Summary
Properties of cancer stem cells involved in drug resistance and relapse have significant effects on clinical outcome. Although tyrosine kinase inhibitors (TKIs) have dramatically improved survival of patients with chronic myeloid leukemia (CML), TKIs have not fully cured CML due to TKI-resistant CML stem cells. Moreover, relapse after discontinuation of TKIs has not been predicted in CML patients with the best TKI response. In our study, a model of CML stem cells derived from CML induced pluripotent stem cells identified ADAM8 as an antigen of TKI-resistant CML cells. The inhibition of expression or metalloproteinase activity of ADAM8 restored TKI sensitivity in primary samples. In addition, residual CML cells in patients with optimal TKI response were concentrated in the ADAM8+ population. Our study demonstrates that ADAM8 is a marker of residual CML cells even in patients with optimal TKI response and would be a predictor of relapse and a therapeutic target of TKI-resistant CML cells.
Keywords: chronic myeloid leukemia, disease specific iPSCs, TKI-resistant CML stem cells
Highlights
-
•
We established a model of CML stem cells derived from CML-iPSCs
-
•
ADAM8 is identified as an antigen of TKI-resistant CML cells
-
•
The inhibition of ADAM8 restored TKI sensitivity in primary samples
-
•
ADAM8 is a marker of residual CML cells in patients with optimal TKI response
The paucity and heterogeneity of CML stem cells are obstacles for analyses. In our study, a model of CML stem cells derived from CML-iPSCs identified ADAM8 as an antigen of TKI-resistant cells. In CML patients, ADAM8+ cells showed TKI resistance and residual CML cells after TKIs-treatment were concentrated in ADAM8+ population, suggesting that ADAM8 is a marker of TKI-resistant CML cells.
Introduction
Cancer stem cells (CSCs) have a significant effect on clinical outcome due to the properties of drug resistance and relapse (Valent et al., 2012). Despite recent advances in cancer therapies, conventional treatments fail to eradicate drug-resistant CSCs in some types of cancers, which requires permanent drug intakes to control diseases and could be a hotbed of relapse (Nguyen et al., 2012).
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm that originates in hematopoietic stem cells (HSCs) by the BCR-ABL chimeric oncogene arising from t(9;22) (q34;q11) chromosomal translocation. BCR-ABL causes increased proliferation of myeloid cells with the capacity for differentiation and progresses CML from the chronic phase (CML-CP), controllable by tyrosine kinase inhibitors (TKIs), into the lethal phase (accelerated phase [AP] and blast crisis [BC]) if untreated (O'Hare et al., 2012). Although imatinib and other TKIs targeting the resultant protein of BCR-ABL have dramatically improved survival (Druker et al., 2001), these drugs do not kill leukemic stem cells (LSCs), the CSCs in leukemia (Graham et al., 2002). Of the best 10%–20% of TKI responders with undetectable expression of BCR-ABL in blood samples by RT-PCR, half of the responders faced molecular relapse after the discontinuation of TKIs (Mahon et al., 2010, Imagawa et al., 2015), meaning that currently established tests for CML could not predict the relapse after the discontinuation of TKIs. The antigen, which marks residual CML cells after treatment with TKIs, would be a therapeutic target and a predictor of relapse.
Although many studies have reported TKI-resistant CML stem cells, CML stem cells were hard to analyze, mainly due to their paucity and heterogeneity. The population in primary CML samples, which were defined as CML stem cells by immunophenotype, was still heterogeneous and the size of the population was, in some cases, not enough for robust and repeated analysis. One of the potent models would be hematopoietic cells (HCs) derived from induced pluripotent stem cells (iPSCs) of CML patient's samples (CML-iPSCs) (Carette et al., 2010, Hu et al., 2011, Bedel et al., 2013), which were more homogeneous than primary samples and were able to expand at the stage of iPSCs. We have previously reported that HCs derived from CML-iPSCs contained a TKI-resistant population (Kumano et al., 2012). Although a survival factor for CML cells has been reported using CML-iPSCs from single patients, the CML-iPSCs had complex chromosomal aberration of four-way translocation (Suknuntha et al., 2015), which was a potential bias in the analysis.
To find out which antigen marked TKI-resistant CML cells, we took advantage of HCs derived from integration-free CML-iPSCs with solo chromosomal translocation of t(9;22) (q34;q11) from two CML-CP patients. Using CML-iPSCs, we revealed that ADAM metallopeptidase domain 8 (ADAM8), also known as CD156, marked TKI-resistant cells, which remained after the treatment with TKIs even in good responders. ADAM8 would be an effective predictor of relapse after TKI discontinuation and a potential therapeutic target of TKI-resistant cells.
Results
Establishment of Integration-free iPSCs from Two CML-CP Patients
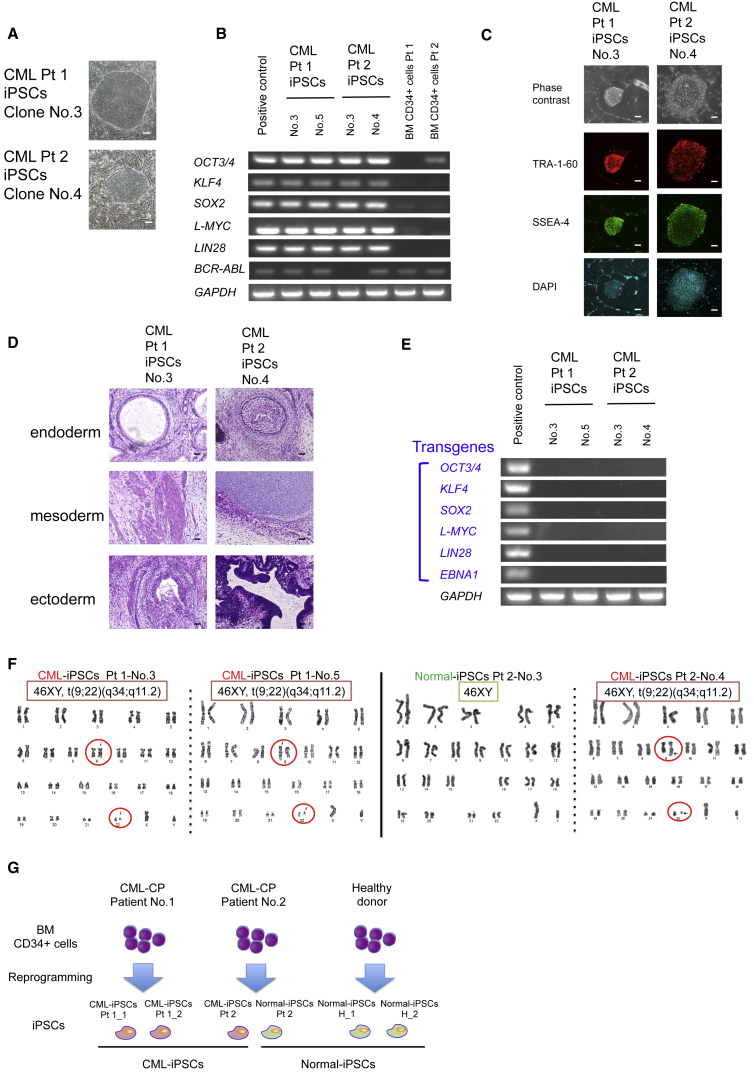
To overcome the paucity and heterogeneity of CML stem cells, we established integration-free iPSCs from the bone marrow (BM) of two CML-CP patients and used hematopoietic differentiated cells (DCs) from CML-iPSCs as a model of TKI-resistant CML stem cells. After the induction of reprograming factors in the CD34+ primary samples of two patients with newly diagnosed CML-CP, we obtained embryonic stem cell (ESC)-like colonies (Figure 1A) in which the expression of stem cell genes and BCR-ABL was confirmed by RT-PCR (Figure 1B), the expression of stem cell marker by immunofluorescence microscopy (Figure 1C), and the pluripotency by teratoma formation (Figure 1D).
Figure 1.
Establishment of Integration-free iPSCs from Two Patients with CML-CP
(A) Morphology of iPSCs from CML-CP patients. Scale bars, 200 μm.
(B) RT-PCR for the expression of endogenous stem cell genes and BCR-ABL in iPSCs from CML-CP patients.
(C) Immunofluorescence microscopy of CML-iPSCs. TRA-1-60 and SSEA-4, both of which are surface marker proteins of embryonic stem cells, are expressed in CML-iPSCs. Scale bars, 200 μm.
(D) Teratoma formation assays of CML-iPSCs. CML-iPSCs showed the reconstitution of multiple lineages. Scale bars, 50 μm.
