Abstract
Pigs are emerging as important large animal models for biomedical research, and may represent a source of organs for xenotransplantation. The MHC is pivotal to the function of the immune system in health and disease, and is particularly important in infection and transplant rejection. Pigs deficient in class I MHC could serve as important reagents to study viral immunity, allograft and xenograft rejection. We report the creation and characterization of class I MHC knockout pigs using the Cas9 nuclease and gRNAs. Pig fetal fibroblasts were genetically engineered using Cas9 and gRNAs, and class I MHC negative cells were then used as nuclear donors for somatic cell nuclear transfer. We produced 3 piglets devoid of all cell surface class I proteins. Though these animals have reduced levels of CD4−CD8+ T cells in peripheral blood, the pigs appear healthy and are developing normally. These pigs are a promising reagent for immunological research.
Keywords: Swine leucocyte antigen class I, MHC, swine, CRISPR/Cas9, Gene knockout, SLA deficiency
Introduction
Classical Class I MHC genes synthesize polymorphic proteins which form a heterotrimeric complex with the β2-microglobulin (β2m) protein and short peptides [1,2]. The polymorphism arises from the existence of multiple class I genes each having many alleles. A groove, formed by the α1 and α2 domains of the protein, contains the majority of variation between different class I polypeptides and has pockets that accommodate specific amino acids and determine peptide anchor positions [3, 4, 5]. Variation in non-anchor residues allows a single class I MHC protein to present a diverse set of peptides.
Rodent models and in vitro assays have provided much of the mechanistic information regarding the role of class I molecules in antigen presentation. The heterotrimeric complex is scanned by antigen receptors on CD8+ T lymphocytes [6, 7]. Some MHC-displayed peptides originating from infectious agents or cancers activate CD8 cells. Despite understanding these processes in great detail, identifying relevant antigens has been difficult. Variations in both the class I MHC protein and in other antigen presentation machinery can independently shape the peptide repertoire [8, 9, 10]. In addition to their role in CD8+ T lymphocyte activation, class I MHC molecules also regulate the activity of other immune effectors such as natural killer cells. Understanding the nuances of how class I MHC regulates immunity in various species will benefit from the development of additional animal models.
To improve the ability to study class I MHC function in pigs, we have used the Cas9 nuclease and gRNA to disrupt class I MHC genes, which are also known as Swine Leukocyte Antigens (SLA). Following somatic cell nuclear transfer (SCNT), we have created cloned animals lacking class I MHC protein expression. The development of class I MHC deficient swine will aid the study of immunity to infectious diseases which cost the agricultural industry hundreds on millions of dollars annually [11]. These novel pigs can also be used in studies of xenotransplantation in an effort to eliminate the human anti-pig immunity that currently prevents the use of pig tissues as replacements for failing human organs.
Materials and Methods
Cell Culture
Fetal fibroblast cells from a cloned pig with known class I SLA alleles were used in this study [12]. FFSC were resuspended and cultured in a SCM (stem cell media) prepared with MEM-α/EGM-MV media (Invitrogen/Lonza, Carlsbad, CA; Switzerland) supplemented with 10% Fetal Bovine Serum (Hyclone, Logan UT), 10% Horse serum (Invitrogen, Carlsbad, CA), 12 mM HEPES (Sigma) and 1% pen/strep (Gibco Grand Island, NY), and cultured in Collagen-I-coated plates (Becton Dickinson, Bedford, MA) at 38.5oC, 5% CO2 and 10% O2. The cells treated with SLA-specific gRNA and Cas9 contained a GGTA1 gene that had been previously inactivated [13]. SLA expressing control cells were derived from GGTA1 deficient animals. The genetic backgrounds of the control and experimental animals are very similar as they were cloned from cells originating from a single donor.
Generation of gRNAs Expression Vector
Plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid 42230, http://www.addgene.org/42230/) was used to clone the designed annealed oligos (Figure 1E, Supplementary Figure 1) to generate guide RNA using the CRISPR associated Cas9 nuclease system [14]. One ug of plasmid pX330 was digested with BbsI (New England Biolabs, Ipswich, MA) for 30 min at 37 °C. Each pair of phosphorylated oligos were annealed using a Veriti themocycler (Applied Biosystems, Grand Island, NY) starting at 37 °C for 30 min, followed by a step at 95 °C for 5 min and then ramp down to 25 °C at 5 °C/min. Digested pX330 was ligated to the annealed par of oligos for 10 min at room temperature. Ligation reaction was used to transform TOP10 competent cells (Invitrogen, Carlsbad, CA), following the manufacture’s protocol. The QIAprep kit (Qiagen, Valencia, CA) was used to isolate plasmid from 15 colonies per treatment. DNA clones were sequenced (DNA Sequencing Core Facility, Department of Biochemistry and Molecular Biology, IUPUI) and used to transfect porcine FFSC cells.
