Abstract
Schizophrenia is a debilitating psychiatric condition often associated with poor quality of life and decreased life expectancy. Lack of progress in improving treatment outcomes has been attributed to limited knowledge of the underlying biology, although large-scale genomic studies have begun to provide insights. We report a new genome-wide association study of schizophrenia (11,260 cases and 24,542 controls), and through meta-analysis with existing data we identify 50 novel associated loci and 145 loci in total. Through integrating genomic fine-mapping with brain expression and chromosome conformation data, we identify candidate causal genes within 33 loci. We also show for the first time that the common variant association signal is highly enriched among genes that are under strong selective pressures. These findings provide new insights into the biology and genetic architecture of schizophrenia, highlight the importance of mutation-intolerant genes and suggest a mechanism by which common risk variants persist in the population.
Schizophrenia is characterized by psychosis and negative symptoms such as social and emotional withdrawal. While onset of psychosis typically does not occur until late adolescence or early adulthood, there is strong evidence from clinical and epidemiological studies that schizophrenia reflects a disturbance of neurodevelopment1. It confers substantial mortality and morbidity, with a mean reduction in life expectancy of 15-30 years2,3. Although recovery is possible, most patients have poor social and functional outcomes4. No substantial improvements in outcomes have emerged since the advent of antipsychotic medication in the mid-twentieth century, a fact that has been attributed to a lack of knowledge of pathophysiology1.
Schizophrenia is both highly heritable and polygenic, with risk ascribed to variants spanning the full spectrum of population frequencies5–7. The relative contributions of alleles of various frequencies are not fully resolved, but recent studies estimate that common alleles, captured by genome-wide association study (GWAS) arrays, explain between one-third and one-half of the genetic variance in liability8. There has been a long-standing debate, from an evolutionary standpoint, as to how common risk alleles persist in the population, particularly given the early mortality and decreased fecundity associated with schizophrenia9. Various hypotheses have been proposed, including compensatory advantage (balancing selection), whereby schizophrenia-associated alleles confer reproductive advantages in particular contexts10,11; hitchhiking, whereby risk-associated alleles are maintained by their linkage to positively selected alleles12; and contrasting theories that attribute these effects to rare variants and gene-environment interaction13. Addressing these competing hypotheses is now tractable given advances from recent studies of common genetic variation in schizophrenia.
The largest published schizophrenia GWAS, that from the Schizophrenia Working Group of the Psychiatric Genomics Consortium (PGC), identified 108 genome-wide significant loci and unequivocally demonstrated the value of increasing sample sizes for discovery in schizophrenia genetics research5. Here we report a large, phenotypically homogeneous GWAS of schizophrenia that, when combined with previously published data, identifies new facets of genetic architecture and biology and demonstrates that the evolutionary process of background selection contributes to the persistence of common risk alleles in the population.
Results
GWAS and meta-analysis
We obtained genome-wide genotype information for schizophrenia cases from the UK (the CLOZUK sample), which we combined with control datasets obtained from public repositories or through collaboration. The final sample size was 11,260 cases and 24,542 controls (5,220 cases and 18,823 controls not in previous schizophrenia GWAS; Methods and Supplementary Figs. 1 and 2). At a genome-wide level, the association statistics indicated that the common variant architecture in the CLOZUK sample was highly correlated with that in an independent sample of 29,415 cases and 40,101 controls from the PGC (genetic correlation = 0.954± 0.030; P = 6.63 × 10−227), and this was further confirmed by polygenic risk score and trend test analyses across the datasets at a range of association P-value thresholds (Methods and Supplementary Tables 1 and 2).
Meta-analysis of the CLOZUK and independent PGC datasets, excluding related and overlapping samples (total of 40,675 cases and 64,643 controls; Supplementary Fig. 3) identified 179 independent genome-wide significant SNPs (P < 5 × 108; Supplementary Table 3) mapping to 145 independent loci (Fig. 1, Methods and Supplementary Table 4). The 145 associated loci included 93 of those that were genome-wide significant in the study of the PGC, the majority of which showed a strengthened association (Supplementary Fig. 4 and Supplementary Table 5). This does not imply that the remaining 15 PGC loci were false positives; rather, this reflects the expected inflation of effect sizes for genome-wide significant SNPs in incompletely powered studies and, as we demonstrate, is consistent with all 108 PGC loci representing true positives (Supplementary Note). Of the 52 loci not identified by the PGC, 2 have been reported as genome-wide significant in other studies: the locus at ZEB214 and a locus on chromosome 8 (38.0-38.3 Mb)15.
Fig. 1. Manhattan plot of schizophrenia GWAS associations.
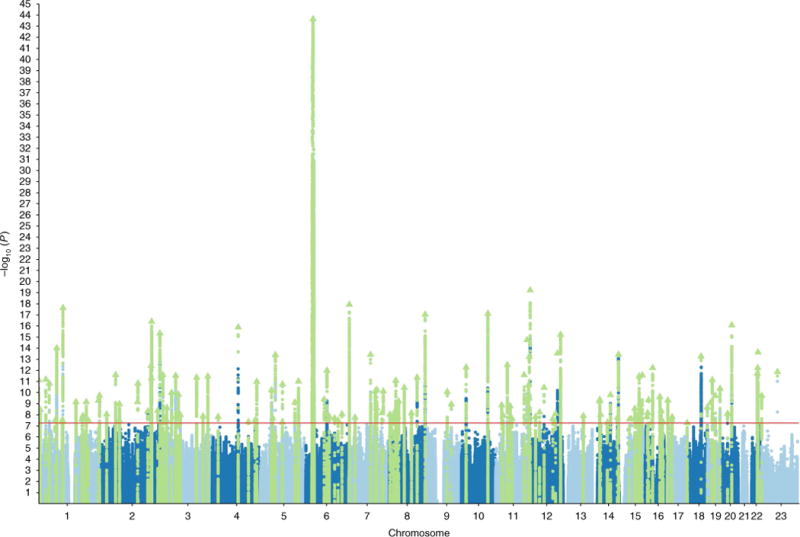
Associations are shown from the meta-analysis of CLOZUK and an independent PGC dataset (n = 105,318; 40,675 cases and 64,643 controls). The 145 genome-wide significant loci are highlighted in green. The red horizontal line indicates the genome-wide statistical significance threshold (P = 5×10−8).
In further independent samples (5,662 cases and 154,224 controls), 43 of the 50 genome-wide significant index SNPs showed the same pattern of allelic association, a level that far surpassed chance (P = 1.05 × 10−7). Despite the modest number of cases in these samples, 18 of the 50 index alleles reached nominal significance (P < 0.05), which again is implausible by chance (P = 1.46 × 10−11). None demonstrated evidence for heterogeneity of effect (Methods and Supplementary Table 6).
Mutation-intolerant genes
Recent studies have shown that mutation-intolerant genes capture much of the rare variant architecture of neurodevelopmental disorders such as autism, intellectual disability and developmental delay, as well as schizophrenia16–19. Here we show that, for schizophrenia, this also holds for common variation. Using gene set analysis in MAGMA20, loss-of-function (LoF)- intolerant genes (n = 3,230) as defined by the Exome Aggregation Consortium (ExAC)21 using their gene-level constraint metric (pLI ≥ 0.9), were enriched for common variant associations with schizophrenia in comparison with all other annotated genes (P = 4.1 × 10−16).
It has been shown that pLI is correlated with gene expression across tissues, including brain21, which raises the possibility that the enrichment for LoF-intolerant genes in schizophrenia may reflect enrichment for signal in genes expressed in the brain. However, LoF-intolerant gene set enrichment was robust to the inclusion of both ‘brain-expressed’ (n = 10,360) and ‘brain-specific’ (n = 2,647) gene sets19 as covariates in the analysis (P = 1.89 × 10−10) or to controlling for FPKM gene expression values in brain22 (P = 1.03 × 10−14).
It has been suggested that clustering of risk alleles in mutationintolerant genes is a hallmark of early-onset traits under natural selection23,24. However, LoF-intolerant genes are known to be enriched for SNPs identified as genome-wide significant in GWAS (as listed in the NHGRI-EBI GWAS Catalog25) and for broad categories of disorders21. To examine whether our finding is a property of polygenic disorders in general, we obtained summary genetic data from a late-onset neuropsychiatric disorder (Alzheimer’s disease), a non-psychiatric disorder (type 2 diabetes) and a psychological trait (neuroticism), each of which has been shown to be under minimal selective pressure (Methods). These other phenotypes showed at best a weak signal for enrichment of the LoF-intolerant gene set in the MAGMA analysis, with the signal not comparable to that seen in schizophrenia (Alzheimer’s disease, P = 0.008; type 2 diabetes, P = 0.016; neuroticism, P = 0.066).