(E) RT-PCR for the expression of transgenes in iPSCs from CML-CP patients. The expression of transgenes is undetectable.
(F) G-banded chromosomal analysis of iPSCs from CML-CP patients. CML-iPSCs have solo chromosomal aberration of t(9;22) (q34;q11). Moreover, iPSCs with normal karyotypes are obtained from CML-CP patient no. 2.
(G) Experimental scheme for establishment of iPSCs from CML-CP patients. Three CML-iPSCs derived from two CML-CP patients and three normal-iPSCs derived from a healthy donor and CML-CP patient no. 2 are used for further analysis.
To minimize the bias due to the reprograming process, we confirmed the silence of exogenous expression by episomal vectors, which do not integrate genomes (Figure 1E), and selected the clones with none other than the t(9;22) (q34;q11) chromosomal translocation, the same karyotype as primary samples by G-banded chromosomal analysis (Figure 1F).
Several clones of iPSCs with normal chromosomal karyotype were established from CML-CP patient no. 2, which were considered to be derived from normal CD34+ cells in BM of the CML-CP patient and we used the clones as normal controls. We also established iPSCs from CD34+ cells in BM of a healthy donor as normal controls. All iPSC clones obtained were listed in Table S1. Finally, we selected three normal-iPSC clones (two from a healthy donor and one from a CML-CP patient) and three CML-iPSC clones (two from CML-CP patient no. 1 and one from CML-CP patient no. 2) and performed further analysis (Figure 1G).
Definition of Pre-HPCs from CML-iPSCs as Immature, Multi-potent, and TKI-Resistant Subpopulations
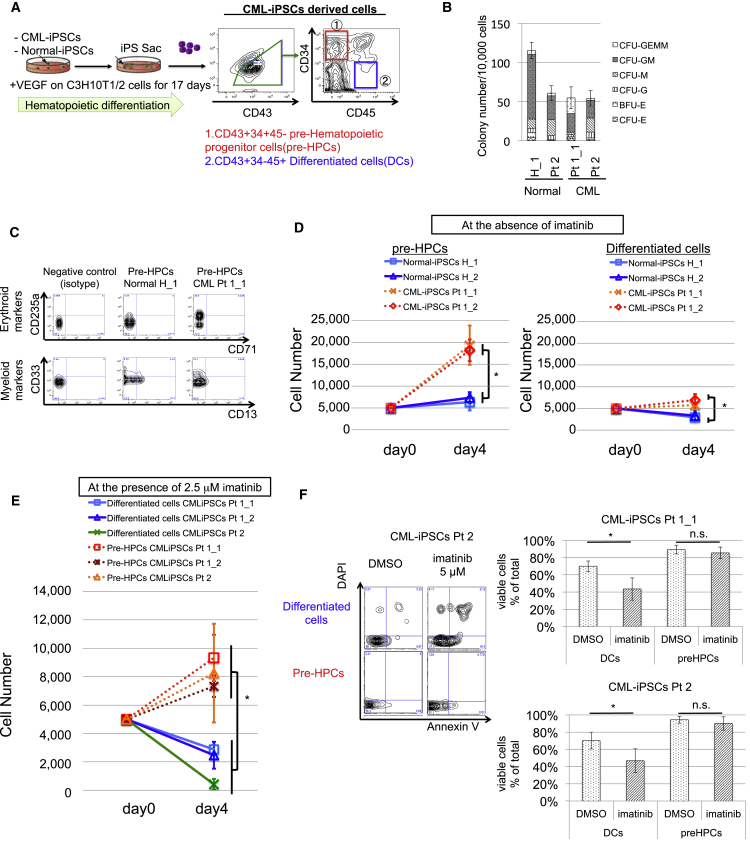
To analyze HCs derived from CML-iPSCs, CML-iPSCs were cultured with C3H10T1/2 stromal cells with vascular endothelial growth factor (VEGF) and then we obtained pre-hematopoietic progenitor cell (HPCs), immature hematopoietic DCs phenotypically defined by CD34+/CD45− and DCs (CD34+/CD45−), more DCs than pre-HPCs among CD43+ HCs (Figure 2A) (Nakajima-Takagi et al., 2013). Colony-forming cell (CFC) assay showed pre-HPCs from CML-iPSCs (CML-pre-HPCs) gave rise to numbers of myeloid and erythroid colonies comparable with those from normal iPSCs (Figure 2B). Flow cytometry (FCM) analysis showed that the myeloid and erythroid colonies from CML-pre-HPCs expressed myeloid and erythroid surface markers including CD13, CD33, and CD71 (Figure 2C).
Figure 2.
Definition of CD34+/CD43+/CD45− Pre-HPCs from CML-iPSCs as Immature, Multi-Potent, and TKI-Resistant Subpopulations
(A) Experimental scheme of hematopoietic differentiation. On day 17, pre-HPCs and differentiated cells (DCs) are obtained.
(B) CFC assay of CML-pre-HPCs. CFC assay revealed the differentiation capacity of CML-pre-HPCs into erythroid and myeloid lineages. Data show means ± SD, n = 3 independent experiments. BFU, burst-forming unit; CFU, colony-forming unit.
(C) Representative FCM data are shown. Analysis for total colonies derived from normal and CML-pre-HPCs on CFC assay. The expression of CD13 and CD33 (myeloid markers), and CD235 (erythroid marker) is detected.
(D) Proliferation assay of CML-pre-HPCs and CML-DCs at the absence of imatinib for 96 hr. Both CML-pre-HPCs and CML-DCs show increased proliferation, compared with normal counterparts. Data show means ± SD, n = 3 independent experiments (p values: two-tailed Student's t test; ∗p < 0.05).
(E) Proliferation assay of CML-pre-HPCs in the presence of 2.5 μM imatinib for 96 hr. CML-pre-HPCs show resistance to imatinib compared with CML-DCs. Data show means ± SD, n = 3 independent experiments (p values: two-tailed Student's t test; ∗p < 0.05).
(F) Apoptosis assay of CML-pre-HPCs. Representative FCM data are shown on the left. Five micromolar imatinib for 48 hr, which significantly induces apoptosis in CML-DCs, does not induce apoptosis in CML-pre-HPCs. Data show means ± SD, n = 3 independent experiments (p values: two-tailed Student's t test; ∗p < 0.05). E, erythroid; G, granulocyte; GEMM, granulocyte/erythroid/macrophage/megakaryocyte; GM, granulocyte/macrophage; M, macrophage.
Proliferation assay in the liquid culture demonstrated that CML-pre-HPCs exhibited increased cell proliferation compared with pre-HPCs from normal iPSCs (normal-pre-HPCs) (Figure 2D), which was canceled by imatinib. Even in the presence of imatinib, however, CML-pre-HPCs kept on growing (Figure 2E) and did not undergo apoptosis (Figure 2F), in marked contrast with DCs from CML-iPSCs (CML-DCs). These findings indicate that CML-pre-HPCs not only recapitulate CML disease phenotype but also show multi-potent capacity and resistance against imatinib, principal features of CML stem cells.
Gene Expression Profiling of Pre-HPCs from CML-iPSCs as a Comparable Model of CML Stem Cells
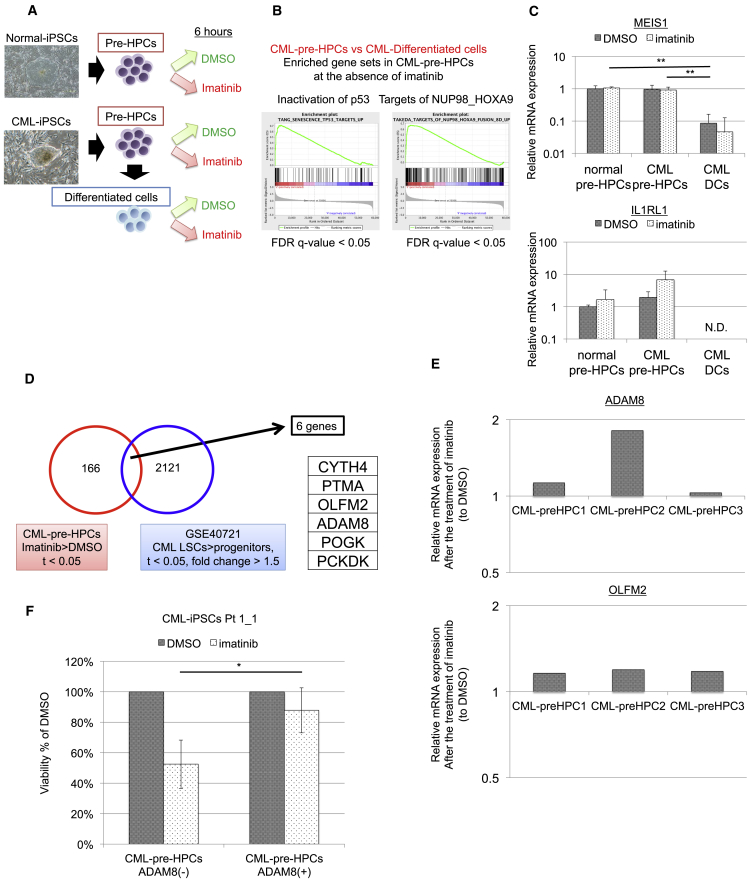
To investigate the genes involved in imatinib resistance, we performed gene expression profiling of CML-pre-HPCs (Figure 3A). qRT-PCR revealed that CML-pre-HPCs showed elevated expression levels of BCR-ABL compared with those of CML-DCs (Figure S1A). Clustering analysis divided two major clusters, one including pre-HPCs from normal-iPSCs and CML-iPSCs and another including DCs from CML-iPSCs, indicating that the expression profiling was reasonable (Figure S1B, Table S2).
