Abstract
Background
Recent studies examining the association between posttraumatic stress disorder (PTSD) and accelerated aging, as defined by DNA methylation-based estimates of cellular age that exceed chronological age, have yielded mixed results.
Methods
We conducted a meta-analysis of trauma exposure and PTSD diagnosis and symptom severity in association with accelerated DNA methylation age using data from 9 cohorts contributing to the Psychiatric Genomics Consortium PTSD Epigenetics Workgroup (combined N = 2,186). Associations between demographic and cellular variables and accelerated DNA methylation age were also examined, as was the moderating influence of demographic variables.
Results
Meta-analysis of regression coefficients from contributing cohorts revealed that childhood trauma exposure (when measured with the Childhood Trauma Questionnaire) and lifetime PTSD severity evidenced significant, albeit small, meta-analytic associations with accelerated DNA methylation age (ps = .028 and .016, respectively). Sex, CD4 T cell proportions, and natural killer cell proportions were also significantly associated with accelerated DNA methylation age (all ps < .02). PTSD diagnosis and lifetime trauma exposure were not associated with advanced DNA methylation age. There was no evidence of moderation of the trauma or PTSD variables by demographic factors.
Conclusions
Results suggest that traumatic stress is associated with advanced epigenetic age and raise the possibility that cells integral to immune system maintenance and responsivity play a role in this. This study highlights the need for additional research into the biological mechanisms linking traumatic stress to accelerated DNA methylation age and the importance of furthering our understanding of the neurobiological and health consequences of PTSD.
Keywords: DNA methylation, traumatic stress, PTSD, accelerated aging, meta-analysis, epigenetic clock
1.1 Introduction
Traumatic stress (e.g., psychiatric symptoms related to traumatic experiences) may precipitate a host of negative outcomes inclusive of psychological and medical conditions (Schnurr et al., 2000; Afari et al., 2014). Theory (Miller & Sadeh, 2014; Lohr et al., 2015; Williamson et al., 2015) and empirical research (Roberts et al., 2017, Li et al., 2017; Wolf et al., 2016, 2017) suggest that traumatic stress may also advance the pace of cellular aging such that it exceeds that of chronological aging and this may potentially lead to, or be a marker for, negative health outcomes (Horvath, 2013).
There are highly reliable age-related changes in DNA methylation (DNAm) throughout the epigenome (Christensen et al., 2009). Recent research has capitalized on these associations and on the substantial information available from state-of-the-art DNAm arrays, capturing methylation levels at hundreds of thousands of CpG (Cytosine-phosphate-Guanine) loci using just a single beadchip and a small amount of DNA, to develop methylation-based estimates of chronological age. Specifically, Hannum et al. (2013) developed a DNAm age algorithm derived from whole blood that was based on 71 probes (89 in the “all data” model) and it correlated with chronological age at r = .96. Independently, Horvath (2013) identified 353 DNAm loci that when combined into a weighted summary score also evidenced very strong correlations (r = .96) with chronological age across multiple tissues. Evidence for the utility and validity of DNAm age is evident in research demonstrating that DNAm age estimates that are higher than expected given chronological age (i.e., “accelerated DNAm age”) are associated with age-related disorders and mortality (Chen et al., 2016; Christiansen et al., 2016; Horvath et al., 2015; Levine et al., 2015; Marioni et al., 2016; Marioni et al., 2015). Collectively, this suggests that accelerated DNAm age may be a biomarker for a generalized pathological cellular aging process, with a variety of environmental conditions and diseases associated with this basic epigenetic “clock.”
Emerging research raises the possibility that traumatic stress may be associated with advanced DNAm age, though results to date have been mixed. Specifically, Wolf et al. (2016; 2017) reported that symptoms of posttraumatic stress disorder (PTSD) were associated with DNAm age acceleration (relative to chronological age) per the Hannum, but not the Horvath (Wolf et al., 2016), algorithm in two samples of predominately male U.S. military veterans (sample sizes ranged from 281 to 339). In contrast, in a Dutch sample of 96 male military veterans, Boks et al. (2015) reported that PTSD was negatively associated with Horvath DNAm age estimates over time, while combat trauma was positively associated with Horvath DNAm age. In that study, the relationship between DNAm age and chronological age was not factored in to the definition of age acceleration. In a largely female civilian sample (n = 392), Zannas et al. (2015) found no evidence of an association between childhood trauma exposure or PTSD and accelerated Horvath DNAm age relative to chronological age. However, the authors did report an association between personal life stressors and advanced Horvath DNAm age, particularly among older participants. The variability in the approach to measuring DNAm age across these studies (e.g., Horvath versus Hannum metrics; inconsistent use of operational definitions that model the relationship between DNAm age and chronological age) and the variability in results across studies to date makes it difficult to discern a clear pattern of association between traumatic stress and accelerated aging in DNAm.
Given this, the aim of this study was to bring the strengths of the Psychiatric Genomics Consortium (PGC) PTSD Epigenetics Workgroup (Ratanatharathorn et al., 2017) to bear on the evaluation of the association between traumatic stress and accelerated DNAm age using data from nine cohorts encompassing 2,186 subjects. The PGC (Sullivan et al., 2017), which began in 2007 and added the PTSD Working Group in 2012, represents the largest consortium in biological psychiatry; this study used a subset of PGC-affiliated datasets with relevant data.
1.2 Study Aims
The specific aims of the study were to: (1) evaluate DNAm age and DNAm age acceleration1 in association with key demographic (i.e., sex, ancestry, age) and cellular variables (i.e., white blood cell proportions); (2) examine associations between trauma exposure and PTSD with accelerated DNAm age; and (3) examine demographic variables that might moderate the association between traumatic stress and accelerated DNAm age. Given that PTSD-related accelerated DNAm age has only been observed with the Hannum et al. DNAm age algorithm (Wolf et al., 2016; 2017), we hypothesized that PTSD diagnosis and severity would be associated with accelerated Hannum et al. DNAm age, but we investigated Horvath DNAm age in parallel. These goals were accomplished by deploying a standardized script and instructions to individual investigators with relevant data who participate in the PGC PTSD Epigenetics Workgroup and then meta-analyzing results across cohorts.
2.1 Method
2.2 Participants
Table 1 lists the individual cohorts included in the meta-analysis and key demographic and methodological details of the studies. There were seven military samples. These included: (1) the National Center for PTSD cohort (NCPTSD; Logue et al., 2013), a sample of 465 white, non-Hispanic trauma-exposed male and female veterans from mixed war eras and a subset of their trauma-exposed spouses;2 (2) Translational Research Center for TBI and Stress Disorders (TRACTS) cohort, a sample of 289 primarily white, non-Hispanic male and female veterans who deployed to Iraq and/or Afghanistan (McGlinchey et al., 2017; Sadeh et al., 2016; Wolf et al., 2016);3 (3) the Marine Resiliency Study (MRS; Baker et al. 2012; Nievergelt et al., 2015), a sample of 126 male mixed-ancestry Marines who were deployed to Afghanistan and who were assessed pre, and at 3- and 6-months post-deployment (PTSD data and DNAm data for this study reflect the time point with the most severe symptoms post-deployment); (4) the Army Study to Assess Risk and Resilience in Service Members (Army STARRS; Ursano et al., 2014; Stein et al., 2016), a sample of 102 male Army service members of primarily European ancestry who were assessed pre- and post-deployment to Afghanistan (data for this study were from the 3-month post-deployment assessment); (5 & 6) the Mid-Atlantic Mental Illness Research Education and Clinical Center PTSD Study (Mid-Atlantic MIRECC; Ashley-Koch et al., 2015), two related cohorts of male and female veterans who deployed to Iraq and/or Afghanistan (one cohort, MIRECC-a, was white, non-Hispanic [n = 176] and the second cohort, MIRECC-b, was black, non-Hispanic [n = 369]); and (7) the Dutch Prospective Research in Stress-related Military Operations (PRISMO) study (Boks et al., 2015), a sample of 62 male Dutch soldiers who were assessed pre- and 6-months post-deployment to Afghanistan (data for this study come from the post-deployment PTSD assessment).4 There were two non-military cohorts: (1) the Detroit Neighborhood Health Study (DNHS; Ruggiero et al., 2003; Uddin et al., 2010), a sample of 179 primarily black male and female urban community members; and (2) the Grady Trauma Project (GTP; Gillespie et al., 2009; Binder et al. 2008), a cohort of 418 primarily black male and female urban community members. The total sample size across all cohorts was 2,186. This included 855 lifetime PTSD cases and 516 lifetime controls and 876 current PTSD cases and 1,228 current controls.5 All participants were adults. Additional details on these cohorts and the PGC PTSD approach to DNAm analyses are available in Ratanatharathorn et al. (2017).
Table 1.