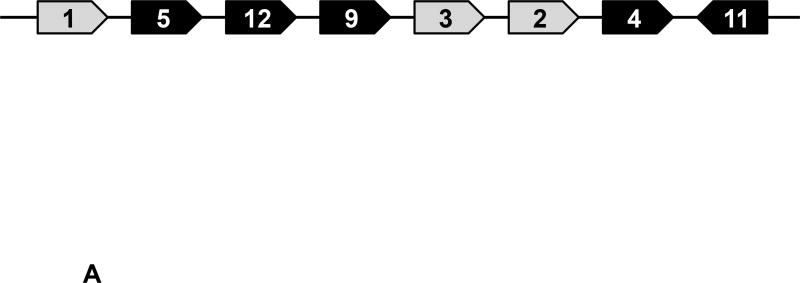
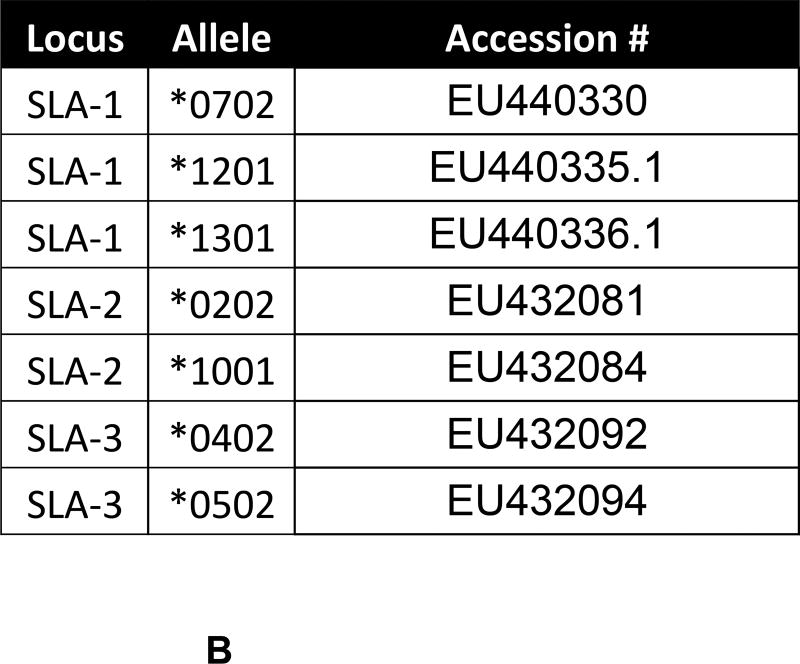
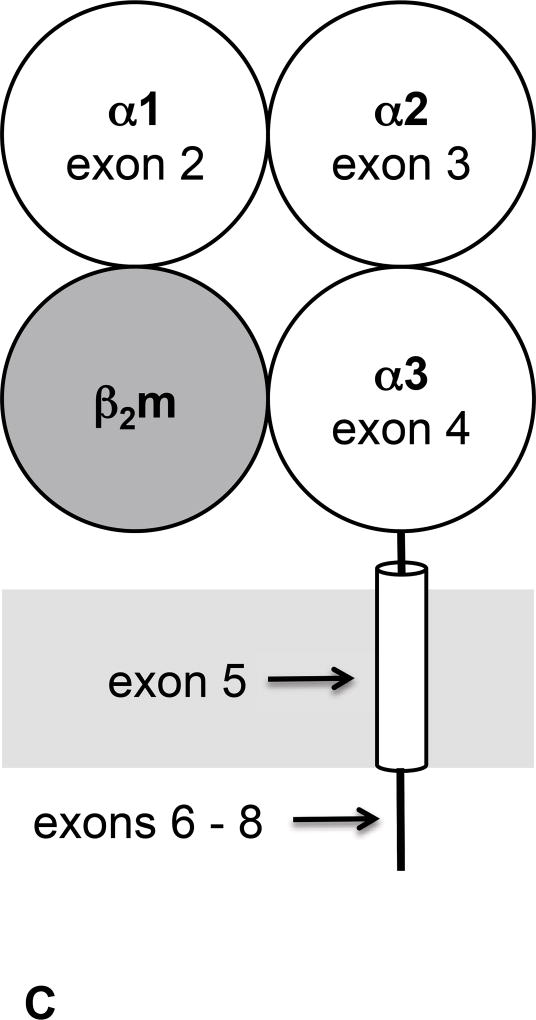
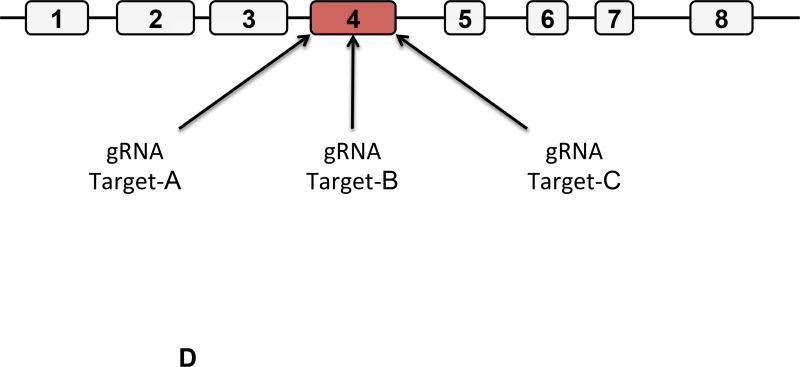
Figure 1. Description of Swine Class I MHC Genes.
A) The class I region of swine MHC contains three classical class I genes (SLA-1, −2, −3;), several pseudogenes (SLA-4, −5, and −9) and two class I like genes (SLA-11 and −12). The approximate size of the entire classical class I MHC region is 130kb. The SLA-1 and −3 loci are separated by approximately 65kb and the SLA-2 and −3 loci are separated by about 17kb. B) NCBI accession numbers are shown that are relevant to the alleles of this study. C) The five protein domains of the class I protein are shown with an indication of which gene exon encodes each specific polypeptide region. The β2m protein is also shown. D) Description of the relative location of the gRNA targets in exon four of the class I gene. E) gRNA target sequences of exon four are shown. Exon four consists of 276 bp in the different alleles, and the class I genes are approximately 3.5 kb long.
Transfection and Identification of Targeted Cells
FFSC cells were seeded in early passage (passage 2) onto 6-well plates 24h before transfection. Cells were harvested and counted, 1 ×106 cells were resuspended in 800 ul of fresh sterile electroporation buffer (75% Cytosalt buffer: 120 mM KCl; 0.15 mM CaCl2; 10 mM K2HPO4, pH 7.6; 5 mM MgCl2) and 25% OPTI-MEM (Gibco, Grand Island, NY). Cells were mixed with two ug of plasmid DNA in 4-mm cuvettes. Transfection was performed using the GenePulser X-cell (Bio-Rad, Hercules, CA) following the manufacture’s protocol for mammalian cells. Treated cells were seeded into 6-well plate and grown until confluent. Cell screening was performed using a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA), using mouse anti-pig SLA class I-FITC antibody (AbD Serotec, Raleigh, NC). Cells with low expression for SLA class I antigen were expanded and fluorescence activated cell sorting (FACS) at least twice, using the FACS Vantage SE at the Indiana University Flow Cytometry Resource Facility. Selected treatments were used for SCNT.
Genotypic Analysis
Genomic DNA was isolated from pig cells using the QIAmp® DNA mini kit (Qiagen, Valencia, CA). RNA samples were isolated using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. RNA quality and quantity were affirmed by Agilent Bioanalyzer analysis (Core Facility of the Department of Biochemistry and Molecular Biology, School of Medicine, IUPUI). RNA samples were reverse transcribed using OneStep RT-PCR Kit (Qiagen, Valencia, CA). PCR products were purified and ligated into the pCR4-TOPO TA (Invitrogen, Carlsbad, CA). Transformed bacteria were plated onto Luria–Bertani agar containing 50 µg/ml Kanamycin for clone selection. Plasmids were isolated using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA, USA). Nucleotide sequences were performed by the Sanger method using custom sequencing service (Elim Biopharm, Hayward, CA). Primers used in this study are shown in Supplementary Figure 1.
Phenotypic analyses
Flow Cytometry
Porcine peripheral blood mononuclear cells (PBMCs) were prepared using Ficoll-Paque Plus as described previously [15]. PBMC were stained with the following antibodies: mouse anti-pig PerCP-Cy5.5 CD3, PE CD4, FITC CD8α, and mouse Isotype control (BD Biosciences, CA, USA). Dead cells were excluded from analysis using Fixable Viability Dye eFluor 660 (eBioscience, Inc. CA, USA). Analysis was performed using an Accuri C6 flow cytometer and CFlow software (Accuri, Ann Arbor, MI, USA) and FlowJo software (Tree Star, Inc., Ashland, OR, USA).