Figure 3.
Gene Expression Profiling of CML Pre-HPCs as a Comparable Model of CML Stem Cells Identifies ADAM8 as a Candidate of Antigen for TKI-Resistant CML Cells
(A) A scheme of gene expression profiling. Normal pre-HPCs, CML-pre-HPCs, and CML-DCs are cultured under the same conditions as proliferation assay for 6 hr in the presence or absence of 2.5 μM imatinib.
(B) GSEA for CML-pre-HPCs compared with CML-DCs in the absence of imatinib. Gene set upregulated by inactivation of p53 and targets of NUP98 HOXA9 are enriched in CML-pre-HPCs (false-discovery rate (FDR) q value < 0.05). Twenty-three gene sets enriched from 3,267 curated gene sets in CML-pre-HPCs (FDR q value < 0.05) are listed in Table S3.
(C) Validation of genes involved in TKI resistance. Expression levels of Meis1 and IL1RL1 are elevated in CML-pre-HPCs, both of which are reported to be involved in TKI resistance of CML cells. Data show means ± SD, n = 3 independent experiments (p values: two-tailed Student's t test; ∗∗p < 0.01).
(D) Investigation of candidate genes involved in TKI resistance of CML cells.
(E) Validation of candidate genes. qRT-PCR demonstrates that expression levels of ADAM8 and OLFM2 are enhanced by treatment with imatinib in all CML-pre-HPCs; CML-pre-HPCs1 from CML-iPSCs Pt.1_1, CML-pre-HPCs2 from CML-iPSCs Pt.1_2, and CML-pre-HPCs3 from CML-iPSCs Pt.2. Data for each clone show n = 1.
(F) Viability assay for purified ADAM8+ cells in CML-pre-HPCs from Pt.1_1. ADAM8+ cells showed TKI resistance. Data show means ± SD, n = 3 independent experiments (p values: two-tailed Student's t test; ∗p < 0.05).
Compared with CML-DCs, gene sets enrichment analysis (GSEA) for CML-pre-HPCs demonstrated that 23 gene sets enriched from 3,267 curated gene sets (false-discovery rate q value < 0.05) included the gene set upregulated by inactivation of p53 and targets of NUP98-HOXA9, both of which were known to be involved in TKI resistance of CML (Figures 3B and S1C; Table S3) (Yamamoto et al., 2000, Dash et al., 2002, Li et al., 2012, Abraham et al., 2016). As the candidates of the genes involved in imatinib resistance, we selected 166 genes, which had elevated expression levels in CML-pre-HPCs by the treatment with imatinib. Among the 166 candidates, IL1RL1, reported as a gene associated with imatinib resistance (Levescot et al., 2014), was the gene of which expression level was elevated most. Both MEIS1, critical targets of NUP98-HOXA9, and IL1RL1 were validated by qRT-PCR, suggesting that CML-pre-HPCs are also a comparable model of TKI-resistant CML cells in terms of gene expression profiling and that the candidate genes are potent (Figure 3C).
The analysis for CML stem cells in published array data for patients with CML (GEO: GSE40721) narrowed down the 166 candidates to six genes, based on enhanced expression levels in TKI-resistant CML stem cell fractions (CD34+/CD38−) more than in TKI-sensitive CML progenitor fractions (CD34+/CD38+) (Figure 3D). In six genes, the expression levels of ADMA8 and OLFM2 were elevated by the treatment with imatinib among all clones of CML-pre-HPCs (Figure 3E) and purified ADAM8+ cells in CML-pre-HPCs showed TKI resistance compared with ADAM8− cells in CML-pre-HPCs (Figure 3F), suggesting that the mechanism of upregulation of ADAM8 in CML-pre-HPCs by imatinib reflected the enhanced accumulation of imatinib-resistant ADAM8+ cells. These findings indicate that ADAM8 would be a potent marker of TKI-resistant CML cells. We then performed further analyses to address the significance of ADAM8.
Gene induction of ADAM8 in CML-BC cell lines by a retroviral vector demonstrated no significant difference in TKI sensitivity (Figure S2). Moreover, in a mouse CML model using retroviral induction of BCR-ABL and BM transplantation, immunofluorescence microscopy revealed that Adam8 was not involved in TKI resistance (Figure S3). These findings suggest that the CML-BC cell line and mouse models might be unsuitable for the evaluation of ADAM8. We then evaluated the significance of ADAM8 as a marker in CML-CP primary samples.
ADAM8 as an Antigen of TKI-Resistant CML Cells in Newly Diagnosed CML-CP Patients
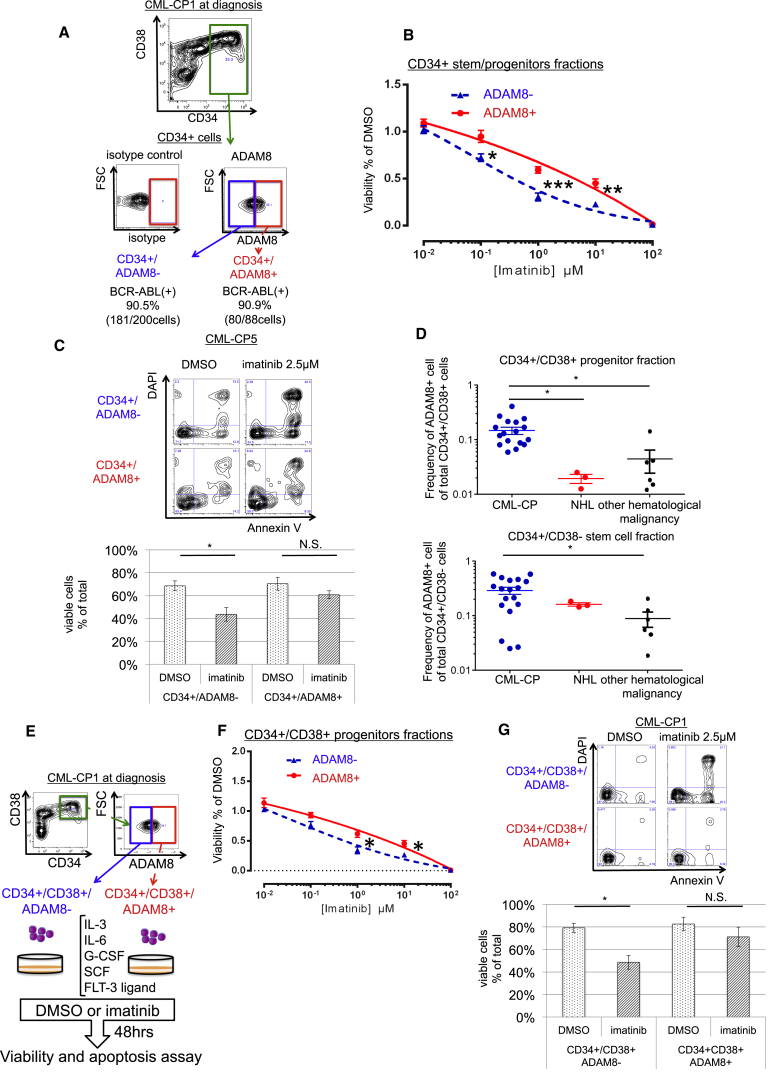
To address the significance of ADAM8 in CMP-CP patients, we evaluated purified ADAM8+ cells by fluorescence-activated cell sorting (FACS) in primary samples (Figure 4A). Morphologically, CD34+/ADAM8− and CD34+/ADAM8+ cells looked similar: round HCs (Figure S4A). Fluorescent in situ hybridization (FISH) revealed that there was no difference in positivity of BCR-ABL between CD34+/ADAM8− and CD34+/ADAM8+ cells (Figure S4B), whereas the expression of BCR-ABL in both mRNA and protein levels was suppressed more in CD34+/ADAM8+ cells than CD34+/ADAM8− cells (Figures S4C and S4D). In addition, CD34+/ADAM8+ cells had higher colony-forming capacity than CD34+/ADAM8− cells (Figure S4E) and accumulated in the G1 phase of the cell cycle (Figure S4F). These findings suggest that CD34+/ADAM8+ cell would be CML cells that had a distinct feature from CD34+/ADAM8− cells.