Study Characteristics of each Cohort Included in the Meta-Analysis
| Study | Total N (% male) |
M Age (SD; Range) |
Ancestry | Sample Description |
Source of PCs for ancestry covariates |
% PTSD Cases |
% Trauma Exposure |
% Ch Trauma Exposure |
LT Trauma Measure |
Ch Trauma Measure |
PTSD DX Measure |
PTSD Severity Measure |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| National Center for PTSD (NCPTSD)1 | 465 (64.7%) | 52.38 (10.64; 23–75) |
|
Trauma-exposed veterans and a subset of their trauma-exposed spouses | 10,000 common SNPs on Illumina OMNI 2.5–8 array | C: 38.7% L: 63.7% |
100% per CAPS | 18.3% | TLEQ11 | TLEQ11 (select items) | CAPS12 (C & L) | CAPS12 (C & L) |
| TRACTS2 | 289 (88.2%) | 32.02 (8.37; 19–58) |
|
Veterans deployed to Iraq and/or Afghanistan | 100,000 common SNPs on Illumina OMNI 2.5–8 array | C: 55.4% L: 72.7% |
79.5% | 23.7% | TLEQ11 | TLEQ11 (select items) | CAPS12 (C & L) | CAPS12 (C & L) |
| Marine Resiliency Study (MRS)3 | 126 (100%) | 22.21 (3.04; 18.80–38.84) |
|
US Marines deployed to Iraq or Afghanistan | 10,000 common SNPs on Illumina Human Omni Express Exome chip in concert with ancestry informative markers | C: 50.0% L: NA |
100.0% | 40.3% | LEC13 | CTQ14 | CAPS12 (C) | CAPS12 (C) |
| Army STARRS4 | 102 (100%) | 23.78 (4.24; 18–42) |
|
US Army soldiers deployed to Afghanistan | 25,000 common SNPs on Illumina Human Omni Express Exome chip | C: 50.0% L: 53.9% |
81.4% | 16.7% | Self-report of trauma exposure | CTQa | CIDI15 (C & L) | PCL16 (C) |
| Mid-Atlantic MIRECC-a5 | 176 (78%) | 34.87 (9.89; 20–64) |
|
Veterans deployed to Iraq or Afghanistan | 104,846 common SNPs on (a) Human Hap650, (b) Human1 Mill Duo, (c) Human Omni 2.5–8 | C: 49.4% L: NA |
99.4% | 40.9% | TLEQ11 | TLEQ11 (select items) | SCID17 (C) | DTS18 (C) |
| Mid-Atlantic MIRECC-b6 | 369 (50%) | 38.37 (9.36; 20–66) |
|
Veterans deployed to Iraq or Afghanistan | 237,080 common SNPs 3 on Illumina beadchips with imputation across them: (a) Human Hap650, (b) Human1 Mill Duo, (c) Human Omni 2.5–8 |
C:49.6% L: NA |
99.7% | 55.6% | TLEQ11 | TLEQ11 (select items) | SCID17 (C) | DTS18 (C) |
| PRISMO7 | 62 (100%) | 27.79 (9.98; 19–54) |
|
Dutch veterans deployed to Afghanistan | Illumina Infinium 450k DNAm beadchip10 | C:50% L: NA |
NA | 85.5% | List of deploy-ment events19 | ETI20 | SRIP21 (C) | N/A |
| Detroit Neighbor-hood Health Study (DNHS)8 | 179 (37%) | 53.89 (13.94; 20–89) |
|
Trauma-exposed (PTSD enriched) participants from Wave 1 or 2 of the DNHS sample (representative of adult Detroit residents) | Illumina Infinium 450k DNAm beadchip10 | C:22.3% L:66.5% |
100.0% | 59.4% | List of traumatic events22 | Items from the CTQ14 & CTS23 | PCL16 (C & L) | N/A |
| Grady Trauma Project (GTP)9 | 417 (28%) | 41.61 (12.66; 18–77) |
|
Subset of GTP participants with relevant data; participants recruited from large, urban, public hospital | 5000 common SNPs on Illumina Human Omni1-Quad beadchip | C: 19.4% L:41.9% |
96.4% | 54.8% | TEI24 | CTQ14 | CAPS12 (C & L) | CAPS12 (C & L) |
Note. All studies used the Illumina HumanMethylation450K Beadchip. TRACTS = Translational Research Center for TBI and Stress Disorders; Army STARRS = Army Study to Assess Risk and Resilience in Service Members; MIRECC = Mid-Atlantic Mental Illness Research Education and Clinical Center PTSD Study; PRISMO = Dutch Prospective Research in Stress-related Military Operations; TLEQ = Traumatic Life Events Questionnaire; TEI = Traumatic Events Inventory; CTQ = Childhood Trauma Questionnaire; CAPS = Clinician Administered PTSD Scale; PCL = PTSD Checklist; SCID = Structured Clinical Interview for DSM-IV; CIDI = Composite International Diagnostic Interview; DTS = Davidson Trauma Scale; SRIP = Self-rating Inventory for Post-traumatic Stress Disorder; ETI = Early Trauma Inventory; LEC = Life Events Checklist; PTSD = posttraumatic stress disorder; C = current; L = lifetime; Ch = childhood; LT = lifetime; DX = diagnosis; WNH = white, non-Hispanic; EA = European ancestry; PC = principal component; SNP = single nucleotide polymorphism; DNAm = DNA methylation.
Army STARRS used a measure that was nearly identical to the CTQ and for the purposes of this study, the measure was scored consistent with the CTQ.
Superscripted numerals refer to the reference for each measure in the reference section of this paper. 1Logue et al., 2013; 2. McGlinchey et al., in press; Wolf et al., 2016; 3. Baker et al. 2012; Nievergelt et al., 2015; 4. Ursano et al., 2014; Stein et al., 2016; 5. Koch et al., 2015; 6. Ashley-Koch et al., 2015; 7. Boks et al., 2015; 8. Ruggiero et al., 2003; Uddin et al., 2010; Breslau et al. 1998; 9. Gillespie et al., 2009; Binder et al., 2008; 10. Barfield et al., 2014; 11. Kubany et al., 2000; 12. Blake et al., 2005; 13. Gray et al., 2004; 14. Bernstein et al., 2003; 15. Andrews & Peters, 1998; 16. Ruggiero et al., 2003; 17. First et al., 1994; 18. Davidson et al., 1997; 19. Van Zuiden et al., 2011; 20. Bremner et al., 2007; 21. Hovens et al., 2000; Hovens et al., 2002; 22. Breslau et al., 1998; 23. Straus, 1979; 24. Ribbe, 1996
2.3 Measures
Given variability in the measures used to assess trauma exposure and PTSD symptoms (Table 1), we provided guidance and instruction on the scoring of these variables in order to harmonize the phenotypes of interest. Total trauma exposure reflected a count of the number of different types of lifetime traumatic experiences (though studies differed with respect to the specific type and number of events that were included in each measure). Measures of childhood trauma reflected a count of the number of different types of traumatic events (e.g., witnessing family violence, sexual abuse, physical abuse, or neglect occurring prior to age 18). Three studies employed the Childhood Trauma Questionnaire (CTQ; Bernstein et al., 2003) or a near identical version of the measure.6 In order to harmonize the approach to scoring this measure and keep the scaling consistent with the other measures of childhood trauma, this scale was scored to reflect a 0–3 count of any exposure to sexual abuse, physical abuse, and/or neglect. Our use of a variable representing the number of different types of traumatic experiences was based on the need to use a consistent definition of trauma across the various cohorts (which utilized different trauma inventories) to aid in interpretation of results. It was also based on evidence that the number of different types of traumatic experiences poses the strongest risk for the development of PTSD and other psychopathology, compared to other metrics of trauma exposure (e.g., Hedtke et al., 2008). Current (past month) and lifetime (worst period of symptoms) DSM-IV PTSD diagnostic determinations were based on the established scoring instructions for each measure employed by each study, and PTSD severity was based on total self-report or interviewer-rated severity (e.g., total score on the PTSD Checklist [Weathers et al., 1993] or total frequency + intensity ratings on the Clinician Administered PTSD Scale [Blake et al., 1995]). As not all studies had data pertaining to each variable of interest (Table 1), the sample size for each meta-analysis varied as detailed below.
2.4 Procedure
The last author, in consultation with the first author, developed an R script to deploy to data analysts at each study. The analysts followed the aforementioned guidance for scoring each variable, executed the script, and sent summarized results to the first and last author for meta-analysis. Results were carefully screened for potential errors (e.g., missing or mis-coded variables, inconsistent sample sizes, etc.) prior to meta-analysis.
DNA was extracted from peripheral blood samples according to the procedures identified in the original publications for each cohort (Table 1). Genotyping was completed on a variety of genome-wide arrays and for this study, is relevant only for the development of principal components (PCs) for use in ancestry evaluation (Table 1). All studies employed the Illumina Infinium Human Methylation BeadChip, with details on methylation procedures for each study available7 in the original publications (Table 1) and the DNAm processing pipeline described in Ratanatharathorn et al. (2017). Each study site obtained local IRB approval and all participants provided written informed consent. The IRB of the VA Boston Healthcare System approved the meta-analytic procedures of the summarized data.
2.5 Statistical Procedures
2.5.1 Ancestry
PCs were calculated to reflect ancestry within each cohort from either the genotyped data (in most cases) or the DNAm data (Table 1). The first three PCs were controlled for in all analyses in GWAS-based ancestry inferences, whereas PCs 2–4 were controlled for in the DNAm-based ancestry analyses as described in Ratanatharathorn et al. (2017). Both sets of PCs reflect ancestry. While SNP-based PCs are more common, as detailed by Ratanatharathorn et al., when DNAm probes located within 1 base pair of a SNP are used to index ancestry (i.e., essentially proxies for the SNP), the resulting PCs are significantly correlated with genome-wide SNP-based PCs. Ratanatharathorn et al. showed that in the PGC PTSD Epigenetics workgroup data that is also featured in this study, PCs 2–4 derived from probes within 1 base pair of SNPs aligned with SNP-based PCs 1–3. These ancestry PCs from DNAm data are distinct from quantification of cell mixtures, batch effects, and white blood cell counts (see below).
