Abstract
The generation of large-scale biomedical data is creating unprecedented opportunities for basic and translational science. Typically, the data producers perform initial analyses, but it is very likely that the most informative methods may reside with other groups. Crowdsourcing the analysis of complex and massive data has emerged as a framework to find robust methodologies. When the crowdsourcing is done in the form of collaborative scientific competitions, known as Challenges, the validation of the methods is inherently addressed. Challenges also encourage open innovation, create collaborative communities to solve diverse and important biomedical problems, and foster the creation and dissemination of well-curated data repositories.
The growth of data in biomedicine is best exemplified by the estimated more than 250,000 human genomes that have been sequenced to date1, compared with the handful of genomes available only a decade ago. Sequencing data are only a small component of the big data deluge. Scientists are generating all types of omics data (including genomics, proteomics and metabolomics data), such as those produced by the Encyclopedia of DNA Elements (ENCODE)2, The Cancer Genome Atlas (TCGA)3, the International Cancer Genome Consortium (ICGC)4 and the Human Protein Atlas5. These projects are just a small portion of the biomedical data that are available6, which include: clinical, imaging, wearables and behavioural data.
In response to the challenges imposed by big data, new approaches to scientific research, such as cloud computing, are evolving to meet the needs of biomedical scientists7. Biomedical research can learn from other scientific fields that routinely deal with big data, such as astronomy8 and meteorology9, the communities of which have already learned how to share data and models as a common resource. Working within an information commons has facilitated the modelling of complex phenomena (including climate, ecology, migration and economics) and will do the same in the life sciences.
In addition to data sharing, a combined approach to data analysis is required. Reproducible analytical workflows of high sophistication are needed to maximize the extraction of hypotheses and, ultimately, knowledge out of the big data. The complexity and pace of data generation goes beyond the capacity and expertise of individuals or classic research groups, and requires the joint effort of a large number of scientists with a diverse set of skills. Only a concerted effort, driven by the scientific community, will accelerate the data-to-knowledge pipeline that will help us to address some of the most important and pressing issues in biomedicine.
An emerging paradigm that brings together large numbers of research scientists to address complex problems is the concept of crowdsourcing: a methodology that uses the voluntary help of large communities to solve problems posed by an organization10. Although the idea is not new11, the current practice of crowdsourcing is truly a product of our times in that it leverages the prompt feedback, ease of access, communication and a participatory culture that is fuelled by the Internet. Over the past 20 years, crowdsourcing has developed rapidly across many academic12 and commercial13 initiatives. In the context of biomedical research, many initiatives have developed in areas ranging from protein structure prediction to disease prognosis12.
In this Review, we begin with a brief introduction and a historical perspective on the use of crowdsourcing with a focus on scientific applications. We then focus on specific forms of crowdsourcing, known as Challenges or collaborative competitions, that are a powerful methodology to rigorously evaluate and vigorously advance the state-of-the-art of methods used to address certain scientific questions. Next, we present the most important elements of organizing a Challenge, which will provide useful information for both prospective organizers as well as participants to understand the motivation and rationale underlying some of the decisions that need to be made in defining Challenges. Finally, we review some of the scientific and sociological insights that this framework has provided and end with perspectives for the future development and applications of crowdsourcing.
What is crowdsourcing?
Coined by Jeff Howe in an article in Wired Magazine14, crowdsourcing combines the bottom-up creative intelligence of a community that volunteers solutions with the top-down management of an organization that poses the problem. The idea of leveraging a community of experts and non-experts to solve a scientific problem has been around for hundreds of years. One early example is the 1714 British Board of Longitude Prize that was awarded to the person who could solve what was arguably the most important technological problem of the time: to determine the longitude of a ship at sea15. After eluding many famous scientists, such as Leonhard Euler, the prize was awarded to a relatively unknown clockmaker named John Harrison for his invention of the marine chronometer. This example underlies two important concepts. The first is that the best solutions to difficult problems may require the knowledge from experts in adjacent fields — in this case, a carpenter and clockmaker. The second key idea is to pose the problem as an open participation challenge, what today is known as crowdsourcing, in order to solicit solutions from a wide range of sources without a priori expectations as to who may be best positioned to solve the problem.
Extrapolating the take home messages from the Longitude Prize to big data analytics, it is highly likely that the methods and breakthroughs that get the most useful signal from big data may reside with groups other than the data generators or the most famous and best published groups in a field. Furthermore, a common theme that arises across crowdsourced efforts is that the ensemble of analytical models that are independently generated by a crowd of experts offers robust predictions that are often better than the best individual predictions in the ensemble.
Crowdsourcing has been used in many contexts, including business (the design of consumer products13), journalism (the collection of information) and peer review (in the evaluation of patent applications). In this Review, we are interested in the application of crowdsourcing to the computational problems in biomedical sciences. Although there are different types of crowdsourcing (BOX 1), we will focus on Challenges.
Box 1. Types of crowdsourcing.
Generally speaking, crowdsourcing can refer to efforts in which the crowd provides data (for example, patients provide their medical information) to be mined by others or, alternatively, to initiatives in which the crowd actively works on solving a problem66. One type of active crowdsourcing is labour-focused crowdsourcing, in which work that needs to be done is proposed to a community willing to take up such a job13. A well-known example of labour-focused crowdsourcing is the ‘Mechanical Turk’ run by Amazon. The Mechanical Turk approach provides an online workforce that allows people to complete work, or ‘human intelligence tasks’, in exchange for a small amount of money67.
A complex problem can be divided into a set of smaller, independent tasks to benefit from crowdsourcing. Crowdsourcing data annotation and curation in bioinformatics can be handled well with this approach. This scheme has also been applied to provide pathway resources68,69, reconstruct the human metabolic network70, annotate molecular interactions in Mycobacterium tuberculosis71 and identify crucial errors in ontologies72.
In contrast to labour-focused forms of crowdsourcing, there are forms of crowdsourcing in which individuals participate because of their interest in the project or cause13. An example of this is the crowdsourced approach taken to develop the popular community encyclopedia Wikipedia. In some instances (such as Wikipedia and the protein structure game Foldit73), participants contribute their time and intellectual capacity, whereas in other examples (such as the Folding@home74 and Rosetta@home75 protein folding projects), participants provide computational power from their personal equipment to help solve the problem.
In some instances, crowdsourcing can be implemented in the form of a game76 to maximize the number of solvers who work on the problem and to increase the likelihood that they will stay engaged. For example, in the Foldit project, the problem of determining protein structure is transformed into an entertaining game. Such ‘ gamification’, in which game-design elements are used to allow an enjoyable experience, has proved a spectacular approach to raise participant numbers and interest. It also leads to results: Foldit’s 57,000 players provided useful results that matched or outperformed algorithmically computed solutions73. Foldit was followed by a similarly popular project, EteRNA77, in which more than 26,000 participants provided an RNA sequence that fits a given shape. The best designs, as chosen by the community, were then tested experimentally73,78. Hence, gamification is a powerful tool to engage massive numbers of volunteer citizen scientists to solve complex problems in which human intuition can outperform computer algorithms, even for abstract problems such as quantum computing79.
Crowdsourcing projects are also effective for collecting new ideas or directions that may be needed to solve a tough problem. These are referred to as ‘ideation’ Challenges, and the British Board of Longitude Prize mentioned in the introduction falls into this category. More recently, the Longitude Prize 2014 (REF. 80) built on the success of its predecessor to address the problem of antibiotic resistance through the creation of point-of-care test kits for bacterial infections. Among other ideation Challenges, the Qualcomm Tricorder XPRIZE81 encourages participants to develop a handheld wireless device that monitors and diagnoses health conditions.
Finally, crowdsourcing has been used in the context of benchmarking new computational methods. In this modality, a Benchmarking Challenge is set up in which data are provided to participants along with the particular question to be addressed. This is often to predict a different data set known only to the organizers (the so-called ‘gold standard’) and requires clear scoring metrics to evaluate the solutions (see Supplementary information S1 (box)). When the benchmarking aim is complemented with a framework that lets participants compete with others for the best solution, and the right incentives are provided to encourage participation, then a collaborative competition, or Challenge, is established, which is the focus of this Review.
Challenges: overview and platforms
A Challenge is a specific form of crowdsourcing that is now very popular among research scientists. These Challenges can be competitions organized by academic groups or by a for-profit company; they use voluntary labour to solve their own problems or those of a third party (typically other for-profit companies).
In the academic setting, the competitive side of a Challenge is usually complemented with an aim to build a community of solvers that work collaboratively to solve a tough scientific problem. Challenge organizers not only broadcast Challenges to a community of potentially interested solvers but also ask for ideas from the ‘crowd’ to address current problems in academic research.
For-profit companies are also leveraging the advantages of crowdsourcing. One of the best known examples of crowdsourcing in the for-profit world is the Netflix Prize, a Challenge that was organized by Netflix from 2006 to 2009 to identify the algorithms that would best determine which movies to suggest to their subscribers. The business of crowdsourcing consists of organizing Challenges as a fee-for-service for other companies that may not have the in-house expertise necessary to give solutions to a specialized task13,16. In such cases, crowds can fill that expertise gap.
The success of the crowdsourcing paradigm has spurred a proliferation of Challenge initiatives and platforms. Wikipedia17 lists more than 150 crowdsourcing projects in very diverse areas, such as design and technology innovation.