Figure 4.
ADAM8 Marks TKI-Resistant CML Cells in BM Samples of Patients with Newly Diagnosed CML-CP
(A) A scheme of viability and apoptosis assay for ADAM8+ cells in CD34 + stem/progenitor fraction.
(B) Viability assay for CD34+/ADAM8+ cells in BM of newly diagnosed CML-CP patients. Data show means ± SEM, n = 6 patients (p values: two-tailed Student's t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(C) Apoptosis assay for CD34+/ADAM8+ cells in BM of newly diagnosed CML-CP patients. Representative FCM data are shown on the left; 2.5 μM imatinib for 48 hr, which significantly induces apoptosis in ADAM8− cells, does not induces apoptosis in ADAM8+ cells. Data show means ± SEM, n = 4 patients (p values: two-tailed Student's t test; ∗p < 0.05).
(D) Frequency of ADAM8+ cells in CD34+/CD38+ progenitor fraction and CD34+/CD38−stem cell fraction. In the progenitor fraction, the frequency of ADAM8+ cells is significantly higher in CML-CP patients (n = 18) than in non-Hodgkin lymphoma (n = 3) and other hematological malignancies (AML, n = 1; acute lymphocytic leukemia, n = 1; myelodysplastic syndrome, n = 1; chronic myelomonocytic leukemia, n = 3). Data show means ± SEM, n = shown above patients (p values: two-tailed Student's t test; ∗p < 0.05).
(E) A scheme of viability and apoptosis assay for ADAM8+ cells in CD34+/CD38+ stem/progenitor fraction.
(F) Viability assay for CD34+/CD38+/ADAM8+ cells in BM of newly diagnosed CML-CP patients. Data show means ± SEM, n = 4 patients (p values: two-tailed Student's t test; ∗p < 0.05).
(G) Apoptosis assay for CD34+/CD38+/ADAM8+ cells in BM of newly diagnosed CML-CP patients. Representative FCM data are shown on the left; 2.5 μM imatinib for 48 hr, which significantly induces apoptosis in ADAM8− cells, does not induce apoptosis in ADAM8+ cells. Data show means ± SEM, n = 6 patients (p values: two-tailed Student's t test; ∗p < 0.05).
Among the CD34+ stem/progenitor fraction, FCM analysis found that ADAM8+ cells were enriched more in BM samples of patients with newly diagnosed CML-CP than normal controls and other types of leukemias (Figure S4G). Cell viability assays showed that ADAM8+ CML cells in a newly diagnosed CML-CP patient exhibited imatinib resistance (Figures 4B and S6) and imatinib-induced apoptosis was suppressed in ADAM8+ CML cells compared with ADAM8− cells (Figure 4C), indicating that ADAM8 is specific for CML-CP and ADMA8+ cells are involved in TKI resistance of CML-CP patients.
Even in the CD34+/CD38+ progenitor fraction, which was previously known as TKI-sensitive fraction, ADAM8+ cells exhibited TKI resistance in both cell viability and apoptosis assay (Figures 4D–4G and S7A), indicating that ADAM8 would be a useful marker of TKI-resistant CML cells.
To investigate the association between the disease progression of CML and the expression of ADAM8, we analyzed the published datasets. The analysis for the expression of ADAM8 in the published array data for patients with CML-CP, CML-AP, and CML-BC (GEO: GSE47927) demonstrated that primary samples from CML-BC statistically significantly expressed ADAM8 in HSCs, common myeloid progenitors, and granulocyte monocyte progenitors (Figure S5A). This finding indicates that primary samples from CML-BC patients may express ADAM8 at the elevated levels.
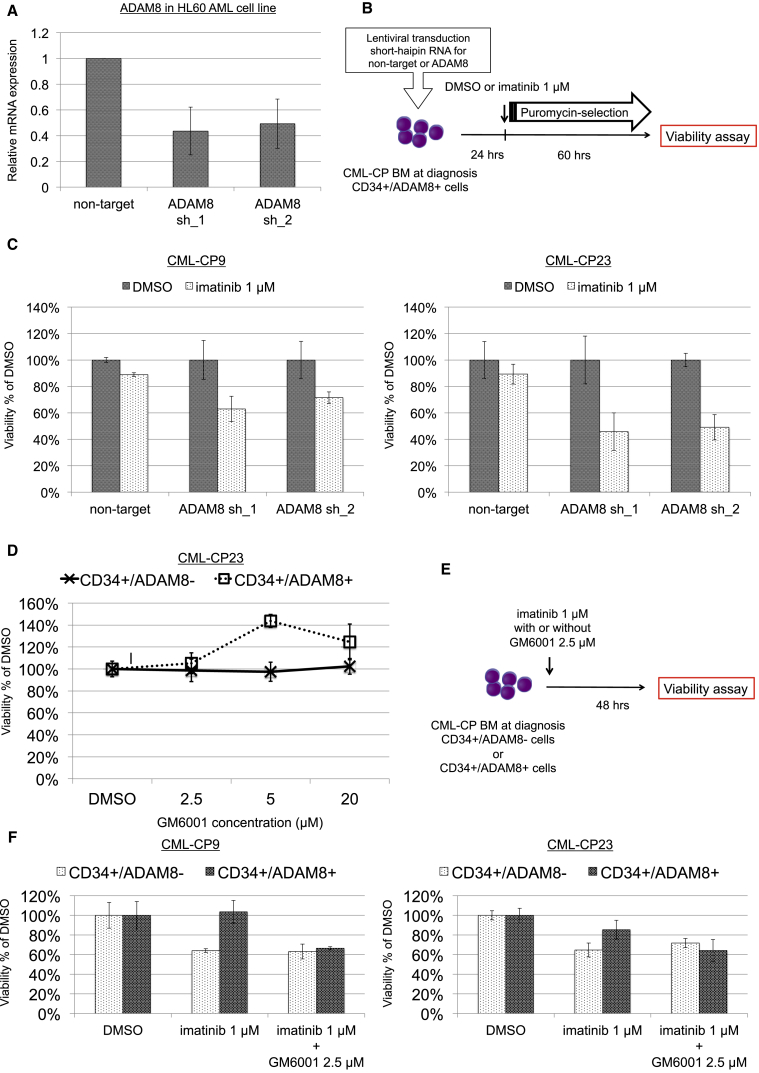
To address the functional role of ADAM8 in TKI resistance, knockdown of ADAM8 expression was performed in primary samples by lentiviral transduction of short hairpin RNA (shRNA) for ADAM8. The efficacy of knockdown for ADAM8 was confirmed in HL60 acute myeloid leukemia (AML) cell lines (Figure 5A). After the lentiviral transduction of CD34+/ADAM8+ cells in primary samples, transfected cells were selected by puromycin for 60 hr (Figure 5B). Viability assay showed that knockdown of ADAM8 canceled the TKI resistance in CD34+/ADAM8+ cells (Figure 5C). Pharmacological inhibition of metalloproteinase activity, one of the critical functions of ADAM8, with GM6001 demonstrated that 2.5 μM GM6001, which had no effect on viability in both CD34+/ADAM8− and CD34+/ADAM8+ cells (Figure 5D), fully canceled the TKI resistance in CD34+/ADAM8+ cells (Figures 5E and 5F). These findings suggest that ADAM8 has a functional role in TKI resistance, mainly via the metalloproteinase activity.
Figure 5.
Functional Role of ADAM8 in Resistance against TKI
(A) Efficiency of knockdown of ADAM8 by shRNAs in the HL60 AML cell line. qRT-PCR showed the effective knockdown of ADAM8 by two shRNAs. Data show means ± SD, n = 3 independent experiments.
(B) Experimental scheme for the transduction of shRNAs and viability assay.
