Abstract
Increasing number of studies have indicated aberrant microRNA (miRNA) expression could affect normal biological progress in non-small cell lung cancer (NSCLC) cells. This study was performed to evaluate the biologic functions of microRNA-224 (miR-224) in NSCLC. Real-time PCR was performed to evaluate the expression of miR-224 and Homeobox D10 (HOXD10) in NSCLC cell lines and tissues. Transwell assays were performed to investigate the function of miR-224 on NSCLC cell migration and invasion. Moreover, western blotting and luciferase assays were used to investigate HOXD10 as miR-224 downstream targets. miR-224 is increased in NSCLC metastatic tissues and cell lines. Increased miR-224 expression promoted NSCLC cell migration and invasion, while low miR-224 expression suppressed NSCLC cell migration and invasion. Furthermore, HOXD10 was targeted directly by miR-224 in NSCLC cells. Moreover, we found that HOXD10 was a functional target and influenced tumour-inductive functions of miR-224 on progression of NSCLC. These findings suggest that miR-224 may be used in the treatment of NSCLC. Targeting this novel strategy, miR-224/HOXD10 axis may be helpful as promising biomarker and therapeutic method to control NSCLC cell metastasis.
Keywords: miR-224, HOXD10, invasion and metastasis, non-small cell lung cancer
Introduction
Lung cancer remains the leading cause of cancer-associated mortality (1). Non-small cell lung cancer (NSCLC) develop as the predominant form, and approximately accounts for 85% of all lung cancer related deaths (2), mainly consisting of three subtypes: adenocarcinoma, large cell cancer, and squamous cell cancer (3,4). Regardless of great improvements in surgical techniques and chemotherapy, the prognosis for NSCLC remains inconclusive, the 5-year survival rate of is approximately 15% (5,6). Therefore, an improved comprehension of the pathophysiological functions and mechanisms involved in the NSCLC development appears to be pivotal for blossoming novel and effective therapeutic approaches.
MicroRNAs (miRNAs) are small non-coding RNAs, ranging in length from 18 to 24 nucleotides, base binding with target mRNAs in the 3′-untranslated region (3′-UTR) (7), causing mRNA degradation, cleavage, translational repression or activation (8, 9). Increasing research has indicated that miRNAs are regulators in cancer, that might function either as suppressors or activators of cancer development by regulating different targets (10). Furthermore, many studies have indicated that the aberrant expression of various miRNAs could affect normal biological progress of NSCLC cells that are involved in tumor invasion and metastasis (11). Moreover, previous studies have reported that miR-224 facilitated tumor progression to enhanced invasion and growth in NSCLC and implicated pathogenesis of lung cancer (12,13). However, the specific functional molecular mechanisms of miR-224 in the mediation of NSCLC cancer process are still elusive, and the potential mechanism of miR-506 as a therapeutic target for NSCLC has not been fully evaluated.
The Homeobox D10 (HOXD10) belongs to the homeobox (HOX) gene family, has been reported to be the main regulator that can directly affect organogenesis and plays a role in maintaining the differentiated tissues functions (14), encodes a sequence-specific transcription factor with a homeobox DNA-binding domain (15,16). Furthermore, Li et al showed that miR-224 could promote hepatocellular carcinoma cell invasion and migration by targeting HOXD10, and HOXD10 was recognized as an essential tumor suppressor (17). Whether HOXD10 could play an important function in the mediation of NSCLC needs further investigation.
In this study, we proved the expression level of HOXD10 could be regulated by miR-224. miR-224 may play a vital part in mediation of malignant disease development, especially NSCLC, by directly targeting HOXD10. These findings may suggest a critical regulatory mechanism for cancer progress or development of NSCLC and give a feasible therapeutic option for exploring NSCLC carcinogenesis.
