ABSTRACT
Middle East respiratory syndrome coronavirus (MERS-CoV) causes a highly lethal pulmonary infection with ∼35% mortality. The potential for a future pandemic originating from animal reservoirs or health care-associated events is a major public health concern. There are no vaccines or therapeutic agents currently available for MERS-CoV. Using a probe-based single B cell cloning strategy, we have identified and characterized multiple neutralizing monoclonal antibodies (MAbs) specifically binding to the receptor-binding domain (RBD) or S1 (non-RBD) regions from a convalescent MERS-CoV-infected patient and from immunized rhesus macaques. RBD-specific MAbs tended to have greater neutralizing potency than non-RBD S1-specific MAbs. Six RBD-specific and five S1-specific MAbs could be sorted into four RBD and three non-RBD distinct binding patterns, based on competition assays, mapping neutralization escape variants, and structural analysis. We determined cocrystal structures for two MAbs targeting the RBD from different angles and show they can bind the RBD only in the “out” position. We then showed that selected RBD-specific, non-RBD S1-specific, and S2-specific MAbs given prophylactically prevented MERS-CoV replication in lungs and protected mice from lethal challenge. Importantly, combining RBD- and non-RBD MAbs delayed the emergence of escape mutations in a cell-based virus escape assay. These studies identify MAbs targeting different antigenic sites on S that will be useful for defining mechanisms of MERS-CoV neutralization and for developing more effective interventions to prevent or treat MERS-CoV infections.
IMPORTANCE MERS-CoV causes a highly lethal respiratory infection for which no vaccines or antiviral therapeutic options are currently available. Based on continuing exposure from established reservoirs in dromedary camels and bats, transmission of MERS-CoV into humans and future outbreaks are expected. Using structurally defined probes for the MERS-CoV spike glycoprotein (S), the target for neutralizing antibodies, single B cells were sorted from a convalescent human and immunized nonhuman primates (NHPs). MAbs produced from paired immunoglobulin gene sequences were mapped to multiple epitopes within and outside the receptor-binding domain (RBD) and protected against lethal MERS infection in a murine model following passive immunization. Importantly, combining MAbs targeting distinct epitopes prevented viral neutralization escape from RBD-directed MAbs. These data suggest that antibody responses to multiple domains on CoV spike protein may improve immunity and will guide future vaccine and therapeutic development efforts.
KEYWORDS: MERS-CoV, protection, RBD, S1, escape mutation, monoclonal antibody
INTRODUCTION
Middle East respiratory syndrome coronavirus (MERS-CoV) is one of four β-coronaviruses in the family Coronaviridae found to infect humans. It has emerged as a highly fatal cause of severe acute respiratory infection since April 2012. As of 10 November 2017, there have been 2,103 confirmed cases of infection, including 733 related deaths from 27 countries (http://www.who.int/emergencies/mers-cov/en/). Although human-to-human transmission of the virus has been identified on several occasions, most outbreaks are hospital related, as exemplified by a large outbreak that occurred in South Korea in 2015 and in Saudi Arabia since 2013 (1, 2). Dromedary camels serve as a principal reservoir for transmission, and future outbreaks of infection in humans are expected (3–6). The high case fatality rate, vaguely defined epidemiology, and absence of prophylactic or therapeutic measures against this novel virus have created an urgent need for effective vaccines and therapeutic options, should outbreaks expand to pandemic proportions.
Administration of neutralizing monoclonal antibodies (MAbs) has become an increasingly attractive option for prophylaxis or therapy of viral infections (7). The homotrimeric spike glycoprotein (S) is the primary target for neutralizing antibodies on coronaviruses. S is a typical class I fusion glycoprotein that undergoes proteolytic cleavage resulting in two subunits: S1, which is distal to the virus membrane, and S2, which includes the amino-terminal hydrophobic fusion peptide, transmembrane domain, and two heptad repeat sequences that mediate membrane fusion required for viral entry (8–11). The S1 subunit contains the receptor-binding domain (RBD) that mediates virus attachment to its host receptor, dipeptidyl peptidase 4 (DPP4) (10, 12). Several research groups have isolated anti-MERS-CoV MAbs from immunized mice, convalescent MERS patients, or naive human antibody phage display libraries (13–19). While most reported MERS-CoV-neutralizing MAbs are RBD specific, two non-RBD S1-specific MAbs have recently been reported (20, 21). Structural studies revealed that three RBD-specific MAbs, D12 and 4C2 (from immunized mice) and MERS-27 (from a naive human phage display library), have highly similar binding patterns in complex with the RBD, mainly interacting with residues centered around W535 in the RBD, while m336 (isolated from a human immunoglobulin library) and MCA1 (from a MERS human survivor) interacted more broadly with RBD contact residues largely overlapping the DPP4-binding site (15, 20, 22–24). Although RBD-specific MAbs can neutralize virus with high potency and have been reported to protect animals against infection in pre- and postexposure models (13, 14, 25, 26), there is a potential risk for virus to escape under selection pressure if only a single site is targeted (27, 28). Therefore, using two or more MAbs targeting different RBD epitopes or conserved non-RBD domains may have advantages for therapeutic applications (27, 29–32). Using hybridoma technology, we previously generated a panel of MAbs from full-length S-immunized mice that target the RBD, S1 (non-RBD portion), and S2. In this study, we used MERS-CoV S-specific probes to sort single B cells and clone the heavy- and light-chain immunoglobulin genes to identify neutralizing MAbs specifically binding to the RBD or S1 (non-RBD) from a MERS-CoV-infected individual and immunized nonhuman primates (NHPs). Six RBD-specific and five S1-specific MAbs were further characterized according to binding specificity, neutralization breadth, shared and unique neutralizing epitopes, and atomic-level structure. We demonstrate that a combination of RBD- and S1-specific MAbs or two RBD-specific MAbs targeting distinct epitopes could prevent emergence of escape mutations in the RBD. In addition, MAbs specific to the RBD, S1, or S2 protected DPP4-transgenic mice against MERS-CoV infection.
RESULTS
Characterization of serum from immunized rhesus macaques and a MERS CoV-infected convalescent human volunteer.
First, we analyzed serum from immunized rhesus macaques and a convalescent MERS patient for antibody binding specificity (see Fig. S1A in the supplemental material) by enzyme-linked immunosorbent assay (ELISA) and a pseudotyped lentivirus reporter neutralization assay (Fig. S1B). Similar to serum from macaques immunized three times with full-length S DNA (3×DNA) or two times with full-length S DNA and once with S1 protein (2×DNA/protein) (20), human serum could bind to the RBD, S1, and S2 of MERS-CoV S. In contrast, two immunizations with S1 protein (2×protein) (20) induced antibodies binding only the RBD and S1, but not S2, as expected. Anti-RBD and -S1 antibody titers in the human donor obtained 3 weeks after infection were lower than those in macaques immunized with 2×DNA/protein or 2×protein (Fig. S1A). Similar results were observed using the pseudotyped reporter virus neutralization assay, perhaps attributable to the time point for sample collection and difference between natural infection and immunization (Fig. S1B). However, there was a higher level of anti-S2 antibody detected in human serum (Fig. S1A), suggesting that natural infection of MERS-CoV or vaccination with full-length S induces antibodies targeting multiple structural domains of S. Since the 2×DNA/protein regimen induced the highest neutralizing activity among the three groups, we isolated MAbs from immunized macaques in addition to those from the human donor.
Isolation and characterization of MAbs.
In this study, we isolated neutralizing MAbs targeting the RBD and S1 from a MERS patient and vaccinated macaques using MERS-CoV specific probes combined with single B cell cloning strategy (33, 34). Peripheral blood mononuclear cells (PBMCs) from the immunized macaques (2 weeks after the last boost) and human donor (3 weeks after onset of illness) were stained with a viability marker (VIVID), antibodies to exclude T cells and monocytes (CD3/CD4/CD8/CD14), antibodies to label B cells (CD20/IgG/IgM for NHP and CD20/IgG for human), and probes for the RBD and S1. IgG+ IgM− probe+ (NHP) or IgG+ probe+ (human) single B cells were sorted and subjected to PCR amplification and cloning of VH/VL genes (Fig. S2A and S2B). A panel of MAbs was generated (Fig. S2C) and selected for further characterization on the basis of binding specificity and neutralization potency. Three MAbs from the macaque JC57 (JC57-11, JC57-13, and JC57-14) were from the same VH germ line (IGHV4-2), as observed in anti-simian immunodeficiency virus (anti-SIV) antibodies (34), but were paired with different VL genes (IGKV3-9, IGKV1-20, and IGKV2S17). A MAb isolated from another macaque (FIB), FIB-H1, used a different VH and VL than JC57-derived MAbs. The somatic mutation rate (nucleotide) ranged from 3% to 17% for VH and 1% to 9% for VL based on currently available data in the IMGT database for rhesus macaques. Two of four MAbs from the human donor (CDC2-C2 and CDC2-A10) utilized IGHV1-69, which is a commonly used allele for neutralizing antibodies against many other viruses, including MERS-CoV (17, 18, 35–37). All four human MAbs used different VL genes. Remarkably, V segments of human MAbs showed very low levels of somatic mutation, ranging from 0% to 5% for VH and 1% for VL, which may be related to the early postinfection collection time point. Both JC57-11 and CDC2-C2 have relatively long VH complementarity-determining region 3 (CDRH3) sequences, 23 and 20 amino acids, respectively (Fig. S2C).
Binding specificity, neutralization potency, and breadth.
