Abstract
Dichloro-diphenyl-trichloroethane (DDT) resistance among arthropod species is a model for understanding the molecular adaptations in response to insecticide exposures. Previous studies reported that DDT resistance may involve one or multiple detoxification genes, such as cytochrome P450 monooxygenases (P450s), glutathione S-transferases (GSTs), esterases, and ATP binding cassette (ABC) transporters, or changes in the voltage-sensitive sodium channel. However, the possible involvement of microRNAs (miRNAs) in the post-transcriptional regulation of genes associated with DDT resistance in the Drosophila melanogaster strain 91-R remains poorly understood. In this study, the majority of the resulting miRNAs discovered in small RNA libraries from 91-R and the susceptible control strain, 91-C, ranged from 16–25 nt, and contained 163 precursors and 256 mature forms of previously-known miRNAs along with 17 putative novel miRNAs. Quantitative analyses predicted the differential expression of ten miRNAs between 91-R and 91-C, and, based on Gene Ontology and pathway analysis, these ten miRNAs putatively target transcripts encoding proteins involved in detoxification mechanisms. RT-qPCR validated an inverse correlation between levels of differentially-expressed miRNAs and their putatively targeted transcripts, which implies a role of these miRNAs in the differential regulation of detoxification pathways in 91-R compared to 91-C. This study provides evidence associating the differential expression of miRNAs in response to multigenerational DDT selection in Drosophila melanogaster and provides important clues for understanding the possible roles of miRNAs in mediating insecticide resistance traits.
Introduction
While chemical insecticidal agents have been developed and widely applied to suppress pest arthropod populations in ongoing efforts to enhance the efficiency of agricultural production and protect human health [1], this intensive use of chemical insecticides has also led to the development of resistance to one or more classes of insecticides [2–4]. The neurotoxic organochlorine insecticide, dichlorodiphenyltrichloroethane (DDT), has been extensively used to control many agricultural insect pests and insects that vector human diseases, but was banned in most countries in the 1980s due to environmental concerns [5]. The genetic basis of DDT resistance in the Drosophila melanogaster (D. melanogaster) provides a model system for studying the evolution of insecticide resistance. Indeed, the low-level DDT resistance phenotype in D. melanogaster is thought to be associated with a single cytochrome P450, Cyp6g1 [6,7]. However, moderate to high-level DDT resistance is polygenic [8] involving modulation in DDT penetrance and excretion and multiple differentially-regulated Phase I, II, and III detoxification genes/enzymes, respectively, including P450s, glutathione S transferases (GSTs), and ATP binding cassette (ABC) transporters. Regarding the P450s, members of the Cyp6 (Cyp6g1, Cyp6g2, Cyp6a2, and Cyp6w1) and Cyp12 (Cyp12d1) subfamilies have been implicated in polygenic DDT resistance [9–13]. Furthermore, the overexpression and structural changes in membrane-spanning ABC transporters were shown to be involved in DDT efflux and contribute to the overall DDT resistance phenotype in D. melanogaster [14]. Full genome re-sequencing identified 13 different regions showing evidence of high nucleotide diversity, directional selection (selective sweeps), and thus putatively associated with the evolution of DDT resistance in the DDT-resistant strain, 91-R, when compared with a susceptible isoline, 91-C [15]. Analyses of this same whole genome re-sequencing data also identified a large panel of fixed amino acid changing mutations between 91-R and 91-C, many of which showed evidence of directional selection [16]. Most recently, a multigenic response to DDT selection was demonstrated within 91-R where transcripts with constitutive differential-expression compared to 91-C were enriched in cell survival, stress response, and neurological functions [17]. Additionally, reduced expression of the putative calcium/lipid binding domain-containing protein from gene model CG10737 is associated with short-term (3–5 hrs) DDT knockdown resistance within the Drosophila Genetic Reference Panel, suggesting a role for ameliorated effects of muscle overstimulation in DDT resistance phenotypes [13]. Despite these lines of evidence, the systemic basis and underlying genetic control of polygenic DDT resistance remains enigmatic.
Accumulating evidence demonstrates that the post-transcriptional regulation (RNA editing and alternative splicing) may contribute to the evolution of insecticide resistant phenotypes [18,19]. The miRNAs are a class of endogenous 18–25 nt non-coding small RNAs that, since their discovery in Caenorhabditis elegans over two decades ago [20], have been identified across several arthropod species including twelve Drosophila species [21]. Biogenesis of miRNAs occurs from transcript-derived stem-loop structures with canonical CNNC downstream elements [22] that are processed into a ~70 bp double-stranded precursor RNAs (pre-RNAs) within the nucleus by the ribonuclease type III enzyme Drosha [23]. After translocation to the cytoplasm, miRNA precursors exhibit canonical stem-loop secondary structures, which usually have two arms called the miRNA-5p arm or -3p arm [24]. The pre-RNAs are further processed into mature 21–22 nt miRNAs by Dicer [25,26]. Following the degradation of the passenger strand [27], the resulting single-stranded guide sequence is loaded into the RNA-induced silencing complex (RISC) [28]. Subsequent base pairing of the miRNA guide “seed sequence” (nucleotides 2–7 at the 5' end of guide RNAs) [29] with the 3'-untranslated region (3'-UTR) of target cellular mRNA transcripts most often targets the mRNA for degradation by RISC [30], either blocking translation initiation factor binding or inhibiting elongation factor progression that leads to premature termination [31–33]. Alternatively, a miRNA binding of a target transcript can result in translational up-regulation [34] within their mRNA resulting in inhibition of translation or mRNA degradation [35]. Overall, miRNAs are recognized as potent regulators of eukaryotic gene expression at the post-transcriptional level [36,37] that regulate cell differentiation [38], migration [39], and neuronal development [40]. Approximately 256 D. melanogaster miRNA precursors have been identified and deposited in the mirBase (Release 21) [24] and have been shown to be involved in regulating biological processes such as development [41], immune response [42], and metabolism [43].
Recent publications implicate a role of specific miRNAs in the regulation of insecticide resistance mechanisms among insect species. Several comparative analyses have estimated significant differences in miRNA expression between chemical insecticide-resistant and -susceptible strains [44,45]. Additionally, miRNAs differentially expressed between insecticidal Bacillus thuringiensis (Bt) toxin-resistant and -susceptible strains of Ostrinia furnacalis were predicted to target potential receptor genes [46]. In Culex pipiens, miRNAs differentially expressed between deltamethrin-sensitive and -resistant strains were proposed to mediate the expression of putative cytochrome P450 target genes [47]. Specifically, Culex pipiens miR-285 and miR-278 were implicated in pyrethroid resistance through the transcriptional regulation of Cyp6n23 and Cyp6ag11 [48,49], as well as a miRNA cluster involved in regulation of Cyp9j35 and Cyp325bg3 [50] and upregulation of miR-932 that regulates transcript levels of the cuticular gene CpCPR5 [51,52].
To date, the authors have no knowledge of any investigation of the contributions of post-transcriptional gene expression regulation on DDT resistance phenotypes. In order to partially address this knowledge gap, the present study estimated significant quantitative miRNA differences between the highly DDT-resistant D. melanogaster strain 91-R compared to the DDT-susceptible strain 91-C. Additionally, correlation between levels of differentially expressed miRNAs and corresponding putative computationally predicted P450 target transcripts were made within 91-R. This study provides insights into the role of miRNAs for the regulation of metabolic resistance to DDT as well as the effects of multigenerational DDT selection on the evolution of miRNA-mediated post-transcriptional regulation in a polygenic D. melanogaster DDT resistance phenotype.
Materials and methods
miRNA library preparation, sequencing, and annotation
The DDT-resistant 91-R and -susceptible 91-C strains were obtained from Dr. Ranjan Ganguly (University of Tennessee-Knoxville) and developed as described previously [53]. Both strains were reared on a commercially available medium (Jazz-Mix Drosophila Food, Fischer Scientific, Cat. No. AS153) under the conditions of 25 ± 1°C, 55–70% relative humidity and a 14 h light /10 h dark cycle. 91-R has been continually selected by maintaining the flies in colony bottles with the presence of a 150 mg DDT impregnated filter paper disk, while 91-C was maintained without any exposure to DDT. Recent topical bioassays estimated that 91-R is ~107-fold more resistant to DDT compared to 91-C [17].
Three biological replicates of one hundred 3–5 day-old virgin female adults were collected from both 91-R and 91-C (n = 6). In order to compare the constitutive expression of miRNAs in subsequent analyses (see below), neither population was exposed to DDT within that generation. The small RNAs (sRNAs) were immediately extracted from live collected flies from each replicate using the Qiagen miRNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Valencia, CA). RNA degradation and contamination were assessed for all samples using an Agilent 2100 Bioanalyzer (Agilent Technologies, Germany), and RNA concentrations were estimated using a NanoDrop One (Thermo, Wilmington, USA). Illumina sRNA libraries were constructed from each pool, and 50-bp single-end (SE50) sequence read data were generated on an Illumina HiSeq 4000 at the Research Technology Support Facility (Michigan State University, East Lancing, MI).