2.5.2 DNAm Age, White Blood Cell Counts
Two metrics of DNAm age were computed. Horvath DNAm age estimates were computed per the instructions on Dr. Horvath’s website (https://labs.genetics.ucla.edu/horvath/dnamage/) on the raw (non-normalized) probe data as the Horvath program includes a normalization step at part of its procedures. The algorithm is based on 353 probes. The Horvath website outputs DNAm age estimates as well as proportional white blood cell (WBC) counts estimated from the methylation data according to the Houseman method (Houseman et al., 2012; Jaffe and Irizarry, 2014.) These WBC estimates (CD8 T cells, CD4 T cells, B cells, natural killer [NK] cells, and monocytes) were retained as covariates in the analyses and their potential associations with DNAm age residuals were evaluated accordingly. Cleaning and imputation of the methylation data at each site followed the protocol for the PGC-PTSD EWAS group. This protocol is described in detail by Ratanatharathorn et al. (2017), and will only be described briefly here. Individual methylation values failing to meet a detection p-value threshold of 0.001 were set to missing. Subjects and probes with more than 10% missing data were excluded. Samples with intensity of less than 50% of the experiment-wide mean or with intensity <2,000 arbitrary units were excluded. Probes that cross hybridize between autosomes and sex chromosomes (Chen et al., 2013) were excluded. Normalization was performed using beta mixture quantile dilation method as implemented in the wateRmelon (Touleimat et al., 2012; Pidsley et al., 2013) Bioconductor package. Missing data were imputed using the impute package with a k nearest neighbor method (http://www.bioconductor.org/packages/release/bioc/html/impute.html). An empirical Bayes batch-correction method (ComBat; Johnson et al., 2007), as implemented in the Bioconductor sva package (Leek et al., 2013), was used to remove chip effects and systematic variation due to chip and sample position on the chip. Hannum et al.’s “all data” algorithm (inclusive of 89 probes) was used to calculate DNAm age on the batch-corrected and normalized probe values. This algorithm reflects the linear weighted combination of methylation levels at the select loci. Across studies, the maximum percentage of subjects with missing data on a given probe in the Hannum et al. algorithm was 3.29%, with 0.11% as the maximum number of imputed probes across subjects and probes in any study.8 One probe (cg25428494) in the Hannum algorithm was excluded from all studies because it was identified as a cross-reactive probe that binds with elements on the sex chromosomes (see above).
2.5.3 Data Analyses
We first evaluated the meta-correlations between Horvath and Hannum DNAm age estimates with chronological age and the correlations between Horvath and Hannum age estimates using the metacor package in R. Each study calculated DNAm age residuals by regressing each DNAm age estimate on chronological age and saving the unstandardized residuals from this equation. As described elsewhere (Wolf et al. 2016), when DNAm age is over-estimated relative to chronological age, this yields positive DNAm age residuals and can be conceptualized as accelerated DNAm age. Likewise, when DNAm age is under-estimated relative to chronological age, this is reflected in negative age residuals and is thought to denote decelerated cellular aging. These residualized variables were the primary dependent variables for use in the analyses. We examined the average age residual across studies and the correlation between the Horvath and Hannum age residuals. Analysts for each cohort then executed a series of multiple regression analyses, with the parameter coefficients from each cohort subsequently meta-analyzed. Specifically, the script included procedures to regress each DNAm age residual on (separately) childhood trauma, total lifetime trauma, current and lifetime PTSD diagnosis, and current and lifetime PTSD severity. In each of these six regressions (per each DNAm age metric), we covaried for WBC proportions, sex, and the three ancestry PCs reflecting either population stratification (in mixed ancestry samples) or population substructure (in homogenous ancestry samples). This allowed us to meta-analyze associations between these key demographic factors and advanced DNAm age. Interaction terms between age and each trauma exposure and PTSD variable and between sex and each trauma exposure and PTSD variable were computed and the interaction term added to each model predicting DNAm age residuals. This step was omitted for samples with no variability in sex and/or age. Regressions were protected F-tests given that all covariates and predictors (and interaction terms) were included in the same analysis; the difference in the exogenous variables across regressions was based on which trauma or PTSD variable was included, as determined by the available data for each cohort. All study-specific results were summarized and sent to the first author for meta-analysis.
Meta-analysis was conducted in R using the rma function from the metafor package (Viechtbauer, 2010). A random effects model was used to meta-analyze across cohorts. All regression parameter coefficients reported are unstandardized. To evaluate potential heterogeneity of effects, we also conducted a meta-analysis of interaction terms for the moderating influence of sex and chronological age on the association between each traumatic stress variable and DNAm age residuals. Finally, to explore potential methodological sources of variation on meta-analytic results, we meta-analyzed across studies that used the same assessment tool (i.e., the most common measure employed to assess each phenotype of interest across studies). For childhood trauma, we meta-analyzed across the subset of studies using the CTQ (n = 487 across 3 studies), as differential effects for the CTQ compared to other childhood trauma measures have previously been reported (Polanczyk et al., 2009). Separately, we also meta-analyzed results across studies that employed select childhood items from the Traumatic Life Events Questionnaire (TLEQ; n = 1,295 across 4 studies). For lifetime trauma exposure, we focused on studies that employed the TLEQ (n = 1,296 across 4 studies) and for current PTSD diagnosis and severity, we focused on studies that employed the gold-standard Clinician Administered PTSD Scale (CAPS; diagnosis: n = 1,216 across 4 studies; severity: n = 1,210 across 4 studies). As the majority of the studies (3 out of 5 for PTSD diagnosis; 3 out of 4 for PTSD severity) with any lifetime PTSD diagnostic and/or severity data used the CAPS, we did not prioritize a CAPS-specific lifetime PTSD analysis in the results, but instead footnote these results for completeness.
Given that parallel tests were conducted for the Horvath and Hannum algorithms, we corrected the p-value threshold in a manner that also took into account the correlation between the two metrics by adjusting for 1.8 tests (adjusted p-value threshold = .028),9 though we note that prior studies examining both metrics have not adjusted for multiple sets of tests (e.g., Chen et al., 2017; Quach et al., 2017, Marioni et al., 2015). For significant effects of interest (i.e., those relating to trauma and PTSD), we report the I2 statistic, an index of the proportion of the variance across studies that is due to true heterogeneity across populations (i.e., not an absolute index of the heterogeneity of effect size; Borenstein et al., 2017). We include this statistic for completeness, but note that caution is warranted in interpreting it because meta-analyses with a small number of contributing cohorts tend to yield inexact and systematically biased values (von Hippel et al., 2015).
3.1 Results
3. 2 Associations between DNAm Age and Chronological Age and across Hannum and Horvath Models
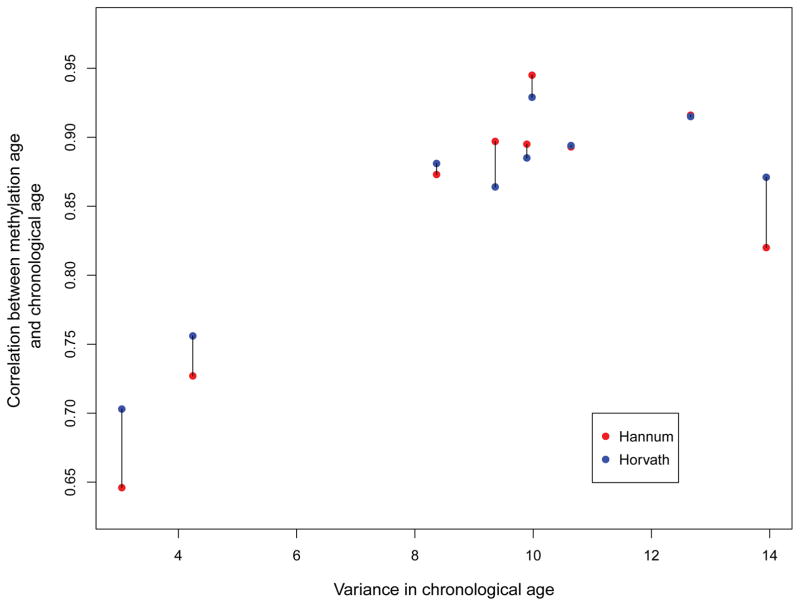
The meta-correlation between Hannum DNAm age and chronological age was r = .87 (SD = .09). The meta-correlation between Horvath DNAm age and chronological age was r = .87 (SD = .07; Table 2). In general, the strength of this correlation was associated with the variance in chronological age in each sample. This effect is shown in Figure 1, which plots the association between the variance in age in each sample and the correlation between DNAm age and chronological age for both DNAm age algorithms (linked via barbell for each study). The Hannum and Horvath age estimates correlated with each other at r = .89 in the meta-analysis. Across studies, the mean Hannum age residual was < .001 years (SD = 4.33 years, range: −12.21 to 15.68 years), and the mean Horvath age residual was < .001 years (SD = 4.33 years, range: −12.98 to 15.71 years). Upon meta-analysis, the Hannum and Horvath age residuals were moderately correlated with each other (r = .56). Details on these statistics are provided in Table 2.
Table 2.
Associations between DNAm Age, Chronological Age, and DNAm Age Residuals and Descriptive Statistics for DNAm Age Residuals
| DNAm Age with Chron. Age | Horvath with Hannum DNAm Age | M DNAm Age Residuals | Horvath with Hannum DNAm Age Residuals | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| Hannum | Horvath | r | Hannum | Horvath | ||||||
|
|
|
|||||||||
| r | r | M | SD | Range | M | SD | Range | r | ||
| NCPTSD | .893 | .894 | .878 | <.001 | 4.84 | −14.37 to 14.72 | <.001 | 4.11 | −15.91 to 25.85 | .392 |
| TRACTS | .873 | .881 | .883 | <.001 | 4.24 | −11.84 to 19.23 | <.001 | 3.82 | −13.00 to 16.99 | .493 |
| MRS | .646 | .703 | .722 | <.001 | 3.04 | −7.97 to 8.67 | <.001 | 3.42 | −9.75 to 8.67 | .493 |
| Army STARRS | .727 | .756 | .756 | <.001 | 3.52 | −9.12 to 9.67 | <.001 | 4.12 | −13.71 to 11.23 | .460 |
| MIRECC-a | .895 | .885 | .932 | <.001 | 3.92 | −9.11 to 12.22 | <.001 | 3.95 | −11.40 to 8.57 | .671 |
| MIRECC-b | .897 | .864 | .928 | <.001 | 4.07 | −11.34 to 11.32 | <.001 | 4.55 | −12.11 to 13.39 | .688 |
| PRISMO | .945 | .929 | .932 | <.001 | 3.23 | −7.89 to 7.89 | <.001 | 3.98 | −9.30 to 11.15 | .448 |
| DNHS | .820 | .871 | .908 | <.001 | 6.81 | −18.85 to 37.04 | <.001 | 5.65 | −17.96 to 24.70 | .690 |
| Grady Trauma Project (GTP) | .916 | .915 | .930 | <.001 | 5.32 | −19.41 to 20.33 | <.001 | 5.03 | −13.68 to 20.81 | .565 |
| Meta stats | .87 (.82 to .90) | .87 (.83 to .90) | .89 (.85 to .92) | < .001 | 4.33 | −12.21 to 15.68 | < .001 | 4.33 | −12.98 to 15.71 | .56 (.47 to .63) |
Note. Study name abbreviations are found in Table 1. Mean Hannum and Horvath DNAm age residuals are < .001 because, on average, estimated DNAm age was very close to chronological age. Chron = chronological; DNAm = DNA methylation.
