Abstract
The 14-3-3 family of proteins has undergone considerable expansion in higher eukaryotes with humans and mice expressing seven isoforms (β, ε, η, γ, θ, ζ, and σ) from seven distinct genes (YWHAB, YWAHE, YWHAH, YWHAG, YWHAQ, YWHAZ, and SFN). Growing evidence indicates that while highly conserved, these isoforms are not entirely functionally redundant as they exhibit unique tissue expression profiles, subcellular localization, and biochemical functions. A key limitation in our understanding of 14-3-3 biology lies in our limited knowledge of cell-type specific 14-3-3 expression. Here we provide a characterization of 14-3-3 expression in whole retina and isolated rod photoreceptors using reverse-transcriptase digital droplet PCR. We find that all 14-3-3 genes with the exception of SFN are expressed in mouse retina with YWHAQ and YWHAE being the most highly expressed. Rod photoreceptors are enriched in YWHAE (14-3-3 ε). Immunohistochemistry revealed that 14-3-3 ε and 14-3-3 ζ exhibit unique distributions in photoreceptors with 14-3-3 ε restricted to the inner segment and 14-3-3 ζ localized to the outer segment. Our data demonstrates that, in the retina, 14-3-3 isoforms likely serve specific functions as they exhibit unique expression levels and cell-type specificity. As such, future investigations into 14-3-3 function in rod photoreceptors should be centered on 14-3-3 ε and 14-3-3 ζ, depending on the subcellular region of question.
Keywords: 14-3-3, retina, photoreceptor, rd1, cone, digital droplet PCR, YWHAE, YWHAZ
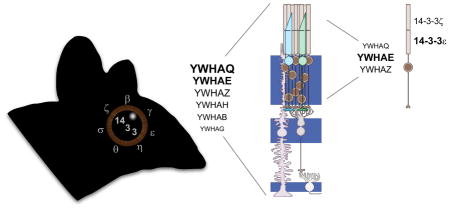
Graphical abstract
INTRODUCTION
The 14-3-3s are a small family of highly conserved proteins found in all eukaryotic cells. They function as adaptors that can modulate the enzymatic activity, structure, and stability of client proteins by inducing or stabilizing a conformational change upon binding or by bridging together domains or proteins in a larger signaling complex (Aitken, 2006; Bridges and Moorhead, 2005; Cornell and Toyo-Oka, 2017). 14-3-3s are small, ~30 kDa, proteins, and they are hockey-stick shaped with the client binding site consisting of a shallow groove on the concave side (Gardino et al., 2006). The 14-3-3 ligand in the client protein is a short linear motif, most often phosphorylated, found in intrinsically disordered regions (Bustos and Iglesias, 2006). 14-3-3s serve thousands of different client proteins, influencing nearly every aspect of cellular biology (Madeira et al., 2015; Tinti et al., 2012; Tinti et al., 2014). Examples include regulation of cell growth and differentiation through modulation of the MAPK pathway via activation of Raf1, negative regulation of apoptosis through multiple mechanisms including cytosolic sequestration of Bad, and cell cycle regulation via modulation of the transcription factors p53, FOXO, and MIZ1 (Fantl et al., 1994; Hermeking and Benzinger, 2006; Zha et al., 1996). Likely to accommodate all of these roles, the family of 14-3-3 proteins has been expanded in higher eukaryotes. There are two 14-3-3 proteins in yeast, seven in mammals, and at least a dozen in many plants (Chandna et al., 2016). 14-3-3 may function as monomers, homodimers, or heterodimers, further expanding the palette of available 14-3-3 in any given cell (Gardino et al., 2006; Sluchanko et al., 2012; Sluchanko and Gusev, 2012).
The various 14-3-3 isoforms exhibit a highly conserved structure with significant sequence similarity; there is 46% sequence identity between the human isoforms which results in functional redundancy. The degree to which this redundancy extends is not completely understood. Historically, it has been assumed that all isoforms are entirely functionally redundant so that most studies investigating 14-3-3 and a given client use a single 14-3-3 isoform as an implicit representative of the entire family. However, emerging evidence calls this assumption into question. 14-3-3 proteins recognize their targets via mode I (RXX[pS/pT]XP), mode II (RX[F/Y]X[pS/pT]XP), or mode III ([pS/T]X1-2COOH) consensus motifs and, a growing number of client proteins have been observed to be preferentially recognized by specific isoforms suggesting that subtle variations of these consensus sites may provide a favorable interaction with one isoform over another (Coblitz et al., 2006; Yaffe et al., 1997). Examples of this include the water channel aquaporin-2 which is selectively recognized by 14-3-3 θ and 14-3-3 ζ isoforms and the potassium channel TASK-3 which is most strongly bound by 14-3-3 γ (Kilisch et al., 2016; Moeller et al., 2016). In addition to this biochemical specificity, there also appears to be specificity in the expression of 14-3-3 isoforms (e.g. Martin et al., 1994; Moreira et al., 2008; Schindler et al., 2006; Watanabe et al., 1993; Watanabe et al., 1994; Wilson et al., 2016). This is most striking for 14-3-3 σ, which is largely restricted to epithelial cells (Leffers et al., 1993).
One type of cell where differential 14-3-3 expression or function is likely to come into play is in the highly specialized mammalian photoreceptor. Photoreceptor outer segments are sensory cilia with a greatly elaborated membrane surface area which allows for maximal light sensitivity. All cilia require microtubule motor-driven intraflagellar transport (IFT) for assembly and maintenance (Satir, 2017). 14-3-3 η has been reported to link the IFT motor KIF3 to a cell polarity complex (Fan et al., 2004) and ciliary-localized β-arrestin 2 which is part of G-protein coupled receptor – MAPK signaling pathway (Molla-Herman et al., 2008). Phototransduction, the signaling cascade in outer segments that converts photon absorption into an electrical signal is regulated by 14-3-3 proteins. Two photoreceptor enriched clients of 14-3-3 that modulate phototransduction are phosducin and GCAP2. Phosducin is a Gβγ binding protein. Light-dependent phosphorylation allows phosducin to interact with 14-3-3, releasing Gβγ thereby facilitating dark adaptation of photoreceptors (Lee et al., 2004; Nakano et al., 2001; Thulin et al., 2001). GCAP2 is a calcium sensor that regulates guanylate cyclase (GC), the enzyme responsible for producing the second messenger, cGMP (Wen et al., 2014). In the dark, GCAP2 is bound to Ca2+ and inhibits GC, preventing toxic accumulation of cGMP. In the light, Ca2+ free GCAP2 activates GC to restore cGMP levels. When Ca2+ free GCAP2 becomes phosphorylated it can interact with 14-3-3 ε, and at least one other 14-3-3 isoform. 14-3-3 binding to GCAP2 sequesters it in the inner segment (Hoyo et al., 2014). Another aspect of photoreceptor function is the production of melatonin for regulation of circadian rhythms. Arylalkylamine N-acetyltransferase (AANAT) regulates melatonin synthesis. 14-3-3 binding to phosphorylated AANAT in the dark protects AANAT from degradation which is required for rhythmic production of melatonin at night (Fukuhara et al., 2004; Pozdeyev et al., 2006).
