Abstract
Expansins are extracellular proteins that facilitate cell wall extension, possibly by disrupting hydrogen bonding between hemicellulosic wall components and cellulose microfibrils. In addition, some expansins are expressed in non-growing tissues such as ripening fruits, where they may contribute to cell wall disassembly associated with tissue softening. We have identified at least three expansin genes that are expressed in tomato (Lycopersicon esculentum Mill.) seeds during germination. Among these, LeEXP4 mRNA is specifically localized to the micropylar endosperm cap region, suggesting that the protein might contribute to tissue weakening that is required for radicle emergence. In gibberellin (GA)-deficient (gib-1) mutant seeds, which germinate only in the presence of exogenous GA, GA induces the expression of LeEXP4 within 12 hours of imbibition. When gib-1 seeds were imbibed in GA solution combined with 100 μm abscisic acid, the expression of LeEXP4 was not reduced, although radicle emergence was inhibited. In wild-type seeds, LeEXP4 mRNA accumulation was blocked by far-red light and decreased by low water potential but was not affected by abscisic acid. The presence of LeEXP4 mRNA during seed germination parallels endosperm cap weakening determined by puncture force analysis. We hypothesize that LeEXP4 is involved in the regulation of seed germination by contributing to cell wall disassembly associated with endosperm cap weakening.
In most seeds, radicle extension through the structures surrounding the embryo is the event that terminates germination and marks the commencement of seedling growth (Bewley, 1997a). In seeds whose embryos are embedded in a rigid endosperm, the micropylar portion of the endosperm, termed the endosperm cap, presents a physical restraint to radicle extension. This restraint must be lessened through the weakening of the endosperm cap to allow radicle emergence (Groot and Karssen, 1987). Endosperm cap weakening is associated with cell wall hydrolysis (Watkins et al., 1985; Sánchez et al., 1990). As Man-containing polysaccharides are a major component of the endosperm cell walls of seeds of tomato (Lycopersicon esculentum Mill.) and other Solanaceae (Sánchez et al., 1990; Dahal et al., 1997), endo-β-mannanase has been regarded as a candidate enzyme to control the weakening process (Groot et al., 1988; Nomaguchi et al., 1995). Increased mannanase activity is consistently associated with radicle emergence (Nonogaki and Morohashi, 1996; Nonogaki et al., 1998, 2000), but there are also conditions where emergence does not occur even though high enzyme activity is present (Toorop et al., 1996; Dahal et al., 1997; Still and Bradford, 1997). Thus, although endo-β-mannanase may be necessary for germination, it does not appear to be sufficient in all cases.
In addition to endo-β-mannanase, other enzymes, including mannosidase, galactosidase, cellulase, pectin methylesterase, polygalacturonase, arabinosidase, xyloglucan endotransglycosylase, β-1,3-glucanase, and chitinase, are also expressed during tomato seed germination (Groot et al., 1988; Leviatov et al., 1995; Downie et al., 1998; Sitrit et al., 1999; Bradford et al., 2000). Since most of these hydrolases are associated with cell separation or cell wall disassembly in other developmental processes such as abscission zones and fruit ripening (Del Campillo and Lewis, 1992; Lashbrook et al., 1994), it is reasonable to expect that they also may be involved in endosperm weakening. However, specific hydrolase activities have not been connected directly with the mechanism of endosperm weakening required for radicle emergence, and additional factors may be involved in controlling this process (Bewley, 1997b; Bradford et al., 2000; Toorop et al., 2000).
Expansin proteins are candidates to be such factors. Expansin was first identified from cucumber hypocotyls by its ability to induce stress relaxation of cell walls in killed tissue segments (McQueen-Mason et al., 1992). Expansins are proposed to function as cell wall loosening factors by disrupting non-covalent linkages, such as hydrogen bonds, at the cellulose-hemicellulose interface thereby relaxing an important constraint to turgor-driven cell expansion (McQueen-Mason and Cosgrove, 1994; Cosgrove, 1998). Expansins have been highly conserved through plant evolution as homologous genes have been identified in gymnosperms and in both monocots and dicots among the angiosperms (Cosgrove, 1998). Expansins occur as multi-gene families in Arabidopsis, rice, cucumber, tomato, and other species where they have been examined in detail (Cosgrove, 1999). The large number of expansin-like genes (e.g. at least 22 in Arabidopsis) suggests multiple developmental or tissue-specific roles for these proteins in addition to cell expansion. Expansins are expressed in growing tissues including cucumber hypocotyls, deepwater rice internodes, shoot meristems, and developing fruits (Cho and Kende, 1997; Fleming et al., 1997; Reinhardt et al., 1998; Brummell et al., 1999). They are also expressed in non-growing tissues such as ripening fruits (Rose et al., 1997; Brummell et al., 1999). During ripening, extensive cell wall degradation and solubilization of wall components results in tissue softening and cell separation (Fischer and Bennett, 1991). Preliminary results with expansin promoters linked to the β-glucuronidase reporter also indicate that expression of specific expansin genes occurs in germinating seeds, in the root cap, and in association with abscission zones or tissues where cell separation will take place (Cosgrove et al., 1998). Thus, in addition to their role in cell growth, specific expansins may also contribute to cell wall processes associated with developmental events such as ripening, abscission, and cell separation (Cosgrove, 1997).
Since cell wall disassembly and cell separation accompany endosperm weakening during tomato seed germination, we investigated the possibility that expansin(s) could be involved. Here we report that a specific expansin gene (LeEXP4), also expressed in tomato flowers and enlarging fruit (Brummell, et al., 1999), is expressed in the endosperm cap region and is regulated by hormonal and environmental factors that control seed germination. The expression of LeEXP4 is consistently associated with endosperm cap weakening, supporting a role for expansins in the cell wall modification of this tissue.
