Significance
The completion of seed germination is an irrevocable event for plants, determining, for most plants, the site of the remainder of their life cycle. One environmental cue important to the completion of seed germination is light, which, in Arabidopsis thaliana, can influence a host of transcription factors, including PHYTOCHROME-INTERACTING FACTOR1 (PIF1), a negative regulator of the completion of germination and seedling de-etiolation. The KELCH F-BOX protein COLD TEMPERATURE GERMINATING10 (CTG10) can recognize and bind to PIF1, negatively influencing PIF1 stability, stimulating the completion of germination, and promoting a de-etiolated seedling morphology. PIF1, in turn, can downregulate CTG10 expression, revealing a complex coregulation orchestrated by light presence and quality that dictates whether the seed completes germination.
Keywords: germination, seed, ubiquitination, light
Abstract
Seeds employ sensory systems that assess various environmental cues over time to maximize the successful transition from embryo to seedling. Here we show that the Arabidopsis F-BOX protein COLD TEMPERATURE-GERMINATING (CTG)-10, identified by activation tagging, is a positive regulator of this process. When overexpressed (OE), CTG10 hastens aspects of seed germination. CTG10 is expressed predominantly in the hypocotyl, and the protein is localized to the nucleus. CTG10 interacts with PHYTOCHROME-INTERACTING FACTOR 1 (PIF1) and helps regulate its abundance in planta. CTG10-OE accelerates the loss of PIF1 in light, increasing germination efficiency, while PIF1-OE lines fail to complete germination in darkness, which is reversed by concurrent CTG10-OE. Double-mutant (pif1 ctg10) lines demonstrated that PIF1 is epistatic to CTG10. Both CTG10 and PIF1 amounts decline during seed germination in the light but reaccumulate in the dark. PIF1 in turn down-regulates CTG10 transcription, suggesting a feedback loop of CTG10/PIF1 control. The genetic, physiological, and biochemical evidence, when taken together, leads us to propose that PIF1 and CTG10 coexist, and even accumulate, in the nucleus in darkness, but that, following illumination, CTG10 assists in reducing PIF1 amounts, thus promoting the completion of seed germination and subsequent seedling development.
Seed germination, senso stricto, begins with imbibition and ends with the protrusion of part of the embryo through the covers (1). Changes in transcription (2, 3) and translation (4) have been documented to influence hormonal quantities and sensitivities (5), but the causal chain of events is complex. Unsurprisingly, there are still discoveries to be made in how signals become integrated into one of the few truly committed programmatic switches experienced by an individual plant—the completion of germination.
Due to the severity of the consequences, evolution has selected for the integration of internal developmental stages with environmental feedback required for successful establishment, ultimately creating the molecular complexity exhibited. One of the diverse environmental stimuli to which many seeds are sensitive is light. Studies on the influence of light quality on lettuce seed germination (6–8) led to the discovery of the phytochrome (Phy) family of bilin-containing photoreceptors (9–11). In addition to influencing seed germination, Phys govern a wide range of developmental responses throughout a plant’s life cycle via their ability to interact with and influence the abundance and activity of the PHYTOCHROME-INTERACTING FACTOR (PIF), basic helix–loop–helix (bHLH) transcription factor family (12–15).
One Arabidopsis PIF in particular, PIF1 (or PIL5), inhibits the completion of seed germination in darkness (16) by preventing radicle protrusion through an indirect repression of bioactive gibberellic acid (GA) accumulation (17, 18). PIF1 also represses the completion of seed germination in darkness by direct transcriptional stimulation of genes encoding the GRAS-domain-containing DELLA proteins REPRESSOR OF ga1-3 (RGA) and GA-INSENSITIVE (GAI) (19) that act to decrease GA sensitivity, thereby repressing the completion of seed germination (17).
Upon illumination of imbibed seeds or etiolated seedlings, PIF1 interacts with one or more photoactivated Phys, triggering PIF1 phosphorylation (20). Binding of PIFs to photoactivated Phys also strips the PIF dimers from their cognate DNA-binding elements (21). In addition, families of HLH proteins are present in plants that interfere with bHLH transcription factor dimerization (necessary for DNA binding) through the formation of non–DNA-binding heterodimers (22–25). One such HLH protein, LONG HYPOCOTYL IN FR LIGHT (HFR)-1 increases in abundance in the light (26) and binds PIF1 to further attenuate its interactions with DNA (27). Through this collective inhibition, light relieves PIF1-mediated suppression without reducing PIF1 nuclear titer.
Upon binding photoactivated Phys, many PIFs undergo phosphorylation and rapid declines in abundance (28, 29) due to polyubiquitination and 26S proteasomal degradation, a system exquisitely suited to mediate in unidirectional, all-or-nothing developmental switches (e.g. the completion of germination) (30). In Arabidopsis, an aberrant member of the base of the proteasomal regulatory particle (rpn10-1), while viable, leads to a plethora of perturbations, including seed germination (31), implicating proteasomal-mediated protein degradation in the normal completion of germination. A defined role for the 26S proteasome in seed germination was acquired from work with the F-BOX protein SLEEPY1 (SLY1) responsible for the degradation of members of the GRAS domain-containing family of transcription factors (namely RGA and GAI) inhibitory to the GA response (32–37).
Ubiquitin-mediated turnover is directed by a collection of E3 ligases that covalently attach chains of ubiquitin onto appropriate targets followed by the recognition and breakdown of these polyubiquitinated intermediates by the 26S proteasome. In Arabidopsis thaliana it had been demonstrated that the REALLY INTERESTING NEW GENE (RING) CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) protein, capable of E3 ubiquitin ligase activity, interacts with members of the family of four SUPPRESSOR OF PHYTOCHROME A-105 (SPA) proteins and associates with the CULLIN4 (CUL4) and DAMAGED DNA BINDING1 (DDB1) proteins to modulate a variety of plant developmental aspects, including both flowering time and photomorphogenesis (38, 39). Furthermore, various complexes of these proteins can recognize and recruit PIF1 in darkness to collectively inhibit photomorphogenesis, partially through enhanced LONG HYPOCOTYL5 (HY5) degradation (40). Upon illumination, PIF1, assisting CUL4COP1-SPA–mediated repression of photomorphogenesis, is, itself, polyubiquitinated by the complex, leading to its degradation (41).
A second influential family of ubiquitin E3 ligases encompasses the Skp1–Cdc53/Cullin–F-BOX protein (SCF) ligase complexes that use a polymorphic collection of F-BOX proteins for target recognition (42, 43). The F-BOX superfamily in plants is diverse, with ∼700 genes in the Arabidopsis genome (44). Genetic studies have linked SCF E3s to a number of cellular processes critical to plants, including the completion of seed germination (45–52).
Previously, we used an unbiased activation-tagging genetic screen to identify candidate Arabidopsis loci whose expression is positively correlated with the completion of germination. From analysis of a collection of mutants that completed germination faster at cold temperatures (53), we identified an F-BOX protein gene COLD TEMPERATURE GERMINATING (CTG)-10 (52). We present evidence that CTG10 interacts with PIF1 in vitro and in vivo, leading to a positive correlation between CTG10 titer and PIF1 destabilization in planta upon illumination.
Results
CTG10 Is Expressed in Mature Seeds and Pollen.
CTG10 (GenBank accession no. DQ666277; At4g19330) is an intronless gene encoding a 383-aa protein with an F-BOX domain of 43–47 residues and two KELCH repeats (52), possible sites of substrate recognition (54). CTG10 expression patterns were assessed using a promoter fragment 1,097-bp upstream of the start codon to drive β-glucuronidase (GUS) expression. GUS expression was detected in mature pollen and mature seeds (Fig. 1 A and B and Fig. S1). Expression was greatest in the lower hypocotyl, with no staining observed in the endosperm or adherent testa (Fig. 1B).
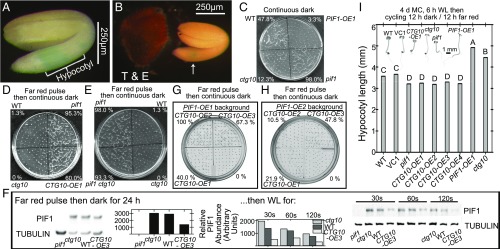
Fig. 1.