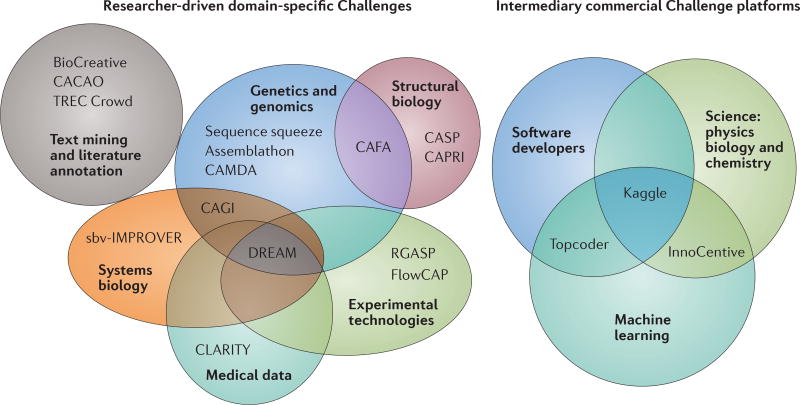
FIGURE 1 highlights some of the most notable crowdsourcing efforts and platforms in life science research. Among the researcher-driven Challenges, the areas that have most profited are: structural biology (Critical Assessment of protein Structure Prediction (CASP)18 and Critical Assessment of PRediction of Interaction (CAPRI)19); genomics (Sequence Squeeze, Assemblathon and Critical Assessment of Massive Data Analysis (CAMDA)); systems biology (Systems Biology Verification combined with Industrial Methodology for Process Verification in Research (sbv-IMPROVER) and Critical Assessment of Genome Interpretation (CAGI)); text mining (BioCreative20, Cross-language Access to Catalogues And On-line libraries (CACAO) and Text REtrieval Conference Crowdsourcing Track (TREC Crowd)); curation and annotation (Critical Assessment of Functional Annotation (CAFA)); medicine (Children’s Leadership Award for the Reliable Interpretation and appropriate Transmission of Your genomic information (CLARITY) and Medical Image Computing and Computer Assisted Intervention (MICCAI)); and emerging technologies in search of benchmarking and new analytical tools (Flow Cytometry Critical Assessment of Population Identification Methods (FlowCAP)21). Challenges also provide a framework to evaluate the ability of software pipelines to process different data types, such as the RNA-seq Genome Annotation Assessment Project (RGASP), which runs a competition to evaluate the software to align partial transcript reads to a reference genome sequence, which is a key step in RNA sequencing (RNAseq) data processing22,23. Other initiatives started with a narrow focus and then broadened their range. For example, the DREAM Challenges originally addressed the problem of inferring gene regulatory networks from experimental data24, hence the name DREAM: Dialogue for Reverse Engineering Assessment and Methods. However, DREAM has evolved to address challenges ranging from regulatory genomics24,25 to translational medicine26. These initiatives are often driven by academic efforts, although companies27 or other institutions — such as health providers (for example, the Heritage Provider Network (HPN) and their Heritage Health Prize Challenge16) and non-profit organizations and disease foundations (for example, the DREAM–Phil Bowen ALS Prediction Prize4Life Challenge and the Prostate Cancer DREAM Challenge in partnership with the Project Data Sphere Initiative) — also take an active part in their organization. The for-profit side of crowdsourcing Challenges is best exemplified by companies such as InnoCentive, Kaggle and Topcoder (FIG. 1).
Figure 1. Challenge platforms and organizations.
The most popular researcher-driven Challenge initiatives in the life sciences (left) and the most popular commercial Challenge platforms (right) are shown. Initiatives, such as DREAM (Dialogue for Reverse Engineering Assessment and Methods), FlowCAP (Flow Cytometry Critical Assessment of Population Identification Methods), CAGI (Critical Assessment of Genome Interpretation) and sbv-IMPROVER (Systems Biology Verification combined with Industrial Methodology for Process Verification in Research), organize several Challenges per year; only the generic project and not the specific Challenges are shown. Among the most popular and successful commercial Challenge platforms are: InnoCentive, which crowdsources Challenges in science and technology (social sciences, physics, biology and chemistry); Topcoder, which serves the software developer community; and Kaggle, which administers Challenges to machine-learning and computer experts, addressing predictive analytics problems in a wide range of disciplines. The figure is not comprehensive, but highlights the most consistent and well-established Challenge initiatives. CAFA, Critical Assessment of Functional Annotation; CACAO, Cross-language Access to Catalogues And On-line libraries; CAMDA, Critical Assessment of Massive Data Analysis; CAPRI, Critical Assessment of PRediction of Interaction; CASP, Critical Assessment of protein Structure Prediction; CLARITY, Children’s Leadership Award for the Reliable Interpretation and appropriate Transmission of Your genomic information; RGASP, RNA-seq Genome Annotation Assessment Project; TREC Crowd, Text REtrieval Conference Crowdsourcing Track.
Steps and components of a Challenge
The scientific question
Challenges often arise from scientific problems for which answers need new method development and validation28, or from the need to benchmark algorithms that yield divergent results and for which an objective evaluation could be appropriate29,30. However, the genesis of a Challenge could also be the emergence of new data repositories, the analysis of which could benefit from the crowdsourcing paradigm31,32,33. In all cases, the starting point is the definition of the scientific question that the Challenge aims to answer (FIG. 2). This question needs to be of fundamental clinical and/or basic research importance and formulated in a way that can be addressed in a collaborative-competition setting, typically in the form of an algorithmic prediction. This step usually involves coordination with a steering committee of experts in the domain area, such as physicians, biologists, toxicologists and genomicists. In addition, the question posed needs to be conceptually clear and attractive to researchers from many fields of study who can apply their specific principles and methods to address the question.
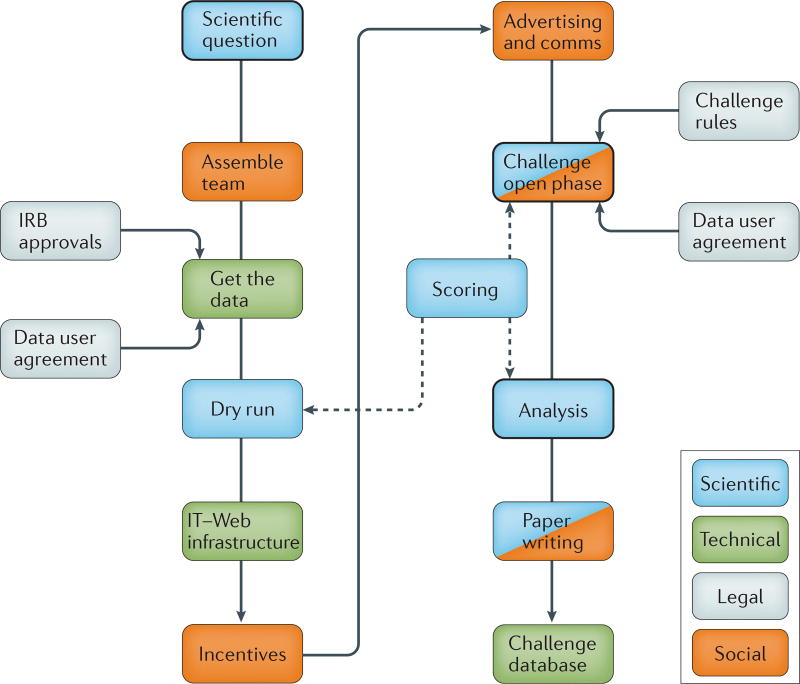
Figure 2. The steps and tasks in the organization of a Challenge.
The main scientific steps of developing a Challenge are: the determination of the scientific question, the pre-processing and curation of the data, the dry run, the scoring and judging, the post-Challenge analysis and the Challenge reporting and paper writing. Technical considerations include: development and maintenance of the IT infrastructure that requires registration, creation of computing accounts, security needed for cloud-based data hosting and development of submission queues, leaderboards and discussion forums. The legal considerations include agreements with the data providers regarding restrictions of data use and the agreement that participants will abide by the Challenge rules. The social dimension includes the creation of an organizing team to plan, run and analyse the Challenge, as well as to determine and put incentives in place for participation, to advertise the Challenge, to moderate the discussion forum and to lead the post-Challenge activities, such as paper writing and conferences. Comms, communications; IRB, Institutional Review Board.
Organizational infrastructure
Running Challenges requires input and expertise from different sets of specialists who all need to work together in a coordinated fashion. It is essential to assemble a team of specialists that includes: scientists, who develop the challenge question or questions; data governance specialists, who manage the data use agreements; data scientists, who perform data analysis tasks; and IT engineers, who support the IT infrastructure. Sometimes participants of previous Challenges can be engaged to help with these tasks. The typical tasks involved in a Challenge comprise four layers of expertise: scientific, technical, legal and social (FIG. 2).
Data procurement, hosting and internal analysis
The appropriate procurement and evaluation of the data needed for a Challenge is essential to the success of the effort. It is highly desirable that a portion of the data be unpublished so that it can be used as the ground truth (‘ gold standard’): that is, as a validation data set against which to score Challenge submissions. The amount of data provided in a Challenge must be sufficient to address the intended question. The underlying data must be of high quality but also have sufficient diversity and complexity that researchers will extract different patterns of signal from the data instead of finding only a subset of the important predictive features.
Having the data organized and packaged as an easy-to-use data set is necessary to reduce the barriers to participation. Adequate data governance has to be in place to ensure that data sharing is conducted in a legal and ethically responsible manner, particularly in the instances of data sets that include human data. This may require legal agreements with data producers and/or Institutional Review Board (IRB) oversight of human data sharing protocols.
A Challenge needs an IT infrastructure and web content. Important parts of such an infrastructure are: a registration system (one that requires participants to agree to the Challenge terms and conditions, including data use terms); a Challenge website that contains a detailed Challenge description, data set storage and the capability to download data sets and upload submissions; leaderboards that provide real-time feedback of performance; and a discussion forum where participants can communicate with organizers and other participants. To address issues around hosting big data and ensuring that algorithms are reusable, a few Challenge platforms (such as Kaggle, Synapse34 and Topcoder) have started to use cloud systems (for example, Amazon Web Services, IBM Softlayer and Microsoft Azure) for the storage of Challenge data and ‘Docker’ containers with the participants’ executable programs, which are ported to the cloud for running on the data. Finally, an archive of open-source Challenge methods in the form of ‘Dockerized’ re-runnable models (Synapse and Kaggle) facilitates ongoing open science research, even after a Challenge has finished.