All raw Illumina sequence data were imported into CLC Genomics Workbench v.9.5 (Qiagen) and all reads were processed to remove low-quality reads, poly A sequences, adapters, reads without 3’ adapter, and sequences shorter than 15 nt. Using the “small RNA analysis” tool in CLC Genomics Workbench, annotation of the trimmed read data from each library (n = 6) was made by comparing against the known miRNA precursors of D. melanogaster in the miRBase R.21 (http://www.mirbase.org/; file hairpin.fa); subsequent generation of relative miRNA counts were made within each library. Only tags matching exactly with the mature 5’ or 3’ regions of previously annotated D. melanogaster miRNAs (miRbase R.21; file mature.fa) were accepted and retained for the further analysis. The sRNA sequencing data with annotated tags were deposited to NCBI Short Read Archive (SRA) with the accession number SRP136631. Additionally, miRDeep2 v.0.0.8 software was used in order to predict novel miRNA candidates [54]. Illumina adapters of sRNA raw sequence reads were trimmed using cutadapt v.1.15 [55]. The trimmed reads were quality-checked and curated using personal perl script. The results were aligned to the D. melanogaster reference genome (Release 6; dmel-all-chromosome-r6.19.fasta.gz at http://flybase.org/) using bowtie v.1.2.2 [56] and also were mapped to D. melanogaster non-coding RNA database (dmel-all-tRNA-6.1/8.fasta and dmel-all-miscRNA-6.1/8.fasta file at flybase.org) in order to filter the small conditional RNAs (scRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs) and transfer RNAs (tRNAs). Novel miRNAs were then identified against known miRNA precursors (hairpin.fa) and previously annotated miRNAs (mature.fa) using default parameters suggested by the developers [57]. The structure and minimal free energy (MFE) of all potential novel miRNAs were predicted using a RNAfold [58] with algorithms described previously [59]. The MFE ≤ -25 kcal mol-1, the randomization test of secondary structure MFE, called randfold P-value ≤ 0.05, and miRDeep2 score ≥ 3 were used as the cutoff level to declare them as potential novel miRNAs [60].
Differential expression of miRNAs
Estimates of miRNA expression level analysis within each replicate library from 91-R (n = 3) and 91-C (n = 3) were performed using the “Annotate and Merge” command on the CLC Genomics Workbench v.9.5 (Qiagen). This procedure used the D. melanogaster miRBase (release 21, http://www.mirbase.org/) as the primary database and the non-coding RNA database (dmel-all-tRNA-6.1/8.fasta and dmel-all-miscRNA-6.1/8.fasta) as the secondary database for annotation. The read counts of miRNAs were first normalized using the tag per million reads (TPM) method: TPM = (number of mapped reads for each miRNA/total number of mapped reads) ×106. Subsequently, the Empirical analysis of Differential Gene Expression (EDGE) algorithm [61] was used to estimate differences in read counts comparing 91-R to 91-C strains, with P-values adjusted for multiple testing calculated using the Benjamini–Hochberg false discovery rate (FDR) procedure [62]. The variance in miRNA levels between the 91-R and 91-C with a log2 fold-change > 1.0 or < -1.0, and a FDR ≤ 0.05, were defined as significant [63].
Target prediction and functional annotation of differentially expressed miRNAs
The potential target transcripts of miRNAs predicted to be differentially expressed between 91-R and 91-C were predicted using three different types of software packages, RNAhybrid [64], DIANA [65], and ComiR [66], using the following criteria: (1) RNAhybrid: the target site prediction was restricted to the 3’-UTR region obtained from the 3’-UTR database of D. melanogaster (dmel-all-three_prime_UTR-r6.19.fasta at http://www.flybase.org) with MFE ≤ -30 kcal mol-1; (2) DIANA: miTG score ≥ 0.8; and (3) ComiR: high threshold ≥ 0.8. Additionally, target gene ontology (GO) and corresponding pathways for all putative target transcripts were retrieved from the FlyMine database (http://www.flymine.org), and the GO terms were classified with CateGOrizer (https://www.animalgenome.org).
Validation and correlated expression between miRNAs and target transcripts
Reverse transcriptase-quantitative PCR (RT-qPCR) was carried out on selected eight known miRNAs for validation of sRNA sequencing estimated differences between the 91-R and 91-C strains. Moreover, the correlation was assessed between the relative expression levels of eight differentially expressed miRNAs and their potential targeted detoxification genes: Cyp6a8, Cyp6g1, Cyp6w1, Cyp4s3, Cyp6g2, Cyp309a2, Cyp313a4, Cyp313b1, Cyp4ae1, Cyp4d2, Cyp4g15, Cyp4p3, Cyp6d5, Cyp6t3, Cyp6u1, Cyp6v1, Cyp49a1, Cyp18a1, Cyp303a1, Cyp4aa1, Cyp4e3, Cyp4g1, Cyp6a19, Cyp6a22, Cyp6a9, Cyp4d20, GstD1, GstS1, GstE6, GstE10, Esterase10, Esterase7, Esterase8, ABC-B7, GABA-R, mAChR-A, GluR-IB, nAcRalpha-A, and Cpr65Ec. Three biological replicates of 15 adult female flies were sampled per strain (91-R and 91-C) and—identically with treatments used in Illumina sRNA sequencing—all samples were not exposed to DDT within the generation used. Both miRNAs and total RNAs were extracted from each biological replicate using a miRNeasy Mini Kit and a RNeasy Mini Kit, with resulting extracts treated with DNAse I (Qiagen) to remove contaminating genomic DNA. The first-strand cDNA was synthesized from mature miRNA and total RNA respectively with a miScript II RT kit (Qiagen) and a SuperScript™ III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. RT-qPCR reactions were performed with a miScript SYBR® Green PCR Kit (Biorad, Hercules, CA) according to the manufacturer’s instructions using miRNA-specific forward primers (S1 Table) with miScript II RT kit (Qiagen) products as a template. Analogously, RT-qPCR reactions for corresponding putative target mRNA transcripts were performed using SYBR® Green Master Mix (Biorad) according to the manufacturer’s instructions using target transcript-specific forward and reverse primers (S1 Table), with products from the SuperScript™ III Reverse Transcriptase kit used as a template. All amplification reactions were performed on a StepOnePlus Real-Time PCR system (Applied Biosystems Inc., Foster City, CA), with three technical replicates across all biological replicates. Normalized miRNA and target transcript expression levels were calculated using the 2−ΔΔC(t) method [67] with the internal references U6 snRNA and 5S rRNA for miRNA normalization and rp49 for mRNA normalization, respectively. A one-way ANOVA was used to examine the significance of expression differences between the two samples using XLSTAT software (Addinsoft, USA). The correlation coefficient was determined by Pearson's correlation analysis between the transcript levels of the selected eight miRNAs and their corresponding putative target genes.
Results
miRNA library preparation, sequencing, and annotation
Six miRNA libraries (three biological replicates from each of 91-R and 91-C strains) were constructed, from which sequencing generated 216.2 million total raw reads (≥ 30.1 million per library; Table 1). After trimming reads (i.e., removal of reads without a 3' adaptor and < 15 nt) 5.1 to 5.8 and 4.4 to 7.7 million reads were retained respectively among triplicates from 91-R and 91-C (Table 1). These were then used in subsequent analyses. Considering trimmed reads across all six libraries, the majority of the sequences (55.5%) were distributed from 16–25 nt (S1 Fig), which is the standard size of described miRNAs [68]. A class of 26–31 nt long RNA sequences accounted for 25.4% of the total reads (34,598,261) and were classified as suspected piwi-interacting RNAs (piRNAs) [69]. Of the trimmed reads, 531,843 to 992,816 miRNA tags were identified across all replicate libraries of 91-R and 91-C (Table 1), and among these between 6,050 and 9,356 matched D. melanogaster miRNA entries in the miRBase R.21 database.
