The regulation of Trypanosoma brucei life stage development remains unclear. Inositol polyphosphate multikinase regulates the development of mammalian bloodforms to insect stages that normally develop in flies. Specific inositol phosphates, perhaps as second messengers, interact with proteins of the regulatory network that controls development.
Abstract
Many cellular processes change during the Trypanosoma brucei life cycle as this parasite alternates between the mammalian host and tsetse fly vector. We show that the inositol phosphate pathway helps regulate these developmental changes. Knockdown of inositol polyphosphate multikinase (IPMK), which phosphorylates Ins(1,4,5)P3 and Ins(1,3,4,5)P4, resulted in changes in bloodstream forms that are characteristic of insect stage procyclic forms. These changes include expression of the procyclic surface coat, up-regulation of RNA-binding proteins that we show to regulate stage-specific transcripts, and activation of oxidative phosphorylation with increased ATP production in bloodstream forms. These changes were accompanied by development of procyclic morphology, which also occurred by the expression of a catalytically inactive IPMK, implying that regulation of these processes entails IPMK activity. Proteins involved in signaling, protein synthesis and turnover, and metabolism were affinity-enriched with the IPMK substrate or product. Developmental changes associated with IPMK knockdown or catalytic inactivation reflected processes that are enriched with inositol phosphates, and chemical and genetic perturbation of these processes affected T. brucei development. Hence, IPMK helps regulate T. brucei development, perhaps by affecting inositol phosphate interactions with proteins of the regulatory network that controls energy metabolism and development.
INTRODUCTION
Life cycle–stage developmental regulation is essential for the transmission of trypanosomatid pathogens and includes morphological, transcriptional, and physiological adaptations for survival in the host and the vector (MacGregor et al., 2012). Trypanosoma brucei is transmitted by tsetse flies and causes human African trypanosomiasis and related cattle diseases in sub-Saharan Africa (Stuart et al., 2008). In the mammalian host, T. brucei proliferates as slender bloodstream forms (BFs) and undergoes antigenic variation by expressing variant surface glycoproteins (VSGs), thus avoiding host antibody clearance (Cestari and Stuart, 2018). BFs rely solely on glycolysis for energy production, and many glycolytic enzymes are in peroxisome-like organelles called glycosomes (Furuya et al., 2002; Haanstra et al., 2008). In the mammalian bloodstream, the slender BFs develop into nonreplicating stumpy BFs, which then develop into procyclic forms (PFs) after transmission to the fly (MacGregor et al., 2012; Figure 1A). The transition entails morphological and metabolic changes, including a switch from glycolysis to oxidative phosphorylation (Bochud-Allemann and Schneider, 2002), and changing the surface coat from VSGs to procyclins (Roditi et al., 1989). Different procyclins appear to be expressed in a particular order after T. brucei infection of tsetse flies (Urwyler et al., 2005), although that may differ between T. brucei strains or in vivo versus in vitro (Roditi et al., 1989; Vassella et al., 2000; Batram et al., 2014). Other developmental changes accompany the life-cycle progression in the fly from PFs to epimastigotes and then to mammalian infective metacyclic trypomastigotes.
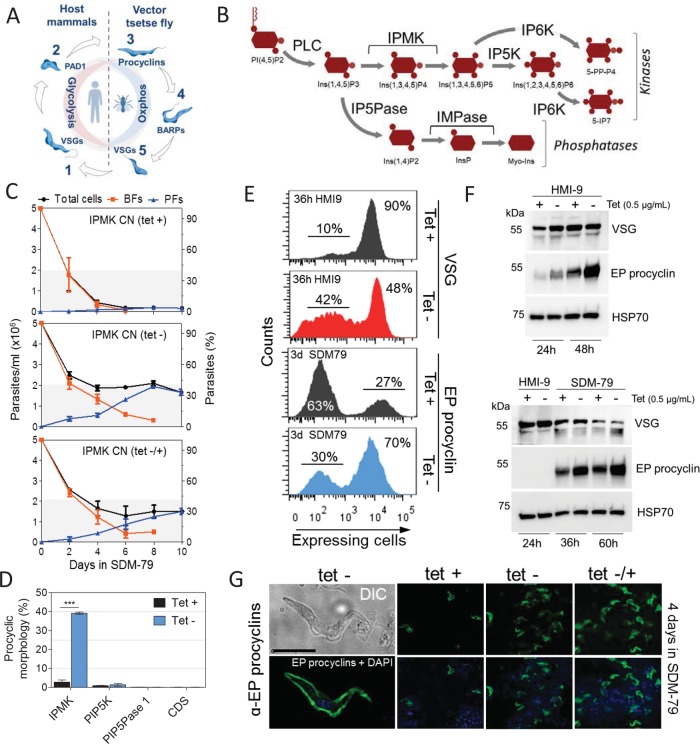
FIGURE 1:
IPMK regulates development of BF T. brucei to PF. (A) Diagram of the T. brucei life cycle stages. Long slender BFs (1) differentiate to short stumpy BFs (2). BFs after ingestion by tsetse flies differentiate to PFs and then to epimastigotes (4) and metacyclics (MFs) (5), which are infectious to mammals. BFs and MFs express VSGs; PFs express procyclins; epimastigotes expresses brucei alanine-rich proteins (BARPs); and stumpy expresses PAD1. (B) Diagram of a portion of the IP pathway. IP5K, inositol pentakisphosphate kinase; IP6K, inositol hexakisphosphate kinase; IP5Pase, inositol polyphosphate 5-phosphatase; IMPase, inositol monophosphatase. See Materials and Methods for gene IDs and additional abbreviations. (C) Morphological quantification of T. brucei BFs and PFs. IPMK was either expressed or knocked down for 36 h in T. brucei BF in HMI-9 at 37°C, followed by transfer to SDM-79 at 27°C (time 0). Top, continuous IPMK expression (tet +); middle, continuous IPMK knockdown (tet –); and bottom, IPMK was reexpressed after 36 h knockdown, at the transfer to SDM-79 at 27°C (tet –/+). Transition forms were counted as BFs. (D) Quantification as a percentage of CN cells that developed PF morphology after gene knockdown. IPMK, PIP5K, PIP5Pase 1, or CDS were either expressed or knocked down, as described in C, and quantification of cells with PF morphology were done at day 8. (E) Flow cytometry analysis of VSG or EP procyclin expression after IPMK knockdown in T. brucei BF in HMI-9 at 37°C for 36 h or SDM-79 for 3 d, respectively. (F) Western blot analysis of VSG or EP procyclin expression after IPMK knockdown in T. brucei BF. Top, BFs were collected in late-log growth in HMI-9 at 37°C for 24 h (1.5 × 106 cells/ml) and 48 h (2.5 × 106 cells/ml), respectively. Bottom, BFs were collected in mid–log growth (8.0 × 105 cells/ml) in HMI-9 at 37°C for 24 h and then transferred to SDM-79 at 27°C for an additional 12 and 36 h. Western blots were run with 1.0 × 107 cells (top) and 5.0 × 106 cells equivalents/lane (bottom). Blots were stripped and reblotted with monoclonal antibodies (mAb) α-HSP70. (G) EP procyclin IF analysis after IPMK knockdown and 4 d in SDM-79 at 27°C as described in C. Differential interference contrast (DIC) image shows T. brucei (tet-) with PF morphology, and IF shows surface EP procyclin expression. The pictures are representative of three independent experiments. Blue, DAPI staining of DNA; green, EP procyclin. Bar, 10 µm. Tet, 0.5 µg/ml. Data are represented as mean ± SEM.
Cell signaling and metabolic changes are implicated in regulating development between life-cycle stages. The AMP-activated protein kinase (AMPK) complex functions as an AMP/ATP sensor and regulates development of stumpy forms (Saldivia et al., 2016). AMP and cAMP analogues, which inhibit TbTOR4 expression, induce development of BF slender to stumpy, as does knockdown of TbTOR4 (Barquilla et al., 2012). In contrast, knockdown of a Nek-related kinase or protein phosphatase 1 inhibits stumpy formation (Mony et al., 2014; Domingo-Sananes et al., 2015). Moreover, expression of metabolic enzymes and mitochondrial enzyme activities are altered during T. brucei development (Vassella et al., 2004; Domingo-Sananes et al., 2015). Tricarboxylic acid (TCA) cycle metabolites citrate and cis-aconitate (CCA) and a temperature shift from 37 to 27°C stimulate development of BFs to PFs and activation of oxidative metabolism (Overath et al., 1986). CCA is transported by the carboxylate transporter proteins associated with differentiation (PAD), which is expressed in stumpy BFs and PFs (Dean et al., 2009). CCA inhibits activation of the protein tyrosine phosphatase TbPTP1, which is a negative regulator of BF-to- PF development, by the protein phosphatase TbPIP39 resulting in BF-to-PF development (Szoor et al., 2010). Other treatments have been shown to stimulate BF-to-PF development, such as mild acid (Rolin et al., 1998) or protease treatment (Yabu and Takayanagi, 1988). Mild acid was shown to affect TbPIP39 phosphorylation, whereas protease treatment appeared to affect an alternate process that is dispensable in tsetse flies (Szoor et al., 2013). The RNA-binding proteins (RBPs) RBP6, RBP7, and RBP10 are also implicated in T. brucei development, likely by regulating stage-specific transcript expression (Mony et al., 2014; Kolev et al., 2012; Wurst et al., 2012). Hence, T. brucei development employs multiple processes including signaling, metabolism, and control of gene expression to coordinate changes in parasite morphology, surface composition, and physiological adaptations to the host and vector.
We showed that the inositol phosphate (IP) pathway controls telomere silencing and allelic exclusion of VSG expression sites (ES) (Cestari and Stuart, 2015), which is developmentally regulated in T. brucei. IPs function as second messengers via interactions with proteins that regulate many metabolic and developmental processes in eukaryotes (Szijgyarto et al., 2011; Chavez et al., 2015; Seeds et al., 2015; Wu et al., 2016). The IP pathway enzymes catalyze the synthesis and phosphate remodeling of IPs and phosphatidylinositols (PIs). A PI-specific phospholipase C (PLC) cleaves PI(4,5)P2 to generate diacylglycerol and Ins(1,4,5)P3. We showed that Ins(1,4,5)P3 is further converted by T. brucei IP multikinase (IPMK) to Ins(1,3,4,5)P4 and Ins(1,3,4,5,6)P5 (Figure 1B; Cestari et al., 2016). We show here that the IP pathway functions in the control of T. brucei development. Specifically, IPMK knockdown in BFs resulted in loss of surface VSGs and expression of a procyclin surface coat, a metabolic switch from glycolysis to oxidative phosphorylation, a shift from the BF to PF stage specific transcriptome, and development of PF morphology. The expression of a kinase-dead IPMK also resulted in BF-to-PF development, indicating that IPMK enzymatic activity, and likely its substrates or products, is involved in these processes. We found that proteins that interact with Ins(1,4,5)P3 or Ins(1,3,4,5)P4 have functions associated with various cellular processes that differ between stages and thus may be controlled by these metabolites. We propose that IPs may be part of a regulatory system that controls T. brucei metabolism and development.