(C) Viability assay for vector-transduced CD34+/ADAM8+ cells with or without 1 μM imatinib. TKI resistance was canceled by shRNAs targeting ADAM8. Data show means ± SD, n = 3 technical replicates.
(D) Viability assay for CD34+/ADAM8− and CD34+/ADAM8+ cells with GM6001; 2.5 μM GM6001 had little effect on the viability of both cells. Data show means ± SD, n = 3 technical replicates.
(E) Experimental scheme for the combination of imatinib with GM6001.
(F) Viability assay with imatinib and GM6001. Combination of imatinib with GM6001 canceled the TKI resistance in CD34+/ADAM8+ cells. Data show means ± SD, n = 3 technical replicates.
ADAM8 as an Antigen of Residual CML Cells in CML-CP Patients with Optimal TKI Response
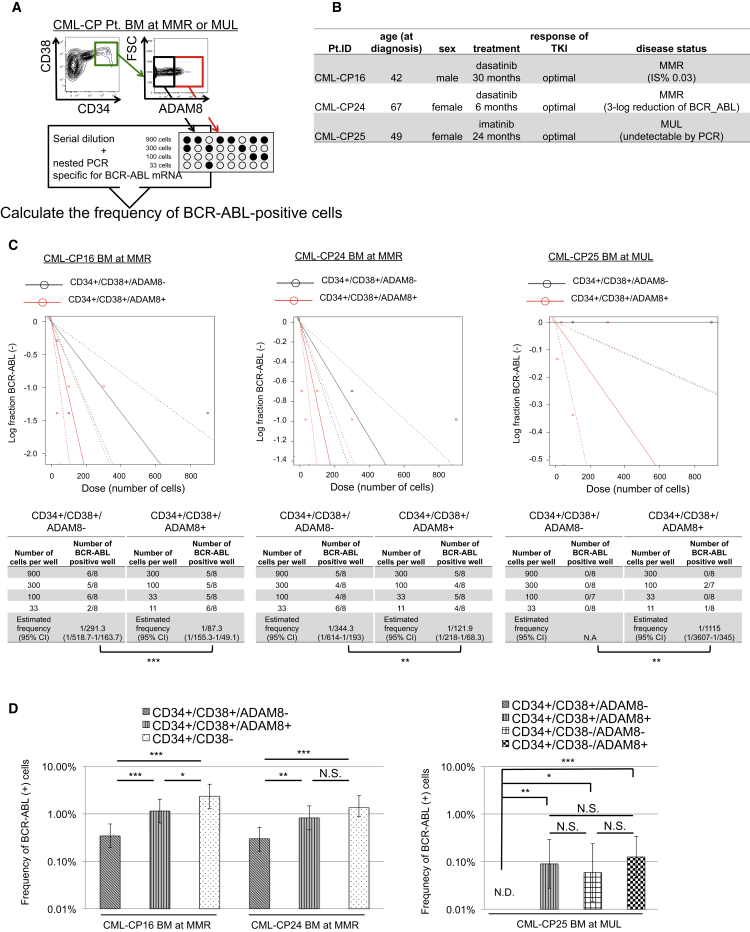
To evaluate the significance of ADAM8 as a marker of TKI-resistant CML cells in CML-CP patients under the treatment with TKIs, we measured the frequency of CML cells in BM samples of CML-CP patients who had achieved major molecular response (MMR; n = 2) or molecular undetectable leukemia (MUL; n = 1) after the administration of TKIs by limiting dilution analysis (Figures 6A, 6B, and S6A).
Figure 6.
ADAM8 Marks TKI-Resistant Residual CML Cells in BM of CML-CP Patients with Optimal TKI Response under Treatment with TKI
(A) A scheme of limiting dilution assay.
(B) Characteristics of CML-CP patients with optimal TKI response under treatment with TKIs.
(C) Limiting dilution assay for CML-CP patients with optimal TKI response under treatment with TKIs. Residual CML cells are concentrated more in the ADAM8+ subpopulation than in the ADAM8− subpopulation. Data for each patient show n = 1. CI, confidence interval.
(D) Frequency of residual CML cells in CML-CP patients with optimal TKI response. Data for each patient show n = 1.
Error bars represent 95% CI. p values: chi-square test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In CML patients with MMR, CML cells remained in CD34+/CD38+/ADAM8+ cells at higher frequency in spite of steep decline of CML cells in CD34+/CD38+/ADAM8− cells. The frequency of CML cells was as high in the CD34+/CD38+/ADAM8+ fraction as in the CD34+/38− CML stem cell fraction. Even in a patient with MUL, residual CML cells were detected in the ADAM8+ population among the CD34+/CD38+ fraction, whereas CML cells were undetectable in the ADAM8− population (Figures 6C and 6D). In the CD34+/38− fraction, the ADAM8+ population had no significant difference in the frequency of CML cells compared with the ADAM8− population, which was compatible with the results of the viability assay of newly diagnosed CML-CP primary samples (Figures S6B and S6C). These findings indicate that ADAM8 works as a marker of TKI-resistant residual CML cells even in CML-CP patients with favorable response of TKIs.
Discussion
By using CML-iPSCs, the present study identifies a significance of ADAM8 as a marker of residual CML cells even in patients with CML-CP who have achieved favorable molecular response. In the CD34+/CD38+ fraction, which was previously known as the TKI-sensitive fraction, ADAM8+ cells showed significantly higher frequency of residual CML cells than ADAM8− cells in CML-CP patients with favorable molecular response. These results were compatible with the result of the viability assay of newly diagnosed CML-CP primary samples, indicating that ADAM8 marks TKI-resistant CML cells even in CML-CP patients under treatment with TKIs.
Several possible explanations could account for the fact that CD34+/ADAM8+ CML cells exhibit TKI resistance. One possibility is that CD34+/ADAM8+ CML cells suppress the expression levels of BCR-ABL, which leads CD34+/ADAM8+ cells to be less dependent on BCR-ABL. The possible explanation is compatible for single-cell analysis of CML stem cells in BC, previously reported (Nukina et al., 2014). However, the relationship between the expression levels of BCR-ABL and TKI sensitivity is still controversial.
Moreover, although ADAM8 plays a role in leukocyte recruitment in terminally differentiated HCs, the functional role of ADAM8 in TKI resistance of CML has remained unknown (Xia et al., 2002, Zarbock et al., 2007, Yago et al., 2010, Domínguez-Luis et al., 2011). The present study showed that both knockdown of ADAM8 by shRNA and pharmacological inhibition of the function of ADAM8 by GM6001, a matrix metalloproteinase inhibitor, canceled the TKI resistance in primary samples. These findings suggest that ADAM8 has a functional role in TKI resistance, mainly via the metalloproteinase activity. One possible reason that ADAM8 overexpression did not affect TKI resistance in CML-BC cell lines is that numerous additional gene alterations accumulated in the cell lines have prevented ADAM8 overexpression from conferring apparent gain of TKI resistance. In other malignancies, previous studies for ADAM8 have reported that ADAM8 is involved in resistance against chemotherapies through the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase pathway (Dong et al., 2015, Schlomann et al., 2015). Further studies of the function in TKI resistance are needed for the assessment of the potential of ADAM8 as a therapeutic target.
The present study demonstrates that, even in CML-CP patients who have achieved optimal TKI response under treatment with TKIs, ADAM8 enhances the sensitivity of PCR specific for BCR-ABL, a well-established test for the clinical assessment of CML-CP patients. However, it is also unclear whether ADAM8+ residual CML cells caused relapse after discontinuation of TKIs. In addition, it remained uncertain whether CD34+/ADAM8+ CML cells exist in the peripheral blood of CML-CP patients with MMR or CMR during treatment with TKIs. Although several studies focusing on the surface marker proteins of CML stem cells have been reported, including CD25 (Kobayashi et al., 2014, Sadovnik et al., 2016), CD26 (Herrmann et al., 2014, Culen et al., 2016) and T cell immunoglobulin mucin-3 (Kikushige et al., 2015), so far, there are no antigens that have significance as a predictor of relapse after discontinuation of TKIs in CML-CP patients with favorable TKI response. Further examination of predictive impact and clinical applicability of these antigens, including ADAM8, are needed.
Using reprograming technology, the present study provides a platform to analyze TKI-resistant CML stem cells as a model of chemo-resistant CSCs. Although CML-iPSCs overcome the heterogeneity and paucity of CML stem cells to some degree, limitations specific for cancer-derived iPSCs still remain. At first, samples obtained are still limited mainly due to the efficiency of tissue-specific differentiation. For example, several protocols refined for hematopoietic differentiation have been reported; however, more accessible and efficient protocols for differentiation are necessary for robust analysis, such as multi-omics studies. Secondly, cancer-derived iPSCs are difficult to establish. To generalize the present approach to CSCs, further studies are needed to clarify the mechanism by which cancers interrupt the reprograming process to iPSCs. Finally, the degree to which the present strategy of CSCs through cancer-derived iPSCs is applicable to other cancers also remains elusive but warrants examination.