Table 1. Small RNA sequences from 91-C and 91-R triplicate libraries read data.
| Category | Analyses of total reads data | |||||
| 91-R-1 | 91-R-2 | 91-R-3 | 91-C-1 | 91-C-2 | 91-C-3 | |
| Raw reads | 35,712,203 | 30,108,348 | 33,996,366 | 43,821,999 | 34,205,253 | 38,365,489 |
| Trimmed reads | 5,801,176 | 5,105,542 | 5,439,260 | 7,738,822 | 6,065,134 | 4,448,327 |
| Annotated with ncRNA (rRNA, tRNA, snRNA, snoRNA and others) | 4,399,524 | 4,072,155 | 4,196,227 | 5,961,313 | 4,553,746 | 3,182,916 |
| D. melanogaster miRNAs | 384,620 | 252,554 | 339,001 | 455,995 | 336,210 | 356,146 |
| Unannotated | 1,017,032 | 780,833 | 904,032 | 1,321,514 | 1,175,178 | 909,265 |
|
Data Processing/ Strains |
Analyses of unique reads | |||||
| 91-R-1 | 91-R-2 | 91-R-3 | 91-C-1 | 91-C-2 | 91-C-3 | |
| Unique miRNA | 992,816 | 920,013 | 944,922 | 900,308 | 894,908 | 531,843 |
| Annotated with ncRNA (rRNA, tRNA, snRNA, snoRNA and others) | 490,904 | 510,823 | 503,922 | 344,222 | 375,672 | 174,207 |
| D. melanogaster miRNAs | 7,643 | 6,050 | 7,079 | 9,356 | 7,947 | 7,942 |
| Unannotated | 494,269 | 403,140 | 433,921 | 546,730 | 511,289 | 349,694 |
Annotation of known miRNAs in the 91-R and 91-C strains identified 163 precursor and 256 mature miRNAs following alignment of trimmed reads to the D. melanogaster precursor/mature miRNAs in the miRBase R.21 (S2 Table). None of the precursor or mature miRNAs were expressed uniquely in either strain. However, we failed to identify any of the 93 known D. melanogaster precursors that are recorded in the miRBase R.21 (S3 Table). In both strains, miR-184-3p miRNA was the most abundant among the means of non-normalized reads across triplicate libraries, with miR-8-3p, miR-276a-3p, bantam-3p, and miR-33-5p as the next most abundant miRNAs in both strains (Table 2). Algorithms in the miRDeep2 package identified 17 potential novel miRNAs in 91-R and 91-C, of which 15 were in common between libraries derived from both strains; putatively novel-miR-3L-18860981 and novel-miR-2R-20583765 were uniquely observed in 91-R and 91-C, respectively (S4 Table). The range of estimated MFE among potential novel miRNAs was between -31.7 and -25.5 kcal mol-1 and their mature lengths from 18–25 nt. These 17 novel miRNAs were named based on the chromosome and position on which the miRNA gene is located in the D. melanogaster genome.
Table 2. The most abundant reads from 91-R and 91-C small RNA libraries corresponding to known Drosophila melanogaster miRNAs in miRBase R.21.
| miRNA |
91-R Read countsa |
91-C Read countsa |
Mature sequence |
|---|---|---|---|
| miR-184-3p | 27,954 | 27,402 | UGGACGGAGAACUGAUAAGGGC |
| miR-8-3p | 24,289 | 25,956 | UAAUACUGUCAGGUAAAGAUGUC |
| miR-276a-3p | 20,174 | 17,849 | UAGGAACUUCAUACCGUGCUCU |
| bantam-3p | 7,498 | 9,036 | UGAGAUCAUUUUGAAAGCUGAUU |
| miR-33-5p | 4,208 | 5,040 | GUGCAUUGUAGUCGCAUUGUC |
| miR-10-5p | 3,464 | 3,954 | ACCCUGUAGAUCCGAAUUUGUU |
| miR-317-3p | 2,908 | 3,292 | UGAACACAGCUGGUGGUAUCCAGU |
| miR-14-3p | 2,603 | 2,611 | UCAGUCUUUUUCUCUCUCCUAU |
| miR-31a-5p | 3,580 | 2,354 | UGGCAAGAUGUCGGCAUAGCUGA |
| miR-312-3p | 1,056 | 2,318 | UAUUGCACUUGAGACGGCCUGA |
| miR-11-3p | 1,659 | 2,275 | CAUCACAGUCUGAGUUCUUGC |
| miR-311-3p | 596 | 2,170 | UAUUGCACAUUCACCGGCCUGA |
| miR-318-3p | 994 | 1,977 | UCACUGGGCUUUGUUUAUCUCA |
| miR-999-3p | 1,604 | 1,660 | UGUUAACUGUAAGACUGUGUCU |
| miR-957-3p | 1,744 | 1,576 | UGAAACCGUCCAAAACUGAGGC |
| miR-276b-3p | 1,846 | 1,553 | UAGGAACUUAAUACCGUGCUCU |
| miR-277-3p | 2,046 | 1,489 | UAAAUGCACUAUCUGGUACGACA |
| miR-995-3p | 542 | 1,275 | UAGCACCACAUGAUUCGGCUU |
| miR-305-3p | 1,000 | 1,235 | CGGCACAUGUUGAAGUACACUCA |
| let-7-5p | 1,094 | 1,208 | UGAGGUAGUAGGUUGUAUAGU |
| miR-986-5p | 3,069 | 139 | UCUCGAAUAGCGUUGUGACUGA |
| miR-958-3p | 1,020 | 899 | UGAGAUUCUUCUAUUCUACUUU |
a Non-normalized reads summed across triplicate libraries
Differential expression analysis and RT-qPCR validation
Comparison of normalized estimates of miRNA quantity (log2 fold-changes; S5 Table) demonstrated an overall parity across replicated libraries derived from DDT resistance strain, 91-R, as compared to the susceptible control, 91-C (counts pooled across replicates within strain; S2 Fig). Exceptions were found among ten known miRNAs that showed significant levels of differential expression between 91-R and 91-C (log2 fold-change ≥ |1|; FDR ≤ 0.05). Specifically, nine miRNAs were significantly down-regulated, and only one (miR-986-5p) was significantly up-regulated in 91-R (25.3-fold) when compared with the susceptible control 91-C (Table 3). Interestingly, four out of these nine down-regulated miRNAs [miR-310, miR-311, miR-312 and miR-313] are clustered miRNAs and belong to the miR-310 family. Overall, most of the differentially expressed miRNAs were down-regulated in the DDT resistant strain 91-R, suggesting that those down-regulated miRNAs may be potentially involved in DDT resistance.
Table 3. Differentially expressed miRNAs between 91-C and 91-R.
| miRNA | p-value | FDRa | Fold-change | Log2 fold-changeb |
miRNA sequence |
|---|---|---|---|---|---|
| miR-986-5p | 3.0E-53 | 7.8E-51 | 25.3 | 4.66 | TCTCGAATAGCGTTGTGACTGA |
|
miR-2a-1-3p// miR-2a-2-3p |
7.0E-04 | 1.3E-02 | -2.01 | -1.01 | TATCACAGCCAGCTTTGATGAGC |
| miR-312-3p | 1.4E-07 | 5.3E-06 | -2.02 | -1.01 | TATTGCACTTGAGACGGCCTGA |
| miR-995-3p | 2.8E-09 | 1.4E-07 | -2.17 | -1.12 | TAGCACCACATGATTCGGCTT |
| miR-286-3p | 2.1E-03 | 3.2E-02 | -2.29 | -1.19 | TGACTAGACCGAACACTCGTGCT |
| miR-92a-3p | 2.9E-10 | 1.9E-08 | -2.34 | -1.23 | CATTGCACTTGTCCCGGCCTAT |
| miR-4919-3p | 1.4E-04 | 3.2E-03 | -2.57 | -1.36 | TAATCCCTGAACGACTTGCAG |
| miR-311-3p | 1.8E-15 | 1.5E-13 | -3.38 | -1.76 | TATTGCACATTCACCGGCCTGA |
| miR-310-3p | 9.1E-16 | 1.2E-13 | -3.94 | -1.98 | TATTGCACACTTCCCGGCCTTT |
| miR-313-3p | 4.4E-07 | 1.4E-05 | -4.89 | -2.29 | TATTGCACTTTTCACAGCCCGA |
a FDR: False discovery rate. Differentially expressed miRNAs were identified at the threshold [FDR < 0.05 and log2(fold change) ≥|1.0|] of 91-C/91-R.
b Fold change was calculated as 91-C/91-R.