Immunolabeling experiments have repeatedly demonstrated expression of 14-3-3 throughout the retina (Buret et al., 2016; Hoyo et al., 2014; Kim et al., 2005; Koseki et al., 2012; Nakano et al., 2001; Pozdeyev et al., 2006; Yang et al., 2008). However, the reported localization of 14-3-3 varies somewhat which may be due to subtle differences in how tissues were processed, or the specificity and sensitivity of the antibodies used. There is no comprehensive analysis of 14-3-3 in the retina taking into account all the distinct isoforms of 14-3-3. The goal of this study was to determine which 14-3-3 isoforms are expressed in the retina, with an emphasis on photoreceptors to provide a platform for further investigations into the role of 14-3-3 in retinal biology. We used quantitative digital-droplet PCR and immunohistochemistry with validated isoform specific 14-3-3 antibodies to detail the expression level and localization of the individual 14-3-3 isoforms.
MATERIALS AND METHODS
Animals
C57BL/6J (wild type, WT) mice of both sexes and at 1–4 months of age were obtained from The Jackson Laboratory. Female C3H/HeJ mice carrying the Pde6brd1 mutation from The Jackson Laboratory were used at 2.5 months of age when degeneration of rods is complete and termed “ΔRods”. RPE65 R91W; Nrl−/− (conefull) mice were a generous gift from Marijana Samardzija and Christian Grimm (Samardzija et al., 2014); males at 1–3 months of age were used. Mice were housed in a central vivarium, maintained on a standard 12/12 hour light/dark cycle, with food and water provided ad libitum in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All procedures adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the University of Iowa IACUC committee.
Antibodies and Immunohistochemistry
Antibodies and lectins used in this study are listed in Table 1. Mouse eyes from at least 3 individual mice were enucleated and the posterior eyecups collected by dissection. Eyecups were fixed in 4% paraformaldehyde at room temperature for 60–90 min, cryoprotected in 30% sucrose, and then frozen in O.C.T (Tissue-Tek, Electron Microscopy Sciences, Hatfield, PA, USA) before collecting 12 μm cryosections. For immunostaining, the sections were permeabilized in PBS containing 0.5% Triton X-100 for 10 min, followed by incubation in 10% goat serum to block nonspecific labeling. Primary antibodies were incubated on sections overnight at 4 °C, washed and secondary antibodies along with WGA and Hoechst were added for 1–2 hours at room temperature. Fluoromount-G was used as the mounting media and No 1.5 cover slips were sealed in place with nail polish. Images were collected using a 20x objective, with digital zoom set to capture the span of the retina from the OS to the GC and at 1 μm steps in the z-axis, with the pinhole set to 1 AU on a Zeiss 710 confocal microscope (Central Microscopy Research Facility, Univ. Iowa). Maximum through stack projections were made using Zen Light 2009 (Carl Zeiss); manipulation of images was limited to rotation, cropping, and adjusting the brightness and contrast levels using Zen Light 2009 or Photoshop CC (Adobe).
Table 1.
Antibodies & Lectins used for immunohistochemistry (IHC) or Western blot (WB)
| Source | Cat# | Conc. | RRID | |
|---|---|---|---|---|
| Rabbit anti-pan 14-3-3 | Abcam | Ab9063 | 1:250 (IHC), 1:300 (WB) | AB_306972 |
| Mouse anti-pan 14-3-3 (H8) | Santa Cruz | sc-1657 | 1:100 (IHC), 1:500 (WB) | AB_626618 |
| Rabbit anti-14-3-3 epsilon | Cell Signaling | 9635 | 1:100 (IHC), 1:1000 (WB) | n/a |
| Mouse anti-14-3-3 zeta (1B3) | Santa Cruz | sc-293415 | 1:100 (IHC), 1:500 (WB) | n/a |
| WGA: Alexa 594 | ThermoFisher | W11262 | 1:500 (IHC) | n/a |
| Goat anti-rabbit IgG: Alexa 647 | ThermoFisher | A21244 | 1:500 (IHC) | AB_141663 |
| Goat anti-mouse IgG: Alexa 488 | ThermoFisher | A11001 | 1:500 (IHC) | AB_2534069 |
| IRDye® 680RD Goat anti-Rabbit IgG | LI-COR | 925-68071 | 1:20000 (WB) | n/a |
| IRDye® 800CW Goat anti-Mouse IgG | LI-COR | 925-32210 | 1:20000 (WB) | n/a |
RNA isolation and RT-ddPCR
Freshly dissected mouse retina was placed in RNAlater (Qiagen, Hilden, Germany) and stored for up to 24 hours at 4 °C. Retinas were mechanically disrupted in a TissueLyser LT (5Hz, 5 min) (Qiagen) and RNA collected using the RNAeasy mini (Qiagen) standard protocol, RNA concentration and quality was measured using a Dropsense16 and an Agilent BioAnalyzer 2100 respectively (The Iowa Institute of Human Genetics, University of Iowa). Synthesis of cDNA from 1 μg RNA (RIN > 8.5) was done using the iScript kit according to manufacturer’s instructions (BioRad, Hercules, CA, USA). PrimeTime qPCR 5′ Nuclease assays with FAM for the 5′ dye and Zen/Iowa Black FQ for the 3′ quencher were purchased from IDT (Table 2). 24 μl PCR reactions containing 500 nM primers, 250 nM probe, and variable amounts of cDNA were assembled with the ddPCR Supermix for Probes (BioRad). From this, 20ul of reaction mix was mixed with Droplet Generation Oil for Probes (BioRad) and the reaction was partitioned into oil droplets using the QX200 droplet generator (BioRad). The oil droplets were then transferred to a 96 well plate that was sealed before subjecting the samples to PCR amplification using a C1000 Touch ™ thermal cycler. PCR amplified cDNA was then analyzed for presence or absence of fluorescence using the Qx200 droplet reader. Results obtained from the droplet reader were then analyzed by QuantaSoft ™ Analysis Pro 1.0.596 (BioRad).
