Reduced Anthocyanins in Petioles (RAP) encodes a glutathione S-transferase anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry.
Keywords: Anthocyanin transporter, foliage coloration, Fragaria vesca, fruit coloration, glutathione S-transferase, mutant analysis, strawberry
Abstract
The red color of the foliage and fruit in strawberry comes from anthocyanins stored in the vacuole; however, how this anthocyanin accumulation is regulated remains unclear. A reduced anthocyanin in petioles (rap) mutant was identified in an N-ethyl-N-nitrosourea (ENU) mutagenized population of YW5AF7, a white-fruited variety of the wild strawberry Fragaria vesca. The causative mutation was identified to be a premature stop codon in a glutathione S-transferase (GST) gene. In addition to the foliage coloration, RAP also mediates fruit pigmentation and acts downstream of the fruit-specific transcription factor FvMYB10. Among all eight GST genes in the same subfamily, RAP is most abundantly expressed in the ripening fruit. Expression analysis and transient expression assays demonstrated that RAP is the principal transporter of anthocyanins among the paralogs. Moreover, domain-swap experiments showed that both the N- and C-terminals of RAP are essential for the binding capability of anthocyanins. In addition, transient knock-down of RAP resulted in reduced fruit coloration in cultivated strawberry. Collectively, our results demonstrate that RAP encodes the principal GST transporter of anthocyanins in the strawberry foliage and fruit, and it could be modified to alter the fruit color in strawberry.
Introduction
Cultivated strawberry (Fragaria ananassa) is one of the major fruit crops grown worldwide. The wild diploid F. vesca has emerged as a model plant for cultivated strawberry, as well as other Rosaceae fruit, because of the fact that it has a small stature, a short life cycle, its genome has been sequenced (~240 Mb; Shulaev et al., 2011), and it is easy to transform (Oosumi et al., 2006). In strawberry, the botanical fruits are the small and multiple achenes adhering to the receptacle surface, while the juicy flesh is derived from the enlarged stem tip that forms the receptacle. In recent years, extensive transcriptome datasets generated from F. vesca during flower and fruit development have provided a valuable resource for studying this unique receptacle and its development (Kang et al., 2013; Hollender et al., 2014; Li et al., 2017). Our own research has frequently utilized three F. vesca accessions: the 7th inbred line of ‘Yellow Wonder’ (YW5AF7), ‘Hawaii 4’ (H4), and ‘Ruegen’. While all three accessions have red petioles, only Ruegen develops red fruit, including red receptacles and red achenes. YW5AF7 and H4, in contrast, bear white receptacles and white achenes. The loss of red color in YW5AF7 and H4 was recently shown to result from a natural mutation in the FvMYB10 gene, which encodes a key transcription factor for anthocyanin biosynthesis (Hawkins et al., 2016).
Anthocyanins are a group of water-soluble flavonoid compounds that perform diverse biological functions, such as attracting pollinators and seed dispersers (Schaefer et al., 2004), conferring stress resistance, and prolonging fruit life span (Zhang et al., 2013). Anthocyanins also give rise to the brilliant red strawberry fruit favored by consumers. Similar to other plant species, anthocyanins are synthesized in strawberry by a well-studied pathway with a series of enzymes using phenylalanine as the precursor (Pillet et al., 2015). At the final step, a UDP-glucosyltransferase, such as UGT71K3 (Song et al., 2016), catalyses the glucosylation of the anthocyanins to increase their stability. These anthocyanin biosynthetic enzymes contribute to fruit coloration in strawberry. Most of their coding genes are differentially expressed between the red- and white-fruited strawberry varieties (Zhang et al., 2015; Hawkins et al., 2017), and knock-down of their expression is able to alter the fruit color (Hoffmann et al., 2006; Fischer et al., 2014).
It has been demonstrated in several plant species that the genes coding for anthocyanin biosynthesis enzymes are subjected to transcriptional regulation by the well-studied ‘MBW’ complex. This consists of one MYB transcription factor, one bHLH transcription factor, and one WD-40 protein (Xu et al., 2015). In strawberry, this complex has also been shown to be a master regulator of pigmentation. Overexpression of FaMYB10 in F. ananassa or FvMYB10 in F. vesca results in an accumulation of anthocyanins in the roots, leaves, and fruit (Lin-Wang et al., 2010, 2014), whereas Fa/FvMYB10-RNAi transgenic lines produce white fruit (Lin-Wang et al., 2014; Medina-Puche et al., 2014). As referred to above, a natural SNP that causes an alteration in the amino acids from W (Trp) to S (Ser) at position 12 of FvMYB10 results in white fruit color in the H4 and YW accessions of F. vesca (Hawkins et al., 2016). Hence, the genotype of H4 and YW is myb10, while the genotype of Ruegen is MYB10. In addition, FaMYB1, a different MYB transcription factor, is a transcriptional repressor of anthocyanin biosynthesis genes that acts in the last few steps of the flavonoid pathway (Aharoni et al., 2001). More regulatory transcription factors have been identified by expression correlation analysis in strawberry (Pillet et al., 2015), indicating that our knowledge of the gene network of this pathway is still incomplete.
Anthocyanins are synthesized at the endoplasmic reticulum, at the side that faces the cytoplasm, and are then transported into the vacuole for storage. Several types of mechanism are known to be responsible for anthocyanin transport in plants, including glutathione S-transferases (GSTs), multidrug and toxic extrusion (MATE), ATP-binding cassette (ABC) proteins, and possibly the allergen Fra a 1 (Goodman et al., 2004; Marinova et al., 2007; Gomez et al., 2009; Muñoz et al., 2010; Zhao et al., 2011; Francisco et al., 2013). Of particular interest in relation to the current study are the GSTs (EC 2.5.1.18), which are dimeric enzymes involved in cellular detoxification by conjugating glutathione (GSH) to a variety of electrophilic compounds (Dixon et al., 2002). The functions of plant GSTs include detoxification of xenobiotics as well as responses to biotic and abiotic stresses (Loyall et al., 2000; Agrawal et al., 2002). GSTs comprise a large gene family in plant species, and are soluble and highly abundant in the cytosol (McGonigle et al., 2000; Wagner et al., 2002; Dixon et al., 2009; Jain et al., 2010). Plant GSTs can be divided into five subfamilies, namely phi, tau, theta, zeta, and lambda (Dixon et al., 2002), with those involved in anthocyanin transport belonging to the plant-specific phi subfamily (Kitamura et al., 2004). A typical GST protein contains a conserved GSH-binding site (G-site) located in the N-terminus domain and a C-terminus substrate-binding domain (H-site), with the two being in proximity of each other in a 3-D structure that forms the catalytic site (Dixon et al., 2002).
Among the anthocyanin transporters, GSTs probably play the most important role as loss of their function causes a phenotype with a visible loss of pigment, such as bz2 (Bronze-2) from maize, an9 (Anthocyanin 9) from petunia, fl3 (Flavoniod 3) from carnation, and tt19 (Transparent Testa 19) from Arabidopsis (Marrs et al., 1995; Alfenito et al., 1998; Larsen et al., 2003; Kitamura et al., 2004; Sun et al., 2012). Several studies have carefully examined the roles of TT19 in anthocyanin accumulation in Arabidopsis and provided strong evidence that TT19 acts as a carrier protein for sequestration of anthocyanins from the cytosol into the vacuole (Kitamura et al., 2004; Li et al., 2011; Sun et al., 2012). In fruit crops, GSTs have been associated with fruit or flower pigmentation, such as LcGST4 from lychee, MdGST from apple, and Riant from peach (Cardoso et al., 2012; Cheng et al., 2015; El-Sharkawy et al., 2015; Hu et al., 2016). However, none of them has been studied using genetic approaches. In our current study, a F. vesca mutant produced as a result of an N-ethyl-N-nitrosourea (ENU) chemical mutagenesis of YW was found to accumulate very little anthocyanin in the leaf petioles and was thus named reduced anthocyanins in petioles (rap). Mapping by sequencing revealed that RAP encodes a GST transporter for anthocyanin. Using a combination of different approaches, this study demonstrates that RAP is the pivotal anthocyanin transporter in the foliage and fruit of strawberry, thus providing a promising candidate gene for improving or manipulating fruit and foliage color in cultivated strawberry.