In conclusion, this study identifies ADAM8 as an antigen of TKI-resistant CML cells, which remains after TKI treatment even in CML-CP patients with favorable molecular response from using CML-iPSCs. This study also provides a powerful platform with CML-iPSCs to investigate the pathophysiology of TKI-resistant CML stem cells. The degree to which these results can be generalized to analysis for chemo-resistance of other CSCs is unclear but warrants examination. ADAM8 would be a promising predictor of relapse after discontinuation of TKIs and a potential therapeutic target against residual CML cells in CML-CP patients with favorable molecular response.
Experimental Procedures
Patient Samples
All the samples from patients with CML-CP that were chosen were determined to have no more than t(9;22) (q34;q11) chromosomal translocation on G-banded chromosomal analysis and were preserved in two or more aliquots from January 2010 to September 2017. Mononuclear cells (MNCs) were isolated by centrifugation through a Ficoll gradient. Frozen samples were incubated overnight in an adequate amount of pre-culturing medium (minimal essential medium [α-MEM] containing 20% fetal calf serum [FCS] supplemented with 50 ng/mL stem cell factor [SCF] and 50 ng/mL thrombopoietin [TPO]) and subsequently were used for further analyses. Characteristics of all patients are described in Table S4.
Animals
C57BL/6 and NOD.Cg-PrkdcscidIl2rgtm1Wjl/Szj (NSG) mice were purchased from Japan SLC and Charles River Laboratories Japan, respectively. All mice we used were aged 8–12 weeks.
Cell Lines
CML cell line K562 was purchased from RIKEN BRC and NCO2 was purchased from JCRB Cell Bank. Imatinib-resistant CML cell lines, KCL22/SR and KU812/SR, were kindly gifted by Dr. Nagai (Ohmine et al., 2003, Miyoshi et al., 2005). The mouse C3H10T1/2 cells were cultured as previously descried (Takayama et al., 2008).
Flow Cytometry
Isolation of each fraction from HCs derived from iPSCs, BM cells from mice, and primary samples from patients with leukemia was performed using FACSAria II and FACSAria III cell sorter (BD Biosciences). Data were analyzed with FlowJo (TreeStar, Ashland, OR). Antibodies are listed in Table S5.
Generation of iPSCs from CML-CP Samples with Episomal Vectors
To establish iPSCs from CML patient samples with optimal TKI response, purified CD34+ cells from BM of patients with newly diagnosed CML-CP, who had achieved MMR after 2-year treatment with TKIs, were expanded for 2 days in α-MEM supplemented with 20% FCS, SCF, ligand for fms-like tyrosine kinase 3 (FL3L), interleukin (IL)-3, IL-6, and TPO, as previously described (Kumano et al., 2012). The plasmid mixture containing pCXLE-hOCT3/4-shp53-F, pCXLE-hSK, pCXLE-hUL, and pCXWB-EBNA1 was electroporated into 2 × 105 CD34+ cells, as described in the literature (Okita et al., 2013). The cells were then cultured with mouse embryonic fibroblast for 20–30 days and we obtained ESC-like colonies from two patients with CML-CP.
RNA Extraction, qRT-PCR, One-Step RT-PCR Analysis
After extraction of total RNA with RNAeasy reagents (QIAGEN), reverse transcription was performed with ReverTra Ace qPCR RT Master Mix (TOYOBO). Primer sequences used for PCR are listed in Table S6. qRT-PCRs were carried out in the LightCycler480 system (Roche) with THUNDERBIRD SYBR qPCR Mix according to the manufacturer's instructions (TOYOBO). Each assay was performed in triplicate and the results were normalized to 18S. Primer sequences are listed in Table S6.
Characterization of CML-iPSCs Clones
Semi-quantitative RT-PCR for stem cell genes was performed with the primers described in the literature (Okita et al., 2011). Immunofluorescence microscopy for SSEA-4 and TRA-1-60 was conducted as previously described (Kumano et al., 2012). Cells were observed with a FLUOVIEW FV10i (Olympus). Teratoma formation assay was performed by the injection of 1.0 × 106 cells in the testis capsule of NSC mice. Ten weeks after injection, developed tumors were harvested and fixed with 3.8% formaldehyde. The fixed tumors embedded in paraffin were stained with H&E for histological analysis. Karyotype was determined by the G-banding method (Nihon Gene Research Laboratories).
Hematopoietic Differentiation from iPSCs
For hematopoietic differentiation from iPSCs, we adopted the protocols described in the literature (Takayama et al., 2010). In brief, clusters of iPSCs were transferred onto C3H10T1/2 cells treated with mitomycin C and co-cultured in the medium for hematopoietic differentiation, which consisted of Iscove's modified Dulbecco medium containing a cocktail of 10 mg/mL human insulin, 5.5 mg/mL human transferrin, 5 ng/mL sodium selenite, 2 mmol/L L-glutamine, 0.45 mmol/L monothioglycerol, 50 mg/mL ascorbic acid, and 15% highly filtered fetal bovine serum in the presence of 20 ng/mL human VEGF. Culture medium was replaced on days 4, 7, 10, 12, 14, and 16. After 17 to 18 days of culture, the iPSC sacs crushed with pipette tips were filtered with a cell strainer, and among the CD43+ HCs, CD34+/CD45− pre-HPCs and CD34-/CD45+ DCs were isolated by FACS, as described above.
Colony-Forming Assay
In each experiment, 10,000 pre-HPCs from iPSCs or 300 primary cells were plated onto MethoCult GF M4434 medium (STEMCELL Technologies). Numbers of colonies in each experiments were scored on day 14 following manufacturer's instructions.
Proliferation Assay
In each experiment, 5,000 pre-HPCs and DCs from iPSCs were cultured in the α-MEM containing 20% FCS supplemented with 100 ng/mL SCF, 10 ng/mL TPO, 100 ng/mL FL3L, 10 ng/mL IL-3, and 100 ng/mL IL-6 in the presence or the absence of imatinib, as previously described (Kumano et al., 2012). Each assay was performed in duplicate.
Microarray Analysis
Gene expression analysis was carried out with the use of the SurePrint G3 Human GE 8x60K v2 Microarray (Agilent). The preparation of amplified cDNA was performed with the Ovation Pico WTA System V2 (NuGEN) according to manufacturer's protocol. We also analyzed publicly available gene expression microarray data on human samples from the Gene Expression Omnibus (GEO) database (GEO: GSE40721, GSE47927) (Cramer-Morales et al., 2013, Herrmann et al., 2014). Raw data were normalized using CLC Genomics Workbench and clustering analysis was performed using the heatmap.2 function in the gplots R package. Clustering analysis was supervised in the top 1,000 genes, which were differentially expressed among samples. To compare expression profiles of pre-HPCs and DCs from iPSCs, normalized data were tested for GSEA using curated gene sets. For screening of genes with elevated expression levels in pre-HPCs compared with DCs from CML-iPSC, the expression values of individual genes were compared between groups. Genes significantly elevated in pre-HPCs from CML-iPSCs compared with DCs as determined by an unpaired Student's t test (p < 0.05) were selected.
Apoptosis Assay
For HCs from iPSCs, culture medium of hematopoietic differentiation in the presence or the absence of 5 μM imatinib was replaced on day 16. For primary samples of patients with newly diagnosed CML-CP, purified CMP-CP samples were cultured in α-MEM containing 20% FCS supplemented with 100 ng/mL SCF, 100 ng/mL granulocyte colony-stimulating factor (G-CSF), 20 ng/mL FL3L, 20 ng/mL IL-3, and 20 ng/mL IL-6 in the presence or the absence of 5 μM imatinib, as previously described (Ma et al., 2014). Forty-eight hours after the exposure of imatinib, cells were stained by the antibody for annexin-V and 4,6-diamidino-2-phenylindole following manufacturer's protocol.