Figure 1.
shows the relationship between variance in each study in chronological age and the magnitude of the correlation between chronological age and Horvath and Hannum DNAm age estimates (linked via barbell for each study).
3.3 Associations between DNAm Age Residuals and Demographic and Cellular Variables
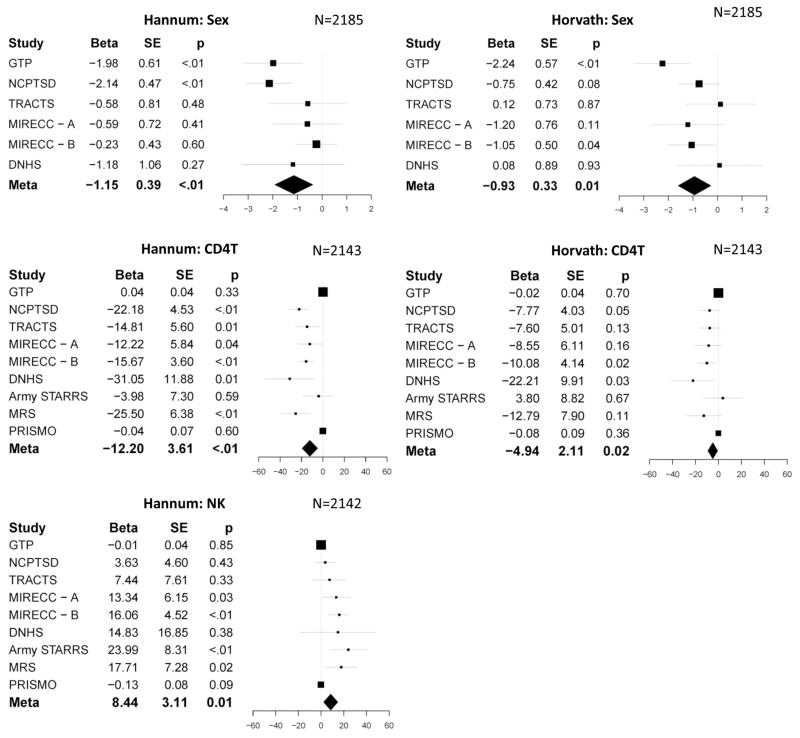
Tables 3 and S1 show the patterns of association between DNAm age residuals and sex, ancestry, and WBC proportions, meta-analyzed from the covariate portion of the regression equations. Of note, at the meta-analytic level (Table 3), there were significant associations between sex and both DNAm age residuals, such that women had decelerated aging relative to men. CD4 T cell proportions were inversely associated with both DNAm age residuals, and NK cell counts were positively associated with Hannum DNAm age residuals. Forest plots for these significant meta-analytic effects are shown in Figure 2.
Table 3.
Meta-analytic associations between Demographic, Cellular, and Traumatic Stress Variables in Association with DNAm Age Residuals
| N | Hannum | Horvath | |||||
|---|---|---|---|---|---|---|---|
| beta | se | p | beta | se | p | ||
| Sex | 2185 | −1.1455 | 0.389282 | 0.003255 | −0.93492 | 0.334843 | 0.005236 |
| PC1 | 2152 | 0.23162 | 2.367165 | 0.922054 | 1.0379 | 2.391098 | 0.664239 |
| PC2 | 2152 | 0.47236 | 2.046801 | 0.817486 | −0.24999 | 2.016886 | 0.901355 |
| PC3 | 2152 | 1.666092 | 2.499842 | 0.505105 | 2.673311 | 2.79451 | 0.338754 |
| CD8T | 2158 | −0.00293 | 0.050745 | 0.954011 | 2.946052 | 2.345547 | 0.209109 |
| CD4T | 2143 | −12.1967 | 3.610022 | 0.000729 | −4.94381 | 2.107359 | 0.018978 |
| Bcell | 2127 | 0.031993 | 0.043265 | 0.459626 | −5.40386 | 4.355332 | 0.214699 |
| NK | 2142 | 8.444542 | 3.10706 | 0.006571 | −0.08119 | 0.083031 | 0.328179 |
| Mono | 2170 | 0.139378 | 0.109426 | 0.20276 | 0.034167 | 0.107404 | 0.750396 |
| Childhood Trauma | 2009 | 0.060947 | 0.096259 | 0.526634 | −0.03471 | 0.110594 | 0.753619 |
| Childhood Trauma (CTQ) | 487 | 0.459596 | 0.209814 | 0.028488 | −0.11822 | 0.237175 | 0.618178 |
| Childhood Trauma (TLEQ) | 1295 | −0.1468 | 0.173309 | 0.396967 | −0.16058 | 0.132627 | 0.225986 |
| Lifetime Trauma | 2024 | 0.017713 | 0.036284 | 0.625429 | −0.05254 | 0.033963 | 0.121898 |
| Lifetime Trauma (TLEQ) | 1296 | −0.02009 | 0.040609 | 0.620864 | −0.05992 | 0.043818 | 0.171457 |
| Current PTSD Dx | 2104 | 0.12899 | 0.189075 | 0.495103 | −0.11764 | 0.193403 | 0.542997 |
| Current PTSD Sev | 2081 | 0.004478 | 0.002969 | 0.131459 | 0.000469 | 0.003043 | 0.877467 |
| Current PTSD Dx (CAPS) | 1216 | 0.341131 | 0.262793 | 0.194254 | 0.010832 | 0.251552 | 0.965654 |
| Current PTSD Sev (CAPS) | 1210 | 0.008573 | 0.004587 | 0.061615 | 0.004787 | 0.004356 | 0.271811 |
| Lifetime PTSD Dx | 1371 | 0.529147 | 0.276291 | 0.05547 | 0.122311 | 0.410977 | 0.766 |
| Lifetime PTSD Sev | 1251 | 0.011099 | 0.004608 | 0.01601 | 0.005694 | 0.006226 | 0.3604 |
Note. Significant effects are shown in bold font. DNAm = DNA methylation; PC = principal component; NK = natural killer cells; mono = monocytes; CTQ = Childhood Trauma Questionnaire; TLEQ = Traumatic Life Events Questionnaire; PTSD = posttraumatic stress disorder; dx = diagnosis; sev = severity; CAPS = Clinician Administered PTSD Scale.
Figure 2.
shows the forest plots for the significant meta-analytic associations between Hannum (left panel) and Horvath (right panel) DNAm age residuals and demographic and cellular variables. Error bars represent 95% confidence intervals. Study abbreviations are defined in Table 1.
3.4 Associations between Trauma, PTSD, and DNAm Age Residuals
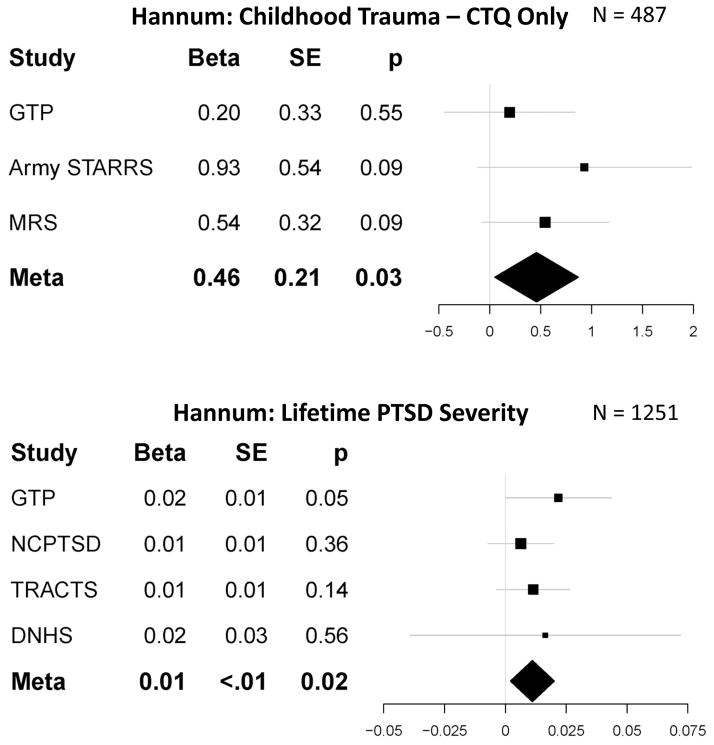
Tables S2 and 3 show the individual and meta-analytic results, respectively, for the regressions of each DNAm age residual on the trauma and PTSD variables. There were no significant meta-analytic effects of childhood (or lifetime) trauma exposure on either DNAm age residual (smallest p = .12) when evaluating all metrics of trauma exposure across all studies. To evaluate potential methodological sources of variability (see above), we then examined associations between childhood trauma as assessed by the CTQ (3 studies) and separately, items on the TLEQ (4 studies). This analysis revealed that CTQ-defined childhood trauma exposure was positively associated with Hannum DNAm age residuals, with overlapping 95% confidence intervals for the magnitude of the effect across cohorts (meta β = .46, p = .028 which was exactly equal to the adjusted threshold, I2 = 0, see Figure 3, Tables 3, S3) while there was no significant effect for studies utilizing the TLEQ (Tables 3, S3). No significant effects emerged when limiting the evaluation of lifetime trauma history to the TLEQ (Tables 3, S3).