Conducting an internal ‘dry run’ among the Challenge organizing team can be very revealing. It provides organizers a preview of the way in which participants will experience the Challenge website information and IT infrastructure, as well as the opportunity to work with the data sets to determine whether the scientific goals laid out in the Challenge can be attained. The typical dry run processes are: first, data sets are split into a training set, a cross-validation set and a test set; second, scoring metrics are selected; third, an estimate of the Challenge’s difficulty is made, considering the data at hand (if a Challenge seems impossible or too easy, then it may be better not to do it); and fourth, a definition is made of a baseline solution that the participants should improve upon.
Participant enrolment
The next step in organizing a Challenge is to define the incentives that will motivate as many participants as possible to take part. Incentives could include an invitation to the best performers to co-author a scientific paper describing the Challenge outcomes and insights, a speaking invitation to conferences and/or monetary awards. Many participants are enticed to just participate in a collaborative effort in which they can work on interesting and unpublished data to address a fundamental problem.
Before launching the Challenge, an aggressive advertising campaign should be in place. Successful marketing approaches include the use of press releases, pre-Challenge commentaries in relevant journals and outreach to researchers in the communities that are most directly connected to the Challenge in question.
Scoring
Challenges offer researchers a unique opportunity to have an objective, unbiased and rigorous performance evaluation of their algorithms and to avoid the traps of self-assessment35. Evaluation of Challenge solutions requires the development of quantitative metrics to compare submissions against the true outcomes, which are known to the organizers but not to the participants. Several scoring metrics can be used in the same Challenge to assess different aspects of the predictions36 (Supplementary information S1 (box)).
It is important to keep in mind that the scores in a Challenge are specific to the gold standard at hand, and the specific performance ranking that results from a Challenge may differ (albeit, not too much) if a different gold standard were used. The choice of a gold standard can be very clear (for example, in cases in which the Challenge is about predicting response to treatment37 or patient survival38) or noisy (such as in cases in which the predictions are compared with measurements containing experimental noise28,29). However, there are cases in which there is no perfect gold standard (often referred to as a ‘copper standard’). In such cases, the organizers can find alternative ways to score the submissions, but it may require the scoring metrics to be kept partially undisclosed. For example, in the HPN–DREAM Breast Cancer Network Inference Challenge39 the aim was to determine a causal signalling network in breast cancer cells from phosphoproteomics data. Because the true network is unknown, the Challenge used a procedure to determine causal links indirectly from experiments in which specific nodes are perturbed39.
In order for a final score to be meaningful, it has to be accompanied with a statistical criterion of how difficult reaching that degree of performance is, typically under a null hypothesis that assumes random predictions or predictions originating from off-the-shelf solutions to the Challenge.
Challenge open phase
After much preparation, the day arrives when the Challenge is launched, the data is crowdsourced and solutions to the scientific problems posed in the Challenge are solicited (TABLE 1). This ‘open phase’ is characterized by the progressive improvement of the algorithmic and mathematical techniques developed to solve the Challenge, which is facilitated by the use of leaderboards that allow participants to monitor their relative ranking with respect to others. A dialogue of ideas and data features can be encouraged by using a discussion forum. The open phase typically lasts from 3 to 6 months, but the specific duration depends on the complexity of the question. Challenge organizers can impose different restrictions about the copyright and intellectual property rights associated with the submissions. In an academic setting, participants are often asked to submit open-source code and a publicly accessible description of the methods used to make predictions in order to promote open and reproducible research. In a for-profit context, a winning participant may be asked to transfer copyrights and intellectual property rights in exchange for monetary awards.
Table 1.
Examples of Challenges*
| Challenge‡ | Challenge question | Gold standard | Winning methodology or algorithm |
Lessons and conclusions |
Legacy |
|---|---|---|---|---|---|
| Gene regulation and signalling network Challenges | |||||
|
Infer a transcription factor-to-target gene regulatory network |
|
|
|
|
|
Model the DNA binding sites of a TF based on PBM data | The measured degree of binding of each TF in the test set in an independent PBM | A method based on a k-mer model |
|
|
|
Predict the expression levels of genes downstream of ribosomal promoters based on the promoter DNA sequence | GFP fluorescence intensity driven by each promoter | SVM with a previous search for the best adapted feature, complemented by a previous physical model of TF and RNA polymerase interaction with DNA | General models to predict promoter expression did not fare well for predicting a specific family of promoters (ribosomal genes) |
|
|
|
|
Maximum likelihood fit of the model parameters given, observed data obtained from in silico experiments and construction of a game tree of possible sequences of the most informative data to use and experiments to perform |
|
|
|
|
|
|
|
|
| Translational and clinical challenges | |||||
|
Classify AML versus normal blood samples from flow cytometry data | Actual diagnosis of healthy versus AML blood samples in the test data set | Not very relevant in this context, as many algorithms had a perfect score | If the signal is clearly contained in the data, the choice of machine learning algorithms is not essential to identify correlates of clinical outcomes in flow cytometry data |
|
|
Predict the progression of patients with ALS from clinical trial data | Slope of change in ALS functional rating scale (a measure of disease status) per unit time | Two teams were identified as winners. One of them used a Bayesian additive random trees, whereas the other used random forest |
|
|
|
Predict the survival of patients with breast cancer on the basis of gene expression data, genomic copy number data and clinical covariates | The actual survival of patients in the test set | A method that used ‘attractor metagenes’ (REF. 88); these are features built by combining the expression of multiple genes using a mutual-information-based iterative algorithm | Copy number and gene expression data provided only an incremental performance improvement over clinical covariates alone, especially for aggressive high-grade tumours. This suggests that additional genomics data may be necessary to capture tumour progression |
|
|
|
|
|
|
|
|
Use genotype information to predict the response to anti-TNF therapy in patients with rheumatoid arthritis | Known response of patients in the test set | Gaussian process regression | Community phase showed that genetic predictors did not significantly contribute to anti-TNF response prediction |
|
| Genotype-to-phenotype prediction Challenges | |||||
|
Rank a panel of breast cancer cell lines from the most sensitive to the most resistant to a set of drugs based on gene expression, mutation, copy number, DNA methylation and protein quantification of the untreated cell lines | The concentration of a drug that inhibits the growth to 50% of the maximum (GI50), measured for 28 drugs across 18 breast cancer cell lines | A novel method that leveraged various machine learning approaches, including Bayesian inference, multitask learning, multiview learning and kernelized regression. This nonlinear, probabilistic model aims to learn and predict drug sensitivities simultaneously from all drugs |
|
|
|
Rank 91 compound pairs (all possible pairs of 14 compounds) from the most synergistic to the most antagonistic in a human lymphoma cell line, using gene expression profiles of cells perturbed with the individual compounds | Excess over Bliss, a measure of the deviation from additivity for all compound pairs | A method that hypothesized that when cells are sequentially treated with two compounds, the transcriptional changes induced by the first contribute to the effect of the second. A synergistic score was calculated by averaging two possible sequential orders of treatment between pairs of compounds |
|
|
|
|
Example data were provided | Recursive feature elimination followed by classification with an artificial neural network consisting of 50 input units, 10 hidden units and 1 output unit with sigmoid activation | The prediction of liver injury in humans using toxicogenomic data from animals is possible, but more data (especially non-toxic drugs) would be necessary to obtain better predictions | Challenge publication91 |
|
|
|
|
|
|
|
Match each of 77 genomes to the corresponding phenotypic profile from a list of 291 profiles (containing 214 ‘decoy’ profiles); each profile consists of 243 phenotypes | Known phenotypes of the subjects, as self-reported in surveys | Bayesian probabilistic model predicting the risk of a dichotomous phenotype using population-level prevalence as a prior, and integrating the contribution of rare and common variant genotypes in an individual | A model using the combination of GWAS hits, low-penetrance genes, high-penetrance genes and high-penetrance variants yields the best performance | Challenge publication63 |
| NGS data analysis | |||||
|
Assemble de novo a simulated diploid genome from short-read sequences | Simulated data | Several of the methods used variants of de Bruijn graphs; the best methods used heuristics for error correction, bubble removal, contig resolution, scaffolding and so on | The best sequence assemblers could reconstruct large sequences of a de novo genome at high coverage and with good accuracy |
|
|
Align RNA-seq reads to reference genomes, identifying loci of origin and reporting alignments with correctly placed introns, mismatches and small indels | RNA-seq from simulated transcriptome data | GSNAP, GSTRUCT, MapSplice and STAR compared favourably to other methods tested |
|
|
|
Identification and quantification of transcript isoforms based on RNA-seq data, assessed against well-curated reference genome annotation | RNA-seq and NanoString data |
|
|
|
|
|
|
Consensus model from the first three simulated data rounds resulted in a ‘meta’ algorithm that is far superior to any single algorithm used in genomic data analysis to date, highlighting the importance of considering a wisdom of crowds approach |
|
|
ALS, amyotrophic lateral sclerosis; AML, acute myeloid leukaemia; CAGI, Critical Assessment of Genome Interpretation; CAMDA, Critical Assessment of Massive Data Analysis; ChIP, chromatin immunoprecipitation; DREAM, Dialogue for Reverse Engineering Assessment and Methods; E. coli, Escherichia coli; FlowCAP, Flow Cytometry Critical Assessment of Population Identification Methods; GLM, generalized linear model; GRN, gene regulatory network; GSNAP, Genomic Short-read Nucleotide Alignment Program; GWAS, genome-wide association study; HPN, Heritage Provider Network; ICGC, International Cancer Genome Consortium; NCATS, US National Center for Advancing Translational Sciences; NCI, US National Cancer Institute; NGS, next-generation sequencing; NIEHS, US National Institute of Environmental Health Sciences; PBM, protein-binding microarray; PGP, Personal Genome Project; PWM, position weight matrix; RGASP, RNA-seq Genome Annotation Assessment Project; RNA-seq, RNA sequencing; S. cerevisiae, Saccharomyces cerevisiae; SNP, single-nucleotide polymorphism; SNV, single-nucleotide variant; STAR, Spliced Transcripts Alignment to a Reference; SubC, SubChallenge; SVM, support vector machine; TCGA, The Cancer Genome Atlas; TF, transcription factor; TNF, tumour necrosis factor; UNC, University of North Carolina.