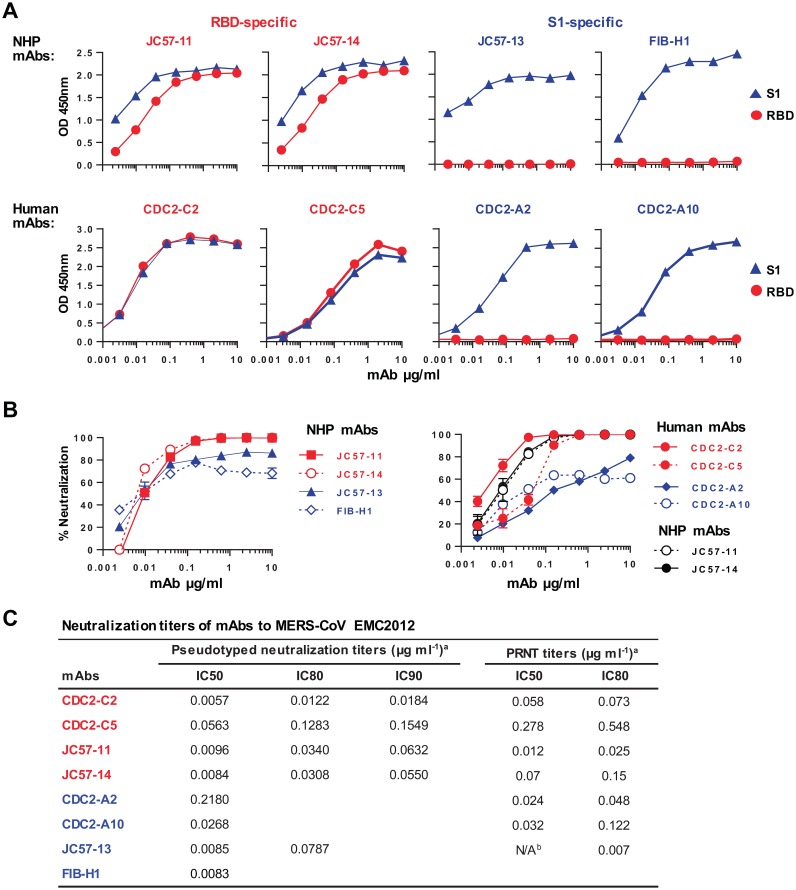
MAbs were first tested for binding to the MERS-CoV S RBD or S1 proteins by ELISA. JC57-11 and JC57-14 from macaques and CDC2-C2 and CDC2-C5 from humans were found to be RBD-specific MAbs, binding to both the RBD and S1. JC57-13 and FIB-H1 from macaques and human CDC2-A2 and CDC2-A10 bound to S1, but not the RBD, suggesting specificity for non-RBD S1 domains (Fig. 1A). The MAbs display nanomolar affinity for S1 similar to those of previously described MERS-specific antibodies (Fig. S3). MAbs were also tested in an S-pseudotyped lentiviral reporter neutralization assay (Fig. 1B and C). Reciprocal IC50 neutralization titers (where IC50 is the concentration resulting in 50% reduction of infectivity) for JC57-11 and JC57-14 against MERS-CoV EMC pseudoparticles were similar (0.0096 and 0.0084 μg/ml, respectively), and these MAbs had higher potency than the human MAb CDC2-C5 (IC50 = 0.0563 μg/ml). CDC2-C2 was the most potent RBD-specific MAb against MERS-CoV EMC, with IC50 and IC80 values of 0.0057 and 0.0122 μg/ml, respectively, in the pseudovirus neutralization assay and 0.058 and 0.073 μg/ml, respectively, in plaque reduction neutralization tests (PRNT) using native MERS-CoV EMC (Fig. 1C). The neutralizing potential of CDC2-C2 is comparable to that of the two most potent MAbs (m336 and REGN3051) described to date (13, 18). All S1 non-RBD-specific MAbs failed to neutralize 100% of the input virus, reaching no more than 90% neutralization. Therefore, these MAbs had high reciprocal IC50 titers and had negative IC90 neutralizing activity. This phenomenon has also been reported for influenza virus and human immunodeficiency virus type 1 (HIV-1) antibodies. Although not as potent as the RBD-specific MAbs, S1-specific MAbs, like G2, previously isolated by our group (20) were different from other reported MAbs against MERS-CoV (13–19) in their ability to neutralize virus despite targeting epitopes outside the RBD (Fig. 1B and C).
FIG 1.
MAb binding specificity and neutralization potency. (A) Binding specificity. MAbs isolated from immunized NHPs and a MERS survivor were assayed by ELISA for binding to soluble receptor-binding domain (RBD) or S1 protein. RBD-specific MAbs (in red) bound to both RBD and S1 proteins, while S1-specific MAbs (in blue) bound only to S1. (B) Neutralization potency. Neutralization activity was measured using a MERS-CoV EMC S-pseudotyped lentivirus neutralization assay. NHP MAbs are shown in the left graph and human MAbs in the right graph (two NHP RBD-specific MAbs in black were included for comparison). Percent neutralization at the different MAb concentrations is shown. Data points represent the means of triplicate replicates with standard errors. (C) IC50, IC80, and IC90 neutralization titers. aIC values represent average results from two technical replicates of neutralization testing using a pseudovirion entry assay or plaque reduction neutralization testing (PRNT). bThe PRNT IC50 for MAb JC57-13 was not available (N/A), as >50% neutralization was obtained at the lowest concentration of MAb tested, 0.0032 μg ml−1.
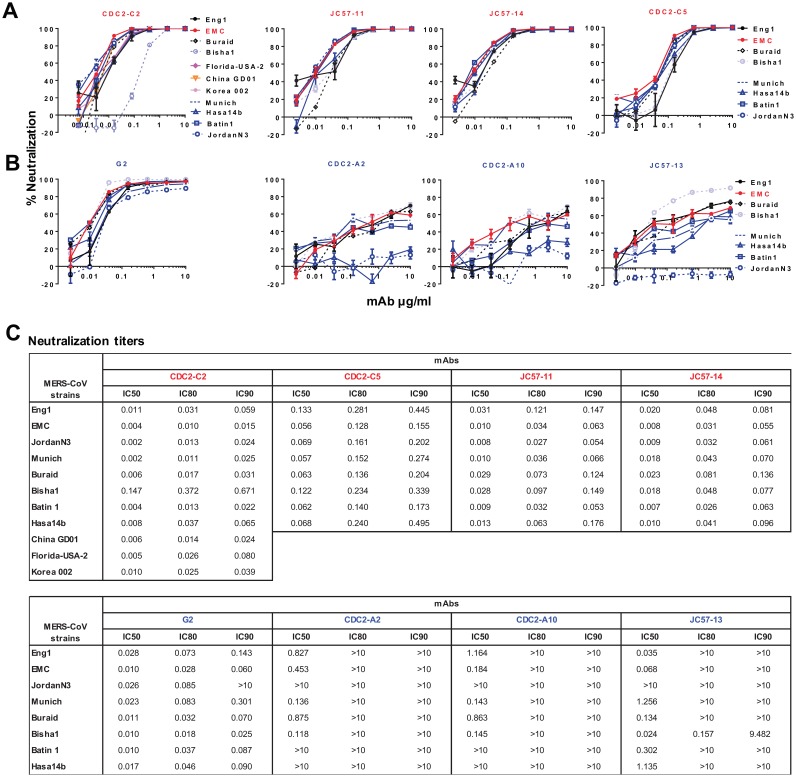
MAbs were tested for cross-neutralization against a panel of 8 to 11 MERS-CoV S-pseudotyped lentiviruses. The most potent RBD-specific MAb, CDC2-C2, neutralized pseudoparticles representing 10 MERS-CoV strains with high potency and partially neutralized the Bisha1 strain (GenBank accession number KF600620.1) (Fig. 2A and C), which differs from other strains by an aspartic acid-to-glycine substitution at residue 509 (D509G). The other three RBD-specific MAbs (JC57-11, JC57-14, and CDC2-C5) neutralized pseudoparticles bearing S from each of eight tested MERS-CoV strains, including Bisha1, but with lower potency than CDC2-C2 (Fig. 2A and C). These MAbs can also neutralize S-pseudotyped particles corresponding to most recent strains from Korea and China (Fig. 2A and C). Notably, all three S1-specific MAbs isolated from macaques and the human donor were unable to neutralize the Jordan N3 strain (GenBank accession number KC776174.1, which contains three differences in the N-terminal domain (NTD) of S: glycine versus valine, histidine versus tyrosine, and arginine versus leucine at positions 94, 194, and 301, respectively. In addition, CDC2-A2 and CDC2-A10 showed no neutralization activity against Hasa14b (GenBank accession number KF600643.1), which differs from EMC by a tyrosine-to-histidine substitution at position 58. These results suggest that the S1-specific MAbs interact with the NTD. Unexpectedly, these MAbs showed lower neutralization potency than G2 (Fig. 2B), a murine MAb previously reported by our group (20), which neutralized S pseudoparticles representing all eight strains, including those containing polymorphisms in the NTD (blue lines). Taken together, these findings indicate that human MAb CDC2-C2 is most potent RBD-specific Mab, with a neutralization IC50 of 0.002 to 0.011 μg/ml against pseudovirion surrogates of 10 MERS-CoV strains, and that murine MAb G2 is the broadest and most potent S1-specific MAb, with an IC50 neutralizing activity of 0.010 to 0.028 μg/ml to S pseudoparticles representative of eight MERS-CoV strains (Fig. 2C).
FIG 2.
MAb neutralization breadth. RBD-specific MAbs (in red) (A) and S1-specific MAbs (in blue) (B) were measured for neutralization activity against 8 to 11 strains of MERS-CoV S-pseudotyped viruses as indicated, including strains identified from 2012 to 2015. CDC2-C2, CDC2-C5, CDC2-A2, and CDC2-A10 were isolated from a human donor, JC57-11, JC57-14, JC57-13, and FIB-H1 from immunized rhesus macaques, and G2 from an immunized mouse (20). Percent neutralization at the different MAb concentrations is shown. Data points represent the means of triplicate replicates with standard errors. (C) IC50, IC80, and IC90 neutralization titers for panels A and B.
Structural analysis of RBD-specific antibodies.
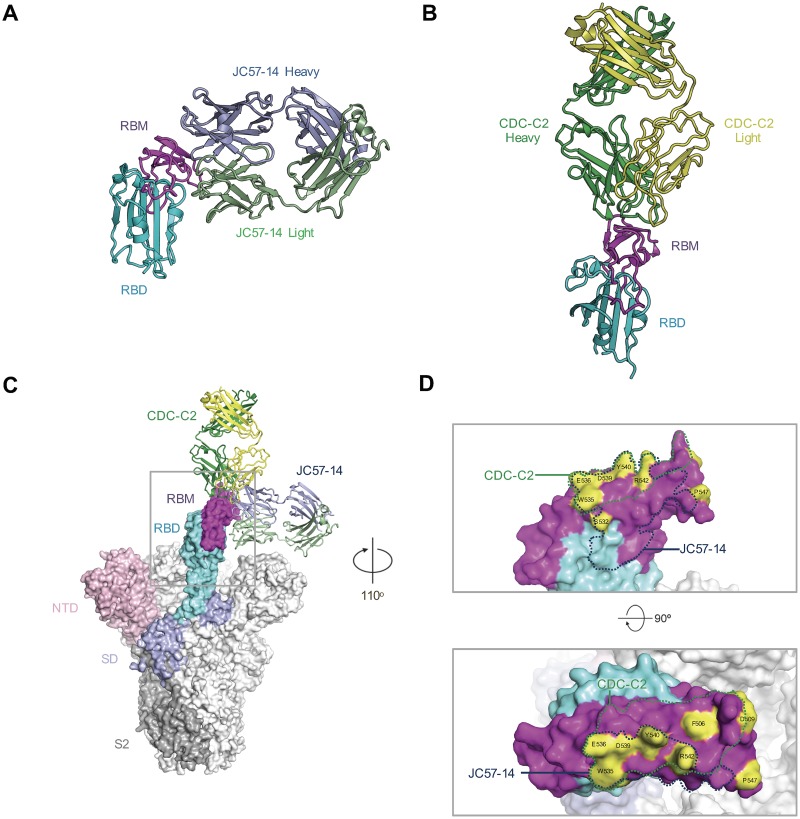
To structurally characterize the RBD-specific antibodies we determined the crystal structure of JC57-14 and CDC2-C2 in complex with the RBD (Fig. 3; see also Fig. S4 and Tables S1 to S5). Consistent with the blocking and neutralization data, JC57-14 binds the RBD similarly to the previously described D12 antibody (Fig. 3A; see also Fig. S4A) (20). JC57-14 binds the RBD utilizing both heavy- and light-chain interactions with W535 at the center of the epitope. Residues that have undergone somatic mutation within the heavy chain do not contact the RBD, while those in the light chain make significant contacts with the RBD, including residues Asn 30 and Tyr 50 (Table S3), forming hydrogen and salt bridge bonds.