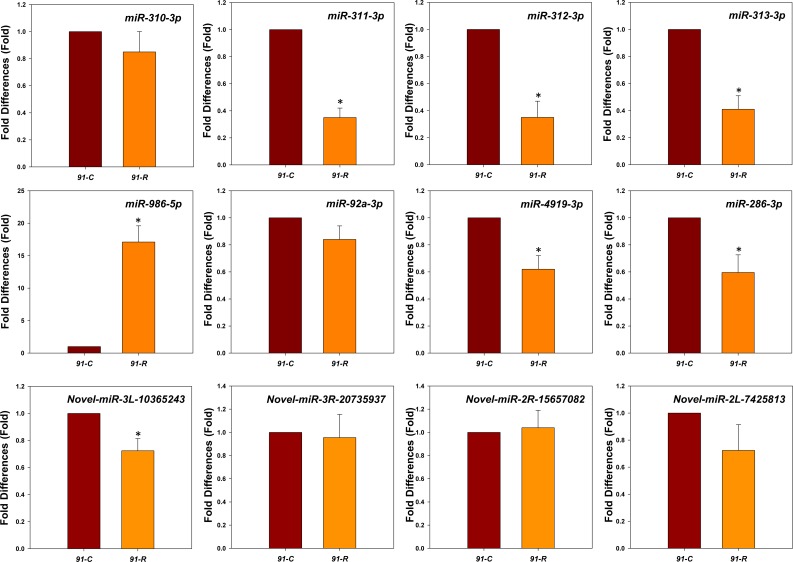
The RT-qPCR validation of the predicted significant quantitative differences in miRNA levels among eight known and four novel miRNAs were amplified showing that the expression levels of miR-986-5p were highly up-regulated in 91-R, whereas miR-286-3p, miR-4919-3p, miR-311-3p, miR-312-3p, and miR-313-3p were significantly down-regulated in 91-R (Fig 1). However, levels of miR-92a-3p and miR-310-3p showed no significant difference between strains (P-value > 0.05). Based on Pearson’s correlation coefficient test, the expression patterns of selected miRNAs showed a similar trend between the results of sRNA sequencing and RT-qPCR (R2 = 0.971, P-value < 0.01), confirming the predicted differential expression between strains. Additionally, the expression level of one putative novel miRNA (novel-miR-3L-10365243) was significantly down-regulated in 91-R, whereas three other putative novel miRNAs showed no significant differences between two strains (Fig 1).
Fig 1. RT-qPCR validation of differentially expressed known and novel miRNAs identified between 91-R and 91-C strains.
The expression levels were normalized by both U6 and 5S rRNA. Statistical significance was analyzed using one-way ANOVA. The asterisks represent significance, where one asterisk indicates P < 0.05.
Target transcript predictions and correlated expression between miRNAs and target transcripts
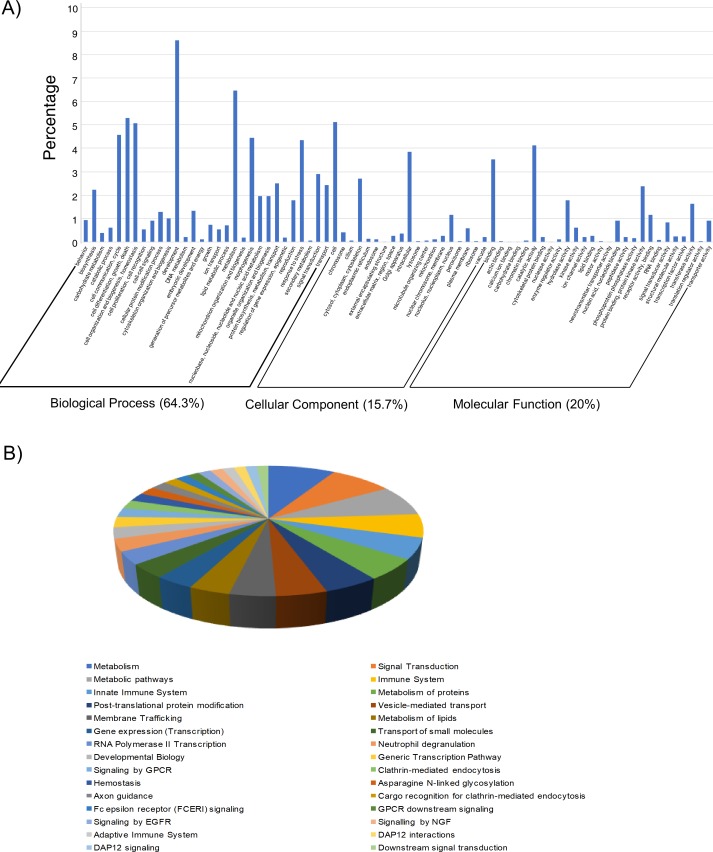
Considering only the ten miRNAs predicted to be differentially expressed between 91-R and 91-C, a total of 46,368 miRNA-target pairs were predicted by the algorithms applied by ComiR (n = 5,577), DIANA (n = 2,548), and RNAhybrid (n = 38,243; S6 Table). Overlap in output occurred among 664 transcript targets predicted by all three algorithms. Functional GO annotations were received for 603 of these 664 transcript targets, with 64.3%, 20%, and 15.7% that respectively received terms in biological process (BP), molecular function (MF), and cellular component (CC) categories (GO level 2; S7 Table). Specifically, two putative functions (development and metabolism) were highly represented in the BP category, and the MF category showed the function of ‘binding’ and ‘catalytic activity’ was most prevalent among target genes. Moreover, cell, intracellular component, cytosol, cytoplasm, and cytoskeleton were largely overrepresented in CC category (Fig 2A). A total of 2,175 biological pathways were assigned to 258 of the 664 predicted transcript targets (38.9%; Fig 2B; S8 Table), and the target Ras85D (CG9375) is associated with regulation of tissue growth and development represented 93 biological pathways. The remaining 405 transcript targets (61.1%) received no pathway annotations. Among these 2,175 biological pathways, 58 (2.7%), 54 (2.5%), and 53 (2.4%) target genes of the known differentially expressed miRNAs, respectively, were assigned to metabolism, signal transduction, and metabolic pathway (Fig 2B).
Fig 2. Gene ontology (GO) analysis and pathway annotation of the most targeted genes of the known differentially expressed miRNAs between 91-R and 91-C strains.
(A) GO analysis. 664 target genes from miRNAs predicted to be differentially expressed were analyzed with FlyMine to obtain the GO terms, and the GO terms were classified with CateGOrizer and separated into three major categories. (B) Pathway annotations. 664 target genes from miRNAs predicted to be differentially expressed were submitted to FlyMine to get the pathway classification. The 30 most abundant pathways were represented.
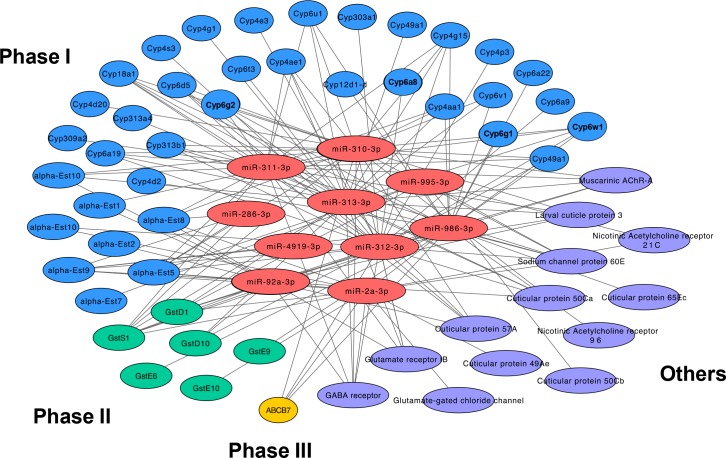
Additional annotation focused on a subset of the 664 transcripts putatively targeted by differentially expressed miRNAs; specifically, those transcripts likely to be involved in xenobiotic metabolism. These putative transcripts were predicted to target phase I, II, and III detoxification pathways such as cytochrome P450s, GSTs, esterases, and ABC transporters (43 target transcripts; Fig 3). Additionally, differentially expressed miRNAs were predicted to regulate other genes associated with cuticular proteins, acetylcholine receptors, nicotinic acetylcholine receptors, and glutamate-gated chloride channels (13 target transcripts; Fig 3). The phase I, cytochrome P450 targets Cyp6a8, Cyp6g1, Cyp6g2, and Cyp6w1, were previously associated with DDT resistance as described in the Introduction. Also of note, the down-regulated miR-310-313 cluster in 91-R strain was predicted to share several P450s, sodium channel proteins, and cuticular proteins encoding target genes (Fig 3). Additionally, sixteen P450 genes (MFE ≤ -25; Cyp18a1, Cyp305a1, Cyp309a2, Cyp312a1, Cyp313b1, Cyp49a1, Cyp4ae1, Cyp4g1, Cyp4g15, Cyp4p2, Cyp4s3, Cyp6a19, Cyp6g1, Cyp6g2, Cyp6v1, and Cyp9h1) were among the predicted targets of 9 of the 17 putative novel miRNAs (S9 Table).
Fig 3. Putative xenobiotic metabolism-related target genes of differentially expressed miRNAs.
The network consists of differentially expressed miRNAs and their corresponding target genes. Red hexagons represent differentially expressed miRNAs; blue circles represent target genes as phase I detoxification genes; green circles represent target genes encoding phase II detoxification genes; yellow circle represents target genes as phase III detoxification gene; and purple circles represent other genes affecting the insecticide insensitivity in insects. The network was generated and visualized in Cytoscape v3.6.0. Cyp6a8, Cyp6g1, Cyp6g2, and Cyp6w1 associated with DDT resistance are in bold.