Materials and methods
Plant material and ENU mutant screening
The 7th generation inbred lines of three F. vesca accessions, namely Yellow Wonder 5AF7 (YW5AF7, white-fruited), Ruegen (Ru F7-4, red-fruited), and Hawaii 4 (PI551572, National Clonal Germplasm Repository, USA, white-fruited), were used as wild-types in this study (Slovin et al., 2009; Hawkins et al., 2016). The plants were cultivated in a growth room under a light intensity of 100 μmol m−2 s−1 with a 16/8 h light/dark photoperiod at 22 °C. For ENU mutagenesis, the seeds of YW5AF7 were first soaked in water for 1 d at 4 °C, then treated with 0.4% ENU (N3385, Sigma-Aldrich) for 8 h at room temperature with gentle shaking, and finally rinsed thoroughly 10 times with water (Caruana et al., 2018). After storage at 4 °C for 2 weeks, the seeds were propagated in a greenhouse. Mutants were screened in the M2 generation.
Identification and isolation of RAP via mapping-by-sequencing
The rap mutant in the M3 generation was backcrossed into the parent YW5AF7 to generate an F2 population. In this population, equal amounts of young leaves were pooled from 27 wild-type plants and 18 mutant plants. DNA extraction for the two groups was performed using a CTAB method (Porebski et al., 1997; Oosumi et al., 2006). A total of 6G paired-end reads at 150 bp were generated for each of the two groups using the Illumina HiSeq X Ten platform (Biomarker Technologies, Beijing). The reads were aligned to the v2.0 genome of F. vesca using Bowtie2 (http://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.2.6/) (Tennessen et al., 2014). The SNPs were called by SAMtools (https://sourceforge.net/projects/samtools/files/samtools/). The following criteria were applied to filter the SNPs: 100% presence in the mutant, <50% in the wild-type, and absent in the wild-type parent YW5AF7. In addition, the candidate SNPs had to be located in the exon and cause non-synonymous or nonsense mutations in the protein product. After passing these filtering criteria, the resulting candidate SNPs were further confirmed by PCR-amplification and Sanger sequencing in each individual rap mutant in the F2 population.
Phylogenetic analysis
The protein sequences were downloaded from PLAZA (Proost et al., 2015) or NCBI (https://www.ncbi.nlm.nih.gov/) according to the accession numbers (see Results). Protein sequences of the six mis-annotated GST-encoding genes in F. vesca (RAP, RAP-L1, RAP-L2, RAP-L4, RAP-L5, and RAP-L7) were obtained from the new F. vesca annotation v2.0.a2 (Supplementary Fig. S3 at JXB online) (Li et al., 2018). The sequence alignment was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). An unrooted phylogenetic tree was constructed using MEGA 7 (http://www.megasoftware.net/) with the neighbor-joining statistical method and bootstrap analysis (1000 replicates).
Plasmid construction
The primers used for plasmid construction are listed in Supplementary Table S1. Genomic DNA or cDNA obtained from fruit or young leaves of Ruegen were used for sequence amplification. For complementation rescue, the genomic sequence of RAP from 1114 bp upstream of the translation start site to 1182 bp downstream of the stop codon (3317 bp in total) was amplified, sub-cloned into pDONR221, and then inserted into the binary vector pMDC99. For overexpression, RAP-RFP, MYB10, RAP-L1-7, and the two chimeric genes of RAP and RAP-L1 (Supplementary Fig. S4B) were cloned into either pDONR221 or pENTR1A and inserted into the binary vector pK7WG2D. For RNAi, the entire coding sequence of RAP from 1 to 512 bp was inserted into the binary vector pK7GWIWG2D. To examine the subcellular localization, coding sequences of RAP and RAP-L1-5 were cloned into the EcoRI- and NdeI-digested binary vector pRI101 to fuse with GFP (green fluorescent protein) using the Gibson cloning method. These constructs were transformed into Agrobacterium tumefaciens strain GV3101 for plant transformation.
Stable transformation in Arabidopsis
Arabidopsis transformation was carried out by the floral-dip method (Clough and Bent, 1998). T1 transgenic seeds were screened on half-strength Murashige and Skoog (MS) medium (M5524, Sigma-Aldrich) with 100 mg l–1 kanamycin.
Transient gene expression in strawberry fruit
Transient expression assays in strawberry fruit were performed as described by Hoffmann et al. (2006). Briefly, a single Agrobacterium colony was selected and grown in 2 ml of liquid LB medium until OD600 reached about 0.8–1.0. The culture was then spun down and resuspended in buffer (1×MS, 2% sucrose) to reach an OD600 of exactly 0.8. Fruit at the white stage were injected using a 10-ml syringe. The color phenotype was examined 1 week after the injection, and at least 10 fruit were used for each construct.
Transient gene expression in tobacco leaves and microscopy
Transient expression in tobacco leaves was performed as described by Sparkes et al. (2006). The fluorescence in transformed cells was observed using a confocal microscope (Leica, SP8). GFP was excited at 488 nm and captured at 500–530 nm. Red (RFP) and orange fluorescent proteins (OFP) were excited at 543 nm and captured at 560–630 nm. Chlorophyll autofluorescence was captured at 650–750nm.
Quantitative RT-PCR
Total RNA was extracted using a Plant Total RNA Isolation Kit (Sangon Biotech, Shanghai, China, No. SK8631) following the manufacturer’s instructions. Approximately 1 μg of total RNA was used for cDNA synthesis using a PrimeScriptTM RT reagent kit (TaKaRa, Shiga, Japan, Cat# RR047A). For qPCR, a total volume of 10 μl reaction mixture was used containing 5 μl of 2×SYBR Green master mix (Cat# 172–5124, BioRad), 1 μl of 5× diluted cDNA, 0.25 μl of each primer, and 3.5 μl ddH2O (Supplementary Table S2). Amplification was performed using a QuantStudio 7 Flex system (Applied Biosystems, USA). The amplification program consisted of one cycle of 50 °C for 2 min and 95 °C for 10 min, followed by 50 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 20 s. The fluorescent product was detected at the third step of each cycle. The expression level of each gene was calculated using the 2−∆∆CT method (Livak and Schmittgen, 2001). All analyses were repeated three times using biological replicates.
Measurement of total anthocyanins
Approximately 0.5 g fresh tissue was ground in liquid nitrogen, added to 5 ml of extraction solution (methanol: H2O: formic acid: trifluoroacetic acid, 70:27:2:1), and kept at 4 °C for 12 h in the dark. The supernatant was transferred to a new tube by filtration. The absorbance was measured at 530 and 657 nm by Hoefer Vision (SP-2001). The anthocyanin content was calculated using the following formula: QAnthocyanins = [A530−(0.25×A657)]/M, where QAnthocyanins is the amount of anthocyanins, A530 and A657 are the absorbance at the indicated wavelengths, and M is the fresh weight of the plant material used for extraction (Zhang et al., 2009). All samples were measured as triplicates in three independent biological replicates.
HPLC analysis of anthocyanins in strawberry petioles and fruit
Approximately 0.5 g fresh tissue was ground in liquid nitrogen, added to 2.5 ml of extraction solution (methanol: H2O: hydrochloric acid, 80:20:0.1), and kept at 4 °C for 12 h in the dark. The mixture was centrifuged at 9000 g for 20 min. The supernatant was filtered through a 0.45-μm millipore membrane. HPLC analysis was performed using a Daojing LC-20AT system. Separation was performed using a Develosil-ODS C18, 5-μm, 4.6 × 250-mm column. The mobile phase was 0.1% formic acid in water (solvent A) and methanol (solvent B) at a flow rate of 0.6 ml min–1. The linear gradient of phase B was as follows: 0–10 min, 10–25%; 10–15 min, 25–30%; 15–50 min, 30–50%; 50–60 min, 50–60%; 60–68 min, 60–10%; 68–70 min, 10%. The UV-visible light detector wavelength was set at 510 nm for detecting anthocyanins. Cyanidin (Cy) 3-gluc (Aladdin, 27661-36-5) was used as the authentic standard.
Statistical analyses
For the segregation ratio test of the F2 population, the χ2 value was calculated manually. Statistical analyses were performed using SPSS (IBM SPSS Statistics v22.0).