FIG 3.
Structural characterization of macaque and human MERS virus-neutralizing antibodies. (A) Crystal structure of JC57-14 antibody bound to MERS England1 RBD. (B) Crystal structure of CDC-C2 bound to MERS England1 RBD. (C) JC57-14 and CDC-C2 are modeled using the MERS spike trimer structure (PDB code 5W9H) bound to a single RBD in the open conformation. (D) Antibody epitopes of JC57-14 and CDC-C2 are displayed on the surface of the RBD structure. Residues highlighted and labeled differing in natural viral variants were tested to characterize antibody epitopes.
CDC-C2 binds to the MERS RBD at an angle 90° rotated from JC57-14 (Fig. 3B; see also Fig. S4B) binding to the RBD predominantly using the antibody heavy chain. The angle of approach is similar to that of the human receptor DPP4, and there is significant overlap with DPP4 contacting residues (Fig. S4D). The CDC-C2 antibody is reminiscent of the previously described m336 using a related VH1–69 gene-encoded heavy chain and containing a noncanonical disulfide bond in the CDR H3. In fact, most residues involved in recognition of the RBD are germ line-encoded, including significant interactions by Phe 54 and Lys 73, with the exception of Ile 31 (Table S4). In contrast to m336, the CDC-C2 CDR H3 is two residues longer than the m336 CDR H3 and does not contain the m336 junction-encoded NRG motif, although it can still make significant contacts with the RBD through Tyr100a and an additional eight residues of the CDR H3.
Structural comparison of the seven structurally characterized RBD-specific antibodies described indicates that to date, there are two major modes of recognition that have been identified. One focuses around W535 as well as contacting residues 390 to 400 (Fig. S4D) seen with JC57-14, D12, 4C2, and MERS27 antibodies and utilizing both heavy and light chains for recognition (Table S3). The second mode of binding targets the DPP4 receptor-binding site contacting residues 500 to 515 as well as W535 and adjacent residues, predominantly using a VH1–69 gene-encoded heavy chain to bind the MERS RBD, as exemplified by antibodies CDC-C2, m336, and MCA1, and to bind to residues largely contacted by the DPP4 receptor (Table S4).
In the context of the recently described MERS spike trimer, all structurally characterized antibodies can bind to the outward-rotated RBD molecule (Fig. 3C). However, the JC57-14 mode of recognition is fully ablated in the closed RBD structure, and in addition, both m336 and MCA1 antibodies clash with a neighboring protomer, indicating that these antibodies would not be able to bind the fully closed trimer molecule (Fig. S4C). Based on the antibody epitope contact residues (Fig. 3D; see also S4D) and mutagenesis data, CDC2-C5 matches well with the JC57-14 mode of binding, while CDC2-C2 is consistent with the m336 binding pattern. The neutralization and binding data indicate that both JC57-11 and F11 have epitopes dissimilar to the structurally characterized antibodies. Further structural analysis of these MAbs in complex with the RBD or the intact S trimer in combination with escape mutation data will be required for a comprehensive understanding of epitopes and binding orientations.
Mapping RBD-specific neutralizing MAbs.
We previously demonstrated that mouse RBD-specific MAbs F11 and D12 make contact with two distinct regions of the RBD at and around residue 509 for F11 and at residues 535 and 536 for D12. Therefore, F11 and D12 can bind to the RBD simultaneously, suggesting the potential for additive neutralization effects and prevention of escape mutations (20).
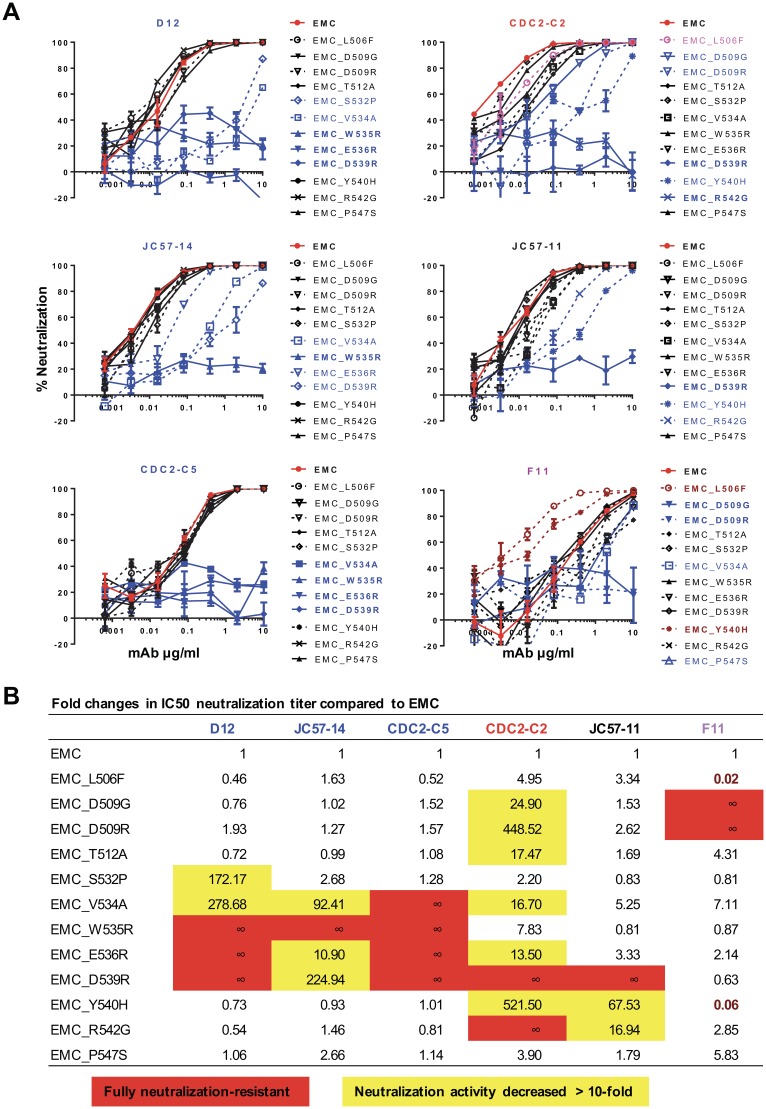
To define the epitopes for macaque and human RBD-specific MAbs, we generated 12 mutations (Fig. 4) within the RBD of MERS-CoV EMC S. These were designed based on published MAb structures and escape mutants (15, 20, 22, 23). Six MAbs were tested for neutralization activity against pseudotyped lentiviral particles containing EMC S and its mutant derivatives. D12, JC57-14, and CDC2-C5 showed similar specificities of neutralization (Fig. 4; see also Fig. S5), dependent on residues 534, 535, 536, and 539. E536R and D539R mutations completely eliminated neutralization by D12 and CDC2-C5 and partially diminished the neutralizing activity of JC57-14, which matches the binding footprint of these MAbs on the RBD (Fig. 3). Mutation of residues between 539 and 542 resulted in reduced sensitivity to neutralization by MAbs JC57-11 and CDC2-C2, while mutations at positions 506 and 509 resulted in loss of neutralizing sensitivity to MAbs F11 and CDC2-C2. Therefore, in agreement with the crystal structure, MAb CDC2-C2 (Fig. S4D) interacts more broadly across epitopes within the RBD, which may explain its higher potency, similar to that of m336 isolated from a nonimmune human immunoglobulin library (23). Notably, the D539R mutation completely or almost completely ablated neutralizing activity by all MAbs except for F11, which was unique in its neutralization specificity, with D509G diminishing and L506F and Y540H enhancing neutralizing sensitivity. In aggregate, we demonstrated that six RBD-specific MAbs have four different patterns of binding to mediate neutralization.
FIG 4.
Neutralization epitopes of RBD-specific MAbs. RBD-specific MAbs neutralize MERS-CoV by interacting with the different residues in the receptor-binding domain. (A) Neutralization curves. Six RBD-specific MAbs (CDC2-C2 and CDC2-C5 from a human, JC57-11 and JC57-14 from NHPs, and F11 and D12 from mice) were tested for neutralizing activity against pseudoviruses displaying MERS-CoV EMC S with engineered mutations in the RBD. EMC is shown in red, mutant constructs fully resistant to neutralization are shown in boldface blue type, mutant constructs partially resistant to neutralization are shown in lightface blue type, and mutant constructs with enhanced neutralization are shown in dark red. Data points represent the means of triplicate replicates with standard errors. The experiment was repeated once to ensure reproducible results; results from one of the two experiments are shown. (B) Fold changes in IC50 neutralization titers corresponding to mutant forms of EMC S. Mutant S IC50 neutralization titers (Fig. S5) are presented relative to native S (set equal to 1). Six MAbs display a total of four unique neutralization patterns, which are color-coded in blue, red, black, and purple.
Competitive antibody binding.