Materials and methods
Tissue specimen collection
Forty paired samples of the NSCLC and adjacent non-tumor tissues were obtained at the Department of Respiratory Medicine, The First Affiliated Hospital of Jiamusi University (Jiamusi, China). Informed consent was obtained from all patients and the present study was approved by the Ethics Committee of The First Affiliated Hospital of Jiamusi University. All tissue specimens were confirmed by pathological diagnoses and no patients received radiotherapy or chemotherapy before surgery. The characteristics of the NSCLC tissues and the corresponding normal tissues are described in Table I.
Table I.
Clinicopathological characteristics of patients with NSCLC.
| miR-224 expression | ||||
|---|---|---|---|---|
| Characteristics | Total cases (n=40) | High | Low | P-value |
| Age (years) | 0.7356 | |||
| ≤60 | 25 | 17 | 8 | |
| >60 | 15 | 9 | 6 | |
| Sex | 0.8843 | |||
| Male | 23 | 13 | 10 | |
| Female | 17 | 10 | 7 | |
| Tumor size (cm) | 0.5628 | |||
| <3 | 28 | 19 | 9 | |
| ≥3 | 12 | 7 | 5 | |
| Metastasis | 0.0262a | |||
| Yes | 21 | 17 | 4 | |
| No | 19 | 9 | 10 | |
| Differentiation | 0.0312a | |||
| Well/Moderate | 14 | 6 | 8 | |
| Poor | 26 | 20 | 6 | |
| TNM stage | 0.0281a | |||
| I+II | 21 | 15 | 6 | |
| III+IV | 19 | 7 | 12 | |
P<0.05.
Cell lines
Three human NSCLC cell lines (H358, 95-D, A549) and one normal human lung epithelial cell line (BEAS-2B) were purchased from Shanghai Institute of Cell Biology (Shanghai, China). The H358 cell line has been reported to be possibly identical to the H358M cell line, of the bronchioloalveolar carcinoma type (http://mct.aacrjournals.org/content/12/5/643). The cells were maintained in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA) at 37°C and 5% CO2.
Cell transfections
Cells were transfected with the negative control miRNA (NC miRNA) and miRNA mimics/inhibitor (100 µM) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) after the recommended instructions. The miR-224 mimics/inhibitors and NC miRNA were synthesized with RiboBio (Guangzhou, China).
RNA isolation and RT-qPCR assays
Total RNA were isolated from cultured cells using the RNA simple Total RNA Extraction kit (Ambion, Carlsbad, CA, USA) for cDNA synthesis. To perform the quantitative detection of miR-224 or mRNA expression, quantitative PCR was carried out with StepOne Plus RT-PCR System. The RT-qPCR data were calculated by the 2−∆∆Cq method and relative to SnRNA U6 or GAPDH.
Luciferase reporter assay
To find the targets of miR-506, we utilized bioinformatics software (Targetscan, miRanda). The pmirGLO vectors with wild-type (WT) or mutant type (MUT) miR-224 binding site in HOXD10 3′-UTR were constructed by GenePharma Co., Ltd. (Shanghai, China). The miR-224 mimics/inhibitor, and miR-224 NC were synthesized and purified by RiboBio; siHOXD10 and anti-siHOXD10 were purchased from RiboBio. Then, 50 ng of WT or MUT luciferase vector and miR-224 mimic/inhibitor or NC miRNA (20 µM) were co-transfected into H358 or A549 cells. Then luciferase activity was determined in two days with Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) after transfection.
Western blotting
RIPA lysis buffer was chosen to extract all proteins, supplemented with a protease inhibitor, and separated by 10% SDS polyacrylamide gels (SDS-PAGE) and were electro-transferred to polyvinylidene difluoride (PVDF) membranes (Life Technologies, Grand Island, NY, USA). Subsequently, the membranes were incubated with a specific primary antibody: rabbit monoclonal anti-HOXD 10 (ab 138508; 1:1,000; Abcam, Cambridge, MA, USA) for 12 h at 4°C, and then horseradish peroxidase-conjugated (HRP, 1:10,000), secondary antibody; goat anti-rabbit IgG-HRP (sc-2,004; 1:3,000; Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA) was incubated for 1 h at 25°C. Finally, the proteins of interest were detected by the enhanced chemiluminescence (ECL) detection system (Sea Biotech, Shanghai, China).