RESULTS
Cloning and Sequence Analysis of Tomato Seed Expansins
Using primers to conserved expansin sequences, an approximately 540-bp cDNA band was amplified by reverse transcriptase (RT)-PCR from germinating tomato seed RNA and cloned into pCR2.1. Subsequent sequence analyses indicated the presence of six expansin homologs (termed TE1 to TE6). These six independent fragments shared high amino acid sequence homology among themselves and with known expansins. The sequence of TE1 was identical to a region of LeEXP1, which is expressed during tomato fruit ripening (Rose et al., 1997), and the sequence of TE2 was identical to a region of LeEXP2, which is expressed in expanding tissues (Reinhardt et al., 1998). TE3 was identical with a region of LeEXP4, which is detected in flowers and expanding fruits (Brummell et al., 1999). TE5 was identical to a region of LeEXP9. The remaining two fragments TE4 and TE6 were unique in the database. Since the colonies were randomly selected for sequencing, it is possible that additional expansin homologs are expressed in germinating tomato seeds.
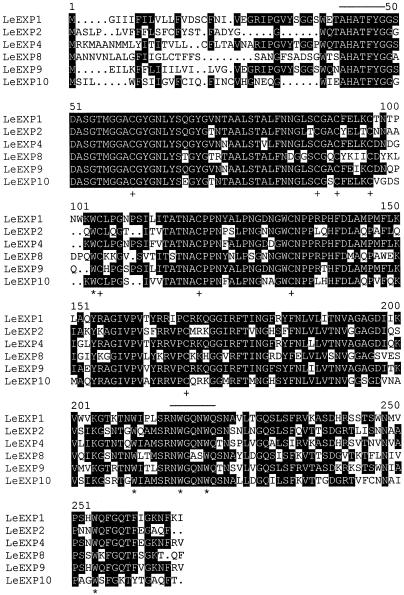
A cDNA library prepared from gibberellin (GA)-treated gib-1 tomato seeds was screened with TE1 to TE6. The nucleotide sequences of the full-length cDNAs identified by TE1, TE2, TE3, and TE5 were confirmed to be identical with LeEXP1, LeEXP2, LeEXP4, and LeEXP9, respectively. The full-length cDNAs corresponding to TE4 and TE6 were named LeEXP8 and LeEXP10, respectively (Cosgrove, 1999). The multiple alignment of the predicted amino acid sequences of these six genes confirms that they are all members of an α-expansin gene family (Fig. 1). The predicted signal sequences (approximately the first 30 N-terminal amino acids) are divergent, but the proposed mature polypeptides shared a high degree of amino acid identity. The predicted amino acid sequences of all six tomato genes exhibit the diagnostic characteristics of expansins, including eight conserved cysteines and conserved tryptophans (Fig. 1; Shcherban et al., 1995).
Figure 1.
Multiple alignment of the deduced amino acid sequences of full-length expansin cDNAs from germinating tomato seeds. LeEXP1 (U82123), LeEXP2 (AF096776), LeEXP4 (AF059488), LeEXP8 (AF184232), LeEXP9 (AJ243340), and LeEXP10 (AF184233) were aligned using the MACDNASIS PRO3.5 program (Hitachi Software, San Bruno, CA). Amino acids identical among at least three of the cDNAs are shaded using the Boxshade program (http://www. isrec.isf). The conserved cysteines and typtophans are indicated by * and +, respectively. The lines above specific sequences indicate the conserved regions for which RT-PCR primers were designed.
Preparation of Gene-Specific Probes and Genomic Analysis of Novel Expansin Genes
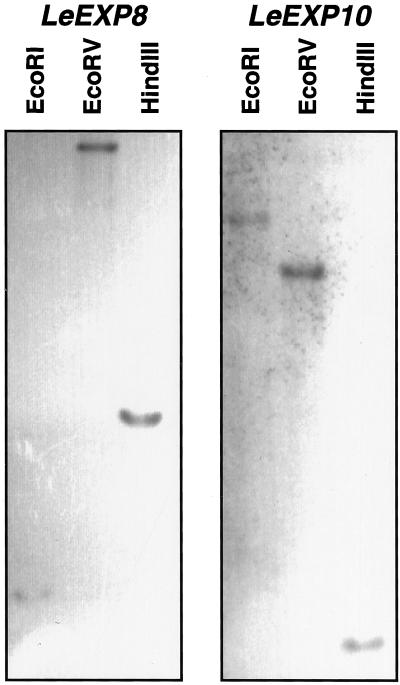
Gene-specific probes were generated by amplification of 3′-terminal untranslated regions of each gene. When the probes were hybridized with all six full-length expansin cDNAs isolated from germinating tomato seeds, each probe hybridized only with its corresponding cDNA (data not shown), demonstrating the specificity of the probes. Because LeEXP8 and LeEXP10 are novel expansin genes, genomic analysis was performed. Each of the gene-specific probes strongly hybridized to a single genomic fragment (Fig. 2), indicating that both LeEXP8 and LeEXP10 are single-copy genes and that the probes are gene-specific. A Southern blot using the gene-specific probe for LeEXP4 also hybridized with single bands identical to those shown for this gene by Brummell et al. (1999) (data not shown).
Figure 2.
Genomic DNA gel-blot analysis of tomato expansin genes. Tomato genomic DNA (10 μg) was digested by EcoRI, EcoRV, and HindIII, respectively, and subjected to gel-blot hybridization using gene-specific cDNA probes amplified by PCR using primers corresponding to the 3′-untranslated regions of LeEXP8 (left) and LeEXP10 (right).
Different Expansins Exhibit Tissue-Specific Expression in Germinating Tomato Seeds
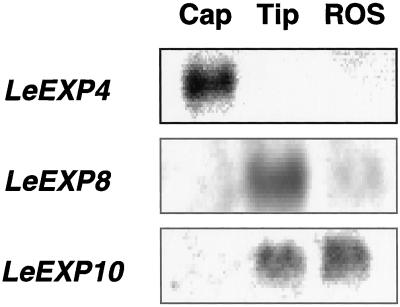
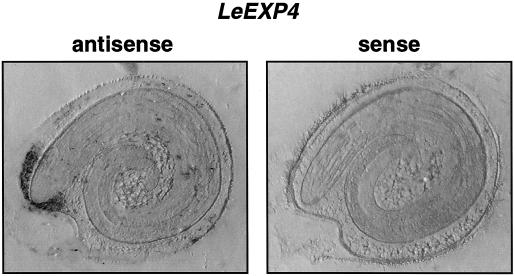
When gene-specific probes were hybridized with total RNA isolated from seeds imbibed for 40 h (just prior to first radicle emergence), no signal or only very weak hybridization occurred using probes for LeEXP1, LeEXP2, or LeEXP9, whereas strong signals were detected for LeEXP4, LeEXP8, and LeEXP10 (data not shown). When seed tissues were isolated and tested separately, LeEXP4 was expressed specifically in the endosperm cap, whereas LeEXP8 was expressed only in the radicle tip and LeEXP10 mRNA was present in both the radicle tip and the rest of the seed (Fig. 3). Localization of LeEXP4 expression to the micropylar region of the endosperm was confirmed by northern tissue printing (Fig. 4). Because our primary interest is in endosperm cap weakening, and LeEXP4 expression is localized to this tissue, further studies focused on LeEXP4.