To quantify the contribution of SNPs within LoF-intolerant genes to schizophrenia SNP-based heritability (h2SNP), we used partitioned linkage disequilibrium score regression (LDSR)26 (Supplementary Table 7). Overall, genic SNPs accounted for 64% of h2SNP, a 1.23-fold enrichment proportional to their SNP content (P = 5.93 × 10−14). Consistent with the analysis using MAGMA, h2SNP was enriched in LoF-intolerant genes (2.01-fold; P = 2.78 × 10−24), which explained 30% of all h2SNP (equating to 47% of all genic h2SNP). In contrast, genes classed as not LoF intolerant (pLI < 0.9) were significantly depleted for h2SNP relative to their SnP content (0.90-fold; P = 5.86 × 10−3), although in absolute terms SNPs in these genes accounted for 34% of h2SNP A finer-scale analysis of the relationship between LoF intolerance scores and enrichment for association showed that enrichment was restricted to genes with a pLI score above 0.9, precisely those defined as ‘LoF intolerant’ (Supplementary Fig. 5).
Common risk alleles in regions under background selection
Our finding that LoF-intolerant genes are enriched for common risk variants raises the question of how such alleles are found at common frequencies in the population. While the contribution of ultra-rare variation in functionally important genes to disorders associated with low fecundity can be accounted for by de novo mutation16,19,27, this cannot explain the persistence of common alleles. To address this question, we used partitioned LDSR to test the relationship between schizophrenia-associated alleles and SNP-based signatures of natural selection. These included measures of positive selection, background selection and Neanderthal introgression. We examined the heritability of SNPs after thresholding them at extreme values for these metrics (top 2%, 1% and 0.5%), including in the baseline model annotation sets such as LoF-intolerant genes and genomic regions with extreme LD patterns (Methods).
We observed strong evidence for schizophrenia h2SNP enrichment in SNPs under strong background selection (BGS), which was consistent across all the thresholds we examined (Table 1). We also found a significant depletion of h2SNP in SNPs subject to positive selection as indexed by the CLR statistic. These two results are mutually consistent, as calculation of the CLR statistic explicitly controls for the effect of BGS28. This suggests that SNPs under positive selection, but under weak or no BGS, are depleted for association with schizophrenia. No significant relationship between h2SNP and other positive selection or Neanderthal introgression measures was found after correction for multiple testing (Table 1). An LDSR analysis treating BGS measures as a quantitative trait rather than as a binary one confirmed that the relationship between BGS and schizophrenia association was not due to the imposition of arbitrary thresholds to define strong BGS (P = 7.73 × 10−11). We also note that the τc statistic of the LDSC model was significant for BGS, in both the binary (P = 0.041) and quantitative (P = 0.023) analyses (Supplementary Table 8). The τc statistic indicates the enrichment of BGS after controlling for all other annotations in the model (including LoF-intolerant genes)26 and thus represents a robust and conservative test for BGS enrichment.
Table 1.
Heritability analysis of natural selection metrics
| Top 2% of scores (genome wide) | Top 1% of scores (genome wide) | Top 0.5% of scores (genome wide) | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Metric | Ref. | Enrichment | 2-sided P value | Enrichment | 2-sided P value | Enrichment | 2-sided P value |
| Background selection (B statistic) | [29] | 1.801 | 0.001 | 2.341 | 9.90×10−4 | 2.365 | 0.002 |
| Positive selection (CLR) | [28] | 0.408 | 6.53×10−5 | 0.173 | 5.80×10−7 | 0.259 | 0.016 |
| Positive selection (CMS) | [88] | 0.054 | 0.001 | −0.037 | 0.006 | −0.039 | 0.007 |
| Positive selection (XP-EEH) | [87] | 0.621 | 0.342 | 0.383 | 0.303 | 0.125 | 0.268 |
| Positive selection (iHS) | [86] | 0.973 | 0.946 | 0.980 | 0.974 | 1.633 | 0.557 |
| Neanderthal posterior probability (LA) | [89] | 0.807 | 0.347 | 0.800 | 0.462 | 0.858 | 0.745 |
Partitioned LDSR regression results for SNPs thresholded by extreme values (defined as top percentiles versus all other SNPs) of each natural selection metric. All tests have been adjusted for 58 ‘baseline’ annotations, which include categories such as LoF intolerant, recombination coldspot and conserved (Methods). Enrichment values below 1 indicate a depletion of h2SNP in an annotation category (less contribution than expected for a given number of SNPs). Negative enrichments should be considered zero (no contribution to h2SNP by these SNPs). Bold values indicate results surviving correction after adjusting for all tests (Bonferroni a = 0.05/18 = 0.0028).
The above analyses account for a possible confounding relationship between LoF intolerance and BGS. To illustrate this more clearly, we binned the BGS intensities into four categories of increasing score and classified SNPs in these bins according to whether they were in LoF-intolerant genes, ‘all other’ gene sets or a non-genic set (Supplementary Fig. 6). Note that the lower boundary of the top bin (BGS intensity > 0.75) corresponds approximately to the top 2% BGS threshold in Table 1 and is equivalent to a reduction in effective population size estimated at each SNP of 75% or more29. We found significant heritability enrichment across all BGS intensity intervals in LoF-intolerant genes that increased progressively with higher intensity scores. Notably, we also found heritability enrichment for SNPs under BGS pressure in genes that were not LoF intolerant, restricted to the highest BGS intensity bin. Indeed, the highest BGS intensity bin in non-LoF-intolerant genes was enriched for heritability at a level roughly equivalent to that for all LoF-intolerant genes. These findings point to BGS and LoF intolerance as making at least partially independent contributions to heritability enrichment in schizophrenia. In contrast, none of the phenotypes we selected on the basis of their minimal impact on fecundity (Alzheimer’s disease, type 2 diabetes and neuroticism) showed significant BGS enrichment for heritability either when using the BGS τc statistic of the LDSR model (minimum P > 0.22; Supplementary Table 8) or when specifically testing regions of high BGS intensity in genes that were tolerant (pLI < 0.9) of functional mutations (minimum P > 0.40).
Systems genomics
Using MAGMA, we undertook a primary analysis of 134 central nervous system (CNS)-related gene sets we have previously shown capture the excess copy number variation (CNV) burden in schizophrenia30. In a GWAS context, we now show that, collectively, this group of gene sets captures a disproportionately high fraction of h2SNP (30% of total heritability, enrichment = 1.63, P = 8.57 × 10−13, 46% of genic heritability; Supplementary Table 7). Of the 134 sets, 54 were nominally significant, of which 12 survived multiple-testing correction (family-wise error rate (FWER) P < 0.05; Supplementary Table 9), with no notable association for gene sets such as the ARC protein complex and the NMDAR protein network, that we have previously implicated in rare variant studies30,31. Stepwise conditional analysis, adjusting sequentially for the more strongly associated gene sets, resulted in six gene sets that were independently associated with schizophrenia (Table 2 and Supplementary Data). These extended from low-level molecular and subcellular processes to broad behavioral phenotypes. The most strongly associated gene set constituted the targets of the fragile X mental retardation protein (FMRP)32. FMRP is a neuronal RNA-binding protein that interacts with polyribosomal mRNAs (the 842 target transcripts of this gene set32) and is thought to act by inhibiting translation of target mRNAs, including many transcripts of pre- and postsynaptic proteins. The FMRP target set has been shown to be enriched for rare mutational burden in exome sequencing studies of de novo variation in autism33 and intellectual disability31. In schizophrenia, it has also been shown to be nominally significantly enriched for association signal in sequencing studies8,31 and GWAS5,8, but has only inconsistently been associated in studies of CNV30,34. Here we provide the strongest evidence thus far for enrichment of this gene set in schizophrenia.
Table 2.
Functional gene set analysis highlights six independent gene sets associated with schizophrenia
| Gene set | Number of genes | Enrichment P value (FWER)a | Conditional P valueb |
|---|---|---|---|
| Targets of FMRP32 | 798 | 1×10−5 | 1.9×10−8 |
| Abnormal behavior (MP:0004924) | 1,939 | 1.8×10−4 | 1.4×10−5 |
| 5-HT2C receptor complex37 | 16 | 0.029 | 0.001 |
| Abnormal nervous system electrophysiology (MP:0002272) | 201 | 0.003 | 0.002 |
| Voltage-gated calcium channel complexes36 | 196 | 0.011 | 0.016 |
| Abnormal long-term potentiation (MP:0002207) | 142 | 0.030 | 0.031 |
MP refers to Mammalian Phenotype Ontology terms of the MGI35, from which gene sets were derived. FMRP, fragile X mental retardation protein.
Westfall-Young family-wise error rate, as implemented in MAGMA20.
From stepwise conditional analysis that adjusts sequentially for ‘stronger’ associated gene sets.