CTG10 expression has a limited tissue distribution, and a phenotypic parity of opposing mutations exists for PIF1 and CTG10. (A and B) GUS staining from pCTG10-GUS expression in mature seeds which were vacuum-infiltrated with substrate and left overnight before washing. The bracket in A and the arrow in B indicate the zone of GUS accumulation in the hypocotyl. GUS staining was never visible in the dead testa (T) tissue or the endosperm (E) of mature quiescent seeds or following imbibition. (Scale bars: 250 µm.) (C–E, G, and H) Completion of germination of defined genotypes of seeds in continuous darkness (C) or in constant darkness after an FRp-D (0.49 µmol⋅m−2⋅s−1) (D, E, G, and H) as scored after 1 wk (D and E) or 6 d (G and H) of imbibition. For scale, all Petri dishes are 9 cm across. The percentage of germination is provided in each image. (F) PIF1 protein abundance in the seeds of four genotypes following exposure to an FRp-D before 24-h darkness. When seeds are then exposed to light for various times, PIF1 protein is more stable in the ctg10 mutant seeds relative to WT, while PIF1 is destabilized in the seeds of a CTG10-OE line. All blots were repeated at least three times. (I) Hypocotyl lengths of defined genotypes after ensuring the completion of germination and growth under 12-h cycles of darkness and FR light (0.49 µmol⋅m−2⋅s−1). Significantly deviating average hypocotyl lengths (±SEM) (experiments were performed twice) were identified using Duncan’s multiple pairwise comparison and are indicated by different uppercase letters. Representative images of seedlings grown under this light regime are provided as Insets. (Scale bar: 1 mm.) VC, empty vector control.
Alteration of CTG10 Influences Germination Percentages.
Two-week afterripened CTG10-OE seeds completed germination to 75% within 5 d compared with only 20% for vector control seeds (Fig. S2A). If CTG10-OE results in faster than usual completion of germination (52, 53), a reduction of CTG10 amounts may detrimentally influence seed germination percentage. An exonic transfer DNA (T-DNA) insertion in CTG10 (SALK_104830; ctg10) was identified [SALK SIGnAL database (55)] and acquired from The Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH). Freshly harvested ctg10 seeds, without moist chilling, completed germination significantly less in continuous darkness than did WT control seeds harvested and treated in the same manner (Fig. 1C).
An extensive survey of the dormancy of variously afterripened seeds of diverse genotypes did not show alterations in seed dormancy. The single exception was the tendency for a greater percentage of partially afterripened, unchilled CTG10-OE2 seeds to complete germination, predicated on the presence of light (Figs. S2 B and C and S3).
Opposing Alterations in CTG10 and PIF1 Expression Produce a Parity of Phenotypes.
The inability of ctg10 seeds to complete germination in darkness to the same extent as WT seeds (Fig. 1C) was reminiscent of PIF1-OE seeds (16). CTG10-OE or pif1 seeds were capable of completing germination in darkness, following a pulse of far-red light before darkness (FRp-D) early during imbibition (Fig. 1D). Neither PIF1-OE nor ctg10 seeds completed germination well if they were kept in constant darkness (Fig. 1C) or were exposed to an FRp-D (see ctg10 in Fig. 1 D and E and PIF1-OE in Fig. 1 G and H).
Except for the pif1 genotype, PIF1 protein was detected in seeds exposed to an FRp-D (24 h in darkness) (Fig. 1F). When 24 h-dark imbibed seeds were subsequently exposed to light for 30, 60, or 120 s, PIF1 amounts declined most rapidly in the CTG10-OE3 line, while PIF1 was relatively more stable in ctg10 than in WT seeds (Fig. 1F). If CTG10 destabilizes PIF1 during seed germination, and CTG10 stimulates the completion of seed germination at 10 °C (53), then perhaps pif1 and/or PIF1-OE1 seeds would also have a phenotype at 10 °C. While there was no stimulation of the speed of seed germination completion for pif1, both ctg10 and PIF1-OE1 seeds lagged significantly behind the WT seeds from 5–7 d after imbibition at 10 °C (Fig. S4A). PIF1 exerts an indirect influence over the transcription of genes encoding abscisic acid (ABA)-synthetic and -metabolizing enzymes (17) and has a role in ABA signaling (56). Exogenous ABA might influence seed germination of the ctg10 mutant or CTG10-OE lines. Seeds of the CTG10-OE strain were no better than WT seeds in completing germination on a variety of ABA concentrations, whereas at 0.3 μM ABA, ctg10 and PIF1-OE seed performance was similarly delayed, relative to WT (Fig. S4B).
Double Mutants Reveal That pif1 Is Epistatic to ctg10.
The repression of radicle protrusion from ctg10 seeds germinated in continuous darkness (Fig. 1C), exacerbated by FRp-D (Fig. 1D), was alleviated in ctg10, pif1 double mutants (Fig. 1E). Simultaneous CTG10-OE PIF1-OE partially reversed PIF1-OE repression of seed germination after an FRp-D (Fig. 1 G and H). Additionally, PIF1-OE repression of the completion of germination under cycles of 12-h far-red (FR) light/12-h darkness was also alleviated in CTG10-OE, PIF1-OE seeds (Fig. S5A).
Another phenotype of pif1 mutants is a shorter hypocotyl than in WT under 12-h cycles of darkness and FR illumination (16). The CTG10-OE lines also had statistically significantly shorter hypocotyls under 12-h darkness/12-h FR light illumination cycles (Fig. 1I), whereas both the PIF1-OE and the ctg10 mutants had longer hypocotyls than WT under this light regime (Fig. 1I). Seedlings of all genotypes were statistically indistinguishable from WT when grown in continuous darkness or under 12-h light/12-h darkness (Fig. S5B).
Yeast Two-Hybrid Identification of PIF1 as a CTG10 Interactor.
A PIF1–CTG10 interaction occurred in yeast two-hybrid assays (Fig. S6). Both the N-terminal and C-terminal halves of PIF1 had lower affinity for CTG10 compared with the full-length PIF1 (Fig. S6B). The inclusion of increasing lengths of the bHLH domain with the N terminus of PIF1 resulted in stronger interaction between these PIF1 fragments and CTG10. Although the C-terminus of PIF1 without the bHLH domain showed no significant interaction with CTG10, its inclusion in the full-length PIF1 protein increased the affinity of CTG10 for PIF1, at least in yeast two-hybrid assays (Fig. S6B).
Association of PIF1 and CTG10 in Planta.
If the observed phenotypic alterations in seed germination and hypocotyl elongation are the result of CTG10–PIF1 interaction, then the two proteins must associate in the nucleus where PIF1 resides, and yet the CTG10 N terminus contains a predicted leucine-rich nuclear exclusion signal (NES) within the C-terminal portion of the F-BOX region (Fig. S7A) (57). Potentially, CTG10 could be excluded from the nucleus (and hence PIF1) until some signaling event (most obviously light) allows CTG10 nuclear access. Subcellular localization was preliminarily examined using a YFP:CTG10 fusion protein transiently expressed in tobacco leaf cells. Regardless of whether the leaf cells were cultured following bombardment for 1 d in darkness or in light, YFP was detectable in the nucleus (Fig. S7 B and C).
To provide evidence in support of CTG10 nuclear localization in Arabidopsis, functional CTG10:GFP fusions (Fig. 2 B and C and Fig. S8A) stably expressed in Arabidopsis seedlings were examined. Etiolated seedlings expressing GFP alone displayed signal throughout the cell but only a weak signal in the nucleus (Fig. 2A, GFP after 15-min light). Etiolated seedlings expressing a CTG10:GFP fusion behind the CaMV35S promoter were mounted and scanned as quickly as possible (darkness) or after 15 min of white light. CTG10:GFP was localized in the nucleus regardless of illumination (see Fig. 2 B and C for representative images). Neither the CTG10 C-terminal fusions with GFP nor the N-terminal HIS-myc fusions with CTG10, used to verify the specificity of the CTG10 antibody, interfered with the capacity of these CTG10-OE lines to promote the completion of germination in continuous darkness following an FRp-D (Fig. S8).
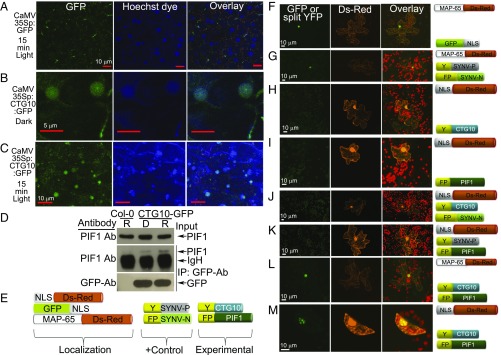
Fig. 2.