Figure 3.
RNA gel-blot analysis of abundance of specific expansin mRNAs in different parts of tomato seeds just prior to radicle emergence (40 h of imbibition). Total RNA (5 μg) from endosperm caps (Cap), radicle tips (Tip), and the rest of the seed (ROS) was separated by electrophoresis and hybridized with gene-specific probes for LeEXP4, LeEXP8, and LeEXP10. Each gene exhibits a distinct expression pattern among seed tissues.
Figure 4.
Tissue printing to localize LeEXP4 mRNA expression. Tomato seeds imbibed for 24 h were bisected and the cut surfaces were printed onto two membranes. One membrane (left) was hybridized with an antisense probe for LeEXP4 and the other one (right) was hybridized with the corresponding sense probe as a control for non-specific binding. LeEXP4 mRNA was detected only in the endosperm cap tissue.
To determine the expression pattern of LeEXP4 in other tissues, RNA gel-blot analyses were conducted using total RNA isolated from tomato roots, stems, leaves, flowers, dry seeds, and ripening fruits. Among these tissues, LeEXP4 mRNA could only be detected in flowers (data not shown), consistent with the results of Brummell et al. (1999).
Hormonal and Environmental Regulation of LeEXP4 Expression and Endosperm Cap Weakening
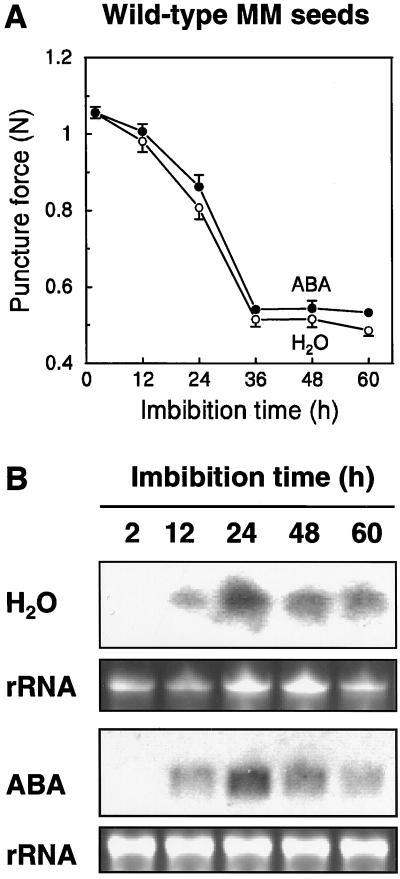
Weakening of the endosperm cap can be measured by puncture force analysis (Groot and Karssen, 1987). Physical weakening of the cap tissue begins within 12 h of imbibition of wild-type seeds in water, and reaches a minimum value in ungerminated seeds by 36 h (Fig. 5A). Whereas mRNA of LeEXP4 is not detectable in seeds soon after imbibition, it begins to accumulate within 12 h and is abundant by 24 h (Fig. 5B), when physical weakening is proceeding rapidly (Fig. 5A). LeEXP4 mRNA abundance then declines with longer imbibition times in seeds that have not completed germination. This expression pattern was essentially unaffected by imbibition in the presence of 100 μm abscisic acid (ABA), which prevents radicle emergence (Fig. 5B). However, ABA also had no significant effect on the decrease in puncture force of the endosperm cap (Fig. 5A). Thus, regardless of the presence of ABA, there was a correspondence between endosperm cap weakening and the expression of LeEXP4.
Figure 5.
A, Puncture force analysis of wild-type MM seeds at different times after imbibition in water (○) or 100 μm ABA (●). Error bars indicate ± se (n = 24) when they exceed the size of the symbols. B, LeEXP4 mRNA abundance in MM seeds after imbibition in water or in 100 μm ABA. Total RNA extracted at various times after imbibition was separated by electrophoresis and hybridized with a LeEXP4-specific cDNA probe. Bottom sections in each pair show ethidium bromide-stained rRNA to indicate the relative RNA loading of each lane.
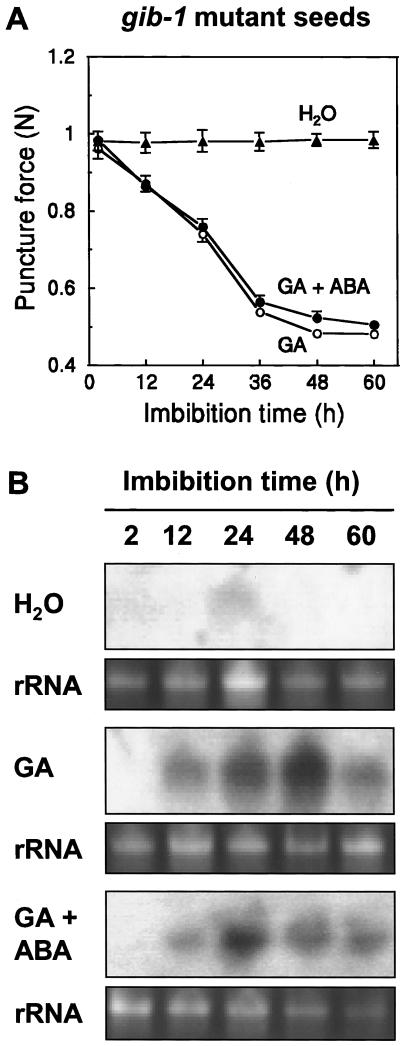
Endosperm cap weakening and radicle emergence of gibberellin-deficient gib-1 tomato seeds are completely dependent on exogenous GA (Groot and Karssen, 1987). Endosperm caps did not weaken when gib-1 seeds were imbibed in water, but weakening did occur when GA4+7 was present in the imbibition solution (Fig. 6A) with a time course similar to that for tomato wild-type cv Moneymaker (MM) seeds (Fig. 5A). No or only slight expression of LeEXP4 mRNA was detected in gib-1 seeds imbibed in water, but in the presence of GA4+7, LeEXP4 mRNA accumulated within 12 h and remained abundant until 48 h of imbibition before declining by 60 h (Fig. 6B). (Approximately 4% of the seeds had completed radicle emergence by 48 h, although only un-germinated seeds were sampled for RNA.) The simultaneous presence of ABA can block the effect of GA on seed germination (Groot and Karssen, 1992) but did not affect GA-induced endosperm cap weakening (Fig. 6A) or the expression of LeEXP4 (Fig. 6B).