We highlight another five gene sets that are independently associated with schizophrenia. Three of these derive from the Mouse Genome Informatics (MGI) database35 and relate to behavioral and neurophysiological correlates of learning: abnormal behavior (MP:0004924), abnormal nervous system electrophysiology (MP:0002272) and abnormal long-term potentiation (MP:0002207). We note that two of these gene sets (MP:0004924 and MP:0002207) were among the five most enriched of the 134 gene sets tested in a recent schizophrenia CNV analysis30. The remaining two independently associated genes sets were voltage-gated calcium channel complexes36 and the 5-HT2C receptor complex37. The calcium channel finding confirms extensive evidence from common and rare variant studies implicating calcium channel genes in schizophrenia5,8, including a new GWAS locus in CACNA1D identified in our meta-analysis. While there is less convergent evidence in support of the involvement of the 5-HT2C receptor complex in schizophrenia, the fact that we identify independent association for this gene set implicates these genes in schizophrenia pathophysiology and potentially rejuvenates a previous avenue of 5-HT2C ligand therapeutic endeavor in schizophrenia research38. However, we interpret this result with caution given the small size of this gene set and the fact that a number of its genes encode synaptic proteins that are structurally related to other receptor complexes37, not only 5-HT2C.
Systems genomics and mutation-intolerant genes
The LoF-intolerant genes and the six conditionally independent (‘significant’) CNS-related gene sets together account for 39% of schizophrenia SNP-based heritability (P = 5.07 × 10−26), equating to 61% of genic heritability (Fig. 2a and Supplementary Table 7). This is likely to be an underestimation of the true effect of these gene sets, as distal non-genic regulatory elements (not included in this analysis) will add to the heritability explained by these genes. In examining the relationship between the LoF-intolerant and CNS-related gene sets (Fig. 2a), genes belonging to both categories were the most highly enriched (2.6-fold, P = 7.90 × 10−15), although LoF-intolerant genes that were not annotated to our significant CNS gene sets still displayed enrichment for SNP-based heritability (1.74-fold, P = 9.77 × 10−10), while genes that were in the significant CNS gene sets but had pLI <0.9 showed more modest enrichment (1.39-fold, P = 6.05 × 10−4). Notably, genes outside these categories were depleted in heritability relative to their SNP content (enrichment = 0.79, P = 1.82 × 10−7).
Fig. 2. Partitioned heritability analysis of gene sets in schizophrenia.
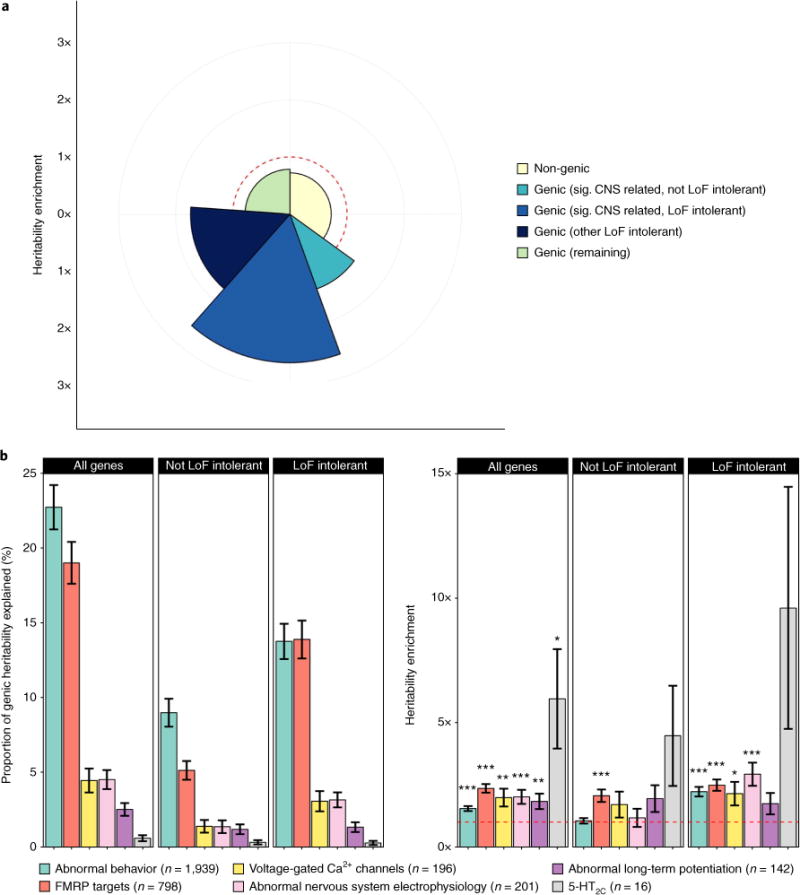
a, Heritability of genomic partitions and the six conditionally independent (‘significant’) gene sets (Table 2). The radius of each segment indicates the degree of enrichment, while the arc (angle of each slice) indicates the percentage of total SNP-based heritability explained. No relative enrichment (enrichment = 1) is shown by the dashed red line (and depletion equates to enrichment <1, inside red line). b, Heritability of the significant CNS gene sets dissected by their overlap with LoF-intolerant genes. Whiskers represent heritability or enrichment standard errors. Asterisks indicate the significance of each heritability enrichment (*P≤0.05, **P≤0.01, ***P≤0.001).
This general pattern remained when we focused on the six significant CNS gene sets individually, in that the enrichment in these gene sets derived primarily from their intersection with LoF-intolerant genes (Fig. 2b). Indeed, only the targets of FMRP showed significant enrichment for SNPs in genes that were not LoF intolerant (2.06-fold, P = 4.23 × 10−5).
Data-driven gene set analysis
To set the systems genomics results in context and to ensure that we were not missing enrichment in other gene sets by our hypothesis-driven approach, we undertook a purely data-driven analysis of a larger comprehensive annotation of gene sets from multiple public databases, totaling 6,677 gene sets (Methods and Supplementary Table 10). Six gene sets survived FWER correction for the full 6,677 gene sets and showed independence through conditional analyses. The LoF-intolerant gene set was the most strongly enriched, followed by the two most strongly associated functional gene sets we had specified in our hypothesis-driven CNS gene set analysis (FMRP targets and MGI abnormal behavior genes). The other three sets were calcium ion import (GO:0070509), membrane depolarization during action potential (GO:0086010) and synaptic transmission (GO:0007268). These are highly overlapping with the independently associated sets from our primary CNS systems genomics analysis. Indeed, if we repeat the data-driven comprehensive gene set analysis while adjusting for the six independently associated CNS gene sets, the only surviving enrichment term is the LoF-intolerant genes. These results are consistent with those from CNV analysis30 in that they do not support annotations other than those related to CNS function and demonstrate that hypothesis-based analysis to maximize power does not substantially impact the overall pattern of results.
Identifying likely candidates within associated loci
To identify SNPs and genes that might be causally linked to the genome-wide significant associations, we used FINEMAP39 to identify credibly causal alleles (those with a cumulative posterior probability for a locus of at least 95%) and functionally annotated these alleles using ANNOVAR40. This identified 6,105 credible SNPs across 144 genome-wide significant loci, excluding the major histocompatibility complex (MHC) region (Methods and Supplementary Table 11). From these, we defined a highly credible set of SNPs (n = 25) as those that were more likely to explain the associations than all other SNPs combined (i.e., with a FINEMAP posterior probability greater than 0.5). Of these, 14 mapped to genes on the basis of putative functionality (exonic SNPs that cause nonsynonymous or splice variations or promoter SNPs; n = 6) or mapped to regions identified as likely regulatory elements (n = 8) through chromosome conformation analysis performed in tissue from the developing brain using Hi-C41 physical interactions (Methods and Supplementary Table 12). One of the implicated alleles was a nonsynonymous variant in the manganese and zinc transporter gene SLC39A8. Nonsynonymous variants in this gene, which lead to SLC39A8 deficiency, have been associated with severe neurodevelopmental disorders putatively through impaired manganese transport and glycosylation42, highlighting a mechanism of therapeutic potential for schizophrenia.
We also applied Summary-data-based Mendelian Randomization (SMR) analysis43 to the data in concert with dorsolateral prefrontal cortex expression quantitative trait locus (eQTL) data from the CommonMind Consortium44, aiming to identify variants that might be causally linked through expression changes in specific genes (Methods and Supplementary Table 13). After applying a conservative threshold (PHEIDI > 0.05) that prioritized colocalized signals due to a single causal variant43, we identified 22 candidates at 19 loci with false discovery rate (FDR) P < 0.05.
In total, the combination of FINEMAP, Hi-C and SMR analyses assigned potentially causal genes at 33 genome-wide significant loci and implicated a single gene at 27 of these loci. However, the analyses intersect for only a single gene, ZNF823, indicating the need for more comprehensive functional genomic annotations in CNS- relevant tissues.