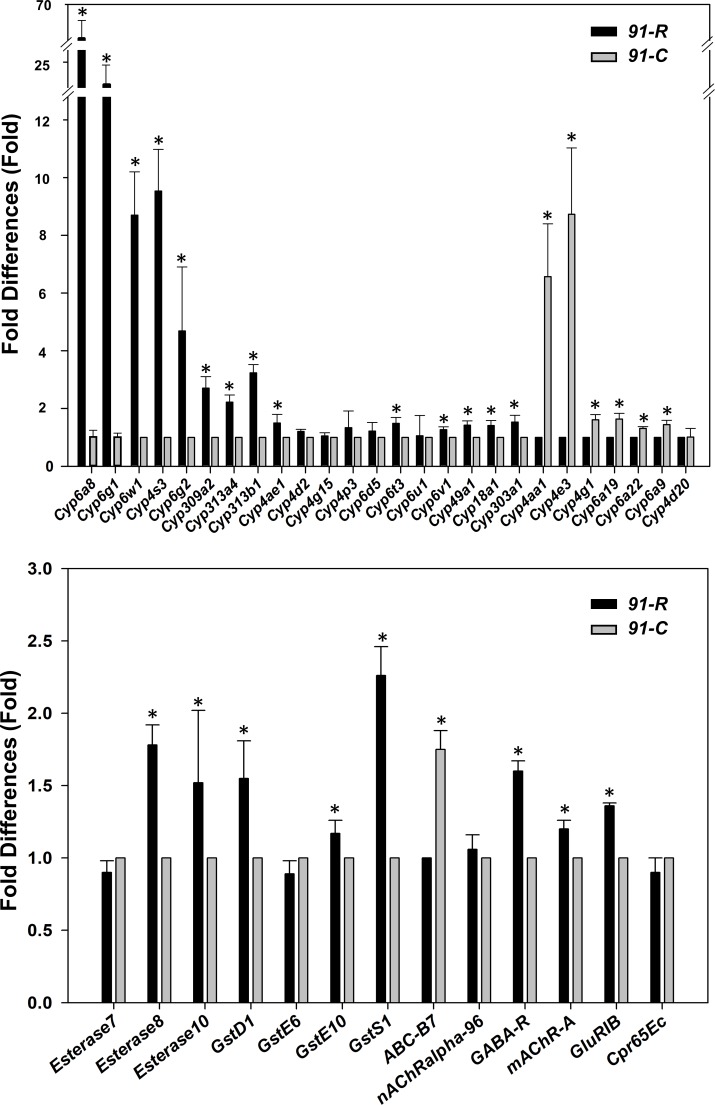
Moreover, the RT-qPCR-estimated quantities of 26 putatively targeted P450 transcripts showed an inverse relationship with the expression level of the corresponding miRNA(s) predicted to target them in 91-R. For example, the relative expression of three miRNAs (miR-311-3p, miR-312-3p, and miR-313-3p) were significantly down-regulated in 91-R strain (Fig 1), while the corresponding predicted targets (Cyp6a8, Cyp4s3, Cyp4ae1, Cyp6g1, Cyp6g2, Cyp6t3, Cyp6v1, Cyp18a1, Cyp49a1, Cyp303a1, Cyp309a2, Cyp313a4, and Cyp313b1) were significantly up-regulated (Fig 4). Furthermore, other corresponding predicted detoxification targets, Esterase8, Esterase10, GstD1, GstE10, GstS1, GABA-R, mAChR-A, nAcRalpha-A and GluR-IB were significantly up-regulated with the down-regulation of miR-286-3p, miR-2a-3p, miR-311-3p, miR-312-3p, and miR-313-3p in 91-R strain (Fig 4). The resulting coefficient of correlation showed that the strong negative correlation between miRNAs and putative target transcripts (R = -0.9558; P-value < 0.01) suggests the possibility that higher levels of detoxification transcripts may be influenced by the reduced levels of corresponding targeting miRNAs within the DDT resistant 91-R strain. The precise role of these detoxification genes in mediating DDT resistance remains unknown, but may include increasing cellular resistance to oxidative stress, cuticular penetrance, or other systemic responses to exposure.
Fig 4. Expression level analysis of potential target detoxification genes of differentially expressed miRNAs between 91-R and 91-C strains.
The expression levels were normalized by rp49. Statistical significance was analyzed using one-way ANOVA. The asterisks represent significance, with a single asterisk indicating P < 0.05.
Discussion
Resistant phenotypes have evolved within arthropod populations that lead to survival when exposed to various chemical or biological insecticidal toxins and seemingly arise to each successively introduced novel mode of action [70]. Although direct correlations have been made between single transposon-based mutations at single genetic loci (an Accord element associated with Cyp6g1) and corresponding insecticide resistance traits [71–73], in many instances the causal mutations and molecular mechanisms involved in resistance are yet to be fully understood. Moreover, definitive linkages between phenotypes and corresponding adaptive mutations remain difficult to define [74], especially in instances where phenotypes arise due to the contribution of multiple genes or gene interactions, or are complexed with variance due to the environment [75]. For instance, DDT resistance was linked to the up-regulation of Cyp6g1 caused by the upstream integration of an Accord transposon at the DDT-R locus in D. melanogaster populations [71] but shown to be independent of DDT-R in the highly DDT resistant strain 91-R [76]. The prediction of thirteen selective sweeps that became fixed in the genome of 91-R during the course of DDT selection implicated a complex polygenic mode of evolution [15], which likely involves a combination of cis- and trans-regulatory mutations that modulate the function of stress response and neurogenic pathways [17]. Regardless of evidence that strongly implicates miRNAs as potent modulators of gene expression at the post-transcriptional level [77], the role of miRNAs in the evolution of differential gene expression in insecticide resistant phenotypes among arthropods remains suggestive or associative in many cases [44,45].
The current study identified a total of 163 precursors with 256 mature known miRNAs and 17 novel miRNAs. This study failed to identify 93 known precursor miRNAs, previously recorded in the miRBase R.21. The extraction of sRNAs from 3–5 day-old virgin adult female samples may likely have biased the number of miRNA types. Previously, miRNAs were shown to be sex-biased in D. melanogaster, where expression was preferentially associated with the reproductive functions [78]. More recent work that compared miRNA expression between mature male and female reproductive organs in Bactrocera dorsalis demonstrated that sex-biased miRNAs are likely involved in sexual differentiation [79]. Furthermore, the expression of miRNAs varied across developmental stages of Xenopus laevis [80] and across different human tissues [81]. This suggests that our use of virgin female adults might have narrowed the pool of potential miRNAs that could be encountered within the resultant sRNA libraries but is justified since adults are exposed to DDT selection within the 91-R laboratory colony. Another hypothesis may reside in the potential saturation of sRNA samples with 2S ribosomal RNA (rRNA). Specifically, D. melanogaster rRNA is composed of four individual rRNAs, 28S, 18S, 5.8S, and 2S, with the 2S rRNA being 30-nt in length [82]. Analysis of our sRNA read data revealed that 32% of non-miRNA sequences matched the D. melanogaster 2S rRNA, suggesting the possibility that the 2S rRNA component of our libraries might have affected the subsequent read depths and the inability to identify 93 known miRNAs from 91-R and 91-C strains if they would have been at low copy number. Neither of the above explanations can be ruled out but require additional investigation. The current study nonetheless predicted the significant quantitative difference in ten miRNA levels between 91-R and 91-C at the adult virgin female stage.
Ten differentially expressed miRNAs and their corresponding putative target transcripts were predicted and received GO and pathway database annotations in this study. Among these ten differentially expressed miRNAs, four members of the miR-310 family, miR-310, miR-311, miR-312 and miR-313, were significantly down-regulated in 91-R when compared with the 91-C strain. The miR-310 family form a cluster on chromosomal arm 2R between the CDS of qsm and Nnf1a (positions 20,583,752 to 20,584,260) [83] and arose via duplication from the ancestral miR-91 [84]. Previous studies showed that the miR-310 family regulates genes in the D. melanogaster Toll-mediated innate immune response pathway [85], via hedgehog signaling [86], and beta-catenin that in turn regulates cell adhesion and outgrowth [87]. Furthermore, the mir-310-313 cluster has been reported to be associated with hypersensitivity to nicotine in D. melanogaster [88]. These authors observed that the expression of the miR-310-313 downregulates escargot (esg) gene expression involved in the development of sensory organs and neurons in the thoracoabdominal ganglion. The overexpression of the miR-310-313 induces an abnormal sensitivity to nicotine by abating esg transcription, then disrupting sensory organs involved in chemical perception and cuticle development. This mechanism may be similar to the DDT resistance of miR-310 family members in 91-R strain. Additionally, miR-310 family members impact synaptic functions through the regulation of neurotransmitter release at neuromuscular junctions of D. melanogaster larvae [89]. These authors observed that miR-310-313 cluster regulates neurotransmitter release at presynaptic terminals by decreasing the expression of Kinesin heavy chain-73, Khc-73, which is involved in early neural development. Interestingly, our prior research validated the significant differential regulation of genes directing neuronal development in 91-R, including Unc-115b, and CG31832 that function in neural growth and development, but detected no significant difference in expression of Khc-73 [17]. Our prior results suggest the possibility that adaptive responses to multigenerational neurotoxic DDT selection in 91-R may affect the function of neuromuscular junctions, although the role of the miR-310 family in the regulation of genes associated with neuronal functions in 91-R remains a hypothesis that needs to be tested experimentally.