CTG10 is present in the nucleus, regardless of illumination, where it interacts with PIF1 as supported by both pulldown and BiFC assays in planta. (A–C) Cotyledon cells of etiolated Arabidopsis seedlings stably transformed with CaMV35S-driven GFP constructs after 15-min white light treatment before Hoechst dye immersion (A) and similar tissue of CaMV35S-driven CTG10:GFP translational fusions without (B) or with (C) 15-min white light treatment. (Scale bars: 5 µm in B and 10 µm in A and C.) (D) Four-day-old dark-grown Col-0 and CTG10:GFP seedlings were pretreated with 200 µM bortezomib for 4 h followed by either darkness (D) or red light (R, 100⋅µmol⋅m−2) treatment. Total protein was extracted in buffer, and CTG10:GFP was immunoprecipitated using the GFP antibody and probed with the PIF1 antibody. (Top) Input protein. (Middle and Bottom) CTG10:GFP immunoprecipitated samples probed with PIF1 (Middle) or GFP (Bottom) antibodies. IgH, Ig heavy chain; IP, immunoprecipitation. (E) Representations of the constructs used to assess the interaction of PIF1 and CTG10 in vivo. Three constructs (NLS:Ds-Red, GFP:NLS, and MAP-65:Ds-Red) were used as indicators of successful cellular bombardment and subcellular location. Two were positive controls of proteins (Y:SYNV-P and FP:SYNV-N) known to interact and fluoresce in the plant nucleus. Two (Y:CTG10 and FP:PIF1) were the respective experimental proteins. CTG10, COLD TEMPERATURE GERMINATING10; Ds-Red, long-wave-length-emission maximum fluorescent protein cloned from coral of the Discosoma species; FP, the C-terminal 67 amino acids of the YFP; GFP, a green fluorescent protein from the jellyfish, Aequorea victoria; MAP-65, microtubule-associating protein-65; NLS, nuclear localization signal; PIF1, the PHYTOCHROME-INTERACTING FACTOR1 protein; SYNV-N, Sonchus yellow net virus nucleocapsid protein; SYNV-P, Sonchus yellow net virus phosphoprotein; Y, the N-terminal 174 amino acids of the YFP. (F) Both the MAP-65:Ds-Red and NLS:GFP localized as expected to the microtubules and the nucleus, respectively. (G) The viral P and N proteins interacted in the plant nucleus and led to the fluorescence of the split YFP under the experimental conditions employed. (H–K) Neither the Y:CTG10 nor the FP:PIF1 fusion protein was capable of fluorescing alone (H and I) or when coupled with an inappropriate binding partner Y:CTG10 FP:SYNV-N (J) or Y:SYNV-P FP:PIF1 (K). (L and M) When Y:CTG10 was cobombarded with FP:PIF1, a strong, nuclear-localized yellow fluorescence was observed. The yellow fluorescence visible in M is not due to bleedover of nuclearly localized Ds-Red into the yellow channel, as it was not observed in H–K. (Scale bars: 10 µm in F–M.)
Transgenic plants expressing functional CTG10-GFP protein were used to perform in vivo coimmunoprecipitation assays. These demonstrated that CTG10 associated with PIF1 both in darkness and in light (Fig. 2D); however, this association was enhanced in response to a light stimulus. These data support the contention that CTG10 and PIF1 associate in planta and that this association is promoted by light.
To further assess the physical association between CTG10 and PIF1 in planta, bimolecular fluorescence complementation (BiFC) experiments were performed using Nicotiana tabacum leaves. YFP fluorescence accumulated in the nuclei 24 h following bombardment with gold particles coated with constructs for fluorescent-positive controls [GFP-nuclear localization signal (NLS)] and for the two nuclear-localized interacting proteins of the Sonchus yellow net nucleorhabdovirus (SYNV): the phosphoprotein (P) and the HLH-containing nucleocapsid protein (N) (Fig. 2 F and G and ref. 58). In no instance did the split YFP constructs for CTG10 and PIF1 provide fluorescent signal when they were introduced into cells without an appropriate partner (Fig. 2 H–K). Only when these two constructs were cobombarded did signal accumulate in the nuclei (Fig. 2 L and M).
Seed CTG10 and PIF1 Proteins Fluctuate Depending on Impinging Light and Hours After Imbibition.
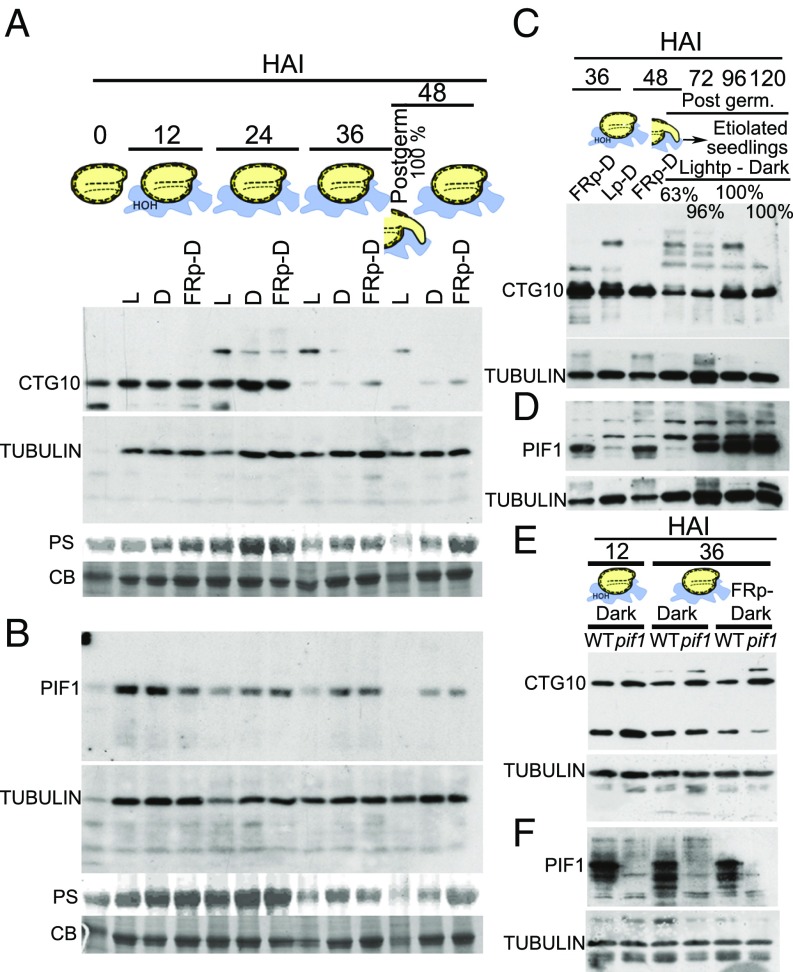
CTG10-specific antibodies (Fig. S8C) demonstrated the presence of CTG10 protein stored in the mature, dehydrated seed (0 time point in Fig. 3A). Western blots revealed that the CTG10 protein was unstable in seeds beyond 24 h after imbibition (HAI) (Fig. 3A and Fig. S9 A and B). In contrast to CTG10, PIF1 protein was present in very low amounts in the mature, dehydrated seed but subsequently increased at 12 HAI, regardless of illumination (Fig. 3B and Fig. S9 A and B). At and beyond 24 HAI, PIF1 declined in abundance predicated on the presence of light conducive to Phy activation; however, PIF1 also declined somewhat over time in darkness following an FRp-D and in continuous darkness, although these seeds still retained detectable amounts of PIF1. Upon completion of germination in the light (48 HAI L) (Fig. 3 A and B and Fig. S9 A and B), neither CTG10 nor PIF1 was abundant.
Fig. 3.