Primary kidney endothelial cells were isolated using 0.025% of collagenase type IV from Clostridium histolyticum (Sigma-Aldrich St. Louis, MO), and cultured for 3 days in RPMI medium supplemented with 10% FBS, 100ug/ml Endothelial cell growth supplement (BD Biosciences, CA, USA).
Fibroblasts were grown under the same conditions used to maintain fetal fibroblasts as described above.
Somatic cell nuclear transfer (SCNT)
SCNT was performed as described previously [16] using in vitro matured oocytes (DeSoto Biosciences Inc, St. Seymour, TN). Cumulus cells were removed from the oocytes by pipetting in 0.1% hyaluronidase. Only oocytes with normal morphology and a visible polar body were selected for SCNT. Oocytes were incubated in manipulation media (Ca-free NCSU-23 with 5% FBS) containing 5 µg/mL bisbenzimide and 7.5 µg/mL cytochalasin B for 15 min. Oocytes were enucleated by removing the first polar body plus metaphase II plate, and one cell was injected into each enucleated oocyte. Couples were fused and activated simultaneously by two DC pulses of 180 V for 50 µsec (BTX cell electroporator, Harvard Apparatus, Hollison, MA, USA) in 280mM Mannitol, 0. 1mM CaCl2, and 0.05mM MgCl2. Activated embryos were placed back in NCSU-23 medium with 0.4% bovine serum albumin (BSA) and cultured at 38.5 °C, 5% CO2 in a humidified atmosphere for less than 1 h, before being transferred into the recipient. Recipients were synchronized occidental gilts on their first day of estrus. Animals used in this study were under protocols approved by the Institutional Biosafety and Institutional Animal Care and Use Committee-IUPUI.
Sequence analysis
Nucleotide sequences were analyzed using the MacVector with Assembler 12.7 (MacVector, Inc. Apex, NC). Overlapping forward and reverse sequence fragments assembled the complete coding sequence of each allele. Multiple sequence alignment was created for each locus. Sequences were confirmed by using The Immune Polymorphism Database (IPD)-MHC SLA sequence database (http://www.ebi.ac.uk/ipd/mhc/sla/).
Results
Description of Swine Class I MHC Genomic Organization and Individual Gene Structure
As depicted in Figure 1A, the class I region consists of three classical MHC genes SLA-1, −2, and −3. This genomic location also contains three pseudogenes, SLA-4, −5, and −9, and two poorly characterized class I-like genes SLA-11 and −12 [17, 18, 19, 20]. The alleles of SLA-1, −2, and −3 present in the cells and animals used in this study are shown in Figure 1B. Three SLA-1 alleles presumably exist as a result of gene duplication in one of the haplotypes [21, 22].
Classical class I MHC proteins are membrane proteins having three extracellular domains, a transmembrane segment, and short cytoplasmic tail, and are encoded by an eight-exon gene (Figures 1C and 1D). Exons two and three are highly polymorphic while the remaining exons have limited variability [20, 23]. Exon four encodes the extracellular α 3 domain which is critical for assembly of the class I molecule with β2m and its transport to the cell surface [24, 25].
The Cas9 endonuclease is now commonly used to inactivate genes [14, 26, 27, 28, 29]. This is accomplished by designing gRNA that recruit Cas9 to target sequences. Cleavage initiates repair mechanisms that are imperfect leading to the creation of insertions and deletions that disrupt gene activity. Three separate gRNA were designed that specified unique locations in exon four (Figures 1D, and 1E). All three targeted sequences are present in the alleles of SLA-1, −2, and −3 that are relevant to this study. Comparison with NCBI sequences indicates the targeted nucleotides are also present in SLA −4, −5, −9, −11, and −12 (pseudo)genes, though the consequences of gRNA treatment on these genes were not examined.
Gene Disruption
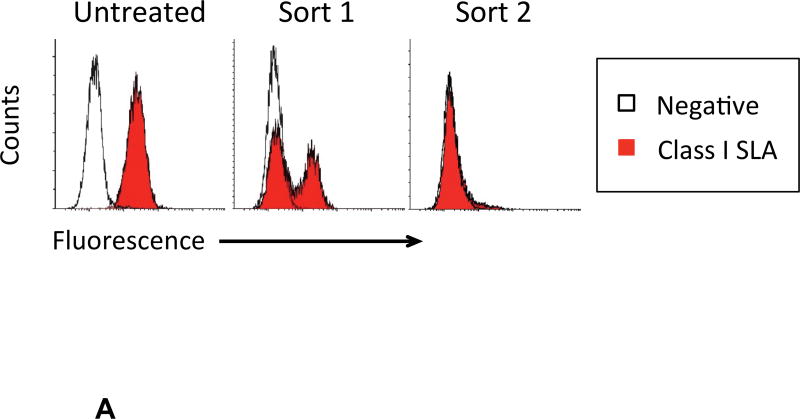
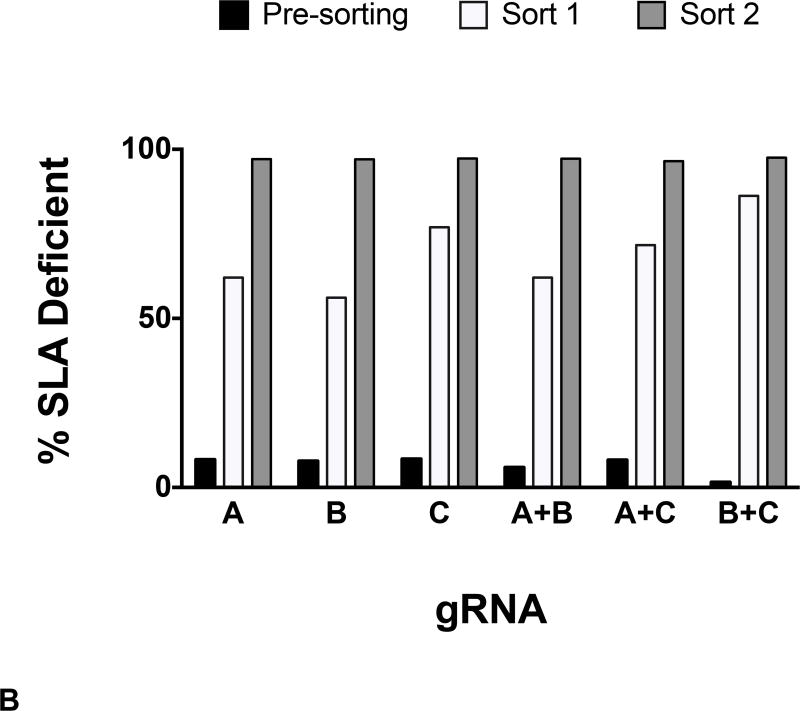
Fibroblasts expressing classical class I SLA molecules (Figure 2A, Untreated) were transfected with gRNA. Following gRNA and Cas9 treatment, expression of SLA-1, −2, and −3 was evaluated with a monoclonal antibody that binds all three MHC proteins. Two rounds of flow cytometry sorting produced an SLA negative population of cells (Figure 2A). The isotype control peak in Sort 2 is difficult to see because of overlap with the class I SLA histogram. All three gRNA successfully disrupted class I SLA genes when used alone and in combination (Figure 2B).