Findings from this study show that the down-regulation of miR-311-3p, miR-312-3p, and miR-313-3p is correlated with the up-regulation of their respective in silico predicted cytochrome P450 target transcripts, Cyp6a8, Cyp4s3, Cyp4ae1, Cyp6g1, Cyp6g2, Cyp6t3, Cyp6v1, Cyp18a1, Cyp49a1, Cyp303a1, Cyp309a2, Cyp313a4, and Cyp313b1 in 91-R. This up-regulation of Cyp6g1 target is predicted to occur via a decreased miR-310-313 expression in 91-R and also corresponds to the cytochrome P450 initially implicated in conferring DDT resistance at the DDT-R locus via integration of the Accord transposon among D. melanogaster populations [71]. Our computational and empirical data suggests that the miR-310 family members may be involved in the posttranscriptional regulation of these cytochrome P450s, which in turn might be directly involved in DDT detoxification or stress response [17]. Thus, the involvement of miR-310-313 could explain prior evidence that even though DDT resistance in 91-R is independent of the Accord insertion [76], significant up-regulation of Cyp6g1, as well as Cyp6a8, Cyp4s3, Cyp4ae1, Cyp6g2, Cyp6t3, Cyp6v1, Cyp18a1, Cyp49a1, Cyp303a1, Cyp309a2, Cyp313a4, and Cyp313b1 occurs constitutively in the strain. Regardless of these strong correlations, additional functional analyses are required to confirm these predicted impacts of miRNA-based posttranscriptional regulation.
The function of the most highly up-regulated miRNA in 91-R strain, miR-986-5p, is currently unknown, but levels in the hemolymph of adult virgin males are known to significantly decrease over time [90]. Interestingly, the miR-986 precursor is located on the second chromosome within the third intron of the Cyp4e2 gene, which is involved in the metabolism of endogenous and exogenous compounds [91]. Our transcript target site predictions suggest that miR-986-5p could interact with transcripts of cytochrome P450s, GSTs, esterases, and superoxide dismutases (SODs). Specifically, the GSTs and SODs are a group of a multifunctional antioxidant enzymes that play an important role in mediating oxidative stress caused by reactive oxygen species (ROS) in insects[92–94]. Therefore, the miR-986 may be involved in the posttranslational regulation of genes that alleviate oxidative stress induced by DDT insecticide exposures. The putative targeted transcripts of miR-986-5p include Cyp6g1 and Cyp6g2. However, the RT-qPCR performed here, as well as prior RNA-seq results [17], indicate that the expression levels of putative targets Cyp6g1 and Cyp6g2 were not decreased. In contrast, both Cyp6g1 and Cyp6g2 were significantly up-regulated in 91-R, suggesting miR-986-5p may have a transcript stabilizing or enhancing effect, as was demonstrated previously [95,96]. Alternatively, algorithms used for in silico prediction of miRNA-transcript target interactions can produce variable results depending on the input database and models that are applied [97,98]. Regardless, in vivo or in vitro validation of these assumptions will be required.
Conclusion
Analyses conducted in this study focused on differentially expressed miRNAs that were predicted to regulate transcript levels of both phase I, II and III detoxification genes previously shown to be associated with the DDT resistance phenotype. Cytochrome P450s that are involved in many cellular processes, including xenobiotic detoxification, have been studied [99] for associations between miRNA levels and corresponding putatively targeted P450 transcripts. For example, a negative relationship was shown between up-regulated miRNAs miR-8534-5p and miR-375-5p and their respective predicted targeted cytochrome P450s, Cyp6b6 and Cyp4g15, in chlorantraniliprole-resistant strains of Plutella xylostella [44]. Several miRNAs down-regulated in deltamethrin-resistant mosquitoes played a role in pyrethroid resistance through the reduced targeting of Cyp325bg3, Cyp6n23, and Cyp9j35 [47,48,50], while the miRNAs (miR-155, miR-216b, miR-499) modulate the abundance of Cyp561d2 transcripts in response to fipronil exposure [100]. Additionally, miR-285 and miR-278 differentially regulate Cyp6n23 and Cyp6ag11 in pyrethroid resistant compared to susceptible Culex pipiens [48,49].
Thus, our implication of down-regulated miR-311-3p, miR-312-3p, and miR-313-3p with the corresponding constitutive up-regulation of in silico predicted targets Cyp6a8, Cyp4s3, Cyp4ae1, Cyp6g2, Cyp6t3, Cyp6v1, Cyp18a1, Cyp49a1, Cyp303a1, Cyp309a2, Cyp313a4 in 91-R may provide yet another example of a miRNA-mediated posttranscriptional modification that contributes to an insecticide resistance trait. Additionally, 4 out of 10 miRNAs predicted to be differentially expressed between 91-R and 91-C (miR-986-5p, miR-995-3p, miR-312-3p, miR-2a-3p) were also predicted to interact with and impact the transcript level of multidrug resistance-associated protein B7 (ABC-B7). The ABC-B subfamily member, MDR49, had been previously been reported to show significant levels of differential expression between 91-R and the DDT-susceptible strain Canton-S [12] but not between 91-R and 91-C [12,17]. It was subsequently shown that the 91-R-derived 91-R-MDR49B allele provided increased DDT tolerance via transgenic expression in susceptible D. melanogaster, implicating structural, as opposed to dosage, effects on the DDT resistance trait [14]. Thus, one could hypothesize that ABC-B7 might be regulated by one or multiple miRNAs that are differentially expressed between 91-R and 91-C. In addition to further empirical study, however, it should also be kept in mind that multiple different mutational mechanisms can contribute to DDT resistance in 91-R.
In this study, we identified a set of differentially expressed known miRNAs and several novel miRNAs from DDT-resistant 91-R and -susceptible 91-C strains. Since 91-R and 91-C strains have a common initial origin (genetic background), and 91-R has been selected for survival when exposed to chronic high levels of DDT for over six decades, changes between the strains are speculated to result from either the effects of random genetic drift or from selection. The experiments described here do not allow the differentiation of any impacts from drift versus selection, but the strong correlation between differential expression of miRNAs and their corresponding in silico predicted target transcripts suggests the potential for the involvement of posttranscriptional regulation of several detoxification genes.
Moreover, the experimental procedures do not allow the formulation that down-regulated miRNAs identified in the present study could lead directly to the overexpression of detoxification genes in the 91-R strain, thus requiring further functional experiments in order to elucidate the mechanisms of miRNAs involvement in DDT resistance in 91-R. As such, the overexpressed P450s putatively targeted by differentially-regulated miRNAs in 91-R strain need to be further evaluated to identify the ones directly involved in the DDT resistance mechanism. Due to the polygenic nature of DDT resistance in 91-R (see Introduction), however, this trait may be the result of interactions or additive/non-additive effects from several distinct genetic factors. This study, for the first time, suggests that these might include impacts mediated by miRNA posttranscriptional regulation. Our results provide valuable information for exploring the mechanisms of miRNAs involved in insecticide resistance and for understanding the evolution of post-transcriptional regulation in response to DDT pressure in D. melanogaster.
Supporting information
(TIF)
Each plot represents a miRNA. The X- and Y-axis show the normalized read counts of miRNAs in the two strains respectively.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
The small RNA sequencing data sets generated during this study are available in the NCBI Sequence Read Archive (SRA) under accession numbers SRP136631 (https://www.ncbi.nlm.nih.gov/sra/ <https://www.ncbi.nlm.nih.gov/sra/>).
Funding Statement
The authors received no specific funding for this work.