Migration and invasion assays
Cells were washed using serum-free medium three times. A total of 5×105 cells (H358), transfected with the miR-224 mimic/inhibitor and NC, were resuspended in 200 µl serum-free medium and added into upper chamber with or without 150 mg Matrigel (BD Biosciences, San Jose, CA, USA). The lower chamber was filled with 600 µl medium containing serum and incubated for 48 h for migration assay and 72 h for invasion assay. Matrix gels were added into upper chamber of the Transwell. At the end of the experiment, cells of the lower chamber were stained with crystal violet for 20 min. Stained cells were visualized and counted under a CKX41 inverted microscope (SZ61; Olympus, Tokyo, Japan).
Statistical analysis
The statistical analyses of experimental results were conducted using SPSS 16.0 statistics software package (SPSS, Inc., Chicago, IL, USA). The results are expressed as the mean ± SD. The Student's t-test and the Chi-square test were used to analyze experimental data. The unpaired two group comparison and multiple comparisons were made with analysis of variance (ANOVA) with Tukey-Kramer post-hoc test. Correlation between mRNA and miRNA expression level were estimated using the Spearman's correlation method. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-224 is significantly upregulated in NSCLC
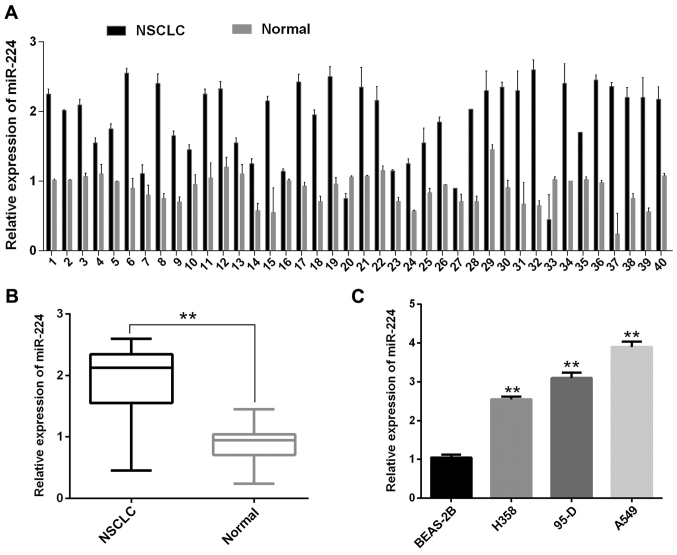
In order to evaluate the expression levels of miR-224 in NSCLC tissues, the RT-qPCR was used in 40 pairs of collected NSCLC tissues and their adjacent non-tumor tissues samples. miR-224 showed a remarkably higher expression in NSCLC tissues than that in adjacent non-tumor lung tissues (Fig. 1A and B). Moreover, the expression levels of miR-224 in three NSCLC cell lines (H358, 95-D, A549) were also higher compared with BEAS-2B (Fig. 1C). These results can help us to understand the correlation between miR-224 and NSCLC, which is characterized by high expression.
Figure 1.
miR-224 expression in NSCLC tissues and cell lines. (A and B) Relative expression of miR-224 in 40 pairs of NSCLC tissues. (C) The miR-224 was significantly higher expressed in NSCLC cell lines (H358, 95-D, A549) than that in BEAS-2B. **P<0.01.