Discussion
In the largest genetic study of schizophrenia thus far, we explore the genomic architecture of and the evolutionary pressures on common variants associated with the disorder. Our study provides the first evidence linking common variation in LoF-intolerant genes to risk of developing schizophrenia and demonstrates that these genes account for a substantial proportion (30%) of the SNP-based heritability for schizophrenia. Systems genomics analysis highlights six gene sets that are independently associated with schizophrenia and point to molecular, physiological and behavioral pathways involved in schizophrenia pathogenesis.
Given that mutation intolerance is due to high selection pressure21,23,24, our finding that schizophrenia risk variants that persist at common allele frequencies are enriched in LoF-intolerant genes might appear counterintuitive. However, new evidence presented here suggests that this can be reconciled by BGS, which is a consequence of purifying selection in regions of low recombination45,46. In such regions, recurrent selection against deleterious variants causes haplotypes to be removed from the gene pool, which reduces genetic diversity in a manner equivalent to a reduction in effective population size47. This in turn impairs the efficiency of the selection process, allowing alleles with small deleterious effects to rise in frequency by drift48. Such a consequence of purifying selection has been shown to be compatible with the genomic architecture of complex human traits49 and to influence phenotypes in model organisms50. We have explicitly modeled this effect (both theoretically and via simulations; Supplementary Note) and provide strong evidence for the feasibility of this effect as explanatory for the effect sizes seen for common alleles in schizophrenia.
We did not find enrichment for any measure of positive selection or Neanderthal introgression. A recent study explained a negative correlation between schizophrenia associations and metrics indicative of a Neanderthal selective sweep as evidence for positive selection or polygenic adaptation in schizophrenia12. We do not find any significant correlation in our model, which addresses the contribution of BGS, and hence our results are not consistent with large contributions of positive selection to the genetic architecture of schizophrenia (Table 1). Indeed, positive selection is not widespread in humans, as reported by other studies that explicitly considered or accounted for BGS28,51. Polygenic adaptation, the co-occurrence of many subtle allele frequency shifts at loci influencing complex traits52, remains an intriguing possibility but has not been implicated in psychiatric phenotypes, including schizophrenia, in recent analyses53,54. In contrast, BGS has been proposed as a mechanism driving human-Neanderthal incompatibilities, as regions with stronger estimated BGS have lower estimated Neanderthal introgression55. We therefore conclude that the bulk of the BGS signal we obtain is unlikely to be influenced by positive selection29, challenging theories of the selective advantage of schizophrenia risk alleles to explain the high population frequencies of these alleles.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41588-018-0059-2.
Methods
GWAS and reporting of independently associated regions
Details of sample collection and genotype quality control are given in the Supplementary Note. The CLOZUK schizophrenia GWAS was performed using logistic regression with imputation probabilities (‘dosages’) adjusted for 11 principal-component analysis (PCA) covariates. These covariates were chosen as those nominally significant (P < 0.05) in a logistic regression for association with the phenotype56. To avoid overburdening the GWAS power by adding too many covariates to the regression model57, only the first 20 principal components were considered and tested for inclusion, as higher numbers only become useful for the analysis of populations that bear strong signatures of complex admixture58. The final set of covariates included the first five principal components (as recommended for most GWAS approaches59) and principal components 6, 9, 11, 12, 13 and 19. Quantile–quantile and Manhattan plots are shown in Supplementary Figs. 7 and 8.
To identify independent signals among the regression results, signals were amalgamated into putative associated loci using the same two-step strategy and parameters as PGC (Supplementary Table 14). In this procedure, regular LD clumping is performed (r2 = 0.1, P < 1×10−4; window size < 3 Mb) to obtain independent index SNPs. Afterward, loci are defined for each index SNP as the genomic region that contains all other imputed SNPs within the region with r2 ≥ 0.6. To avoid inflating the number of signals in gene-dense regions or in those with complex LD, all loci within 250 kb of each other were annealed.
Meta-analysis with PGC
A total of 6,040 cases and 5,719 controls from CLOZUK were included in the recent PGC study5. We reanalyzed the PGC data after excluding all these cases and controls, obtaining a sample termed ‘INDEPENDENT PGC’ (29,415 cases and 40,101 controls). Adding the summary statistics from this independent sample to the CLOZUK GWAS results allowed for a combined analysis of 40,675 cases and 64,643 controls (without duplicates or related samples). This meta-analysis was performed using the fixed-effects procedure in METAL60 with weights derived from standard errors. For consistency with the PGC analysis, additional filters (INFO > 0.6 and MAF > 0.01) were applied to the CLOZUK and INDEPENDENT PGC summary statistics, leaving 8 million markers in the final meta-analysis results. Quantile–quantile and Manhattan plots are shown in Supplementary Fig. 3 and Fig. 2. The same procedure as above was used to report independent loci from this analysis (Supplementary Tables 3 and 4). As raw PGC genotypes were not available for the LD clumping procedure, phase 3 of the 1000 Genomes Project (1KGPp3) was used as a reference.
Replication of new GWAS loci
To validate the association signals from the CLOZUK + PGC meta-analysis, we amalgamated data contributed by other schizophrenia genetics consortia (total of 5,762 cases and 154,224 controls; details in the Supplementary Note). We sought GWAS summary statistic data for the index SNPs from the 50 new genome-wide significant loci (Supplementary Table 4). These summary statistics were subjected to meta-analysis in METAL using the fixed-effects procedure to obtain replication and heterogeneity statistics (Supplementary Table 6).
Estimation and assessment of a polygenic signal
Association signals caused by the vast polygenicity underlying complex traits can be hard to distinguish from confounders related to sample relatedness and population stratification. To effectively disentangle this issue, we used the software LD Score v1.0 to analyze the summary statistics of our association analyses and estimate the contribution of confounding biases to our results by LDSR61. An LD reference was generated from 1KGPp3 after restricting this dataset to strictly unrelated individuals and retaining only markers with MAF >0.01. To improve accuracy, the summary statistics used as input were refined by discarding all indels and restricting SNPs to those with INFO >0.9 and MAF >0.01, a total of 5.16 million SNPs. The resulting LD score intercept for the CLOZUK GWAS was 1.085 ± 0.010, which compared to a mean χ2 of 1.417 indicates a polygenic contribution of at least 80%. For the CLOZUK + PGC meta-analysis, the LD score intercept was 1.075 ± 0.014 (mean χ2 = 1.960), which supports more than 90% of the signal being driven by polygenic architecture. Both of these figures are in line with those for other well-powered GWAS of complex human traits64, including schizophrenia5. This analysis was also used to calculate SNP-based heritability (h2SNP) for our three datasets (CLOZUK, INDEPENDENT PGC and the CLOZUK + PGC meta-analysis), which we transformed to a liability scale using a population prevalence of 1% (registry-based lifetime prevalence62). For reference and compatibility with epidemiological studies of schizophrenia, prevalence estimates of 0.7% (lifetime morbid risk63) and 0.4% (point prevalence63, more akin to treatment-resistant schizophrenia prevalence (appropriate for CLOZUK)) were used for additional liability-scale h2SNP calculations (Supplementary Table 15).
The LDSR framework allowed us to compare the genetic architecture of CLOZUK and INDEPENDENT PGC, by calculating the correlation of their summary statistics64. A genetic correlation coefficient of 0.954±0.030 was obtained, with a P value of 6.63×10−227. We also examined the independent SNPs that reached a genome-wide significant level in the INDEPENDENT PGC dataset, of which there were 76 after excluding the extended major histocompatibility complex (xMHC) region. In the CLOZUK sample, 76% (n = 57) of these genome-wide significant SNPs were nominally significant (P < 0.05). Using binomial sign tests based on clumped subsets of SNPs65, we found that all but 1 (98.6%) of these 76 genome-wide significant SNPs were associated with the same direction of effect in the CLOZUK sample, a result highly unlikely to reflect chance (P = 2.04 × 10−21; Supplementary Table 1). Moreover, of the 1,160 SNPs with an association P value less than 1 × 10−4 in the INDEPENDENT PGC sample, 82% showed enrichment in the CLOZUK cases (P = 3.44 × 10−113), confirming that very large numbers of true associations will be discovered among these SNPs with increased sample sizes. Additionally, the new sample introduced in this study (CLOZUK2) was compared by the same methods with the PGC dataset and showed results consistent with the full CLOZUK analysis, providing molecular validation of this sample as a schizophrenia sample (Supplementary Table 1).