Table 2.
qPCR assays used for ddPCR
| Assay ID | Gene | Ref Seq # | Exon location |
|---|---|---|---|
| Mm.PT.58.10072972.gs | YWHAB | NM_018753 | 4-6 |
| Mm.PT.58.43656403 | YWHAE | NM_009536 | 5-6 |
| Mm.PT.58.42727760 | YWHAG | NM_018871 | 1-2 |
| Mm.PT.58.42310160 | YWHAH | NM_011738 | 1-2 |
| Mm.PT.58.29642684.g | YWHAQ | NM_001739 | 5-5 |
| Mm.PT.58.31794574 | YWHAZ | NM_001253807 | 7-9 |
| Mm.PT.58.42262048.g | SFN | NM_018754 | 1-1 |
Retina dissociation and FACS sorting
Rod perikarya were isolated from C57Bl6/J mouse retina as described (Feodorova et al., 2015). Briefly, four freshly dissected mouse retina were digested using the papain dissociation system (Worthington Biochemical Corporation, Lakewood, NJ, USA). Dissociated retinas were filtered through a 70μm nylon mesh (Falcon, Durham, NC USA) and sorting of rod perikarya was done using an Becton Dickinson Aria II, equipped with a 70μm nozzle, with standard forward and sideward scatter settings (Flow Cytometry Facility, Univ. Iowa). 20,000 sorted cells (~100ul volume) were collected in 500ul RLT buffer for RNA isolation using the RNeasy micro (Qiagen) standard protocol, which was used for RT-ddPCR as described above.
Plasmids
A pPROEX-HTb vector for expression of human 14-3-3 ζ with an N-terminal His6-tag was a gift from Dr. Christian Ottmann (Stevers et al., 2016). Human and mouse 14-3-3 ζ only differ at one amino acid (A112 in human, P112 in mouse); for expression of mouse 14-3-3 ζ, the A112P mutation in human YWHAZ was generated using overlap extension primers. Mouse YWHAB, YWHAE, YWHAG, YWHAH, YWHAQ, and SFN cDNA clones were purchased from Origene (Rockville, MD USA) and subcloned into pPROEX-HTb at the BamHI and NotI sites. All primers were purchased from IDT (Coralville, IA USA). All inserts and mutations were verified by Sanger sequencing (Iowa Institute of Human Genetics).
Protein purification and Western blotting
His-tagged 14-3-3 isoforms were expressed in BL21(DE3) E. coli, induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at mid-log phase. Induced cells were lysed using B-PER Bacterial Protein Extraction Reagent (ThermoFischer Scientific, Waltham, MA USA) and purified using HisPur Ni-NTA resin (ThermoFisher Scientific). Equal amounts of 14-3-3 proteins (determined by absorbance at 280nm) were separated using SDS-PAGE and transferred to PVDF for staining with REVERT-total protein stain (LI-COR, Lincoln, NE USA) and immunoblotting with the antibodies listed in Table 1. Blots were imaged using a LI-COR Odyssey FC and analyzed with Image Studio (v5) software. Signal intensities were normalized to that of total protein and at least 2 blots per antibody were analyzed.
Phylogenetic tree
Mouse 14-3-3 protein sequences (UniProt ID: Q9CQV8, β; P62259, ε; P61982, γ; P68510, η; P68254, θ; O70456, σ; P63104, ζ) were aligned using the T-Coffee multiple sequence alignment web server (Notredame et al., 2000), the neighbor-joining tree without distance corrections was displayed as a cladogram using iTOL (Letunic and Bork, 2016).
Statistical Analysis
Statistical differences were determined using GraphPad Prism software (v7). Statistical significance was defined using an alpha of 0.05. Normality was assessed by the Shapiro-Wilk test and parametric data were analyzed by ANOVA.
RESULTS AND DISCUSSION
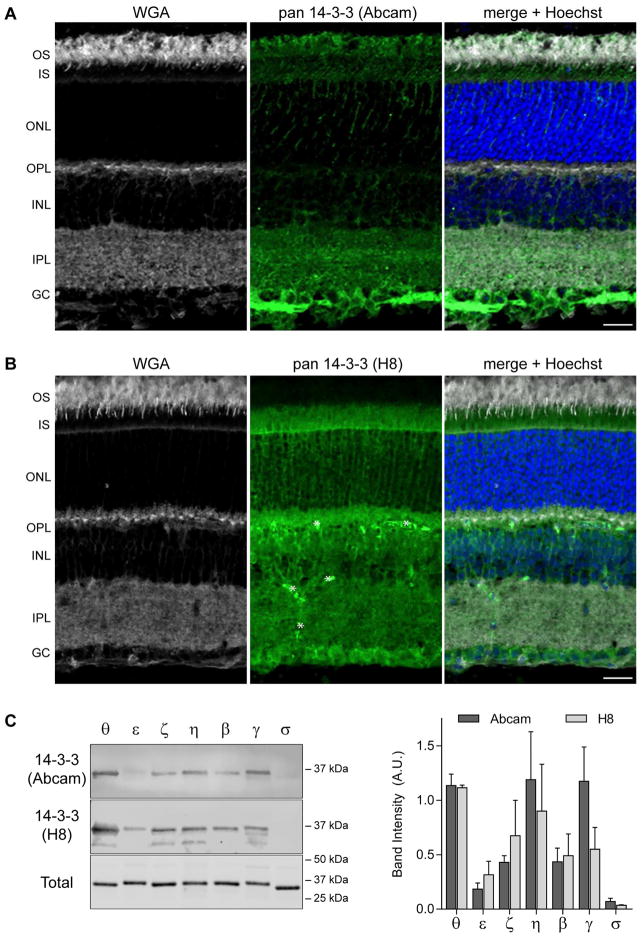
The expression of 14-3-3 in mouse retina was examined by immunohistochemistry with wheat germ agglutinin (WGA) as a marker of the plexiform (synaptic) layers and photoreceptor outer segments. One pan 14-3-3 antibody robustly labeled the nerve fiber layer, followed by the inner plexiform layer and photoreceptor outer segments (Figure 1A). A pattern of streaks running through the outer nuclear layer indicative of Muller glia labeling was also noted. When a second pan 14-3-3 antibody from a different commercial source was used a different labeling pattern was obtained (Figure 1B). The second pan-14-3-3 antibody labeled ganglion cell bodies, the outer plexiform layer, inner plexiform layer, and inner segments. The labeling pattern in the inner segments seems the brightest at the outer limiting membrane. That pattern is consistent with either labeling of photoreceptor Golgi or Muller glia microvilli. The discrepancy in labeling for 14-3-3, particularly in the inner and outer segments of photoreceptors, was unexpected.