Figure 6.
A, Puncture force analysis of gib-1 seeds at different times after imbibition in water (▴), 100 μm GA (○), or 100 μm GA plus 100 μm ABA (●). Error bars indicate ± se (n = 24) when they exceed the size of symbols. B, LeEXP4 mRNA abundance in gib-1 seeds after imbibition in water, 100 μm GA, or 100 μm GA plus 100 μm ABA. Total RNA extracted at various times after imbibition was separated by electrophoresis and hybridized with a LeEXP4-specific probe. Bottom sections in each pair show ethidium bromide-stained rRNA to indicate the relative RNA loading of each lane.
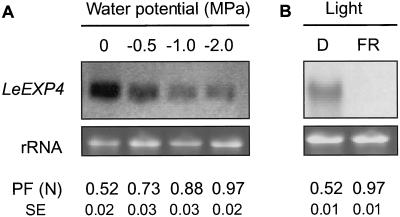
Low water potential can delay or prevent tomato seed germination (Dahal and Bradford, 1990). When MM tomato seeds were imbibed for 40 h in polyethylene glycol (PEG) solutions maintaining osmotic potentials of −0.5, −1.0, or −2.0 MPa, the abundance of LeEXP4 mRNA decreased as the water potential decreased (Fig. 7A). The decline in LeEXP4 mRNA abundance corresponded to an increase in the puncture force of the endosperm caps (Fig. 7A).
Figure 7.
LeEXP4 expression in wild-type MM seeds imbibed at different water potentials or under far-red light. A, Total RNA isolated from seeds imbibed for 40 h in water or in PEG 8,000 solutions of −0.5, −1.0, or −2.0 MPa was hybridized with a LeEXP4-specific probe. B, Total RNA isolated from seeds imbibed for 40 h in the dark (D) or under continuous far-red (FR) light was hybridized with a LeEXP4-specific probe. Lower sections in each pair show ethidium bromide stained rRNA to indicate the relative RNA loading of each lane. In A and B, the puncture force (PF) values (and se, n = 24) below each lane were recorded for endosperm caps of each treatment at the time that RNA samples were collected (40 h of imbibition).
Far-red light can also inhibit tomato seed germination (Downie et al., 1999). When MM seeds were imbibed for 40 h under continuous far-red light, no expression of LeEXP4 mRNA was detected, in contrast to the accumulation observed in seeds imbibed in the dark (Fig. 7B). Far-red light also prevented any weakening of the endosperm cap (Fig. 7B).
DISCUSSION
Expansins comprise a large superfamily of proteins sharing amino acid sequences conserved both within and among species. It is becoming evident that different expansin genes are expressed in unique tissue-specific and developmentally regulated patterns (Cosgrove, 1998). The plant body is composed of many organs, tissues, and cell types, each of which requires a characteristic and highly precise pattern of cell enlargement or cell wall modification with differential control by various developmental signals, hormones, and environmental conditions. For example, four expansin genes from rice have different expression patterns in leaves, stems, and roots (Cho and Kende, 1997), and six expansin genes show differential expression during tomato fruit development and ripening (Rose et al., 1997; Brummell et al., 1999). A number of expansin promoters from Arabidopsis exhibit varied and tissue-specific expression based upon reporter gene assays (Cosgrove et al., 1998).
We report here that this is also the case during tomato seed germination. At least six expansins may be expressed at some level in tomato seeds prior to radicle emergence, based upon RT-PCR amplification and subsequent screening of an imbibed tomato seed cDNA library (Fig. 1). The transcripts of three of these expansin genes accumulate sufficiently to be detected by gel hybridization, and each of these exhibits a distinct pattern of localization: LeEXP4 expression is localized to the endosperm cap; LeEXP8 is confined to the radicle tip; and LeEXP10 accumulates in both the radicle tip and the rest of seed (Fig. 3). As the cell wall composition of the embryo is quite distinct from that of the mannan-rich endosperm cell walls, and smaller differences are discernable even between the endosperm cap and the remaining lateral endosperm (Dahal et al., 1997), it is perhaps not surprising that specialized expansins might be required to interact with each type of cell wall. The fates and functions of the cell walls also differ among tissues. In the embryo, cell expansion is the primary event accompanying protrusion of the radicle from the seed, requiring extension of the cell walls. In both the endosperm cap and the lateral endosperm, the cells do not expand and the cell walls instead constitute a major carbohydrate reserve that is mobilized during germination. However, the thinner-walled cells of the endosperm cap begin to degrade prior to radicle emergence, presumably resulting in tissue weakening to allow radicle emergence, whereas the thicker cell walls of the lateral endosperm are degraded only during seedling growth following radicle emergence (Nonogaki et al., 1998; Toorop et al., 1998, 2000). In addition to expressing different expansin genes, each of these tissues also expresses distinct isoforms of endo-β-mannanase (Nonogaki et al., 1995; Nonogaki and Morohashi, 1996; Voigt and Bewley, 1996), which is due to the activation of different genes (Nonogaki et al., 2000). Thus, although the biochemical processes involved in cell wall disassembly in both parts of the endosperm may be similar, these processes are clearly regulated differentially with specific expansin and endo-β-mannanase genes being expressed solely in the endosperm cap prior to radicle emergence (Fig. 3; Nonogaki et al., 1998, 2000).