A set of nineteen Challenges organized in the past six years (see also the additional case studies in the main text). Challenges are classified according to the research area. Challenge participants generally had a quantitative background from disciplines such as bioinformatics, computational biology, mathematics, statistics, physics, engineering and computer science; however, there were participants coming from the biological and medical sciences. An expanded version of this table, including information on the scoring metrics and the solvability of the problems from the supplied data, is provided as Supplementary information S2 (table).
Challenge overview, including name, reference, active years of Challenge and participation numbers.
Evaluation and analysis
When the open phase of the Challenge finishes, the analysis phase begins, in which submissions are evaluated to determine the best performers. In addition, meta-analyses of the submissions may be conducted to extract global insights into aspects of the Challenge, such as the scientific problem and the methods used (BOX 2).
Box 2. Lessons from Challenges.
Algorithms and methodological lessons
Simple is often better
Because a Challenge’s crowdsourcing attracts participants from many disciplines, the methodologies applied are very diverse. Often fairly simple methods, such as regression-based approaches, perform very well across many different domains, as they depend less on unverified hypotheses and are thus good starting points.
Prior knowledge
Integration of domain-specific prior knowledge about the problem under consideration seems to provide advantages in algorithm development. For example, in a Challenge to predict gene expression from promoter sequences, the best-performing team used machine learning without the use of additional biological knowledge. However, adding a posteriori information on the binding sites of a transcription factor significantly boosted the performance21,82. Similarly, one of the outcomes of the Heritage Provider Network (HPN)–Dialogue for Reverse Engineering Assessment and Methods (DREAM) Breast Cancer Network Inference Challenge was that the use of prior knowledge on signalling networks, even if obtained from different cellular contexts, boosted the performance in predicting causal interactions between signalling proteins39.
The wisdom of crowds
Another recurrent theme is that there is wisdom in the crowds39,43. The aggregation of solutions proposed by different teams is routinely as good as, and often better than, any of the single solutions29,60. This community wisdom gives real meaning to the notion of collaboration by competition (FIG. 3). As it is uncertain a priori which algorithm is going to perform best in any given problem, an aggregation of multiple methods is a robust strategy to attain good results.
Multitask learning boosts performance
Many problems in systems biology require the prediction of the response to a set of perturbations of the same system, such as the sensitivity of a panel of cell lines to different drugs or toxic compounds, or the determination of the essentiality of genes across a given set of cell lines. Predictors that learn jointly from perturbations that can have similar response rather than independently from each perturbation generally perform better29,60.
Challenge organization lessons
The organization of a Challenge requires scientific, technological, legal, financial and social considerations.
Scoring strategies generally need to be made transparent, which often, but not always, means disclosing the evaluation metric. However, there are cases in which the organizers may prefer to keep aspects of the metric undisclosed until the end of the Challenge, to prevent participants from focusing on optimizing their submissions to the metric rather than focusing on solving the scientific problem at hand. In such cases, organizers may disclose just the areas that will be evaluated in a general sense without giving the specific scoring criteria.
There is the risk that the focus on winning a Challenge may lead some participants to tweak existing approaches so as to maximize the score rather than develop innovative approaches that may not be competitive to well-studied ones in the first implementation.
Organizers need to find ways to prevent data leakage and overfitting83, such as by limiting the number of submissions to the leaderboard or limiting the information revealed by the leaderboard84.
It may be wise not to provide any information about the test set, as it can provide unintended information to the participants. Instead, participants should submit code.
The advantage of having many participants in a Challenge creates the problem of multiple testing during scoring, which may diminish the statistical significance of the results.
It is important to determine that the data are of good quality before the Challenge and whether it is going to be too easy or too difficult, or whether there is sufficient statistical power in the data. This is typically accomplished during the dry runs (FIG. 2). It might be better not to launch promising Challenges that, after close inspection in the dry run, have a high probability of being unsolvable. At the same time, hard Challenges might be worth running for fundamental questions, as they provide a sound assessment of the current state-of-the-art methodologies that scientists can build upon.
Sociological lessons
A major consideration in using the Challenge framework is the question of how best to incentivize participation. The most typical incentives are monetary awards, the possibility to co-author a high-profile paper reporting on a Challenge, an invitation to present the best-performing method at a conference or the desire to access and analyse the data sets provided in the Challenge.
Given that meaningful participation requires a substantial time investment from each team, a ‘winner-takes-all’ approach for selecting top performers can limit the diversity and depth of involvement, whereas intermediate awards can directly motivate participants to exert costly effort.
Unsportsmanlike behaviour — in which participants register under different identities in order to send more predictions to the leaderboard than allowed — has been observed, but fortunately this is rare and not difficult to detect.
Many teams welcome the opportunity to come together as a community to compare approaches and share the lessons from a Challenge, in the form of leaderboards, webinars, forums, e-mail lists or hackathons85. Recent DREAM Challenges have included a post-competition collaborative phase in which top teams are brought together to further improve on solutions or to address post-hoc analytical questions.
Is there a strategy to win in Challenges?
Each Challenge has several specific features. Hence, it is hard to extract a general strategy as to what it takes to perform well in a Challenge.
Aspects that seem to lead to decreased performance include making technical mistakes, such as overfitting a model to the training data, or not using prior knowledge or biological thinking to guide model development.
There is also no obvious general pattern of what is the best composition of a team. The best-performing teams can be composed of many researchers with different backgrounds or consist of a single individual with very specific expertise (typically in machine learning).
Generally, success in solving computational biology problems (such as the ones presented as Challenges) depends on teams, methods and data. In the absence of the right data, even the most proficient experts using the best methods will not be able to solve the Challenge. Likewise, if a cutting-edge method is used by an inexperienced team not using best practices, the resulting solutions may be less powerful than they could have been.
Challenge outputs and legacy
The outputs of a Challenge are manifold. One important legacy includes the large number of methodologies used to solve the Challenge. Although the best-performing approach is normally highlighted, the true value of a Challenge is the large collection of methods that, although individually may not be particularly predictive, collectively provide a robust solution (the concept of the ‘ wisdom of crowds’ (BOX 2; FIG. 3)).
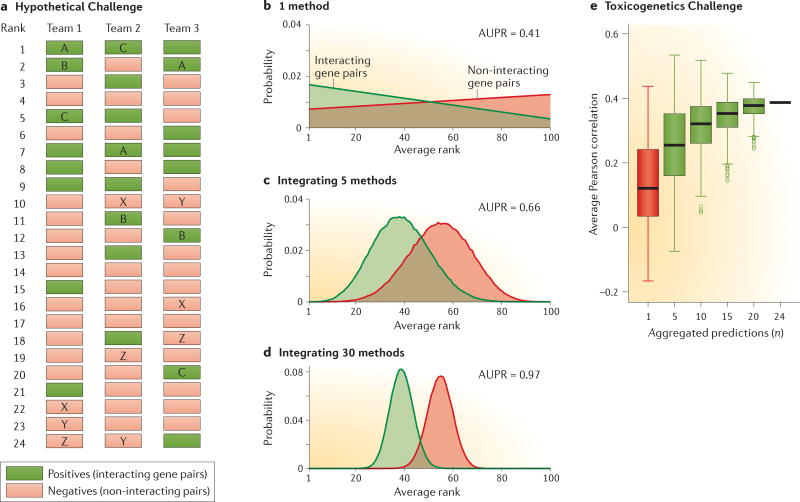
Figure 3. The wisdom of crowds in theory and in practice.
Two case studies in the context of a hypothetical Challenge43 or the NIEHS–NCATS–UNC DREAM Toxicogenetics Challenge (a collaboration between the US National Institute of Environmental Health Sciences (NIEHS), the US National Center for Advancing Translational Sciences (NCATS) and the University of North Carolina (UNC))60. a–d | The hypothetical example shows three of the predictions that will be integrated into an aggregate ranked list. Two sufficient conditions for integration to outperform individual inference methods are: first, each of the inference methods must have better than random predictive power (that is, on average, items in the positive set are assigned better (lower) ranks than items in the negative set), and second, predictions of different inference methods must be statistically independent. In part b, we show the probability that a given method places a positive or negative item at a given rank. Positive items are assigned lower ranks on average, yet there is still some considerable probability of giving a low rank to a negative item. The area under the precision-recall curve (AUPR) of this method is only 0.41; for a random prediction with these parameters, we would expect an AUPR of 0.3. Suppose now that the integrated solution is computed for each item as the average of the assigned ranks to that item by each method. If, for the sake of simplicity, we assume that all methods have the same probability and the assigned ranks are independently chosen for the positive and negative sets, then the central limit theorem establishes that the average rank probability will approach a Gaussian distribution, with its variance shrinking as more methods are integrated. In this way, the probability of a positive to have lower ranks than negatives increases (parts c and d), resulting in an AUPR that tends to 1 (perfect prediction) as the number of integrated inference methods increases. e | An equivalent trend is seen in the Toxicogenetics Challenge using a different metric (Pearson correlation). The Pearson correlation is shown for all 24 methods submitted, and the box-plot for n randomly chosen predictions out of the 24. The median correlation of the aggregates increases as the number of aggregated methods increases. Parts a–d are adapted from REF. 43, Nature Publishing Group. Part e is adapted from REF. 60, Nature Publishing Group.