miR-224 is overexpressed following transfection with miR-224-mimic
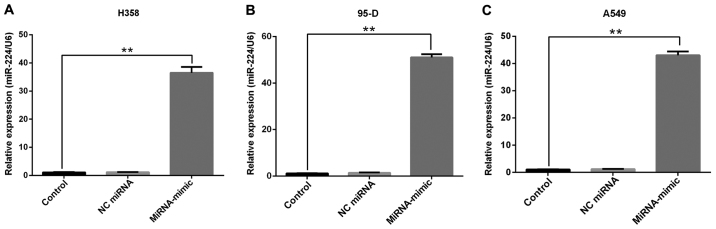
To investigate the causal role of miR-224 in tumor progression and development, the transfected miR-224 mimics were transfected into the three different NSCLC cell lines (H358, 95-D, A549). The results as shown in Fig. 2, after transfection with miR-224 mimics, miR-224 showed an average of 35±3-fold expression in H358 cells (Fig. 2A), 50±2-fold expression in A549 cells (Fig. 2B), and 42±2-fold expression in 95-D cells (Fig. 2C), suggested that the expression level of miR-224 was markedly increased with miR-224 mimics compared with NC miR-224 and the control group.
Figure 2.
miR-224 expression level after transfection with miRNA-mimics. The miR-224 was overexpressed via transfection with miRNA-mimics, but still unchanged in control and NC miRNA group in H358 (A) 95-D (B) and A549 (C) cells. **P<0.01.
HOXD10 acts as an oncogene and inversely correlates with miR-224
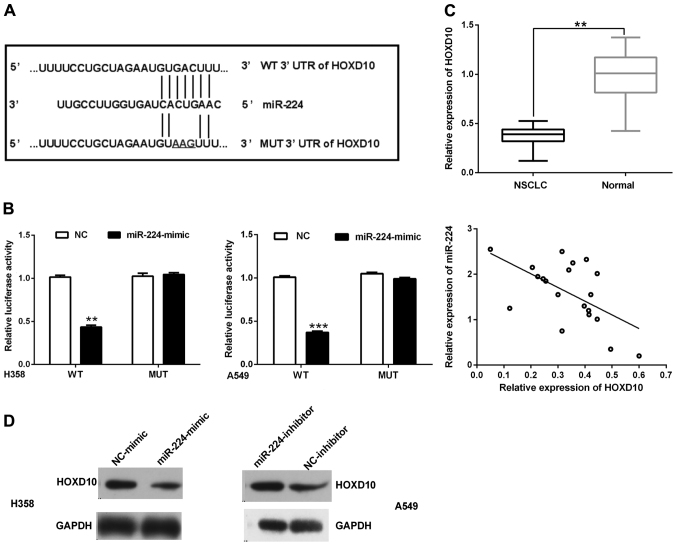
To find the putative downstream target of miR-224c, bioinformatic tools, such as TargetScan and miRanda, were used. By the online prediction, we obtained the outcome that HOXD10 was selected as a potential target gene of miR-224 and has a highly conserved binding site to miR-224 at 152–158 bp in the 3′-UTR (Fig. 3A). In order to further verify the predicted binding site, dual luciferase reporter assay was carried out using two NSCLC cell lines (H358, A549). The results indicated that the relative luciferase activity transfected with HOXD10-3′UTR-WT reporter and miR-224 mimics was significantly decreased, on the other hand, it had no suppressive effect on the mutant reporter in H358 and A549 cells (Fig. 3B).
Figure 3.
miR-224 downregulates HOXD10 expression by targeting its 3′UTR. (A) An illustration of WT and MUT 3′-UTR of HOXD10 showing a putative miR-224 target site. (B) The luciferase activities of HOXD10-WT, and HOXD10-MUT in H358 and A549 cells transfected with miR-224 mimic. (C) HOXD10 mRNA level was remarkably lower examined by qRT-PCR and it was inversely correlated with miR-224 level in NSCLC tissues (20 paired tissues). (D) Protein level of HOXD10 and GAPDH in H358 and A549 cells. **P<0.01, ***P<0.001.
The mRNA expression levels of HOXD10 were also tested in 40 pairs of samples using the approach of qRT-PCR. The outcome suggested that the expression of HOXD10 was obviously lower in NSCLC tissues. Furthermore, a correlation was performed among mRNA levels of HOXD10 and miR-224 by Spearman's correlation analysis. The statistical analysis suggested that the expression of HOXD10 was significantly inversely correlated with miR-224 levels in the NSCLC tissues (Fig. 3C).