Results
Identification of a reduced-color mutant rap in F. vesca and RAP gene isolation
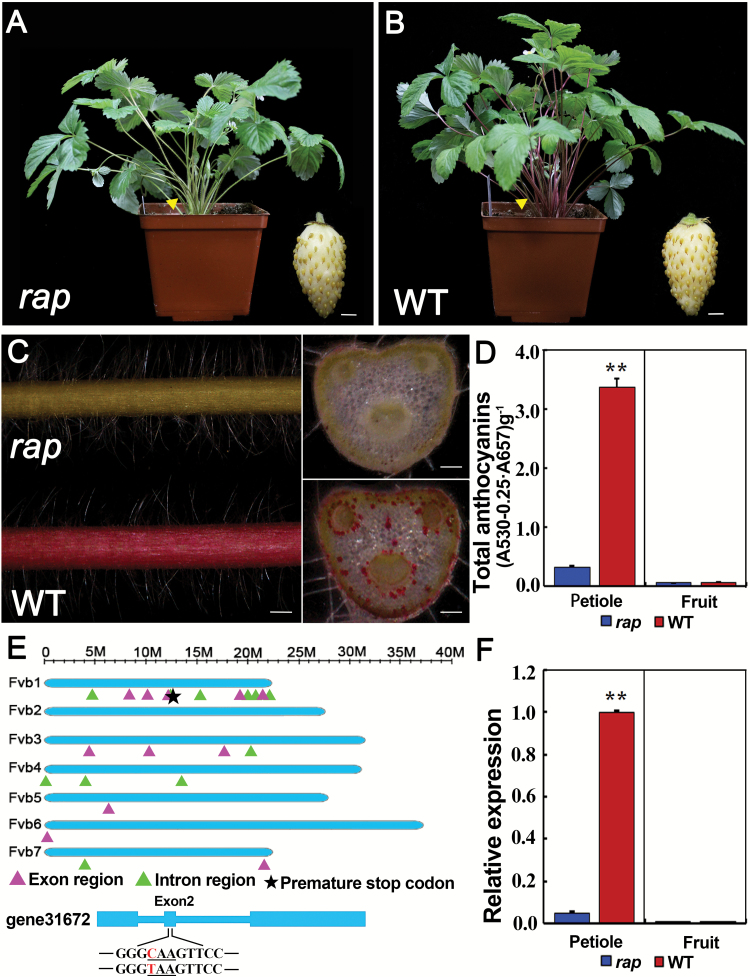
To identify genes essential for the regulation of fruit development in F. vesca, we mutagenized YW5AF7, the 7th inbred line of the strawberry variety Yellow Wonder, using the chemical mutagen ENU (Justice, 2000). In the M2 generation, a mutant with green petioles and leaves was identified and named as rap (reduced anthocyanins in petioles) (Fig. 1A, B). Closer observation showed that the pigmentation of epidermal cells was greatly reduced in the rap mutant, in contrast to the bright-red color in leaf petioles of the wild-type (YW5AF7). Similarly, cross-sections of leaf petioles showed the same change in some cortex cells, especially around the vasculature (Fig. 1C). Consistent with the reduced-pigment phenotype, the total anthocyanin content was remarkably reduced in the leaf petioles of rap (P<0.01, Student’s t-test), and very low in the fruit of both rap and the YW5AF7 wild-type control (Fig. 1D).
Fig. 1.
Phenotypes and gene isolation of the rap mutant. Images of the plant and mature fruit (inset) of (A) the rap mutant and (B) the wild-type (WT) YW5AF7. (C) Images showing the epidermis and cross-sections of a petiole of rap and YW5AF7 (WT). (D) Total anthocyanin contents in the petioles of mature leaves and in mature fruit of rap and YW5AF7 (WT). **, P<0.01, Student’s t-test. (E) Diagram showing the locations of high-quality exonic and intronic SNPs. The chromosome length is indicated at the top. SNPs located in exons or introns are indicated. The star indicates the SNP in gene31672 that causes a premature stop codon. In the model of gene31672, the three underlined nucleotides indicate the codon affected, and the SNP (C to T) concerned is in red. (F) Expression levels of RAP in the petioles of mature leaves and in mature fruit of rap and YW5AF7 (WT) as analysed by qRT-PCR. Gene11892 was used as the internal control. Data are means (±SD) obtained from three technical replicates. **, P<0.01, Student’s t-test. The experiment was repeated three times with similar results. Scale bars: (A, B) 3 mm; (C) 1 mm (left), and 0.3 mm (right).
To examine whether a known gene in the anthocyanin pathway was mutated, the expression levels of catalytic enzyme genes (including CHS, FSH, ANS) and regulatory genes (MYB10, MYB1, bHLH33, and bHLH3) were compared between rap and YW5AF7 in the leaf petioles by qRT-PCR (Supplementary Fig. S1A). A majority of these genes were either similarly or more highly expressed in rap; however, the expression of LAR and MYB10 was significantly reduced (Supplementary Fig. S1B). The coding sequences of MYB10 and LAR were subsequently examined, but neither was found to harbor any mutation. These results indicated that none of the examined genes had the causative mutation for rap.
To identify the causative mutation, the rap mutant was backcrossed with YW5AF7 to generate the F2 mapping population. In this population, the ratio of mutant to wild-type plants was 48:137, which is close to 1:3 (χ2=0.045; χ20.05=3.84). Young leaves from the F2 mutant group (18 plants) and the F2 wild-type group (27 plants) were respectively pooled for whole-genome resequencing. Totals of 43.3 million and 48.5 million paired-end reads at 150 bp were obtained for the mutant and wild-type groups, respectively. We found that 93.7% of the mutant reads and 86.8% of the wild-type reads were aligned to the updated F. vesca genome (Fvb) (Tennessen et al., 2014). SNP calling and filtering (see Methods) identified a total of 64 high-quality SNPs, of which 41 were in the intergenic regions, 11 in introns, three in UTRs, and nine in coding sequences. The locations of the exonic and intronic SNPs in each chromosome are indicated in Fig. 1E. It was notable that more SNPs were located in Chromosome 1 than in other chromosomes. Among the SNPs in the coding region, eight caused either a synonymous or a missense mutation, whilst only one SNP (C to T) in the 2nd exon of gene31672 resulted in a premature stop codon (Fig. 1E, indicated by a star in Chromosome 1). Out of the 31 mapped reads from the genome resequencing data of the mutant group, this position had 31 Ts, i.e. a SNP index of 100%, while the wild-type group had 5 Ts out of the 24 mapped reads, a SNP index of 20.8% (Supplementary Fig. S2). We then examined this SNP individually in 50 F2rap mutants by PCR-amplification and Sanger sequencing, and found that all 50 were homozygous for the mutation. Moreover, the expression level of RAP was greatly reduced in the petioles of rap (Fig. 1F), indicating nonsense-mediated decay for the rap mutant mRNA. Taking the results together, gene31672 was identified as the primary candidate for RAP.