Because endosperm cap weakening is the critical process governing radicle emergence in tomato seeds, we have further examined the regulation of expression of LeEXP4 in the endosperm cap. In wild-type MM seeds, the expression of LeEXP4 can be detected within 12 h after imbibition (Fig. 5A). This is consistent with the physical weakening of the endosperm caps of imbibed MM seeds, which also begins by this time (Fig. 5B), and with the initial appearance of endo-β-mannanase (LeMAN2) mRNA and activity (Nonogaki et al., 2000). In gib-1 seeds, which require GA to complete germination, GA induces accumulation of LeEXP4 mRNA within 12 h of imbibition (Fig. 6B), concomitant with the initiation of endosperm cap weakening (Fig. 6A). Even though the entire seed is in contact with the imbibition solution containing GA, LeEXP4 mRNA is detected only in the endosperm cap (Bradford et al., 2000), indicating that its promoter is sensitive to regulation by both cell type and GA. It is also possible that the sensitivity of cells to GA is greater in the micropylar (proximal) part of the seed as has been shown for barley aleurone layer cells (Ritchie et al., 1999).
Both low water potential and far-red light delay or inhibit germination of wild-type seeds, and both factors reduce or prevent expression of LeEXP4 (Fig. 7). Furthermore, the extent of endosperm cap weakening was quantitatively proportional to the abundance of LeEXP4 mRNA across all hormonal and environmental conditions (Figs. 5–7). Low water potential can also reduce endo-β-mannanase activity (Toorop et al., 1998). Thus, a proportional down-regulation of genes involved in endosperm cap weakening may account for the characteristic inverse relationship between seed water potential (in excess of a threshold) and time to completion of radicle emergence (Bradford, 1995). Far-red light also inhibits expression of the germinative endo-β-mannanase (Nomaguchi et al., 1995), which is GA-dependent and localized to the endosperm cap region (Nonogaki et al., 2000). In lettuce and Arabidopsis seeds, red and far-red (FR) light control germination through the action of phytochrome by regulating gibberellin biosynthesis (Toyomasu et al., 1998; Yamaguchi et al., 1998). This could also be the case with tomato seeds where FR light might inhibit GA synthesis that is required to trigger expression of genes involved in endosperm cap weakening (Fig. 7).
The relationship of LeEXP4 expression to ABA at first appears rather anomalous. ABA effectively inhibits germination, and in isolated endosperm caps it also blocked GA-induced weakening (Groot and Karssen, 1992). However, in intact seeds (both MM seeds imbibed in ABA solution and gib-1 seeds imbibed in GA+ABA solution), ABA did not prevent LeEXP4 expression (Figs. 5B and 6B) and had no effect on endosperm weakening (Figs. 5A and 6A). The latter result is in agreement with the results of Toorop et al. (2000), and ABA did not inhibit the appearance of endo-β-mannanase mRNA and activity in the endosperm cap following imbibition (Toorop et al., 1996; Dahal et al., 1997; Nonogaki et al., 2000). Thus ABA does not appear to inhibit germination by blocking the expression of genes associated with endosperm cap weakening (Bradford et al., 2000) in contrast with earlier hypotheses (Ni and Bradford, 1993). It has been suggested that there is a second phase of weakening required for radicle emergence and that ABA inhibits this second phase (Toorop et al., 2000). Alternatively, ABA may act on the growth potential of the embryo (Schopfer and Plachy, 1985; Ni and Bradford, 1992), reducing it below the level required to penetrate even the weakened endosperm cap. Since the expression of LeEXP8 was localized to radicle tips and LeEXP10 to radicle tips and the rest of the seed (Fig. 3), we are currently studying whether ABA and/or GA regulate their expression. Further studies will also be required to understand the differences in hormonal responses between intact and excised endosperm caps. The importance of the embryo in regulating physiological events in the endosperm cap has been clearly demonstrated in Datura ferox seeds (Sánchez and de Miguel, 1997).
A key question is how expansin might interact with cell walls or wall-hydrolyzing enzymes to induce weakening or enhance disassembly of wall polymers. We have expressed LeEXP4 protein in Escherichia coli but have been unable to demonstrate any activity of the fusion protein in the endosperm cap puncture force assay (data not shown). However, the expressed LeEXP4 protein also did not exhibit activity in the cucumber hypocotyl extension assay (D. Cosgrove, personal communication), as has generally been the case with bacterially expressed expansins (McQueen-Mason and Rochange, 1999). Better results have been obtained recently with plant and insect expression systems (Sheih and Cosgrove, 1998; McQueen-Mason and Rochange, 1999), and we are pursuing those approaches to obtain active LeEXP4 protein for further studies of its function and mode of action in endosperm caps. It is intriguing that the same LeEXP4 expansin is apparently involved both in the early stages of tomato fruit growth (Brummell et al., 1999) and in endosperm cap weakening, despite the differences between the two tissues in cell wall composition. For example, tomato fruit cell wall hemicelluloses are low in Man content (Gross and Wallner, 1979), whereas cell walls of tomato endosperm caps contain 60% Man (Dahal et al., 1997). However, tissue-specific expression of the same expansin promoter in multiple locations in the plant was also evident in Arabidopsis (Cosgrove et al., 1998).
We have demonstrated that three distinct expansin gene family members (LeEXP4, LeEXP8, and LeEXP10) are expressed in tomato seeds prior to radicle emergence. Expansin gene LeEXP4 is expressed specifically in the endosperm cap of imbibed tomato seeds, as well as in flowers and expanding fruits. Expression of LeEXP4 is initiated within 12 h of seed imbibition, is regulated by factors that affect germination, and is quantitatively correlated with the extent of weakening of the endosperm cap tissues. Together, these results support the hypothesis that LeEXP4, most likely in conjunction with cell wall hydrolases, is involved in the cell wall degradation associated with tissue weakening and cell separation in the endosperm cap. If LeEXP4 protein is required to loosen hemicellulosic bonds and/or to facilitate access of hydrolases to the polymer matrix, regulation of its expression could be a critical control point in the germination process. At the same time, we recognize that our inferences are based on mRNA abundance, not protein amounts or functional assays. Ultimately it will be necessary to localize and quantify expansin proteins and demonstrate expansin activity and function. Antibodies against individual expansins and transgenic plants to modify LeEXP4 expression are under development to determine the expression patterns of expansin proteins and the consequences for seed germination of altered expansin expression.