Many Challenge platforms (such as CASP, CAMDA and DREAM) organize a post-Challenge conference to discuss take-home lessons and to encourage participants to meet and learn from each other’s experience. When the results are adequately interesting, the organizers coordinate efforts with participants to write a paper describing the results of the Challenge and the lessons learned.
The legacy of a Challenge may also include a database containing the Challenge data, leaderboards, submissions and, sometimes, the source code and documentation of participants, for future use in education, research and subsequent benchmarking (TABLE 1), which can also be supported by tools for offline scoring36.
What have Challenges taught us?
Many Challenges have been crowdsourced over the past two decades. The collective wisdom resulting from these Challenges yields a wealth of scientific, methodological, epistemological, sociological and organizational lessons (BOX 2). In this section, we highlight a few case studies that represent a non-exhaustive list of successful Challenges that have been held in the field of genetics, genomics and systems biology, with an emphasis on the scientific and algorithmic insights gained. Summaries of a wider range of Challenges are listed in TABLE 1 and Supplementary information S2 (table).
The wisdom of crowds produces the most robust regulatory network inference results
Challenges on inference and modelling of gene regulatory networks were the main focus in early editions of the DREAM Challenges40–45. The aim of the transcriptional network inference Challenges was to predict causal regulatory interactions between transcription factors (TFs) and target genes on the basis of gene expression data of a particular cell of interest. Different types of gene expression data were given to participants to solve these Challenges, including single measurements as well as time series measurements for genetic, drug and environmental cell perturbations. Rigorous evaluation of gene network inference methods is non-trivial because the underlying gold standards (in this case, the ‘true’ networks) are generally not known. Different strategies were used to circumvent this problem: simulated expression data, which enabled systematic evaluation based on the underlying in silico gene networks40–42,45–47; an in vivo synthetic network of five genes that had been engineered in Saccharomyces cerevisiae44,45; and microarray compendia from model organisms (Escherichia coli and S. cerevisiae), in which predictions could be evaluated based on experimentally supported TF–target gene interactions (for example, by chromatin immunoprecipitation43,45).
These Challenges enabled for the first time direct comparison of a broad range of inference methods across multiple networks, giving valuable insights for both method development and application. Regression-based methods, information-theoretical methods and meta-predictors that combine multiple inference approaches each performed well, especially when combined with data resampling techniques to improve robustness, whereas probabilistic graphical models, such as Bayesian networks — a popular network inference approach in the literature — never achieved top performance. TF knockouts were the most informative experiment, whereas dynamics in time series data proved difficult to leverage for inferring transcriptional interactions. An important lesson was that no single inference method performed robustly across diverse networks. Moreover, different types of inference approaches captured complementary features of the underlying networks. Consequently, the integration of predictions from multiple inference methods resulted in more robust and accurate networks, achieving top performance in several Challenges40–43,45–47. In the DREAM5 Challenge, this approach was used to construct robust, community-based networks for E. coli and Staphylococcus aureus, thus leveraging the wisdom-of-crowds phenomenon (BOX 2; FIG. 3), not only for method assessment but also to gain new biological insights43,45. As part of the legacy of this Challenge, the top-performing inference methods and the tools to integrate predictions across methods were made available on a web-based platform (GenePattern (GP)–DREAM)48.
It is important to emphasize that in these Challenges, network inference methods were successful only when applied to large expression compendia (those comprising hundreds of different conditions and perturbations) from either in silico networks or bacterial organisms. By contrast, performance was poor for S. cerevisiae, suggesting that additional inputs besides expression data are needed to accurately reconstruct transcriptional networks for eukaryotes43,45. As rich data sets (such as epigenetic and chromatin conformation data) are becoming available for human cell types and tissues, integrative methods are being developed to reconstruct fine-grained regulatory circuits connecting TFs, enhancers, promoters and genes49. Consequently, there will be a need for novel benchmarks and Challenges to rigorously assess these methods on human regulatory circuits.
Benchmarking of TF–DNA binding motif prediction methods showed that position weight matrix models perform well for most TFs but fall short in specific cases
The TF–DNA Binding Motif Recognition Challenge25 aimed to benchmark algorithms and models for describing the DNA-binding specificities of TFs; this is a central problem in regulatory genomics. For example, many disease-associated genetic variants occur in non-coding regions of the genome50,51, suggesting that some variants might act by modulating binding sites for TFs. The major paradigm in modelling TF sequence specificity is the position weight matrix (PWM) model. However, it has been increasingly recognized that the shortcomings of PWMs, such as their inability to model gaps, to capture dependencies between the residues in the binding site, or to account for the fact that TFs can have more than one DNA-binding interface, can make them inaccurate52– 54. Alternative models that address some of the shortcomings of PWMs have been developed55–57, but before this Challenge, their relative efficacies had not been rigorously compared. A major difficulty in predicting TF–DNA binding interactions had been the scarcity of data about the relative preference of a TF to a wide range of individual sequences, as such data are needed to train the models. This limitation was overcome with the introduction of the universal protein-binding microarray (PBM)58, which provides information about the relative affinity of a given TF to each of the 32,896 possible 8-base sequences in the PBM.
The PBM data set released for this Challenge describes the binding preferences of 86 mouse TFs (representing a wide range of TF families). Two independent probe sequence designs were used to generate two PBMs for assaying each TF. For 20 TFs, data were provided from both PBMs, for ‘practice’ and method calibration; the remaining 66 TFs were used in the Challenge. For each TF, participants were asked to predict the probe intensities of one type of PBM, given the probe intensities of the other. In total, 14 groups from around the world participated. Five evaluation criteria were used to assess the ability of an algorithm to either predict probe sequence intensities or assign high ranks to preferred 8-base sequences. The top-performing method was based on a k-mer model59, which captured short-range interdependencies between nucleotides by making use of longer nucleotide sub-sequences (known as k-mers) rather than mononucleotide-based PWM models. A web server has been released that allows anyone to submit their predictions and compare the performance of their method25. Among the key findings were: first, the simple PWM-based model performs well for ~90% of the TFs examined, with advanced models generally being required for specific families (such as C2H2 zinc fingers); second, the methods that perform well in the in vitro comparisons also tended to perform well in distinguishing binding sites from random sequences in vivo; and third, the best PWMs tended to have low information content, consistent with high degeneracy in eukaryotic TF binding specificities. In summary, the results of this community-based effort have led to multiple new insights into TF function and have provided a suite of new computational methods for predicting (and evaluating) TF binding.
Predicting toxic-compound effects from basal genomic features is difficult but possible
The NIEHS–NCATS–UNC DREAM Toxicogenetics Challenge60 was a collaboration between the US National Institute of Environmental Health Sciences (NIEHS), the US National Center for Advancing Translational Sciences (NCATS) and the University of North Carolina (UNC). It was designed to assess the capabilities of current methodologies to address two crucial issues in the context of chemical safety testing: first, the use of genetic information to predict cellular toxicity in response to environmental compounds across cell lines with different genetic backgrounds; and second, the use of compound structure information to predict population-level cellular toxicity in response to new environmental compounds. The data set used for the Challenge was unique in terms of size and scope, containing cytotoxicity measurements for 884 lymphoblastoid cell lines (derived from the 1000 Genomes Project) in response to 156 environmental compounds. Genotype, transcriptional data and chemical attributes were also provided to Challenge participants. A portion of the cytotoxicity data was given as a training set, and a portion was kept to assess the performances of the methods.
Objective assessment using the Challenge framework demonstrated that predictions from participants’ models of cytotoxicity that were based on genetic background were overall modest (although top-performing predictions were significantly better than random), suggesting that genetic data is insufficient to meaningfully address the Challenge question. The availability of transcriptomics data (from RNA-seq), which were provided for only a subset of the cell lines, was shown to significantly improve the overall accuracy of the predictions, suggesting that additional molecular characterization could improve the predictability. Larger training data sets are also expected to improve predictability by using the state-of-the-art approaches developed to solve the Challenge.
By contrast, a subset of participants’ predictive models that were based on compound structure performed well, as they were accurate and robustly better than random, indicating that this Challenge question was difficult but solvable using a subset of current methodologies. Being able to predict not only the average toxic effect of an environmental compound in the population, but also the variability in the population response, plays a crucial part in assessing exposure risk in silico. Challenge results showed that it is indeed possible to effectively rank chemicals by toxicity based on their chemical structure alone, and methods developed to solve the Challenge could thus be used to prioritize the tested compounds for chemical safety.
Integrating over multiple omics data types is best for predicting drug response, but gene expression or phosphoproteomics are the most informative individual data types
Similar results to the NIEHS–NCATS–UNC DREAM Toxicogenetics Challenge were obtained in the US National Cancer Institute (NCI)–DREAM Drug Sensitivity Prediction Challenge29 to predict drug response on a panel of breast cancer cell lines (TABLE 1). Here again, the Challenge revealed that although there is signal in the data, the models showed far from optimal performance. Participants were given 6 omics training data sets from 35 cell lines that were each treated with 28 drugs. Given these data, the Challenge was to predict the response for 18 other cell lines to each of the 28 drugs.