Figure 3.
shows the forest plots for the significant meta-analytic associations between childhood trauma (top panel) and lifetime PTSD severity (bottom panel) and Hannum DNAm age residuals. Error bars represent 95% confidence intervals. Study abbreviations are defined in Table 1.
Meta-analyses revealed a near significant association between lifetime PTSD diagnosis and Hannum DNAm age residuals (meta β = .53, p = .06, total n = 1,371) and a significant association between lifetime PTSD severity and Hannum DNAm age residuals (meta β = .01, p = .016, I2 = 0, total n = 1,317, see Tables 3, S2, Figure 3), with overlapping 95% confidence intervals across the cohorts.10 There were no significant associations between current PTSD diagnosis or severity and DNAm age residuals (smallest p = .13; Tables 3, S2). The CAPS-specific association between current PTSD severity and Hannum DNAm age residuals just missed the unadjusted threshold for statistical significance (p = .06); there was no significant association for current PTSD diagnosis when limiting the analysis to studies using the CAPS (Tables 3, S3).
All significant trauma and PTSD effects noted above were with the Hannum DNAm age residuals. There were no significant associations between any of the trauma or PTSD variables and Horvath DNAm age residuals (Tables 3, S2, S3). There were also no significant meta-analytic interaction effects to suggest a moderating influence of sex or chronological age on the associations between trauma and PTSD with either set of DNAm age residuals (smallest p = .10; Table S4).
4.1 Discussion
This was the largest and most demographically heterogeneous evaluation of the associations between trauma exposure, PTSD, and accelerated aging in DNAm, spanning nine studies and over 2,000 participants. Results of meta-analyses suggested that both childhood trauma exposure (when assessed with the CTQ) and lifetime PTSD severity (assessed with the CAPS for the majority of studies with lifetime PTSD severity data), were associated with accelerated epigenetic aging. Associations between lifetime PTSD diagnosis and current PTSD severity (when assessed with the CAPS) and DNAm age residuals just failed to meet the unadjusted threshold for statistical significance, though the direction of the effect was consistent with those for lifetime PTSD severity and CTQ-measured childhood trauma. Though there was variability in the p-values associated with each cohort, collectively, results suggest an overlapping pattern of association between traumatic stress and acceleration of the pace of the epigenetic clock, albeit the relationship was modest in magnitude. Each additional exposure to a new type of childhood trauma on the CTQ was associated with nearly a half-year of age acceleration; the difference with respect to age acceleration between someone with a score of 0 on the CAPS versus a highly symptomatic patient with a score of 100 would be expected to be about 1.1 years. Small effects in DNAm studies are commonplace and it is critical to next examine the functional effects of such differences, which may be considerable with respect to gene expression, and to model the potential cumulative effects of advanced DNAm over time (Breton et al., 2017).
As many as one-third of patients with PTSD exhibit a chronic form of the disease that persists for years (Kessler, 2000) and the chronicity and severity of such symptoms would be expected to magnify the negative health correlates of PTSD. Indeed, one study that modeled PTSD burden as defined by both symptom severity and duration (e.g., chronicity) found that the burden of the disease evidenced stronger negative associations with cortical thickness than did PTSD severity alone (Lindemer et al., 2013). Based on this, it is possible that effect size estimates in this study underestimate the association between traumatic stress and cellular age because they do not account for the burden of traumatic stress across time. Future research may benefit from quantifying the burden of PTSD as a function of both disorder severity and chronicity. Consistent with this interpretation, we found that childhood, but not lifetime, trauma exposure was associated with accelerated aging and this could suggest: (a) that there are critical windows during childhood in which the effects of trauma exposure are particularly damaging; and/or (b) that childhood trauma could be a marker for a more prolonged period of psychiatric distress that contributes to overall greater burden. Longitudinal studies are necessary to evaluate these possibilities.
Associations between traumatic stress and advanced epigenetic age were observed for the Hannum et al. algorithm but not for the Horvath metric. To our knowledge, prior positive associations between PTSD and epigenetic age have only been shown for the Hannum et al. metric (Wolf et al., 2016, 2017), while positive effects for general life stressors, violence, and trauma exposure have previously been reported for the Horvath index (Zannas et al., 2015; Boks et al., 2015; Jovanovic et al., 2017). It is remarkable that the two algorithms show differential patterns of association given that they are highly correlated with each other and with chronological age. Despite these strong correlations, the components of each age algorithm that do not index chronological age, as captured by the age residuals, appear to be fairly distinct from each other: the two age residuals shared approximately 25% of the variance and evidenced differential patterns of association with the WBCs in this study. The two algorithms have just 6 loci and 11 genes in common (Wolf et al., 2016) and together with the results of this study, this implies that each algorithm may be sensitive to different pathogenic environmental and biological processes, though more research is needed to test this. The strong meta-analytic correlations between both DNAm age estimates and chronological age in the context of the relationship between variance in age in each sample and the strength of the DNAm age/chronological age correlation (Figure 1) are also informative. This supports the accuracy of the DNAm age estimates and raises the possibility that in instances in which the correlation between DNAm age and chronological age is weaker than expected, that this may be due to limited age variance in the sample, and may not necessarily reflect a failure in the algorithm.
Results of this study underscore the importance of identifying the biological mechanisms that link traumatic stress to age acceleration. Zannas et al. (2015) examined the responsivity of the loci included in the Horvath algorithm to the glucocorticoid receptor agonist dexamethasone and found that approximately 31% of the DNAm loci were responsive to dexamethasone while over 80% of genes located near the DNAm loci evidenced dexamethasone-related changes in gene transcription. Consistent with this, about a quarter of the DNAm loci were shown to be localized to glucocorticoid response elements. As well, Davis et al. (2017) found advanced Horvath DNAm age to be correlated with diurnal cortisol levels. This pattern of results is consistent with a central role of stress hormones in PTSD (Yehuda, 2009). Dexamethasone-regulated genes also showed enrichment in gene networks associated with age-related diseases (i.e., coronary artery disease, arteriosclerosis, leukemia, etc.; Zannas et al., 2015). This raises the possibility that accelerated cellular aging in DNAm may link traumatic stress to increased risk for pre-mature disease onset.
Complementary research on the loci included in the Hannum algorithm has yet to be conducted and comparing the responsivity of these loci to those in the Horvath algorithm is critical for understanding differential patterns of association across the two metrics. As well, evaluation of the sensitivity of the loci to other dynamic biological processes implicated in PTSD, such as pro-inflammatory cytokines (Passos et al., 2015), and catecholamines (Highland et al., 2015), would help to elucidate the biological mechanisms involved in traumatic-stress related accelerated aging. This type of evaluation is also important for determining whether epigenetic age acceleration is a mechanism for, or simply a biomarker of, early health decline. Behavioral pathways to epigenetic aging are also critical to evaluate further, given evidence that insomnia (which is also a symptom of PTSD) and obesity (which is highly comorbid with PTSD; Pagoto et al., 2012) are associated with accelerated DNAm age (Carroll et al., 2017; Nevalainen et al., 2017).
We also observed meta-analytic effects suggesting that women, on average, had decelerated aging relative to men (using both indices). This has been reported previously in blood and brain tissue (Horvath et al., 2016; Hannum et al. 2013) but the interpretation of the effect is unclear. This finding could reflect an underlying differential rate of epigenetic aging in men versus women, and/or differential susceptibility to factors that cause epigenetic age to deviate from chronological age across the sexes. These explanations are consistent with the common finding that women tend to outlive men (Beltrán-Sánchez et al., 2015), perhaps because their pace of cellular aging is attenuated. On the other hand, the finding could indicate that the age algorithms are simply more accurate in one sex over the other. There is preliminary evidence to suggest that differential age acceleration across the sexes does not alter the strength of the association between epigenetic age and important clinical correlates, including traumatic stress in this study, and time to death in prior work (Chen et al., 2016). This suggests that regardless of the reason for the differential age acceleration across sexes, the predictive power of accelerated epigenetic age is not diminished in one sex compared to the other.
We also observed a negative relationship between estimates of CD4 T cells and both DNAm age residuals and a positive association between NK cells and Hannum age residuals. Similar patterns of association were reported in meta-analyses of 13,000 individuals by Chen et al. (2016) and in over 4,600 individuals by Marioni et al. (2015). These findings raise the possibility that cells that are integral to immune system maintenance and responsivity play a role in altering the pace of the epigenetic clock so that it loses synchronicity with the pace of chronological aging. Alternatively, Chen et al. and Horvath et al. (2016) combined information from age residuals and estimated WBC counts and found that this metric was a better predictor of time to death (Chen et al. 2016) and metabolic and inflammatory markers (Horvath et al., 2016) compared to the DNAm age residuals alone. The combined index was referred to as “extrinsic epigenetic age acceleration” and conceptualized as a marker of the biological age of the immune system in blood (Chen et al. 2016), however, it was not clear which components of this combined metric were responsible for the increased predictive strength of time till death. Studies of the responsivity of DNAm age probes to inflammatory and anti-inflammatory agents could shed light on how inflammation is associated with the DNAm age clock.