Both CTG10 and PIF1 proteins are present in seeds before the completion of germination, and their amounts attenuate before radicle protrusion but reaccumulate if seedlings are etiolated. (A and B) Western blots of mature, dehydrated WT seeds or WT seeds exposed to light (L), kept in darkness (D), or exposed to an FRp-D (0.49 µmol⋅m−2⋅s−1) before being placed in darkness for 12, 24, 36, or 48 HAI. All blots were repeated at least three times. (A) Blots were probed with CTG10 antibody and reprobed with tubulin antibody. (B) Blots of the protein extracts in A probed with PIF1antibody and reprobed with tubulin antibody. CB, Coomassie Blue; PS, Ponceau S. (C and D) The time course of WT seed germination and seedling establishment on water, designed to permit completion of germination and subsequent etiolated seedling generation after various light treatments. Western blots of proteins from WT seeds exposed to a light pulse and then incubated in darkness for 72, 96, or 120 h. The seeds had all completed germination (Post germ.), and extracts were from these etiolated seedlings. All blots were repeated at least three times. The blots were probed with either CTG10 antibody (C) or PIF1 antibody (D) and then were reprobed with tubulin antibodies. The percentage of germination is provided when it had occurred. (E and F) WT or pif1 seeds were kept in darkness for 12 or 36 h or were exposed to an FRp-D (0.49 µmol⋅m−2⋅s−1) before being kept in darkness for 36 h. Protein extracts were taken from these seeds at these times and were separated and blotted. Immunoblots were probed with CTG10 antibody (E) or PIF1 antibody (F), and both blots were reprobed with tubulin antibody. CTG10 and PIF1 band volumes in A and B at T0 (low tubulin signal) were normalized by first making PS-stained bands at T0 and T12-h light relative to each other (T0/T12). This relative quantity was then multiplied by the T12-h light tubulin relative value, and the T0 tubulin band volume was represented by this value (Fig. S9).
Embryo-to-Seedling Transition: CTG10 and PIF1 Reaccumulate in Etiolated Seedlings.
Neither CTG10 nor PIF1 was abundant in the seeds that had completed germination following 48 h in the light (Fig. 3 A and B and Fig. S9 A and B). This is perplexing, because seeds completing germination in the dark produce etiolated seedlings in this developmental continuum. PIF1 is responsible for a plethora of phenotypes associated with seedling etiolation (59, 60). Then, following the completion of germination, PIF1 must reaccumulate in etiolated seedlings if it is to orchestrate seedling etiolation. An additional question was whether CTG10 also increased in abundance during seedling etiolation. This would also determine if the reduction in CTG10 protein was due to the completion of germination per se (and therefore was permanently switched off after this developmental event) or if it, too, could reaccumulate following the completion of germination if light was removed to produce an etiolated seedling.
All seeds that had been exposed to a pulse of white light during germination before darkness (Lp-D) were capable of completing germination at 72 HAI, while those exposed to an FRp-D were not (Fig. 3 C and D). When comparing FRp-D with Lp-D at 36 and 48 HAI, it is evident that both PIF1 and CTG10 proteins are declining as seeds approach completion of germination (Fig. 3 C and D and Fig. S9 C and D). However, the overall decline in the abundance of both CTG10 and PIF1 proteins in seeds up to 48 HAI when treated with Lp-D is reversed in etiolated seedlings following the completion of germination in darkness (72–120 HAI in Fig. 3 C and D and Fig. S9 C and D). The reaccumulation of both CTG10 and PIF1 provides the seedlings with PIF1 for proper etiolation responses as well as an F-BOX protein capable of helping target PIF1 for degradation upon illumination of the seedlings.
PIF1 Represses CTG10.
The fact that one of the two proteins reaccumulating in tandem in etiolated seedlings was a transcription factor suggested that PIF1 was stimulating CTG10 expression. However, it was possible that CTG10, when targeting PIF1 for polyubiquitination and degradation, was also destroyed, allowing it to reaccumulate under conditions where PIF1 was stable. To deconvolute PIF1 stimulation of CTG10 transcription from PIF1-coupled CTG10-concurrant degradation, light environments in which CTG10 and PIF1 coexist (continuous darkness and FRp-D) were chosen, and WT or pif1 seeds were exposed to these conditions. Under conditions in which PIF1 was present and stable (WT seeds), CTG10 amounts were less than in the pif1 mutant, particularly later during the seed germination time course (Fig. 3 E and F), suggesting that CTG10 expression was influenced by PIF1.
PIF1 Is Lost More Rapidly from WT than from ctg10 Etiolated Seedlings Exposed to Light.
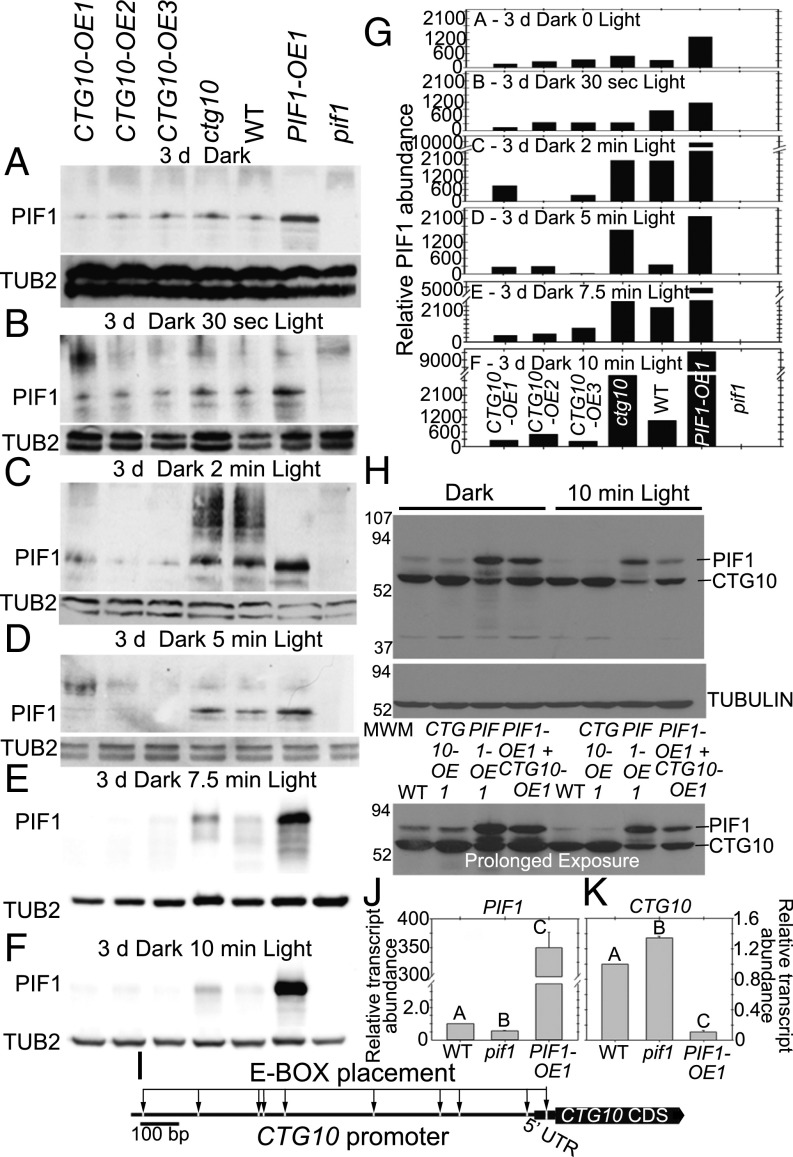
While CTG10 and PIF1 amounts change over the course of many hours during seed germination (Fig. 3) (60, 61), changes in PIF1 abundance are known to be very rapid (within minutes) in etiolated seedlings exposed to illumination (62). The rapid loss of PIF1 protein from etiolated seedlings upon illumination was judged to be the most sensitive light-dependent transition with which to monitor PIF1 stability and its control over CTG10 expression. Both the completion of seed germination in darkness and seedling hypocotyl elongation were influenced in the ctg10 mutant (Fig. 1 C and H), and so enhanced PIF1 stability should be evident using the ctg10 mutant. PIF1 protein was detectable in seedlings etiolated for 3 d in all genotypes examined except pif1 (Fig. 4 A and G). While WT seedlings retained detectable amounts of PIF1 even after 10-min exposure to light, PIF1 protein abundance was considerably reduced in the CTG10-OE lines within this period (Fig. 4 A–G). Both PIF1-OE and ctg10 seedlings retained more PIF1 than WT seedlings for the last 5 min of illumination (Fig. 4 D–G). The abundance of PIF1 among genotypes was quantified relative to tubulin amounts within each blot and, following background subtraction (pif1 lane), was graphically depicted to further document the retention of PIF1 in the ctg10 and PIF1-OE seedlings compared with the WT and CTG10-OE lines (Fig. 4G).