RESULTS
IPMK regulates BF-to-PF development in T. brucei
To identify the role of IPs in T. brucei, we generated a conditional null (CN) IPMK cell line by removing both endogenous IPMK alleles and inserting a tetracycline (tet)-regulatable IPMK allele into the ribosomal DNA spacer (Cestari and Stuart, 2015). Knockdown of IPMK resulted in BFs developing to PFs (Figure 1C). IPMK was knocked down by withdrawal of tet, and after 36 h, a time point in which cell viability were >90% (Supplemental Figure S1, A–C), the cells were transferred from BF (HMI-9 at 37°C) to PF (SDM-79 at 27°C) growth conditions, and parasite survival and development to PFs were monitored by cell morphology and procyclin expression. The IPMK knockdown resulted in nearly half of the cells surviving under PF conditions, whereas essentially all IPMK-expressing cells remained as BFs and eventually died (Figure 1C). In addition, ∼40% of the surviving cells developed into morphological PFs over time (Figure 1, C and D). During this period of time, the BF cell population gradually decreased concomitant with an increase in the PF cell population. IPMK knockdown under BF growth conditions for 36 h resulted in loss of VSG2 expression in ∼42% of the cell population (Figure 1E). In addition, the proportion of cells expressing PAD1, which increased from 45% to 60% between 24 and 36 h when IPMK was expressed, declined from 49% to 23% in IPMK knockdown cells (Supplemental Figure S1D). Western blot analysis showed a gradual increase in EP procyclin expression in BFs growing at late log (24 h) and stationary (48 h) phases, especially after IPMK knockdown (Figure 1F, top panel), but not in BFs at mid–log growth (24 h; Figure 1F, bottom panel). The IPMK knockdown also increased EP procyclin expression after transfer to PF conditions, which was accompanied by a decrease in VSG expression (Figure 1F, bottom panel). After 3 d, ∼70% of the population expressed EP procyclins (Figure 1E), and immunofluorescence (IF) analysis showed EP procyclin surface expression by cells with PF morphology (Figure 1G). IPMK reexpression after 36 h knockdown under BF conditions followed by transfer to PF conditions resulted in ∼30% of cells with PF morphology compared with 40% when IPMK was continuously knocked down (Figure 1C). The data indicate that IPMK knockdown resulted in cells committed to develop into PFs. However, overexpression of IPMK had no effect on T. brucei BF development (Supplemental Figure S1E). Knockdown of other genes of the IP pathway, PI 5-kinase (PIP5K), PI 5-phosphatase (PIP5Pase 1), or CDP-diacylglycerol synthase (CDS), all of which have growth kinetics similar to that of IPMK CN (Cestari and Stuart, 2015), did not result in development of BF to PFs (Figure 1D), which suggests that the development phenotype is related to perturbation of a specific step of the pathway, the IPMK step, and not nonspecific effects due to gene knockdown, viability, or tet. Notably, the parental cell line SM427 differentiates into PFs that survive in PF culture for ∼21 d but for unknown reasons are not continuously cultivatable. Exclusive expression of T. cruzi or L. major IPMK orthologues, which rescued T. brucei from the lethal IPMK knockdown in BFs (Cestari et al., 2016), only partially reverted development rates (Supplemental Figure S1F), likely due to IPMK sequence differences among these species. Our data suggest that IPMK regulate development of T. brucei BF to PF.
Development of BFs to PFs depends on IPMK catalytic activity
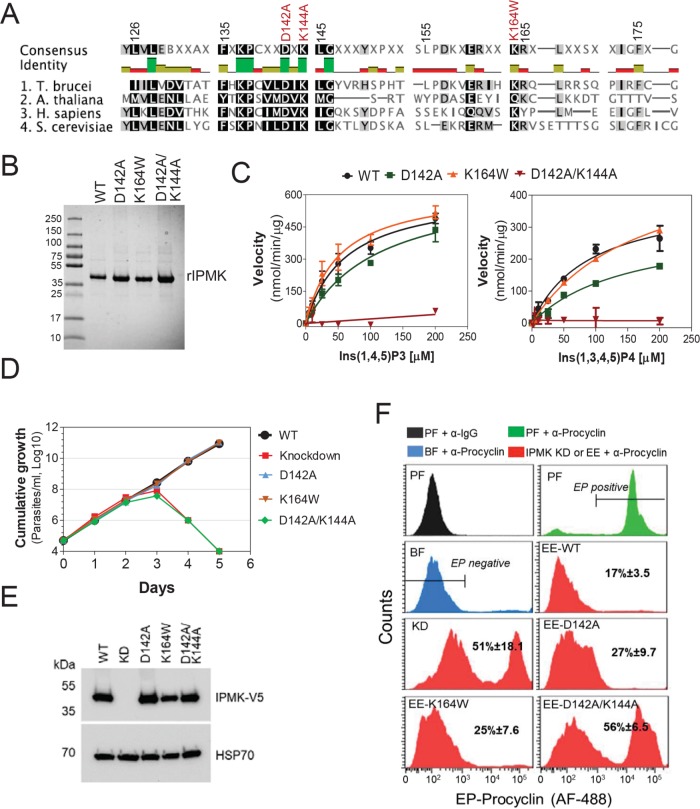
To determine whether IPMK catalytic activity is required for T. brucei BF development to PF, we generated mutations in the predicted IPMK catalytic site to eliminate its activity. T. brucei IPMK has some amino acids (aa) within the catalytic site that are conserved with IPMKs from yeast, humans, and Arabidopsis (Figure 2A), despite the overall low aa conservation (aa identities are 23.3%, 22.6%, and 24.9%, respectively). The D131A and/or K133A mutations within the yeast IPMK catalytic site have been shown to eliminate its activity (Dubois et al., 2000; Bosch and Saiardi, 2012), whereas a K121W mutation in Arabidopsis thaliana IPMK was shown to selectivity eliminate its 6-kinase activity (Endo-Streeter et al., 2012). Hence, we expressed in Escherichia coli and purified the following his-tagged recombinant versions of T. brucei IPMK: wild type (WT), D142A (equivalent to yeast D131A), D142/D144A (equivalent to yeast D131A/K133A), and K164W (equivalent to A. thaliana K121W; Figure 2B). Enzymological analysis showed that the D142A mutation reduced but did not eliminate IPMK activity. Specifically, the Km value for Ins(1,4,5)P3 was increased from 61.4 to 113.0 µM and the Km value for Ins(1,3,4,5)P4 was increased from 91.4 to 153.5 µM (Figure 2C and Table 1). While the K164W mutation did not significantly affect Ins(1,4,5)P3 phosphorylation, it increased the Km value for Ins(1,3,4,5)P4 from 91.4 to 169.9 µM, implying that this mutation reduced 6-kinase activity. Importantly, the D142A/K144A double mutation eliminated IPMK activity on both Ins(1,4,5)P3 and Ins(1,3,4,5)P4 substrates. Exclusive expression of T. brucei V5-tagged WT, D142A, or K164W rescued the growth of the T. brucei IPMK CN, whereas exclusive expression of V5-tagged D142A/K144A IPMK did not (Figure 2, D and E), indicating that IPMK catalytic activity is essential for T. brucei growth. Exclusive expression of the D142A/K144A mutant also resulted in a significant increase in BF to PF development compared with that for cells expressing WT IPMK (Figure 2F), whereas expression of either the D142A or K167W mutant only slightly affected development. Hence, development of BF to PF results from the loss of IPMK catalytic activity as well as the elimination of IPMK protein.
FIGURE 2:
Development of BFs to PFs depends on IPMK catalytic activity. (A) Alignment of catalytic sites of IPMKs of T. brucei, A. thaliana (gene ID: AED91147.1), Homo sapiens (gene ID: NP_689416.1), and Saccharomyces cerevisiae (gene ID: PJP09331.1). Mutated sites are shown in red. (B) Recombinant T. brucei IPMK WT or mutants (37 kDa) resolved on a 4–20% SDS–PAGE gel stained with Coomassie Imperial Staining (Pierce). (C) Activity analysis of rIPMK WT and mutants with Ins(1,4,5)P3 and Ins(1,3,4,5)P4 substrates. See Table 1 for kinetic parameters of each rIPMK enzyme and substrates. Growth curve (D) and Western blot analysis (E) of T. brucei IPMK CN exclusively expressing V5-tagged IPMK WT, mutants D142A, K164W, D142A/K144A, or no IPMK gene (knockdown). Cells were grown in the absence of tet for exclusive expression of introduced V5-tagged WT, mutant allele, or IPMK knockdown. Blots were stripped and reblotted with mAb 78, which recognizes mitochondrial HSP70. (F) Flow cytometry analysis and quantification of T. brucei expressing EP procyclin after IPMK knockdown or exclusive expression (EE) of WT or mutant IPMK alleles for 36 h in HMI-9 at 37°C in BF followed by 6 d in SDM-79 at 27°C.
TABLE 1:
Kinetic parameters of WT and mutant T. brucei rIPMK.
| Wild type | D142A | K164W | D142A/K144A | |
|---|---|---|---|---|
| Ins(1,4,5)P3 | ||||
| Km (µM) | 61.41 (±15.37) | 113.0 (±31.93) | 51.04 (±13.84) | ND |
| Vmax (nmol/min per µg) | 617.0 (±62.64) | 660.2 (±92.97) | 629.6 (±65.16) | ND |
| Kcat/Km (S–1M–1) | 1.67 × 105 | 9.73 × 104 | 2.06 × 105 | ND |
| Ins(1,3,4,5)P4 | ||||
| Km (µM) | 91.44 (±52.01) | 153.5 (±33.97) | 169.9 (±20.39) | ND |
| Vmax (nmol/min per µg) | 400.8 (±52.01) | 315.2 (±38.48) | 540.3 (±36.96) | ND |
| Kcat/Km (S–1M–1) | 7.31 × 104 | 3.42 × 104 | 5.23 × 104 | ND |
ND, not determined. The D142A/K144A mutation eliminates enzyme activity.