4.2 Study Limitations
These results should be considered in light of a number of limitations. First, this was a cross-sectional study and, as such, we cannot infer causal associations between traumatic stress and accelerated DNAm age. Second, given that the contributing studies had diverse available data, there were different samples comprising each meta-analysis. This also meant that we were unable to discern the relative effects of childhood trauma exposure versus lifetime PTSD severity as only one study (Grady Trauma Project) had both the CTQ childhood trauma exposure variable and lifetime PTSD severity. For the same reasons, we were unable to include childhood trauma and adult trauma exposure in the same model or to evaluate the influence of chronic interpersonal violence across the lifetime. Thus, as this line of work is developed and new data become available, it will be important to disentangle the relative effects of trauma exposure versus PTSD on accelerated aging. As this meta-analysis was based on a relatively nascent field with fewer than 10 contributing studies, statistical power was also a potential limitation. However, power for meta-analyses based on summary data across individual studies has been shown to be similar to power for analyses in which data from distinct studies are pooled to form one large sample (Olkin & Sampson, 1998; Mathew & Nordstrom, 1999; Lin & Zeng, 2010) and this applies to genetic association studies as well (Lin & Zeng, 2010; Sung et al., 2014). Our pooled sample size should have provided adequate power in this context, though inclusion of more cohorts into the meta-analysis would strengthen results. As well, while we attempted to look for sources of demographic variation in the results, there are undoubtedly other variables that we did not have access to that might moderate the primary associations (or otherwise serve as important covariates). This concern is offset by the strength of the meta-analytic approach which yields improved statistical power and generalizability of results across populations relative to an individual cohort.
4. 3 Conclusions
Evaluation of epigenetic cellular aging is in its infancy, with the vast majority of research to date focused on the utility of the DNAm age calculators to predict mortality and health outcome. This study was the largest to evaluate traumatic stress-related advanced epigenetic age, and to our knowledge, was the largest examination of any psychiatric symptom domain in association with DNAm age to date. By deploying a uniform data analytic script and harmonizing critical variables, our analytic approach was able to eliminate some sources of methodological noise compared to more traditional meta-analyses that are based on published summary statistics. Results suggested that childhood trauma (when assessed with a detailed instrument) and lifetime PTSD severity were both associated with advanced DNAm age, though the magnitude of the effect was small and additional research is needed to determine how the chronicity of psychiatric symptoms might contribute to accelerated aging. As genome-wide and epigenome-wide testing move from the research to the clinical domains, assessment of epigenetic age may become a practical approach for tracking an individual’s cellular age, and testing the utility of interventions designed to slow the pace of cellular aging. In the future, we may develop traumatic-stress specific epigenetic age profiles that contribute to our understanding of the neurobiological consequences of trauma and PTSD.
Supplementary Material
Acknowledgments
The Psychiatric Genomics Consortium (PGC) PTSD Epigenetics Workgroup is supported by the U.S. Army Medical Research and Materiel Command and the National Institute of Mental Health (NIMH; R01MH108826).
This work was supported in part by: Merit Review Award Number I01 CX-001276-01 (EJW) from the United States (U.S.) Department of Veterans Affairs Clinical Sciences R&D (CSR&D) Service, National Institute On Aging of the National Institutes of Health under Award Number R03AG051877 (EJW), Presidential Early Career Award for Scientists and Engineers to EJW as administered by U.S. Department of Veterans Affairs Office of Research and Development (PECASE 2013A).
Nicole Nugent’s effort on this project was supported by NIMH R01MH105379 & R01MH108641.
Funding for the TRACTS study was supported in part by NIMH grant R21MH102834 “Neuroimaging Genetics of PTSD” to MWM and the Translational Research Center for TBI and Stress Disorders (TRACTS), a VA Rehabilitation Research and Development Traumatic Brain Injury Center of Excellence (B9254-C). This research is the result of work supported with resources and the use of facilities at the Pharmacogenomics Analysis Laboratory, Research and Development Service, Central Arkansas Veterans Healthcare System, Little Rock, Arkansas.
Funding for the NCPTSD cohort was supported by U.S. Department of VA CSRD Merit Review award 5I01CX000431-02 and by U.S. Department of VA Biomedical Laboratory Research & Development Program award 1I01BX002150-01, both to MWM. This project was also supported by NIMH RO1 MH079806 to MWM.
Funding for the Mid-Atlantic MIRECC cohorts was provided in part by grant #I01 BX002577 from the Biomedical and Laboratory Research and Development (BLR&D) Service of the Department of Veterans Affairs Office of Research and Development (VA ORD), grant #IK2CX000525 and #11S-RCS-009 from the CSR&D Service of VA ORD, and the VA Mid-Atlantic Mental Illness Research, Education, and Clinical Center (MIRECC).
Funding for MRS was provided by the Marine Corps, Navy Bureau of Medicine and Surgery (BUMED), VA Health Research and Development (HSR&D) and NIH R01MH093500. Acknowledged are Mark A Geyer Ph.D (VA San Diego Healthcare System & UCSD), Daniel T. O’Connor (UCSD) and all MRS investigators and staff. The authors also thank the Marine and Navy Corpsmen volunteers for military service and participation in MRS.
Funding for DNHS was provided by NIH R01DA022720 and RC1MH088283. We thank the many Detroit residents who chose to participate in the DNHS, as well as the technical and support staff who assisted with the study.
The Grady Trauma Project, which appreciates the technical support of all of the staff, volunteers and participants, is supported by the National Institutes of Mental Health (MH096764 and MH071537).
The VA Mid-Atlantic MIRECC Workgroup contributors for this publication include: Mira Brancu, Patrick S. Calhoun, Kathleen P. Decker, Eric Dedert, Eric B. Elbogen, John A. Fairbank, Kimberly T. Green, Robin A. Hurley, Jason D. Kilts, Angela Kirby, Christine E. Marx, Gregory McCarthy, Scott D. McDonald, Marinell Miller-Mumford, Scott D. Moore, Rajendra A. Morey, Jennifer C. Naylor, Treven C. Pickett, Jared Rowland, Jennifer J. Runnals, Cindy Swinkels, Steven T. Szabo, Katherine H. Taber, Larry A. Tupler, Elizabeth E. Van Voorhees, H. Ryan Wagner, Richard D. Weiner, and Ruth E. Yoash-Gantz.
Army STARRS was sponsored by the Department of the Army and funded under cooperative agreement number U01MH087981 with the U.S. Department of Health and Human Services, National Institutes of Health, National Institute of Mental Health (NIH/NIMH).
The contents of this article do not represent the views of the U.S. Department of Veterans Affairs, the National Institutes of Health, or the United States Government.
Footnotes
While the primary variable of interest in this study captured the extent to which DNAm age deviated from chronological age along a spectrum ranging from under-predicted age to over-predicted age, for the sake of simplicity, we refer here to the over-predicted/accelerated end of this spectrum.
Examination of this cohort by Wolf et al. (2017) focused on a subset of 339 veterans, employed a different definition of PTSD than that used here, and did not evaluate the Horvath index.
Examination of this cohort by Wolf et al. (2016) employed a different definition of PTSD than that used here.
Examination of this cohort by Boks et al. (2015) was based on a larger sample, did not include evaluation of the Hannum DNAm age index, and was based on a different operational definition of the association between DNAm age and chronological age than that employed here.
The number of controls differs as a function of which datasets had current and/or lifetime PTSD diagnostic data. For analyses involving lifetime PTSD, we only included cohorts that specifically assessed history of the disorder (worst period of symptoms) as it is unknown if current controls had a history of PTSD and we had no index of the severity of past symptoms unless the study explicitly assessed lifetime symptoms.
One study (DNHS) combined items across the CTQ with another measure (Table 1) and therefore was not included in CTQ-specific analyses. Army STARRS used a near-identical version of the CTQ and was included in CTQ-specific analyses.
Details concerning the use of the Infinium beadchip in the DNHS cohort are not yet available in the published literature, thus the references listed in Table 1 for this cohort are included to provide a more general overview of the study methodology. An overview of the DNAm processing pipeline employed for all cohorts included in this meta-analysis are provided in the methods section of this paper and in Ratanatharathorn et al. (2017).
Information on missing probe values is not output by the Horvath website and thus we were only able to obtain this information for the Hannum et al. algorithm.
Specifically, using a permutation testing procedure for use in raw data (see Miller et al., 2015), we determined the effective number of tests given the correlation across the Hannum and Horvath age residuals as evaluated in the TRACTS data cohort (raw data from that cohort were available to the first and last author and this procedure can only be performed on raw, not summarized, data). This revealed that the adjusted p-value represented 1.8 tests based on the r =.49 DNAm age residual association in that dataset.
When these two analyses were restricted to the three studies that employed the CAPS for lifetime PTSD assessment, results were largely unchanged: meta β for lifetime PTSD dx = .58, p = .066; meta β for lifetime PTSD severity = .01, p = .019.
Disclosures
In the past 3 years, Dr. Kessler received support for his epidemiological studies from Sanofi Aventis; was a consultant for Johnson & Johnson Wellness and Prevention, Sage Pharmaceuticals, Shire, Takeda; and served on an advisory board for the Johnson & Johnson Services Inc. Lake Nona Life Project. Kessler is a co-owner of DataStat, Inc., a market research firm that carries out healthcare research.
All other authors report no financial conflicts of interest relevant to this work.