Figure 2. gRNA-Cas9 Treatment and Flow Sorting of Class I SLA Negative Fibroblast Cells.
Following gRNA treatment, two successive rounds of flow cytometry sorting yielded class I negative SLA cells. A representative example of enrichment is shown (A). When used singly or in combination, all three gRNA targeting exon four were capable of producing cells deficient in class I SLA expression (B).
Generation of Multi-Gene Mutant Pig by SCNT
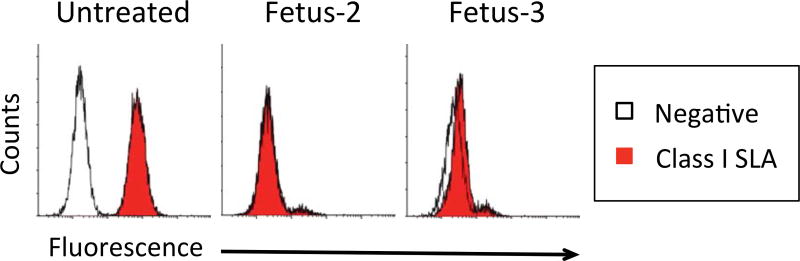
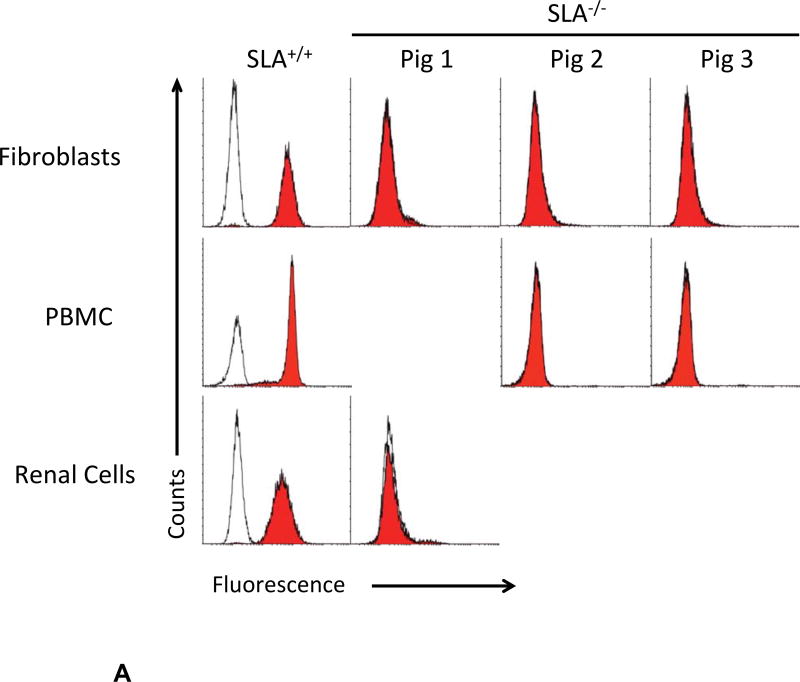
Class I SLA-negative cells from gRNA treatment A+C were used for SCNT (Table I). 211 cloned embryos were transferred to two recipients; one became pregnant. Fetuses were collected at day 32 of gestation. Fibroblast cells from well formed fetuses 2 and 3 were grown in culture and retested for phenotypic expression of class I SLA by flow cytometry (Figure 3). Though cells from fetus-3 appeared to express a small quantity of MHC protein, fetus-2 cells remained negative and were re-cloned, producing two other pregnancies. One sow spontaneously aborted at day 45 and the other produced three clonal piglets (Table I). One animal was sacrificed for cell collection. At the time of this report the other two have been healthy for 3.5 months. Renal cells from piglet 1, PBMC from piglets 2 and 3, and fibroblasts from all three animals were negative for class I SLA cell surface expression (Figure 4A).
Table 1.
Cloning Statistics
| Recipient | Donor Cells |
Transferred Embryos |
Pregnancy | Fetuses | Piglets | Cloning Efficiency (%) |
|---|---|---|---|---|---|---|
| 1 | FFSC | 113 | No | 0 | 0 | 0 |
| 2 | FFSC | 98 | Yes | 3 | 0 | 3.1 |
| 3 | FFSC-R2 | 144 | Yes | Aborted day 45 | 0 | 0 |
| 4 | FFSC-R2 | 121 | Yes | 0 | 3 | 2.5 |
| TOTAL | 476 | 75.0 | 3 | 3 | 1.4 |
Figure 3. Selection of Class I SLA negative Fetal Fibroblast Cells.
SCNT of fibroblasts isolated in figure 2 were used to create embryos. 32 days after impregnating a sow with these embryos, three fetuses were collected. Two of the fetuses were well formed and used to create fibroblast cultures. The fibroblasts were stained with a negative isotype control or with and antibody specific for class I SLA. Fetus-3 expressed low levels of SLA protein. Cells derived from Fetus-2 were devoid of class I SLA proteins.
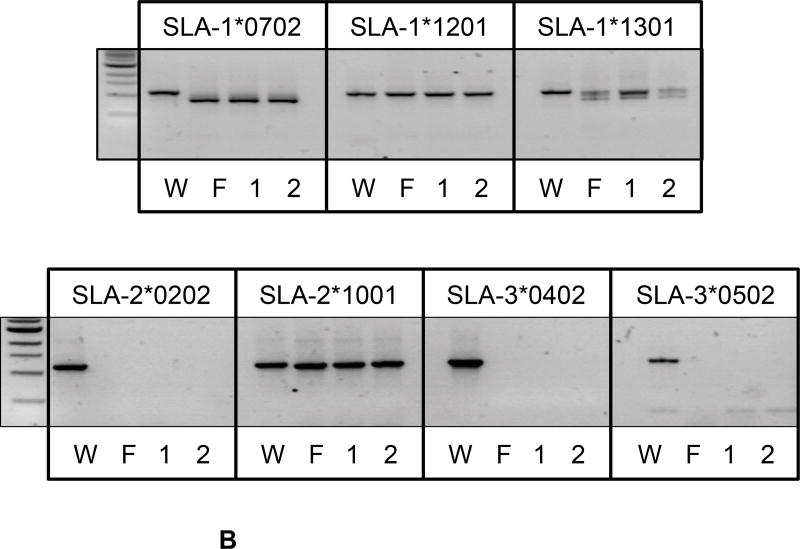
Figure 4. Phenotypic and cDNA Analyses of Class I SLA Deficient Piglets.