Fig. 4.
In seedlings, PIF1 is stabilized upon illumination in the ctg10 mutant relative to the WT or CTG10-OE lines, and these two proteins negatively influence each other through different mechanisms. (A–F) After 72 h darkness etiolated WT, OE, and mutant seedlings were illuminated for various times over a 10-min time period. PIF1 amounts in the seedlings were assessed using PIF1 antibody after 0 (A), 0.5- (B), 2- (C), 5- (D), 7.5- (E), and 10-min (F) exposure to white light. Blots were reprobed with tubulin antibody demonstrating equal loading/transfer. (G) PIF1 band volumes were normalized within blots for tubulin amounts (equal loading). Following background subtraction (PIF1 band volume in pif1), the relative amount of PIF1 for each genotype was displayed graphically for each exposure time to further emphasize PIF1 stability within ctg10. Relative band volume is depicted on the ordinate axis. (H) PIF1 and CTG10 protein abundance in etiolated seedlings before and after exposure of various genotypes (WT, PIF1-OE, CTG10-OE, and PIF1-OE + CTG10-OE) to light for 10 min. The blot was reprobed with tubulin antibody. Two exposures to film are provided for the PIF1- and CTG10-probed blot. (I) Schematic of the promoter, 5′ UTR, and CDS of CTG10 indicating all 10 E-BOX motifs (denoted by arrows and white gaps). (Scale bar: 100 bp.) (J and K) Real-time qPCR of cDNA from mRNA extracted from WT, pif1, and PIF1-OE 72-h etiolated seedlings using primers for PIF1 (J), CTG10 (K), or ACTIN2 (control). Average expression amounts (±SEM) relative to ACTIN2 are presented (n = 3). Significantly deviating average relative expression levels were identified using Duncan’s multiple pairwise comparison and are represented by different uppercase letters.
CTG10 and PIF1 Protein Abundance Negatively Influence Each Other in the Light.
Seeking to clarify the consequences of PIF1 influence on CTG10 amounts, we examined protein blots from seedlings of WT, CTG10-OE, PIF1-OE, and a PIF1-OE CTG10-OE line, all etiolated for 5 d and then provided light for 0 or 10 min before extraction. The results demonstrate that PIF1 amounts were stable in dark-grown seedlings, regardless of CTG10 amounts (Fig. 4H, dark versus 10-min light comparison for all genotypes tested; obvious in prolonged exposure). However, the amount of CTG10 present in the etiolated seedlings (dark in Fig. 4H) was reduced by PIF1-OE but only when CTG10 transcription was driven solely by its endogenous promoter (Fig. 4H, Upper; PIF1-OE compared with WT). Upon transfer to the light for 10 min, PIF1 amounts declined (Fig. 4H, 10-min light compared with dark for all genotypes). As well, with greater amounts of CTG10 present in the seedlings (Fig. 4H, Upper, dark, CTG10-OE and double-OE compared with WT and PIF1-OE, respectively), less PIF1 protein was present following 10-min illumination (Fig. 4H, Upper and Lower, 10-min light, CTG10-OE and double-OE compared with WT and PIF1-OE, respectively). However, the converse was also true, because with more PIF1 present in etiolated seedlings (Fig. 4H, Upper, dark, CTG10-OE compared with double-OE), the less CTG10 remained following 10-min illumination (Fig. 4H, Upper, 10-min light, CTG10-OE compared with double-OE).
There are no G-BOXES (CACGTG) in the CTG10 promoter, but there are nine E-BOXES (CANNTG) in the CTG10 promoter and one in the 5′ UTR (Fig. 4I). To examine possible CTG10 regulation by PIF1 we used RT-qPCR analysis of PIF1 or CTG10 transcript amounts in WT, pif1, and PIF1-OE etiolated seedlings. As anticipated for PIF1 transcript amounts, when examined in these genetic backgrounds, less and more transcript, respectively, was evident in 3-d etiolated pif1 and PIF1-OE seedlings relative to WT seedlings (Fig. 4J). There was a slight but significant increase in CTG10 transcript abundance in the pif1 seedlings and a considerable reduction of CTG10 in PIF1-OE etiolated seedlings, relative to WT (Fig. 4K).
Discussion
Physiological and genetic evidence from seed and seedling growth-habit alterations influenced by mutation or OE and the behavior of double-mutant and double-OE lines indicates that CTG10 helps regulate PIF1 amounts. Enhanced light-mediated degradation of PIF1 in CTG10-OE lines and PIF1 stabilization in the ctg10 mutant, combined with a demonstration of the physical interaction of CTG10 with PIF1 through yeast two-hybrid, BiFC, and coimmunoprecipitation assays, constitutes biochemical and molecular evidence that corroborates the role of CTG10 in the regulation of PIF1.
Factors up-regulated in the cold-temperature germinating (CTG) screen resulting in more rapid completion of germination at 10 °C (53) were predicted to have a positive role in normal seed germination. This was affirmed by the identification of CTG10 as an F-BOX protein promoting the destruction of PIF1, a bHLH transcription factor negatively influencing the completion of germination (16). Moreover, the phenotype of CTG10-OE seeds indicates that the abundance of the F-BOX protein can be a bottleneck in PIF1 degradation by the 26S proteasome, despite the efficiency with which CUL4cop1-spa acts (41). It was intriguing that the seed tissue most effectively expressing the GUS reporter from the CTG10 promoter was the lower hypocotyl (Fig. 1 A and B). This region has been demonstrated to be the first region of the Arabidopsis embryo to commence elongation, resulting in radicle protrusion, the culmination of germination (63). This tissue region, known as the “photosensitive site,” is also the only one in the positively photoblastic lettuce (Lactuca sativa) embryo known to be necessary to perceive impinging red light and thereby orchestrate the completion of germination in this species (64).
Why would CTG10 be identified in a CTG screen? The bHLH transcription factor SPATULA (SPT) has been recognized as a light-stable repressor of seed germination in dormant Arabidopsis seeds whose influence can be removed by either moist chilling or dry afterripening (65). The CTG screen was designed with this in mind, as Arabidopsis (Col-0) dormancy is alleviated at 10 °C (53), thus removing the completion of germination from SPT control. Still, the response to cold stress is light dependent (specifically through Phy), signifying that light- and cold-signal transduction are inexorably linked (66–68). Indeed, seeds of both the ctg10 and PIF1-OE1 lines were delayed in completing germination when imbibed at 10 °C. For some species, brief red illumination is a more potent stimulus of subsequent completion of germination if it is provided at low temperature (69). Furthermore, the identity of the Phy most instrumental for stimulating the completion of germination changes in Arabidopsis and is based on the temperature under which the seeds are germinating (70). A series of requirements for PIF destruction upon seedling illumination have been documented (71), any or all of which may be similar in seeds and have slower kinetics at lower temperatures, leading to conditions allowing the identification of CTG10 overexpression at low temperatures. Alternatively, PIF1 is known to exert part of its negative influence over the completion of germination in darkness by directly enhancing the transcription of genes encoding RGA and GAI (17), two DELLA proteins that are themselves repressive of GA responses (72). Additionally, PIF1 modifies bioactive GA production (18) by its indirect influence over genes encoding enzymes involved in GA synthesis and degradation (17). Consistent with PIF1’s role as a primary deterrent of the completion of germination in darkness, it was found to stabilize ABA titer in the seed via its indirect influence over the transcription of genes encoding ABA-synthetic and -metabolizing enzymes (17). In a detailed examination of four dormancy-alleviating treatments in Arabidopsis ecotype Cape Verde Islands, it was hypothesized that cold temperature stimulated the production of GA precursors, whereas light enhanced their use in the synthesis of bioactive GA (73). Thus PIF1 is indirectly involved in (i) repressing bioactive GA production and sensitivity while (ii) maintaining seed ABA amounts. This may explain why, in a line hyperexpressing CTG10, seed germination under continuous light at low temperature (imposing more rapid PIF1 degradation relative to WT) would result in a system that can amplify minor temporal differences in PIF1 degradation (due to the overall slower rate of germination at 10 °C) to the extent that it was a visible, selectable phenotype.