References
- 1.Afari N, Ahumada SM, Wright LJ, Mostoufi S, Golnari G, Reis V, Cuneo JG. Psychological trauma and functional somatic syndromes: a systematic review and meta-analysis. Psychosom Med. 2014;76:2. doi: 10.1097/PSY.0000000000000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews G, Peters L. The psychometric properties of the composite international diagnostic interview. Soc Psychiatry Psychiatr Epidemiol. 1998;33:80–8. doi: 10.1007/s001270050026. [DOI] [PubMed] [Google Scholar]
- 3.Ashley-Koch AE, Garrett ME, Gibson J, Liu Y, Dennis MF, Kimbrel NA, Beckham JC, Hauser MA Veterans Affairs Mid-Atlantic Mental Illness Research, Education, and Clinical Center Workgroup. Genome-wide association study of posttraumatic stress disorder in a cohort of Iraq-Afghanistan era veterans. J Affect Disord. 2015;184:225–34. doi: 10.1016/j.jad.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker DG, Nash WP, Litz BT, Geyer MA, Risbrough VB, Nievergelt CM, O’Connor DT, Larson GE, Schork NJ, Vasterling JJ, Hammer PS, Webb-Murphy JA MRS Team. Predictors of risk and resilience for posttraumatic stress disorder among ground combat Marines: methods of the Marine Resiliency Study. Prev Chronic Dis. 2012;9:E97. doi: 10.5888/pcd9.110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R, Klengel T, Mehta D, Binder EB, Epstein MP, Ressler KJ, Conneely KN. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38:231–241. doi: 10.1002/gepi.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrán-Sánchez H, Finch CE, Crimmins EM. Twentieth century surge of excess adult male mortality. Proc Natl Acad Sci U S A. 2015;112:8993–8. doi: 10.1073/pnas.1421942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T, Stokes J, Handelsman L, Medrano M, Desmond D, Zule W. Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl. 2003;27:169–90. doi: 10.1016/s0145-2134(02)00541-0. [DOI] [PubMed] [Google Scholar]
- 8.Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, Schwartz AC, Cubells JF, Ressler KJ. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM. The development of a Clinician-Administered PTSD Scale. J Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- 10.Boks MP, van Mierlo HC, Rutten BP, Radstake TR, De Witte L, Geuze E, Horvath S, Schalkwyk LC, Vinkers CH, Broen JC, Vermetten E. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology. 2015;51:506–12. doi: 10.1016/j.psyneuen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Borenstein M, Higgins JP, Hedges LV, Rothstein HR. Basics of meta-analysis: I2 is not an absolute measure of heterogeneity. Res Synth Methods. 2017;8:5–18. doi: 10.1002/jrsm.1230. [DOI] [PubMed] [Google Scholar]
- 12.Bremner JD, Bolus R, Mayer EA. Psychometrics properties of the Early Trauma Inventory-Self Report. J Nerv Ment Dis. 2007;195:211–8. doi: 10.1097/01.nmd.0000243824.84651.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breslau N, Kessler RC, Chilcoat HD, Schultz LR, Davis GC, Andreski P. Trauma and posttraumatic stress disorder in the community: the 1996 Detroit Area Survey of Trauma. Arch Gen Psychiatry. 1998;55:626–32. doi: 10.1001/archpsyc.55.7.626. [DOI] [PubMed] [Google Scholar]
- 14.Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, Herbstman J, Holland N, LaSalle JM, Schmidt R, Yousefi P, Perera F, Joubert BR, Taylor M, Yang IV, Chen R, Hew KM, Freeland DM, Miller R, Murphy SK. Small-magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: the children’s environmental health and disease prevention research center’s epigenetics working group. Environ Health Perspect. 2017;125:511–26. doi: 10.1289/EHP595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll JE, Irwin MR, Levine M, Seeman TE, Absher D, Assimes T, Horvath S. Epigenetic aging and immune senescence in women with insomnia symptoms: findings from the Women’s Health Initiative Study. Biol Psychiatry. 2017;81:136–144. doi: 10.1016/j.biopsych.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, Roetker NS, Just AC, Demerath EW, Guan W, Bressler J, Fornage M, Studenski S, Vandiver AR, Moore AZ, Tanaka T, Kiel DP, Liang L, Vokonas P, Schwartz J, Lunetta LL, Murabito JM, Bandinelli S, Hernandez DG, Melzer D, Nalls M, Pilling LC, Price TR, Singleton AB, Gieger C, Holle R, Kretschmer A, Kronenberg F, Kunze S, Linseisen J, Meisinger C, Rathmann W, Waldenberger M, Visscher PM, Shah S, Wray NR, McRae AF, Franco OH, Hofman A, Uitterlinden AG, Absher D, Assimes T, Assimes T, Levine ME, Lu AT, Feinberg AP, Levy D, Baccarelli A, van Meurs J, Bell JT, Peters A, Deary IJ, Pankow JS, Ferrucci L, Horvath S. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YA, Lemire M, Choufani S, Butcher DS, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, Christensen K. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–54. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis EG, Humphreys KL, McEwen LM, Sacchet MD, Camacho MC, MacIsaac JL, Lin DTS, Kobor MS, Gotlib IH. Accelerated DNA methylation in adolescent girls: associations with elevated diurnal cortisol and reduced hippocampal volume. Transl Psychiatry. 2017;7:e1223. doi: 10.1038/tp.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson JR, Book SW, Colket JT, Tupler LA, Roth S, David D, Hertzberg M, Mellman T, Beckham JC, Smith RD, Davison RM, Katz R, Feldman ME. Assessment of a new self-rating scale for post-traumatic stress disorder. Psychol Med. 1997;27:153–60. doi: 10.1017/s0033291796004229. [DOI] [PubMed] [Google Scholar]
- 22.First MB, Spitzer RL, Gibbon M, Williams JB. Structured clinical interview for Axis I DSM-IV disorders. New York: Biometrics Research; 1994. [Google Scholar]
- 23.Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, Weiss T, Schwartz AC, Cubells JF, Ressler KJ. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry. 2009;31:505–14. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray MJ, Litz BT, Hsu JL, Lombardo TW. Psychometric properties of the life events checklist. Assessment. 2004;11:330–41. doi: 10.1177/1073191104269954. [DOI] [PubMed] [Google Scholar]
- 25.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Kajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–67. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedtke KA, Ruggiero KJ, Fitzgerald MM, Zinzow HM, Sauders BE, Resnick HS, Kilpatrick DG. A longitudinal investigation of interpersonal violence in relation to mental health and substance use. J Consult Clin Psychol. 2008;76:633–47. doi: 10.1037/0022-006X.76.4.633. [DOI] [PubMed] [Google Scholar]
- 27.Highland KB, Costanzo M, Jovanovic T, Norrholm SD, Ndiongue R, Reinhardt B, Rothbaum B, Roy MJ. Biomarkers of post-deployment resilience among military service members. Neurobiol Stress. 2015;2:62–6. doi: 10.1016/j.ynstr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, Ritz BR, Chen B, Lu AT, Rickabaugh TM, Jamieson BD, Sun D, Li S, Chen W, Quintana-Murci L, Fagny M, Kobor MS, Tsao PS, Reiner AP, Edlefsen KL, Absher D, Assimes TL. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath S, Pirazzini C, Bacalini MG, Gentilini D, Di Blasio AM, Delledonne M, Mari D, Arosio B, Monti D, Passarino G, De Rango F, D’Aguila P, Giuliani C, Marasco E, Collino S, Descombes P, Garagnani P, Franceschi C. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany NY) 2015;7:1159–70. doi: 10.18632/aging.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hovens JE, Bramsen I, van der Ploeg HM. Self-rating inventory for posttraumatic stress disorder: review of the psychometric properties of a new brief Dutch screening instrument. Percept Mot Skills. 2002;94:996–1108. doi: 10.2466/pms.2002.94.3.996. [DOI] [PubMed] [Google Scholar]
- 33.Hovens JE, Bramsen I, van der Ploeg HM. Self Report Measure for PTSD Symptoms: SRIP Manual. Lisse; 2000. [Google Scholar]
- 34.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-side association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 36.Jovanovic T, Vance LA, Cross D, Knight AK, Kilaru V, Michopoulos V, Klengel T, Smith AK. Exposure to violence accelerates epigenetic aging in children. Sci Rep. 2017;7:8962. doi: 10.1038/s41598-017-09235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessler RC, Ustün TB. The World Mental Health (WMH) Survey Initiative version of the World Health Organization (WHO) Composite International Diagnostic Interview (CIDI) Int J Methods Psychiatr Res. 2004;13:93–121. doi: 10.1002/mpr.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessler RC. Posttraumatic stress disorder: the burden to the individual and to society. J Clin Psychiatry. 2000;61:4–12. [PubMed] [Google Scholar]
- 39.Kubany ES, Haynes SN, Leisen MB, Owens JA, Kaplan AS, Watson SB, Burns K. Development and preliminary validation of a brief broad-spectrum measure of trauma exposure: the Traumatic Life Events Questionnaire. Psychol Assess. 2000;12:210–224. doi: 10.1037//1040-3590.12.2.210. [DOI] [PubMed] [Google Scholar]
- 40.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. R package version 3. 2013. sva: Surrogate variable analysis. [Google Scholar]
- 41.Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY) 2015;7:690–700. doi: 10.18632/aging.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Wang J, Zhou J, Huang P, Li J. The association between post-traumatic stress disorder and shorter telomere: a systematic review and meta-analysis. J Affect Disord. 2017;218:322–326. doi: 10.1016/j.jad.2017.03.048. [DOI] [PubMed] [Google Scholar]