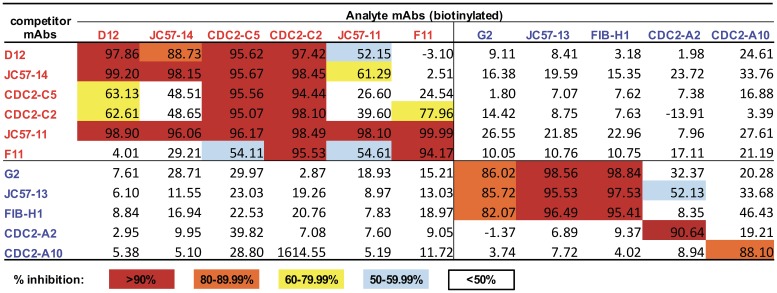
To determine whether MAbs recognize similar or different epitopes, binding competition assays by ELISA and biolayer interferometry (BLI) were performed. Percent inhibition of analyte MAb binding by competing untagged MAbs is shown in Fig. 5 and in Fig. S6 in the supplemental material and competition curves are shown in Fig. S7. F11 binding to S1 was completely blocked by JC57-11. D12 and JC57-14 showed a similar pattern, competing with each other and also with JC57-11. CDC2-C2 was blocked by all RBD-specific MAbs, and CDC2-C5 exhibited a similar pattern, except for reduced competition by F11. Conversely, CDC2-C2 but not CDC2-C5 could block F11 binding (77.96% inhibition by CDC2-C2). Notably, JC57-11 could block all RBD-specific MAbs, while CDC2-C2 could be competed by all RBD-specific MAbs. These results indicate that the six RBD-specific MAbs represent four distinct binding patterns represented by (i) D12, JC57-14, and CDC2-C5, (ii) CDC2-C2 (m336-like), (iii) JC57-11, and (iv) F11, which is generally consistent with neutralization escape mapping data (Fig. 4). Therefore, the combined data demonstrate that RBD-specific MAbs have 2 major modes of recognition but four different patterns of binding to mediate neutralization.
FIG 5.
Competition map of MAb binding by ELISA. RBD-specific MAbs (in red) and S1-specific MAbs (in blue) were biotinylated and used for competition binding to MERS-CoV S1 with MAbs listed in the leftmost column. Percent inhibition of analyte (biotinylated)-MAb binding by competitor (unlabeled) MAbs is indicated by color as shown in the color key. Competition binding curves are shown in Fig. S7A and S7B.
The five non-RBD S1-specific MAbs showed three binding patterns. G2, JC57-13, and FIB-H1 competed with each other but did not compete with CDC2-A2 or CDC2-A10. CDC2-A2 and CDC2-A10 did not compete with each other despite similar neutralization patterns (neither was able to neutralize JordanN3 S pseudovirions), suggesting that additional non-RBD S1 domains outside the NTD, such as SD1 and SD2, may be targets by S1-specific MAbs. Results of competition analysis by ELISA and BLI for S1-specific MAbs were in full agreement, while some discrepancies between the two assays were observed for RBD-specific MAbs (Fig. 5; see also Fig. S6).
Passive administration of MAbs protects mice against MERS-CoV infection.
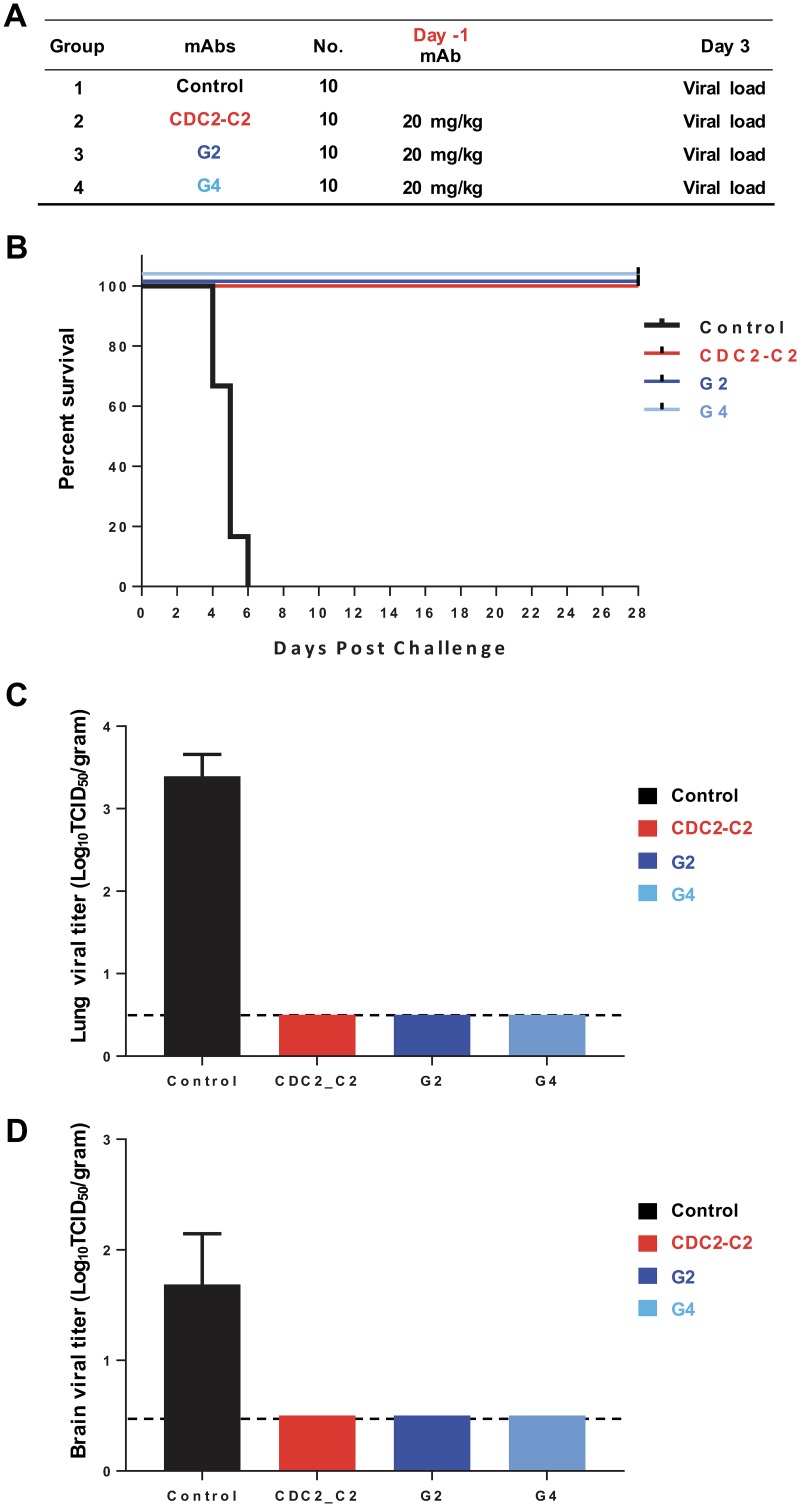
To test whether MAbs targeting different regions of S could protect against lethal MERS-CoV infection, DPP4-transgenic mice were intraperitoneally injected with 20 mg/kg (of body weight) of antibody 24 h prior to intranasal infection with 106 50% tissue culture infective doses (TCID50) of MERS-CoV (strain EMC/2012) (Fig. 6A). All control animals died within 6 days postchallenge, while the animals that received single RBD-specific MAb CDC2-C2, S1-specific MAb G2 (20), or S2-specific MAb G4 (20) survived during 28 days of observation (Fig. 6B). Infectious virus was undetectable in lungs of MAb-treated groups 3 days postchallenge (Fig. 6C). None of the treated animals and 1 out of 4 control animals had detectable virus in brain tissue at day 3 postchallenge (Fig. 6D). We did not collect brain samples at the postmortem time point when virus is typically present in untreated animals. These results show that MAbs targeting the RBD, non-RBD S1, or S2 of S can block MERS-CoV replication in the lower airways and lungs of mice and prevent severe disease following lethal MERS-CoV challenge.
FIG 6.
Passive transfer of RBD-, S1-, and S2-specific neutralizing MAbs protect against MERS-CoV infection. (A) Groups of 10 human DPP4-transgenic (hDPP4) mice were administered MAb CDC2-C2 (RBD specific), G2 (S1 specific), or G4 (S2 specific) (20) via the intraperitoneal route 1 day prior to challenge with MERS-CoV EMC. hDPP4 mice were challenged intranasally with 106 TCID50 of MERS-CoV (strain EMC/2012). At 3 days postinfection (dpi), four animals were sacrificed and lungs were collected for analyses. The remaining six animals were observed for 28 days for survival. (B) Survival curves of the different groups (6 hDPP4 mice per group). After challenge, hDPP4 mice were euthanized due to the severity of disease signs or at 28 dpi. (C and D) MERS-CoV viral titers in the lower respiratory tract (C) and in the brain (D) of hDPP4 mice at 3 dpi. Mean values ± SD were calculated. The dashed line indicates the cutoff limit of the TCID50 assay.
Combining MAbs to delay escape mutations.