We went on to conduct polygenic risk score analysis. Polygenic scores for CLOZUK were generated from INDEPENDENT PGC as a training set, using the same parameters for risk profile score (RPS) analysis in PGC5, arriving at a high-confidence set of SNPs for RPS estimation by removing the xMHC region and indels, and applying INFO > 0.9 and MAF > 0.1 cutoffs. Scores were generated from the autosomal imputation dosage data, using a range of P-value thresholds for SNP inclusion66 (5 × 10−8, 1 × 10−5, 0.001, 0.05 and 0.5). In this way, we can assess the presence of a progressively increasing signal-to-noise ratio in relation to the number of markers included67. As in the PGC study, we found the best P-value threshold for discrimination to be 0.05 and report highly significant polygenic overlap between the INDEPENDENT PGC and CLOZUK samples (P < 1 × 10−300, Nagelkerke r2 = 0.12; Supplementary Table 2), confirming the validity of combining the datasets. For comparison with other studies, we also report polygenic variance on the liability scale68, which amounted to 5.7% for CLOZUK at the 0.05 P-value threshold (Supplementary Table 2). As in the PGC study, the limited r2 and area under the receiver operating characteristic curve (AUROC) obtained by this analysis restrict the current clinical utility of these scores in schizophrenia.
Gene set analysis
To assess the enrichment of sets of functionally related genes, we used MAGMA v1.0320 on the CLOZUK + PGC meta-analysis summary statistics. From these, we excluded the xMHC region for its complex LD and the X chromosome given its smaller sample size. In the resulting data, gene-wide P values were calculated by combining the P values of all SNPs inside genes after accounting for LD and outliers. This was performed allowing for a window of 35 kb upstream and 10 kb downstream of each gene to capture the signal of nearby SNPs that could fall in regulatory regions69,70. Next, we calculated competitive gene set P values on the gene-wide P values after accounting for gene size, gene set density and LD between genes. For multiple-testing correction in each gene set collection, an FWER71 was computed using 100,000 resamplings.
We performed sequential analyses using the following approaches:
LoF-intolerant genes. We tested the enrichment of the LoF-intolerant genes described by ExAC21. This set comprised all genes defined in the ExAC database as having a probability of LoF intolerance (pLI) statistic higher than 90%. Although these genes do not form part of cohesive biological processes or phenotypes, they have previously been found to be highly expressed across tissues and developmental stages21. Also, they are enriched for hub proteins72, which makes them interesting candidates for involvement in the ‘evolutionary canalization’ processes that have been proposed to lead to pleiotropic, complex disorders73.
CNS-related genes. These gene sets were compiled in our recent study30 and include 134 gene sets related to different aspects of CNS function and development. These include, among others, gene sets that have been implicated in schizophrenia by at least two independent large-scale sequencing studies8,31: targets of FMRP32, constituents of the N-methyl-D-aspartate receptor (NMDAR74) and activity-regulated cytoskeleton-associated protein complexes (ARCs75,76), as well as CNS and behavioral gene sets from MGI database version 635.
Genes identified by data-driven analysis. The final systems genomic analysis was designed as an ‘agnostic’ approach, with the aim of integrating a large number of gene sets from different public sources, not necessarily conceptually related to psychiatric disorders, as this has been successful elsewhere70,77. We conducted this analysis to test whether additional gene sets were associated in addition to those from the 134 CNS-related gene sets. For this, first we merged together the LoF-intolerant gene set and the 134 sets in the CNS-related collection. Second, we selected additional gene set sources to encompass a comprehensive collection of biochemical pathways and gene regulatory interaction networks: 2,693 gene sets with direct experimental evidence and a size of 10–200 genes70 were extracted from Gene Ontology (GO78) database release 01/02/2016; 1,787 gene sets were extracted from the fourth ontology level of MGI database version 635; 1,585 gene sets were extracted from REACTOME79 version 55; 290 gene sets were extracted from KEGG80 release 04/2015; and 187 gene sets were extracted from OMIM81 release 01/02/2016.
The total number of gene sets included was 6,677.
Detailed results of the analyses of the CNS-related and data-driven collection are given in Supplementary Tables 9 and 10. Reported numbers of genes in each gene set are those with available data in the meta-analysis. This may differ from the original gene set description, as some genic regions had null or poor SNP coverage. Following the data-driven gene set analysis as described, we also conducted analysis adjusting for our CNS-related gene sets to determine whether the data-driven analysis was contributing additional findings.
Partitioned heritability analysis of gene sets
It is known that the power of a gene set analysis is closely related to the total heritability of the phenotype and the specific heritability attributable to the tested gene set82. To assess the heritability explained by the genes carried forward after the main gene set analysis, LD Score was again used to compute a partitioned heritability estimate of CLOZUK + PGC using the gene sets as SNP annotations. As in the MAGMA analysis, the xMHC region was excluded from the summary statistics. These were also trimmed to contain no indels and only markers with INFO >0.9 and MAF >0.01, for a total of 4.64 million SNPs. As a recognized caveat of this procedure is that model misspecification can inflate the partitioned heritability estimates26, all gene sets were annotated twice: once using their exact genomic coordinates (extracted from the NCBI RefSeq database83) and another time with putative regulatory regions taken into account using the same upstream/downstream windows as in the MAGMA analyses. Additionally, all SNPs not directly covered by our gene sets of interest were explicitly included into other annotations (‘non-genic’, ‘genic but not LoF intolerant’) on the basis of their genomic location. Finally, the ‘baseline’ set of 53 annotations from Finucane et al.26, which recapitulates important molecular properties such as presence of enhancers or phylogenetic conservation, was also incorporated in the model. All of these annotations were then tested jointly for heritability enrichment. We note that using exact genic coordinates or adding regulatory regions made little difference to the estimated enrichment of our gene sets; thus, throughout the manuscript, we report the latter for consistency with the gene set analyses (Fig. 2 and Supplementary Table 8).
Natural selection analyses
We aimed to explore the hypothesis that some form of natural selection is linked to the maintenance of common genetic risk in schizophrenia12,84,85. To do this, for all SNPs included in the CLOZUK + PGC meta-analysis summary statistics, we obtained four different genome-wide metrics of positive selection (iHS86, XP-EEH87, CMS88 and CLR28), one of background selection (B statistic29, postprocessed by Huber et al.28) and one of Neanderthal introgression (average posterior probability LA89). The use of different statistics is motivated by the fact that each of them is tailored to detect a particular selective process that acted on a particular timeframe (see Vitti et al.51 for a review). For example, iHS and CMS are based on the inference of abnormally long haplotypes and thus are better powered to detect recent selective sweeps that occurred during the last ~30,000 years88, such as those linked to lactose tolerance or pathogen response90. On the other hand, CLR incorporates information about the spatial pattern of genomic variability (the site frequency spectrum91) and corrects explicitly for evidence of BGS, thus being able to detect signals from 60,000 to 240,000 years ago28. The B statistic uses phylogenetic information from other primates (chimpanzee, gorilla, orangutan and rhesus macaque) to infer the reduction in allelic diversity that exists in humans as a consequence of purifying selection on linked sites over evolutionary time frames92. As the effects of background selection on large genomic regions can mimic those of positive selection46, it is possible that the B statistic might amalgamate both, although the rather large diversity reduction that it infers for the human genome as a whole suggests that any bias due to positive selection is likely to be minor93. Finally, XP-EEH is a haplotype-based statistic that compares two population samples, and its power is thus increased for alleles that have suffered differential selective pressures since those populations diverged90. Although methodologically different, LA has a similar rationale by comparing human and Neanderthal genomes89, to infer the probability of each human haplotype having been the result of an admixture event with Neanderthals.
For this work, CLR, CMS, the B statistic and LA were retrieved directly from their published references and lifted over to GRCh37 genomic coordinates if required using the Ensembl LiftOver tool94,95. As the available genome-wide measures of iHS and XP-EEH were based on HapMap 3 data96, both statistics were recalculated with the HAPBIN97 software directly on the EUR superpopulation of the 1KGPp3 dataset, with the AFR superpopulation used as the second population for XP-EEH. Taking advantage of the fine-scale genomic resolution of these statistics (between 1-10 kb), all SNP positions present in CLOZUK + PGC were assigned a value for each measure, either directly (if the position existed in the lifted-over data) or by linear interpolation. To simplify the interpretation of our results, all measures were transformed before further analyses to a common scale, in which larger values indicate stronger effect of selection or increased probability of introgression. For example, the BGS B statistic, for which values of zero indicate the strongest effect (see Charlesworth45 for its theoretical derivation), was included in all our analyses as 1 - B, which we termed ‘BGS intensity’.
Heritability enrichment of these statistics was tested by the LD Score partitioned heritability procedure. We derived binary annotations from the natural selection metrics by dichotomizing at extreme cutoffs defined by the top 2%, 1% and 0.5% of the values of each metric in the full set of SNPs. This approach is widely used in evolutionary genomics, owing to the difficulty of setting specific thresholds to define regions under selection28,51. Consistent with the previously described LDSR partitioned heritability protocol, enrichment was estimated with all binary annotations included in a model with multiple categories that represent important genomic features. This model included the 3 main categories of our set-based analysis (‘non-genic’, ‘genic’ and ‘LoF intolerant’), 2 categories based on genomic regions with outlying LD patterns (recombination hotspots and coldspots)98 and the 53 ‘baseline’ categories of Finucane et al.26.