A total of 44 predictions were evaluated that covered a range of methods, from a simple correlation-based method, which finished third overall, to a novel Bayesian multitask, multiple kernel learning (MKL) model, which was the top-performing model. In addition to the method assessment, an extensive evaluation of the underlying data was conducted in a post-challenge analysis. Using the Bayesian multitask MKL and an elastic net, predictors were built using all possible combinations of omics data; results showed that integrating five or six of the data types consistently had the best performance, but gene expression microarrays provided the single best data type to use with the Bayesian multitask MKL method, and reverse phase protein array (RPPA) data were best using the elastic net. Other observations made from the Challenge results are that methods using prior biological knowledge, such as pathway information, outperformed methods that did not use prior information, and nonlinear models tended to perform better than linear methods.
Clinical outcomes can be more accurately predicted with clinical data than with molecular data
Although examination of molecular mechanisms that underlie clinical outcomes is an important scientific step for disease research, experience from several Challenges indicates that the types of molecular and genetic data used in these Challenges provide less predictive information than do clinical measures. Two Challenges have established community efforts to build predictive models based on single-nucleotide polymorphism (SNP) data, including the prediction of clinical non-response following anti-TNF (tumour necrosis factor) treatment in patients with rheumatoid arthritis37 or the prediction of Alzheimer disease diagnosis61. The outcomes of these Challenges demonstrated that the genetic contribution to overall performance was minimal, suggesting that current methodologies are not able to identify and compile genetic signals given existing sample collections.
An alternative approach for capturing complex genetic signals in predictive models is to incorporate downstream phenotypic measures that are themselves influenced by genetic variation. Clinical measures of disease state that represent the complex interactions of human biology aggregated across multiple genetic and non-genetic factors tend to provide the greatest contribution to predictions. In the Alzheimer’s Disease Big Data DREAM Challenge61, cognitive measures of brain function greatly outperformed SNP genotypes for predicting disease status. In the Rheumatoid Arthritis Responder Challenge37, clinical measures of pretreatment disease severity had the greatest contribution to prediction of anti-TNF treatment response. Similarly, in the Sage Bionetworks–DREAM Breast Cancer Prognosis Challenge38, the use of genomics information (in this case, gene expression and copy number variation) increased the predictive ability by a modest 8% with respect to clinical covariates only (TABLE 1; Supplementary information S2 (table)). Additional work with large molecular data sets is needed to further understand what makes a certain size and type of data useful for predictive analytics.
In genome-interpretation Challenges, tailored approaches typically perform best
CAGI is a very successful community effort to objectively assess computational methods for predicting the phenotypic effects of genomic variation. Participants are provided with genetic variants and are invited to make predictions of resulting phenotypes. These predictions are evaluated against experimental characterizations by independent assessors.
Each year, CAGI includes approximately ten different Challenges, addressing different scales and aspects of the relationship between genotype and phenotype. At one extreme are predictions of biochemical activity. For example, the Cystathionine beta-Synthase (CBS) Challenge62 sought to understand the biochemical effects of CBS mutations, which underlie clinical homocystinuria. In this Challenge, participants were given individual amino acid substitutions in the CBS protein and asked to predict the biochemical activity as measured through a yeast growth assay. Participants typically trained their models on numerous different non-synonymous variants and their impacts, although some focused training on other available mutation data in the CBS gene. Two very different evolutionary methods worked particularly well, whereas biophysical approaches performed poorly. The performance of the most popular methods was generally in the middle of the ranking. Overall, this Challenge revealed that the phenotype prediction methods embody a rich representation of biological knowledge, making statistically significant predictions. However, the accuracy of prediction on the phenotypic effect of any specific variant was unsatisfactory and of questionable clinical utility.
At the other extreme of the CAGI Challenges are the genotype-to-phenotype Challenges. An insightful example is the prediction of phenotypic traits of public genomes in the Personal Genome Project (PGP). The Challenge consisted of matching each of 77 given human genomes to the right phenotypic profiles among 291 possible profiles, of which 214 were decoys. Each phenotype profile consisted of 243 binary traits comprising 239 traits that were self-reported by the PGP participants and supplemented with blood groups extracted from electronic health records. The Challenge was assessed by counting the number of correct genotype-to-phenotype assignments. This Challenge ran from 2012 to 2013 and had 16 submissions. The top performer63 used a Bayesian probabilistic model to predict clinical phenotypic traits from genome sequence and population prevalence.
Overall, CAGI Challenges showed that the most effective predictions came from methods honed to the precise Challenge.
Conclusions and perspectives
As we face the challenges of data analysis that are emerging from the scale and complexity of the growing body of biological data, we must explore different modes of research to advance science. Crowdsourced Challenges present a different way of doing science. This is not to say that Challenges are better than traditional approaches, but they provide an alternative way to engage researchers and make valuable data open to the community. A key requirement for this is data sharing. Even though the idea of data sharing has obvious societal and scientific advantages, its implementation is less straightforward than it might seem at first sight. This is, at least in part, due to the fact that some data producers are reluctant to share data, either because they want to publish the data for their own benefit before it becomes public or because they misunderstand the benefits of crowdsourcing64. Conversely, new frameworks are required that carefully balance the needs for security and ethics with desires for broad data reuse65 and education. Reflective of striking this balance, open computational platforms — such as Synapse — are emerging to provide Challenges with IRB-approved data hosting services as well as a social layer and working environment that makes it easy for Challenge teams to work together.
Traditional training of research scientists can also be enriched with the use of scientific Challenges. There are students who use Challenges in their dissertation work as sources of data to test their computational approaches and compare their performance relative to the best solutions that result from the Challenges. In addition, instructors in different disciplines (such as biology, bioinformatics and computational systems biology) can use past or ongoing Challenges as modules to introduce computational methodologies along with best practices for rigorous validation and reproducibility. Perhaps more importantly, students can learn to collaborate on a global stage with fellow researchers in the pursuit of solutions to specific problems, while they develop their skills by participating in ongoing Challenges.
Crowdsourcing research problems has the potential to accelerate research manifold owing to the sheer amount of work that can be focused on one Challenge question in a short period of time. As an illustration, the NCI–DREAM Drug Sensitivity Challenge29 ran in 2012 for a period of 5 months and had 127 participants (Challenges can often recruit even more participants than this). Assuming that each researcher worked on average 100 hours on the Challenge, this represents ~127,000 hours (~14 person-years) of research effort dedicated to addressing one question. Even if a single researcher were able to dedicate this amount of time to address a single question, it is unlikely that this individual would have the cross-disciplinary knowledge of 127 participants; thus, a much smaller sampling of methods would be explored. Hence, the value of Challenges resides not only in the acceleration, but just as importantly, in the diversification of approaches used to attack a problem. By engaging multiple groups with different backgrounds and ideas, various solutions can be integrated to add on the benefits of the wisdom of crowds (BOX 2; FIG. 3). Compared to individual solutions, integrated solutions are much more robust to the specific composition of the data used to answer a Challenge question and often yield results that are better than the best individual solution. In addition, crowdsourced Challenges produce rigorous, unbiased benchmarked data and methods that have been subjected to a rigorous vetting that can be used to aid peer review (BOX 3).
Box 3. Challenge-assisted peer review.
The wide availability of genetics and genomics data has encouraged the development of many statistical methodologies and algorithms to analyse and interpret those data. Under ideal conditions, the performance of these algorithms should be soundly assessed by the method developers in the first instance, followed by evaluation by peer reviewers when these methods are sent for publication. However, it has been documented that there is a natural tendency towards leniency when scientists evaluate their own research35, and peer reviewers are often unable to thoroughly evaluate claims of good performance of all the complex and involved algorithmic pipelines reported in a publication. The consequences of this state of affairs are a lack of rigour in the characterization of the performance of algorithms and a proliferation of positive results that fail reproducibility86,87.
One possible solution to the enforcement of best practices in the evaluation of computational methods before publication could be to have Challenge organizers and journal editors work together on the assessment of method performance. This could be done by using blind Challenges as an aid to the traditional peer review system. This hybrid review system, which we have called ‘Challenge-assisted peer review’, would leverage the rigour in method evaluation provided by blind Challenges with the assessment of clarity, originality and other aspects properly handled in the traditional peer review process. Similarly, a Challenge assessment would also address the potential lack of reproducibility issues, as the code submitted to a Challenge is typically re-run by the organizers to verify that the submitted results are reproducible. To be clear, the goal of a Challenge-assisted peer review is not to forcefully identify the single best method for publication, but rather to flesh out the strengths and weaknesses of the different methods in a controlled evaluation protocol. In a Challenge-assisted peer review scenario, a journal editor could coordinate the organization of a Challenge to test and broadcast a specific scientific question of interest to the journal. Alternatively, Challenge organizers could contact a journal editor and propose to publish, after proper peer review, the rigorously evaluated results of a Challenge. For example, the best-performing algorithm in the Sage Bionetworks–Dialogue for Reverse Engineering Assessment and Methods (DREAM) Breast Cancer Prognosis Challenge38 was published following a previous agreement with the journal editor and peer review88; this Challenge provided a common platform for data access and blinded evaluation of the accuracy of 1,400 submitted models in predicting the survival of 184 patients with breast cancer using gene expression, copy number data and clinical covariates from 1,981 patients. Several publications resulting from the DREAM Challenges have followed similar approaches26,28,29,89.
In addition, the partnership between Challenge organizers and journal editors allows the Challenge organizers to announce that the journal is interested in considering the paper resulting from the Challenge. The possibility of contributing to a top-tier publication can be a strong incentive for researchers to participate in a Challenge. Furthermore, the publication of the results of a Challenge in a high-profile journal makes the results, algorithms and analyses of the participants’ submissions widely available and provides, through the Challenge-assisted evaluation, a true seal of quality. In summary, Challenge-assisted peer review could be a useful tool to enhance the peer review system for publications with strong computational biology and bioinformatics content.