IPMK knockdown affects stage-specific patterns of gene expression
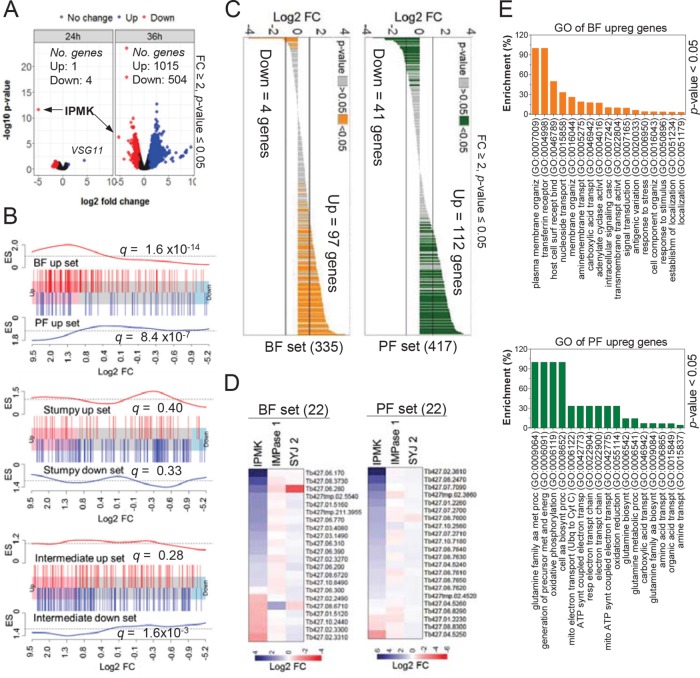
To investigate how IPMK affects T. brucei development, we collected RNA at 24 and 36 h after IPMK knockdown in BF conditions, times during which cells are growing and viable (Supplemental Figure S1, A–C), and performed transcriptomic analysis by RNA sequencing (RNAseq). While many genes were differentially regulated at 24 h, the most significant changes were at 36 h, that is, within four to six cell divisions, with ∼1500 genes differentially regulated (Figure 3A and Supplemental Dataset 1; fold change ≥2, p value ≤ 0.05). We performed gene set enrichment analysis (GSEA) of the 36-h RNAseq data using sets of genes that are differentially expressed between T. brucei stages (Capewell et al., 2013). We found a significant enrichment in gene sets from BF (q = 1.6 × 10–14), from PF (q = 8.4 × 10–7), and to a lesser extent from intermediate forms (q = 1.6 × 10–3; Figure 3B), implying changes in the expression of stage-specific genes. In addition, we found that ∼1⁄3 of the top ∼400 genes that had been shown by ribosome profiling to be up-regulated in BFs or PFs (Jensen et al., 2014) were also up-regulated after 36 h of IPMK knockdown (Figure 3C). We randomly selected a subset of 44 genes from the BF and PF gene sets and validated their differential expression by real-time PCR after IPMK knockdown (Figure 3D). Knockdown of SYJ 2 or IMPase 2 enzymes, which are predicted to dephosphorylate IP or PI metabolites, did not affect expression of these gene sets, indicating that their regulation is dependent on IPMK expression (Figure 3D). Gene ontology (GO) analysis of the 97 BF and 112 PF up-regulated genes (Figure 3C) revealed that BF genes were associated with adenylate cyclase and signal transduction, surface receptors, response to stimulus, and antigenic variation (Figure 3E). In contrast, PF genes were associated with amino acid and carboxylic acid transporters and metabolism, oxidative phosphorylation, and ATP biosynthesis (Figure 3E). IPMK knockdown also affected expression of protein kinases and phosphatases, including those involved in development (Supplemental Dataset 1). An increase in VSG11 expression was also detected at 24 h after IPMK knockdown (Figure 3A) followed by up-regulation of other telomeric and nontelomeric VSG genes at 36 h (Supplemental Figure S2). This suggests that, in addition to changes from VSG to procyclins (Figure 1), some cells in the population either lost expression site regulation or switched VSG expression. Given the role of the IP pathway in regulating VSG allelic exclusion and antigenic switching (Cestari and Stuart, 2015), it is likely that IPMK knockdown affected IP steady state levels and thus VSG regulation. Notably, knockdown of PIP5K or PIP5Pase 1 (Cestari and Stuart, 2015), both of which are involved in the regulation of VSG expression, did not result in BF-to-PF development (Figure 1D), and hence IPMK effects on cell development is unlikely to be due to ES derepression or to lethal effects resulting from knockdown of these essential genes.
FIGURE 3:
IPMK knockdown alters stage-specific gene expression in T. brucei. (A) Volcano plot shows differentially expressed (DE) genes after IPMK knockdown at 24 and 36 h by RNAseq. See Supplemental Dataset S1 for RNAseq data. Arrows indicate IPMK. RNAseq data of knockdown (tet –) IPMK cells were compared with tet + cells at their respective time points (24 or 36 h). (B) Barcode plot shows GSEA of genes up-regulated in BF or PF (top) or up- and down-regulated in stumpy (middle) or intermediates (bottom) in the 36-h RNAseq data. Each bar indicates a gene of the set and its location on the x-axis is its fold change. ES, enrichment score (see Supplemental Information text for details on gene sets). False discovery rates (q) are indicated for each gene set analysis. (C) Expression profile in the 36-h IPMK knockdown RNAseq data of the top ∼400 translated genes of BF or PF (Jensen et al., 2014). (D) Real-time PCR analysis of 44 genes selected from BF or PF gene sets after 30 h knockdown of IPMK, SYJ 2, or IMPase 2 in T. brucei BFs. (E) Gene ontology (GO) enrichment analysis of the 97 BF and 112 PF up-regulated genes shown in C. Tet, 0.5 µg/ml. Enrichment represents the percentage of genes in BF or PF sets that are associated with a GO process divided by the percentage of genes of the same process in the genome. Data are represented as the mean.
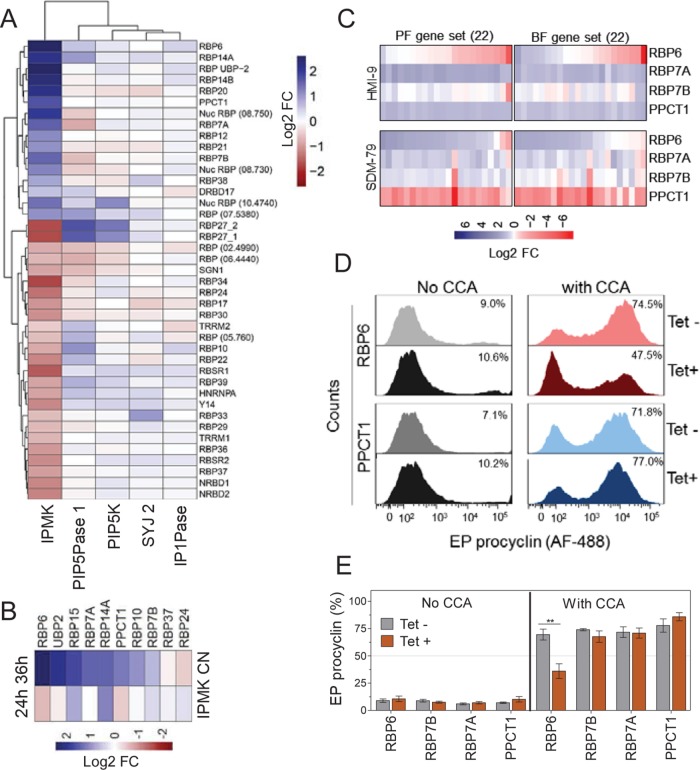
IPMK knockdown also affected the expression of many RBP mRNAs, such as up-regulation of RBP6, RBP7A and B, and PPCT1 (Figure 4A). Real-time PCR analysis confirmed RBPs’ differential expression (Figure 4B), whereas knockdown of other IP pathway genes did not similarly affect their expression. Overexpression of RBP6-V5 in BFs down-regulated the expression of gene sets under BF conditions for 24 h, but up-regulated their expression if BFs were cultured under PF conditions for 36 h (Figure 4C). Moreover, overexpression of PPCT1-V5 or RBP7A-V5 in BFs resulted in differential expression of the genes sets in a manner dependent on growth conditions. Hence, these RBPs affect expression of genes that are differentially expressed between BFs and PFs, and their effects on transcript abundances are dependent on growth conditions. Overexpression of RBP6, RBP7A, RBP7B, or PPCT1 in BFs did not result in development of BFs to PFs (Figure 4, D and E). However, EP procyclin expression decreased by ∼35% when RBP6-V5 was overexpressed, with concomitant induction of BF-to-PF development by CCA (Figure 4, D and E). This suggests that RBP6 may function as a negative regulator of BF-to-PF development, in addition to its overexpression resulting in PF-to-MF development (Kolev et al., 2012). Hence, these RBPs may play various complementary roles in T. brucei development that are affected by IPMK knockdown. Overall, IPMK knockdown alters expression of genes involved in metabolism, signaling, control of gene expression, and development in T. brucei.
FIGURE 4:
RBPs associated with stage specific gene regulation. (A) Differential expression of RBPs after knockdown of IPMK (36 h) or other IP pathway genes (24 h). (B) Real-time PCR analysis of 10 selected RBPs after 24 and 36 h IPMK knockdown. (C) Real-time PCR analysis of 44 genes from BF and PF gene sets after tet-regulatable overexpression of C-terminally V5-tagged RBPs in T. brucei BFs cultured in HMI-9 at 37°C for 24 h or SDM-79 at 27°C for 36 h. (D) Flow cytometry analysis of EP procyclin expression in T. brucei BFs overexpressing RBP6-V5 or PPCT1-V5. Cells were incubated in SDM-79 at 27°C for 3 d. (E) Flow cytometry quantification of EP procyclin expression after RBPs overexpression in T. brucei BFs (as described in D). Tet, 1 µg/ml. Data are represented as mean (heat maps) or mean ± SEM (bar graphs).
Ins(1,4,5)P3- and Ins(1,3,4,5)P4-interacting proteins function in cell signaling, metabolism, and regulation of protein synthesis
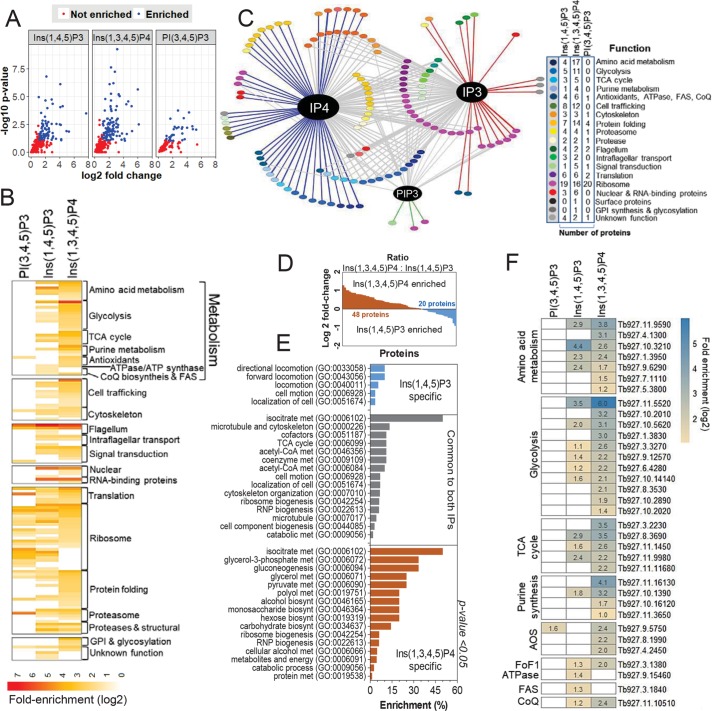
Because IPMK phosphorylates Ins(1,4,5)P3 to generate Ins(1,3,4,5)P4 initially (Figure 1B; Cestari et al., 2016), we sought to identify proteins that might interact with these IPs by affinity purification of T. brucei BF proteins using IPs or PIs conjugated with agarose beads that are commercially available or with nonconjugated beads as a control (Jungmichel et al., 2014; Wu et al., 2016). Out of ∼1200 proteins detected by mass spectrometry, 129 (fold change ≥ 2, p value < 0.1) were identified by label-free quantitative analysis associated with each metabolite in multiple experiments when compared with nonconjugated beads (Figure 5, A and B; Supplemental Dataset 2). Eighty-five proteins interacted with Ins(1,4,5)P3, 109 with Ins(1,3,4,5)P4, and 68 of these with both IPs, although the enrichment values of these 68 proteins were higher with Ins(1,3,4,5)P4 than with Ins(1,4,5)P3 (Figure 5, C and D). Other true interactions may not have been identified due to the stochastic nature of mass spectrometry analysis, which often results in variable detection of less abundant proteins, or less frequently detected peptides, deemed not significant in statistical analysis (Clough et al., 2012); or due to other factors that may affect protein interactions after cell lysis, such as changes in protein conformation or protein complex stability. Nevertheless, the statistical analysis provides a list of potential IP–protein interactions. Affinity purification with biotin-conjugated Ins(1,4,5)P3 or Ins(1,3,4,5)P4 followed by Western blot analysis confirmed the interactions of V5-tagged cysteinyl-tRNA synthetases (CysRS), phosphoglycerate kinase C (PGKC), peroxisomal membrane protein 14 (PEX14), heat shock protein 70 (HSP70), and universal minicircle sequence–binding protein (UMSBP1) with Ins(1,3,4,5)P4, and the interactions of V5-tagged intraflagellar transport proteins 54/57 (IFT54/57), HSP70, and UMSBP1 with Ins(1,4,5)P3 (Supplemental Figure S3). We also performed affinity purification and mass spectrometry to identify proteins that bound to agarose-conjugated PI(3,4,5)P3 to control for potential nonspecific IP–protein binding due to, for example, IP charges. The IP configuration of PI(3,4,5)P3 is similar to that of Ins(1,3,4,5)P4, except that phosphate 1 of the inositol ring is linked to a diacylglycerol moiety. Thirty-three proteins (fold change ≥ 2, p value < 0.1) were found to interact with PI(3,4,5)P3 when compared with the set interacting with agarose beads alone, 22 of which also interacted with Ins(1,4,5)P3 and 25 with Ins(1,3,4,5)P4 (Figure 5, A–D). The detection of a protein interacting with more than one IP (Figure 5C) may be due to flexible protein domain requirements for IP binding (Lemmon, 2008), which may involve specific protein regions and which may be affected in vivo by subcellular concentrations and by locations and interactions of the proteins and substrates. The annotated functions of these proteins identified processes including metabolism, translation, protein folding and degradation, cytoskeleton, trafficking, flagellum organization, and nuclear function, and only ∼6% of the 129 proteins had been annotated as hypothetical (unknown function; Figure 5, B and C). GO analysis indicated that the Ins(1,4,5)P3-specific proteome was enriched in proteins with functions associated with cell motility, which were also present in the common subset of 68 proteins, whereas that of Ins(1,3,4,5)P4 was enriched in proteins with metabolic functions some of which were also in the common subset (Figure 5E). More proteins with metabolic functions were associated with Ins(1,3,4,5)P4 than with Ins(1,4,5)P3 or PI(3,4,5)P3, which included proteins involved in glycolysis, amino acid and purine metabolism, the TCA cycle, and antioxidants (AOS; Figure 5F). Hence, Ins(1,4,5)P3 and Ins(1,3,4,5)P4 binding are associated with cell signaling, metabolism, and protein synthesis and turnover and are potentially involved in regulating these processes.