We then derived the τc coefficient26 (and associated P value) of the significantly enriched natural selection annotations (i.e., the background selection metric), This represents the enrichment of an annotation over and above the enrichment of all other annotations, which is a conservative approach, as most of the categories in our model are partially overlapping. To increase our power and for additional validation, we noted that LD Score allows testing of the full range of quantitative metrics, in an extension of the partitioned heritability framework. Results of this analysis are reported in Supplementary Table 8.
Analysis of other phenotypes
To explore the specificity of our natural selection results, we retrieved data from other well-powered GWAS of complex traits. We selected three phenotypes for which (i) the genome-wide summary statistic data were publicly available, (ii) the sample size was larger than 50,000 individuals, (iii) the phenotype has minimal impact on fecundity99–101 (and hence the traits behave as neutral or approximately neutral to selection) and (iv) summary statistics were considered adequate for LD Score analysis based on baseline z scores >426,102 (Supplementary Table 8). The phenotypes chosen were Alzheimer’s disease103, neu roticism104 and type 2 diabetes105. For the LD Score analyses, as the public release of these statistics did not include imputation INFO scores at the time of this study, we restricted the set of SNPs to those included in the HapMap 3 project96, as recommended61. To facilitate comparison with the schizophrenia results, we also restricted our schizophrenia summary statistic data to these SNPs and repeated the analyses above using BGS as a binary (top 2%) and quantitative trait.
We also employed MAGMA on the summary statistics of these additional phenotypes to examine whether the LoF-intolerant gene set enrichment displayed specificity to schizophrenia, after excluding the xMHC and APOE regions.
Fine-mapping, Hi-C and SMR
Accurately locating causal genes (‘fine-mapping’) for complex disorders is a challenge to GWAS and usually requires multiple approaches105. To highlight credibly causal variants, we used FINEMAP v1.139 at each of the 145 identified loci (Supplementary Table 3), selecting variants with a cumulative posterior probability of 95%. These were then annotated with ANNOVAR40 release 2016Feb1 (Supplementary Table 11). We mapped the SNPs with a FINEMAP posterior probability higher than 0.5 to the developing brain Hi-C data generated by Won et al.41, following the methodology described therein, which allowed us to implicate genes by chromatin interactions instead of solely chromosomal position (Supplementary Table 12). We compiled results from the eQTL analysis of the CommonMind Consortium post-mortem brain tissues44. This included 15,782 genes, which were curated to remove any genes with FPKM = 0 across >10% of individuals. All the SNPs from the meta-analysis data were mapped to the eQTL data using rs numbers, position and allele matching. Both datasets were analyzed together using SMR43, which resulted in 4,276 genes showing eQTLs with overlapping SNPs and genome-wide significant P values (Supplementary Table 13).
URLs
CLOZUK + PGC2 meta-analysis summary statistics, http://walters.psycm.cf.ac.uk/; CRESTAR Consortium, http://www.crestar-project.eu/; Wellcome Trust Case Control Consortium, http://www.wtccc.org.uk/; People of the British Isles project, http://www.peopleofthebritishisles.org/; Mouse Genome Informatics (MGI), http://www.informatics.jax.org/; Psychiatric Genomics Consortium, http://www.med.unc.edu/pgc/; 1000 Genomes IBD segment sharing within and between populations, http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/ibd_by_pair/.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
The gene content of the CNS-related gene sets that survived conditional analysis (significant) is given in MAGMA format in the Supplementary Data. Summary statistics from the CLOZUK + PGC2 GWAS are available for download (see URLs).
Supplementary Material
Acknowledgments
General. This project has received funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement 279227 (CRESTAR Consortium). The work at Cardiff University was funded by the Medical Research Council (MRC) Centre (MR/L010305/1), a program grant (G0800509) and a project grant (MR/L011794/1) and by the European Community’s Seventh Framework Programme HEALTH-F2-2010-241909 (project EU-GEI). U.D. received funding from the German Research Foundation (DFG, grant FOR2107 DA1151/5-1; SFB-TRR58, project C09) and the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant Dan3/012/17). E.M.B. and N.R.W received salary funding from the National Health and Medical Research Council (NHMRC; 1078901, 105363). E. Santiago and A.C. received funding from the Agencia Estatal de Investigación (AEI; CGL2016-75904-C2-1-P), Xunta de Galicia (ED431C 2016-037) and Fondo Europeo de Desarrollo Regional (FEDER). The iPSYCH and GEMS2 teams acknowledge funding from the Lundbeck Foundation (grants R102-A9118 and R155-2014-1724), the Stanley Medical Research Institute, an advanced grant from the European Research Council (project 294838), the Danish Strategic Research Council and grants from Aarhus University to the iSEQ and CIRRAU centers.
Case data. We thank the participants and clinicians who took part in the CardiffCOGS study. For the CLOZUK2 sample, we thank Leyden Delta for supporting the sample collection, anonymization and data preparation (particularly M. Helthuis, J. Jansen, K. Jollie and A. Colson), Magna Laboratories, UK (A. Walker) and, for CLOZUK1, Novartis and the Doctor’s Laboratory staff for their guidance and cooperation. We acknowledge L. Bates, C. Bresner and L. Hopkins, at Cardiff University, for laboratory sample management. We acknowledge W. Lawrence and M. Einon, at Cardiff University, for support with the use and setup of computational infrastructures.
Control data. A full list of the investigators who contributed to the generation of the Wellcome Trust Case Control Consortium (WTCCC) data is available from its website. Funding for the project was provided by the Wellcome Trust under award 076113. The UK10K project was funded by Wellcome Trust award WT091310. Venous blood collection for the 1958 Birth Cohort (NCDS) was funded by UK MRC grant G0000934, peripheral blood lymphocyte preparation was funded by the Juvenile Diabetes Research Foundation (JDRF) and the Wellcome Trust, and cell line production, DNA extraction and processing were funded by Wellcome Trust grant 06854/Z/02/Z. Genotyping was supported by the Wellcome Trust (083270) and the European Union (ENGAGE: HEALTH-F4-2007-201413). The UK Blood Services Common Controls (UKBS-CC collection) was funded by the Wellcome Trust (076113/C/04/Z) and by a National Institute for Health Research (NIHR) programme grant to the NHS Blood and Transplant authority (NHSBT; RP-PG-0310-1002). NHSBT also made possible the recruitment of the Cardiff Controls, from participants who provided informed consent. Generation Scotland (GS) received core funding from the Chief Scientist Office of the Scottish government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR03006). Genotyping of the GS:SFHS samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, Edinburgh, Scotland, and was funded by the MRC and Wellcome Trust (grant 10436/Z/14/Z). The Type 1 Diabetes Genetics Consortium (T1DGC; EGA dataset EGAS00000000038) is a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Institute of Allergy and Infectious Diseases (NIAID), the National Human Genome Research Institute (NHGRI), the National Institute of Child Health and Human Development (NICHD) and JDRF. The People of the British Isles project (POBI) is supported by the Wellcome Trust (088262/Z/09/Z). TwinsUK is funded by the Wellcome Trust, MRC, European Union, NIHR-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. Funding for the QIMR samples was provided by the Australian NHMRC (241944, 339462, 389875, 389891, 389892, 389927, 389938, 442915, 442981, 496675, 496739, 552485, 552498, 613602, 613608, 613674, 619667), the Australian Research Council (FT0991360, FT0991022), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254) and the US National Institutes of Health (NIH; AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206, DA12854, DA019951) and the Center for Inherited Disease Research (Baltimore, MD, USA). TEDS is supported by a program grant from the MRC (G0901245-G0500079), with additional support from the NIH (HD044454, HD059215). In the GERAD1 Consortium, Cardiff University was supported by the Wellcome Trust, the MRC, Alzheimer’s Research UK (ARUK) and the Welsh government. King’s College London acknowledges support from the MRC. The University of Belfast acknowledges support from ARUK, the Alzheimer’s Society, Ulster Garden Villages, the Northern Ireland R&D Office and the Royal College of Physicians/Dunhill Medical Trust. Washington University was funded by NIH grants, the Barnes Jewish Foundation, and the Charles and Joanne Knight Alzheimer’s Research Initiative. The Bonn group was supported by the German Federal Ministry of Education and Research (BMBF), Competence Network Dementia and Competence Network Degenerative Dementia and by the Alfried Krupp von Bohlen und Halbach-Stiftung.