Figure 1.
14-3-3 proteins are expressed throughout the retina. A–B) mouse retina labeled with WGA (left), pan 14-3-3 (center, green), with merged images including Hoechst labeling for nuclei on the right. A polyclonal pan 14-3-3 antibody (Abcam) was used in A, and a monoclonal pan-14-3-3 (H8) in B. Asterisks in (B) indicate labeling of blood vessels by the secondary antibody. Scale bar, 20 μm and abbreviations are: OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GC, ganglion cell layer. C) Western blots of recombinant 14-3-3 proteins probed with the antibodies used in A–B; blots stained with REVERT to label total protein. Quantitation of the band intensities, data represent mean + SD, reveals no statistically significant differences between the two antibodies.
A possible explanation for the discrepancy is that the individual 14-3-3 isoforms are not distributed equally across the retina and the pan-antibodies do not have equal sensitivity for the individual isoforms. To test this idea, the pan 14-3-3 antibodies were blotted against purified recombinant 14-3-3 θ, ε, ζ, η, β, γ, and σ. The pan-antibodies had a similar pattern in terms of both specificity and sensitivity. 14-3-3 θ, η, and γ were detected equally. The sensitivity for 14-3-3 β and ζ was reduced approximately by half, and by half again for 14-3-3 ε. Only trace amounts of 14-3-3 σ could be detected (Figure 1C). Since the two antibodies generated similar Western blotting results, the variability in immunostaining remains uncertain. We decided to investigate this issue by first quantifying the expression of 14-3-3 transcripts.
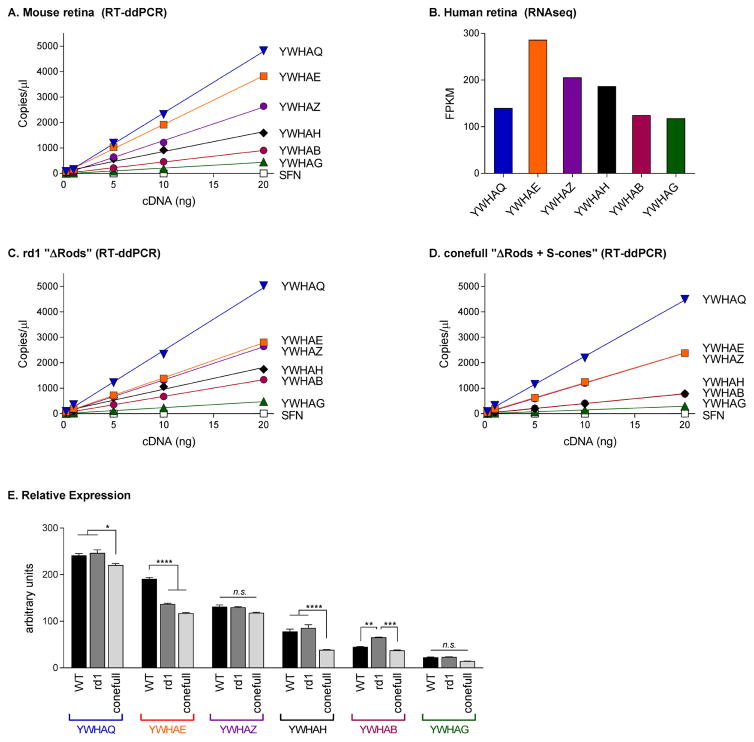
We used Reverse Transcriptase-digital droplet PCR (RT-ddPCR) to investigate the relative expression levels of all seven 14-3-3 isoforms. In RT-ddPCR, TaqMan probe based PCR amplification of cDNA takes place in a single sample partitioned into ~10,000–20,000 individual droplets; the proportion of droplets generating a fluorescent signal are fit to a Poisson distribution to calculate the absolute initial copy number of the target cDNA. This is currently the most accurate and sensitive technique available for quantitating RNA expression levels (Hindson et al., 2013; Racki et al., 2014; Sanders et al., 2013; Taylor et al., 2015). We performed five ddPCR reactions per gene using a cDNA concentration ranging from 0.25–20ng. The results were fit with a simple linear regression model; r2 of the fits was 0.99. The abundance of the 14-3-3 isoforms in RNA isolated from mouse retina was as follows: YWHAQ (14-3-3 θ) > YWHAE (14-3-3 ε) ≫ YWHAZ (14-3-3 ζ) > YWHAH (14-3-3 η) > YWHAB (14-3-3 β) > YWHAG (14-3-3 γ); Sfn (14-3-3 σ) was only detected in trace amounts (Figure 2A). We queried the expression levels of the 14-3-3 genes from a previously published RNAseq dataset of human retina and found a similar pattern of expression (Figure 2B, (Whitmore et al., 2014)). With the exception of YWHAQ, the mouse and human data are in agreement with YWHAE being more abundant than YWHAZ, YWHAH, YWHAB, and YWHAG in that order and SFN undetectable from background. RNAseq is very precise when comparing the expression levels of a single gene across samples. However, quantifying expression among genes within a sample is challenging due to imprecise transcript specifications, amplification bias during library construction, and non-unique mapping of reads (Li and Dewey, 2011; Wagner et al., 2012). While it is possible that the expression of YWHAQ is different in mouse versus human retina, we think it is more likely that the RNAseq data is underreporting the abundance of YWHAQ in human retina. The large range of expression levels for the different 14-3-3 transcripts supports the idea that there may be cell type dependent differential expression of the individual isoforms.
Figure 2.