ATP-Luciferase Viability Assay
Purified HCs derived from iPSCs were suspended in the same medium as in the proliferation assay. Purified samples of patients with CML-CP were suspended in the same medium as in the apoptosis assay. The suspensions of each cell were placed into a 384-well plate (Greiner Bio-One) and were incubated at 37°C in a 5% CO2 incubator for 48 to 60 hr in the presence of imatinib and GM6001 (Merck Millipore) at the concentration described. Subsequently, ATP-luciferase (TOYO INK) was added in each well following manufacturer's protocol and the luminescence from each well was measured in triplicate in relative light units (RLU) on a TriSter2 LB942 (BEERTHOLD). The cell viability was calculated by the formula as follows: cell viability (%) = 100 × (RLU from each well – RLU from background)/(RLU from control (DMSO) – RLU from background).
FISH
FISH staining was performed (BIO MEDICAL LABORATORIES). Briefly, FISH analysis for t(9;22) translocation was performed on interphase, using the LSI-BCR-ABL dual color extra signal probe (Vysis) (Primo et al., 2003). LSI-ES probe for ABL (red) hybridizes fusion der(22), der(9), and nonrearranged 9. LSI-ES probe for BCR (green) hybridizes fusion der(22) and nonrearranged 22.
Plasmids
Plasmids containing the sequence of flag-tagged human ADAM8 was kindly gifted by Dr. Jörg W. Bartsch (Schlomann et al., 2015). Plasmids encoding proteins of flag-tagged human ADAM8 were subcloned into the EcoR1/BamH1 site of pBleuscript II. To produce protein-expressing retrovirus, we used the following plasmids: pMXs-ADAM8-flag-neo. All of the PCR products were verified by DNA sequencing.
Lentivirus Production and Knockdown of ADAM8 Expression by shRNA
To obtain lentivirus supernatants, HEK293T cells were transiently transfected with pLKO.1-puro-CMV-tGFP containing shRNA sequences (Sigma-Aldrich Japan), pMISSION GAG POL (Sigma-Aldrich Japan), and pMISSION VSV-G (Sigma-Aldrich Japan). Forty-eight hours later, the viral supernatant was collected and utilized for infection. The vector-transduced cells were selected by medium containing puromycin (1.0 μg/mL for HL60 and 2.5 μg/mL for primary samples) and subjected to assays in vitro. Non-target shRNA was a nonfunctional construct purchased from Sigma-Aldrich Japan. The target sequences, from 5′ to 3′, were GGGCCTGGAGATTTGGAATAG (ADAM8sh_1) and GGAATAGTCAGGACAGGTTCC (ADAM8sh_2).
Limiting Dilution Analysis
In the subpopulation of CD34+ and CD34+/CD38+, ADAM8+ cells were plated on a 96-well plate at 300, 100, 33, and 11 cells/well and ADAM8− cells at 900, 300, 100, 33 cells/well with eight replicates. Then we performed one-step RT-PCR for BCR-ABL and β-actin in each well (Chu et al., 2014) following the pre-amplification of target genes and calculated the absolute frequency of BCR-ABL-expressing cells using ELDA software (Hu and Smyth, 2009). Primers for pre-amplification and one-step RT-PCR are described in the literature (Chu et al., 2014, Sato et al., 2014).
Statistics
The estimated frequency of CML cells was generated using the ELDA software (http://bioinf.wehi.edu.au/software/elda/index.html). Statistical significance of differences between groups was assessed with chi-square test in limiting dilution assay. Except for limiting dilution assay, statistical significance of differences between different groups was assessed with a two-tailed unpaired t test. Differences were considered statistically significant at a p value < 0.05.
Study Approval
Primary samples from BM of all patients were obtained after informed consent. All studies using human cells were reviewed and approved by the institutional review boards of the University of Tokyo. All animal experiments adhered to the guidelines for animal experiments of the University of Tokyo.
Author Contributions
M.M., J.K., K. Kataoka, A.Y., S.A., K. Kumano, and M.K. designed the research; M.M., S.Y., A.H., and T.K. performed the experiments; M.M., J.K., A.Y., and M.K. wrote the paper.
Acknowledgments
We thank T. Kitamura for Plat-E and Plat-A packaging cells; H. Nakauchi for pGCDNsam-IRES-EGFP retroviral vector; T. Nagai for imatinib-resistant CML cell lines; K. Okita for pCXLE-hOCT3/4-shp53-F, pCXLE-hSK, pCXLE-hUL, and pCXWB-EBNA1; J.W. Bartsch for pCMV6-ADAM8-flag; Yoko Hokama and Keiko Tanaka for expert technical assistance; and Kyowa Hakko Kirin for recombinant human G-CSF. This work was supported in part by Advanced Research & Development Programs for Medical Innovation (AMED-CREST), the Japan Society for the Promotion of Science (JSPS) KAKENHI (no. 14J02953 and no. 16K19568), and a grant from the Foundation for Promotion of Cancer Research, Okinaka Memorial Institute for Medical Research and the Tokyo Biochemical Research Foundation. Since the current study started, M.K. has received an honorarium from Shionogi; M.K. has received an honorarium from the Japan Science and Technology Agency; M.K. has received research funding from Astellas, Bristol-Myers Squibb, Novartis, Pfizer, and Takeda; S.A. has received research funding from Bristol-Myers Squibb and Novartis.
Published: February 8, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and six tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.015.
Accession Numbers
All microarray data have been deposited in the NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GEO: GSE92624.
Supplemental Information
References
- Abraham S.A., Hopcroft L.E.M., Carrick E., Drotar M.E., Dunn K., Williamson A.J.K., Korfi K., Baquero P., Park L.E., Scott M.T. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedel A., Pasquet J.-M., Lippert E., Taillepierre M., Lagarde V., Dabernat S., Dubus P., Charaf L., Beliveau F., de Verneuil H. Variable behavior of iPSCs derived from CML patients for response to TKI and hematopoietic differentiation. PLoS One. 2013;8:e71596. doi: 10.1371/journal.pone.0071596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Pruszak J., Varadarajan M., Blomen V.A., Gokhale S., Camargo F.D., Wernig M., Jaenisch R., Brummelkamp T.R. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., McDonald T., Lin A., Chakraborty S., Huang Q., Snyder D.S., Bhatia R. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood. 2014;118:5565–5573. doi: 10.1182/blood-2010-12-327437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer-Morales K., Nieborowska-Skorska M., Scheibner K., Padget M., Irvine D.A., Sliwinski T., Haas K., Lee J., Geng H., Roy D. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122:1293–1304. doi: 10.1182/blood-2013-05-501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culen M., Borsky M., Nemethova V., Razga F., Smejkal J., Jurcek T., Dvorakova D., Zackova D., Weinbergerova B., Semerad L. Quantitative assessment of the CD26+ leukemic stem cell compartment in chronic myeloid leukemia: patient-subgroups, prognostic impact, and technical aspects. Oncotarget. 2016;7:33016–33024. doi: 10.18632/oncotarget.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash A.B., Williams I.R., Kutok J.L., Tomasson M.H., Anastasiadou E., Lindahl K., Li S., Van Etten R.A., Borrow J., Housman D. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc. Natl. Acad. Sci. USA. 2002;99:7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Luis M., Lamana A., Vazquez J., García-Navas R., Mollinedo F., Sánchez-Madrid F., Díaz-González F., Urzainqui A. The metalloprotease ADAM8 is associated with and regulates the function of the adhesion receptor PSGL-1 through ERM proteins. Eur. J. Immunol. 2011;41:3436–3442. doi: 10.1002/eji.201141764. [DOI] [PubMed] [Google Scholar]