GERAD1 Consortium
Denise Harold47,48, Rebecca Sims47, Amy Gerrish47, Jade Chapman47, Valentina Escott-Price1, Richard Abraham47, Paul Hollingworth47, Jaspreet Pahwa47, Nicola Denning47, Charlene Thomas47, Sarah Taylor47, John Powell49, Petroula Proitsi49, Michelle Lupton49, Simon Lovestone49,50, Peter Passmore51, David Craig51, Bernadette McGuinness51, Janet Johnston51, Stephen Todd51, Wolfgang Maier52, Frank Jessen52, Reiner Heun52, Britta Schurmann52,53, Alfredo Ramirez52, Tim Becker54, Christine Herold54, André Lacour54, Dmitriy Drichel54, Markus Nothen55, Alison Goate56, Carlos Cruchaga56, Petra Nowotny56, John C. Morris56, Kevin Mayo56, Peter Holmans1, Michael O’Donovan1, Michael Owen1 and Julie Williams47
CRESTAR Consortium
Evanthia Achilla57, Esben Agerbo21,22, Cathy L. Barr58, Theresa Wimberly Böttger59, Gerome Breen7,8, Dan Cohen60, David A. Collier7,44, Sarah Curran61,62, Emma Dempster63, Danai Dima7, Ramon Sabes-Figuera57, Robert J. Flanagan64, Sophia Frangou65, Josef Frank66, Christiane Gasse59,67, Fiona Gaughran4, Ina Giegling45, Jakob Grove21,23,24,32, Eilis Hannon63, Annette M. Hartmann45, Barbara Heißerer68, Marinka Helthuis69, Henriette Thisted Horsdal59, Oddur Ingimarsson70, Karel Jollie69, James L. Kennedy71, Ole Köhler33, Bettina Konte45, Maren Lang66, Sophie E. Legge1, Cathryn Lewis7, James MacCabe4, Anil K. Malhotra72, Paul McCrone57, Sandra M. Meier59, Jonathan Mill7,63, Ole Mors21,34, Preben Bo Mortensen21,22,23, Markus M. Nöthen55, Michael C. O’Donovan1, Michael J. Owen1, Antonio F. Pardiñas1, Carsten B. Pedersen21,22, Marcella Rietschel66, Dan Rujescu45,46, Ameli Schwalber68, Engilbert Sigurdsson70, Holger J. Sørensen35, Benjamin Spencer73, Hreinn Stefansson39, Henrik Støvring67, Jana Strohmaier66, Patrick Sullivan74,75, Evangelos Vassos7, Moira Verbelen7, James T. R. Walters1 and Thomas Werge21,41,42
47MRC Centre for Neuropsychiatric Genetics and Genomics, Neurosciences and Mental Health Research Institute, Department of Psychological Medicine and Neurology, School of Medicine, Cardiff University, Cardiff, UK. 48Neuropsychiatric Genetics Group, Department of Psychiatry, Trinity Centre for Health Sciences, St James’s Hospital, Dublin, Ireland. 49Institute of Psychiatry, Department of Neuroscience, King’s College London, London, UK. 50Department of Psychiatry, University of Oxford, Oxford, UK. 51Ageing Group, Centre for Public Health, School of Medicine, Dentistry and Biomedical Sciences, Queen’s University, Belfast, UK. 52Department of Psychiatry, University of Bonn, Bonn, Germany. 53Institute for Molecular Psychiatry, University of Bonn, Bonn, Germany. 54Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Bonn, Germany. 55Department of Genomics, Life & Brain Center, University of Bonn, Bonn, Germany. 56Departments of Psychiatry, Neurology and Genetics, Washington University School of Medicine, St. Louis, MO, USA. 57Centre for Economics of Mental and Physical Health, Health Service and Population Research Department, Institute of Psychiatry, King’s College London, London, UK. 58Toronto Western Research Institute, University Health Network, Toronto, Ontario, Canada. 59National Centre for Register-Based Research, Department of Economics and Business, School of Business and Social Sciences, Aarhus University, Aarhus, Denmark. 60Department of Community Mental Health, Mental Health Organization North-Holland North, Heerhugowaard, the Netherlands. 61Department of Child and Adolescent Psychiatry, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK. 62Brighton and Sussex Medical School, University of Sussex, Brighton, UK. 63University of Exeter Medical School, RILD, University of Exeter, Exeter, UK. 64Toxicology Unit, Department of Clinical Biochemistry, King’s College Hospital NHS Foundation Trust, London, UK. 65Clinical Neurosciences Studies Center, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 66Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim/Heidelberg University, Mannheim, Germany. 67Centre for Integrated Register-based Research, CIRRAU, Aarhus University, Aarhus, Denmark. 68Concentris Research Management, Fürstenfeldbruck, Germany. 69Leyden Delta, Nijmegen, the Netherlands. 70Department of Psychiatry, Landspitali University Hospital, Reykjavik, Iceland. 71Centre for Addiction and Mental Health, Toronto, Ontario, Canada. 72Division of Psychiatry Research, Zucker Hillside Hospital, Northwell Health System, Glen Oaks, NY, USA. 73Department of Psychological Medicine, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK. 74Center for Psychiatric Genomics, Department of Genetics, University of North Carolina, Chapel Hill, NC, USA. 75Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
Footnotes
Author contributions
A.F.P. curated and processed genetic data, performed statistical analyses, contributed to the interpretation of results and participated in the primary drafting of the manuscript. P.H., A.J.P., V.E.-P., A.C. and E. Santiago performed statistical analyses, contributed to the interpretation of results and participated in the primary drafting of the manuscript. S.R. curated and processed genetic data and participated in the primary drafting of the manuscript. N.C. and M.L.H. contributed to the interpretation of results and participated in the primary drafting of the manuscript. S.E.L., S.B. and A.L. participated in the recruitment of participants for the study and curated and managed their phenotypic information. D.C., J.H., L.H., E.R. and G.K. contributed and curated data used in the statistical analyses. K.M. managed the laboratory and genotyping procedures at Cardiff University. J.H.M., D.A.C. and D.R. supervised the recruitment of the participants for the study. S.A.M. managed the genotyping of samples for the study. N.R.W contributed genotypes of control samples and participated in the primary drafting of the manuscript. Control data were obtained from the GERAD1 Consortium; as such, the investigators within the GERAD1 Consortium contributed to the design and implementation of GERAD1 and/or provided control data but did not participate in analysis or writing of this report. D.H.G., L.M.H., D.M.R., P.S., E.A.S. and H.W. performed statistical analyses and contributed to the interpretation of results. M.J.O. and M.C.O’D. conceived and supervised the project, contributed to the interpretation of results and participated in the primary drafting of the manuscript. J.T.R.W. conceived and supervised the project, led the recruitment of the participants and sample acquisition for the study, performed statistical analysis, contributed to the interpretation of results and participated in the primary drafting of the manuscript. All other authors contributed genotypes of control samples or summary statistics of replication samples. All authors had the opportunity to review and comment on the manuscript, and all approved the final manuscript.
Competing interests
D.A.C. is a full-time employee and stockholder of Eli Lilly and Company. The remaining authors declare no conflicts of interest.