How can both CTG10 and PIF1 be present in the seed at the same time and place? Western blot results demonstrated that both CTG10 and PIF1 are present in mature, quiescent seeds, with PIF1 increasing in abundance early during germination (Fig. 3B), so a PIF1–CTG10 interaction early during seed germination is not prevented through a temporal control of transcription, translation, or CTG10 stability. Neither is there evidence that the predicted NES in the CTG10 F-BOX N terminus functions to compartmentalize CTG10 (Fig. S7) from its nuclear-localized substrate PIF1, at least not in Arabidopsis seedlings (Fig. 2 A–C) or tobacco leaves (Fig. 2 L and M). Pertaining to seed germination 12 HAI in the light, it was surprising that PIF1 amounts increased relative to mature, dehydrated seeds despite the presence of CTG10 (Fig. 3 A and B). Arabidopsis seeds placed on water for 12 h would be fully hydrated at this time in the lag phase of germination (seed expansion from water uptake is complete after ∼10 h) (74). They would also be capable of responding to incident light because PhyB, at least, is also present at this time (75), and PIF1 has affinity for PhyB (62), which should permit proper light-signal transduction into the nucleus and ultimately PIF1 recognition by CTG10 (or any other E3 ligase assembled at the time). Furthermore, PIF1 amounts decline only slightly 24 HAI in light despite the presence of CTG10 (Fig. 3). Only at 36 HAI and beyond in light do PIF1 (and CTG10) amounts decline considerably. GA signaling has been described as a de-repressible system (76), and the same could be said for seed germination with respect to the events required to permit its completion, which must act to remove previously imposed impediments. It is in keeping with this concept that PIF1 protein abundance may increase initially after imbibition to set up PIF1-mediated repression, being somehow impervious to SCFCTG10 (or any other E3, e.g., CUL4cop1-spa)-mediated degradation during this period, regardless of illumination. Only subsequently would the environmental conditions (in this case, light) determine whether this obstruction is retained or removed, and apparently 24 h in continuous light is barely sufficient time to initiate all the systems required for light-mediated reduction of PIF1 amounts. A similar scenario exists for the DELLA protein RGL2. This repressor of seed germination accumulates during prolonged (6-d) moist chilling in seeds of the Ler and Ws ecotypes (77) or from 0–12 HAI in unchilled Col ecotype seeds (78), only to be removed in WT seeds if conditions favor GA accumulation and ABA reduction.
PIF1 remained detectable much longer in seeds imbibed in the light or shown a pulse of white light (Fig. 3 B and D) than it did in etiolated seedlings, where it is almost below the level of detectability after 10-min illumination (Fig. 4 A–G). Slower PIF1 degradation in seeds relative to seedlings has been demonstrated previously (60, 61) and has been confirmed in an extensive germination time course here. This could be due to more efficient targeting of PIF1 by CTG10 in seedlings if the CTG10 titer is greater than in seeds early during germination. PIF1 protein amounts decreased more rapidly in etiolated CTG10-OE seedlings than in WT seedlings over 10-min exposure to light (Fig. 4 A–G). Yeast two-hybrid attempts to identify a single PIF1 degron recognized by CTG10 were unsuccessful (Fig. S6B). Both N- and C-terminal PIF1 moieties resulted in significant CTG10 interactions in yeast two-hybrid assays (Fig. S6B), suggesting that CTG10 may recognize multiple PIF1 degrons and providing a reason why CTG10 hyperproduction may target PIF1 for elimination more efficiently (i.e., via avidity increases rather than affinity for any one site). Indeed, both the PIF3 N- and C-terminal regions are required for its recognition by the light-response Bric-a-Brac/Tramtrack/Broad (LRB2) complex assisting polyubiquitination of this PIF (71). Furthermore, PIF1 requires phosphorylation before degradation, and the protein has been shown to be phosphorylated in multiple locations by multiple kinases (29, 79).
The results presented here imply only that CTG10 has PIF1 as one of its targets (and not necessarily a direct target), not that PIF1 is the sole target of CTG10 or that the germination phenotype documented for seeds from CTG10-OE plants is caused solely by more efficient (rapid) PIF1 destruction. Similarly, PIF1 is known to be the target of another E3 ubiquitin ligase (i.e., CUL4cop1-spa) leading to its destruction (41), a situation hypothesized for PIF3 after the observation that, even in triple mutants (which remove all three family members) of the LRB family, PIF3 slowly declines in abundance (71), suggesting that other E3 ligases may participate in its degradation.
It is certainly intriguing that PIF1 can somehow (directly or indirectly) transcriptionally repress CTG10 (Fig. 4K), the promoter of which contains no G-BOX elements (Fig. 4I). Although CTG10 was not recognized as a PIF1 target in a genome-wide survey of PIF1-binding sites in Arabidopsis, this does not preclude direct PIF1 binding to CTG10, because the state of the art at the time utilized arrays that did not include CTG10 (19). PIF1 is known to form heterodimers with a variety of bHLH proteins (80), and the 10 E-BOXES present in the CTG10 promoter 5′ UTR (Fig. 4I) implies that, should PIF1 exert its repressive influence directly, it may do so either as a homodimer, similar to PIF3 (81), or as a member of a bHLH heterodimer. This raises the question of whether any such participating bHLH proteins may also be CTG10 targets. Certainly, the contention that bHLH heterodimers may have greater affinity for E-boxes than homodimers in Arabidopsis has precedents in other light-regulated systems (82) and during seed development (83).
It could be expected that, in constant light, CTG10 targeting of PIF1 during germination would result in the low abundance of PIF1, permitting the completion of germination, and that subsequent photomorphogenic seedling development would not require CTG10 protein because (i) no PIF1 was present, or (ii) active PhyB and/or HFR1 were sequestering PIF1 away from its DNA targets (21, 27), or (iii) CUL4cop1-spa-mediated PIF1 recognition and (upon illumination) degradation is capable of efficiently eliminating the transcription factor (41). However, seeds completing germination in darkness produce etiolated seedlings, for which PIF1 is necessary (20, 84). In etiolated seedlings PIF1 reaccumulates, but given PIF1’s repressive influence on CTG10 transcription (Fig. 4K), it was unexpected that CTG10 should also increase in abundance in darkness concomitant with PIF1. Although we have not yet tested this assumption, the lack of an hypothesized PIF1 bHLH-binding partner when seeds complete germination in darkness may remove the E-BOX–containing CTG10 promoter from PIF1 repression, allowing the concurrent increase of both PIF1 and CTG10 during etiolation. Hence, CTG10 is not irrevocably eliminated upon the completion of germination (Fig. 3C) and is available to assist in a proper de-etiolation response upon illumination.
CTG10 is one F-BOX protein interacting with the bHLH transcription factor PIF1, mediating PIF1’s degradation during seed germination and during seedling de-etiolation. Nevertheless, PIF1 is impervious to proteolysis early during germination, regardless of light. PIF1 can somehow down-regulate the expression of CTG10. Despite PIF1 reaccumulation in etiolated seedlings, it fails to repress CTG10 protein accumulation in them.
Methods
Plant material and vector construction are described in SI Methods.
Double-OE and Double-Mutant Generation.
Two PIF1-OE lines (kindly provided by Giltsu Choi, Korea Advanced Institute of Science and Technology, Daejeon, South Korea) were crossed with three CTG10-OE lines (52). Double-homozygous (PIF1-OE CTG10-OE) plants were identified by BASTA foliar application (CTG10-OE) and by kanamycin resistance (PIF1-OE lines). The ctg10 line (52) was crossed with the pil5-1 line (16) (kindly provided by Giltsu Choi), and homozygous mutant plants were identified in the F3 generation by PCR analysis.
CTG10 Promoter-GUS.
To determine the localization of CTG10 expression, various plant parts from pCTG10:GUS lines were submerged in a solution containing 5-bromo-4-chloro-3-indolyl glucuronide (85) in microcentrifuge tubes. The tubes were placed in a desiccator, and a vacuum was applied and released three times (for 1 min each at 1-min intervals) to infiltrate the tissue with the substrate. Thereafter, the tubes were closed and placed in the dark at 37 °C overnight. The following day the substrate was removed and replaced with 70% EtOH twice to wash the tissue and then, when beneficial, left in 95% EtOH for 24 h to clear the tissue. Histochemical evidence of GUS activity relative to untransformed controls was recorded using a Zeiss Stemi SV11 or Axioplan 2 microscope attached to an AxioCam MRc 5 digital camera using AxioVision 4.8 software.
Phenotypic Analysis of CTG10 and PIF1 Single and Double Mutants, and OE Lines.
Germination assay and plant growth.