MATERIALS AND METHODS
Plant Materials
Tomato (Lycopersicon esculentum Mill.) seeds from either Moneymaker (MM) plants grown in the field or homozygous gibberellin-deficient (gib-1) mutant plants grown in a greenhouse in 1997 were used throughout the study. The gib-1 mutant and its isogenic parent line were originally obtained from Dr. Cees Karssen (Wageningen University, The Netherlands). Mutant plants were sprayed three times per week with 100 μm GA4+7 to revert the dwarf habit and allow more vigorous growth and fertility. After fruits were harvested in the fall of 1997, seeds were extracted, treated with 0.25 m HCl, dried to 6% moisture content (fresh basis), and stored at −20°C until used (Ni and Bradford, 1993).
Germination Conditions
Approximately 500 seeds were incubated at 25°C in the dark in 9-cm diameter Petri dishes on top of two layers of blotter paper moistened with 12 mL of deionized water, 100 μm GA4+7, 100 μm ABA, or PEG 8,000 solutions having water potentials of −0.5, −1.0, or −2.0 MPa. For FR light treatment, seeds were imbibed at 25°C for 40 h under continuous FR illumination in a custom-made FR chamber (Lagarias et al., 1997) where peak transmittance, half-band pass, and fluence rate were 760 nm, 85 nm and 22 μmol m−2 s−1, respectively, at the level of the seed as measured by a LI-COR LI-8,000 portable spectroradiometer (LI-COR, Lincoln, NE).
RNA Isolation, PCR Amplification, and cDNA Library Screening
Samples of 500 MM seeds imbibed for 24 h were pulverized in liquid nitrogen and the frozen material transferred to 2 mL of extraction buffer (10 mm Tris-HCl, pH 8.2, 100 mm LiCl, 1 mm EDTA, 1% [w/v] SDS, 25 mm dithiothreitol) in a ground glass homogenizer on ice. Extraction followed a modification of the phenol/SDS method of Ausubel et al. (1987). Purified total RNA (1 μg) was used as template for RT-PCR. Degenerate 5′ [G(GC)(N) CA(TC) GC(N) AC(N) TT(CT) TA(CT) GG(N) G] and 3′ [(TC) TGCCA(AG) TT(TC) TG(N) CCCCA(AG) TT] (n = A, T, G, or C) PCR primers were designed based on two conserved domains according to the alignment of deduced amino acid sequences of known expansins (Shcherban et al., 1995; Cho and Kende, 1997; Rose et al., 1997). After amplification for 36 cycles (94°C for 1 min, 50°C for 1.5 min, and 72°C for 1.5 min), the amplified fragments were cloned into pCR2.1 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). DNA sequences were determined with universal primers (T3 and M13-forward) using an Applied Biosystems model 377 sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA) with dye termination chemistry and AmpliTaq DNA polymerase FS (Perkin-Elmer Applied Biosystems). The PCR fragments were used to screen a cDNA library prepared from whole gib-1 seeds imbibed in 100 μm GA4+7 for 24 h. The cDNAs were labeled with enhanced chemiluminescence (ECL) nucleic acid labeling reagents (ECL kit, Amersham Life Science, Arlington Heights, IL) and were added to prehybridization solution at a final concentration of 10 ng/mL. Prehybridization was for 30 min at 42°C and hybridization was for 3 h at 42°C. Following hybridization, the membranes were washed twice at 42°C with 6 m urea, 0.5% (w/v) SDS (low stringency), or 0.2% (w/v) SDS (high stringency) and then washed twice for 5 min each with 2× SSC at room temperature. Hybridization was visualized using the chemiluminescent reagents in the ECL kit and exposure to x-ray film. Independent inserts in the library vector pBK-CMV were sequenced. Six distinct full-length cDNAs were identified, four of which were previously reported in the GenBank database (LeEXP1 [U821223], LeEXP2 [AF096776], LeEXP4 [AF059488], LeEXP9 [AJ243340]) and two of which were novel (LeEXP8 [AF184232] and LeEXP10 [AF184233]).
DNA Gel-Blot Analyses
To generate gene-specific probes, DNA fragments were amplified by PCR from primarily the 3′-untranslated regions of the genes. The PCR products were composed of (nucleotide positions are relative to the translation start ATG): for LeEXP1, nucleotides (nt) 739 to 1,001; for LeEXP2, nt 729 to 1,047; for LeEXP4, nt 690 to 1,011; for LeEXP8, nt 784 to 1,022; for LeEXP9, nt 741 to 1,002; and for LeEXP10, nt 750 to 1,061. Labeling of the PCR products was as mentioned above for cDNA library screening. For cDNA gel-blot analysis, 5 ng of PCR product was obtained from each of the six target genes using T3/T7 primers to amplify the full-length sequence from a library vector. The cDNAs were subjected to electrophoresis and transferred to a nylon membrane (Hybond N+, Amersham). Prehybridization, hybridization, washing, and chemiluminescent visualization of the membrane were performed as described for cDNA library screening using ECL, except that hybridization was for 16 h. Six identical blots were each hybridized with one of the PCR products described above.
For Southern blotting, genomic DNA was isolated from young MM tomato leaves as described by Murray and Thompson (1980) and modified by Bernatzky and Tanksley (1986). Aliquots (10 μg) were digested with restriction enzymes, fractionated on a 0.8% (w/v) agarose gel, and transferred to Hybond N+ membranes. Probe amplification and labeling, prehybridization, hybridization, washing, and chemiluminescent detection of the blots were performed as described for cDNA gel-blot analysis.