A) Three piglets, recloned from the SLA negative fetal fibroblast cells isolated in Figure 3, were examined for cell surface expression of class I SLA proteins on the surface of fibroblasts, (all three animals), PBMC (piglets-2 and −3), or cells isolated from the kidney (piglet-1). Corresponding class I SLA positive cells are shown for comparison. Relative binding of class I specific SLA antibodies and an irrelevant isotype control are shown. B) cDNA, prepared from fetus-2 and piglets-1 and −2, were subjected to PCR with primers designed to amplify individual alleles of class I SLA. Sample W represents an identical analysis of the untreated parental, SLA expressing, fibroblasts. The length of the predicted full-length transcript is indicated below each allele name. Samples F, 1, and 2 represent the fetus, and cloned animals 1 and 2 respectively.
cDNA from Fetus-2 and piglets-1 and −2 were evaluated by allele-specific PCR to determine why class I MHC proteins were absent (Figures 4B). PCR of cDNA from engineered animals failed to amplify SLA-2*0202, SLA-3*0402, and SLA-5*0502 transcripts. Apparently full-length transcripts representing SLA-1*1201 and SLA-2*1001 were obtained. These modified animals also contained multiple sizes of SLA-1*0702 and −1*1301 transcripts. The parental unmodified cells were also analyzed for comparison and yielded primarily full-length transcripts (Figure 4B). The lengths of the full-length transcripts were: SLA-1*0702 (1109 bp), SLA-1*1201 (1109 bp), SLA-1*1301 (1147 bp), SLA-2*0202 (1118 bp), SLA-2*1001 (1118 bp), SLA-3*0402 (1106 bp), and SLA-3*0502 (1109bp).
Because the cDNA analyses did not explain the loss of every classical class I molecule, allele specific PCR amplifications of cDNA were inserted into vectors. Multiple cDNA clones for each allele were sequenced in an effort to find inactivating insertions and deletions (Table II). These analyses were performed on untreated parental cells and on the modified cells isolated from fetus-2 and two SLA deficient piglets. The majority of cDNA clones derived from the parental cells contained transcripts capable of encoding functional class I SLA proteins. A small number of cDNA clones indicated that some alleles SLA-1*0702 and −1*1201 expressed transcripts lacking 276 bases. In contrast, neither fetus-2 nor the cloned piglets contained full-length transcripts capable encoding functional class I SLA proteins. Transcripts lacking 276 bases became the predominant SLA-1*0702 species in Piglets-1 and −2. In fetus-2, the SLA-1*0702 transcripts lacked either 276 or four bases. Neither of these transcript variants should encode functional protein because lacking 276 bases eliminates the α3 domain, and the four-base deletion creates a frameshift mutation.
Table 2.
Sequencing results: Numbers in each column represent the counts of individual cDNA clones having a particular sequence.
| SLA-1*0702 | SLA-1*1201 | SLA- 1*1301 |
|||
|---|---|---|---|---|---|
|
| |||||
| FL | (−)276 bp | (−)4bp | FL | (−)276 bp | FL |
| 0 | 8 | 11 | 0 | 0 | 0 |
| 0 | 23 | 0 | 0 | 0 | 0 |
| 0 | 23 | 0 | 0 | 0 | 0 |
| 96 | 6 | 0 | 31 | 3 | 31 |
| SLA-2*0202 | SLA-2*1001 | SLA-3*0402 | SLA- 3*0502 |
|---|---|---|---|
|
| |||
| FL | FL | FL | FL |
|
| |||
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 32 | 39 | 19 | 46 |
| RECOMBINANTS | ||
|---|---|---|
|
| ||
| *1301/*0702 | *1301/*1001 | *1001/SLA-12 |
|
| ||
| (−)276 bp | (−)2 bp | (+)1bp |
|
| ||
| 0 | 41 | 14 |
| 0 | 27 | 20 |
| 6 | 25 | 20 |
| 0 | 0 | 0 |
Though PCR analyses generated products for SLA-1*1201, SLA-1*1301, and SLA-2*1001, these were not confirmed in the sequence analyses. Instead these cDNA revealed multiple recombinant molecules (Table II). Alleles SLA-1*1301 and SLA-2*1001, SLA-1*1301 and SLA-1*0702, and SLA-2*1001 and SLA-12 recombined in close proximity to the location of the gRNA binding sites. These recombinants were incapable of encoding functional class 1 SLA molecules as a consequence of frameshifts arising from a 2 base deletion or 1 base insertion, or they lacked 276 bases of exon four (Supplementary Figure 2).
Functional Consequences of Class I SLA Deficiency
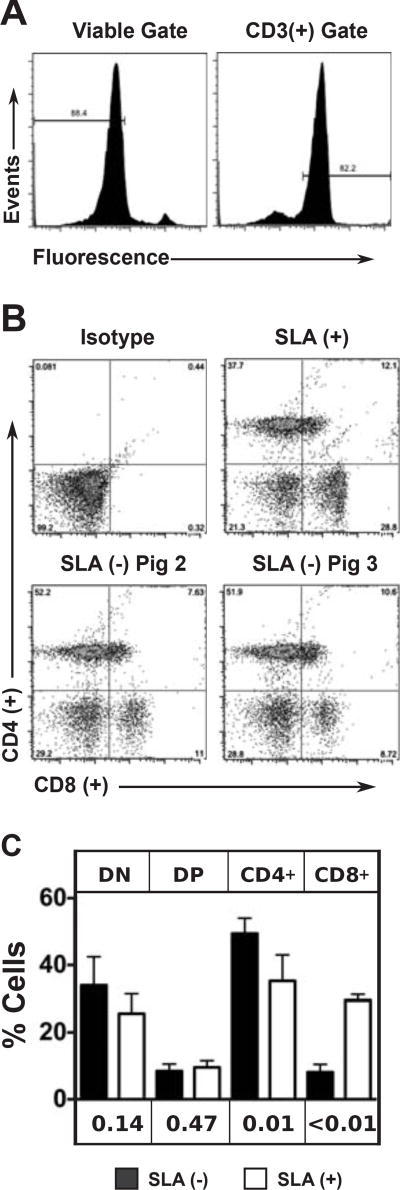
Because class I MHC molecules are needed for the efficient development of mature T lymphocytes expressing CD3 and CD8 markers, a T cell subset analysis was performed (Figure 5). PBMC were gated for viability and expression of the CD3 protein (Figure 5A) and then analyzed for expression of CD4 and CD8 (Figure 5B). This analysis was performed on four separate blood draws from an SLA positive animal, and on a total of five blood draws from two cloned SLA deficient animals (Figure 5C). The lymphocyte subsets in the control animal closely match what has been reported for other class I SLA expressing pigs. The SLA deficient animals showed a significant reduction in CD3+/CD8+ mature T lymphocytes and an increase in CD3+/CD4+ T cells.