Unless otherwise stated, germination assays were at 25 °C. Seed germination (completed as soon as the radicle protruded beyond the testa) assays comprised three replications of 50 seeds each placed on two layers of Whatman no. 1 filter paper (Whatman) moistened with 2 mL distilled, deionized water. For seed germination assays on sterile medium, three replications (unless otherwise stated) of 50 seeds each were surface sterilized and plated on aqueous agar medium [0.6% (wt/vol) agar, 10 mM MES (pH 5.7)], which in one experiment also contained filter-sterilized ABA at defined concentrations. Depending on the experiment, plates were immediately wrapped in three layers of aluminum foil (darkness), moist chilled at 4 °C for 3 d or not, and then were treated with a 5-min FRp-D or with 12-h FR/12-h darkness cycles at 0.49 µmol⋅m−2⋅s−1) or were provided with white light as a pulse or continuously [photosynthetic active radiation (PAR) of 135 μmol⋅m−2⋅s−1]. Plants grown to produce seeds were also grown at 25 °C and PAR of 135 μmol⋅m−2⋅s−1. Seed germination percentage was assessed each day or 6 or 7 d after imbibition.
Hypocotyl elongation assay.
To assess the effect of FR light on hypocotyl length, seeds from CTG10-OE, ctg10, pif1, or PIF1-OE plants, a plant containing the empty vector control, and WT Col-0 plants were surface sterilized, plated on MS agar [half-strength MS; 38 mM MES (pH 5.7), 0.6% (wt/vol) agar] and imbibed for 4 d at 4 °C in the dark. Moist chilled seeds were then placed at 25 °C and provided white light for 6 h before the plates were incubated for five additional days in (i) continuous darkness, or (ii) under a 12-h FR light/12-h darkness cycle, or (iii) under a 12-h white light/12-h darkness cycle. For each genotype, 50 establishing seedlings were randomly selected, and the hypocotyl length for the longest 30 seedlings for each genotype was recorded (16). These experiments were repeated twice. White light was provided at 135 μmol⋅m−2⋅s−1 PAR.
Yeast two-hybrid assay.
A possible interaction between CTG10 and PIF1 (At2g20180) based on a parity of phenotypes between opposite mutants was explored using the GAL4BD CTG10 coding sequence (CDS) fusion and PIF1 expressed as a C-terminal fusion to GAL4AD in pAD-GAL4-2.1 (Stratagene Agilent Technologies Division). Both bait and prey constructs were cotransformed into an YRG-2 yeast strain and selected on synthetic complete (SC) medium lacking leucine and tryptophan (Becton Dickinson Biosciences Clontech). Protein interaction was assayed by histidine resistance and β-galactosidase liquid assay using o-nitrophenyl β-D-galactopyranoside (ONPG) as a substrate (86, 87). Protein concentrations were determined using the Bradford (88) protein assay (Bio-Rad Laboratories). Attempts to switch vectors to verify the CTG10/PIF1 interaction were unsuccessful due to the ability of PIF1 to act as a transcriptional activator when it contained a DNA-binding domain.
Examination of the regions of the PIF1 protein with which the CTG10 was interacting was performed by amplifying (Table S1) and cloning various regions of PIF1 into pAD-GAL4-2.1 and assessing the strength of their interaction with CTG10 relative to the full-length PIF1 by liquid ONPG assay (86, 87).
BiFC analysis.
BiFC analysis used transient expression of CTG10 fused with the N-terminal portion (174 amino acids) of the EYFP reporter (Y:CTG10), and PIF1 fused to the C-terminal moiety (67 amino acids, FP:PIF1) was orchestrated through Gateway technology (Invitrogen; Thermo Fisher Scientific) into pSITE-BiFC vectors (89). Negative controls included Y:CTG10 or FP:PIF1 alone or mismatched with inappropriate, nuclearly localized binding partners, Y:CTG10 + FP:SYNV-N and Y:SYNV-P + FP:PIF1. Cells receiving plasmid-coated gold particles and their transcriptional/translational capacity was confirmed by including a plasmid constitutively expressing nuclearly localized Ds-Red, nuclearly localized GFP, or the tubulin-targeting MICROTUBULE-ASSOCIATED PROTEIN65-1 (MAP65-1:Ds-Red) (90) on the particles for each bombardment.
Transient and stable expression analysis of CTG10 subcellular localization using confocal imaging.
Transient YFP:CTG10/AtZFP11:dsRED assays were conducted by the introduction of Nanogold particles (Bio-Rad Laboratories) coated with both plasmids into tobacco (Nicotiana tabacum L. cultivar KY160) leaf cells using a Biolistic PDS-1000/He particle-delivery system (Bio-Rad Laboratories).
Leaf cells were cultured for at least 24 h on medium (91) either under low light (light) or wrapped in two layers of aluminum foil (darkness). The cells were quickly mounted on slides under very dim light, and emission from the various reporter genes and, in some instances, chlorophyll autofluorescence, was viewed using an Olympus FV1000 laser-scanning confocal microscope (Olympus America, Inc.). Assays of CTG10 residence also used etiolated seedlings stably expressing CTG10:GFP fusions under the control of the CaMV35S promoter, mounted in Hoechst’s dye, that had or had not been exposed to light before visualization.
Antibody preparation and Western blotting.
Epitope Informatics, Ltd. was contracted to examine the CTG10 and PIF1 for unique regions predicted to be of high antigenicity. Peptides (21-mers including a noncoded C-terminal cysteine; CTG10: NH2-MAYLSFKSNMERTPRESNTPC-COOH; PIF1: NH2-EKTNVDDRKRKEREATTTD EC-COOH) were synthesized (United Biochemical Research, Inc.) to the most unique antigenic region, and ∼5 mg was linked to Keyhole limpet hemocyanin, and an additional ∼10 mg was linked to agarose gel (Strategic Diagnostics Inc., now SDIX, LLC). Two New Zealand White rabbits were immunized for each antibody, and boosters for both antigens were provided. Serum was prepared, and antibody was affinity-purified over the appropriate agarose-linked peptide. Aliquots of the affinity-purified antibody were stored frozen until use. Tubulin and hexahistidyl-epitope antibodies were purchased from Sigma-Aldrich Co. and Qiagen, respectively.
Seeds/seedlings subjected to a variety of light regimes before protein extraction were ground in liquid nitrogen and homogenized in extraction buffer [50 mM Tris⋅Cl (pH 8), 10 mM EDTA, 50 mM NaCl, 0.5% (wt/vol) SDS, 10 mM β-mercaptoethanol (Fisher Scientific), and Sigma-Aldrich plant protease inhibitor mixture (1:100)]. The homogenate was centrifuged at 8,100 × g for 10 min at 4 °C; the supernatant was collected, and aliquots were made for Western blot analysis. Proteins were quantified using the Bradford assay according to the manufacturer’s protocol (Pierce). Protein extracts were size fractionated using 10% SDS/PAGE and transferred to nitrocellulose membrane (Protran BA85; Whatman International, Ltd.) using a mini Trans-Blot Electrophoritic transfer cell (Bio-Rad) with transblot buffer [25 mM Tris, 192 mM glycine, and 20% (vol/vol) methanol]. The Western blot was performed according to published procedures (92) using the KPL LumiGLO Chemiluminescent Kit (Kirkegaard & Perry Laboratories, Inc.) and X-ray film. CTG10, PIF1, or hexahistidyl primary antibodies were all used at 1:25,000 dilutions in blocking solution, except for PIF1 for 24 h dark-imbibed seeds, and de-etiolating seedlings at 7.5- and 10-min light, where it was applied at 1/12,500 dilution. Enzyme-conjugated secondary antibody was applied at 1:30,000. As loading controls, tubulin was detected using anti-tubulin primary monoclonal antibodies [1:5,000 for seeds (Sigma-Aldrich Co. catalog no. T9028) or 1:1,000 for seedlings (Sigma-Aldrich Co. catalog no. T6074)] following the same procedure. Blots of seed proteins were also stained with Ponceau S Red Stain, and occasionally, a duplicate gel was run and stained with Coomassie Brilliant Blue. The tubulin antibody reacted poorly with protein from mature, dehydrated seeds. Ponceau S dye was used to visualize seed-storage proteins to demonstrate equal loading at this early time point. The storage proteins were degraded at later time points, especially in seeds about to complete or which had completed germination (see the Coomassie Blue-stained image of a gel of the same extracts used on the blot; Fig. 3 A and B), necessitating the use of the tubulin antibody immunoblots at later times to demonstrate equal loading.