FIGURE 5:
Proteomic analysis of inositol phosphate interacting proteins. (A) Volcano plots indicate proteins that interact with Ins(1,4,5)P3 or Ins(1,3,4,5)P4 compared with nonconjugated beads (enrichment cutoff ≥ 2-fold and p value < 0.1). See Supplemental Dataset S2 for list of proteins. (B) Fold enrichment analysis of proteins that interact with each IP by functional categories. (C) Network analysis of protein IP interactions. Blue line, Ins(1,3,4,5)P4 interactions; red line, Ins(1,3,4)P3 interaction; gray line, proteins that interact with both IPs. (D) Ratio of fold enrichment of proteins identified as interacting with Ins(1,3,4,5)P4 vs. Ins(1,4,5)P3. (E) GO enrichment analysis of proteins interacting with Ins(1,4,5)P3, Ins(1,3,4,5)P4, or both. (F) Heat map of metabolic proteins that interact with each IP (subset indicated by brackets in B). Fold enrichment values are indicated for each protein.
IPMK regulates oxidative phosphorylation inhibitor–sensitive ATP production in BFs
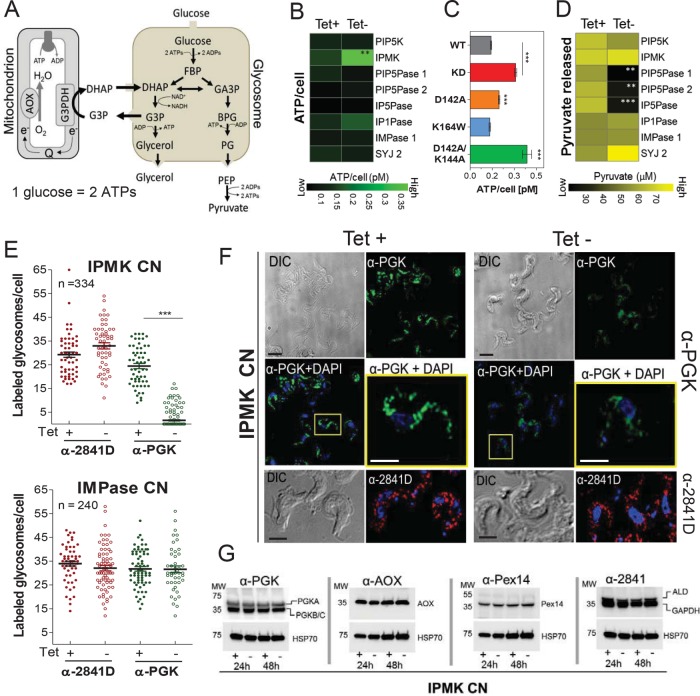
The affinity enrichment of proteins involved in energy metabolism by Ins(1,3,4,5)P4 suggests that the IPMK knockdown may affect, among other processes, cellular energy metabolism. T. brucei BF relies solely on glycolysis for energy production, consuming glucose to generate ATP and excreting pyruvate (Figure 6A). Knockdown of IPMK in BFs (under BF growth conditions) resulted in an increase in ATP levels that was 2.5-fold by 48 h, whereas knockdown of eight other IP pathway genes did not significantly affect ATP levels (Figure 6B). Exclusive expression of the catalytically inactive version of IPMK harboring the D142A/K144A mutations or the partially active D142A mutant also resulted in increased ATP levels, whereas exclusive expression of the K164W mutant (which partially affected 6-kinase activity) did not affect ATP levels (Figure 6C). Notably, a decrease rather than an increase in ATP level is associated with loss of viability (Sykes and Avery, 2009) and thus the ATP accumulation may result from changes in energy metabolism in these cells. In contrast, the amounts of pyruvate released into culture medium after 48 h IPMK knockdown were not significantly affected (Figure 6D). The decrease in pyruvate release after PIP5Pase 1 knockdown correlates with its early growth defect (Cestari and Stuart, 2015). However, there was also a decrease in pyruvate release after knockdown of PIP5Pase 2 or IP5Pase and an increase after knockdown of SYJ 2 (Figure 6C). Because these genes are not essential for T. brucei growth (Supplemental Figure S4; Cestari et al., 2016), the effect of their knockdown may relate directly to perturbations of the IP pathway.
FIGURE 6:
IPMK regulation of T. brucei energy metabolism. (A) Diagram of T. brucei BF glycolysis. Some steps have been omitted for simplicity. FBP, fructose 1,6-biphosphate; DHAP, dihydroxyacetone phosphate; G3P, glycerol-3-phosphate; G3AP; glyceraldehyde 3-phosphate; BPG, 2,3-bisphosphoglyceric acid, PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; AOX, alternative oxidase; G3PDH, glyceraldehyde 3-phosphate dehydrogenase (or GAPDH). (B–D) Measurements of ATP produced per cell (B, C) or pyruvate released into medium (D) after 48 h knockdown of IP pathway genes (B, D) or exclusive expression of IPMK mutant alleles (C). (E) Quantification and (F) IF analysis of the number of BF glycosomes labeled with antibodies against PGK, aldolase, and GAPDH after 48 h knockdown of IPMK or IMPase. α-2481D recognizes glycosomal aldolase and GAPDH. The pictures are representative of six independent experiments. n, number of cells counted. (G) Western blot analysis of T. brucei BF lysates after 24 and 48 h IPMK knockdown. Polyclonal antibodies (pAb) α-PGK detect all three PGK isoforms. Blots were stripped and reblotted with mAb α-HSP70. Tet, 0.5 µg/ml. Data are represented as means. *p < 0.05; **p < 0.01, ***p < 0.005 comparing tet- vs. tet+ conditions or IPMK mutant vs. WT alleles by t test.
Owing to the lack of feedback regulation of glycolytic enzymes in trypanosomes, ATP net production is controlled by compartmentalization of some glycolytic enzymes into glycosomes, and hence loss of glycosomes is toxic to BFs (Furuya et al., 2002; Haanstra et al., 2008). Glycosomes of BFs were analyzed by three-dimensional (3D) deconvolution IF microscopy of glycosomal PGK, aldolase, and GAPDH enzyme (Sommer and Wang, 1994). IPMK knockdown for 48 h under BF conditions resulted in a decrease of PGK-labeled glycosomes per cell, and PGK had a somewhat dispersed rather than a punctate localization (Figure 6, E and F). However, IPMK knockdown did not affect the expression levels of other glycosomal proteins or the overall number of glycosomes per cell (Figure 6, E–G). Knockdown of IMPase 1, which did not affect pyruvate production (Figure 6D), also did not affect PGK localization or glycosome numbers (Figure 6E). T. brucei expresses glycosomal PGKA (56 kDa) in BF and PF, whereas the PGKC (47kDa) and PGKB (45 kDa) are developmentally regulated and expressed in the glycosomes of BF and in the cytosol of PF, respectively (Blattner et al., 1998). Although this α-PGK antibody cannot distinguish PGK isoforms and size differences between PGKB and C cannot be resolved by Western blotting (Figure 6G), real-time PCR showed down-regulation of PGKC mRNA levels (Supplemental Figure S5). Thus, the excess of ATP production and PGK localization is not due to loss of glycosomes but may be a consequence of metabolic and developmental changes in BFs.
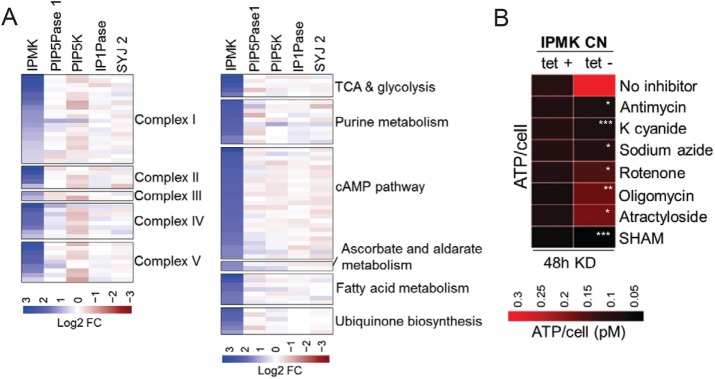
Because of the elevated ATP levels (Figure 6), we explored whether IPMK knockdown altered mitochondrial ATP production. RNAseq analysis after 36 h of IPMK knockdown revealed up-regulation of several genes encoding enzymes of the TCA cycle, fatty acid and ubiquinone metabolism, and the respiratory chain, including those encoding complexes II, III, and IV which are specific from T. brucei PFs (Figure 7A). Knockdown (under BF growth conditions) of four other genes of the IP pathway, PIP5K, PIP5Pase 1, IP1Pase, and SYJ 2, did not significantly affect expression of these genes (Figure 7A). Remarkably, the excess ATP produced after IPMK knockdown in BFs reverted to its normal levels, that is, no knockdown, in the presence of inhibitors of oxidative phosphorylation (Figure 7B), including rotenone, antimycin, cyanide and sodium azide, and oligomycin, which inhibit complexes I, III, IV, and V, respectively. ATP production was also sensitive to SHAM, which indicates that these cells still depend on the alternative oxidase (Figure 7B). The growth of T. brucei CN IPMK BF was unaffected by antimycin, K cyanide, or rotenone whether they express IPMK or not (Supplemental Figure S6), likely because these cells still have a functional glycolytic pathway for energy production. Hence, IPMK knockdown alters T. brucei ATP levels, likely by production of an active oxidative phosphorylation system in BFs, and possibly affects other metabolic pathways, all of which occurred in BFs and hence preceded their development to PFs.