Figure 5. Lymphocyte Subset Analysis of SLA Expressing and SLA Deficient Pigs.
PBMC were isolated from a class I SLA positive animal and two cloned pigs devoid of class I SLA molecules. Cells were incubated with a fluorescent viability dye, and antibodies specific for CD3, CD4, and CD8 molecules. A) Representative histograms demonstrating the gating strategy to select for viable CD3 positive cells. B) CD4 and CD8 expression levels are shown to reveal each T cell subset. An isotype control staining was used to set the gates defining each subset. C) The analysis of panel B was repeated on four separate PBMC isolations from the SLA positive animal and five separate PBMC isolations from the cloned animals (twice for Pig 2 and three times for Pig 3). The means and standard deviations are shown for the various lymphocyte subsets (DN: CD4CD8, DP: CD4+CD8+, CD4: CD4+CD8, CD8: CD4CD8+). Unpaired t tests were used to compare the frequencies of each cell type in SLA expressing and SLA deficient animals. P values are shown beneath the graph for comparison of the frequency of each subset between SLA positive and SLA negative animals.
DISCUSSION
We describe the creation of class I MHC deficient pigs by simultaneously disrupting seven alleles of classical class I SLA genes. At the time of this description, the animals have been healthy and growing well for 14 weeks. All cell types tested from these animals exhibit total loss of cell surface class I MHC proteins. Inactivation of a single gene, β2-microglobulin, also prevents expression of class I MHC heterotrimers at the cell surface. This disruption was not attempted because mice lacking β2m lose the ability to regulate iron homeostasis [30, 31, 32]. The high efficiency of the gRNA/Cas9 technology enabled the rapid production of animals deficient in class I MHC. The relative ease of this approach may enable the rapid production of many novel species lacking class I MHC gene activity.
Multigene families can be difficult to analyze because of functional redundancies. Simultaneous inactivation of all related genes minimizes these challenges by creating a null background that enables the study of one gene at a time. Research into the class I MHC gene family highlights these issues. The development of cell lines with defective expression of class I MHC gene products was crucial to various aspects of their biosynthesis and immune regulatory functions [6, 33, 34, 35, 36, 37, 38]. Animals lacking expression of class I MHC proteins have provided the opportunity to study the function of these molecules at the organismal level. This type of analysis is challenging in humans because of the rare opportunity to identify and study naturally occurring defects in class I MHC biology [39]. Mouse models have been far more productive in this regard because genetic engineering enabled the production of animals with a variety of gene defects preventing biosynthesis of class I MHC proteins [40, 41, 42, 43, 44]. Manipulation of larger mammals using the genome engineering tools applied to murine models has been difficult because they reproduce slowly.
Targeted nucleases have simplified the alteration mammalian genes. Zinc Finger Nucleases (ZFN) and Transcription-Activator LikE Nucleases (TALENs) consist of the nucleolytic portion of the Fok I enzyme covalently attached to a protein domain that can be modified to bind to unique DNA sequences [28, 45, 46]. An RNA-based technology employs the Cas9 endonuclease and a guide RNA (gRNA) to achieve site-specific activity [13, 26, 27, 28, 29]. The generation of cleaved DNA stimulates at least two cellular repair mechanisms. Either can be used to complete the genome editing process. 1) Nonhomolgous end joining of DNA fragments is imperfect and leads to random modifications such as point mutations, insertions, and deletions. 2) Exogenous DNA templates transfer specific genetic modifications to the genome in a process relying on homology directed repair [28].
Though, gRNA can direct Cas9-mediated cleavage of unintended genomic locations even in the presence of sequence mismatches [47, 48, 49], the results reported here arise as a direct consequence of modifying class I MHC genes. We were able to eliminate class I MHC protein expression by treating cells with three unique gRNA (Figure 2). The only common target of the gRNA is exon 4 of classical class I genes and class I-related pseudogenes in the MHC complex. Though it is likely any of the gRNA/Cas9 treatments would have generated SLA deficient animals, cells treated simultaneously with gRNA A and C were chosen for SCNT in an attempt to increase the frequency of larger deletions to simplify analyses. Determining the true effects of simultaneously using two gRNA requires additional analyses.
The combination of high polymorphism and identity between each classical class I MHC gene makes sequence analysis of this region difficult. PCR-amplification of cDNA from these loci introduces point mutations, recombinations, insertions, and deletions [50]. Therefore, we evaluated numerous cDNA clones to ensure accurate sequence analyses of each allele in the unmodified parental cells and in gRNA/Cas9 treated samples. The majority of parental cell cDNA were determined to be full length (Table II), and a few cDNA lacking 276 bases of exon 4 were found for SLA-1*0702 and SLA-1*1201. No transcripts capable of encoding functional class I proteins were found in the SLA deficient animals. SLA-2*0202, SLA-3*0402, or SLA-3*0502 were completely absent. The fetus and piglets all expressed three unique recombinant transcripts not found in the unmodified parental line. These transcripts do not encode functional protein because of frameshifts in two of the recombinants, and the deletion of 276 bp from the third. The only transcript common to the unmodified parental cells and gRNA/Cas9 treated cells originated from SLA-1*0702, but a 276 base deletion in this transcript eliminates the α3 domain of the class I protein.
The reduced frequency of CD4−CD8+ T cells in the SLA deficient animals mimics what has been seen in class I MHC deficient mice, suggesting that many aspects of T cell development are common between the two species [51,52]. In addition, the T Cell subset analysis of CD4 and CD8 expression on peripheral lymphocytes found four populations (double negative, double positive, CD4+CD8−, and CD4−CD8+) similarly to what others have reported [53, 54, 55]. The apparently normal levels of CD4+CD8+ double positive cells in class I SLA deficient animals indicates that engagement of classical class I MHC molecules with either the T cell receptor or CD8 molecule is not essential for the production or maintenance of these cells. The presence reduced levels of CD4−CD8+ T-cells may provide some protection from infection through a mechanism that does not involve class I MHC. This could explain the animals’ apparent good health over several months. However, the fact that the pseudorabies virus has evolved to inhibit presentation of antigenic peptides suggests that class I SLA-antigens are key to viral control in pigs [56]. Longer-term evaluation will be needed to determine the susceptibility of these animals to infectious diseases and cancer.
The enhancements provided by targeted nuclease technologies have dramatically expanded the scope of genome engineering. The ease with which the gRNA-Cas9 system has allowed simultaneous disruption of seven class I MHC alleles in swine suggests this technology should be easily translated to other complex organisms. The ability to simultaneously disrupt several genes in an organism promises to enhance the studies of gene families with redundant functions and will enable the creation of polygenic disease models.