- 43.Lin DY, Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genet Epidemiol. 2010;34:60–6. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin DY, Zeng D. On the relative efficiency of using summary statistics versus individual-level data in meta-analysis. Biometrika. 2010;97:321–332. doi: 10.1093/biomet/asq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindemer ER, Salat DH, Leritz EC, McGlinchey RE, Milberg WP. Reduced cortical thickness with increased lifetime burden of PTSD in OEF/OIF Veterans and the impact of comorbid TBI. Neuroimage Clin. 2013;2:601–11. doi: 10.1016/j.nicl.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide association study of posttraumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol. Psychiatry. 2013;18:937–42. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohr JB, Palmer BW, Eidt CA, Aailaboyina S, Mausbach BT, Wolkowitz OM, Thorp SR, Jeste DV. Is post-traumatic stress disorder associated with premature senescence? A review of the literature. Am J Geriatr Psychiatry. 2015;23:709–25. doi: 10.1016/j.jagp.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marioni RE, Harris SE, Shah S, McRae AF, von Zglinicki T, Martin-Ruiz C, Wray NR, Visscher PM, Deary IJ. The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int J Epidemiol. 2016;45:424–432. doi: 10.1093/ije/dyw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, Pattie A, Corley J, Murphy L, Martin NG, Montgomery GW, Feinberg AP, Fallin MD, Multhaup ML, Jaffe AE, Joehanes R, Schwartz J, Just AC, Lunetta KL, Murabito JM, Starr JM, Horvath S, Baccarelli AA, Levy D, Visscher PM, Wray NR, Deary IJ. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathew T, Nordstrom K. On the equivalence of meta-analysis using literature and using individual patient data. Biometrics. 1999;55:1221–3. doi: 10.1111/j.0006-341x.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 51.McGlinchey RE, Milberg WP, Fonda JR, Fortier CB. A methodology for assessing deployment trauma and its consequences in OEF/OIF/OND veterans: the TRACTS longitudinal prospective cohort study. Int J Methods Psychiatr Res. 2017 doi: 10.1002/mpr.1556. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller MW, Sadeh N. Traumatic stress, oxidative stress and post-traumatic stress disorder: neurodegeneration and the acclerated-aging hypothesis. Mol Psychiatry. 2014;19:1156–62. doi: 10.1038/mp.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller MW, Wolf EJ, Sadeh N, Logue M, Spielberg JM, Hayes JP, Sperbeck E, Schichman SA, Stone A, Carter WC, Humphries DE, Milberg W, McGlinchey R. A novel locus in the oxidative stress-related gene ALOX12 moderates the association between PTSD and thickness of the prefrontal cortex. Psychoneuroendocrinology. 2016;62:359–65. doi: 10.1016/j.psyneuen.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nevalainen T, Kananen L, Marttila S, Jylhävä J, Mononen N, Kähönen M, Raitakari OT, Hervonen A, Jylhä M, Lehtimäki T, Hurme M. Obesity accelerates epigenetic aging in middle-aged but not in elderly individuals. Clin Epigenetics. 2017;9:20. doi: 10.1186/s13148-016-0301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nievergelt CM, Maihofer AX, Mustapic M, Yurgil KA, Schork NJ, Miller MW, Logue MW, Geyer MA, Risbrough VB, O’Connor DT, Baker DG. Genomic predictors of combat stress vulnerability and resilience in U.S. Marines: A genome-wide association study across multiple ancestries implicates PRTFDC1 as a potential PTSD gene. Psychoneuroendocrinology. 2015;51:459–71. doi: 10.1016/j.psyneuen.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 56.Olkin I, Sampson A. Comparison of meta-analysis versus analysis of variance of individual patient data. Biometrics. 1998;54:317–22. [PubMed] [Google Scholar]
- 57.Pagoto SL, Schneider KL, Bodenlos JS, Appelhans BM, Whited MC, May Y, Lemon SC. Association of post-traumatic stress disorder and obesity in a nationally representative sample. Obesity (Silver Spring) 2012;20:200–5. doi: 10.1038/oby.2011.318. [DOI] [PubMed] [Google Scholar]
- 58.Passos IC, Vasconcelos-Moreno MP, Costa LG, Kunz M, Brietzke E, Quevedo J, Salum G, Magalhães PV, Kapczinski F, Kauer-Sant’Anna M. Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. Lancet Psychiatry. 2015;2:1002–12. doi: 10.1016/S2215-0366(15)00309-0. [DOI] [PubMed] [Google Scholar]
- 59.Pidsley R, Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450k methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polanczyk G, Caspi A, Williams B, Price TS, Danese A, Sugden K, … Moffitt TE. Protective Effect of CRHR1 Gene Variants on the Development of Adult Depression Following Childhood Maltreatment: Replication and Extension. Archives of General Psychiatry. 2009;66:978–985. doi: 10.1001/archgenpsychiatry.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ratanatharathorn A, Boks MP, Maihofer AX, Aiello AE, Amstadter AB, Ashley-Koch AE, Baker DG, Beckham JC, Bromet E, Dennis M, Garrett ME, Geuze E, Guffman G, Hauser MA, Kilaru V, Kimbrel NA, Koenen KC, Kuan P, Logue MW, Luft BJ, Miller MW, Mitchell C, Nugent NR, Ressler KJ, Rutten BPF, Stein MB, Vermetten E, Vinkers CH, Youssef NA, Uddin M, Nievergelt CM, Smith AK VA Mid-Atlantic MIRECC Workgroup, PGC PTSD Epigenetics Workgroup. Epigenome-wide association of PTSD from heterogeneous cohorts with a common multi-site analysis pipeline. Am J of Med Genetics Part B: Neuropsychiatric Genetics. 2017;174:619–30. doi: 10.1002/ajmg.b.32568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribbe D. Psychometric review of traumatic event screening instrument for children (TESI-C) Measurement of stress, trauma, and adaptation. 1996:386–7. [Google Scholar]
- 63.Roberts AL, Koenen KC, Chen Q, Gilsanz P, Mason SM, Prescott J, Ratanatharathorn A, Rimm EB, Sumner JA, Winning A, De Vivo I, Kubzansky LD. Posttraumatic stress disorder and accelerated aging: PTSD and leukocyte telomere length in a sample of civilian women. Depress Anxiety. 2017;34:391–400. doi: 10.1002/da.22620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruggiero KJ, Del Ben K, Scotti JR, Rabalais AE. Psychometric properties of the PTSD Checklist-Civilian Version. J Trauma Stress. 2003;16:495–502. doi: 10.1023/A:1025714729117. [DOI] [PubMed] [Google Scholar]
- 65.Sadeh N, Spielberg JM, Logue MW, Wolf EJ, Smith AK, Lusk J, Hayes JP, Sperbeck E, Milberg WP, McGlinchey RE, Salat DH, Carter WC, Stone A, Schichman SA, Humphries DE, Miller MW. SKA2 Methylation is associated with decreased prefrontal cortical thickness and greater PTSD severity among trauma-exposed veterans. Mol Psychiatry. 2016;21:357–63. doi: 10.1038/mp.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnurr PP, Spiro A, 3rd, Paris AH. Physician-diagnosed medical disorders in relation to PTSD symptoms in older male military veterans. Health Psychol. 2000;19:91–97. doi: 10.1037//0278-6133.19.1.91. [DOI] [PubMed] [Google Scholar]
- 67.Sprang G. The traumatic experiences inventory (TEI): A test of psychometric properties. J Psychopathol Behav Assess. 1997;19:257–71. [Google Scholar]
- 68.Stein MB, Chen CY, Ursano RJ, Cai T, Gelernter J, Heeringa SG, Jain S, Jensen KP, Maihofer AX, Mitchell C, Nievergelt CM, Nock MK, Neale BM, Polimanti R, Ripke S, Sun X, Thomas ML, Wang Q, Ware EB, Borja S, Kessler RC, Smoller JW Army Study to Assess Risk and Resilience in Servicemembers (STARRS) Collaborators. Genome-wide Association Studies of Posttraumatic Stress Disorder in 2 Cohorts of US Army Soldiers. JAMA Psychiatry. 2016;73:695–705. doi: 10.1001/jamapsychiatry.2016.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Straus MA. Measuring intrafamily conflict and violence: the Conflict Tactics (CT) Scales. J Marriage Fam. 1979;41:75–78. [Google Scholar]
- 70.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, Cichon S, Edenberg HJ, Faraone SV, Gelernter J, Mathews CA, Nievergelt CM, Smoller J, O’Donovan MC. Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry. 2017 doi: 10.1176/appi.ajp.2017.17030283. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sung YJ, Schwander K, Arnett DK, Kardia SL, Rankinen T, Bouchard C, Boerwinkle E, Hunt SC, Rao DC. An empirical comparison of meta-analysis and mega-analysis of individual participant data for identifying gene-environment interactions. Genet Epidemiol. 2014;38:369–78. doi: 10.1002/gepi.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Touleimat N, Tost J. Complete pipeline for Infinium(®) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–41. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 73.Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Goldmann E, Galea S. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci. 2010;107:9470–5. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ursano RJ, Colpe LJ, Heeringa SG, Kessler RC, Schoenbaum M, Stein MB Army STARRS collaborators. The army study to assess risk and resilience in Servicemembers (Army STARRS) Psychiatry. 2014;77:107–119. doi: 10.1521/psyc.2014.77.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Zuiden M, Geuze E, Willemen HL, Vermetten E, Maas M, Heijnen CJ, Kavelaars A. Pre-existing high glucocorticoid receptor number predicting development of posttraumatic stress symptoms after military deployment. Am J Psychiatry. 2011;168:89–96. doi: 10.1176/appi.ajp.2010.10050706. [DOI] [PubMed] [Google Scholar]
- 76.Viechtbauer W. Conducting meta-analyses in R with the metafor package. Journal of Statistical Software. 2010;36:1–48. [Google Scholar]
- 77.Von Hippel PT. The heterogeneity statistic I2 can be biased in small meta-analyses. BMC Med Res Methodol. 2015;15:35. doi: 10.1186/s12874-015-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weathers FW, Litz BT, Herman DS, Huska JA, Keane TM. The PTSD Checklist (PCL): Reliability, validity, and diagnostic utility. Poster presented at the annual meeting of the International Society for Traumatic Stress Studies; San Antonio, TX. Oct 24; 1993. [Google Scholar]
- 79.Williamson JB, Porges EC, Lamb DG, Porges SW. Maladaptive autonomic regulation in PTSD accelerates physiological aging. Front Psychol. 2015;5:1571. doi: 10.3389/fpsyg.2014.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wolf EJ, Logue MW, Hayes JP, Sadeh N, Schichman SA, Stone A, Salat DH, Milberg W, McGlinchey R, Miller MW. Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–62. doi: 10.1016/j.psyneuen.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolf EJ, Logue MW, Stoop TB, Schichman SA, Stone A, Sadeh N, Hayes JP, Miller MW. Accelerated DNA methylation age: associations with PTSD and mortality. Psychosom Med. 2017 doi: 10.1097/PSY.0000000000000506. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yehuda R. Status of glucocorticoid alterations in post-traumatic stress disorder. Ann N Y Acad Sci. 2009;1179:56–69. doi: 10.1111/j.1749-6632.2009.04979.x. [DOI] [PubMed] [Google Scholar]
- 83.Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, Nemeroff CB, Smith AK, Bradley B, Heim C, Menke A, Lange JF, Brückl T, Ising M, Wray NR, Erhardt A, Binder EB, Mehta D. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.