Characterization of RAP and its paralogs in F. vesca
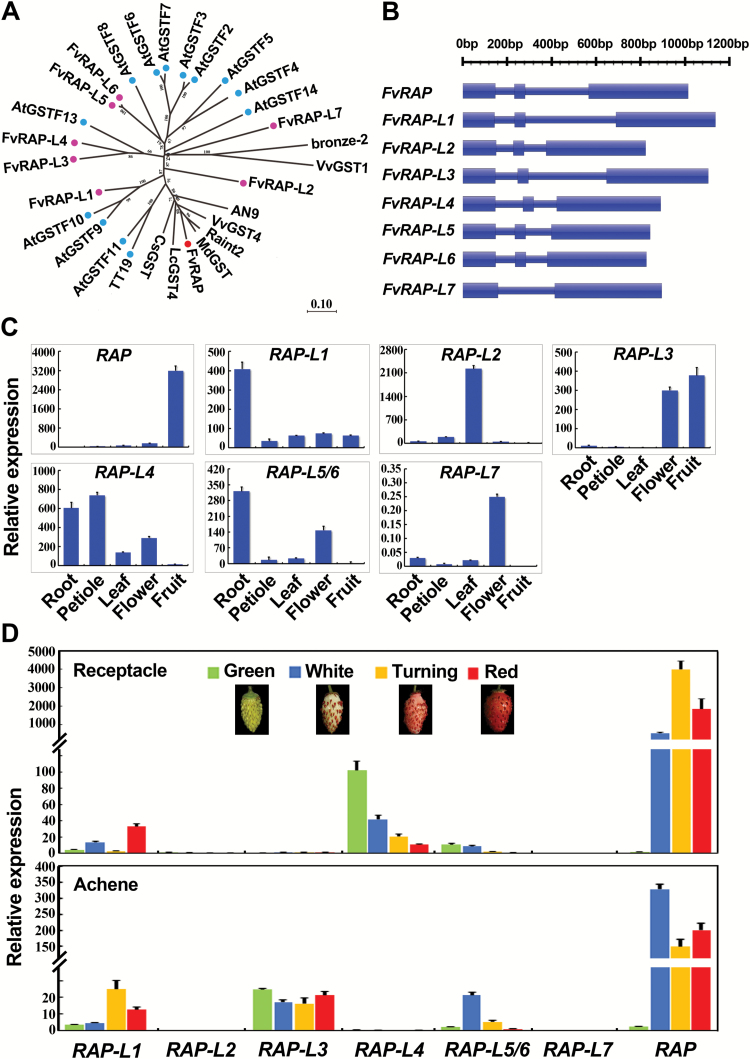
Sequence analysis indicated that RAP encodes a glutathione S-transferase (GST). A BLAST search identified seven other GSTs (Supplementary Fig. S3) in the F. vesca genome that share a high level of similarity to RAP, spanning the entire coding sequence (Supplementary Fig. S4A). They were named as RAP-L1 to RAP-L7 (RAP-Like 1–7). As shown in the phylogenetic tree (Fig. 2A), the eight F. vesca GSTs were closely related to the 13 Arabidopsis GSTs in the phi subfamily (Dixon and Edwards, 2010). The closest homologs of RAP were Riant2 from peach and MdGST from apple, two species in the Rosacea family. RAP-L5 to RAP-L7 were next to each other in Chromosome 2 (Fvb2), perhaps due to gene duplications. The gene models of the RAP paralogs were quite similar: seven of them possessed three exons, with the exception of RAP-L7 that had two exons (Fig. 2B).
Fig. 2.
Phylogenetic analyses and expression patterns of RAP and its paralogs. (A) Phylogenetic tree of RAP and its homologs. A neighbor-joining tree was constructed based on protein sequences of RAP and its homologs from F. vesca, Arabidopsis, and several other species. Gene IDs are shown for genes from F. vesca and Arabidopsis, while the accession numbers in NCBI are shown for others: Riant2 (KT312848), MdGST (AEN84869), AN9 (Y07721), bronze-2 (AAV64226), VvGST4 (AAX81329), VvGST1 (AAN85826), LcGST4 (KT946768), CsGST (ABA42223), FvRAP (gene31672), FvRAP-L1 (gene28763), FvRAP-L2 (gene08595), FvRAP-L3 (gene22014), FvRAP-L4 (gene10549), FvRAP-L5 (gene10550), FvRAP-L6 (gene10551), FvRAP-L7 (gene10552), AtGSTF2 (At4G02520), AtGSTF3 (At2G02930), AtGSTF4 (At1G02950), AtGSTF5 (At1G02940), AtGSTF6 (At1G02930), AtGSTF7 (At1G02920), AtGSTF8 (At2G47730), AtGSTF9 (At2G30860), AtGSTF10 (At2G30870), AtGSTF11 (At3G03190), AtGSTF12/TT19 (At5G17220), AtGSTF13 (At3G62760), and AtGSTF14 (At1G49860). The numbers indicate the bootstrap values calculated from 1000 replicate analyses. (B) Gene models of RAP and its paralogs. Thick bars indicate exons, and thin bars indicate introns. The sequence length is shown at the top. (C) Expression patterns of RAP and its paralogs in the tissues of red-fruited Ruegen as analysed by qRT-PCR. (D) Expression patterns of RAP and its paralogs in fruit receptacles and achenes of Ruegen at four developmental stages, as analysed by qRT-PCR. Gene11892 was used as the internal control in (C) and (D). Data are means (±SD) obtained from three technical replicates. The experiment was repeated for three times with similar results.
The expression patterns of the RAP and RAP-Like genes were examined by qRT-PCR using gene-specific primers (Supplementary Table S2) in the roots, petioles of mature leaves, unfolded leaves, flowers (pooled from entire flower buds at different developmental stages), and mature fruit of the red-fruited Ruegen. The genes exhibited a great diversity in expression patterns (Fig. 2C). RAP was predominantly expressed in fruit, RAP-L1 was more abundant in roots, RAP-L2 was leaf specific, and RAP-L3 was expressed in flowers and fruit. The expression level of RAP-L4 was higher in roots and petioles. RAP-L5/6, the combined expression of RAP-L5 and RAP-L6 due to their high sequence similarity, was expressed in roots and flowers. RAP-L7 was expressed at low levels compared to the other RAP-Like genes, but was found to be more abundant in flowers.
To explore their possible contributions to fruit coloration, the expression trends of RAP and its paralogs in fruit receptacles and achenes were investigated during fruit ripening. Four developmental stages (green, white, turning, and red) were examined. In the fruit receptacles, RAP-L4 was most abundant at the green stage. In the subsequent stages, coinciding with the coloration period, only RAP increased in expression, and this increase was large (Fig. 2D). In the achenes, RAP was also the most abundantly expressed gene during the white, turning, and red stages, while the other RAP-like genes remained at low expression levels at all four stages (Fig. 2D). These results suggested that RAP probably plays more important roles than its homologs in the coloration of fruit receptacles and achenes during F. vesca fruit development.
RAP is the ortholog of TT19 in Arabidopsis
TT19 is the homolog of RAP in Arabidopsis, and has been demonstrated to be an anthocyanin transporter (Sun et al., 2012). To test the role of RAP in anthocyanin transport, 35S::RAP-RFP was transformed into the Arabidopsis mutant tt19-7. A total of 14 independent transgenic lines were obtained with similar phenotypes, and two of them (Line5 and Line10) with high expression levels of RAP validated by qRT-PCR (Supplementary Fig. S5) were chosen for careful characterization. Seeds of wild-type, tt19-7, and the two lines of 35S::RAP-RFP; tt19-7 were germinated on MS medium supplemented with 5% sucrose. At 7 d post germination, hypocotyls of tt19-7 were green, while the 35S::RAP-RFP; tt19-7 seedlings had red hypocotyls identical to the wild-type (Fig. 3A). Consistently, both stems and leaves of the transgenic plants accumulated more anthocyanins than tt19-7 when grown in the growth room without any treatment (Fig. 3B, C). Since RAP can complement tt19-7, this suggested that RAP is an anthocyanin transporter. However, the brown color of seed coats was not rescued in the 35S::RAP-RFP transgenic lines (Fig. 3D), suggesting that RAP may have distinct functions from TT19 during seed-coat pigmentation.
Fig. 3.
Phenotypes of the 35S::RAP-RFP transgenic lines in Arabidopsis tt19-7. (A) Images of 7-d-old seedlings grown on MS medium supplemented with 5% sucrose. (B) Images of the adult plants at bolting. (C) Images of leaves and stems from the adult plants in (B). (D) Images of fresh seeds. Images for the wild-type (WT, Col), tt19-7, and two transgenic lines of 35S::RAP-RFP (L5, L10) in the tt19-7 background are shown. Scale bars: (A–C) 5 mm; (D) 1 mm.