- Dong F., Eibach M., Bartsch J.W., Dolga M.A., Schlomann U., Conrad C., Schieber S., Schilling O., Biniossek M.L., Culmsee C. The metalloprotease-disintegrin ADAM8 contributes to temozolomide chemoresistance and enhanced invasiveness of human glioblastoma cells. Neuro Oncol. 2015;17:1474–1485. doi: 10.1093/neuonc/nov042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker B.J., Sawyers C.L., Kantarjian H., Resta D.J., Reese S.F., Ford J.M., Capdeville R., Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Graham S.M., Jørgensen H.G., Allan E., Pearson C., Alcorn M.J., Richmond L., Holyoake T.L. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- Herrmann H., Sadovnik I., Cerny-Reiterer S., Rülicke T., Stefanzl G., Willmann M., Hoermann G., Bilban M., Blatt K., Herndlhofer S. Dipeptidylpeptidase IV (CD26) defines leukemic stem cells (LSC) in chronic myeloid leukemia. Blood. 2014;123:3951–3962. doi: 10.1182/blood-2013-10-536078. [DOI] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Hu K., Yu J., Suknuntha K., Tian S., Montgomery K., Choi K.-D., Stewart R., Thomson J.A., Slukvin I.I. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa J., Tanaka H., Okada M., Nakamae H., Hino M., Murai K., Ishida Y., Kumagai T., Sato S., Ohashi K. Discontinuation of dasatinib in patients with chronic myeloid leukaemia who have maintained deep molecular response for longer than 1 year (DADI trial): a multicentre phase 2 trial. Lancet Haematol. 2015;2:e528–e535. doi: 10.1016/S2352-3026(15)00196-9. [DOI] [PubMed] [Google Scholar]
- Kikushige Y., Miyamoto T., Yuda J., Jabbarzadeh-Tabrizi S., Shima T., Takayanagi S.I., Niiro H., Yurino A., Miyawaki K., Takenaka K. A TIM-3/Gal-9 autocrine stimulatory loop drives self-renewal of human myeloid leukemia stem cells and leukemic progression. Cell Stem Cell. 2015;17:341–352. doi: 10.1016/j.stem.2015.07.011. [DOI] [PubMed] [Google Scholar]
- Kobayashi C.I., Takubo K., Kobayashi H., Nakamura-Ishizu A., Honda H., Kataoka K., Kumano K., Akiyama H., Sudo T., Kurokawa M. The IL-2/CD25 axis maintains distinct subsets of chronic myeloid leukemia-initiating cells. Blood. 2014;123:2540–2549. doi: 10.1182/blood-2013-07-517847. [DOI] [PubMed] [Google Scholar]
- Kumano K., Arai S., Hosoi M., Taoka K., Takayama N., Otsu M., Nagae G., Ueda K., Nakazaki K., Kamikubo Y. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–6242. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- Levescot A., Flamant S., Basbous S., Jacomet F., Féraud O., Anne Bourgeois E., Bonnet M.-L., Giraud C., Roy L., Barra A. BCR-ABL-induced deregulation of the IL-33/ST2 pathway in CD34+ progenitors from chronic myeloid leukemia patients. Cancer Res. 2014;74:2669–2676. doi: 10.1158/0008-5472.CAN-13-2797. [DOI] [PubMed] [Google Scholar]
- Li L., Wang L., Li L., Wang Z., Ho Y., McDonald T., Holyoake T.L., Chen W., Bhatia R. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Shan Y., Bai R., Xue L., Eide C.A., Ou J., Zhu L.J., Hutchinson L., Cerny J., Khoury H.J. A therapeutically targetable mechanism of BCR-ABL-independent imatinib resistance in chronic myeloid leukemia. Sci. Transl. Med. 2014;6:252ra121. doi: 10.1126/scitranslmed.3009073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon F.-X., Réa D., Guilhot J., Guilhot F., Huguet F., Nicolini F., Legros L., Charbonnier A., Guerci A., Varet B. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11:1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- Miyoshi T., Nagai T., Ohmine K., Nakamura M., Kano Y., Muroi K., Komatsu N., Ozawa K. Relative importance of apoptosis and cell cycle blockage in the synergistic effect of combined R115777 and imatinib treatment in BCR/ABL-positive cell lines. Biochem. Pharmacol. 2005;69:1585–1594. doi: 10.1016/j.bcp.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Nakajima-Takagi Y., Osawa M., Oshima M., Takagi H., Miyagi S., Endoh M., Endo T.A., Takayama N., Eto K., Toyoda T. Role of SOX17 in hematopoietic development from human embryonic stem cells. Blood. 2013;121:447–458. doi: 10.1182/blood-2012-05-431403. [DOI] [PubMed] [Google Scholar]
- Nguyen L.V., Vanner R., Dirks P., Eaves C.J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- Nukina A., Kagoya Y., Watanabe-Okochi N., Arai S., Ueda K., Yoshimi A., Nannya Y., Kurokawa M. Single-cell gene expression analysis reveals clonal architecture of blast-phase chronic myeloid leukaemia. Br. J. Haematol. 2014;165:414–416. doi: 10.1111/bjh.12726. [DOI] [PubMed] [Google Scholar]
- O'Hare T., Zabriskie M.S., Eiring A.M., Deininger M.W. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat. Rev. Cancer. 2012;12:513–526. doi: 10.1038/nrc3317. [DOI] [PubMed] [Google Scholar]
- Ohmine K., Nagai T., Tarumoto T., Miyoshi T. Analysis of gene expression profiles in an imatinib-resistant cell line, KCL22/SR. Stem Cells. 2003;21:315–321. doi: 10.1634/stemcells.21-3-315. [DOI] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Primo D., Tabernero M.D., Rasillo A., Sayagués J.M., Espinosa A.B., Chillón M.C., Garcia-Sanz R., Gutierrez N., Giralt M., Hagemeijer A. Patterns of BCR/ABL gene rearrangements by interphase fluorescence in situ hybridization (FISH) in BCR/ABL+ leukemias: incidence and underlying genetic abnormalities. Leukemia. 2003;17:1124–1129. doi: 10.1038/sj.leu.2402963. [DOI] [PubMed] [Google Scholar]
- Sadovnik I., Hoelbl-Kovacic A., Herrmann H., Eisenwort G., Cerny-Reiterer S., Warsch W., Hoermann G., Greiner G., Blatt K., Peter B. Identification of CD25 as STAT5-dependent growth regulator of leukemic stem cells in Ph+ CML. Clin. Cancer Res. 2016;22:2051–2061. doi: 10.1158/1078-0432.CCR-15-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Goyama S., Kataoka K., Nasu R., Tsuruta-Kishino T., Kagoya Y., Nukina A., Kumagai K., Kubota N., Nakagawa M. Evi1 defines leukemia-initiating capacity and tyrosine kinase inhibitor resistance in chronic myeloid leukemia. Oncogene. 2014;33:5028–5038. doi: 10.1038/onc.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlomann U., Koller G., Conrad C., Ferdous T., Golfi P., Garcia A.M., Höfling S., Parsons M., Costa P., Soper R. ADAM8 as a drug target in pancreatic cancer. Nat. Commun. 2015;6:6175. doi: 10.1038/ncomms7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suknuntha K., Ishii Y., Tao L., Hu K., McIntosh B.E., Yang D., Swanson S., Stewart R., Wang J.Y.J., Thomson J. Discovery of survival factor for primitive chronic myeloid leukemia cells using induced pluripotent stem cells. Stem Cell Res. 2015;15:678–693. doi: 10.1016/j.scr.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama N., Nishikii H., Usui J., Tsukui H., Sawaguchi A., Hiroyama T., Eto K., Nakauchi H. Generation of functional platelets from human embryonic stem cells in vitro via ES-sacs, VEGF-promoted structures that concentrate hematopoietic progenitors. Blood. 2008;111:5298–5306. doi: 10.1182/blood-2007-10-117622. [DOI] [PubMed] [Google Scholar]
- Takayama N., Nishimura S., Nakamura S., Shimizu T., Ohnishi R., Endo H., Yamaguchi T., Otsu M., Nishimura K., Nakanishi M. Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J. Exp. Med. 2010;207:2817–2830. doi: 10.1084/jem.20100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent P., Bonnet D., De Maria R., Lapidot T., Copland M., Melo J.V., Chomienne C., Ishikawa F., Schuringa J.J., Stassi G. Cancer stem cell definitions and terminology: the devil is in the details. Nat. Rev. Cancer. 2012;12:767–775. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- Xia L., Sperandio M., Yago T., McDaniel J.M., Cummings R.D., Pearson-White S., Ley K., McEver R.P. P-selectin glycoprotein ligand-1-deficient mice have impaired leukocyte tethering to E-selectin under flow. J. Clin. Invest. 2002;109:939–950. doi: 10.1172/JCI14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yago T., Shao B., Miner J.J., Yao L., Klopocki A.G., Maeda K., Coggeshall K.M., McEver R.P. E-selectin engages PSGL-1 and CD44 through a common signaling pathway to induce integrin alphaLbeta2-mediated slow leukocyte rolling. Blood. 2010;116:485–494. doi: 10.1182/blood-2009-12-259556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Nakamura Y., Saito K., Furusawa S. Expression of the NUP98/HOXA9 fusion transcript in the blast crisis of Philadelphia chromosome-positive chronic myelogenous leukaemia with t(7;11)(p15;p15) Br. J. Haematol. 2000;109:423–426. doi: 10.1046/j.1365-2141.2000.02003.x. [DOI] [PubMed] [Google Scholar]
- Zarbock A., Lowell C.A., Ley K. Spleen tyrosine kinase Syk is necessary for E-selectin-induced alpha(L)beta(2) integrin-mediated rolling on intercellular adhesion molecule-1. Immunity. 2007;26:773–783. doi: 10.1016/j.immuni.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.