Furthermore, the re-expression of miR-224 significantly suppressed endogenous HOXD10 expression at protein levels in H358 cells. We consistently found that the silencing of miR-224 can increase the protein levels of endogenous HOXD10 in A549 cells (Fig. 3D and E). Our results indicate that both of the binding sites may take part in miR-224 regulation, and miR-224 binds to the 3′-UTR of HOXD10 to affect its expression.
HOXD10 was functionally important for the biological function of miR-224
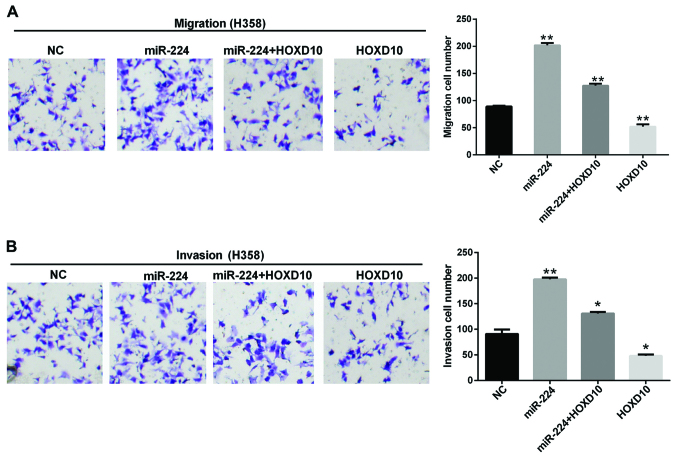
For further determining whether HOXD10 takes part in the regulation of NSCLC cells, migration and invasion assays suggested that silencing of HOXD10 obviously promoted miR-224-induced migration and invasion within H358 cells, and overexpression of HOXD10 could significantly reverse the tumour-inductive functions of miR-224 on the migration and invasion in H358 cells (Fig. 4A and B). These results demonstrated that miR-224 plays a significant role in the migration and invasion of NSCLC cells by targeting HOXD10.
Figure 4.
miR-224 promotes the NSCLC cell migration and invasion. Transwell assay was used to detect the migratory (A) or invasive (B) capacity of H358 cells. *P<0.05; **P<0.01.
Discussion
Lung cancer remains the leading cause of cancer-associated mortality (1), 80% of which are due to NSCLC (2). In recent years, great improvements have been made in comprehension of the pathophysiological functions and finding potential therapeutic methods for NSCLC. miRNAs belong to small endogenous non-coding RNAs, and they may mediate the expression of related gene and/or induce the degradation at the post-transcriptional process (18). Increasing evidence suggests that miRNAs play essential part in the development of cancers such as NSCLC through regulation of target genes (19,20).
Moreover, growing number of evidence has indicated that miR-224 functioned either as tumor oncogene or suppressor. Previous research has indicated that miR-224 was increased in a wide class of solid tumors, such as ovarian cancer (21), hepatocellular carcinoma (22–24), and glioma (25), gastric cancer (26) and may have diagnostic, therapeutic and prognostic values by targeting KLLN, pAKT, EP300, HepG2 and mTOR. In addition, miR-224 acts as a tumor-suppressor in breast cancer and prostate cancer (27,28) by targeting frizzled 5 and TPD52, respectively. Furthermore, various studies suggested that upregulation miR-224 had significant correlation with NSCLC metastasis, and overexpression level of miR-224 enhanced the H1573, H460 cells and H1299 cell abilities of migration, invasion, and proliferation in NSCLC (12). Moreover, Cui et al reported that CASP7 and CASP3 were identified as downstream targets of miR-224, which are functionally important in the miR-224 mediated lung cancer progression (13). Our result showed that miR-224 expression was upregulated in the NSCLC tissues and H358, 95-D, A549 cell lines (Fig. 1A and B), which was consistent with the previous study. Upon this finding, another experiment was performed where miR-224 was overexpressed in three different NSCLC cell lines (H358, A549 and 95D) via transfection (Fig. 2) and the results suggested overexpression of miR-224 may enhance the migration and invasion abilities of NSCLC cells (Fig. 4A and B).