RNA Gel-Blot Analyses
For RNA extraction from seed parts, imbibed seeds were first dissected into the endosperm cap, the radicle tip (removed from within the endosperm cap), and the rest of the seed (lateral endosperm and remainder of the embryo; for a diagram of seed anatomy and dissection, see Cooley et al., 1999). Three pools containing 1,000 seed parts were used for RNA isolation. Total RNA from each sample (5 μg) was subjected to electrophoresis on 1% (w/v) agarose/10% (v/v) formaldehyde denaturing gels, transferred to Hybond N+ membrane and UV cross-linked. Gene-specific probes were generated as described previously except that digoxigenin (DIG)-labeled nucleotides were incorporated during the PCR amplification. The labeling efficiency was estimated according to the manufacturer's instructions (Boehringer Mannheim, Indianapolis). Gene-specific DNA probes were added to the hybridization buffer at a final concentration of 25 ng/mL. Because a DNA probe was used for RNA detection, high SDS buffer (7% [w/v] SDS) was used for hybridization at 42°C. Washing (60°C) and detection followed the recommended method (Boehringer Mannheim). The membranes were blocked for 1 h with 5% (w/v) nonfat milk in 0.1 m maleic acid buffer, pH 7.5, containing 0.15 m NaCl, 0.3% (v/v) Tween 20 (buffer A), and incubated with alkaline phosphatase conjugated anti-DIG antibody (1:15,000 dilution) for 1 h at 25°C. After washing with buffer A, the membranes were incubated with the chemiluminescence substrate CPSD (disodium 3-(4-methoxy-spiro{1,2-dioxetane-3,2′-(5′-chloro) tricyclo [3.3.1.13,7]decan}- 4-yl) phenyl phosphate) (Boehringer Mannheim) and exposed to x-ray film. Exposure times were from 10 min to 2 h depending on the strength of the signal.
Tissue Printing
Tissue prints were prepared as described by Nonogaki et al. (2000). After 24 h of imbibition, seeds were bisected using a razor blade and the cut surfaces were pressed onto a positively charged Hybond N+ membrane (Amersham-Pharmacia Biotech, Piscataway, NJ) for approximately 10 to 15 s. Duplicate membranes were cross-linked using UV light and hybridized with either sense or antisense RNA probes under the same conditions used for northern blots. Hybridization of DIG-labeled probes was as described above, except that the signal was colorimetrically detected with 0.18 m Tris-HCl buffer, pH 8.8, containing 0.025 mg/mL 5-bromo-4-chloro-3-indolyl-phosphate, 0.1 mg/mL nitroblue tetrazolium, and 2 mm MgCl2.
Puncture Force Measurements
The force required to puncture the micropylar endosperm and testa surrounding the radicle tip was analyzed for both MM and gib-1 seeds following various treatments. The micropylar region was sliced from the seed and the radicle tip teased out of the embryo cavity. A food texture analyzer (Stable Micro Systems, Texture Technologies, Scarsdale, NY) fitted with a custom-made probe (0.5-mm diameter) was used to determine puncture force. The endosperm cap with the radicle tip removed was placed on the texture analyzer probe and the test conducted at an inching speed of 10 mm/min (Downie et al., 1999). In each test, the background resistance generated by the probe against the side of the cap was subtracted from the peak resistance to puncture using XT.RA Dimension version 3.7F software supplied by the manufacturer (Stable Micro Systems). Twenty-four individual seeds were measured at each time point and means were expressed as the puncture force in newtons (N).
ACKNOWLEDGMENTS
We express our appreciation to Dr. Joss Rose and Dr. Alan Bennett for providing the degenerate primers used in the initial RT-PCR experiment, to Dr. Clark Lagarias for use of the equipment for far-red light treatment, and to Dr. Daniel Cosgrove for testing for the activity of LeEXP4 protein in the hypocotyl extension assay. We also thank Dr. Peetambar Dahal for providing a membrane for an initial northern blot and Dr. Hiroyuki Nonogaki for helpful discussions. DNA sequencing was performed by the University of California Davis Advanced Plant Genetics Facility.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–9722978 to K.J.B.).
LITERATURE CITED
- Ausubel FM, Brent R, Kinston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 1987. [Google Scholar]
- Bernatzky R, Tanksley SD. Genetics of actin-related sequences in tomato. Theor Appl Genet. 1986;72:314–321. doi: 10.1007/BF00288567. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997a;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci. 1997b;2:464–469. [Google Scholar]
- Bradford KJ. Water relations in seed germination. In: Kigel J, Galili G, editors. Seed Development and Germination. New York: Marcel Dekker; 1995. pp. 351–396. [Google Scholar]
- Bradford KJ, Chen F, Cooley MB, Dahal P, Downie B, Fukunaga KK, Gee OH, Gurusinghe S, Mella RA, Nonogaki H, Wu C-T, Yim K-O. Gene expression prior to radicle emergence in imbibed tomato seeds. In: Black M, Bradford KJ, Vazquez-Ramos J, editors. Advances and Applications in Seed Biology. Wallingford, UK: CAB International; 2000. pp. 231–251. [Google Scholar]
- Brummell DA, Harpster MH, Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol. 1999;39:161–169. doi: 10.1023/a:1006130018931. [DOI] [PubMed] [Google Scholar]
- Cho HT, Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997;9:1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley MB, Yang H, Dahal P, Mella RA, Downie B, Haigh AM, Bradford KJ. Vacuolar H+-ATPase is expressed in response to gibberellin during tomato seed germination. Plant Physiol. 1999;121:1339–1347. doi: 10.1104/pp.121.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Creeping walls, softening fruit, and penetrating pollen tubes: the growing roles of expansins. Proc Natl Acad Sci USA. 1997;94:5504–5505. doi: 10.1073/pnas.94.11.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (1999) Expansin home page. http://www. bio.psu.edu/expansins (December 1999)
- Cosgrove DJ, Scherban TY, Durachko DM. Highly specific and distinct expression patterns for two α-expansin genes in Arabidopsis. Plant Physiol. 1998;Suppl A1 75:57. [Google Scholar]
- Dahal P, Bradford KJ. Effects of priming and endosperm integrity on seed germination rates of tomato seeds: II. Germination at reduced water potential. J Exp Bot. 1990;41:1441–1453. [Google Scholar]
- Dahal P, Nevins DJ, Bradford KJ. Relationship of endo-β-mannanase activity and cell wall hydrolysis in tomato endosperm to germination rates. Plant Physiol. 1997;113:1243–1252. doi: 10.1104/pp.113.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campillo E, Lewis LN. Identification and kinetics of accumulation of proteins induced by ethylene in bean abscission zones. Plant Physiol. 1992;98:955–961. doi: 10.1104/pp.98.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie B, Dirk LMA, Hadfield KA, Wilkins TA, Bennett AB, Bradford KJ. A gel diffusion assay for quantification of pectin methylesterase activity. Anal Biochem. 1998;264:149–157. doi: 10.1006/abio.1998.2847. [DOI] [PubMed] [Google Scholar]