Six of the seven 14-3-3 isoforms are expressed in retina. A) RT-ddPCR results from mouse retina plotted as a function of starting cDNA concentration. Note, error bars are smaller than the symbol size. B) Human retina RNAseq data replotted from (Whitmore et al., 2014). RT-ddPCR results from rd1 (C) or conefull (D) mouse retina. E) Comparison of the slope of the linear regression for each gene from A, C, and D. P-values are represented as n.s., p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
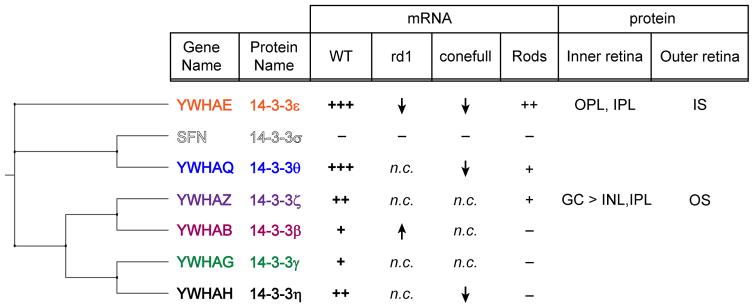
To determine if any particular 14-3-3 isoform is enriched in photoreceptors, we measured the expression levels of 14-3-3 RNA in mouse retinas depleted of rods due to either degeneration or developmental failure. For the degeneration model, we used two and half month old C3H/HeJ mice, a strain that carries the Pde6brd1 allele. The naturally occurring Pde6brd1 mutation causes early onset degeneration resulting in the complete loss of rods well before two months of age (Paquet-Durand et al., 2014). All six 14-3-3 isoforms found in healthy C57Bl/6J “WT” retinas were present in the rd1 or “ΔRods” retina (Figure 2C). Relative expression was determined by comparing the slope of the line fit to each gene within each sample (Figure 2E, black vs gray bars). Two statistically significant changes were observed in ΔRods compared to WT retinas, an increase in the expression of YWHAB (mean diff = 20.3, 95% CI [5.0, 35.6], adj. p = 0.004) and a larger decrease in YWHAE (mean diff = 53.7, 95% CI [38.4, 69.0], adj. p < 0.0001).
We used a mouse line developed for the study of cones for the complimentary ΔRods model. Samardzija and colleagues crossed the NRL KO mouse with a line carrying a hypomorphic mutation in RPE65. In these conefull mice, only S-cone photoreceptors are present and the confounding formation of neural rosettes associated with excessive levels of retinal are attenuated (Mears et al., 2001; Samardzija et al., 2014). We used this line as a “ΔRods + S-cones” model and found that, as in the ΔRods retina, the largest decrease compared to WT was with YWHAE (mean diff = 73.8, 95% CI [58.5, 89.1], adj. p < 0.0001). YWHAQ and YWHAH were also decreased in conefull retina compared to WT (mean diff = 21.2, 95% CI [5.8, 36.5], adj. p = 0.003 and mean diff = 39.3, 95% CI [23.9, 54.6], adj. p < 0.0001 respectively; Figure 2D and 2E, black vs light gray bars).
Several inferences can be drawn from this comparison. None of the 14-3-3 isoforms are exclusively expressed in rods; if that were the case expression should have been lost in both of the Δrods retinas. Since levels of YWHAE were significantly reduced in both models, we conclude that YWHAE is expressed in the inner retina but comparatively enriched in rods. YWHAH is decreased in the conefull but not the degenerated retina where we expected similar changes; given that the cones that are made in the NRL KO are most similar to S-cones, YWHAH could be enriched in M-cones. In both of these Δrods retinas there are more changes occurring than the loss of rods, e.g. gliosis, so interpreting the smaller changes observed for YWHAQ and YWHAB is not straightforward. Next we sought to validate the interpretation that YWHAE is expressed in rods.
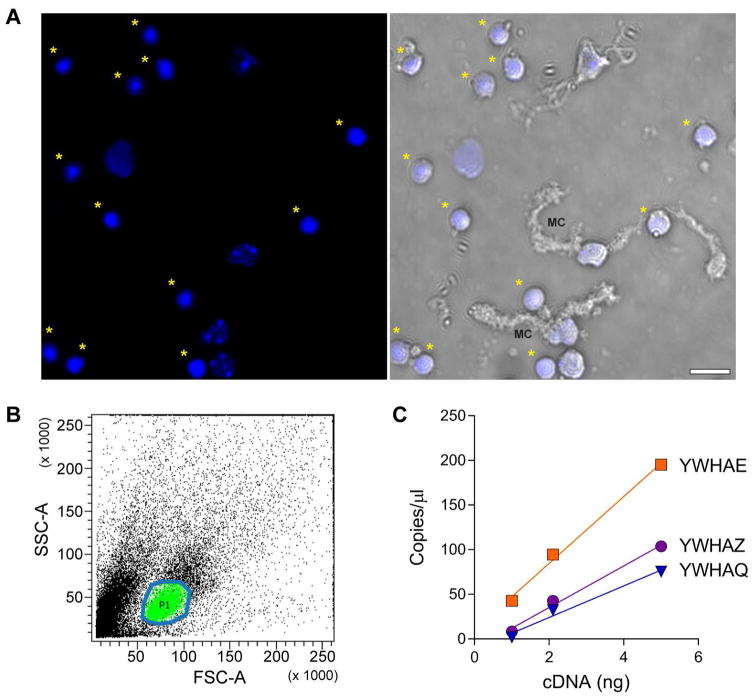
We isolated rods based on the distinctive condensation of heterochromatin in the center of the nucleus (Feodorova et al., 2015; Solovei et al., 2009) and measured the amounts of 14-3-3 transcripts. Papain treatment of the retina released rod perikarya along with fragments of other retinal cells (Figure 3A). The population of living rod perikarya was FACS sorted from the other cells (Figure 3B) and used immediately for RNA extraction. The yield of RNA from this preparation was significantly lower than from whole retina so only three ddPCR reactions were used per gene with a cDNA concentration ranging from 1–5ng. Three of the six 14-3-3 genes were detected: YWHAE was the most abundant, followed by YWHAZ, and YWHAQ (Figure 3C). The amount of YWHAE and YWHAZ detected in isolated rod perikarya was approximately 15% of that detected in intact retina, or 5% for YWHAQ. It is not possible to conclude that the other isoforms are absent from rods, only that YWHAE is the most abundant of 14-3-3s detected in this population, which is consistent with the results from the Δrods retinas. Since some Muller glial cell fragments could remain in association with rod perikarya after sorting (Figure 3A) it is not possible to be absolutely certain of the purity of this preparation. Therefore, we focused on just the two most abundant 14-3-3 isoforms for subsequent validation of rod expression.