References
- 1.Casida JE, Quistad GB. Golden age of insecticide research: past, present, or future? Annu Rev Entomol. 1998;43: 1–16. doi: 10.1146/annurev.ento.43.1.1 [DOI] [PubMed] [Google Scholar]
- 2.Roush RT, McKenzie JA. Ecological genetics of insecticide and acaricide resistance. Annu Rev Entomol. 1987;32: 361–380. doi: 10.1146/annurev.en.32.010187.002045 [DOI] [PubMed] [Google Scholar]
- 3.Mallet J. The evolution of insecticide resistance: Have the insects won? Trends Ecol Evol. 1989;4: 336–340. doi: 10.1016/0169-5347(89)90088-8 [DOI] [PubMed] [Google Scholar]
- 4.Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 2000;45: 371–391. doi: 10.1146/annurev.ento.45.1.371 [DOI] [PubMed] [Google Scholar]
- 5.Organization WH. Indoor residual spraying. Use of indoor residual spraying for scaling up global malaria control and elimination. Geneva, Switzerland (WHO/HTM/MAL/20061112); 2006.
- 6.Daborn P, Boundy S, Yen J, Pittendrigh B, Ffrench-Constant R. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol Genet Genomics. 2001;266: 556–563. doi: 10.1007/s004380100531 [DOI] [PubMed] [Google Scholar]
- 7.Le Goff G, Hilliou F. Resistance evolution in Drosophila: the case of CYP6G1. Pest Manag Sci. 2017;73: 93–499. [DOI] [PubMed] [Google Scholar]
- 8.Strycharz JP, Lao A, Li H, Qiu X, Lee SH, Sun W, et al. Resistance in the highly DDT-resistant 91-R strain of Drosophila melanogaster involves decreased penetration, increased metabolism, and direct excretion. Pestic Biochem Physiol. 2013;107: 207–217. [Google Scholar]
- 9.Pedra JH, McIntyre LM, Scharf ME, Pittendrigh BR. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc Natl Acad Sci U S A. 2004;101: 7034–7039. doi: 10.1073/pnas.0400580101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Festucci-Buselli RA, Carvalho-Dias AS, de Oliveira-Andrade M, Caixeta-Nunes C, Li HM, Stuart JJ, et al. Expression of Cyp6g1 and Cyp12d1 in DDT resistant and susceptible strains of Drosophila melanogaster. Insect Mol Biol. 2005;14: 69–77. doi: 10.1111/j.1365-2583.2005.00532.x [DOI] [PubMed] [Google Scholar]
- 11.Daborn PJ, Lumb C, Boey A, Wong W, Ffrench-Constant RH, Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol. 2007;37: 512–519. doi: 10.1016/j.ibmb.2007.02.008 [DOI] [PubMed] [Google Scholar]
- 12.Gellatly KJ, Yoon KS, Doherty JJ, Sun W, Pittendrigh BR, Clark JM. RNAi validation of resistance genes and their interactions in the highly DDT-resistant 91-R strain of Drosophila melanogaster. Pestic Biochem Physiol. 2015;121: 107–115. doi: 10.1016/j.pestbp.2015.01.001 [DOI] [PubMed] [Google Scholar]
- 13.Schmidt JM, Battlay P, Gledhill-Smith RS, Good RT, Lumb C, Fournier-Level A, et al. Insights into DDT resistance from the Drosophila melanogaster Genetic Reference Panel. Genetics. 2017;207: 1181–1193. doi: 10.1534/genetics.117.300310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seong KM, Sun W, Clark JM, Pittendrigh BR. Splice form variant and amino acid changes in MDR49 confers DDT resistance in transgenic Drosophila. Sci Rep. 2016;6: 23355 doi: 10.1038/srep23355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steele LD, Coates B, Valero MC, Sun W, Seong KM, Muir WM, et al. Selective sweep analysis in the genomes of the 91-R and 91-C Drosophila melanogaster strains reveals few of the 'usual suspects' in dichlorodiphenyltrichloroethane (DDT) resistance. PLoS One. 2015;10: e0123066 doi: 10.1371/journal.pone.0123066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steele LD, Muir WM, Seong KM, Valero MC, Rangesa M, Sun W, et al. Genome-wide sequencing and an open reading frame analysis of dichlorodiphenyltrichloroethane (DDT) susceptible (91-C) and resistant (91-R) Drosophila melanogaster laboratory populations. PLoS One. 2014;9: e98584 doi: 10.1371/journal.pone.0098584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seong KM, Coates BS, Sun W, Clark JM, Pittendrigh BR. Changes in neuronal signaling and cell stress response pathways are associated with a multigenic response of Drosophila melanogaster to DDT selection. Genome Biol Evol. 2017;9: 3356–3372. doi: 10.1093/gbe/evx252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger M, Puinean AM, Randall E, Zimmer CT, Silva WM, Bielza P, et al. Insecticide resistance mediated by an exon skipping event. Mol Ecol. 2016;25: 5692–5704. doi: 10.1111/mec.13882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong K, Du Y, Rinkevich F, Nomura Y, Xu P, Wang L, et al. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem Mol Biol. 2014;50: 1–17. doi: 10.1016/j.ibmb.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75: 843–854. [DOI] [PubMed] [Google Scholar]
- 21.Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, et al. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17: 1865–1879. doi: 10.1101/gr.6593807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152: 844–858. doi: 10.1016/j.cell.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425: 415–419. doi: 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- 24.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42: D68–73. doi: 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116: 281–297. [DOI] [PubMed] [Google Scholar]
- 26.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6: 127–138. doi: 10.1038/nrm1568 [DOI] [PubMed] [Google Scholar]
- 27.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123: 607–620. doi: 10.1016/j.cell.2005.08.044 [DOI] [PubMed] [Google Scholar]
- 28.Wostenberg C, Lary JW, Sahu D, Acevedo R, Quarles KA, Cole JL, et al. The role of human Dicer-dsRBD in processing small regulatory RNAs. PLoS One. 2012;7: e51829 doi: 10.1371/journal.pone.0051829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X. Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics. 2014;30: 1377–1383. doi: 10.1093/bioinformatics/btu045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466: 835–840. doi: 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102: 16961–16966. doi: 10.1073/pnas.0506482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21: 533–542. doi: 10.1016/j.molcel.2006.01.031 [DOI] [PubMed] [Google Scholar]
- 33.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320: 1185–1190. doi: 10.1126/science.1159151 [DOI] [PubMed] [Google Scholar]
- 34.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318: 1931–1934. doi: 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 35.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5: 522–531. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 36.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310: 1817–1821. doi: 10.1126/science.1121158 [DOI] [PubMed] [Google Scholar]
- 37.Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol. 2013;43: 388–397. doi: 10.1016/j.ibmb.2012.10.005 [DOI] [PubMed] [Google Scholar]
- 38.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14: 475–488. doi: 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- 39.Liu Q, Wang W, Yang X, Zhao D, Li F, Wang H. MicroRNA-146a inhibits cell migration and invasion by targeting RhoA in breast cancer. Oncol Rep. 2016;36: 189–196. doi: 10.3892/or.2016.4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460: 642–646. doi: 10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W, Liu Z, Li T, Zhang R, Xue Y, Zhong Y, et al. Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat Commun. 2014;5: 5549 doi: 10.1038/ncomms6549 [DOI] [PubMed] [Google Scholar]
- 42.Xiong XP, Kurthkoti K, Chang KY, Li JL, Ren X, Ni JQ, et al. miR-34 Modulates innate immunity and ecdysone signaling in Drosophila. PLoS Pathog. 2016;12: e1006034 doi: 10.1371/journal.ppat.1006034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gendron CM, Pletcher SD. MicroRNAs mir-184 and let-7 alter Drosophila metabolism and longevity. Aging Cell. 2017;16: 1434–1438. doi: 10.1111/acel.12673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu B, Li X, Liu Y, Gao X, Liang P. Global identification of microRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.). Sci Rep. 2017;7: 40713 doi: 10.1038/srep40713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Xu Z, Wu Q, Peng M, Liu Y, Liu X, et al. Identification of differentially expressed microRNAs between the fenpropathrin resistant and susceptible strains in Tetranychus cinnabarinus. PLoS One. 2016;11: e0152924 doi: 10.1371/journal.pone.0152924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu LN, Ling YH, Wang YQ, Wang ZY, Hu BJ, Zhou ZY, et al. Identification of differentially expressed microRNAs between Bacillus thuringiensis Cry1Ab-resistant and -susceptible strains of Ostrinia furnacalis. Sci Rep. 2015;5: 15461 doi: 10.1038/srep15461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong S, Guo Q, Wang W, Hu S, Fang F, Lv Y, et al. Identification of differentially expressed microRNAs in Culex pipiens and their potential roles in pyrethroid resistance. Insect Biochem Mol Biol. 2014;55: 39–50. doi: 10.1016/j.ibmb.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian M, Liu B, Hu H, Li X, Guo Q, Zou F, et al. MiR-285 targets P450 (CYP6N23) to regulate pyrethroid resistance in Culex pipiens pallens. Parasitol Res. 2016;115: 4511–4517. doi: 10.1007/s00436-016-5238-4 [DOI] [PubMed] [Google Scholar]