- Downie B, Gurusinghe S, Bradford KJ. Internal anatomy of individual tomato seeds: relationship to abscisic acid and germination physiology. Seed Sci Res. 1999;9:117–128. [Google Scholar]
- Fischer RL, Bennett AB. Role of cell wall hydrolases in fruit ripening. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:675–703. [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansin. Science. 1997;276:1415–1418. [Google Scholar]
- Groot SPC, Karssen CM. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta. 1987;171:525–531. doi: 10.1007/BF00392302. [DOI] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. Dormancy and germination of abscisic acid-deficient tomato seeds: studies with the sitiens mutant. Plant Physiol. 1992;99:952–958. doi: 10.1104/pp.99.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Kieliszewska-Rokicka B, Vermeer E, Karssen CM. Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seed prior to radicle protrusion. Planta. 1988;174:500–504. doi: 10.1007/BF00634479. [DOI] [PubMed] [Google Scholar]
- Gross KC, Wallner SJ. Degradation of cell wall polysaccharides during tomato fruit ripening. Plant Physiol. 1979;63:117–120. doi: 10.1104/pp.63.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarias DM, Crepeau MW, Maines MD, Lagarias JC. Regulation of photomorphogenesis by expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Cell. 1997;9:675–688. doi: 10.1105/tpc.9.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Gonzalez-Bosch C, Bennett AB. Two divergent endo-β-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell. 1994;6:1484–1493. doi: 10.1105/tpc.6.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviatov S, Shoseyov O, Wolf S. Involvement of endomannanse in the control of tomato seed germination under low temperature conditions. Ann Bot. 1995;76:1–6. [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Rochange F. Expansins in plant growth and development: an update on an emerging topic. Plant Biol. 1999;1:19–25. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B-R, Bradford KJ. Quantitative models characterizing seed germination responses to abscisic acid and osmoticum. Plant Physiol. 1992;98:1057–1068. doi: 10.1104/pp.98.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B-R, Bradford KJ. Germination and dormancy of abscisic acid- and gibberellin-deficient mutant tomato (Lycopersicon esculentum) seeds: sensitivity of germination to abscisic acid, gibberellin, and water potential. Plant Physiol. 1993;101:607–617. doi: 10.1104/pp.101.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M, Nonogaki H, Morohashi Y. Development of galactomannan-hydrolyzing activity in the micropylar endosperm tip of tomato seed prior to germination. Physiol Plant. 1995;94:105–109. [Google Scholar]
- Nonogaki H, Gee OH, Bradford KJ. A germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol. 2000;123:1235–1245. doi: 10.1104/pp.123.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Morohashi Y. An endo-β-mannanase develops exclusively in the micropylar endosperm of tomato seeds prior to radicle emergence. Plant Physiol. 1996;110:555–559. doi: 10.1104/pp.110.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Nomaguchi M, Morohashi Y. Endo-β-mannanases in the endosperm of germinated tomato seeds. Physiol Plant. 1995;94:328–334. [Google Scholar]
- Nonogaki H, Nomaguchi M, Okumoto N, Kaneko Y, Matsushima H, Morohashi Y. Temporal and spatial pattern of the biochemical activation of the endosperm during and following imbibition of tomato seeds. Physiol Plant. 1998;102:236–242. [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized up-regulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998;10:1427–1438. doi: 10.1105/tpc.10.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S, McCubbin A, Ambrose G, Kao T, Gilroy S. The sensitivity of barley aleurone tissue to gibberellin is heterogeneous and may be spatially determined. Plant Physiol. 1999;120:361–370. doi: 10.1104/pp.120.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez RA, de Miguel L. Phytochrome promotion of mannan-degrading enzyme activities in the micropylar endosperm of Datura ferox seeds requires the presence of the embryo and gibberellin synthesis. Seed Sci Res. 1997;7:27–33. [Google Scholar]
- Sánchez RA, Sunell L, Labavitch JM, Bonner BA. Changes in the endosperm cell walls of two Datura species before radicle protrusion. Plant Physiol. 1990;93:89–97. doi: 10.1104/pp.93.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P, Plachy C. Control of seed germination by abscisic acid: III. Effect on embryo growth potential (minimum turgor pressure) and growth coefficient (cell wall extensibility) in Brassica napus L. Plant Physiol. 1985;77:676–686. doi: 10.1104/pp.77.3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins: a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheih M, Cosgrove DJ. Expression of α- and β-expansins in recombinant systems. Plant Physiol. 1998;Suppl A1 72:57. [Google Scholar]
- Sitrit Y, Hadfield K, Bennett AB, Bradford KJ, Downie AB. Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol. 1999;121:419–428. doi: 10.1104/pp.121.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still DW, Bradford KJ. Endo-β-mannanase activity from individual tomato endosperm caps and radicle tips in relation to germination rates. Plant Physiol. 1997;113:21–29. doi: 10.1104/pp.113.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorop PE, Bewley JD, Hilhorst WM. Endo-β-mannanase isoforms are present in the endosperm and embryo of tomato seeds but are not essentially linked to the completion of germination. Planta. 1996;200:153–158. [Google Scholar]
- Toorop PE, van Aelst AC, Hilhorst HWM. Endosperm cap weakening and endo-β-mannanase activity during priming of tomato (Lycopersicon esculentum cv. Moneymaker) seeds are initiated upon crossing a threshold water potential. Seed Sci Res. 1998;8:583–491. [Google Scholar]
- Toorop PE, van Aelst AC, Hilhorst HWM. The second step of the biphasic endosperm cap weakening that mediates tomato (Lycopersicon esculentum) seed germination is under control of ABA. J Exp Bot. 2000;51:1371–1379. [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B, Bewley JD. Developing tomato seeds when removed from the fruit produce multiple forms of germinative and post-germinative endo-β-mannanase: responses to desiccation, abscisic acid, and osmoticum. Planta. 1996;200:71–77. [Google Scholar]
- Watkins JT, Cantliffe DJ, Huber DJ, Nell TA. Gibberellic acid stimulated degradation of endosperm in pepper. J Am Soc Hortic Sci. 1985;110:61–65. [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun TP. Phytochrome regulation and differential expression of gibberellin 3-β-hybroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]