Figure 3.
YWHAE and YWHAZ are expressed in rods. A) Cell fragments released from retina after papain treatment stained with Hoechst, and in the right panel overlaid with the transmitted light channel. Asterisks indicate rod perikarya; MC is a fragment of a Muller glia cell. B) Profile of sorted cells, area circled in blue containing the cells marked green is the population of rod perikarya collected for RNA extraction. C) RT-ddPCR results from rod perikarya.
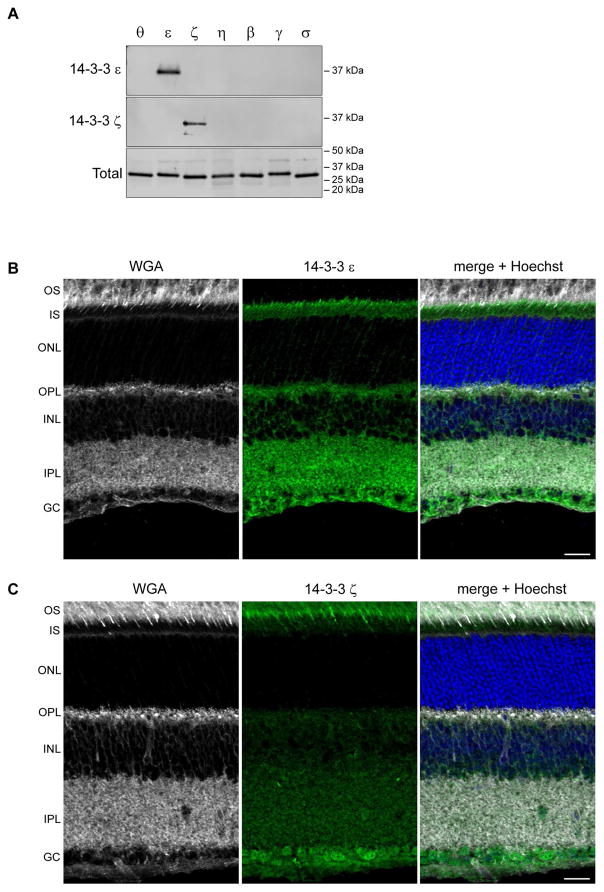
We screened commercially available 14-3-3 antibodies to find an antibody specific for 14-3-3 ε and 14-3-3 ζ respectively (Figure 4A). Returning to immunostaining of whole retina sections, we found that 14-3-3 ε labeled photoreceptor inner segments, with the stain more concentrated at the apical end near the connecting cilia (Figure 4B). 14-3-3 ε also labeled the outer and inner plexiform layers consistent with its reduced but not absent mRNA expression in the Δrods retina models. 14-3-3 ζ labeled photoreceptor outer segments, with the signal being most intense in the basal outer segments and an apparent concentration in cone outer segments. Punctate labeling was present throughout the inner segment. In addition, the ganglion cell bodies were labeled with a weaker signal throughout the inner plexiform and inner nuclear layers (Figure 4C). The striking outer segment signal for 14-3-3 ζ raises the question why reduced YWHAZ transcripts were not observed in the Δrods retinas. A possible explanation is that the amount of transcripts in the cones masked the loss of transcripts from rods since both Δrods retinas contain cones. Returning to the original conundrum of inconsistent immunostaining patterns with the pan-14-3-3 antibodies for inner and outer segments, we suggest that may be explained by competition for antibody binding to other isoforms, subtle differences in the sensitivity for 14-3-3 ε versus ζ, or epitope availability. Regardless, we recommend that future immunostaining experiments use validated isoform-specific antibodies. Especially since the differential subcellular 14-3-3 labeling indicates that 14-3-3 ε and 14-3-3 ζ are not functionally redundant in terms of photoreceptor biology.
Figure 4.
14-3-3 ε and ζ are differentially localized in photoreceptors. A) Western blots of recombinant 14-3-3 proteins probed with 14-3-3 ε (upper) or 14-3-3 ζ (lower) antibodies; blots stained with REVERT to label total protein. Mouse retinas labeled with WGA and either 14-3-3 ε (C) or 14-3-3 ζ (D); scale and abbreviations as in Figure 1.
Comparing our results to the phylogenetic relationship of the 14-3-3 isoforms provides context (Figure 5). The 14-3-3 proteins fall into 3 clades: ε/θ (with σ as an offshoot of θ), ζ/β, and γ/η. The ε/θ clade is most abundantly expressed in retina with 14-3-3 θ likely compensating for any functions that are unique to 14-3-3 σ in other tissues. Within the other two clades, one of the isoforms is most abundant in retina so that it is possible that β and γ are redundant to ζ and η, respectively. The two isoforms with differential subcellular localization in photoreceptors represent two different clades so they may have distinct localization or functions in other tissues as well.
Figure 5.
Correlation between phylogeny and retina expression of 14-3-3. Cladogram of 14-3-3 isoforms adjacent to a summary of the findings regarding mRNA and protein expression in the retina, n.c. indicates no change, information corresponding to the blank rows in the protein columns was not determined.
CONCLUSIONS
The central finding of this study is that 14-3-3 isoforms are differentially expressed in the retina both in terms of overall abundance and at least, in some cases, in terms of cell type and subcellular localization. Future studies of 14-3-3 in photoreceptor biology should focus on 14-3-3 ε and 14-3-3 ζ.
Supplementary Material
Highlights.
Expression of 14-3-3 isoforms in retina was quantified using digital-droplet PCR
Only 14-3-3 σ was not found in retina
14-3-3 antibodies were validated against recombinant proteins
14-3-3 ε and 14-3-3 ζ were detected in photoreceptors
14-3-3 ε and 14-3-3 ζ were found enriched in inner vs. outer segments, respectively
Acknowledgments
Funding: This work was supported by the National Institutes of Health R01, EY027054 (SAB) and P30, EY025580 (Iowa), and the Roy J Carver, Jr. Chair in Bioinformatics and Computational Biology (TES).
We thank Dawn Farruggio and Brandon McKethan from BioRad, Mary Boes and Garry Hauser from the Genomics Division of the Iowa Institute of Human Genetics, and Justin Fishbaugh, Heath Vignes, and Michael Shey from the Univ. of Iowa Flow Cytometry Facility for technical advice. This work utilized the Zeiss LSM710 confocal in the University of Iowa Central Microscopy Research Facilities that was purchased with funding NIH SIG S10 RR025439. Data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility at the University of Iowa. The Facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran’s Administration Medical Center. Data presented herein were obtained at the Genomics Division of the Iowa Institute of Human Genetics which is supported, in part, by the University of Iowa Carver College of Medicine.