FIGURE 7:
IPMK regulates oxidative phosphorylation inhibitor sensitive ATP production in BFs. (A) RNAseq analysis of metabolic genes after knockdown of IPMK (36 h) or other IP pathway genes (24 h). RNAseq data of knockdown (tet-) cells were compared with data on their respective nonknockdown (tet+) cells. (B) Measurements of ATP produced per cell after 48 h IPMK knockdown in absence or presence of oxidative phosphorylation inhibitors. Antimycin (8 mM), potassium cyanide (K cyanide, 50 µM), sodium azide (0.5 mM), rotenone (5 µM), oligomycin (50 ng/ml), atractyloside (200 µM), and SHAM (100 µM) were added to the culture after 24 h IPMK knockdown. Tet, 0.5 µg/ml. Data are represented as means. *p < 0.05; **p < 0.01, ***p < 0.005 comparing tet- conditions treated with inhibitors vs. tet- cells not treated (No inhibitor) by t test.
Regulation of energy metabolism is essential for BF-to-PF development
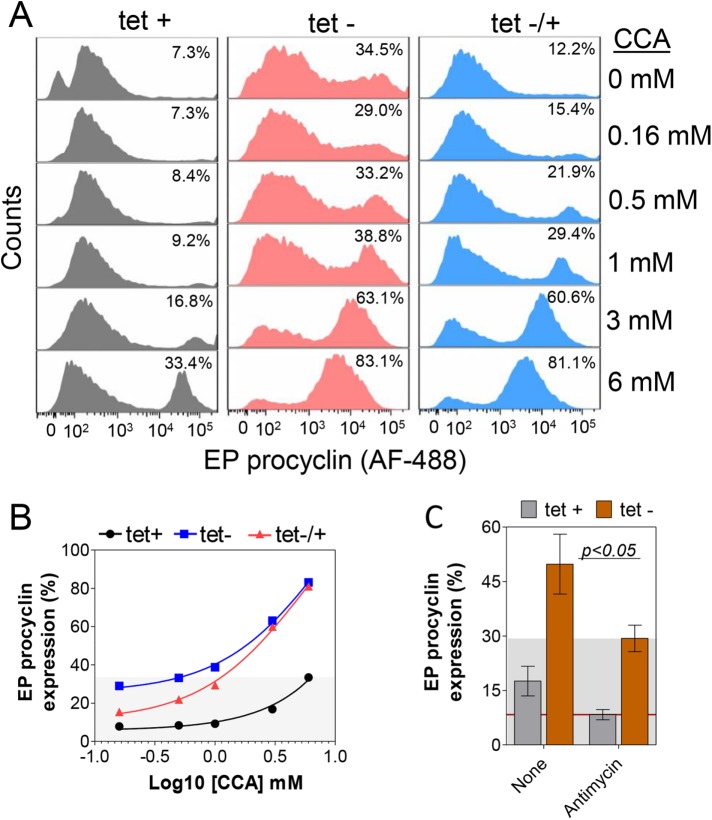
Because the metabolic and transcriptional changes in T. brucei BF after IPMK knockdown preceded development to PFs, we hypothesized that competence for BF-to- PF development is determined, at least in part, by T. brucei BFs attaining a metabolic state that favors the transition between the two stages. We tested this hypothesis by genetic and exogenous perturbation of T. brucei BF energy metabolism followed by analysis of cell development. After 36 h of IPMK knockdown under BF conditions, we transferred BF cells to SDM-79 at 27°C in the presence of various concentrations of CCA and monitored EP procyclin expression by flow cytometry after 3 d. About 35% of the IPMK knockdown T. brucei cells expressed EP procyclin even in the absence of CCA, and the proportion expressing procyclins increased steadily, reaching 83% in 6 mM CCA (Figure 8, A and B). Reexpression of IPMK after BF transfer to PF conditions resulted in a slightly lower rate of EP expression (also noted in Figure 1C), but also, 81% of the cells expressed EP procyclin in 6 mM of CCA. Not only did these cells express procyclins, but also, most of them had a PF morphology. In contrast, continuous expression of IPMK resulted in a lower proportion of T. brucei expressing EP procyclin, reaching only ∼33% in 6 mM of CCA. Thus, IPMK knockdown increased the proportion of T. brucei BFs that are competent to develop to PFs in response to CCA.
FIGURE 8:
Regulation of T. brucei energy metabolism is essential for development competence. (A) Flow cytometry analysis and quantification of cells expressing EP procyclin after IPMK knockdown in BF for 36 h in HMI-9 at 37°C followed by 3 d in SDM-79 at 27°C in the presence of (B) CCA (0.1–6 mM) or (C) antimycin (8 mM). Tet, 0.5 µg/ml. Data are represented as mean ± SEM.
Other TCA cycle intermediates, such as malate, α-ketoglutarate, fumarate or succinate, or myo-inositol, had no significant effect on the development of T. brucei SM427 (parental line) or IPMK CNs (Supplemental Figure S7). The effect of CCA is perhaps due to their uptake by specific transporters (Dean et al., 2009) and/or CCA activation of proteins involved in development (Szoor et al., 2010). In contrast, addition of antimycin, which inhibits oxidative phosphorylation, decreased T. brucei EP procyclin expression by 41% (Figure 8C). This indicates that production of an active oxidative phosphorylation system is essential for BF T. brucei to develop to PF, in agreement with previous data showing that mitochondrial activity is required for development (Timms et al., 2002). Hence, IPMK knockdown altered T. brucei energy metabolism and increased the competence of BFs to develop into PFs, and this was decreased by perturbation of oxidative phosphorylation.
DISCUSSION
We show here that IPMK functions in regulating T. brucei development. IPMK gene knockdown in BFs results in loss of VSGs and expression of procyclins, alteration of glycosomes, production of an active mitochondrial oxidative phosphorylation system, and subsequently morphological changes to PFs. The development is enhanced by addition of TCA cycle metabolites, accompanied by differential expression of RBPs and a PF transcriptome profile. In addition, proteins with roles in cell signaling, metabolism, and protein synthesis and turnover are affinity-enriched by Ins(1,4,5)P3 and Ins(1,3,4,5)P4, the substrate and product of IPMK. A possible explanation is that IPMK function within the complex regulatory network that controls T. brucei development, perhaps via interactions of IPs with metabolic and/or regulatory proteins. Perturbation of the network, such as by changes in IPMK expression (or enzyme activity/interactions), may alter IP metabolite levels and perhaps their subcellular locations. These alterations may impact specific IP binding by proteins of the network, which may affect their functions, such as enzymatic activity, protein interactions, and/or subcellular location. Such “second messenger regulation” has been reported in other systems (Szijgyarto et al., 2011; Seeds et al., 2015; Wu et al., 2016). These changes can result in a cascade of events such as activation of signaling proteins and changes in gene expression, such as mRNA turnover, that further remodels cell metabolism, such as production of an active oxidative phosphorylation system, to establish a physiological state that favors BF-to-PF development.
The regulation of BF-to-PF development entails processes that may be coordinated by successive steps of the IP pathway. The IPMK substrate Ins(1,4,5)P3 is the product of PLC. Overexpression of PLC or knockdown of PIP5K or PIP5Pase 1, which respectively produce and eliminate PLC's substrate (Figure 1B), dysregulate monoalellic expression of telomeric VSG genes and VSG switching (Cestari and Stuart, 2015). Regulation of VSG expression revolves around PI phosphorylation at position 5 (Cestari and Stuart, 2015). However, BF-to-PF development appears to involve IPMK, which phosphorylates Ins(1,4,5)P3 at position 3 and/or 6 (Cestari et al., 2016). Our previous enzymology analysis showed that T. brucei IPMK phosphorylates Ins(1,4,5)P3 to generate Ins(1,3,4,5)P4 and also Ins(1,3,4,5,6)P5 (Cestari et al., 2016), which was confirmed by others (Cordeiro et al., 2017). T. brucei IPMK has low amino acid–sequence homology with human IPMK but conserves the catalytic residues, which are also conserved among yeast, Arabidopsis, Plasmodium, and Drosophila (Figure 2A; Cestari et al., 2016). The results from the IMPK D142A/K144A double mutant, which was unable to phosphorylate Ins(1,4,5)P3 and Ins(1,3,4,5)P4, demonstrate that the role of IPMK in T. brucei development involves its catalytic activity. Although further metabolomics analysis is needed to determine the in vivo consequences of IPMK knockdown for IP metabolites and their fluxes, the data suggest that IPMK expression and its effects on IP phosphorylation are involved in the regulation of BF-to-PF developmental processes. It is possible that IPMK may have other noncatalytic functions, such as protein associations, that are involved in the regulation of energy metabolism and development in T. brucei. Human IPMK interacts with TOR complex 1 to regulate amino acid signaling (Kim et al., 2011), and glucose induces phosphorylation of IPMK as well as its interaction with and down-regulation of AMPK (Bang et al., 2012). T. brucei AMPK and TOR complexes are involved in BF slender to stumpy development in response to AMP and oxidative stress (Barquilla et al., 2012; Saldivia et al., 2016), although it is not known whether T. brucei IPMK interacts with these complexes. IPMK also regulates the switching between glycolysis and oxidative phosphorylation in yeast and mammalian cells (Szijgyarto et al., 2011) via its interaction with a partner of the Arg80-Mcm1 transcriptional complex that controls Pol II transcription of some metabolic genes. Although T. brucei lacks Pol II transcriptional control, it regulates mRNA levels posttranscriptionally (Clayton, 2016), and IPMK and IPs may regulate mRNA nuclear export, as has been shown in yeast and mammalian cells (York et al., 1999; Wickramasinghe et al., 2013). Hence, IPMK catalytic and perhaps noncatalytic roles are involved in the regulation of T. brucei developmental processes.
While typically a very small proportion of cells develop into PF (∼1–3%), IPMK knockdown and its inactivation both resulted in nearly 40% of the cells developing into PFs (Figure 1, C and D). The knockdown of other IP pathway genes, PIP5K, PIP5Pase 1, or CDS, did not result in BF-to-PF development. Thus, it appears that specific perturbation of the IP pathway via IPMK function results in development of PFs. Also, addition of CCA after IPMK knockdown increases the proportion of cells that develop into PFs to 83% (Figure 8). The addition of CCAs, which are TCA–cycle intermediates, may affect the intracellular abundance of metabolites (Overath et al., 1986) or activate signaling proteins (Szoor et al., 2010) and further impact cell development. Although a substantial proportion of cells developed into PFs and survived for nearly 3 wk, they could not be cultivated continuously. This is likely because the cell line used in these studies is derived from the 427 strain, which can develop into PFs if stimulated by CCA, but is unable to be continuously cultivated for unknown reasons (Overath et al., 1986). The manipulation of the IPMK gene may have affected the cell's regulatory network that controls development and thus the increased cell's sensitivity to CCA.