RAP is essential for fruit coloration and acts downstream of FvMYB10 in F. vesca
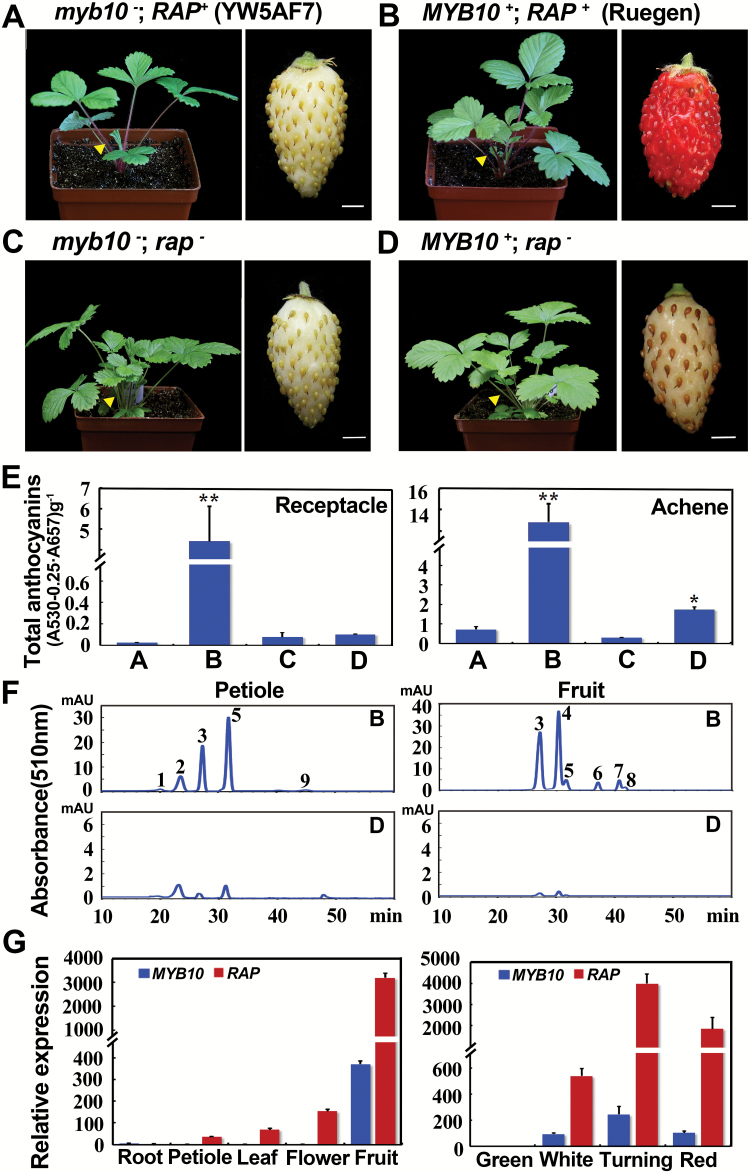
The rap mutant was isolated from the white-fruited and red-petiole parent YW5AF7 (genotype: myb10−) and hence rap is actually a myb10−rap− double-mutant (Fig. 4A, C). This makes it impossible to determine the role of RAP in fruit coloration. To examine whether RAP is involved in fruit pigmentation, we sought to isolate the single rap mutant. rap was crossed with the red-fruited Ruegen (genotype: MYB10+RAP+) (Fig. 4B). In the F2 population, the plants with green petioles, like those of rap (Fig. 4C), were selected. Among these green-petiole plants, the genotype of myb10 was identified via sequencing of the SNP in FvMYB10/gene31413 (Hawkins et al., 2016). Most plants were still myb10−rap− due to the linkage of these two genes on Chromosome 1, and only one MYB10+rap− plant was identified out of 66 green-petiole F2 plants. The isolation of the rap single mutant (MYB10+rap−) enabled us to characterize the effects of RAP without the interference of the background myb10 mutation.
Fig. 4.
The function of RAP in fruit coloration. Images showing the plant and mature fruit of (A) YW5AF7, (B) Ruegen, (C) rap, and (D) MYB10+rap−. The genotypes are indicated above each set of images. Yellow arrowheads point to petioles. (E) Total anthocyanin contents in fruit receptacles and achenes collected from the plants shown in (A–D). The values of B–D are compared to A: *, P<0.05, **, P<0.01, Student’s t-test. (F) HPLC chromatograms of anthocyanins in petioles of mature leaves and mature fruit of Ruegen (as shown in B) and MYB10+rap− (as shown in D). The y-axis shows the absorbance at 510 nm. Peak 1, cyanidin-3,5-diglucoside; peak 2, peonidin-3,5-diglucoside; peak 3, cyanidin-3-glucoside; peak 4, pelargonidin-3-glucoside; peak 5, peonidin-3-glucoside; peak 6, cyanidin-3-malonylglucoside; peak 7, pelargonidin-3-acetylhexoside; peak 8, peonidin-3-malonylglucoside; peak 9, unknown. (G) Co-expression of RAP and MYB10 in different tissues (left) and in fruit receptacles at four developmental stages of Ruegen (right) as analysed by qRT-PCR. Gene11892 was used as the internal control. Data are means (±SD) obtained from three technical replicates. The experiment was repeated three times with similar results. Scale bars in (A–D) are 3 mm.
Mature fruit of MYB10+rap− had white receptacles that were identical to YW5AF7, while the mature achenes were light pink, which was much less coloration than we observed in the achenes of Ruegen (Fig. 4B, D). This suggested that rap alone can affect fruit coloration both in the receptacle and the achenes. To characterize the phenotypes quantitatively, we measured the total anthocyanin contents of fruit receptacles and achenes of the MYB10+rap− plant. The content in the receptacles of MYB10+rap− was as low as that in myb10−rap−, but the content in the achenes of MYB10+rap− was slightly higher than that of myb10−rap− (P<0.05, Student’s t-test). Nevertheless, the anthocyanin contents in the receptacle and achenes of MYB10+rap− were significantly reduced when compared with Ruegen (MYB10+RAP+) (Fig. 4E). Previous studies of Ruegen and YW5AF7 have shown that anthocyanins in petioles and fruit contain a total of nine prominent compounds, eight of which have been identified (Xu et al., 2014). Using similar methods, we were able to observe and identify the same number of HPLC peaks in Ruegen and found that all the peaks were significantly reduced in MYB10+rap− (Fig. 4F).
This genetic experiment indicated that both MYB10 and RAP are required for fruit coloration. To determine the relationship between these two genes, we examined the expression of RAP and MYB10 using qRT-PCR and found that RAP expression nicely correlated with MYB10 expression in different tissues/organs and at different stages of fruit development (Fig. 4G). Moreover, proper expression of RAP relied on the wild-type MYB10, as shown in the three F. vesca varieties. Specifically, YW5AF7 and H4 produced white fruit with nearly no anthocyanin, while Ruegen had red fruit with high anthocyanin content (Supplementary Fig. S6A). The qRT-PCR results showed that RAP had low expression levels in YW5AF7 and H4, the two white-fruited varieties containing the myb10 mutation, and was highly expressed in Ruegen, the red-fruited variety with the wild-type MYB10 (Supplementary Fig. S6B), suggesting that MYB10 probably regulates RAP expression in fruit either directly or indirectly. Of note, the expression level of RAP was comparable in the petioles of YW5AF7, H4, and Ruegen (Supplementary Fig. S6B), suggesting the existence of a different MYB that may act in the petiole for RAP expression.
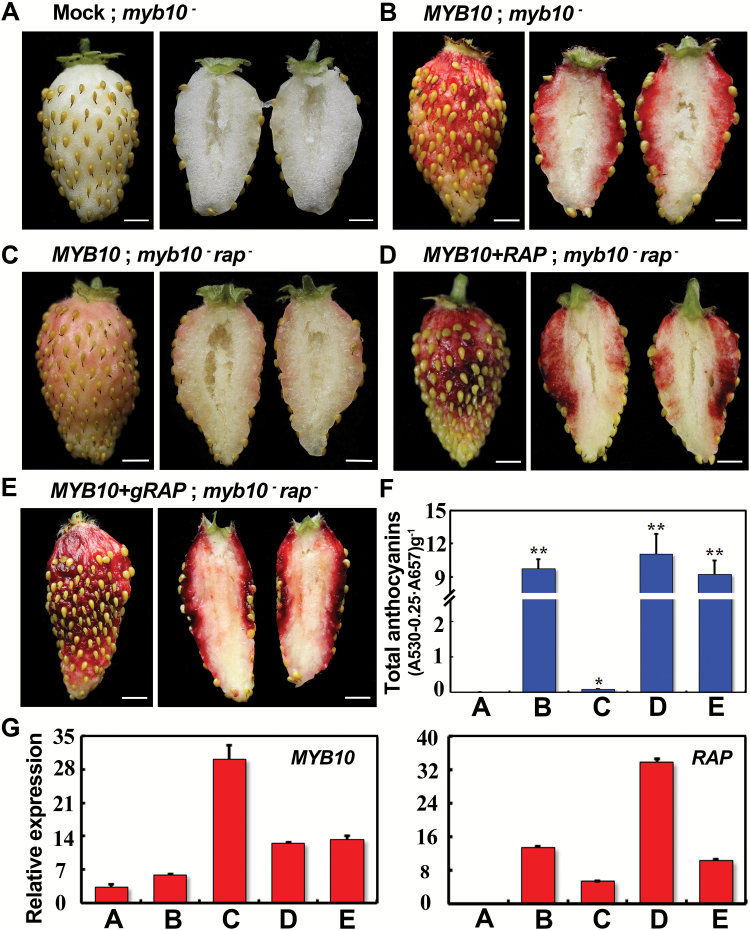
Expression of RAP rescued fruit pigmentation of rap in a transient assay
It has previously been shown that fruit pigmentation in YW5AF7 (genotype: myb10) can be rescued by overexpressing a functional FvMYB10 via agro-infiltration (Hawkins et al., 2016), providing a quick assay to test gene function in anthocyanin accumulation. Using this assay, we found that 35S::FvMYB10 indeed reliably restored fruit coloration (Fig. 5A, B). Accordingly, 35S::FvMYB10 was infiltrated into the rap fruit, but they only exhibited a light-pink color (Fig. 5C), indicating that a functional RAP was necessary to restore the coloration. When 35S::RAP-RFP and 35S::MYB10 were simultaneously infiltrated into the rap fruit, they turned bright red (Fig. 5D). When the genomic fragment of RAP (gRAP::RAP) was transiently transformed into the rap fruis combined with 35S::MYB10, they also turned red (Fig. 5E). The anthocyanin contents in these infiltrated fruit consistently correlated with the color phenotype (Fig. 5F). The expression levels of MYB10 and RAP in these infiltrated fruit were confirmed by qRT-PCR (Fig. 5G). Of note, RAP was significantly induced by overexpression of MYB10 (Fig. 5G, B versus A). Overall, the complementation test proved that gene31672/GST is indeed the RAP gene.