Supplementary information is available for this paper at https://doi.org/10.1038/s41588-018-0059-2.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thornicroft G. Physical health disparities and mental illness: the scandal of premature mortality. Br J Psychiatry. 2011;199:441–442. doi: 10.1192/bjp.bp.111.092718. [DOI] [PubMed] [Google Scholar]
- 3.Olfson M, Gerhard T, Huang C, Crystal S, Stroup TS. Premature mortality among adults with schizophrenia in the United States. JAMA Psychiatry. 2015;72:1172–1181. doi: 10.1001/jamapsychiatry.2015.1737. [DOI] [PubMed] [Google Scholar]
- 4.Morgan C, et al. Reappraising the long-term course and outcome of psychotic disorders: the AESOP-10 study. Psychol Med. 2014;44:2713–2726. doi: 10.1017/S0033291714000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh T, et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–577. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rees E, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Power RA, et al. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry. 2013;70:22–30. doi: 10.1001/jamapsychiatry.2013.268. [DOI] [PubMed] [Google Scholar]
- 10.Huxley J, Mayr E, Osmond H, Hoffer A. Schizophrenia as a genetic morphism. Nature. 1964;204:220–221. doi: 10.1038/204220a0. [DOI] [PubMed] [Google Scholar]
- 11.Shaner A, Miller G, Mintz J. Schizophrenia as one extreme of a sexually selected fitness indicator. Schizophr Res. 2004;70:101–109. doi: 10.1016/j.schres.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan S, et al. Genetic markers of human evolution are enriched in schizophrenia. Biol Psychiatry. 2016;80:284–292. doi: 10.1016/j.biopsych.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uher R. The role of genetic variation in the causation of mental illness: an evolution-informed framework. Mol Psychiatry. 2009;14:1072–1082. doi: 10.1038/mp.2009.85. [DOI] [PubMed] [Google Scholar]
- 14.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 2011;43:1224–1227. doi: 10.1038/ng.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosmicki JA, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese G, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith NGC, Eyre-Walker A. Human disease genes: patterns and predictions. Gene. 2003;318:169–175. doi: 10.1016/s0378-1119(03)00772-8. [DOI] [PubMed] [Google Scholar]
- 24.Blekhman R, et al. Natural selection on genes that underlie human disease susceptibility. Curr Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takata A, Ionita-Laza I, Gogos JA, Xu B, Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron. 2016;89:940–947. doi: 10.1016/j.neuron.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber CD, DeGiorgio M, Hellmann I, Nielsen R. Detecting recent selective sweeps while controlling for mutation rate and background selection. Mol Ecol. 2016;25:142–156. doi: 10.1111/mec.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pocklington AJ, et al. Novel findings from CNVs implicate inhibitory and excitatory signaling complexes in schizophrenia. Neuron. 2015;86:1203–1214. doi: 10.1016/j.neuron.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szatkiewicz JP, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller CS, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci USA. 2010;107:14950–14957. doi: 10.1073/pnas.1005940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bécamel C, et al. Synaptic multiprotein complexes associated with 5-HT2C receptors: a proteomic approach. EMBO J. 2002;21:2332–2342. doi: 10.1093/emboj/21.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, et al. Prediction of efficacy of vabicaserin, a 5-HT2C agonist, for the treatment of schizophrenia using a quantitative systems pharmacology model. CPT Pharmacometrics Syst Pharmacol. 2014;3:e111. doi: 10.1038/psp.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benner C, et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Won H, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JH, et al. SLC39A8 deficiency: a disorder of manganese transport and glycosylation. Am J Hum Genet. 2015;97:894–903. doi: 10.1016/j.ajhg.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 44.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charlesworth B. The effects of deleterious mutations on evolution at linked sites. Genetics. 2012;190:5–22. doi: 10.1534/genetics.111.134288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charlesworth B, Betancourt AJ, Kaiser VB, Gordo I. Genetic recombination and molecular evolution. Cold Spring Harb Symp Quant Biol. 2009;74:177–186. doi: 10.1101/sqb.2009.74.015. [DOI] [PubMed] [Google Scholar]
- 47.Comeron JM, Williford A, Kliman RM. The Hill–Robertson effect: evolutionary consequences of weak selection and linkage in finite populations. Heredity. 2008;100:19–31. doi: 10.1038/sj.hdy.6801059. [DOI] [PubMed] [Google Scholar]
- 48.Charlesworth B. Background selection 20 years on: the Wilhelmine E. Key 2012 Invitational Lecture. J Hered. 2013;104:161–171. doi: 10.1093/jhered/ess136. [DOI] [PubMed] [Google Scholar]
- 49.North TL, Beaumont MA. Complex trait architecture: the pleiotropic model revisited. Sci Rep. 2015;5:9351. doi: 10.1038/srep09351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in Celegans. Science. 2010;330:372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annu Rev Genet. 2013;47:97–120. doi: 10.1146/annurev-genet-111212-133526. [DOI] [PubMed] [Google Scholar]
- 52.Stephan W. Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol Ecol. 2016;25:79–88. doi: 10.1111/mec.13288. [DOI] [PubMed] [Google Scholar]
- 53.Field Y, et al. Detection of human adaptation during the past 2000 years. Science. 2016;354:760–764. doi: 10.1126/science.aag0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Key FM, Fu Q, Romagné F, Lachmann M, Andrés AM. Human adaptation and population differentiation in the light of ancient genomes. Nat Commun. 2016;7:10775. doi: 10.1038/ncomms10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris K, Nielsen R. The genetic cost of Neanderthal introgression. Genetics. 2016;203:881–891. doi: 10.1534/genetics.116.186890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peloso GM, Lunetta KL. Choice of population structure informative principal components for adjustment in a case–control study. BMC Genet. 2011;12:64. doi: 10.1186/1471-2156-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pirinen M, Donnelly P, Spencer CCA. Including known covariates can reduce power to detect genetic effects in case-control studies. Nat Genet. 2012;44:848–851. doi: 10.1038/ng.2346. [DOI] [PubMed] [Google Scholar]
- 58.Bouaziz M, Ambroise C, Guedj M. Accounting for population stratification in practice: a comparison of the main strategies dedicated to genome-wide association studies. PLoS One. 2011;6:e28845. doi: 10.1371/journal.pone.0028845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tucker G, Price AL, Berger B. Improving the power of GWAS and avoiding confounding from population stratification with PC-Select. Genetics. 2014;197:1045–1049. doi: 10.1534/genetics.114.164285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilier CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perälä J, et al. Lifetime prevalence of psychotic and bipolar I disorders in a general population. Arch Gen Psychiatry. 2007;64:19–28. doi: 10.1001/archpsyc.64.1.19. [DOI] [PubMed] [Google Scholar]
- 63.McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- 64.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tansey KE, et al. Common alleles contribute to schizophrenia in CNV carriers. Mol Psychiatry. 2015;21:1085–1089. doi: 10.1038/mp.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dudbridge F. Polygenic epidemiology. Genet Epidemiol. 2016;40:268–272. doi: 10.1002/gepi.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012;36:214–224. doi: 10.1002/gepi.21614. [DOI] [PubMed] [Google Scholar]
- 69.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 70.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox DD, Lee JS. Pointwise testing with functional data using the Westfall–Young randomization method. Biometrika. 2008;95:621–634. [Google Scholar]
- 72.Batada NN, Hurst LD, Tyers M. Evolutionary and physiological importance of hub proteins. PLoS Comput Biol. 2006;2:e88. doi: 10.1371/journal.pcbi.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pocklington AJ, Cumiskey M, Armstrong JD, Grant, S. G. N The proteomes of neurotransmitter receptor complexes form modular networks with distributed functionality underlying plasticity and behaviour. Mol Syst Biol. 2006;2(2006):0023. doi: 10.1038/msb4100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fernández E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pers TH, et al. Comprehensive analysis of schizophrenia-associated loci highlights ion channel pathways and biologically plausible candidate causal genes. Hum Mol Genet. 2016;25:1247–1254. doi: 10.1093/hmg/ddw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fabregat A, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Leeuw CA, Neale BM, Heskes T, Posthuma D. The statistical properties of gene-set analysis. Nat Rev Genet. 2016;17:353–364. doi: 10.1038/nrg.2016.29. [DOI] [PubMed] [Google Scholar]
- 83.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Dongen J, Boomsma DI. The evolutionary paradox and the missing heritability of schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:122–136. doi: 10.1002/ajmg.b.32135. [DOI] [PubMed] [Google Scholar]
- 85.Xu K, Schadt EE, Pollard KS, Roussos P, Dudley JT. Genomic and network patterns of schizophrenia genetic variation in human evolutionary accelerated regions. Mol Biol Evol. 2015;32:1148–1160. doi: 10.1093/molbev/msv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grossman SR, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 91.Ronen R, Udpa N, Halperin E, Bafna V. Learning natural selection from the site frequency spectrum. Genetics. 2013;195:181–193. doi: 10.1534/genetics.113.152587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nordborg M, Charlesworth B, Charlesworth D. The effect of recombination on background selection. Genet Res. 1996;67:159–174. doi: 10.1017/s0016672300033619. [DOI] [PubMed] [Google Scholar]
- 93.Fu W, Akey JM. Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. 2013;14:467–489. doi: 10.1146/annurev-genom-091212-153509. [DOI] [PubMed] [Google Scholar]
- 94.Zhao H, et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maclean CA, Chue Hong NP, Prendergast JG. hapbin: an efficient program for performing haplotype-based scans for positive selection in large genomic datasets. Mol Biol Evol. 2015;32:3027–3029. doi: 10.1093/molbev/msv172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hussin JG, et al. Recombination affects accumulation of damaging and disease-associated mutations in human populations. Nat Genet. 2015;47:400–404. doi: 10.1038/ng.3216. [DOI] [PubMed] [Google Scholar]
- 99.Ptok U, Barkow K, Heun R. Fertility and number of children in patients with Alzheimer’s disease. Arch Womens Ment Health. 2002;5:83–86. doi: 10.1007/s00737-002-0142-6. [DOI] [PubMed] [Google Scholar]
- 100.Whitworth KW, Baird DD, Stene LC, Skjaerven R, Longnecker MP. Fecundability among women with type 1 and type 2 diabetes in the Norwegian Mother and Child Cohort Study. Diabetologia. 2011;54:516–522. doi: 10.1007/s00125-010-2003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jokela M. Birth-cohort effects in the association between personality and fertility. Psychol Sci. 2012;23:835–841. doi: 10.1177/0956797612439067. [DOI] [PubMed] [Google Scholar]
- 102.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith DJ, et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatry. 2016;21:749–757. doi: 10.1038/mp.2016.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene content of the CNS-related gene sets that survived conditional analysis (significant) is given in MAGMA format in the Supplementary Data. Summary statistics from the CLOZUK + PGC2 GWAS are available for download (see URLs).