IPs can function as second messengers via binding to proteins, which regulates their functions (Lemmon, 2008). BF proteins that were affinity-enriched with the IPMK substrate or product are associated with various processes (Figure 6C), as seen in other eukaryotes (Wu et al., 2016; Jungmichel et al., 2014). Some of the enriched proteins have IP- or PI-binding domains, such as PH or Phox domains (Supplemental Dataset S2), but others lack such domains, perhaps reflecting the limited number of such characterized domains or low sequence conservation in T. brucei proteins. We confirmed some of the IP-protein interactions which included proteins involved in metabolism, DNA binding, flagellar function, protein folding and translation. Further validations of these interactions, such as by mutational analysis of potential interacting domains and IP binding assays, will be important to assess their functions in T. brucei. Although further studies are necessary to comprehensively identify IP interactomes, our data indicate potential roles for protein interactions and IP metabolites. Generally, T. brucei proteins enriched by Ins(1,4,5)P3 are associated with cell motility, flagellum and cytoskeleton organization whereas those enriched by Ins(1,3,4,5)P4 are associated with metabolism, such as glycolysis, amino acid metabolism, purine synthesis, and the TCA cycle, namely processes that differ between BF and PF. Thus IP-protein interactions may function in the control of developmental processes in T. brucei, perhaps by affecting protein activity and/or interactions.
T. brucei energy metabolism is developmentally regulated, with BFs relying on glycolysis and PFs on oxidative phosphorylation. BFs only have complexes I (NADH: ubiquinone oxidoreductase) and V (F0F1 ATPase/ATP synthase), and the latter hydrolyzes rather than generates ATP (Schnaufer et al., 2005). IPMK knockdown in BFs results in increased ATP production that is sensitive to oxidative phosphorylation inhibitors of complexes I, III (coenzyme Q: cytochrome c oxidoreductase), IV (cytochrome c oxidase), and V. The production of ATP by these inhibitor-sensitive complexes correlated with the up-regulation of genes encoding respiratory chain proteins expressed by PFs, which also requires the PF type of editing of mRNAs that are encoded in mitochondrial DNA (Feagin et al., 1988). Although IPMK knockdown had little effect on pyruvate production or glycosome numbers, PGK signal was somewhat diffuse in the cytosol, probably due to down-regulation of the glycosomal PGKC. Other glycosomal proteins appeared unaffected, suggesting that protein sorting to this organelle was not affected. These changes in mitochondrial and glycosome content and function that occur in BFs after IPMK knockdown resemble the differences between the BF and PF stages. The metabolic changes may explain the lethality of IPMK knockdown to T. brucei BFs, given that perturbation of glycosomal function is toxic to BFs (Blattner et al., 1998; Furuya et al., 2002). Nevertheless, other factors independent of oxidative phosphorylation or carbon metabolism may also contribute to ATP changes. Further metabolomics studies would be necessary to unravel all metabolic changes associated with IPMK knockdown, given the broad role of IPMK in regulating metabolism (Szijgyarto et al., 2011; Bang et al., 2012), gene expression and nuclear mRNA export (York et al., 1999; Bosch and Saiardi, 2012) and polyphosphate production (Freimoser et al., 2006; Cordeiro et al., 2017).
The transcriptome changes that follow IPMK knockdown also parallel differences between BF and PF cells, including those associated with glucose and carboxylic acid transport, pyruvate and acetyl-coA metabolism, ATP synthesis and cAMP signaling, and surface proteins. T. brucei gene expression is primarily regulated posttranscriptionally and RBPs play a role in this process (Kolev et al., 2012; Wurst et al., 2012; Najafabadi et al., 2013). Overexpression of RBP6, RBP7A or PPCT1 affect expression of BF and PF gene sets that we found were also upregulated after IPMK knockdown. The mechanism by which IPMK affects gene expression in T. brucei is unknown. IPMK may regulate gene expression by direct association with RNAs or alternatively by production of IPs that may regulate processes involved in RNA posttranscriptional control; for example, IPMK is involved in regulating nuclear mRNA export in yeast and mammalian cells (York et al., 1999; Wickramasinghe et al., 2013). Alternatively, T. brucei may have a regulatory system that integrates cell metabolism and RBP function, which controls mRNA transport, stability, or degradation, perhaps analogously to riboswitches in prokaryotes (Garst et al., 2011). The RBP effects on PF and BF gene sets are dependent on growth conditions, such as medium composition and temperature, suggesting that changes in cell physiology may impact RBP function and gene expression. Such types of regulation were shown by the knockdown of mitochondrial metabolic enzymes or hypoxia, which affect T. brucei gene expression (Vassella et al., 2000), and by the identification of a 25nt glycerol-responsive element in the 3′-UTR of GPEET procyclin mRNAs that regulates its developmental expression (Vassella et al., 2004). We did not find conserved nucleotide sequences in the 3′ or 5′-UTRs of genes that were differentially regulated after IPMK knockdown, although structural conformations of these UTRs may affect their interactions with RBPs or regulatory proteins.
Only a portion of the BF population developed to PFs after IPMK knockdown, perhaps due to heterogeneity within the cell population, that is, asynchrony of cell cycle, transcriptome, or metabolome, which suggests that a particular physiological state may be needed for T. brucei BFs to develop into PFs. This may be the case for stumpy BFs, which are apparently preadapted to develop into PFs (MacGregor et al., 2012). The changes in energy metabolism and transcriptome resulting from IPMK knockdown may have affected the proportion of cells that are competent to develop. This was illustrated by an increase in development of IPMK knockdown cells in the presence of CCA or a decrease by inhibition of oxidative phosphorylation. Such a physiological state is also reflected by dramatic changes in expression levels of metabolic proteins during T. brucei BF-to-PF development (Timms et al., 2002; Domingo-Sananes et al., 2015; Dejung et al., 2016). Hence, the competence for development appears to be affected by the cell's physiological state, which can be altered by IPMK knockdown. This competence is likely a temporal condition that anticipates cells’ commitment to development. Reexpression of IPMK after its knockdown did not prevent BF-to-PF development, which implies that some cells committed to this process. This commitment may result from a combination of factors such as signal transduction, metabolic changes, proteins turnover, and changes in gene expression (Wurst et al., 2012; Domingo-Sananes et al., 2015). IPMK overexpression did not affect development, perhaps due to some feedback regulation or posttranslational modification that maintains control of IPMK function. IPMK expression does not appear to change during development to PFs (Jensen et al., 2014), but its activity, which is critical for BF-to-PF development, could be posttranslationally regulated.
The early evolutionary divergence of T. brucei (Excavata) from opisthokonts suggests that the IPMK role in energy metabolism and development, which seems conserved among T. brucei, yeast, and mammals, likely appeared early in evolution. However, the divergent mechanisms of T. brucei gene regulation, which depends on posttranscriptional processes for control of genes transcribed by RNA polymerase II, and its reliance on RNA polymerase I for transcription of certain protein coding genes suggest that aspects of the processes that regulate the development may also be divergent in T. brucei, as they may be in other parasitic protozoa with complex life cycles, such as Leishmania, Plasmodium, and Toxoplasma.
MATERIALS AND METHODS
Cell culture, transgenic cell lines, and growth curves
T. brucei SM427, null, CN, and V5-tagged cell lines were generated and maintained in HMI-9 at 37°C with 5% CO2 as previously described (Cestari and Stuart, 2015; Cestari et al., 2016). Gene IDs and product descriptions are Tb927.9.12470, inositol polyphosphate multikinase (IPMK; Cestari and Stuart, 2015); Tb927.4.1620, phosphatidylinositol 5-kinase 1 (PIP5K 1; Cestari and Stuart, 2015); Tb927.11.6270, phosphatidylinositol (4,5)/(3,4,5)-phosphate 5-phosphatase (PIP5Pase 1; Cestari and Stuart, 2015), Tb927.7.220, CDP-diacylglycerol synthase (Cestari and Stuart, 2015); Tb927.9.5680; phosphatidylinositol 5-phosphatase (PIP5Pase 2; Cestari et al., 2016); Tb927.10.5510, inositol polyphosphate 5-phosphatase (IP5Pase; Cestari et al., 2016); Tb927.8.7170, inositol polyphosphate 1-phosphatase (IP1Pase; Supplemental Figure S4); Tb927.9.6350, inositol-1(or -4)-monophosphatase 2 (IMPase 2; Cestari et al., 2016); and Tb927.7.3490, synaptojanin (IP or PI 5-phosphatase) 2 (SYJ 2; Supplemental Figure S4). For generation of IPMK CN exclusively expressing WT or mutant versions (D142A, K164W, or D142A/K144A) of IMPK, the IPMK genes were amplified from pET-29-IPMK WT or mutants (see Recombinant protein and enzymatic activity for details on the generation of mutated IPMK) using the forward and reverse primers, respectively CCCGGTCTCAAGCTTATGTTAAATATTTGCCAAAACTTGTCTTC and CCCGGTCTCGGATCCTGAAAGAAGAAAAATAATTTTTTCCA, and cloned into pHD-tub-3V5 vector using the HindIII and BamHI sites. Plasmids were integrated into the constitutive tubulin locus by cell electroporation and puromycin resistance selection. Cumulative growth curve analyses were performed as previously described (Cestari and Stuart, 2015). Briefly, T. brucei BFs diluted daily to 5.0 × 104 parasites/ml were grown in the presence or absence of tet (0.5 µg/ml) and counted daily using a cell counter (Beckman), and the procedure was repeated for five to seven consecutive days.
Viability analysis
Viability assays were performed as previously described (Cestari and Stuart, 2013). Briefly, compound stocks were prepared at a 10 mM–1 M concentration in dimethyl sulfoxide (DMSO) or ethanol. T. brucei BF (100 µl at 2.0 × 104 parasites/ml) was plated in 96-well plates and mixed with 100 µl of compounds at twofold serial dilutions: SHAM (100–0.2 mM), oligomycin (10–0.02 µg/ml), sodium azide (40–0.08 µM), antimycin (40–0.08 µM), rotenone (100–0.2 µM), and potassium cyanide (100–0.2 µM), all diluted in HMI-9 medium with 10% fetal bovine serum (FBS). Parasites not treated with compounds were also plated as controls. After 36 h of incubation at 37°C and 5% CO2, 20 µl of alamarBlue (Invitrogen) was added, and the assays were developed for 4 h. Fluorescence measurements were obtained using a SpectraMax M2 microplate reader (Molecular Devices) with excitation at 544 nm and emission at 590 nm (590-nm cutoff). Data were analyzed using GraphPad Prism for Windows.
IF and Western blot analysis
For IF, mid–log phase T. brucei were fixed with 2% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), adhered to a poly-l-lysine–treated 2-mm cover glass (Fisher), permeabilized with 0.2% NP40 in PBS for 5 min, and blocked for 1 h with 3% BSA in PBS. Cells were incubated for 2 h at room temperature (RT) with pAb α-PGK (1789G), α-gGAPDH/Aldolase (2481D), or mAbs α-EP procyclin, clone tbrp1/247 (Cedarlane Laboratories), followed by goat α-rabbit or goat α-mouse immunoglobulin G (IgG) (H+L)–Alexa Fluor 488 or 568 (Molecular Probes); see Supplemental Table S1 for all antibody dilutions. DNA was stained with 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma) and slides mounted with ProLong Gold Antifade Reagent (Life Technologies). Images were acquired with a Deltavision 3D deconvolution microscope (Olympus IX70) and analyzed with Softworks software (Applied Precision). Western blots were performed as described previously (Cestari and Stuart, 2015). Briefly, lysates of T. brucei BF in 1% Triton X-100 in PBS with a protease inhibitors cocktail (Roche) were separated in 4–20% Mini-PROTEAN TGX gel (Bio-Rad) and transferred to immobilon-P PVDF (Millipore). Membranes were probed overnight (ON) at 4°C with abs α-PGK, α-gGAPDH/Aldolase, α-HSP70, α-AOX, α-Pex14, α-procyclins, or α-VSGs, followed by goat α-rabbit or goat α-mouse IgG (H+L) conjugated to horseradish peroxidase (HRP) and developed by chemiluminescence.
Recombinant protein and enzymatic activity
The T. brucei gene encoding IPMK was cloned into pET-29a (Novagen) and expressed and purified from E. coli DE3 pLysS Rosetta cells (Novagen) as previously reported (Cestari et al., 2016). Site-directed mutagenesis of IPMK was performed using the pET-29-IPMK construct and a Q5 site-directed mutagenesis kit (New England Biolabs) according to the manufacturer's instructions. The forward and reverse primers used in site-direct mutagenesis reactions were as follows: D142A: TTGTGTGCTTGCTATCAAACTTG and GGTTTATGAAATGTCGCG, K164W: GCGCATACATTGGAGGCAGCTTC and TCCACCTTGTCGGGTAAT, and D142A/K144A: GCTTGCTATCGCACTTGGATATGTG and ACACAAGGTTTATGAAATGTC. Protein activity was assayed in 30-µl reactions in 96-well plates in a buffer consisting of 20 mM HEPES, 150 mM NaCl, and 2 mM MgCl2 (pH 7.5). The reactions also contained 300 nM rTbIPMK WT or a mutant version thereof, 5–200 mM Ins(1,4,5)P3 or Ins(1,2,4,5)P4 (Echelon Biosciences), and 200 µM of ATP. Reactions were incubated for 60 min at 37°C, after which IMPK activity was measured using the ADP luciferase assay (Promega; Cestari et al., 2016) and a Glomax plate reader (Promega). Kinetic analyses were calculated by nonlinear regression using GraphPad Prism for Windows 5.03 (GraphPad Software).
IP interactions and proteomic analysis and quantification
A sample of 2.0 × 109 mid–log phase T. brucei BFs was harvested from 2 l of culture by centrifugation at 4000 rpm for 10 min at RT, resuspended in 1% Triton X-100 in 50 mM Tris, 150 mM NaCl, and 0.2% NP40 with a protease inhibitor cocktail (Roche) and rotated for 15 min at 4°C for complete cell lysis. The resultant lysates were cleared by centrifugation at 14,000 × g at 4°C and then incubated with agarose beads conjugated with Ins(1,4,5)P3, Ins(1,3,4,5)P4, or PI(3,4,5)P3 or nonconjugated beads in binding buffer 50 mM Tris, 150 mM NaCl, 0.2% NP40, 5 mM MgCl2, pH 7.4, and protease inhibitors (Roche). IPs immobilized on agarose beads were purchased from Echelon Biosciences. Each 1 ml of agarose beads contains 10 nmol of bound IP metabolites as described (Jungmichel et al., 2014; Wu et al., 2016). For confirmation of IP–protein binding, lysates of 5.0 × 107 mid–log growth T. brucei BFs were incubated with biotin-Ins(1,4,5)P3 or biotin-Ins(1,3,4,5)P4 (Echelon Biosciences; biotin linked through the 1-position on the inositol ring) in binding buffer (as above) followed by incubation with 50 µl of Dynabeads M-270 Streptavidin (ThermoFisher Scientific). After 2 h rotation at 4°C the beads were washed with 50 mM Tris, 300 mM NaCl, 0.2% NP40, and 5 mM MgCl2, pH 7.4, and fractions were eluted with Laemmli sample buffer with 5% β-mercaptoethanol. Samples were separated in 4–20% SDS–PAGE and transferred for Western blot analysis or precipitated with cold acetone and processed for mass spectrometry as described (Cestari et al., 2013). Mass spectrometry analyses were performed at the Fred Hutchinson Cancer Research Center using an LTQ-OrbiTrap. MaxQuant was used for peptide and protein identification with mass tolerance of ±10 ppm, fragment mass tolerance of ±0.5 Da, monoisotopic mass values, C13 = 2, against the T. brucei v5.0 predicted protein sequence database. Label-free quantitative analysis was performed using the R package MSstats by comparing each IP–protein interaction against agarose beads–protein interactions as described by Clough et al. (2012). Data include three biological replicates for affinity purifications with nonconjugated agarose beads; three biological replicates for Ins(1,4,5)P3-agarose conjugate beads; two biological replicates for Ins(1,3,4,5)P4-agarose conjugated beads; and two biological replicates for PI(3,4,5)P3-agarose conjugated beads. See Supplemental Dataset 2 for a list of all proteins and peptides identified. The proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD005907.
RNA analysis and sequencing
Mid–log phase CN cell lines were grown with or without 0.5 µg/ml tet for 24 and 36 h and harvested at RT (10 min, 1300 × g) and RNAs were extracted using TRIzol (Life Technologies). Real-time PCR was performed as described by Cestari and Stuart (2015). Supplemental Table S2 lists the primers. RNAseq of poly-A enriched RNAs was performed as described by Kolev et al. (2015). Libraries were sequenced at the Department of Genome Sciences, University of Washington, on two biological replicates for each experiment (see Supplemental Table S3 for a detailed description of library sequences and a biological replicate correlation analysis). Reads alignment and gene ID mapping were done using Bowtie2 and HTSeq, respectively, against the T. brucei 427 genome tritrypDB version 9. Read counts were obtained with Get_ReadCount.py and differential gene expression using edgeR (Robinson et al., 2010) using a likelihood ratio test (McCarthy et al., 2012). A minimum of one count per million (CPM) per gene, that is, five counts per gene in the smallest library, and counts represented in at least two biological replicate libraries were required for a gene to be considered for analysis. Genes were considered differentially expressed when fold changes were ≥2 and p values ≤ 0.05 (Supplemental Dataset S1). Data have been deposited to the SRA repository with the data set identifiers SRR5314671, SRR5314672, SRR5315219, and SRR5315220. GSEA was performed using the phenoTest package in R (Subramanian et al., 2005; Planet, 2013). Gene sets were constructed with genes that are differentially expressed in BF slender, intermediate, stumpy, and PFs (Capewell et al., 2013), and include genes that are up-regulated (72 genes) or down-regulated (254 genes) in BF stumpy compared with BF slender; and up-regulated (130 genes) or down-regulated (178 genes) in BF intermediate compared with BF slender. It also includes sets of genes that are up-regulated in BF compared with PF (157 genes) or PF compared with BF (70 genes). Gene sets were analyzed using 1000 permutation tests by comparing gene sets against the RNAseq data of IPMK knockdown at 36 h. False discovery rates (q) were calculated by the Benjamini–Hochberg procedure and significance indicated by q-value <0.05. Gene sets were represented using a barcode plot using the Limma package in R (Ritchie et al., 2015).
Metabolic analysis
IP pathway genes were knocked down for 24 and 48 h, and cells (100 µl) were incubated with a 100 µl CellTiter-Glo luminescent cell viability assay (Promega) to measure ATP using a GloMax 96-microplate lLuminometer (Promega). In parallel, cell supernatants were quantified using a pyruvate assay kit, both according to manufacturer's instructions (Eton Bioscience). Antimycin (8 mM), K cyanide (50 µM), sodium azide (0.5 mM), rotenone (5 µM), oligomycin (50 µg/ml), atractyloside (200 µM), and SHAM (100 µM) were incubated with CN IPMK 24 h after knockdown and ATP was measured at 48 h as described above.
Development assays and flow cytometry analysis
T. brucei IPMK, PIP5K, PIP5Pase 1, or CDS CNs were grown (∼1.5 × 106 parasites/ml) with or without tet (0.5 µg/ml) for 36 h in 40 ml HMI-9 medium at 37°C and 5.0 × 107 parasites were transferred to 2 or 10 ml SDM-79 and cultured for 3 or 10 d at 27°C. Cells were counted every 2 d by monitoring BF or PF morphology under light microscopy in Neubauer chambers. Aliquots of 1 ml of cells were collected at days 3 and 4 for EP procyclin expression analysis by fluorescence microscopy (see above) or by flow cytometry (details below). T. brucei IPMK CN BFs or SM427 were also incubated in CCA (0.16, 0.5, 1, 3, and 6 mM) or 8 mM antimycin, malate (10 mM), α-ketoglutarate (10 mM), succinate (10 mM), or myo-inositol (10 mM) upon transfer to SDM-79 medium at 27°C; then procyclin expression were analyzed after 3 d by flow cytometry. For procyclin analysis after IPMK overexpression, T. brucei BF that express tet-regulatable C-terminally V5-tagged IPMK were grown at late log phase (∼1.5 × 106 parasites/ml) without tet in HMI-9 at 37°C for 24 h. Afterward, cells were transferred to 24-well plates at 2.0 × 107 parasites/ml without or with tet (1 µg/ml) and cultured in SDM-79 at 27°C for 3 d in the presence of various concentrations of CCA (0.16, 0.5, 1, 3, and 6 mM). Aliquots of 1 ml of culture were fixed in 2% PFA, blocked for 1 h with 3% BSA in PBS, and incubated for 2 h with mAbs α-EP procyclin (Cedarlane Laboratories; see Supplemental Table S1 for antibody dilutions). These cells were washed three times with PBS, incubated for 1 h with mAbs α-mouse IgG–Alexa Fluor 488 (Molecular Probes; see Supplemental Table S1), and washed three times with PBS. Ten thousand cells in PBS were analyzed by flow cytometry using a BD LSR II flow cytometer (BD Biosciences) and FlowJo software (Flowjo). For exclusive expression of Trypanosoma cruzi or Leishmania major IPMK, T. brucei IPMK CN constitutively expressing L. major or T. cruzi IPMK allele introduced in the tubulin locus (Cestari et al., 2016) were grown at late log phase (∼1.5 × 106 parasites/ml) with tet (0.5 µg/ml) for 36 h in 40 ml HMI-9 medium at 37°C. Afterward, cells were washed and 2 ml aliquots were transferred to 24-well plates at 2.0 × 107 parasites/ml without or with tet (1 µg/ml) and cultured in SDM-79 at 27°C for 3 d. Cells were fixed and stained with α-EP procyclin antibodies and analyzed by flow cytometry (all as described above). For PAD1 analysis, T. brucei IPMK CN were grown at late log phase (∼1.5 × 106 parasites/ml) with or without tet (0.5 µg/ml) for 24 and 36 h in 40 ml HMI-9 medium at 37°C. Then 10-ml aliquots were fixed in 1% PFA and blocked in PBS 10% FBS (vol/vol) for 1 h at RT. Cells were washed three times in PBS, incubated in PBS 5% FBS (vol/vol) with pAb α-PAD1 (see Supplemental Table S1) for 2 h at RT, washed three times in PBS, and then incubated with goat α-rabbit IgG (H+L)–Alexa Fluor 568 (Molecular Probes; see Supplemental Table S1) as indicated above. Cells were washed three times in PBS and analyzed by flow cytometry as described.
Data presentation and statistical analysis
Data are shown as means ± SEMs of at least three biological replicates unless otherwise stated. Comparisons among groups were made by a two-tailed t test using GraphPad Prism. P values <0.05 with a confidence interval of 95% were considered statistically significant unless otherwise stated. Graphs were prepared using GraphPad Prism (GraphPad Software) or RStudio (RC Team, 2015).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant R01AI078962 to K.S. and Supplement R01AI014102-37S1 to K.S. and I.C. and by the Center for Infectious Disease Research. We thank Marilyn Parsons for comments on the manuscript and for providing antibodies against Pex14, PGK, GAPDH, Aldolase, and PAD1, Anna Wrem for administrative support, and Lindsay N. Carpp for technical editing of the manuscript.
Abbreviations used:
- BF
bloodstream form
- CCA
citrate and cis-aconitate
- CN
conditional null
- IP
inositol phosphate
- IPMK
inositol polyphosphate multikinase
- PF
procyclic form
- PI
phosphatidylinositol
- RBP
RNA-binding proteins
- TCA
tricarboxylic acid
- VSGs
variant surface glycoproteins
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-08-0515) on March 5, 2018.
REFERENCES
- Bang S, Kim S, Dailey MJ, Chen Y, Moran TH, Snyder SH, Kim SF. (2012). AMP-activated protein kinase is physiologically regulated by inositol polyphosphate multikinase. Proc Natl Acad Sci USA , 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A, Saldivia M, Diaz R, Bart JM, Vidal I, Calvo E, Hall MN, Navarro M. (2012). Third target of rapamycin complex negatively regulates development of quiescence in Trypanosoma brucei. Proc Natl Acad Sci USA , 14399–14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batram C, Jones NG, Janzen CJ, Markert SM, Engstler M. (2014). Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife , e02324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner J, Helfert S, Michels P, Clayton C. (1998). Compartmentation of phosphoglycerate kinase in Trypanosoma brucei plays a critical role in parasite energy metabolism. Proc Natl Acad Sci USA , 11596–11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochud-Allemann N, Schneider A. (2002). Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem , 32849–32854. [DOI] [PubMed] [Google Scholar]
- Bosch D, Saiardi A. (2012). Arginine transcriptional response does not require inositol phosphate synthesis. J Biol Chem , 38347–38355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capewell P, Monk S, Ivens A, Macgregor P, Fenn K, Walrad P, Bringaud F, Smith TK, Matthews KR. (2013). Regulation of Trypanosoma brucei Total and polysomal mRNA during development within its mammalian host. PLoS One , e67069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Haas P, Moretti NS, Schenkman S, Stuart K. (2016). Chemogenetic characterization of inositol phosphate metabolic pathway reveals druggable enzymes for targeting kinetoplastid parasites. Cell Chem Biol , 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Kalidas S, Monnerat S, Anupama A, Phillips MA, Stuart K. (2013). A multiple aminoacyl-tRNA synthetase complex that enhances tRNA-aminoacylation in African trypanosomes. Mol Cell Biol , 4872–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Stuart K. (2013). Inhibition of isoleucyl-tRNA synthetase as a potential treatment for human African trypanosomiasis. J Biol Chem , 14256–14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Stuart K. (2015). Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc Natl Acad Sci USA , E2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Stuart K. (2018). Transcriptional regulation of telomeric expression sites and antigenic variation in trypanosomes. Curr Genomics , 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez M, Ena S, Van Sande J, de Kerchove d'Exaerde A, Schurmans S, Schiffmann SN. (2015). Modulation of ciliary phosphoinositide content regulates trafficking and sonic hedgehog signaling output. Dev Cell , 338–350. [DOI] [PubMed] [Google Scholar]
- Clayton CE. (2016). Gene expression in kinetoplastids. Curr Opin Microbiol , 46–51. [DOI] [PubMed] [Google Scholar]
- Clough T, Thaminy S, Ragg S, Aebersold R, Vitek O. (2012). Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics (Suppl 16), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro CD, Saiardi A, Docampo R. (2017). The inositol pyrophosphate synthesis pathway in Trypanosoma brucei is linked to polyphosphate synthesis in acidocalcisomes. Mol Microbiol , 319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S, Marchetti R, Kirk K, Matthews KR. (2009). A surface transporter family conveys the trypanosome differentiation signal. Nature , 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejung M, Subota I, Bucerius F, Dindar G, Freiwald A, Engstler M, Boshart M, Butter F, Janzen CJ. (2016). Quantitative Proteomics Uncovers Novel Factors Involved in Developmental Differentiation of Trypanosoma brucei. PLoS Pathog , e1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Sananes MR, Szoor B, Ferguson MA, Urbaniak MD, Matthews KR. (2015). Molecular control of irreversible bistability during trypanosome developmental commitment. J Cell Biol , 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E, Dewaste V, Erneux C, Messenguy F. (2000). Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett , 300–304. [DOI] [PubMed] [Google Scholar]
- Endo-Streeter S, Tsui MK, Odom AR, Block J, York JD. (2012). Structural studies and protein engineering of inositol phosphate multikinase. J Biol Chem , 35360–35369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin JE, Abraham JM, Stuart K. (1988). Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell , 413–422. [DOI] [PubMed] [Google Scholar]
- Freimoser FM, Hurlimann HC, Jakob CA, Werner TP, Amrhein N. (2006). Systematic screening of polyphosphate (poly P) levels in yeast mutant cells reveals strong interdependence with primary metabolism. Genome Biol , R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, Kessler P, Jardim A, Schnaufer A, Crudder C, Parsons M. (2002). Glucose is toxic to glycosome-deficient trypanosomes. Proc Natl Acad Sci USA , 14177–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT. (2011). Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol , a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haanstra JR, van Tuijl A, Kessler P, Reijnders W, Michels PA, Westerhoff HV, Parsons M, Bakker BM. (2008). Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc Natl Acad Sci USA , 17718–17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BC, Ramasamy G, Vasconcelos EJ, Ingolia NT, Myler PJ, Parsons M. (2014). Extensive stage-regulation of translation revealed by ribosome profiling of Trypanosoma brucei. BMC Genomics , 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Sylvestersen KB, Choudhary C, Nguyen S, Mann M, Nielsen ML. (2014). Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry. Cell Rep , 578–591. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, Chakraborty A, Koldobskiy MA, Cha SH, Barrow R, et al. (2011). Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab , 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. (2012). Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science , 1352–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ullu E, Tschudi C. (2015). Construction of Trypanosoma brucei Illumina RNA-Seq libraries enriched for transcript ends. Methods Mol Biol , 165–175. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. (2008). Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol , 99–111. [DOI] [PubMed] [Google Scholar]
- MacGregor P, Szoor B, Savill NJ, Matthews KR. (2012). Trypanosomal immune evasion, chronicity and transmission: an elegant balancing act. Nat Rev Microbiol , 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res , 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mony BM, MacGregor P, Ivens A, Rojas F, Cowton A, Young J, Horn D, Matthews K. (2014). Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature , 681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Lu Z, MacPherson C, Mehta V, Adoue V, Pastinen T, Salavati R. (2013). Global identification of conserved post-transcriptional regulatory programs in trypanosomatids. Nucleic Acids Res , 8591–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overath P, Czichos J, Haas C. (1986). The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei. Eur J Biochem , 175–182. [DOI] [PubMed] [Google Scholar]
- Planet E. (2013). phenoTest: Tools to Test Association between Gene Expression and Phenotype in a Way That Is Efficient, Structured, Fast and Scalable. We Also Provide Tools to Do GSEA (Gene Set Enrichment Analysis) and Copy Number Variation. Bioconductor R Package, Version 1.23.1.
- RC Team (2015). R: A Language and Environment for Statistical Computing, Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res , e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics , 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roditi I, Schwarz H, Pearson TW, Beecroft RP, Liu MK, Richardson JP, Buhring HJ, Pleiss J, Bulow R, Williams RO, et al. (1989). Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol , 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolin S, Hancocq-Quertier J, Paturiaux-Hanocq F, Nolan DP, Pays E. (1998). Mild acid stress as a differentiation trigger in Trypanosoma brucei. Mol Biochem Parasitol , 251–262. [DOI] [PubMed] [Google Scholar]
- Saldivia M, Ceballos-Perez G, Bart JM, Navarro M. (2016). The AMPKalpha1 pathway positively regulates the developmental transition from proliferation to quiescence in Trypanosoma brucei. Cell Rep , 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. (2005). The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J , 4029–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeds AM, Tsui MM, Sunu C, Spana EP, York JD. (2015). Inositol phosphate kinase 2 is required for imaginal disc development in Drosophila. Proc Natl Acad Sci USA , 15660–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer JM, Wang CC. (1994). Targeting proteins to the glycosomes of African trypanosomes. Annu Rev Microbiol , 105–138. [DOI] [PubMed] [Google Scholar]
- Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. (2008). Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest , 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA , 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes ML, Avery VM. (2009). A luciferase based viability assay for ATP detection in 384-well format for high throughput whole cell screening of Trypanosoma brucei brucei bloodstream form strain 427. Parasit Vectors , 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. (2011). Influence of inositol pyrophosphates on cellular energy dynamics. Science , 802–805. [DOI] [PubMed] [Google Scholar]
- Szoor B, Dyer NA, Ruberto I, Acosta-Serrano A, Matthews KR. (2013). Independent pathways can transduce the life-cycle differentiation signal in Trypanosoma brucei. PLoS Pathog , e1003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szoor B, Ruberto I, Burchmore R, Matthews KR. (2010). A novel phosphatase cascade regulates differentiation in Trypanosoma brucei via a glycosomal signaling pathway. Genes Dev , 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms MW, van Deursen FJ, Hendriks EF, Matthews KR. (2002). Mitochondrial development during life cycle differentiation of African trypanosomes: evidence for a kinetoplast-dependent differentiation control point. Mol Biol Cell , 3747–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S, Vassella E, Van Den Abbeele J, Renggli CK, Blundell P, Barry JD, Roditi I. (2005). Expression of procyclin mRNAs during cyclical transmission of Trypanosoma brucei. PLoS Pathog , e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Den Abbeele JV, Butikofer P, Renggli CK, Furger A, Brun R, Roditi I. (2000). A major surface glycoprotein of Trypanosomabrucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev , 615–626. [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Probst M, Schneider A, Studer E, Renggli CK, Roditi I. (2004). Expression of a major surface protein of Trypanosoma brucei insect forms is controlled by the activity of mitochondrial enzymes. Mol Biol Cell , 3986–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe VO, Savill JM, Chavali S, Jonsdottir AB, Rajendra E, Gruner T, Laskey RA, Babu MM, Venkitaraman AR. (2013). Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol Cell , 737–750. [DOI] [PubMed] [Google Scholar]
- Wu M, Chong LS, Perlman DH, Resnick AC, Fiedler D. (2016). Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc Natl Acad Sci USA , E6757–E6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst M, Seliger B, Jha BA, Klein C, Queiroz R, Clayton C. (2012). Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol Microbiol , 1048–1063. [DOI] [PubMed] [Google Scholar]
- Yabu Y, Takayanagi T. (1988). Trypsin-stimulated transformation of Trypanosoma bruceigambiense bloodstream forms to procyclic forms in vitro. Parasitol Res , 501–506. [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. (1999). A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science , 96–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.