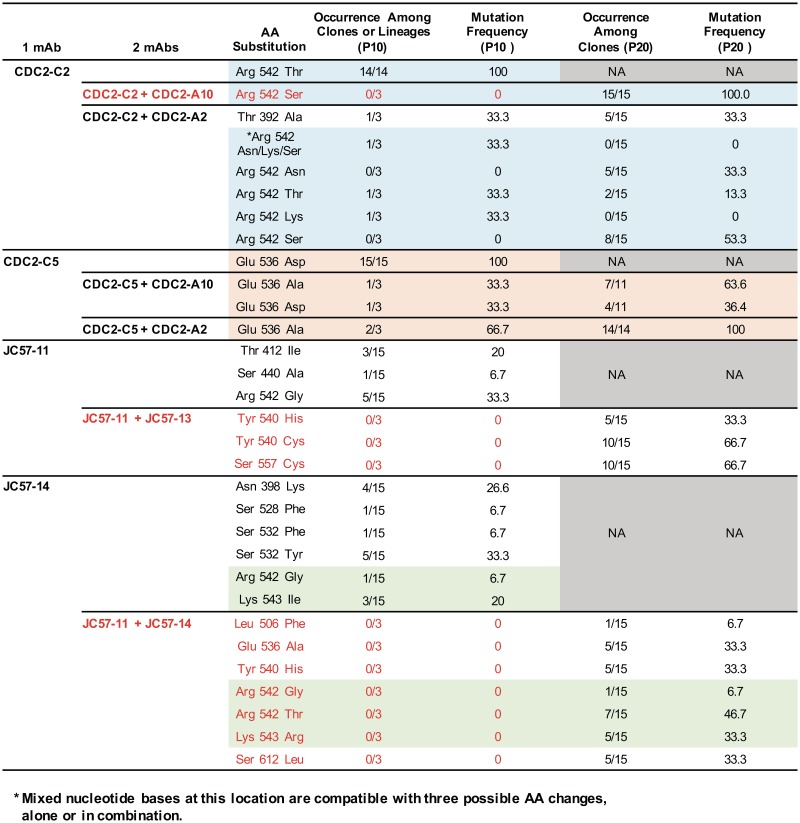
To investigate whether MERS-CoV could evolve strategies for escaping neutralization by MAbs individually or in combination, we performed in vitro selection for MERS-CoV escape mutants using single RBD-specific MAbs and pairs of RBD- plus S1-specific or two RBD-specific MAbs with distinct binding patterns. At passage 10 (P10), escape mutants selected by RBD-specific MAbs (CDC2-C2, CDC2-C5, JC57-11, and JC57-14) contained one or more amino acid substitutions within the RBD (Fig. 7). Consistent with our neutralization mapping data (Fig. 4), the CDC2-C2-, CDC2-C5-, and JC57-11-selected escape mutants contained arginine-to-threonine, glutamic acid-to-aspartic acid, and arginine-to-glycine substitutions at amino acids 542, 536, and 542, respectively. Escape mutations were not detected when CDC2-C2 (RBD specific) and CDC2-A10 (S1 specific), JC57-11 (RBD specific) and JC57-13 (S1 specific), or JC57-11 (RBD specific) and JC57-14 (RBD specific) were combined. In contrast, CDC2-C2 (RBD specific) combined with CDC2-A2 (S1 specific) and CDC2-C5 (RBD specific) combined with CDC2-A2 (S1 specific) or CDC2-A10 (S1 specific) yielded escape mutations, suggesting that the effects of a second MAb on emergence of escape mutant viruses is determined by the specificities of paired MAbs rather than neutralization potency of each MAb. Extending the analysis to the 20th passage (P20) in the presence of protective double-MAb combinations did eventually select for escape mutations (Fig. 7). In general, escape mutations occurred at expected sites in the RBD (e.g., Asn 398 to Lys in the case of JC57-14). The exception was the mutation of Ser 612 to Leu that occurred in the JC57-11/JC57-14 combination. This residue is not surface exposed but does occur at a pivot point at the base of the RBD, and we hypothesize that this may affect the orientation or ratio of outward-facing RBD molecules, thus indirectly reducing RBD-directed neutralization. Achieving viral resistance to two-MAb combinations was more difficult than selection of escape mutant viruses using a single RBD-specific MAb. Shallower concentration ramps of paired MAbs than of single MAbs were necessary to prevent viral extinction under similar time- and cytopathic-effect (CPE)-delimited passage conditions. At P10, MAb concentrations under dual-MAb selection were 1.8- to 6-fold lower than their respective concentrations under single-MAb conditions (Fig. S8). Using two MAbs, virus passage beyond P10 was required to attain MAb concentrations equivalent to those in P10 single-MAb cultures, and in some cases, the final (P20) MAb concentration was substantially lower in the two-MAb passage than in the single-MAb experiment (e.g., JC57-13 alone versus JC57-11 plus JC57-13 [Fig. S8]). Additional empirical modulation of key experimental parameters, such as incubation interval and inoculum volumes, might alter the kinetics of viral adaptation to a second MAb and manifest as a shift in the single- and two-MAb concentration curves. However, taken together, these results support the conclusion that two MAbs targeting distinct epitopes on S can prevent or substantially delay emergence of escape mutations depending on the MAb combination.
FIG 7.
S mutations associated with viral escape from neutralizing MAbs. EMC was serially passaged in the presence of the indicated human and NHP neutralizing anti-S MAbs, singly or combined, and S mutations associated with antibody escape in the 10th (P10) or 20th (P20) passage were determined by S gene sequence analysis of 11 to 15 plaque isolates (P10 single MAb and P20 double MAb) or three independent population virus cultures (P10 double MAb). Percentages of viral isolates or cultures containing the indicated mutation are shown. MAb combinations that prevented the emergence of RBD mutations are shown in red.
DISCUSSION
The continuous increase in MERS cases with high mortality (http://www.who.int/emergencies/mers-cov/en/) and potential for future outbreaks or a pandemic highlight the need for developing effective prophylactic and therapeutic countermeasures against MERS-CoV. Viral S mediates viral attachment and virus-cell fusion and is a key target for antiviral agents and neutralizing antibodies. Several anti-RBD MAbs have been reported to neutralize MERS-CoV in vitro and protect animals against infection in pre- and postexposure models of antibody administration (13, 14, 25, 26). Although RBD-specific MAbs can neutralize virus with high potency, there is a potential risk for virus to escape under selection pressure if only a single site is targeted (27, 28). It is interesting that despite low sequence divergence in isolated MERS viruses, mutations in the RBD have been seen in the Bisha1 strain (506L and 509G) and the more recent Korea002 strain (530L). These mutations map to the two regions on the RBD molecule associated with the two modes of antibody recognition defined by structural analysis. This is consistent with a recent report showing that a stabilized MERS spike protein (S-2P) induced high neutralization titers against all MERS strains, except for Bisha1 and Korea002 (38). Therefore, the ability to use two or more MAbs targeting different epitopes on the RBD or conserved non-RBD domains of S may have advantages for reducing the potential for escape mutations and enabling therapeutic applications (27, 29–32). Most MAbs published to date are RBD specific. The effect of combinations of two MAbs on prevention of escape mutations and treatment of MERS-CoV infection in vivo has not been previously reported.
In this report, we describe the generation of MAbs targeting the RBD and non-RBD S1 subunit of MERS-CoV S from a convalescent patient and vaccine-immunized NHPs using a probe-based single B cell sorting and cloning strategy. RBD-specific MAbs showed higher neutralization potency than non-RBD S1-specific MAbs. Six RBD-specific MAbs and five non-RBD S1-specific MAbs recognize, in seven distinct ways, the RBD and non-RBD S1 regions of MERS-CoV S based on results of competition-binding and neutralization assays. Our results demonstrate that combinations of RBD-specific MAb plus S1-specific MAb or two RBD-specific MAbs targeting different epitopes on S prevented the emergence of escape mutations in the RBD in vitro. In addition, RBD-, S1-, and S2-specific MAbs inhibited MERS-CoV pulmonary infection and prevented lethal disease when delivered 1 day before challenge. Since individual MAbs completely protected mice from infection, it was not possible to assess the value of combination MAbs in vivo. This may take a series of dose titration experiments. Overall, this report describes MAbs targeting both the RBD and non-RBD epitopes of MERS-CoV S that have potential clinical applications in the prevention and/or treatment of disease.
Sera from infected human and immunized NHPs bound the MERS-CoV RBD, S1, and S2, suggesting that natural MERS-CoV infection and vaccination with full-length S induce antibodies targeting multiple structural domains of S. Passive transfer of polyclonal IgG from convalescent infected humans, marmosets, or camels (39–41) effectively reduced signs of clinical disease as well as viral load in lung tissues. The combination of two or more MAbs having unique epitope targets on S, and potentially acting through different mechanisms to achieve neutralization, may function like polyclonal antibodies in suppression of viral infection and with fewer concerns about escape mutations. This premise has been explored in the treatment of other viral infections, such as ZMapp for Ebola virus and neutralizing antibodies targeting the CD4-binding site, the V1, V2, or V3 loop, and membrane-proximal external region (MPER) for HIV-1 (31, 42–44). However, to date, most reported neutralizing antibodies to MERS-CoV target epitopes within the RBD and cross-compete. We and one other group recently have reported non-RBD S1-specific MAbs, G2 and 5F9, and an S2-specific MAb, G4 (20, 21, 38). Because we have isolated a panel of MAbs targeting the RBD, S1, and S2 from mouse, NHP, and human sources, the opportunity now exists to explore combinations of noncompeting MAbs for preventing or treating MERS CoV infection and their impact on disease and escape mutations in vivo.
CDC2-C2 is our most potent RBD-specific MAb comparable to the most potent published MAbs, m336 and REGN3051 (13, 18). CDC2-C2 can cross-neutralize 10 of 11 MERS-CoV strains isolated in 2012 to 2015 with high potency and partially neutralize the Bisha1 strain. In contrast, S1-specific MAbs isolated from a human and NHPs, CDC2-A2, CDC2-A10, and JC57-13, displayed lower neutralizing potency than mouse S1-specific MAb G2 (20). NHP and human S1 MAbs failed to neutralize the JordanN3 strain, which contains amino acid changes within the NTD. These three MAbs did not compete with each other in ELISA and BLI assays, suggesting that in addition to epitopes in the NTD, they may also interact with SD1 or SD2 in the C-terminal aspect of S1 (38, 45). Interestingly, we observed that addition of CDC2-A10, but not CDC2-A2, to CDC2-C2 prevented the emergence of escape mutation at the position 542. Other MAb combinations, JC57-11 with JC57-13 (one RBD-specific MAb with one S1-specific MAb) and JC57-11 with JC57-14 (two RBD-specific MAbs), also prevented escape mutations. Crystal structures of MAbs complexed with the RBD and HR1/HR2 6-helix bundle have helped elucidate neutralizing mechanisms for severe acute respiratory syndrome (SARS)-CoV and other viruses (46, 47). More structural data are needed to determine precise atomic details of epitopes and the orientation and angle of approach for neutralizing MAbs to understand the basis of antibody neutralization and the mechanisms by which combining MAbs can delay viral escape. The availability of stable spike trimers and well-defined epitopes will allow the development of B cell probes to further interrogate vaccine- and infection-induced immune responses and identify novel neutralizing antibodies.
We selected our most potent MAbs from each major specificity, including RBD-specific and S1-specific MAbs, CDC2-C2 and G2, respectively, and S2-specific MAb G4 (38), to test efficacy in vivo. All individual MAbs completely protected against MERS-CoV EMC challenge; 100% of prophylactically treated mice survived a lethal challenge, with undetectable viral load in lung tissues. While there are caveats for interpreting these results in a nonphysiological mouse model, these mice are highly permissive to infection. The data suggest that not only RBD-specific MAbs but also S1- and S2-specific MAbs can play a role in protection against MERS-CoV infection. Further studies to evaluate the combination of two or more MAbs in protection against virus escape mutations in animal models will be needed to define the ability to delay in vivo escape mutations vis-à-vis in vitro escape (Fig. 7).
In conclusion, a panel of murine, NHP, and human MAbs targeting diverse regions of the MERS-CoV S RBD, non-RBD S1, and S2 are valuable reagents for development of passive immunoprophylactic and immunotherapeutic strategies against MERS for supporting studies of structure-function relationships in S and for understanding mechanisms of CoV neutralization. In addition, atomic-level structures of S in its trimeric prefusion conformation have recently been solved for several human CoVs, including MERS-CoV, SARS-CoV, and human CoV HKU1 (45, 46, 48). The definition of neutralizing antibody binding determinants and atomic-level structures will help guide rational antigen design for vaccines against recognized and preemergent human CoVs.
MATERIALS AND METHODS
Samples from macaques and the human subject.