- 49.Lei Z, Lv Y, Wang W, Guo Q, Zou F, Hu S, et al. MiR-278-3p regulates pyrethroid resistance in Culex pipiens pallens. Parasitol Res. 2015;114: 699–706. doi: 10.1007/s00436-014-4236-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Q, Huang Y, Zou F, Liu B, Tian M, Ye W, et al. The role of miR-2~13~71 cluster in resistance to deltamethrin in Culex pipiens pallens. Insect Biochem Mol Biol. 2017;84: 15–22. doi: 10.1016/j.ibmb.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 51.Liu B, Tian M, Guo Q, Ma L, Zhou D, Shen B, et al. MiR-932 Regulates pyrethroid resistance in Culex pipiens pallens (Diptera: Culicidae). J Med Entomol. 2016;53: 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Guo L, Zhou X, Gao X, Liang P. miRNAs regulated overexpression of ryanodine receptor is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Sci Rep. 2015;5: 14095 doi: 10.1038/srep14095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merrell DJ, Underhill JC. Selection for DDT resistance in inbred, laboratory and wild stocks of Drosophila melanogaster. J Econ Entomol. 1956;49: 300–306. [Google Scholar]
- 54.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40: 37–52. doi: 10.1093/nar/gkr688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17: 10–12. doi: 10.14806/ej.17.1.200 [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26: 407–415. doi: 10.1038/nbt1394 [DOI] [PubMed] [Google Scholar]
- 58.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20: 2911–2917. doi: 10.1093/bioinformatics/bth374 [DOI] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26: 139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57: 289–300. [Google Scholar]
- 63.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7: 986–995. [DOI] [PubMed] [Google Scholar]
- 64.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10: 1507–1517. doi: 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41: W169–173. doi: 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coronnello C, Benos PV. ComiR: Combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013;41: W159–164. doi: 10.1093/nar/gkt379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 68.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294: 853–858. doi: 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 69.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12: 246–258. doi: 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- 70.Forgash AJ. History, evolution, and consequences of insecticide resistance. Pestic Biochem Physiol. 1984;22: 178–186. [Google Scholar]
- 71.Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297: 2253–2256. doi: 10.1126/science.1074170 [DOI] [PubMed] [Google Scholar]
- 72.Ffrench-Constant R, Daborn P, Feyereisen R. Resistance and the jumping gene. Bioessays. 2006;28: 6–8. doi: 10.1002/bies.20354 [DOI] [PubMed] [Google Scholar]
- 73.Mateo L, Ullastres A, Gonzalez J. A transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet. 2014;10: e1004560 doi: 10.1371/journal.pgen.1004560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stapley J, Reger J, Feulner PGD, Smadja C, Galindo J, Ekblom R, et al. Adaptation genomics: the next generation. Trends Ecol Evol. 2010;25: 705–712. doi: 10.1016/j.tree.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 75.Wu R. Predicting the genotype-phenotype map of complex traits. J Biomet Biostat. 2012;3: e109. [Google Scholar]
- 76.Kuruganti S, Lam V, Zhou X, Bennett G, Pittendrigh BR, Ganguly R, et al. High expression of Cyp6g1, a cytochrome P450 gene, does not necessarily confer DDT resistance in Drosophila melanogaster. Gene. 2007;388: 43–53. doi: 10.1016/j.gene.2006.09.019 [DOI] [PubMed] [Google Scholar]
- 77.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16: 421–433. doi: 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 78.Marco A. Sex-biased expression of microRNAs in Drosophila melanogaster. Open Biol. 2014;4: 140024 doi: 10.1098/rsob.140024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peng W, Tariq K, Xie J, Zhang H. Identification and characterization of sex-biased microRNAs in Bactrocera dorsalis (Hendel). PLoS One. 2016;11: e0159591 doi: 10.1371/journal.pone.0159591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, et al. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579: 318–324. doi: 10.1016/j.febslet.2004.11.067 [DOI] [PubMed] [Google Scholar]
- 81.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44: 3865–3877. doi: 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tautz D, Hancock JM, Webb DA, Tautz C, Dover GA. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol Biol Evol. 1988;5: 366–376. doi: 10.1093/oxfordjournals.molbev.a040500 [DOI] [PubMed] [Google Scholar]
- 83.Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 2017;45: D663–D671. doi: 10.1093/nar/gkw1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu J, Fu Y, Kumar S, Shen Y, Zeng K, Xu A, et al. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol Biol Evol. 2008;25: 929–938. doi: 10.1093/molbev/msn040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Y, Li S, Li R, Xu J, Jin P, Chen L, et al. Genome-wide miRNA screening reveals miR-310 family members negatively regulate the immune response in Drosophila melanogaster via co-targeting Drosomycin. Dev Comp Immunol. 2017;68: 34–45. doi: 10.1016/j.dci.2016.11.014 [DOI] [PubMed] [Google Scholar]
- 86.Cicek IO, Karaca S, Brankatschk M, Eaton S, Urlaub H, Shcherbata HR. Hedgehog signaling strength is orchestrated by the mir-310 cluster of microRNAs in response to diet. Genetics. 2016;202: 1167–1183. doi: 10.1534/genetics.115.185371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kapoor AJ. The role of mircoRNA 310/13 in regulating cellular adhesion in Drosophila. FASEB J. 2013;27: lb433–lb433. [Google Scholar]
- 88.Sanchez-Díaz I, Rosales-Bravo F, Reyes-Taboada JL, Covarrubias AA, Narvaez-Padilla V, Reynaud E, et al. The Esg gene is involved in nicotine sensitivity in Drosophila melanogaster. PLoS One. 2015;10: e0133956 doi: 10.1371/journal.pone.0133956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsurudome K, Tsang K, Liao EH, Ball R, Penney J, Yang JS, et al. The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction. Neuron. 2010;68: 879–893. doi: 10.1016/j.neuron.2010.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dhahbi JM, Atamna H, Li R, Yamakawa A, Guerrero N, Lam HT, et al. MicroRNAs circulate in the hemolymph of Drosophila and accumulate relative to tissue microRNAs in an age-dependent manner. Genomics Insights. 2016;9: 29–39. doi: 10.4137/GEI.S38147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chahine S, O'Donnell MJ. Interactions between detoxification mechanisms and excretion in Malpighian tubules of Drosophila melanogaster. J Exp Biol. 2011;214: 462–468. doi: 10.1242/jeb.048884 [DOI] [PubMed] [Google Scholar]
- 92.Li HM, Buczkowski G, Mittapalli O, Xie J, Wu J, Westerman R, et al. Transcriptomic profiles of Drosophila melanogaster third instar larval midgut and responses to oxidative stress. Insect Mol Biol. 2008;17: 325–339. doi: 10.1111/j.1365-2583.2008.00808.x [DOI] [PubMed] [Google Scholar]
- 93.Vontas JG, Small GJ, Hemingway J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem J. 2001;357: 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lomate P, Sangole K, Sunkar R, Hivrale V. Superoxide dismutase activities in the midgut of Helicoverpa armigera larvae: Identification and biochemical properties of a manganese superoxide dismutase. Open Access Insect Physiol. 2015;5: 13–20. [Google Scholar]
- 95.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105: 1608–1613. doi: 10.1073/pnas.0707594105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131: 1097–1108. doi: 10.1016/j.cell.2007.10.032 [DOI] [PubMed] [Google Scholar]
- 97.Peter MS, Zsofia T, Viktor M, Andras F, Karoly R, Igaz P. MicroRNA target prediction: problems and possible solutions. Curr Bioinform. 2010;5: 81–88. [Google Scholar]
- 98.Van Peer G, De Paepe A, Stock M, Anckaert J, Volders PJ, Vandesompele J, et al. miSTAR: miRNA target prediction through modeling quantitative and qualitative miRNA binding site information in a stacked model structure. Nucleic Acids Res. 2017;45: e51 doi: 10.1093/nar/gkw1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berge JB, Feyereisen R, Amichot M. Cytochrome P450 monooxygenases and insecticide resistance in insects. Philos Trans R Soc Lond B Biol Sci. 1998;353: 1701–1705. doi: 10.1098/rstb.1998.0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou Y, Huang H, Zhang K, Ding X, Jia L, Yu L, et al. miRNA-216 and miRNA-499 target cyb561d2 in zebrafish in response to fipronil exposure. Environ Toxicol Pharmacol. 2016;45: 98–107. doi: 10.1016/j.etap.2016.05.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Each plot represents a miRNA. The X- and Y-axis show the normalized read counts of miRNAs in the two strains respectively.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The small RNA sequencing data sets generated during this study are available in the NCBI Sequence Read Archive (SRA) under accession numbers SRP136631 (https://www.ncbi.nlm.nih.gov/sra/ <https://www.ncbi.nlm.nih.gov/sra/>).