HOXD10, an important regulator of cell-maintainence on differentiated phenotype, has been suggested to take part in tumorigenesis of various human malignancies (29). HOXD10 was also detected as the cancer suppressor target regularly lost on chromosome 10q23 and a previous study reported that HOXD10 might play a significant part as a cancer suppressor of invasive growth, cell migration, differentiation and morphogenesis (30,31). Another study validated the HOXD10 functioned as a direct downstream target gene of miR-224 in human hepatocellular carcinoma and indicated that the HOXD10 functioned as a crucial inhibitor of cancer cell migration and invasion (17). In addition, in this study, at the molecular level for the first time, luciferase activity assay and western blot suggested that miR-224 targets the 3′-UTR of HOXD10 to regulate its expression in NSCLC (Fig. 3). Furthermore, Transwell assays showed that silencing or overexpression of HOXD10 has different benefits on tumour-inductive influences of miR-224 on the cancer cell migration and invasion (Fig. 4A and B), which indicated miR-224 inhibited expression by targeting 3′-UTR of HOXD10 in NSCLC.
In summary, miR-224 overexpression can enhance the migration and invasion abilities of NSCLC cells. Moreover, HOXD10 was detected to be a downstream target of miR-224, and to regulate the influence on cell migration and invasion of NSCLC. A further study to evaluate the potential function of miR-224/HOXD10 axis on tumorigenesis and progression of NSCLC is needed.
Acknowledgements
Not applicable.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
SL and JZ contributed to the conception of the study; YZ contributed significantly to data analysis and manuscript preparation; FW performed the data analyses and wrote the manuscript; YC and XF helped perform the analysis with constructive discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the of the Ethics Committee of The First Affiliated Hospital of Jiamusi University (Jiamusi, China). Informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Tiseo M, Bordi P, Bortesi B, Boni L, Boni C, Baldini E, Grossi F, Recchia F, Zanelli F, Fontanini G, et al. Bio-FAST trial group: ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer. 2013;108:1695–1703. doi: 10.1038/bjc.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caramori G, Casolari P, Cavallesco GN, Giuffrè S, Adcock I, Papi A. Mechanisms involved in lung cancer development in COPD. Int J Biochem Cell Biol. 2011;43:1030–1044. doi: 10.1016/j.biocel.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Buttery RC, Rintoul RC, Sethi T. Small cell lung cancer: The importance of the extracellular matrix. Int J Biochem Cell Biol. 2004;36:1154–1160. doi: 10.1016/S1357-2725(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 5.Ettinger DS. Ten years of progress in non-small cell lung cancer. J Natl Compr Cancer Netw. 2012;10:292–295. doi: 10.6004/jnccn.2012.0029. [DOI] [PubMed] [Google Scholar]
- 6.Su S, Hu Y, Ouyang W, Ma Z, Li Q, Li H, Wang Y, Wang X, Li T, Li J, et al. Might radiation therapy in addition to chemotherapy improve overall survival of patients with non-oligometastatic Stage IV non-small cell lung cancer?: Secondary analysis of two prospective studies. BMC Cancer. 2016;16:908. doi: 10.1186/s12885-016-2952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saumet A, Lecellier CH. microRNAs and personalized medicine: Evaluating their potential as cancer biomarkers. Adv Exp Med Biol. 2015;888:5–15. doi: 10.1007/978-3-319-22671-2_2. [DOI] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 10.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 11.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 12.Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan M, Jeon YJ, Li B, Vicentini C, Peng Y, et al. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci USA. 2015;112:E4288–E4297. doi: 10.1073/pnas.1502068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui R, Kim T, Fassan M, Meng W, Sun HL, Jeon YJ, Vicentini C, Tili E, Peng Y, Scarpa A, et al. MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget. 2015;6:21802–21815. doi: 10.18632/oncotarget.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 15.Hedlund E, Karsten SL, Kudo L, Geschwind DH, Carpenter EM. Identification of a Hoxd10-regulated transcriptional network and combinatorial interactions with Hoxa10 during spinal cord development. J Neurosci Res. 2004;75:307–319. doi: 10.1002/jnr.10844. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, Maesawa C. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int J Oncol. 2013;43:63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, Ding C, Chen C, Zhang Z, Xiao H, Xie F, Lei L, Chen Y, Mao B, Jiang M, et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29:835–842. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Alonso DF, Ripoll GV, Garona J, Iannucci NB, Gomez DE. Metastasis: Recent discoveries and novel perioperative treatment strategies with particular interest in the hemostatic compound desmopressin. Curr Pharm Biotechnol. 2011;12:1974–1980. doi: 10.2174/138920111798377076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 21.Hu K, Liang M. Upregulated microRNA-224 promotes ovarian cancer cell proliferation by targeting KLLN. In Vitro Cell Dev Biol Anim. 2017;53:149–156. doi: 10.1007/s11626-016-0093-2. [DOI] [PubMed] [Google Scholar]
- 22.Yu L, Zhang J, Guo X, Li Z, Zhang P. MicroRNA-224 upregulation and AKT activation synergistically predict poor prognosis in patients with hepatocellular carcinoma. Cancer Epidemiol. 2014;38:408–413. doi: 10.1016/j.canep.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Wang G, Shan JL, Yang ZX, Wang HZ, Feng J, Zhen JJ, Chen C, Zhang ZM, Xu W, et al. MicroRNA-224 is upregulated in HepG2 cells and involved in cellular migration and invasion. J Gastroenterol Hepatol. 2010;25:164–171. doi: 10.1111/j.1440-1746.2009.05971.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Toh HC, Chow P, Chung AY, Meyers DJ, Cole PA, Ooi LL, Lee CG. MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J. 2012;26:3032–3041. doi: 10.1096/fj.11-201855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Upregulation of microRNA-224 confers a poor prognosis in glioma patients. Clin Transl Oncol. 2013;15:569–574. doi: 10.1007/s12094-012-0972-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Li CF, Ma LJ, Ding M, Zhang B. MicroRNA-224 aggrevates tumor growth and progression by targeting mTOR in gastric cancer. Int J Oncol. 2016;49:1068–1080. doi: 10.3892/ijo.2016.3581. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Liu Y, Shen J, Zhang G, Han J. MicroRNA-224 inhibits proliferation and migration of breast cancer cells by down-regulating Fizzled 5 expression. Oncotarget. 2016;7:49130–49142. doi: 10.18632/oncotarget.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto Y, Nishikawa R, Kojima S, Chiyomaru T, Enokida H, Inoguchi S, Kinoshita T, Fuse M, Sakamoto S, Nakagawa M, et al. Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett. 2014;588:1973–1982. doi: 10.1016/j.febslet.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 29.de Barros E, Bueno Lima R, Ramão A, Pinheiro DG, Alves CP, Kannen V, Jungbluth AA, de Araújo LF, Muys BR, Fonseca AS, Plaça JR, et al. HOX genes: Potential candidates for the progression of laryngeal squamous cell carcinoma. Tumour Biol. 2016;37:15087–15096. doi: 10.1007/s13277-016-5356-8. [DOI] [PubMed] [Google Scholar]
- 30.Vardhini NV, Rao PJ, Murthy PB, Sudhakar G. HOXD10 expression in human breast cancer. Tumour Biol. 2014;35:10855–10860. doi: 10.1007/s13277-014-2324-z. [DOI] [PubMed] [Google Scholar]
- 31.Carrio M, Arderiu G, Myers C, Boudreau NJ. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005;65:7177–7185. doi: 10.1158/0008-5472.CAN-04-1717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.