Human PBMCs and serum were collected and cryopreserved by the U.S. CDC from the laboratory-confirmed Florida/USA-2 MERS patient (https://www.cdc.gov/coronavirus/mers/us.html#florida) at week 3 post-symptom onset. The CDC determined these activities to be encompassed by Public Health Practice and Public Health Response guidelines, and thus Investigational Review Board review was not required. Rhesus macaques were immunized with three different vaccine regimens—3×DNA, 2×DNA/protein, or 2×protein—as described previously (20). Sera and PBMCs were collected 2 weeks after the last injection.
DNA and protein vector constructs.
We synthesized cDNAs encoding S from the following 11 MERS-CoV strains (GenBank accession numbers are in parentheses): England1 (AFY 13307), Batin1 (KF600628), Bisha1 (KF600620), Buraidah1 (KF600630), EMC (AFS88936), Hasa14b (KF600643), JordanN3 (KC776174), Munich (KF192507), Florida/USA-2 (KJ829365), ChinaGD01 (KT006149), and KOR/KNIH/002 (KT029139). We also synthesized 12 EMC-based S genes with RBD mutations for neutralization mapping studies (Fig. 4). All MERS-CoV S genes used for pseudovirus production were cloned into the mammalian expression vector VRC8400 (49, 50) and confirmed by sequencing. S truncations, the RBD, S1, and S2 were synthesized by PCR using a full-length S template, cloned into plasmid vector VRC8400, and sequence confirmed for protein production as described previously (20). S constructs used for ELISA binding contained a C-terminal 6×His tag for protein purification. Constructs used for making S-based probes contained an Avi tag for biotinylation followed by a His tag, and proteins were expressed by transfection of vectors into the Expi293 cell line. Transfected cell culture supernatants were collected and purified on HisTrap HP Hiload 16/60 Superdex columns (GE Healthcare, Piscataway, NJ).
Pseudovirus neutralization assay.
S-containing lentiviral pseudovirions were produced by cotransfection of three plasmids (packaging plasmid pCMVDR8.2, transducing plasmid pHR′ CMV-Luc, and CMV/R-MERS-CoV S plasmid) into 293T cells using Fugene 6 transfection reagent (Promega, Madison, WI) (20, 51, 52). We generated peudoviruses from 11 strains of MERS-CoV S for the cross-neutralization assay (Fig. 2) and 12 MERS-CoV EMC S mutants for neutralization mapping (Fig. 4). Huh7.5 cells, provided by Deborah R. Taylor of the U.S. FDA, were plated into 96-well white/black Isoplates (PerkinElmer, Waltham, MA) at 10,000 per well the day before infection. Serial dilutions of serum or MAbs were mixed with different strains of titrated pseudovirus, incubated for 30 min at room temperature, and added to Huh7.5 cells in triplicate. Following 2 h of incubation, wells were replenished with 100 μl of fresh medium. Cells were lysed 72 h later, and luciferase activity was measured. Percent neutralization and neutralization IC50s were calculated from luminometry data.
Isolation of MAbs by single B cell sorting.
Cryopreserved human or rhesus macaque PBMCs were thawed and stained with LIVE/DEAD fixable violet dead cell stain (Life Technologies). After washing, cells were stained with a cocktail of anti-human antibodies, including CD3 (clone SP34-2; BD Biosciences), CD4 (clone OKT4; BioLegend), CD8 (clone RPA-T8; BioLegend), CD14 (clone M5E2; BioLegend), CD20 (clone 2H7; BioLegend), IgG (G18-145; BD Biosciences), and IgM (clone G20-127; BD Biosciences) and subsequently stained with fluorescently labeled RBD and S1 probes. Probe-positive single B cells were sorted into 96-well plates containing lysis solution as previously described (34, 53, 54). Immunoglobulin heavy- and light-chain mRNAs were reverse-transcribed and amplified by nested PCR using published primers (33). Paired heavy- and light-chain cDNA sequences were cloned into expression vectors containing constant regions of human or rhesus macaque immunoglobulin heavy (γ) and light (κ or λ) chains. IgG was expressed by cotransfecting Expi-293 cells with equal amounts of paired heavy- and light-chain plasmids, followed by antibody purification using protein A Fast Flow (GE Healthcare) according to the manufacturer's instructions. Antibody heavy- and light-chain sequences were compared to human or rhesus monkey immunoglobulin germ line sequences using IMGT/V-QUEST (55, 56).
ELISA.
ELISA plates were coated with MERS-CoV RBD, S1, or S2 protein at 1 μg ml−1 in phosphate-buffered saline (PBS) at 4°C overnight. After standard washes and blocks, plates were incubated with serial dilutions of serum or MAbs. Anti-human IgG-horseradish peroxidase (HRP) conjugates (Jackson Laboratory, Bar Harbor, ME) were used as secondary antibodies, and tetramethylbenzidine (TMB) (KPL, Gaithersburg, MD) was used as the substrate for signal generation.
Competition ELISA.
Detailed methods for competition ELISAs have been published elsewhere (34, 57). Briefly, MAbs were biotinylated using an EZ-Link sulfo-NHS-biotinylation kit (Thermo Fisher Scientific, Waltham, CA) and titrated on MERS-CoV S1-coated plates. Avidin D-HRP conjugate (Vector Laboratories, Burlingame, CA) and TMB (KPL, Gaithersburg MD) were used for color development. The optical density at 450 nm (OD450) was determined with SpectraMax Plus (Molecular Devices, Sunnyvale, CA). The concentration of biotinylated MAb in the linear range of the titration curve was chosen for competition ELISA. Unlabeled competitor MAbs were serially diluted and added to the S1-coated plate. Following incubation for 30 min at room temperature, biotinylated MAbs were added, and OD readings were recorded using biotinylated MAb alone as a binding control. Percent inhibition of binding was calculated as follows: 100 − (reading with biotin-MAb in the presence of competing MAb)/reading with biotin-MAb alone) ×100.
Binding studies using biolayer interferometry.
Binding kinetics of MERS-CoV S molecules to MAbs were carried out using a FortéBio Octet HTX instrument. Assays with agitation set to 1,000 rpm in PBS buffer supplemented with 1% bovine serum albumin (BSA; 50 μl per well) were performed at 30°C in solid black tilted-bottom 384-well plates (Greiner Bio-One). For direct binding studies, MAbs (40 μg ml−1) were used to load anti-human IgG Fc capture (AHC) probes for 300 s to capture levels of 1 to 1.5 nm. Biosensor tips were then equilibrated for 60 s in PBS–1% BSA buffer prior to binding assessment of MERS-CoV S1 protein for 300 s, followed by dissociation for 300 s. For competition binding studies, His-tagged S1 protein was loaded on anti-Penta-His probes for 300 s. Biosensor tips were then equilibrated for 60 s in PBS–1% BSA buffer prior to binding the saturated competitor MAbs (saturated amount of MAb binding to S1 protein was determined prior to competition assay) for 300 s, followed by binding of analyte MAb for 300 s. Data analysis and curve fitting were performed with Octet software, version 9.0.
X-ray crystallography for JC57-14 and CDC2-C2.
RBD England1 (residues 367 to 606) with a C-terminal HRV3C cleavage site and His6 purification tag was produced in GnTi cells for crystallization with JC57-14 as previously described (20). The RBD was purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. The JC57-14 antigen-binding fragment (Fab) was prepared using the Pierce Fab fragment preparation kit. The RBD molecule was mixed with the JC57-14 Fab fragment in a 1:1.2 molar ratio and allowed to sit for 30 min at room temperature. All proteins were purified by size exclusion chromatography (Superdex S200) and concentrated to ∼5 to 8 mg ml−1. Crystallization screening was carried out using a Mosquito crystallization robot, using the hanging-drop vapor diffusion method at 20°C by mixing 0.1 μl of protein complex with 0.1 μl of reservoir solution, followed by manual optimization. The JC57-14 Fab crystals were grown in 0.23 M ammonium sulfate and 22% polyethylene glycol 8000 (PEG 8000). The JC57-14 Fab-RBD England1 complex crystals were grown in 90 mM CHES (pH 9.5) and 18% PEG 8000. Both crystals were cryocooled in liquid nitrogen using mother liquor containing 22% ethylene glycol as a cryoprotectant. Data were collected at a wavelength of 1.00 Å at SER-CAT beamlines ID-22 and BM-22 (Advanced Photon Source, Argonne National Laboratory).
A gene encoding MERS-CoV S1 RBD (England1 strain, residues 367 to 589) with a C-terminal HRV3C cleavage site and human IgG1 Fc fragment was inserted into the eukaryotic expression vector pαH. Three hours after transient transfection of the plasmid into FreeStyle 293-F cells, kifunensine was added to a final concentration of 5 μM. After 6 days, the supernatant was passed over a protein A agarose column, and deglycosylation was conducted on-column by adding endoglycosidase H (Endo H; New England BioLabs) (10% [wt/wt]) at room temperature. After 12 h, the column was washed with PBS and the RBD was eluted by incubating the resin with HRV3C (1% [wt/wt]). The RBD was further purified over a Superdex 75 column (GE Healthcare Biosciences). A gene encoding the CDC2-C2 heavy chain with an HRV3C cleavage site between the first and second constant domains was cloned into the eukaryotic expression vector pVRC8400. This plasmid, along with a similar plasmid encoding the CDC2-C2 light chain, was cotransfected into FreeStyle 293-F cells (Invitrogen). After 6 days, the supernatant was passed over a protein A agarose column, and the Fab was eluted by incubating the resin with HRV3C (1% [wt/wt]). The Fab fragment was further purified over a Superdex 75 column. Purified RBD was mixed with a 1.2-fold molar excess of CDC2-C2 Fab. After incubation on ice for 1 h, the complex was separated from excess Fab fragment on a Superdex 200 column (GE Healthcare Bioscience). Purified CDC2-C2 Fab fragment bound to the RBD was concentrated to 10.5 mg/ml in TBS (2 mM Tris [pH 8.0] and 200 mM NaCl) for crystallization. Crystals were produced at room temperature using the sitting-drop vapor-diffusion method by mixing 0.1 μl of protein with 0.1 μl of reservoir solution containing 0.1 M Tris (pH 6.5), 10% (vol/vol) 2-propanol, and 10% (wt/wt) PEG 3350. Crystals were soaked in reservoir solution supplemented with 20% (vol/vol) ethylene glycol and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the SBC beamline 19-ID (Advanced Photon Source, Argonne National Laboratory).