Fig. 5.
Rescue of the fruit coloration in rap in a transient assay. (A) A fruit of YW5AF7 (genotype: myb10−) infiltrated with buffer. (B) A fruit of YW5AF7 overexpressing MYB10 by agro-infiltration. (C) A fruit of rap overexpressing MYB10 by agro-infiltration. (D) A fruit of rap overexpressing both MYB10 and RAP by agro-infiltration. (E) A fruit of rap expressing both gRAP::RAP and 35S::MYB10 by agro-infiltration. In each case, one representative example is shown from at least 10 infiltrated fruit. (F) Total anthocyanin contents in the fruit shown in (A–E). The values of B–E are compared to A: *, P<0.05, **, P<0.01, Student’s t-test. (G) Expression levels of MYB10 and RAP in the fruit shown in (A–E) as analysed by qRT-PCR. Gene11892 was used as the internal control. Data are means (±SD) obtained from three technical replicates. The experiment was repeated three times with similar results. Scale bars in (A–E) are 3 mm.
Comparison of gene functions between RAP and its paralogs during fruit coloration
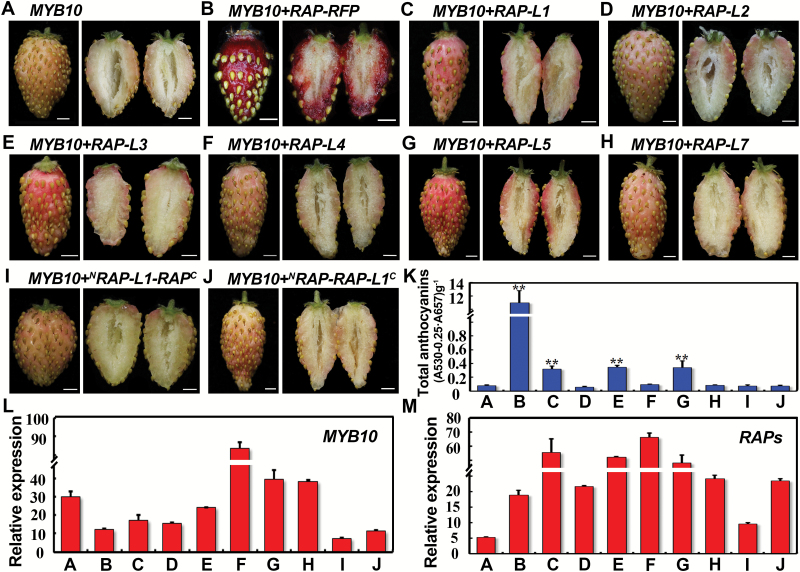
Expression analysis showed that RAP was much more abundantly expressed than its other paralogs during fruit ripening in F. vesca (Fig. 2D). As RAP and its paralogs share a high level of sequence similarity spanning the entire gene (Supplementary Fig. S4A), it is possible that these RAP paralogs have the same anthocyanin binding and transport capacities as RAP. To test this hypothesis, RAP-L1-5 and RAP-L7 driven by the 35S constitutive promoter were each infiltrated together with 35S::MYB10 into the rap fruit. The protein sequence of RAP-L6 is very similar to that of RAP-L5 (only 9 out of 214 amino acids are different), and hence no results are shown for RAP-L6. 35S::MYB10 only served as the negative control and 35S::RAP-RFP together with 35S::MYB10 served as the positive control (Fig. 6A, B). RAP-L1, 3, and 5 induced a deeper pink color compared to the negative control, while the other three RAP-Ls had no obvious effect (Fig. 6C–H). Nevertheless, none of the RAP-Like genes gave the red color to the same extent as RAP (Fig. 6K). To distinguish which part of RAP is more important in determining anthocyanin-binding capacities, the N- and C-terminals were switched between RAP and RAP-L1 (the closest paralog) to create two chimeric proteins (Supplementary Fig. S4B). Transient assays showed that neither of them could promote anthocyanin accumulation (Fig. 6I–K), suggesting that both terminals are necessary for the anthocyanin-binding capacity of RAP. The expression levels of MYB10 and the RAPs were confirmed by qRT-PCR (Fig. 6L, M).
Fig. 6.
Transient expression of RAP and its paralogs in fruit of rap. (A) A fruit of rap overexpressing MYB10 by agro-infiltration. (B–J) Fruits of rap transiently overexpressing MYB10 together with RAP, RAP-Like genes, or chimeric genes between RAP and RAP-L1 (Supplementary Fig. S4B), as indicated. At least 10 fruit were infiltrated for each construct with similar results. (K) Total anthocyanin contents in the fruit shown in (A–J). The values of B–J are compared to A: **, P<0.01, Student’s t-test. (L) Expression levels of MYB10 in the fruit shown in (A–J) as analysed by qRT-PCR. (M) Expression levels of RAPs in the fruit shown in (A–J) as analysed by qRT-PCR. For (A) and (B), RAP was examined; for (C–J), each overexpressed RAP-Like gene was examined. Gene11892 was used as the internal control. Data are means (±SD) obtained from three technical replicates. The experiment was repeated for three times with similar results. Scale bars in (A–J) are 3 mm.
It was shown previously that TT19 is soluble and is localized in the cytosol and on the tonoplast (Sun et al., 2012). To examine the subcellular localization of RAP and its paralogs, RAP and RAP-L1-5 were fused with GFP and transiently expressed in tobacco leaves. We found that RAP, RAP-L1, and RAP-L3-5 (data not shown) were localized in the cytosol and nuclei, but they did not overlap with the endoplasmic reticulum marker (Supplementary Fig. S7A, C) (Nelson et al., 2007). In addition, RAP was partially co-localized with the tonoplast marker CBL6-OFP (Supplementary Fig. S7B) (Batistic et al., 2010). Among the proteins examined, RAP-L2 was notable in that it was primarily localized in chloroplasts (Supplementary Fig. S7D). Both the transient functional assay and the subcellular localization highlight the functional divergence of RAP and its paralogs in F. vesca.
Transient knock-down of RAP reduces fruit coloration in cultivated strawberry
To test whether RAP is essential for fruit coloration in F. vesca, transient RNAi was performed to knock-down RAP in the Ruegen fruit, and it resulted in white fruit receptacles (Supplementary Fig. S8A). As a control, RNAi of gene30464, which encodes a transcription factor not expressed in fruit, did not affect the fruit color (Supplementary Fig. S8B). To determine whether RAP is also important for anthocyanin accumulation in cultivated strawberry (F. ananassa), the two constructs were each transiently infiltrated into the fruit of Sweet Charlie, a popular strawberry cultivar. Similar to the results in Ruegen (F. vesca), transient knock-down of RAP also dramatically reduced fruit coloration in the cultivated strawberry (Supplementary Fig. S8C, D). The anthocyanin content was much lower in RAP-RNAi fruit than controls in both the wild and cultivated strawberry (Supplementary Fig. S8E, F), which correlated with significantly reduced RAP transcript levels (Supplementary Fig. S8G, H). Therefore, RAP is both a good candidate gene for genetic manipulation and a candidate marker for breeding aimed at improving fruit color in cultivated strawberry.