Although Challenges have proved to be a powerful tool in scientific research, not all research questions can be posed as a Challenge. For example, a successful Challenge requires enough data for training and the availability of an unpublished gold standard. If these data do not contain sufficient information to address the scientific questions, the Challenge may be unsolvable. Alternatively, if the questions posed in a Challenge are too easily solved from the data, then a crowdsourced approach is not necessary. A Challenge also has to have sufficient scientific or clinical impact to entice the community to participate. When important problems do not fulfil these criteria, crowdsourcing modalities other than Challenges (BOX 1) can be used. One such type of crowdsourcing is referred to as an ‘ideation’ Challenge, in which organizers solicit new ideas and directions that are conducive to obtaining insights into a problem, even if the solution is unknown.
As community Challenges increase in popularity, the research community may start to feel some degree of Challenge fatigue, and hence organizers will have to evolve different strategies to encourage participation and will need to carefully choose the questions for the community to address.
Challenge funding is also a strategic consideration. Most of the Challenges discussed in this Review (TABLE 1) leveraged the voluntary efforts of participants and organizers. Having volunteers organize Challenges is unsustainable in the long run, particularly if we want to develop and maintain robust platforms and Challenge resources that do not depend on the free time of organizers. To increase the impact of big data and at the same time nurture young computational scientists into collaborative work, it is crucial for funding agencies to create mechanisms to support these scientific crowdsourcing initiatives.
Community efforts can have a major role in defining state-of-the-art solutions to current unsolved problems. For example, ongoing Challenges in the reconstruction of phylogeny in a heterogeneous tumour, detection of RNA transcript fusions or the distinction of driver from passenger mutations from next-generation sequencing data could bring the maturation of data production and analysis that are necessary to develop applications of precision medicine in cancer. Other areas that are ripe for Challenges, but that have not fully benefited from them, include: the identification of patients that will benefit from cancer immunotherapy, the phenotype–genotype mapping for antibiotic resistance and the identification of targets for drug combinations in malaria.
The creativity of a multi-talented community of solvers can be a true innovation engine that brings us one step closer to the solution of today’s most pressing problems in biomedicine. It is precisely because curious and ambitious students, researchers, technologists and citizen scientists find value in contributing to community efforts that Challenges exist. Taken to the next level, we envision community efforts that both generate new data and run a Challenge to address a question in a shorter timeframe than even the best-funded research institutions can attain. If harnessed, we can achieve an extraordinary increase in the speed and depth with which biomedical problems are solved.
Supplementary Material
Acknowledgments
The authors thank N. Aghaeepour, M. Bansal, P. Bertone, E. Bilal, P. Boutros, S. E. Brenner, J. Dopazo, D. Earl, F. Eduati, L. Heiser, S. Hill, P.-R. Loh, D. Marbach, J. Moult, M. Peters, S. Sieberts, J. Stuart, M. Weirauch and N. Zach for information on the crowdsourcing efforts they organized. The authors also thank the DREAM Challenges community, who taught them everything about Challenges that they have tried to share in this Review.
Glossary
- Cloud computing
An internet-based infrastructure to perform computational tasks remotely.
- Crowdsourcing
A methodology that uses the voluntary help of large communities to solve problems posed by an organization.
- Challenges
(Also known as collaborative competitions). Calls to a wide community to submit proposed solutions to a specific problem. These solutions are evaluated by a panel of experts using diverse criteria, and the best performer or winner is selected.
- Gamification
The abstraction of a problem in such a way that working towards its solution feels like playing a computer game.
- Benchmarking Challenge
A Challenge used to determine the relative performance of the methodologies used to solve a particular problem in which a known solution is available to the organizers but not the participants. The organizers compare the proposed solutions to the solution that is only available to them (that is, the gold standard). It is expected that the good solutions will generalize to instances of the problem for which the solution is unknown.
- Gold standard
In allusion to the abandoned system of assigning the true value of a currency, the gold standard in a Challenge is the true solution to the posed problem in one particular instance of that problem.
- Leaderboards
Tables that provide real-time feedback of performance and scores of the proposed solutions to a Challenge, allowing participants to monitor their ranking.
- Training set
In general, this is the portion of the data used to train (fit) a computational model. In a Challenge, this is the data given to the participants to build their models. It normally encompasses most of the data.
- Cross-validation set
A procedure whereby a participant uses subsets of the training data to adjust model parameters based on how well they predict this data set.
- Test set
The subset of data that is separate from the training set and the cross-validation set (that is, the data that participants never have access to in any sort of way). The test set is used to do a final assessment of the predictive power of the models.
- Wisdom of crowds
The collective wisdom that emerges when the solutions to a problem that are proposed by a large pool of people are aggregated. The aggregate solution is often better than the best individual solution.
- Hackathons
Events in which specialists in a topic, normally related to computation, get together to work on a specific problem.
Footnotes
Competing interests statement
The authors declare no competing interests.
See online article: S1 (box), S2 (table)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Stephens ZD, et al. Big Data: astronomical or genomical? PLoS Biol. 2015;13:e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Cancer Genome Consortium et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 6.Toga AW, et al. Big biomedical data as the key resource for discovery science. J. Am. Med. Inform. Assoc. 2015;22:1126–1131. doi: 10.1093/jamia/ocv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snijder B, Kandasamy RK, Superti-Furga G. Toward effective sharing of high-dimensional immunology data. Nat. Biotechnol. 2014;32:755–759. doi: 10.1038/nbt.2974. [DOI] [PubMed] [Google Scholar]
- 8.Henneken E. Unlocking and sharing data in astronomy. Bul. Am. Soc. Info. Sci. Tech. 2015;41:40–43. [Google Scholar]
- 9.World Meteorological Organization. Climate data, management and exchange. WMO. 2009 http://www.wmo.int/pages/themes/climate/climate_data_management_exchange.php.
- 10.Brabham DC. Crowdsourcing. MIT Press; 2013. [Google Scholar]
- 11.Nesta. A guide to historical Challenge prizes. Nesta. 2014 May 13; http://www.nesta.org.uk/news/guide-historical-challenge-prizes.
- 12.Costello JC, Stolovitzky G. Seeking the wisdom of crowds through challenge-based competitions in biomedical research. Clin. Pharmacol. Ther. 2013;93:396–398. doi: 10.1038/clpt.2013.36. [DOI] [PubMed] [Google Scholar]
- 13.Boudreau KJ, Lakhani KR. Using the crowd as an innovation partner. Harv. Bus. Rev. 2013;91:60–69. [PubMed] [Google Scholar]
- 14.Howe J. The rise of crowdsourcing. Wired Magazine. 2006;14:1–4. This article coined the term crowdsourcing and highlighted its potential. [Google Scholar]
- 15.Sobel D. Longitude: The True Story of a Lone Genius Who Solved the Greatest Scientific Problem of His Time. Bloomsbury Publishing; 2007. [Google Scholar]
- 16.Heritage Provider Network Health Prize. Improve healthcare, win $3,000,000. WebCite. 2011 May 4; http://www.webcitation.org/65IuEDAsc.
- 17.Wikipedia. List of crowdsourcing projects. [updated 16 Jun 2016];Wikipedia. https://en.wikipedia.org/wiki/List_of_crowdsourcing_projects.
