potential vectors. Acta Trop. 2015;143:112–120. doi: 10.1016/j.actatropica.2014.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Morphologically identified mosquito species, cox1 (fragment size of 428 bp) and ITS2 sequences (fragment sizes are separately listed for each species) generated from them, and the GenBank accession numbers for relevant submissions. Details of the closest publicly available sequences are also presented for comparison. (DOCX 19 kb)
Data Availability Statement
All data generated during this study are included in this published article. The datasets generated during the current study are available in the GenBank database.