Supplementary Material
Acknowledgments
The authors thank MRI (Methodist Research Institute) and LARC (Laboratory animal research center) staff for taking care of the animals, and Chris Burlak Ph.D. for teaching us the FlowJo software. This investigation utilized a facility constructed with support from Research Facilities Improvement Program grant number C06RR10601-01 from the National Center for Research Resources, National Institute of Health.
This study was funded by Lung Biotechnology LLC, Indiana University Health Transplant Institute, and Indiana University Health. The investigations utilized a facility constructed with support from Research Facilities Improvement Program grant number C06RR10601-01. From the National Center for Research Resources, National Institute of Health. Joe Tector has applied for patents related to the genetic engineering of pigs for use in xenotransplantation and has founded the company Xeno-Bridge llc. R.A. Sidner serves as a consultant to businesses that operate in the field of xenotransplantation.
Abbreviations
- CRSPR/Cas9
Clustered regularly interspaced short palindromic repeats/Cas9 nuclease
- FFSC
Fetal fibroblasts cultured in Steam Cell media
- gRNA
guide RNA
- Hp
Haplotype
- NHEJ
Nonhomolgous end joining
- MHC
Major Histocompatibility Complex
- PBMC
Peripheral blood mononuclear cell
- SLA
Swine Leucocyte Antigen
- SCNT
Somatic cell nuclear transfer
- SLAKO
SLA gene knockout
- β2m
β2-microglobulin
Footnotes
Other authors report no conflicts.
References
- 1.Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annual review of biochemistry. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 2.Rammensee HG, Falk K, Rötzschke O. Peptides naturally presented by MHC class I molecules. Annual review of immunology. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 3.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 4.Madden DR, Gorga JC, Strominger JL, Wiley DC. The three-dimensional structure of HLA-B27 at 2.1 Å resolution suggests a general mechanism for tight peptide binding to MHC. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- 5.Saper MA, Bjorkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. Journal of molecular biology. 1991;219(2):277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 6.Salter RD, Benjamin RJ, Wesley PK, Buxton SE, Garrett TPJ, Clayberger C, Krensky AM, Norment AM, Littman DR, Parham P. A binding site for the T-cell co-receptor CD8 on the α3 domain of HLA-A2. Nature. 1990;345:41–46. doi: 10.1038/345041a0. [DOI] [PubMed] [Google Scholar]
- 7.Garcia KC, Scott CA, Brunmark A, Carbone FR, Peterson PA, Wilson IA, Teyton L. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature. 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 8.Pamer E, Cresswell P. Mechanisms of MHC class I-restricted antigen processing. Annual review of immunology. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 9.Powis SJ, Young LL, Joly E, Barker PJ, Richardson L, Brandt RP, Melief CJ, Howard JC, Butcher GW. The Rat cim Effect: TAP Allele-Dependent Changes in a Class I MHC Anchor Motif and Evidence Against C-Terminal Trimming of Peptides in the ER. Immunity. 1996;4:159–165. doi: 10.1016/s1074-7613(00)80680-9. [DOI] [PubMed] [Google Scholar]
- 10.Morel S, Lévy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin A, Monsarrat B, et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 11.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. Journal of the American Veterinary Medical Association. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 12.Reyes LM, Blosser RJ, Smith RF, Miner AC, Paris LL, Blankenship RL, Tector MF, Tector AJ. Characterization of swine leucocyte antigen alleles in a crossbred pig to be used in Xenotransplant studies. Tissue Antigens. 2014 doi: 10.1111/tan.12430. in press. [DOI] [PubMed] [Google Scholar]
- 13.Li P, Estrada JL, Burlak C, Montgomery J, Butler JR, Santos RM, Wang ZY, Paris LL, Blankenship RL, Downey SM, Tector M, Tector AJ. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate expression. Xenotransplantation. 2014 doi: 10.1111/xen.12131. in press. [DOI] [PubMed] [Google Scholar]
- 14.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutz AJ, Li P, Estrada JL, Sidner RA, Chihara RK, Downey SM, Burlak C, Wang ZY, Reyes LM, Ivary BF, Yin F, Blankenship RL, Paris LL, Tector AJAJ. Double knockout pigs deficient in N-glycolylneuraminic acid and galactose alpha-1,3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation. 2013;20:27–35. doi: 10.1111/xen.12019. [DOI] [PubMed] [Google Scholar]
- 16.Estrada J, Sommer J, Collins B, Mir B, Martin A, York A, Petters RM, Piedrahita JA. Swine generated by somatic cell nuclear transfer have increased incidence of intrauterine growth restriction (IUGR) Cloning Stem Cells. 2007;9:229–36. doi: 10.1089/clo.2006.0079. [DOI] [PubMed] [Google Scholar]
- 17.Lunney JK, Ho C-S, Wysocki M, Smith DM. Molecular genetics of the swine major histocompatibility complex, the SLA complex. Developmental & Comparative Immunology. 2009;33:362–374. doi: 10.1016/j.dci.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Ho C-S, Lunney JK, Franzo-Romain MH, Martens GW, Lee Y-J, Lee J-H, Wysocki M, Rowland RRR, Smith DM. Molecular characterization of swine leucocyte antigen class I genes in outbred pig populations. Animal genetics. 2009;40:468–478. doi: 10.1111/j.1365-2052.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- 19.Renard C, Vaiman M, Chiannilkulchai N, Cattolico L, Robert C, Chardon P. Sequence of the pig major histocompatibility region containing the classical class I genes. Immunogenetics. 2001;5:490–500. doi: 10.1007/s002510100348. [DOI] [PubMed] [Google Scholar]
- 20.Chardon P, Renard C, Vaiman M. The major histocompatibility complex in swine. Immunological reviews. 1999;167:179–192. doi: 10.1111/j.1600-065x.1999.tb01391.x. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka-Matsuda M, Ando A, Rogel-Gaillard C, Chardon P, Uenishi H. Difference in number of loci of swine leukocyte antigen classical class I genes among haplotypes. Genomics. 2009;93:261–273. doi: 10.1016/j.ygeno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Ho CS, Franzo-Romain MH, Lee YJ, Lee JH, Smith DM. Sequence-based characterization of swine leucocyte antigen alleles in commercially available porcine cell lines. International journal of immunogenetics. 2009;36:231–234. doi: 10.1111/j.1744-313X.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- 23.Parham P, Lomen CE, Lawlor DA, Ways JP, Holmes N, Coppin HL, Salter RD, Wan AM, Ennis PD. Nature of polymorphism in HLA-A,-B, and-C molecules. Proceedings of the National Academy of Sciences. 1988;85:4005–4009. doi: 10.1073/pnas.85.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Tector M, Salter RD. Calnexin recognizes carbohydrate and protein determinants of class I major histocompatibility complex molecules. Journal of Biological Chemistry. 1995;270:3944–3948. doi: 10.1074/jbc.270.8.3944. [DOI] [PubMed] [Google Scholar]