- 18.Kryshtafovych A, et al. Challenging the state of the art in protein structure prediction: highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10. Proteins. 2014;82:26–42. doi: 10.1002/prot.24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janin J, et al. CAPRI: a Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 20.Arighi CN, et al. BioCreative-IV virtual issue. Database. 2014;2014:bau039. doi: 10.1093/database/bau039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aghaeepour N, et al. Critical assessment of automated flow cytometry data analysis techniques. Nat. Methods. 2013;10:228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engström PG, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steijger T, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods. 2013;10:1177–1184. doi: 10.1038/nmeth.2714. References 22 and 23 describe RGASP as an early Benchmarking Challenge for RNA-seq data analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stolovitzky GA, Monroe D, Califano A. Dialogue on reverse-engineering assessment and methods. Ann. NY Acad. Sci. 2007;1115:1–22. doi: 10.1196/annals.1407.021. [DOI] [PubMed] [Google Scholar]
- 25.Weirauch MT, et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Küffner R, et al. Crowdsourced analysis of clinical trial data to predict amyotrophic lateral sclerosis progression. Nat. Biotechnol. 2015;33:51–57. doi: 10.1038/nbt.3051. A Challenge with direct clinical implications. [DOI] [PubMed] [Google Scholar]
- 27.Bentzien J, Muegge I, Hamner B, Thompson DC. Crowd computing: using competitive dynamics to develop and refine highly predictive models. Drug Discov. Today. 2013;18:472–478. doi: 10.1016/j.drudis.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Bansal M, et al. A community computational challenge to predict the activity of pairs of compounds. Nat. Biotechnol. 2014;32:1213–1222. doi: 10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costello JC, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat. Biotechnol. 2014;32:1208–1212. doi: 10.1038/nbt.2877. A Challenge to benchmark methods for precision medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boutros PC, et al. Global optimization of somatic variant identification in cancer genomes with a global community challenge. Nat. Genet. 2014;46:318–319. doi: 10.1038/ng.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green AK, et al. The project data sphere initiative: accelerating cancer research by sharing data. Oncologist. 2015;20:464–e20. doi: 10.1634/theoncologist.2014-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdallah K, Hugh-Jones C, Norman T, Friend S, Stolovitzky G. The Prostate Cancer DREAM Challenge: a community-wide effort to use open clinical trial data for the quantitative prediction of outcomes in metastatic prostate cancer. Oncologist. 2015:459–460. doi: 10.1634/theoncologist.2015-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atassi N, et al. The PRO-ACT database: design, initial analyses, and predictive features. Neurology. 2014;83:1719–1725. doi: 10.1212/WNL.0000000000000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omberg L, et al. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nat. Genet. 2013;45:1121–1126. doi: 10.1038/ng.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norel R, Rice JJ, Stolovitzky G. The self-assessment trap: can we all be better than average? Mol. Syst. Biol. 2011;7:537. doi: 10.1038/msb.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cokelaer T, et al. DREAMTools: a Python package for scoring collaborative challenges [version2; referees: 1 approved, 2 approved with reservations] F1000Res. 2015;4:1030. doi: 10.12688/f1000research.7118.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plenge RM, et al. Crowdsourcing genetic prediction of clinical utility in the Rheumatoid Arthritis Responder Challenge. Nat. Genet. 2013;45:468–469. doi: 10.1038/ng.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margolin AA, et al. Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer. Sci. Transl. Med. 2013;5:181re1. doi: 10.1126/scitranslmed.3006112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill SM, et al. Inferring causal molecular networks: empirical assessment through a community-based effort. Nat. Methods. 2016;13:310–318. doi: 10.1038/nmeth.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marbach D, Schaffter T, Mattiussi C, Floreano D. Generating realistic in silico gene networks for performance assessment of reverse engineering methods. J. Comput. Biol. 2009;16:229–239. doi: 10.1089/cmb.2008.09TT. [DOI] [PubMed] [Google Scholar]
- 41.Marbach D, et al. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl Acad. Sci. USA. 2010;107:6286–6291. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prill RJ, et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS ONE. 2010;5:e9202. doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nat. Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. This paper introduces the wisdom-of-crowds concept in computational biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantone I, et al. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell. 2009;137:172–181. doi: 10.1016/j.cell.2009.01.055. [DOI] [PubMed] [Google Scholar]
- 45.Stolovitzky G, Prill RJ, Califano A. Lessons from the DREAM2 Challenges. Ann. NY Acad. Sci. 2009;1158:159–195. doi: 10.1111/j.1749-6632.2009.04497.x. [DOI] [PubMed] [Google Scholar]
- 46.Mendes P, Sha W, Ye K. Artificial gene networks for objective comparison of analysis algorithms. Bioinformatics. 2003;19(Suppl 2):ii122–ii129. doi: 10.1093/bioinformatics/btg1069. [DOI] [PubMed] [Google Scholar]
- 47.Schaffter T, Marbach D, Floreano D. GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics. 2011;27:2263–2270. doi: 10.1093/bioinformatics/btr373. [DOI] [PubMed] [Google Scholar]
- 48.Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 49.Marbach D, et al. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat. Methods. 2016;13:366–370. doi: 10.1038/nmeth.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benos PV. Additivity in protein–DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 55.Zhao X, Xiaoyue Z, Haiyan H, Speed TP. Finding short DNA motifs using permuted Markov models. J. Comput. Biol. 2005;12:894–906. doi: 10.1089/cmb.2005.12.894. [DOI] [PubMed] [Google Scholar]
- 56.Sharon E, Eilon S, Shai L, Eran SA. Feature based approach to modeling protein–DNA interactions. PLoS Comput. Biol. 2008;4:e1000154. doi: 10.1371/journal.pcbi.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He X, et al. A biophysical model for analysis of transcription factor interaction and binding site arrangement from genome-wide binding data. PLoS ONE. 2009;4:e8155. doi: 10.1371/journal.pone.0008155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Annala M, Laurila K, Lähdesmäki H, Nykter M. A linear model for transcription factor binding affinity prediction in protein binding microarrays. PLoS ONE. 2011;6:e20059. doi: 10.1371/journal.pone.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eduati F, et al. Prediction of human population responses to toxic compounds by a collaborative competition. Nat. Biotechnol. 2015;33:933–940. doi: 10.1038/nbt.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allen GI, et al. Crowdsourced estimation of cognitive decline and resilience in Alzheimer’s disease. Alzheimers Dement. 2016;12:645–653. doi: 10.1016/j.jalz.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Critical Assessment of Genome Interpretation. Cystathionine beta-Synthase (CBS) single amino acid mutations. [updated 3 Nov 2010];CAGI. http://cagi2010.org/content/CBS.
- 63.Chen Y-C, et al. A probabilistic model to predict clinical phenotypic traits from genome sequencing. PLoS Comput. Biol. 2014;10:e1003825. doi: 10.1371/journal.pcbi.1003825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Longo DL, Drazen JM. Data Sharing. N. Engl. J. Med. 2016;374:276–277. doi: 10.1056/NEJMe1516564. [DOI] [PubMed] [Google Scholar]
- 65.Wilbanks J, Friend SH. First, design for data sharing. Nat. Biotechnol. 2016;34:377–379. doi: 10.1038/nbt.3516. [DOI] [PubMed] [Google Scholar]
- 66.Khare R, Good BM, Leaman R, Su AI, Lu Z. Crowdsourcing in biomedicine: challenges and opportunities. Brief. Bioinform. 2015;17:23–32. doi: 10.1093/bib/bbv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodman JK, Cryder CE, Cheema A. Data collection in a flat world: the strengths and weaknesses of Mechanical Turk samples. J. Behav. Decis. Mak. 2013;26:213–224. [Google Scholar]
- 68.sbvIMPROVER project team. On crowd-verification of biological networks. Bioinform. Biol. Insights. 2013;7:307–325. doi: 10.4137/BBI.S12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kutmon M, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2015;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thiele I, et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vashisht R, et al. Crowd sourcing a new paradigm for interactome driven drug target identification in Mycobacterium tuberculosis. PLoS ONE. 2012;7:e39808. doi: 10.1371/journal.pone.0039808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mortensen JM, et al. Using the wisdom of the crowds to find critical errors in biomedical ontologies: a study of SNOMED CT. J. Am. Med. Inform. Assoc. 2015;22:640–648. doi: 10.1136/amiajnl-2014-002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cooper S, et al. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larson SM, Snow CD, Shirts M, Pande VS. Folding@Home and Genome@Home: using distributed computing to tackle previously intractable problems in computational biology. arXiv. 2009 https://arxiv.org/abs/0901.0866.
- 75.Das R, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69(Suppl. 8):118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- 76.Good BM, Su AI. Games with a scientific purpose. Genome Biol. 2011;12:135. doi: 10.1186/gb-2011-12-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Treuille A, Das R. Scientific rigor through videogames. Trends Biochem. Sci. 2014;39:507–509. doi: 10.1016/j.tibs.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 78.Lee J, et al. RNA design rules from a massive open laboratory. Proc. Natl Acad. Sci. USA. 2014;111:2122–2127. doi: 10.1073/pnas.1313039111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sørensen JJWH, et al. Exploring the quantum speed limit with computer games. Nature. 2016;532:210–213. doi: 10.1038/nature17620. [DOI] [PubMed] [Google Scholar]
- 80.Rees MA. Longitude Prize for the twenty-first century. Nature. 2014;509:401. doi: 10.1038/509401a. [DOI] [PubMed] [Google Scholar]
- 81.Chandler DL. A doctor in the palm of your hand: how the Qualcomm Tricorder X-Prize could help to revolutionize medical diagnosis. IEEE Pulse. 2014;5:50–54. doi: 10.1109/MPUL.2013.2296803. [DOI] [PubMed] [Google Scholar]
- 82.Meyer P, et al. Inferring gene expression from ribosomal promoter sequences, a crowdsourcing approach. Genome Res. 2013;23:1928–1937. doi: 10.1101/gr.157420.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dwork C, et al. STATISTICS. The reusable holdout: preserving validity in adaptive data analysis. Science. 2015;349:636–638. doi: 10.1126/science.aaa9375. [DOI] [PubMed] [Google Scholar]
- 84.Blum A, Hardt M. The Ladder: a reliable leaderboard for machine learning competitions. arXiv. 2015 https://arxiv.org/abs/1502.04585.
- 85.Möller S, et al. Community-driven development for computational biology at Sprints, Hackathons and Codefests. BMC Bioinformatics. 2014;15:S7. doi: 10.1186/1471-2105-15-S14-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dahlin JL, Inglese J, Walters MA. Mitigating risk in academic preclinical drug discovery. Nat. Rev. Drug Discov. 2015;14:279–294. doi: 10.1038/nrd4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyer P, et al. Verification of systems biology research in the age of collaborative competition. Nat. Biotechnol. 2011;29:811–815. doi: 10.1038/nbt.1968. [DOI] [PubMed] [Google Scholar]
- 88.Cheng W-Y, Ou Yang T-H, Anastassiou D. Development of a prognostic model for breast cancer survival in an open challenge environment. Sci. Transl. Med. 2013;5:181ra50. doi: 10.1126/scitranslmed.3005974. [DOI] [PubMed] [Google Scholar]
- 89.Boutros PC, Margolin AA, Stuart JM, Califano A, Stolovitzky G. Toward better benchmarking: challenge-based methods assessment in cancer genomics. Genome Biol. 2014;15:462. doi: 10.1186/s13059-014-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meyer P, et al. Network topology and parameter estimation: from experimental design methods to gene regulatory network kinetics using a community based approach. BMC Syst. Biol. 2014;8:13. doi: 10.1186/1752-0509-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Uehara T, et al. The Japanese toxicogenomics project: application of toxicogenomics. Mol. Nutr. Food Res. 2010;54:218–227. doi: 10.1002/mnfr.200900169. [DOI] [PubMed] [Google Scholar]
- 92.Earl D, et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2011;21:2224–2241. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bradnam KR, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Earl D, et al. Alignathon: a competitive assessment of whole-genome alignment methods. Genome Res. 2014;24:2077–2089. doi: 10.1101/gr.174920.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ewing AD, et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat. Methods. 2015;12:623–630. doi: 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.