JC57-14 (with or without RBD) diffraction data were processed with the HKL2000 suite (58). CDC-C2 diffraction data were processed using the CCP4 software suite. Data were indexed and integrated in iMOSFLM (59) and scaled and merged with AIMLESS (60). All structures were solved by molecular replacement using PHASER (61). For the JC57-14 Fab structure, four Fab molecules were identified in the asymmetric unit, using the D12 Fab structure as a search model. For the JC57-14–RBD England1 complex, a solution was obtained using PDB code 4ZPT molecule S as a search model for the RBD, the JC57-14 Fab fragment was used as a search model for the Fab variable domain, and the Fab constant domain searched for separately. The CDC-2–RBD complex was solved using PDB code 4XAK as a search model. A cross-validation test set using 5% of the data was used to assess the model refinement process, with structure model validation carried out using MolProbity (62). All structures were built manually in Coot (63) and refined using PHENIX (64).
The JC57-14 Fab structure gave a final R factor value of 17.9% and Rfree value of 21.3%. The JC57-14–RBD England1 complex gave a structure model with a final R factor value of 22.0% and Rfree value of 27.0%. The CDC-C2–RBD England1 complex gave a structure model with a final R factor value of 19.1% and Rfree value of 22.8%. All structures had 100% residues in the favored region of the Ramachandran plot. Data collection and refinement statistics are presented in Table S1.
PRNT.
Fivefold serial dilutions of MAbs ranging from 10 to 0.0032 μg/ml were combined with 26 to 44 PFU of MERS-CoV EMC/2012 in a total volume of 200 μl of gelatin saline (0.3% [wt/vol] gelatin in phosphate-buffered saline supplemented with CaCl2 and MgCl2), and the mixture was applied to confluent Vero 81 cells in 6-well (10-cm2) plates for 1 h at 37°C. Monolayers were overlaid with Dulbecco's modified Eagle's medium (DMEM) containing 1% agar, and plaques were enumerated at 72 h postinfection (hpi). The fraction of residual plaques was plotted as a function of log10 MAb concentration, and the relationship was fit to a four-parameter dose-response curve using GraphPad PRISM 7.00 (R2 values, 0.96 to 1). Using PRISM tools, plaque reduction neutralization testing (PRNT) titers were interpolated from the dose-response curves as MAb concentrations resulting in 50% or 80% plaque reduction. PRNT assays were performed in duplicate for each MAb, and PRNT titers from the two experiments were averaged to generate the final value used in subsequent calculations.
Selection and sequence analysis of antibody escape mutant viruses.
A P0 stock of recombinant MERS-CoV EMC/2012 recovered from an infectious clone (65) was serially passaged in Vero 81 cell (25-cm2 flasks) cultures supplemented with increasing concentrations of individual MAbs or double-MAb combinations. Three parallel passage series were performed for each single- and double-MAb experimental condition. Passages were initiated at viral multiplicities of infection (MOI) ranging from approximately 0.001 to 0.3 PFU per cell and MAb concentrations ranging from 0.12× to 4.2× (single-MAb selection) and 0.06× to 1.9× (double-MAb selection) of their respective PRNT IC80s. Culture supernatants were passed onto fresh cells when monolayer involvement by viral CPE approached 50 to 60%. Viral inoculum volume and MAb concentrations were empirically coadjusted between passage steps to produce the target CPE level of 50 to 60% at approximately 48 hpi. MAb concentrations at the terminal passage level ranged from 3.6 to 105 times (single-MAb selection, P10) and 1.9 to 69 times (double-MAb selection, P20) their respective PRNTs. Clonal escape mutant viruses were isolated from terminal-culture lysates (obtained by freeze-thaw) via plaque purification on Vero 81 cells in the presence of MAbs at their respective final concentrations. Viral plaques were expanded in Vero 81 cells (25-cm2 flasks) without MAb supplementation to generate P0 cultures, from which supernatant virus was preserved and cell-associated virus was harvested in TRIzol reagent (Invitrogen). To identify changes in S associated with antibody escape, TRIzol-extracted viral RNA from cell monolayers, culture lysates (200 μl), or P0 supernatants of plaque expansions (200 to 400 μl) served as template to generate overlapping cDNA amplicons spanning the entire S gene open reading frame (ORF) by reverse transcription-PCR (RT-PCR) using Superscript III reverse transcriptase (Invitrogen) and Easy-A high-fidelity thermostable DNA polymerase (Agilent Technologies) or Q5 high-fidelity thermostable DNA polymerase (New England BioLabs). PCR products were subjected to dideoxy sequencing, and reads were aligned to the native EMC/2012 S gene sequence (GenBank accession number JX869059.2) using MacVector Assembler tools to identify differences from the parental sequence. To identify cell culture-adaptive changes in S resulting from serial MERS-CoV passage, three parallel lineages of antibody-free P10 and P20 EMC/2012 cultures were examined for S mutations by RT-PCR as described above. These mutations were omitted from analyses of changes in S identified in clonal isolates and populations of antibody escape mutant viruses.
Animal studies.
Approval of animal experiments was obtained from the Institutional Animal Care and Use Committee of the Rocky Mountain Laboratories. The performance of experiments was done following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) by certified staff in an AAALAC-approved facility, following the guidelines and basic principles in the Public Health Service Policy on Humane Care and Use of Laboratory Animals (66) and the Guide for the Care and Use of Laboratory Animals (67). Work with infectious MERS-CoV strains under biosafety level 3 (BSL3) conditions was approved by the Institutional Biosafety Committee (IBC). Inactivation and removal of samples from high containment were performed according to IBC-approved standards.
Six- to eight-week-old DPP4-transgenic mice (68, 69) were injected intraperitoneally, while anesthetized by inhalation of isoflurane, with a total of 20 mg/kg of MAbs or sterile saline as a control 1 day prior to infection. Mice were anesthetized via inhalation of isoflurane, followed by intranasal inoculation with 1 × 106 TCID50 of MERS-CoV (EMC/2012) diluted in a total volume of 50 μl. Ten mice per group were used. Four mice per group were euthanized at day 3 postinfection, and lungs were harvested for analysis of MERS-CoV replication. The remaining six animals per group were observed for 28 days.
Infectious virus titration.
Lung tissue samples in 1 ml of 2% DMEM were homogenized, and MERS-CoV was titrated in quadruplicate on VeroE6 cells. Tenfold serial dilutions of tissue homogenates were applied to cells, followed by 1 h of incubation at 37°C and 2 washes with PBS. Cytopathic effect was scored at 5 days postinfection. TCID50 values were adjusted for tissue weight and calculated by the Spearman-Karber method (39).
Statistical analysis.
We used Prism software (version 7.03; GraphPad, La Jolla, CA) to calculate IC50s, IC80s, and IC90s for each MAb from the dose-response curves as MAb concentrations resulting in 50%, 80%, or 90% reduction of infectivity. Log rank (Mantel-Cox) test was used to calculate P value for survival curves using the Prism software. Statistically significant differences met a threshold (α) of 0.05. Statistical variation within each data set is represented as the standard error in each of the figures.
Accession number(s).
Atomic coordinates and structure factors of structures for JC57-14 Fab, JC57-14, and CDC2-C2 in complex with the RBD have been deposited in the Protein Data Bank under accession codes 6C6X, 6C6Y, and 6C6Z.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Viral Pathogenesis and Virology Laboratories at the Vaccine Research Center for insightful comments and helpful discussions. Special thanks are extended to Xiaotao Lu (Vanderbilt) and Erica Andres (Vanderbilt) for assistance with selection and sequence analysis of antibody escape mutant viruses. We thank Carole Hickman (CDC) for technical assistance with processing the human PBMCs. We also thank Brenda Hartman and Kaitlyn Morabito for their assistance in preparing the figures and in the paper submission process.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention
Support for this work was provided by the Intramural Research Program of the Vaccine Research Center (National Institute of Allergy and Infectious Diseases, NIH), the NIAID Division of Intramural Research and NIH under contract HHSN261200800001E (agreement no. 16 × 142 to M.R.D.). Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center (SBC) at the Advanced Photon Source. SBC-CAT is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. Use of sector 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
L.W., W.S., M.G.J., M.K., K.M., W.-P.K., J.R.M., and B.S.G. are inventors on a U.S. patent application describing the use of S glycoprotein as a vaccine antigen and monoclonal antibodies as potential therapeutics and aids in vaccine design. J.D.C. is an inventor on U.S. patent US 9,249,195 B2, Reovirus Vaccines and Methods of Use Therefor.
L.W., W.S., M.G.J., W.-P.K., K.M., J.R.M., and B.S.G. planned the study and experiments. L.W. isolated and characterized MAbs. W.S. and M.C. purified proteins and MAbs. M.G.J., J.G.V.G., N.W., J.S.M., and P.D.K. performed crystallization studies. Y.Z. and K.S.C. performed the pseudovirus neutralization assays. L.W. and M.K. carried out biolayer interferometry measurements. M.M.B., J.D.C., and M.R.D. planned and performed the escape mutation experiments and analyses. R.D.M. and K.O.S. helped with B cell sorting. T.Z. assisted in designing mutations. V.J.M., N.V.D., and R.F. performed the challenge studies. K.M.T. and L.M.H. isolated PBMCs from human donors. L.W., J.R.M., and B.S.G. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02002-17.