- 25.Salter RD. Mutant HLA-A201 heavy chains with lowered affinity for β2m are transported after growth at reduced temperatures. Human immunology. 1992;35:40–49. doi: 10.1016/0198-8859(92)90093-3. [DOI] [PubMed] [Google Scholar]
- 26.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Mali P, Kim-Kiselak C, Church G. CRISPR-Cas-Mediated Targeted Genome Editing in Human Cells. Methods Mol Biol. 2013;1114:245–267. doi: 10.1007/978-1-62703-761-7_16. [DOI] [PubMed] [Google Scholar]
- 28.Carroll D. Genome Engineering with Targetable Nucleases. Annual review of biochemistry. 2014 doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 29.Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proceedings of the National Academy of Sciences. 2013;110:16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos M, Schilham MW, Rademakers LH, Marx JJ, De Sousa M, Clevers H. Defective iron homeostasis in beta 2-microglobulin knockout mice recapitulates hereditary hemochromatosis in man. The Journal of experimental medicine. 1996;184:1975–1985. doi: 10.1084/jem.184.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothenberg BE, Voland JR. beta2 knockout mice develop parenchymal iron overload: A putative role for class I genes of the major histocompatibility complex in iron metabolism. Proceedings of the National Academy of Sciences. 1996;93:1529–1534. doi: 10.1073/pnas.93.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Sousa M, Reimão R, Lacerda R, Hugo P, Kaufmann SHE, Porto G. Iron overload in β2-microglobulin-deficient mice. Immunology letters. 1994;39:105–111. doi: 10.1016/0165-2478(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 33.Salter RD, Cresswell P. Impaired assembly and transport of HLA-A and-B antigens in a mutant TxB cell hybrid. The EMBO journal. 1986;5:943. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storkus WJ, Howell DN, Salter RD, Dawson JR, Cresswell P. NK susceptibility varies inversely with target cell class I HLA antigen expression. The Journal of Immunology. 1987;138:1657–1659. [PubMed] [Google Scholar]
- 35.Spies T, DeMars R. Restored expression of major histocompatibility class I molecules by gene transfer of a putative peptide transporter. Nature. 1991;35:323–324. doi: 10.1038/351323a0. [DOI] [PubMed] [Google Scholar]
- 36.Powis SJ, Townsend AARM, Deverson EV, Bastin J, Butcher GW, Howard JC. Restoration of antigen presentation to t.he mutant cell line RMA-S by an MHC-linked transporter. Nature. 1991;354:528–531. doi: 10.1038/354528a0. [DOI] [PubMed] [Google Scholar]
- 37.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Peter Cresswell. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 38.Kaer LuV, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4− 8+ T cells. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 39.de La Salle H, Hanau D, Fricker D, Urlacher A, Kelly A, Salamero J, Powis SH, Donato L, Bausinger H, Laforet M. Homozygous human TAP peptide transporter mutation in HLA class I deficiency. Science. 1994;265:237–241. doi: 10.1126/science.7517574. [DOI] [PubMed] [Google Scholar]
- 40.Bevan MJ. The earliest knockouts. The Journal of Immunology. 2010;184:4585–4586. doi: 10.4049/jimmunol.1090023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vugmeyster Y, Glas R, Pérarnau B, Lemonnier FA, Eisen H, Ploegh H. Major histocompatibility complex (MHC) class I KbDb−/− deficient mice possess functional CD8+ T cells and natural killer cells. Proceedings of the National Academy of Sciences. 1998;95:12492–12497. doi: 10.1073/pnas.95.21.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Béatrice P, Saron MF, San Martin BR, Bervas N, Ong H, Soloski MJ, Smith AG, Ure JM, Gairin J, Lemonnier FA. Single H2Kb, H2Db and double H2KbDb knockout mice: peripheral CD8+ T cell repertoire and antilymphocytic choriomeningitis virus cytolytic responses. European journal of immunology. 1999;29:1243–1252. doi: 10.1002/(SICI)1521-4141(199904)29:04<1243::AID-IMMU1243>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.Das G, Janeway CA. Development of CD8α/α and CD8α/β T cells in major histocompatibility complex class I-deficient mice. The Journal of experimental medicine. 1999;190:881–884. doi: 10.1084/jem.190.6.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sousa AO, Mazzaccaro RJ, Russell RG, Lee FK, Turner OC, Hong S, Van Kaer L, Bloom BR. Relative contributions of distinct MHC class I-dependent cell populations in protection to tuberculosis infection in mice. Proceedings of the National Academy of Sciences. 2000;97:4204–4208. doi: 10.1073/pnas.97.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): A highly efficient and versatile tool for genome editing. Biotechnology and bioengineering. 2013;110:1811–1821. doi: 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- 46.Gersbach CA, Gaj T, Barbas CF., III Synthetic Zinc Finger Proteins: The Advent of Targeted Gene Regulation and Genome Modification Technologies. Accounts of Chemical Research. 2014 doi: 10.1021/ar500039w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon DK, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013 doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ennis PD, Zemmour J, Salter RD, Parham P. Rapid cloning of HLA-A, B cDNA by using the polymerase chain reaction: frequency and nature of errors produced in amplification. Proceedings of the National Academy of Sciences. 1990;87:2833–2837. doi: 10.1073/pnas.87.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. β2-microglobulin deficient mice lack CD4− 8+ cytolytic T cells. 1990:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 52.Kaer LV, Ashton-Rickardt PG, Ploegh L, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4−8+ T cells. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 53.Zuckermann FA, Husmann RJ. Functional and phenotypic analysis of porcine peripheral blood CD4/CD8 double-positive T cells. Immunology. 1996;87:500. [PMC free article] [PubMed] [Google Scholar]
- 54.Saalmüller A, Reddehase MJ, Bühring HJ, Jonjić S, Koszinowski UH. Simultaneous expression of CD4 and CD8 antigens by a substantial proportion of resting porcine T lymphocytes. European journal of immunology. 1987;17:1297–1301. doi: 10.1002/eji.1830170912. [DOI] [PubMed] [Google Scholar]
- 55.Lunney JK, Pescovitz MD. Phenotypic and functional characterization of pig lymphocyte populations. Veterinary immunology and immunopathology. 1987;17:135–144. doi: 10.1016/0165-2427(87)90134-6. [DOI] [PubMed] [Google Scholar]
- 56.Ambagala APN, Hinkley S, Srikumaran S. An early pseudorabies virus protein down-regulates porcine MHC class I expression by inhibition of transporter associated with antigen processing (TAP) The Journal of Immunology. 2000;164:93–99. doi: 10.4049/jimmunol.164.1.93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.