Densitometric assessment.
Images of Western blots of PIF1 or CTG10 from a variety of genotypes and experiments were loaded into a GelAnalyzer 2010 (Lazor Software) along with images of the blots probed using tubulin antibody or stained with Ponceau S stain. The densities of the immunoreactive bands, following background subtraction, were normalized to constant tubulin amounts for each lane to account for loading variation and are presented as bar graphs.
Coimmunoprecipitation assay.
Four-day-old, CTG10-GFP–expressing dark-grown seedlings (∼400 mg) were pretreated with 200 µM of bortezomib (LC Laboratories) for 4 h. Pretreated seedlings then were either kept in the dark or were treated with red light (100 µmol/m2) and were immediately ground in extraction buffer [100 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.1% Tween20, 10% (wt/vol) glycerol, 1× protease inhibitor mixture (catalog no. P9599; Sigma-Aldrich Co.), 1 mM PMSF, 100 μM bortezomib, 25 mM β-glycerophosphate, 10 mM NaF, and 2 mM Na orthovanadate]. Crude extracts were subjected to centrifugation for 10 min at 16,000 × g, and supernatants were transferred to fresh tubes containing 20 µL of Dynabeads Protein A (catalog no. 10002D; Life Technologies Co.) bound to 1 µg of anti-GFP (catalog no. A11120; Thermo Fisher Scientific). Tubes were incubated in the dark for 2 h on a rotary tube mixer at 4 °C. After incubation, beads were washed three times with extraction buffer (without bortezomib) and were dissolved in 1× SDS loading buffer and heated at 65 °C for 5 min to elute proteins from beads. Proteins were separated using 8% SDS/PAGE and blotted onto PVDF membrane. Blots were challenged using PIF1 (see above) or GFP (catalog no. SC9996; Santa Cruz Biotechnology) antibodies.
Real-time qRT-PCR.
Primers for real-time qPCR were made to ACTIN2, PIF1, and CTG10 (Table S1) cDNA using IDT PrimerQuest (Integrated DNA Technologies, Inc.) and the Primer3 program (93, 94) and were analyzed using NetPrimer (PREMIER Biosoft; www.premierbiosoft.com/netprimer/). A Bio-Rad Laboratories iCycler permitted measurement of SYBR Green fluorescence to determine abundance of the PIF1 and CTG10 transcripts relative to ACTIN2.
Statistical Analysis.
Comparisons were performed among the CTG10-OE lines and vector controls. These comparisons included pif1-mutant or PIF1-OE lines (e.g., RT-qPCR transcript abundances). Differences in percentage of germination at representative time points after imbibition, assessed at 25 °C in the light, or after an FRp-D followed by darkness, or in continuous darkness, as well as hypocotyl lengths, transcript abundances, and Miller units of activity from yeast two-hybrid interactions and the fluorescent β-galactosidase (MUG) assay, were all subjected to ANOVA (SAS Institute Inc.) (95). If the ANOVA indicated that there were significant differences among means, Dunnett’s mean separation test was used to distinguish between WT (control) and mutants at α = 0.05. For comparisons among multiple empty vector and OE lines, or Miller units, Duncan’s test was used at α = 0.05.
Supplementary Material
Acknowledgments
Ms. Amy Crume provided the tobacco plants used for BiFC experiments. Prof. Giltsu Choi (Korea Advanced Institute of Science and Technology) kindly provided the pil5-1 and PIF1-OE lines. A modified pRTL2 vector (double CaMV35S promoter, Tobacco etch virus translational enhancer, NotI sites introduced 5′ and 3′ to the cassette) was the kind gift of Gulvadee Chaiyaprasithi. Prof. Rup K. Kar (Visva-Bharati University) provided useful ideas and encouragement at the inception of this project. Dr. Sharyn Perry (Plant Science Department, University of Kentucky) permitted the use of her growth chamber, and Dr. Tomokazu Kawashima (Plant Science Department, University of Kentucky) allowed the use of his imaging system. Mr. David N. Martin and Mr. Kim Schäfermeyer provided excellent technical assistance in aspects of the project. Funding support included a pilot project and research grant from the Kentucky Tobacco Research and Development Center at the University of Kentucky (to A.B.D.), National Science Foundation Division of Integrative Organismal Systems Collaborative Research Grant 0849230 (to A.B.D. and E.H.), NIH Grant 1R01 GM-114297 (to E.H.), National Science Foundation Supplement 0849230 (to T.D.L. and A.B.D.), an American Society of Plant Biologists Summer Undergraduate Research Fellowship (to T.D.L.), US Department of Agriculture–National Institute of Food and Agriculture Seed Grant 2011-04375 (to A.B.D.), and Kentucky Agricultural Experiment Station Grant KY011038 (to A.B.D. and L.M.A.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711919115/-/DCSupplemental.
References
- 1.Bewley JD, Black M. Seeds: Germination, structure, and composition. In: Bewley JD, Black M, editors. Seeds: Physiology of Development and Germination. 2nd Ed. Plenum; New York: 1994. pp. 1–34. [Google Scholar]
- 2.Winter D, et al. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassel GW, et al. Elucidating the germination transcriptional program using small molecules. Plant Physiol. 2008;147:143–155. doi: 10.1104/pp.107.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galland M, Job D, Rajjou L. The seed proteome web portal. Front Plant Sci. 2012;3:98. doi: 10.3389/fpls.2012.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weitbrecht K, Müller K, Leubner-Metzger G. First off the mark: Early seed germination. J Exp Bot. 2011;62:3289–3309. doi: 10.1093/jxb/err030. [DOI] [PubMed] [Google Scholar]
- 6.Flint LH, McAlister ED. 1935. Wave lengths of radiation in the visible spectrum inhibiting the germination of light-sensitive lettuce seed. Smithsonian Miscellaneous Collections (Smithsonian Institution, Washington, DC), Vol. 4, Publication 3334, pp 1–11.
- 7.Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK. A reversible photoreaction controlling seed germination. Proc Natl Acad Sci USA. 1952;38:662–666. doi: 10.1073/pnas.38.8.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borthwick HA, Hendricks SB, Toole EH, Toole VK. Action of light on lettuce-seed germination. Bot Gaz. 1954;115:205–225. [Google Scholar]
- 9.Butler WL, Norris KH, Siegelman HW, Hendricks SB. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc Natl Acad Sci USA. 1959;45:1703–1708. doi: 10.1073/pnas.45.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegelman HW, Firer EM. Purification of phytochrome from oat seedlings. Biochemistry. 1964;3:418–423. doi: 10.1021/bi00891a019. [DOI] [PubMed] [Google Scholar]
- 11.Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 12.Fankhauser C, et al. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- 13.Colón-Carmona A, Chen DL, Yeh KC, Abel S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000;124:1728–1738. doi: 10.1104/pp.124.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leivar P, Monte E. PIFs: Systems integrators in plant development. Plant Cell. 2014;26:56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosado D, et al. Phytochrome Interacting Factors (PIFs) in Solanum lycopersicum: Diversity, evolutionary history and expression profiling during different developmental processes. PLoS One. 2016;11:e0165929. doi: 10.1371/journal.pone.0165929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh E, et al. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16:3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh E, et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19:1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh E, et al. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- 19.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- 21.Park E, et al. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012;72:537–546. doi: 10.1111/j.1365-313X.2012.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, et al. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- 23.Hao Y, Oh E, Choi G, Liang Z, Wang ZY. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol Plant. 2012;5:688–697. doi: 10.1093/mp/sss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massari ME, Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, et al. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H, et al. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell. 2013;25:3770–3784. doi: 10.1105/tpc.113.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Bu Q, et al. Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J Biol Chem. 2011;286:12066–12074. doi: 10.1074/jbc.M110.186882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smalle J, et al. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell. 2003;15:965–980. doi: 10.1105/tpc.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGinnis KM, et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki A, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 34.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun TP. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154:567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dill A, Thomas SG, Hu J, Steber CM, Sun TP. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004;16:1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyler L, et al. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135:1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernhardt A, et al. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 2006;47:591–603. doi: 10.1111/j.1365-313X.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X, et al. PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell. 2014;26:1992–2006. doi: 10.1105/tpc.114.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu L, et al. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat