Introduction
One key characteristic of many cancers, including breast cancer, is substantial intertumor and intratumor heterogeneity since it confounds the diagnosis and treatment plan. Over the past several decades of research, the depth of knowledge has deepened from recognizing the presence of tumor evolution and heterogeneity (1) to characterizing it at the level of variation in mutational aberrations and gene expression patterns that result in altered cell signaling, growth factor deregulation, and distinct metabolic pathways (2). These diverse events impact an array of oncogenic networks within a single tumor. It has long been recognized that such networks provide the leverage to rely on redundant signaling arms necessary for cellular modification to survive and progress under various conditions (3). Therefore, the presence of heterogeneity poses challenges when choosing a therapeutic intervention to eradicate the tumor without development of drug-resistance, metastasis or tumor relapse. Indeed, it is essential to stratify breast cancer at a genomic and molecular level to decipher tumor heterogeneity and develop robust targeting strategies to improve patient outcome. By using a genomic strategy to predict therapy, personalized medicine can be tailored to individual tumors.
Breast Cancer Classification
To address tumor heterogeneity, breast cancer can be broken down into subgroups in terms of histology, molecular biomarkers, genomic and genetic profiles. Based on the status of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) expression in tumors, breast cancer is classified as hormone receptor positive (HR+), HER2+ or triple-negative in absence of all three receptors. Based on tumor morphology, growth and architecture patterns, breast cancer can be classified into distinct histological subtypes. The most common histological subtype is invasive ductal carcinoma, followed by classic lobular invasive carcinoma. The histopathological classification provides prognostic value. In general, tubular or mucinous carcinoma is associated with excellent prognosis while metaplastic carcinoma has an unfavorable outcome. To gain greater insight into the gene expression patterns that underlie the histological, ER/PR/HER status, prognostic and outcome features of breast cancer, a study was undertaken where global gene expression patterns were analyzed. This study characterized several so-called intrinsic subtypes of breast cancer based on high/low expression of specific sets of genes. This was refined through several critical manuscripts and evolved to intrinsic subtypes including luminal A, luminal B, HER2-enriched, basal-like, claudin-low and normal-like tumors (4–10).
Sequencing the genome of breast cancer has revealed less than 100 driver mutations, most in genes that have previously been widely studied (11). This is consistent with prior reports with several genes that are frequently mutated with other supporting genes that are less commonly altered (12). However, the juxtaposition of sequence data with the intrinsic subtypes reinforces a critical need in breast cancer. That is to say, the ER/PR/HER2-ve (triple negative breast cancer or TNBC which largely overlaps with the basal subtype of breast cancer lacks oncogenic drivers at a high frequency. This is an urgent problem given that on average these tumors have the worst prognosis in breast cancer and typically affect younger women. However, it should be noted that when the subtypes of TNBC are stratified, there are clear distinctions in survival between the various subtypes of TNBC (13, 14).
Targeted therapy for HR+ breast cancer
Approximately 70% of breast cancers are identified as hormone receptor positive (HR+. +) and most of these tumors can be divided into Luminal A and B intrinsic subtypes. Endocrine therapy is the mainstay of treatment for all women with HR+ and is centered around targeting estrogen receptor to suppress its downstream functions. There are three main HR targeting strategies: inhibiting estrogen binding to ER by using selective estrogen receptor modulators such as tamoxifen; degrading ER by using selective estrogen receptor degraders such as fulvestrant; or depleting estrogen production by using aromatase inhibitor or ovarian suppression. The HR+ tumors can be further divided into luminal A and B intrinsic subtypes. Morphologically, luminal A tumors are well differentiated carcinomas and include classical lobular carcinoma, tubular carcinoma, and mucinous carcinoma(15). Tumors of luminal B subtype are less differentiated and consist mostly of invasive ductal carcinoma. Despite the effectiveness of endocrine therapy in HR+ breast tumors, treatment challenges remain in clinical practice. Clinical studies reported that luminal A tumors are more sensitive to therapy while luminal B tumors more often exhibit primary (intrinsic) resistance and may not respond to endocrine therapy alone (16). Conversely, luminal A tumors may respond to single endocrine therapy initially but may recur later due to secondary (acquired) resistance (17). To counteract primary and secondary endocrine resistance, combination strategies of endocrine therapy with cyclin-dependent kinase (CDK) 4/6 inhibitors or PI3K/mTOR/Akt pathway inhibitors have been evaluated. These inhibitors in combination with endocrine therapies have improved progression free survival and disease free survival in women with metastatic breast cancer (18–21). The success of these clinical trials illustrate the need to develop selective targeted therapies based on tumor mechanisms to utilize combinatorial approaches to effectively treat tumors.
Targeted therapy for HER2 positive breast cancer
HER2 is a member of the epidermal growth factor receptor family and is well recognized for its role in cellular differentiation and proliferation. HER2/neu amplification or overexpression is noted in 20% of human breast cancer, and was first observed to be an independent prognostic factor in lymph node positive breast cancer patients (22). Historically it has been seen as a negative prognostic marker associated with increased risk or recurrent disease and worse prognosis. A key mechanism for studying the role of HER2 was the generation of transgenic mice expressing activated c-neu under the control of the mouse mammary tumor virus (MMTV) promoter enhancer. These mice developed multifocal mammary tumors after a brief latency (23).
The development of a humanized HER2/neu monoclonal antibody, trastuzumab, resulted in the first FDA approved targeted therapy for HER2 positive breast cancer patients(24). Multiple clinical trials demonstrated that trastuzumab treatment for the HER2+ breast cancer patients reduced the recurrence risk and improved overall survival (25–27). Currently, there are several HER2 targeting therapeutics utilized in clinical settings: monoclonal antibodies such as trastuzumab and pertuzumab, which bind to the extracellular domain of HER2; tyrosine kinase inhibitors such as lapatinib and neratinib, which bind to the intracellular portion of protein kinase domain of HER2; and toxic drug conjugates such as T-DM1, in which a cytotoxic molecule is linked to the HER2 antibody (28–30). Dual HER2 targeting therapy, a combination of chemotherapy with monoclonal HER2 antibodies, and a combination of endocrine therapy with HER2 targeted agents have been proven to be successful in overcoming breast cancer resistance. HER2+ breast cancer is an excellent example of how identification of a molecular target can lead to effective treatment and improved patient outcomes.
Triple Negative Breast Cancer
TNBC lacks expression of ER, PR, and HER2 receptors, accounts for 10–20% of all reported breast cancer, and largely overlaps with the basal intrinsic subtype. Epidemiologically, TNBC patients are over-represented for young (<50 years), African American and Hispanic women (31). Morphologically, most TNBCs are invasive ductal carcinoma; others include metaplastic carcinoma, carcinoma with medullary features, adenoid cystic carcinoma, and secretory carcinoma (B). Compared to other intrinsic subtypes, TNBC has been clinically observed to be aggressive in nature based on a high grade, advanced stage at diagnosis and a higher risk of metastasis. These tumors tend to recur within three years of diagnosis and are associated with increased mortality within 5 years, resulting in the worst prognosis of all subtypes (32). According to gene expression patterns, TNBC can be subdivided into basal-like subtype, claudin-low breast cancer subtype, and luminal androgen receptor subtype. From a therapeutic standpoint, the basal subtype is most sensitive to chemotherapy while the luminal androgen receptor subtype is least sensitive. Thus, the lack of targeted therapy in TNBC, coupled with the poor outcome and drawbacks of chemotherapy illustrate a critical need to identify the driving factors of TNBC and adapt a personalized medicine approach to effectively treat these patients.
The carriers of BRCA1 germline mutation are more likely to develop TNBC (33). Tumors with BRCA mutations are deficient in homologous recombination repair (HRR). As such, these BRCA mutated TNBC rely on poly (ADP-ribose) polymerase 1 (PARP-1) for single strand DNA break repair, and therefore DNA damaging agents, such as platinum chemotherapy or PARP inhibitors, can induce synthetic lethality in HRR deficient cancer cells. Indeed, PARP inhibitor, olaparib, is currently FDA approved for the treatment for relapsed BRCA-mutated ovarian cancer (34). A recent phase III study reported increased progression free survival in metastatic breast cancer patients with a germline BRCA mutation treated with olaparib as compared to standard therapy (35). Clinical trials of PARP inhibitors in early stage breast cancer with germline BRCA mutation are ongoing.
TNBC is associated with a high number of tumor infiltrating lymphocytes (TILs) and indicate better sensitivity to chemotherapy (36). Furthermore, patients with high TILs may benefit from therapeutics that enhance antitumoral immune responses. Interest in the combination of chemotherapy with immune checkpoint inhibitors is growing and is reflected in a number of active clinical trials.
Luminal androgen receptor (LAR) breast cancer subtype is characterized by luminal gene expression. Preclinical models demonstrated that tumor growth is driven by androgen pathway activation and is estrogen independent (37). The androgen receptor (AR) is an emerging target in TNBC treatment. Androgen blockade in AR+ ER- PR- HER2- breast cancer is being evaluated in clinical trials.
The claudin-low breast cancer subtype is enriched with epithelial to mesenchymal transition (EMT) markers, stem-like features, and immune cell infiltration (38). Currently, there is no targeted treatment shown to be effective in this tumor subtype and preclinical studies are needed to identify actionable targets.
Current Directions
Models for breast cancer
To study breast cancer in a mammalian system, numerous murine models have been developed. The mouse mammary tumor virus (MMTV) promoter enhancer and the Whey Acidic promoter (WAP) have been widely used to drive the expression of specific gene in the mammary epithelium of transgenic mice. These promoters have been used to establish well-known oncogenic mouse models such as MMTV-Myc (39), MMTV-Ras (40), MMTV-Neu (23) and many others. Tumor suppressors have been studied in mouse models through mammary specific knockouts using MMTV-Cre, resulting in mammary specific loss of genes including BRCA and p53 (41, 42). Importantly, many models have now been compared to human breast cancer at a gene expression level, reflecting many of the broad patterns observed in human breast cancer (43–45). When gene expression data from these mouse models was used to predict chromosomal gain and loss, conserved events were noted in subtypes of cancer in specific models that mimicked human breast cancer (46).
Although these mouse models have played critical roles in establishing the mechanisms in breast cancer development and progression, their utility is tempered by limitations for translation to clinical trials. For instance, oncogenes driven by MMTV alleviates the mutation selection pressure and alters natural pattern of genomic stability (46). In addition, human breast cancer routinely metastasizes to liver and bone while most mouse models demonstrate predominately lung metastasis (47). One approach to overcome these restrictions is the use of human breast cancer cell lines. Orthotopic injection of cultured human cell lines in immunocompromised mice allows for in vivo tumor growth and can be used for drug screening. However, cell lines maintained in culture accumulate additional mutations and have gene expression patterns divergent from human breast cancer.
Patient-derived xenograft (PDX) model systems use direct implantation of primary human tumors to the mammary fat pad of immunocompromised Nude/SCID mouse. PDX models eliminate in vitro culture and resulting tumors have been demonstrated to have strong similarities to the original primary tumor at both a histological and gene expression level (48, 49). Indeed, studies with PDX models have been argued to offer a translatable preclinical platform (50). The availability of gene expression data from the PDX models opens the realm of developing personalized medicine for testing potential drugs under preclinical settings, followed by clinical trials for an accelerated transition to develop individualized therapy.
Preclinical trials for personalized medicine
Given that human breast cancer, mouse model systems and PDX tumor samples have extensive gene expression data that has been deposited in public repositories, there is now an opportunity to use the various model systems in preclinical trials. Importantly, the gene expression data allows one to ensure that similar mechanisms to those used in human breast cancer can be tested. We have used a method that predicts pathway activation status from gene expression data to make predictions in both mouse and human. Essentially this method uses gene expression data from training datasets where pathways were activated in human mammary epithelial cells in comparison to non-activated controls. Once signaling pathway signatures were built and validated (13, 51, 52), they could be used to interrogate any breast cancer gene expression dataset. This method has previously been used to make cell signaling pathway predictions in both human breast cancer (13, 52–55) and mouse models (43, 56–62).
Recently we used signaling pathway signatures to predict signaling pathway activation status in MMTV-Myc transgenic mice. While we previously noted that Myc induced tumors with similar histological subtypes shared gene expression patterns (57) and chromosomal gain or deletion events (46), in our recent work we noted key differences in signaling pathways between papillary and epithelial to mesenchymal transition (EMT) tumors (63). Indeed, Myc, Stat3 and AKT signaling were predicted to be strongly activated in papillary tumors while EGFR, RAS and TGFβ activity was upregulated in the EMT tumors. Importantly, as a control we use immunoblotting to validate the gene expression based pathway predictions for each signaling pathway.
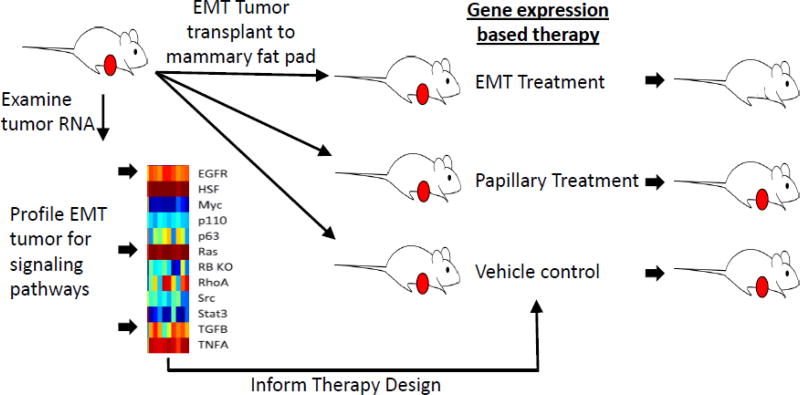
To begin to demonstrate the preclinical application of personalized medicine, we sought to use pathway signature predictions to direct therapeutic design, initially in the MMTV-Myc mouse model system (Figure 1). By selecting therapeutic compounds that are already FDA approved or under various phases of trials for individual signaling pathway, we sought to accelerate the pace for potential transition to clinic. The proof-of-concept experiment demonstrated that combination therapy of three drugs simultaneously targeting the Myc, Stat3, and Akt pathways inhibited papillary tumor growth compared to vehicle controls in a dose dependent manner (63). Importantly, the same combination therapy that prevented papillary tumor failed to arrest growth of EMT tumors, emphasizing the importance of designing treatment regimen based on gene expression patterns for specificity.
Figure 1. Summary of personalized therapy design.
Using the MMTV-Myc mouse model, EMT and Papillary tumors were used in a proof of principle experiment to predict therapy. For all tumors RNA was collected and gene expression was surveyed. Signaling pathway predictions (red on, blue off) for key signaling pathways were calculated based on gene expression signatures. EMT tumors were then transplanted to the mammary glands of syngenic mice. Once tumors reached 6 mm in the largest dimension, therapy for EMT, control therapy based on papillary tumors and vehicle control therapy was initiated in each cohort of mice. EMT treatment was found to be effective and specific for EMT tumors. Papillary treatment was also shown to be effective and specific for papillary tumors (not shown).
In the described experiment, we noted the initial response of the papillary tumors to combination treatment. However, resistance quickly developed and tumors began to grow. Examining global patterns of gene expression from these resistant tumors with signaling pathway signatures, we predicted elevated probabilities of EGFR and Ras pathway activation. This was confirmed through phosphor-ERK immunohistochemistry, and we speculated that the cause of recurrence perhaps is not drug resistance but rather a clonal selection from tumors that were initially heterogenous due to the speed that resistance emerged. With the regrowth of the tumors with other pathways that were targetable, our findings suggest that a repeated biopsy in breast cancer patients to examine tumor cells for changes in characteristics following courses of treatments may provide critical information needed for subsequent treatment decisions.
In much the same manner as the papillary specific treatment, we designed a three drug combination to target EMT tumors arising in the transgenic mice. Targeting EGFR, Ras, and TGFB pathways in combination prevented EMT tumor growth compared to both vehicle control treatment in papillary tumors, again demonstrating that gene expression information could direct an effective and specific therapy. The EMT experiment was critical since prior comparisons to human cancer revealed a similarity of the EMT mouse tumors to the TNBC / Claudin low subtype (43).
To extend the signaling pathway probability method to treating human breast cancer, we applied the same signaling pathway predictions to a collection of human breast cancer samples from numerous datasets and included PDX samples. Limiting the predictions to the basal subset of breast cancer it was readily apparent that many PDX samples were highly similar from the standpoint of which cell signaling pathways were active (63). Choosing several PDX lines, including one that was similar to the MMTV-Myc EMT samples, we implanted the PDX samples into the mammary fat pad and initiated treatment once the tumors reached 6 mm. In a critical finding, we noted that cell signaling pathway inhibitors that were chosen based on gene expression data were capable of shrinking the PDX tumor samples. Together these data demonstrated that gene expression data from both mouse models and human breast cancer samples could be interrogated to predict the optimal therapy. Together these pre-clinical experiments demonstrated that a gene expression based approach could be used to direct effective and specific therapeutic interventions.
5. Future Directions
While there are numerous ongoing studies where molecular profiles are used to direct therapy, the future of personalized medicine in cancer will be tied to information gathered from individual patient tumors. Indeed, the transition to the clinic in the NCI-MATCH trial (https://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/nci-match) has already begun and is reliant upon genomic sequencing of individual tumors to choose drugs, many of which are already FDA approved. In the future, a combination of standard biomarkers (ER/PR/HER2), as well as genomic sequence and gene expression information will provide an opportunity to tailor treatment for each individual breast cancer patient. A future challenge is to develop effective combination strategies. Further preclinical studies may identify resistance mechanisms of targeted therapies and inform clinical trial design of rationale combination strategies.
Acknowledgments
This work was supported with NIH R01CA160514 and Worldwide Cancer Research WCR - 14-1153 to E.R.A.
References
- 1.Fidler IJ, Hart IR. Biological diversity in metastatic neoplasms: origins and implications. Science. 1982;217(4564):998–1003. doi: 10.1126/science.7112116. [DOI] [PubMed] [Google Scholar]
- 2.Network TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heppner GH. Tumor heterogeneity. Cancer Res. 1984;44(6):2259–65. [PubMed] [Google Scholar]
- 4.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96(16):9212–7. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 13.Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010;107(15):6994–9. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–98. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang P, Tse GM. Immunohistochemical Surrogates for Molecular Classification of Breast Carcinoma: A 2015 Update. Arch Pathol Lab Med. 2016;140(8):806–14. doi: 10.5858/arpa.2015-0133-RA. [DOI] [PubMed] [Google Scholar]
- 16.Ades F, Zardavas D, Bozovic-Spasojevic I, Pugliano L, Fumagalli D, de Azambuja E, et al. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol. 2014;32(25):2794–803. doi: 10.1200/JCO.2013.54.1870. [DOI] [PubMed] [Google Scholar]
- 17.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 19.Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N Engl J Med. 2016;375(18):1738–48. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 20.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn RS, Martin M, Rugo HS, Jones S, Im SA, Gelmon K, et al. Palbociclib and Letrozole in Advanced Breast Cancer. N Engl J Med. 2016;375(20):1925–36. doi: 10.1056/NEJMoa1607303. [DOI] [PubMed] [Google Scholar]
- 22.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 23.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54(1):105–15. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 24.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20(3):719–26. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 25.Moja L, Tagliabue L, Balduzzi S, Parmelli E, Pistotti V, Guarneri V, et al. Trastuzumab containing regimens for early breast cancer. Cochrane Database Syst Rev. 2012;(4):CD006243. doi: 10.1002/14651858.CD006243.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gianni L, Eiermann W, Semiglazov V, Lluch A, Tjulandin S, Zambetti M, et al. Neoadjuvant and adjuvant trastuzumab in patients with HER2-positive locally advanced breast cancer (NOAH): follow-up of a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet Oncol. 2014;15(6):640–7. doi: 10.1016/S1470-2045(14)70080-4. [DOI] [PubMed] [Google Scholar]
- 27.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–83. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams CW, Allison DE, Flagella K, Presta L, Clarke J, Dybdal N, et al. Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, pertuzumab. Cancer Immunol Immunother. 2006;55(6):717–27. doi: 10.1007/s00262-005-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loibl S, Jackisch C, Schneeweiss A, Schmatloch S, Aktas B, Denkert C, et al. Dual HER2-blockade with pertuzumab and trastuzumab in HER2-positive early breast cancer: a subanalysis of data from the randomized phase III GeparSepto trial. Ann Oncol. 2017;28(3):497–504. doi: 10.1093/annonc/mdw610. [DOI] [PubMed] [Google Scholar]
- 30.Joensuu H. Escalating and de-escalating treatment in HER2-positive early breast cancer. Cancer Treat Rev. 2017;52:1–11. doi: 10.1016/j.ctrv.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 32.Tischkowitz M, Brunet JS, Begin LR, Huntsman DG, Cheang MC, Akslen LA, et al. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;7:134. doi: 10.1186/1471-2407-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17(5):1082–9. doi: 10.1158/1078-0432.CCR-10-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 35.Robson M, Goessl C, Domchek S. Olaparib for Metastatic Germline BRCA-Mutated Breast Cancer. N Engl J Med. 2017;377(18):1792–3. doi: 10.1056/NEJMc1711644. [DOI] [PubMed] [Google Scholar]
- 36.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32(27):2959–66. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barton VN, D'Amato NC, Gordon MA, Lind HT, Spoelstra NS, Babbs BL, et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Mol Cancer Ther. 2015;14(3):769–78. doi: 10.1158/1535-7163.MCT-14-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107(35):15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38(3):627–37. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 40.Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49(4):465–75. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 41.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 42.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22(1):37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 43.Hollern DP, Andrechek E. A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to human breast cancer. Breast Cancer Research. 2014;16(R59) doi: 10.1186/bcr3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013;14(11):R125. doi: 10.1186/gb-2013-14-11-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rennhack J, To B, Wermuth H, Andrechek ER. Mouse Models of Breast Cancer Share Amplification and Deletion Events with Human Breast Cancer. J Mammary Gland Biol Neoplasia. 2017 doi: 10.1007/s10911-017-9374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73(15):4885–97. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98(20):11462–7. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 53.Bild AH, Parker JS, Gustafson AM, Acharya CR, Hoadley KA, Anders C, et al. An integration of complementary strategies for gene-expression analysis to reveal novel therapeutic opportunities for breast cancer. Breast Cancer Res. 2009;11(4):R55. doi: 10.1186/bcr2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46(10):1051–9. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163(2):506–19. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrechek ER. HER2/Neu tumorigenesis and metastasis is regulated by E2F activator transcription factors. Oncogene. 2015;34(2):217–25. doi: 10.1038/onc.2013.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A, et al. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci U S A. 2009;106(38):16387–92. doi: 10.1073/pnas.0901250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrechek ER, Mori S, Rempel RE, Chang JT, Nevins JR. Patterns of cell signaling pathway activation that characterize mammary development. Development. 2008;135(14):2403–13. doi: 10.1242/dev.019018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujiwara K, Yuwanita I, Hollern DP, Andrechek ER. Prediction and Genetic Demonstration of a Role for Activator E2Fs in Myc-Induced Tumors. Cancer Res. 2011;71(5):1924–32. doi: 10.1158/0008-5472.CAN-10-2386. [DOI] [PubMed] [Google Scholar]
- 60.Hollern DP, Yuwanita I, Andrechek ER. A mouse model with T58A mutations in Myc reduces the dependence on KRas mutations and has similarities to claudin-low human breast cancer. Oncogene. 2012 doi: 10.1038/onc.2012.142. [DOI] [PubMed] [Google Scholar]
- 61.Jhan JR, Andrechek ER. Stat3 accelerates Myc induced tumor formation while reducing growth rate in a mouse model of breast cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turpin J, Ling C, Crosby EJ, Hartman ZC, Simond AM, Chodosh LA, et al. The ErbB2DeltaEx16 splice variant is a major oncogenic driver in breast cancer that promotes a pro-metastatic tumor microenvironment. Oncogene. 2016 doi: 10.1038/onc.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jhan JR, Andrechek ER. Effective personalized therapy for breast cancer based on predictions of cell signaling pathway activation from gene expression analysis. Oncogene. 2017 doi: 10.1038/onc.2016.503. [DOI] [PubMed] [Google Scholar]