Footnotes
Contributions: Experiments were designed, executed, and analyzed by SMI, CKL, JGL, GN, and SAB with bioinformatic analysis by NT, SSW, and TES. SMI, CKL, and SAB wrote the manuscript and all authors approved the final article.
Conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2005;2005:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- Buret L, Rebillard G, Brun E, Angebault C, Pequignot M, Lenoir M, Do-Cruzeiro M, Tournier E, Cornille K, Saleur A, Gueguen N, Reynier P, Amati-Bonneau P, Barakat A, Blanchet C, Chinnery P, Yu-Wai-Man P, Kaplan J, Roux AF, Van Camp G, Wissinger B, Boespflug-Tanguy O, Giraudet F, Puel JL, Lenaers G, Hamel C, Delprat B, Delettre C. Loss of function of in mice induces deafness and cochlear outer hair cells’ degeneration. Cell death discovery. 2016;2:16017. doi: 10.1038/cddiscovery.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos DM, Iglesias AA. Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins. 2006;63:35–42. doi: 10.1002/prot.20888. [DOI] [PubMed] [Google Scholar]
- Chandna R, Augustine R, Kanchupati P, Kumar R, Kumar P, Arya GC, Bisht NC. Class-Specific Evolution and Transcriptional Differentiation of 14-3-3 Family Members in Mesohexaploid Brassica rapa. Frontiers in plant science. 2016;7:12. doi: 10.3389/fpls.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coblitz B, Wu M, Shikano S, Li M. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 2006;580:1531–1535. doi: 10.1016/j.febslet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Cornell B, Toyo-Oka K. 14-3-3 Proteins in Brain Development: Neurogenesis, Neuronal Migration and Neuromorphogenesis. Frontiers in molecular neuroscience. 2017;10:318. doi: 10.3389/fnmol.2017.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Hurd TW, Liu CJ, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14:1451–1461. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Fantl WJ, Muslin AJ, Kikuchi A, Martin JA, MacNicol AM, Gross RW, Williams LT. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- Feodorova Y, Koch M, Bultman S, Michalakis S, Solovei I. Quick and reliable method for retina dissociation and separation of rod photoreceptor perikarya from adult mice. MethodsX. 2015;2:39–46. doi: 10.1016/j.mex.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara C, Liu C, Ivanova TN, Chan GC, Storm DR, Iuvone PM, Tosini G. Gating of the cAMP signaling cascade and melatonin synthesis by the circadian clock in mammalian retina. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:1803–1811. doi: 10.1523/JNEUROSCI.4988-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardino AK, Smerdon SJ, Yaffe MB. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol. 2006;16:183–192. doi: 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyo NL, Lopez-Begines S, Rosa JL, Chen J, Mendez A. Functional EF-hands in neuronal calcium sensor GCAP2 determine its phosphorylation state and subcellular distribution in vivo, and are essential for photoreceptor cell integrity. PLoS Genet. 2014;10:e1004480. doi: 10.1371/journal.pgen.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilisch M, Lytovchenko O, Arakel EC, Bertinetti D, Schwappach B. A dual phosphorylation switch controls 14-3-3-dependent cell surface expression of TASK-1. Journal of cell science. 2016;129:831–842. doi: 10.1242/jcs.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Kim YS, Kang SS, Noh HS, Kim HJ, Cho GJ, Choi WS. Expression of 14-3-3 zeta and interaction with protein kinase C in the rat retina in early diabetes. Diabetologia. 2005;48:1411–1415. doi: 10.1007/s00125-005-1774-7. [DOI] [PubMed] [Google Scholar]
- Koseki N, Kitaoka Y, Munemasa Y, Kumai T, Kojima K, Ueno S, Ohtani-Kaneko R. 17beta-estradiol prevents reduction of retinal phosphorylated 14-3-3 zeta protein levels following a neurotoxic insult. Brain Res. 2012;1433:145–152. doi: 10.1016/j.brainres.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Lee BY, Thulin CD, Willardson BM. Site-specific phosphorylation of phosducin in intact retina. Dynamics of phosphorylation and effects on G protein beta gamma dimer binding. Journal of Biological Chemistry. 2004;279:54008–54017. doi: 10.1074/jbc.M405669200. [DOI] [PubMed] [Google Scholar]
- Leffers H, Madsen P, Rasmussen HH, Honore B, Andersen AH, Walbum E, Vandekerckhove J, Celis JE. Molecular cloning and expression of the transformation sensitive epithelial marker stratifin. A member of a protein family that has been involved in the protein kinase C signalling pathway. J Mol Biol. 1993;231:982–998. doi: 10.1006/jmbi.1993.1346. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic acids research. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Tinti M, Murugesan G, Berrett E, Stafford M, Toth R, Cole C, MacKintosh C, Barton GJ. 14-3-3-Pred: improved methods to predict 14-3-3-binding phosphopeptides. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Rostas J, Patel Y, Aitken A. Subcellular localisation of 14-3-3 isoforms in rat brain using specific antibodies. J Neurochem. 1994;63:2259–2265. doi: 10.1046/j.1471-4159.1994.63062259.x. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nature genetics. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Moeller HB, Slengerik-Hansen J, Aroankins T, Assentoft M, MacAulay N, Moestrup SK, Bhalla V, Fenton RA. Regulation of the Water Channel Aquaporin-2 via 14-3-3theta and -zeta. The Journal of biological chemistry. 2016;291:2469–2484. doi: 10.1074/jbc.M115.691121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla-Herman A, Boularan C, Ghossoub R, Scott MG, Burtey A, Zarka M, Saunier S, Concordet JP, Marullo S, Benmerah A. Targeting of beta-arrestin2 to the centrosome and primary cilium: role in cell proliferation control. PloS one. 2008;3:e3728. doi: 10.1371/journal.pone.0003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira JM, Shen T, Ohlsson G, Gromov P, Gromova I, Celis JE. A combined proteome and ultrastructural localization analysis of 14-3-3 proteins in transformed human amnion (AMA) cells: definition of a framework to study isoform-specific differences. Mol Cell Proteomics. 2008;7:1225–1240. doi: 10.1074/mcp.M700439-MCP200. [DOI] [PubMed] [Google Scholar]