REFERENCES
- 1.Durai P, Batool M, Shah M, Choi S. 2015. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med 47:e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, Alabdullatif ZN, Assad M, Almulhim A, Makhdoom H, Madani H, Alhakeem R, Al-Tawfiq JA, Cotten M, Watson SJ, Kellam P, Zumla AI, Memish ZA, Team KM-CI. 2013. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med 369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, Makhdoom HQ, Zumla AI, Memish ZA. 2013. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breban R, Riou J, Fontanet A. 2013. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet 382:694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cauchemez S, Fraser C, Van Kerkhove MD, Donnelly CA, Riley S, Rambaut A, Enouf V, van der Werf S, Ferguson NM. 2014. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis 14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah AA, Corman VM, Sieberg A, Makhdoom HQ, Assiri A, Al Masri M, Aldabbagh S, Bosch BJ, Beer M, Muller MA, Kellam P, Drosten C. 2014. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis 20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham BS, Ambrosino DM. 2015. History of passive antibody administration for prevention and treatment of infectious diseases. Curr Opin HIV AIDS 10:129–134. doi: 10.1097/COH.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanagh D. 1983. Coronavirus IBV: structural characterization of the spike protein. J Gen Virol 64(Part 12):2577–2583. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, Collins PL. 2004. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A 101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. 2013. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mou H, Raj VS, van Kuppeveld FJ, Rottier PJ, Haagmans BL, Bosch BJ. 2013. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol 87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascal KE, Coleman CM, Mujica AO, Kamat V, Badithe A, Fairhurst J, Hunt C, Strein J, Berrebi A, Sisk JM, Matthews KL, Babb R, Chen G, Lai KM, Huang TT, Olson W, Yancopoulos GD, Stahl N, Frieman MB, Kyratsous CA. 2015. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A 112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti D, Zhao J, Pedotti M, Simonelli L, Agnihothram S, Fett C, Fernandez-Rodriguez B, Foglierini M, Agatic G, Vanzetta F, Gopal R, Langrish CJ, Barrett NA, Sallusto F, Baric RS, Varani L, Zambon M, Perlman S, Lanzavecchia A. 2015. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci U S A 112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Wan Y, Liu P, Zhao J, Lu G, Qi J, Wang Q, Lu X, Wu Y, Liu W, Zhang B, Yuen KY, Perlman S, Gao GF, Yan J. 2015. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res 25:1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang L, Wang N, Zuo T, Shi X, Poon KM, Wu Y, Gao F, Li D, Wang R, Guo J, Fu L, Yuen KY, Zheng BJ, Wang X, Zhang L. 2014. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med 6:234ra259. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 17.Tang XC, Agnihothram SS, Jiao Y, Stanhope J, Graham RL, Peterson EC, Avnir Y, Tallarico AS, Sheehan J, Zhu Q, Baric RS, Marasco WA. 2014. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci U S A 111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying T, Du L, Ju TW, Prabakaran P, Lau CC, Lu L, Liu Q, Wang L, Feng Y, Wang Y, Zheng BJ, Yuen KY, Jiang S, Dimitrov DS. 2014. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol 88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du L, Zhao G, Yang Y, Qiu H, Wang L, Kou Z, Tao X, Yu H, Sun S, Tseng CT, Jiang S, Li F, Zhou Y. 2014. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol 88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Shi W, Joyce MG, Modjarrad K, Zhang Y, Leung K, Lees CR, Zhou T, Yassine HM, Kanekiyo M, Yang ZY, Chen X, Becker MM, Freeman M, Vogel L, Johnson JC, Olinger G, Todd JP, Bagci U, Solomon J, Mollura DJ, Hensley L, Jahrling P, Denison MR, Rao SS, Subbarao K, Kwong PD, Mascola JR, Kong WP, Graham BS. 2015. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun 6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Lu S, Jia H, Deng Y, Zhou J, Huang B, Yu Y, Lan J, Wang W, Lou Y, Qin K, Tan W. 2017. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg Microbes Infect 6:e60. doi: 10.1038/emi.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu X, Zhang S, Jiang L, Cui Y, Li D, Wang D, Wang N, Fu L, Shi X, Li Z, Zhang L, Wang X. 2015. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci Rep 5:13133. doi: 10.1038/srep13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying T, Prabakaran P, Du L, Shi W, Feng Y, Wang Y, Wang L, Li W, Jiang S, Dimitrov DS, Zhou T. 2015. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun 6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Bao L, Chen C, Zou T, Xue Y, Li F, Lv Q, Gu S, Gao X, Cui S, Wang J, Qin C, Jin Q. 2017. Human neutralizing monoclonal antibody efficiently inhibits MERS-CoV replication in common marmoset. J Infect Dis 215:1807–1815. doi: 10.1093/infdis/jix209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson RF, Bagci U, Keith L, Tang X, Mollura DJ, Zeitlin L, Qin J, Huzella L, Bartos CJ, Bohorova N, Bohorov O, Goodman C, Kim do H, Paulty MH, Velasco J, Whaley KJ, Johnson JC, Pettitt J, Ork BL, Solomon J, Oberlander N, Zhu Q, Sun J, Holbrook MR, Olinger GG, Baric RS, Hensley LE, Jahrling PB, Marasco WA. 2016. 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology 490:49–58. doi: 10.1016/j.virol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houser KV, Gretebeck L, Ying T, Wang Y, Vogel L, Lamirande EW, Bock KW, Moore IN, Dimitrov DS, Subbarao K. 2016. Prophylaxis with a Middle East respiratory syndrome coronavirus (MERS-CoV)-specific human monoclonal antibody protects rabbits from MERS-CoV infection. J Infect Dis 213:1557–1561. doi: 10.1093/infdis/jiw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sui J, Deming M, Rockx B, Liddington RC, Zhu QK, Baric RS, Marasco WA. 2014. Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J Virol 88:13769–13780. doi: 10.1128/JVI.02232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coughlin MM, Prabhakar BS. 2012. Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential. Rev Med Virol 22:2–17. doi: 10.1002/rmv.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elshabrawy HA, Coughlin MM, Baker SC, Prabhakar BS. 2012. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One 7:e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong R, Louder MK, Wagh K, Bailer RT, deCamp A, Greene K, Gao H, Taft JD, Gazumyan A, Liu C, Nussenzweig MC, Korber B, Montefiori DC, Mascola JR. 2015. Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. J Virol 89:2659–2671. doi: 10.1128/JVI.03136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M Jr, Lifson JD, Dimitrov DS, Nussenzweig MC, Martin MA. 2013. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundling C, Phad G, Douagi I, Navis M, Karlsson Hedestam GB. 2012. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J Immunol Methods 386:85–93. doi: 10.1016/j.jim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Mason RD, Welles HC, Adams C, Chakrabarti BK, Gorman J, Zhou T, Nguyen R, O'Dell S, Lusvarghi S, Bewley CA, Li H, Shaw GM, Sheng Z, Shapiro L, Wyatt R, Kwong PD, Mascola JR, Roederer M. 2016. Targeted isolation of antibodies directed against major sites of SIV Env vulnerability. PLoS Pathog 12:e1005537. doi: 10.1371/journal.ppat.1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, Wei CJ, Nabel GJ. 2012. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prabakaran P, Zhu Z, Chen W, Gong R, Feng Y, Streaker E, Dimitrov DS. 2012. Origin, diversity, and maturation of human antiviral antibodies analyzed by high-throughput sequencing. Front Microbiol 3:277. doi: 10.3389/fmicb.2012.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong WP, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Doremalen N, Falzarano D, Ying T, de Wit E, Bushmaker T, Feldmann F, Okumura A, Wang Y, Scott DP, Hanley PW, Feldmann H, Dimitrov DS, Munster VJ. 2017. Efficacy of antibody-based therapies against Middle East respiratory syndrome coronavirus (MERS-CoV) in common marmosets. Antiviral Res 143:30–37. doi: 10.1016/j.antiviral.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luke T, Wu H, Zhao J, Channappanavar R, Coleman CM, Jiao JA, Matsushita H, Liu Y, Postnikova EN, Ork BL, Glenn G, Flyer D, Defang G, Raviprakash K, Kochel T, Wang J, Nie W, Smith G, Hensley LE, Olinger GG, Kuhn JH, Holbrook MR, Johnson RF, Perlman S, Sullivan E, Frieman MB. 2016. Human polyclonal immunoglobulin G from transchromosomic bovines inhibits MERS-CoV in vivo. Sci Transl Med 8:326ra321. doi: 10.1126/scitranslmed.aaf1061. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J, Perera RA, Kayali G, Meyerholz D, Perlman S, Peiris M. 2015. Passive immunotherapy with dromedary immune serum in an experimental animal model for Middle East respiratory syndrome coronavirus infection. J Virol 89:6117–6120. doi: 10.1128/JVI.00446-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, Pegu A, Rao E, Doria-Rose N, Beninga J, McKee K, Lord DM, Wei RR, Deng G, Louder M, Schmidt SD, Mankoff Z, Wu L, Asokan M, Beil C, Lange C, Leuschner WD, Kruip J, Sendak R, Do Kwon Y, Zhou T, Chen X, Bailer RT, Wang K, Choe M, Tartaglia LJ, Barouch DH, O'Dell S, Todd JP, Burton DR, Roederer M, Connors M, Koup RA, Kwong PD, Yang ZY, Mascola JR, Nabel GJ. 2017. Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science 358:85–90. doi: 10.1126/science.aan8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Bardelli M, Espinosa DA, Pedotti M, Ng TS, Bianchi S, Simonelli L, Lim EXY, Foglierini M, Zatta F, Jaconi S, Beltramello M, Cameroni E, Fibriansah G, Shi J, Barca T, Pagani I, Rubio A, Broccoli V, Vicenzi E, Graham V, Pullan S, Dowall S, Hewson R, Jurt S, Zerbe O, Stettler K, Lanzavecchia A, Sallusto F, Cavalli A, Harris E, Lok SM, Varani L, Corti D. 2017. A human bi-specific antibody against Zika virus with high therapeutic potential. Cell 171:229–241.e215. doi: 10.1016/j.cell.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. 2017. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. 2017. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilman MS, Moin SM, Mas V, Chen M, Patel NK, Kramer K, Zhu Q, Kabeche SC, Kumar A, Palomo C, Beaumont T, Baxa U, Ulbrandt ND, Melero JA, Graham BS, McLellan JS. 2015. Characterization of a prefusion-specific antibody that recognizes a quaternary, cleavage-dependent epitope on the RSV fusion glycoprotein. PLoS Pathog 11:e1005035. doi: 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. 2016. Pre-fusion structure of a human coronavirus spike protein. Nature 531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barouch DH, Yang ZY, Kong WP, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, Letvin NL, Nabel GJ. 2005. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J Virol 79:8828–8834. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Catanzaro AT, Roederer M, Koup RA, Bailer RT, Enama ME, Nason MC, Martin JE, Rucker S, Andrews CA, Gomez PL, Mascola JR, Nabel GJ, Graham BS. 2007. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 25:4085–4092. doi: 10.1016/j.vaccine.2007.02.050. [DOI] [PubMed] [Google Scholar]
- 51.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A 93:11382–11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang ZY, Werner HC, Kong WP, Leung K, Traggiai E, Lanzavecchia A, Nabel GJ. 2005. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci U S A 102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, NISC Comparative Sequencing Program, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brochet X, Lefranc MP, Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giudicelli V, Brochet X, Lefranc MP. 2011. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 57.Kong R, Li H, Georgiev I, Changela A, Bibollet-Ruche F, Decker JM, Rowland-Jones SL, Jaye A, Guan Y, Lewis GK, Langedijk JP, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2012. Epitope mapping of broadly neutralizing HIV-2 human monoclonal antibodies. J Virol 86:12115–12128. doi: 10.1128/JVI.01632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67(Part 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69(Part 7):1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40(Part 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(Part 1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Part 12):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58(Part 11):1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 65.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Office of Laboratory Animal Welfare. 2015. Public Health Service policy on humane care and use of laboratory animals. NIH, Bethesda, MD. [Google Scholar]
- 67.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 68.Agrawal AS, Garron T, Tao X, Peng BH, Wakamiya M, Chan TS, Couch RB, Tseng CT. 2015. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol 89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Doremalen N, Munster VJ. 2015. Animal models of Middle East respiratory syndrome coronavirus infection. Antiviral Res 122:28–38. doi: 10.1016/j.antiviral.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.