Discussion
Fruit color is an important trait of fruit quality, and color of foliage is also valuable for ornamental plants. Accumulation of anthocyanins not only gives rise to bright colors in plants, but also benefits human health; hence the regulation of anthocyanin biosynthesis is of great interest. It has been shown that GSTs play important roles in anthocyanin accumulation as a result of the characterization of their loss-of-function mutants in maize, petunia, and Arabidopsis (Marrs et al., 1995; Mueller et al., 2000; Kitamura et al., 2004). Hence considerable efforts have been made in exploring the functions of GSTs with regards to fruit pigmentation in grape, apple, and lychee (Conn et al., 2008; El-Sharkawy et al., 2015; Hu et al., 2016). One GST named Riant in peach was found to be responsible for the variegated coloration of petals (Cheng et al., 2015). However, none of these GSTs in fruit crops have been thoroughly studied through genetic approaches, with research being hindered by the difficulties in genetic manipulation of fruit trees. Here, we isolated one green-foliage mutant in wild diploid strawberry, F. vesca, which was shown to be caused by a defective GST gene. This rap mutant is valuable for studying anthocyanin transport in fruit crops.
GSTs form a large gene family. For example, there are 53 GSTs in Arabidopsis (Sappl et al., 2009), 79 in rice (Jain et al., 2010), and 90 in tomato (Islam et al., 2017). GSTs are grouped into several subfamilies. A total of 13 belong to the phi subfamily in Arabidopsis, which is the second most numerous subfamily. Seven FvGSTs together with RAP constitute the phi subfamily in F. vesca. Since a single rap mutation dramatically lowered the anthocyanin level in the fruit and petiole of F. vesca, there is clearly a lack of functional redundancy among the GST genes with roles in anthocyanin transportation in strawberry. Our transient expression results using 35S-driven RAP and RAP family members indicated that the coding sequences are important to their different functions. The differences in coding sequence may also cause distinct subcellular localization that contributes to their functional divergence.
FvMYB10 codes for a master regulatory transcription factor in anthocyanin biosynthesis in strawberry fruit (Lin-Wang et al., 2010, 2014; Medina-Puche et al., 2014). Recently, one naturally occurring SNP in FvMYB10 was found to be responsible for the fruit color of different F. vesca varieties (Hawkins et al., 2016). Noticeably, both red- and white-fruited F. vesca varieties have red petioles, suggesting a minor role of FvMYB10 in anthocyanin accumulation in petioles. Consistent with this observation, the petioles of H4, YW5AF7, and Ruegen have comparable anthocyanin contents and similar profiles of anthocyanin compounds when examined by HPLC (Xu et al., 2014). Moreover, RNAi mediated knock-down of FvMYB10 in red-fruited varieties leads to white fruit but does not change the petiole color (Lin-Wang et al., 2014). Taken together, this suggests that FvMYB10 only plays crucial roles in fruit pigmentation rather than in foliage coloration. The MBW complex is a regulatory paradigm of anthocyanin biosynthesis, and other MYB transcription factors responsible for anthocyanin accumulation in strawberry petioles are yet to be identified.
RAP was significantly induced by overexpression of MYB10 and dramatically reduced in the fruit of myb10 varieties (YW5AF7 and H4) (Fig. 5, Supplementary Fig. S6; Lin-Wang et al., 2014; Härtl et al., 2017), suggesting that RAP expression is regulated by MYB10. Interestingly, the myb10−rap− double-mutant infiltrated with 35S::MYB10 still could not develop red color, indicating that the function of MYB10 in fruit pigmentation depends on wild-type RAP. Based on these results, we conclude that not only is RAP transcriptionally regulated by MYB10 but also that it acts downstream of MYB10 to mediate its effect in fruit coloration. This is consistent with the role of RAP/GST in pigment transport and that of MYB10 in pigment gene biosynthesis. Earlier work in lychee showed that LcGST4 could be up-regulated by LcMYB1 in the dual luciferase assay in tobacco (Hu et al., 2016). Stronger evidence for a direct transcriptional activation of the GST transporter by MYB10 requires further experiments, such as testing the binding of MYB10 to the promoter sequence of RAP containing the C1 motif/MYB binding site (TAACTG, Supplementary Fig. S9) (Bodeau and Walbot, 1996).
In this study we found that RAP was developmentally regulated at the transcriptional level during fruit coloration. As anthocyanin levels are up-regulated in response to different stresses, the GSTs involved in anthocyanin accumulation may also be responsive to both internal and external factors. Consistent with this idea, jasmonic acid elicits an obvious increase of anthocyanin content together with induction of GSTs in grape cell suspension cultures (Conn et al., 2008). LcGST4 is significantly induced by ABA treatment and removing fruit from bagging (Hu et al., 2016). In apple, a transcription factor in light signaling (MdHY5) can directly bind to the G-box motif (CACGTG) in the promoter of MdMYB10 (An et al., 2017), indicating the importance of the G-box in light-mediated regulation of gene expression during anthocyanin production. We found that the promoter of RAP also possesses a G-box motif (238 bp upstream of the translational start codon, Supplementary Fig. S9). These findings suggest that GSTs may be a common downstream target of different regulatory pathways in anthocyanin accumulation.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Anthocyanin biosynthetic genes in F. vesca and their expression levels in rap.
Fig. S2. Integrative Genomics Viewer (IGV; http://software.broadinstitute.org/software/igv/) depiction of the causative SNP of rap in gene31672.
Fig. S3. Genomic sequences of RAP and its paralogs.
Fig. S4. Amino acid sequence alignment of RAP and its paralogs.
Fig. S5. Genotyping of 35S::RAP-RFP transgenic lines in Arabidopsis tt19-7.
Fig. S6. Total anthocyanin contents and expression levels of RAP in petioles and fruit of the three F. vesca varieties.
Fig. S7. Subcellular localization of RAP and its paralogs in tobacco epidermal cells.
Fig. S8. Transient knock-down of RAP reduces fruit coloration in F. vesca and F. ananassa.
Fig. S9. Promoter sequence of RAP.
Table S1. List of primers used for making constructs.
Table S2. List of primers used for qRT-PCR.
Author contributions
HL, CK, and ZL conceived and designed the experiments; HL, CD, YL, and JF performed the experiments; CK, ZL, and HL wrote the paper. All the authors have read and approved the paper.
Acknowledgements
The authors would like to thank Dr Courtney Hollender and Ms Aviva Geretz for the ENU mutagenesis at the University of Maryland, Dr Jirong Huang for providing Arabidopsis tt19-7 seeds, Dr Pengwei Wang for providing the endoplasmic reticulum marker, Dr Zhiyong Pan for providing the tonoplast marker, Dr Shuzhen Yang for technical support with HPLC, and Dr Yujin Hao and Dr Guogui Ning for helpful discussions and suggestions. This work was supported by the National Natural Science Foundation of China (31572098 and 31772274 to CK), US NSF grant (MCB0923913 to ZL), the Scientific & Technological Self-innovation Foundation of Huazhong Agricultural University (2014RC005 to ZL and 2014RC017 to CK), and the Fundamental Research Funds for the Central Universities (2662015BQ031 to CD).