- Nakano K, Chen J, Tarr GE, Yoshida T, Flynn JM, Bitensky MW. Rethinking the role of phosducin: light-regulated binding of phosducin to 14-3-3 in rod inner segments. Proc Natl Acad Sci U S A. 2001;98:4693–4698. doi: 10.1073/pnas.071067198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Paquet-Durand F, Sahaboglu A, Dietter J, Paquet-Durand O, Hitzmann B, Ueffing M, Ekstrom PA. How long does a photoreceptor cell take to die? Implications for the causative cell death mechanisms. Advances in experimental medicine and biology. 2014;801:575–581. doi: 10.1007/978-1-4614-3209-8_73. [DOI] [PubMed] [Google Scholar]
- Pozdeyev N, Taylor C, Haque R, Chaurasia SS, Visser A, Thazyeen A, Du Y, Fu H, Weller J, Klein DC, Iuvone PM. Photic regulation of arylalkylamine N-acetyltransferase binding to 14-3-3 proteins in retinal photoreceptor cells. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:9153–9161. doi: 10.1523/JNEUROSCI.1384-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki N, Morisset D, Gutierrez-Aguirre I, Ravnikar M. One-step RT-droplet digital PCR: a breakthrough in the quantification of waterborne RNA viruses. Anal Bioanal Chem. 2014;406:661–667. doi: 10.1007/s00216-013-7476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samardzija M, Caprara C, Heynen SR, Willcox DeParis S, Meneau I, Traber G, Agca C, von Lintig J, Grimm C. A mouse model for studying cone photoreceptor pathologies. Invest Ophthalmol Vis Sci. 2014;55:5304–5313. doi: 10.1167/iovs.14-14789. [DOI] [PubMed] [Google Scholar]
- Sanders R, Mason DJ, Foy CA, Huggett JF. Evaluation of digital PCR for absolute RNA quantification. PloS one. 2013;8:e75296. doi: 10.1371/journal.pone.0075296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. CILIA: before and after. Cilia. 2017;6:1. doi: 10.1186/s13630-017-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler CK, Heverin M, Henshall DC. Isoform- and subcellular fraction-specific differences in hippocampal 14-3-3 levels following experimentally evoked seizures and in human temporal lobe epilepsy. J Neurochem. 2006;99:561–569. doi: 10.1111/j.1471-4159.2006.04153.x. [DOI] [PubMed] [Google Scholar]
- Sluchanko NN, Artemova NV, Sudnitsyna MV, Safenkova IV, Antson AA, Levitsky DI, Gusev NB. Monomeric 14-3-3zeta has a chaperone-like activity and is stabilized by phosphorylated HspB6. Biochemistry. 2012;51:6127–6138. doi: 10.1021/bi300674e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluchanko NN, Gusev NB. Oligomeric structure of 14-3-3 protein: what do we know about monomers? FEBS Lett. 2012;586:4249–4256. doi: 10.1016/j.febslet.2012.10.048. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Stevers LM, Lam CV, Leysen SF, Meijer FA, van Scheppingen DS, de Vries RM, Carlile GW, Milroy LG, Thomas DY, Brunsveld L, Ottmann C. Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR. Proc Natl Acad Sci U S A. 2016;113:E1152–1161. doi: 10.1073/pnas.1516631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SC, Carbonneau J, Shelton DN, Boivin G. Optimization of Droplet Digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: Clinical implications for quantification of Oseltamivir-resistant subpopulations. J Virol Methods. 2015;224:58–66. doi: 10.1016/j.jviromet.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Thulin CD, Savage JR, McLaughlin JN, Truscott SM, Old WM, Ahn NG, Resing KA, Hamm HE, Bitensky MW, Willardson BM. Modulation of the G protein regulator phosducin by Ca2+/calmodulin-dependent protein kinase II phosphorylation and 14-3-3 protein binding. The Journal of biological chemistry. 2001;276:23805–23815. doi: 10.1074/jbc.M101482200. [DOI] [PubMed] [Google Scholar]
- Tinti M, Johnson C, Toth R, Ferrier DE, Mackintosh C. Evolution of signal multiplexing by 14-3-3-binding 2R-ohnologue protein families in the vertebrates. Open Biol. 2012;2:120103. doi: 10.1098/rsob.120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinti M, Madeira F, Murugesan G, Hoxhaj G, Toth R, Mackintosh C. ANIA: ANnotation and Integrated Analysis of the 14-3-3 interactome. Database (Oxford) 2014;2014:bat085. doi: 10.1093/database/bat085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory in biosciences = Theorie in den Biowissenschaften. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Isobe T, Ichimura T, Kuwano R, Takahashi Y, Kondo H. Developmental regulation of neuronal expression for the eta subtype of the 14-3-3 protein, a putative regulatory protein for protein kinase C. Brain Res Dev Brain Res. 1993;73:225–235. doi: 10.1016/0165-3806(93)90142-w. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Isobe T, Ichimura T, Kuwano R, Takahashi Y, Kondo H, Inoue Y. Molecular cloning of rat cDNAs for the zeta and theta subtypes of 14-3-3 protein and differential distributions of their mRNAs in the brain. Brain Res Mol Brain Res. 1994;25:113–121. doi: 10.1016/0169-328x(94)90285-2. [DOI] [PubMed] [Google Scholar]
- Wen XH, Dizhoor AM, Makino CL. Membrane guanylyl cyclase complexes shape the photoresponses of retinal rods and cones. Frontiers in molecular neuroscience. 2014;7:45. doi: 10.3389/fnmol.2014.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore SS, Wagner AH, DeLuca AP, Drack AV, Stone EM, Tucker BA, Zeng S, Braun TA, Mullins RF, Scheetz TE. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp Eye Res. 2014;129c:93–106. doi: 10.1016/j.exer.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RS, Swatek KN, Thelen JJ. Regulation of the Regulators: Post-Translational Modifications, Subcellular, and Spatiotemporal Distribution of Plant 14-3-3 Proteins. Frontiers in plant science. 2016;7:611. doi: 10.3389/fpls.2016.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Yang X, Luo C, Cai J, Pierce WM, Tezel G. Phosphorylation-dependent interaction with 14-3-3 in the regulation of bad trafficking in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2008;49:2483–2494. doi: 10.1167/iovs.07-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.