References
- Agrawal G, Jwa N-S, Rakwal R. 2002. A pathogen-induced novel rice (Oryza sativa L.) gene encodes a putative protein homologous to type II glutathione S-transferases. Plant Science 163, 1153–1160. [Google Scholar]
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP. 2001. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal 28, 319–332. [DOI] [PubMed] [Google Scholar]
- Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V. 1998. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. The Plant Cell 10, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JP, Qu FJ, Yao JF, Wang XN, You CX, Wang XF, Hao YJ. 2017. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticulture Research 4, 17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J. 2010. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. The Plant Journal 61, 211–222. [DOI] [PubMed] [Google Scholar]
- Bodeau JP, Walbot V. 1996. Structure and regulation of the maize Bronze2 promoter. Plant Molecular Biology 32, 599–609. [DOI] [PubMed] [Google Scholar]
- Cardoso S, Lau W, Eiras Dias J, Fevereiro P, Maniatis N. 2012. A candidate-gene association study for berry colour and anthocyanin content in Vitis vinifera L. PLoS ONE 7, e46021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana JC, Sittmann JW, Wang W, Liu Z. 2018. Suppressor of runnerless encodes a DELLA protein that controls runner formation for asexual reproduction in strawberry. Molecular Plant 11, 230–233. [DOI] [PubMed] [Google Scholar]
- Cheng J, Liao L, Zhou H, Gu C, Wang L, Han Y. 2015. A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. Journal of Experimental Botany 66, 7227–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conn S, Curtin C, Bézier A, Franco C, Zhang W. 2008. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. Journal of Experimental Botany 59, 3621–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. 2010. Glutathione transferases. The Arabidopsis Book 8, e0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Hawkins T, Hussey PJ, Edwards R. 2009. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. Journal of Experimental Botany 60, 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. 2002. Plant glutathione transferases. Genome Biology 3, reviews3004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy I, Liang D, Xu K. 2015. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. Journal of Experimental Botany 66, 7359–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer TC, Mirbeth B, Rentsch J, Sutter C, Ring L, Flachowsky H, Habegger R, Hoffmann T, Hanke MV, Schwab W. 2014. Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria × ananassa). New Phytologist 201, 440–451. [DOI] [PubMed] [Google Scholar]
- Francisco RM, Regalado A, Ageorges A, et al. . 2013. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. The Plant Cell 25, 1840–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C, Terrier N, Torregrosa L, et al. . 2009. Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiology 150, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, Casati P, Walbot V. 2004. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. The Plant Cell 16, 1812–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtl K, Denton A, Franz-Oberdorf K, Hoffmann T, Spornraft M, Usadel B, Schwab W. 2017. Early metabolic and transcriptional variations in fruit of natural white-fruited Fragaria vesca genotypes. Scientific Reports 7, 45113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C, Caruana J, Li J, Zawora C, Darwish O, Wu J, Alkharouf N, Liu Z. 2017. An eFP browser for visualizing strawberry fruit and flower transcriptomes. Horticulture Research 4, 17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C, Caruana J, Schiksnis E, Liu Z. 2016. Genome-scale DNA variant analysis and functional validation of a SNP underlying yellow fruit color in wild strawberry. Scientific Reports 6, 29017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W. 2006. RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: a rapid assay for gene function analysis. The Plant Journal 48, 818–826. [DOI] [PubMed] [Google Scholar]
- Hollender CA, Kang C, Darwish O, Geretz A, Matthews BF, Slovin J, Alkharouf N, Liu Z. 2014. Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiology 165, 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Zhao J, Lai B, Qin Y, Wang H, Hu G. 2016. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Reports 35, 831–843. [DOI] [PubMed] [Google Scholar]
- Islam S, Rahman IA, Islam T, Ghosh A. 2017. Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: gaining an insight to their physiological and stress-specific roles. PLoS ONE 12, e0187504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Ghanashyam C, Bhattacharjee A. 2010. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genomics 11, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ. 2000. Capitalizing on large-scale mouse mutagenesis screens. Nature Reviews Genetics 1, 109–115. [DOI] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z. 2013. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. The Plant Cell 25, 1960–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A. 2004. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. The Plant Journal 37, 104–114. [DOI] [PubMed] [Google Scholar]
- Larsen ES, Alfenito MR, Briggs WR, Walbot V. 2003. A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell Reports 21, 900–904. [DOI] [PubMed] [Google Scholar]
- Li X, Gao P, Cui D, Wu L, Parkin I, Saberianfar R, Menassa R, Pan H, Westcott N, Gruber MY. 2011. The Arabidopsis tt19-4 mutant differentially accumulates proanthocyanidin and anthocyanin through a 3′ amino acid substitution in glutathione S-transferase. Plant, Cell & Environment 34, 374–388. [DOI] [PubMed] [Google Scholar]
- Li Y, Dai C, Hu C, Liu Z, Kang C. 2017. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. The Plant Journal 90, 164–176. [DOI] [PubMed] [Google Scholar]
- Li Y, Wei W, Feng J, Luo H, Pi M, Liu Z, Kang C. 2018. Genome re-annotation of the wild strawberry Fragaria vesca using extensive illumina- and SMRT-based RNA-seq datasets. DNA Research 25, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. 2010. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biology 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, McGhie TK, Wang M, Liu Y, Warren B, Storey R, Espley RV, Allan AC. 2014. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Frontiers in Plant Science 5, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Loyall L, Uchida K, Braun S, Furuya M, Frohnmeyer H. 2000. Glutathione and a UV light-induced glutathione S-transferase are involved in signaling to chalcone synthase in cell cultures. The Plant Cell 12, 1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M. 2007. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. The Plant Cell 19, 2023–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V. 1995. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. [DOI] [PubMed] [Google Scholar]
- McGonigle B, Keeler SJ, Lau SM, Koeppe MK, O’Keefe DP. 2000. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiology 124, 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, Hoffmann T, Ring L, Rodríguez-Franco A, Caballero JL, Schwab W, Muñoz-Blanco J, Blanco-Portales R. 2014. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. Journal of Experimental Botany 65, 401–417. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Goodman CD, Silady RA, Walbot V. 2000. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiology 123, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz C, Hoffmann T, Escobar NM, Ludemann F, Botella MA, Valpuesta V, Schwab W. 2010. The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Molecular Plant 3, 113–124. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Oosumi T, Gruszewski HA, Blischak LA, Baxter AJ, Wadl PA, Shuman JL, Veilleux RE, Shulaev V. 2006. High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta 223, 1219–1230. [DOI] [PubMed] [Google Scholar]
- Pillet J, Yu HW, Chambers AH, Whitaker VM, Folta KM. 2015. Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. Journal of Experimental Botany 66, 4455–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter 15, 8–15. [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K. 2015. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Research 43, D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Harvey Millar A, Singh KB. 2009. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. The Plant Journal 58, 53–68. [DOI] [PubMed] [Google Scholar]
- Schaefer HM, Schaefer V, Levey DJ. 2004. How plant–animal interactions signal new insights in communication. Trends in Ecology & Evolution 19, 577–584. [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, et al. . 2011. The genome of woodland strawberry (Fragaria vesca). Nature Genetics 43, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovin JP, Schmitt K, Folta KM. 2009. An inbred line of the diploid strawberry Fragaria vesca f. semperflorens for genomic and molecular genetic studies in the Rosaceae. Plant Methods 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Zhao S, Hong X, Liu J, Schulenburg K, Schwab W. 2016. A UDP-glucosyltransferase functions in both acylphloroglucinol glucoside and anthocyanin biosynthesis in strawberry (Fragaria × ananassa). The Plant Journal 85, 730–742. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li H, Huang JR. 2012. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Molecular Plant 5, 387–400. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Govindarajulu R, Ashman TL, Liston A. 2014. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biology and Evolution 6, 3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F. 2002. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Molecular Biology 49, 515–532. [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends in Plant Science 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Xu W, Peng H, Yang T, Whitaker B, Huang L, Sun J, Chen P. 2014. Effect of calcium on strawberry fruit flavonoid pathway gene expression and anthocyanin accumulation. Plant Physiology and Biochemistry 82, 289–298. [DOI] [PubMed] [Google Scholar]
- Zhang W, Ning G, Lv H, Liao L, Bao M. 2009. Single MYB-type transcription factor AtCAPRICE: a new efficient tool to engineer the production of anthocyanin in tobacco. Biochemical and Biophysical Research Communications 388, 742–747. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, De Stefano R, et al. . 2013. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Current Biology 23, 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li W, Dou Y, Zhang J, Jiang G, Miao L, Han G, Liu Y, Li H, Zhang Z. 2015. Transcript quantification by RNA-Seq reveals differentially expressed genes in the red and yellow fruits of Fragaria vesca. PLoS ONE 10, e0144356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huhman D, Shadle G, He XZ, Sumner LW, Tang Y, Dixon RA. 2011. MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. The Plant Cell 23, 1536–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.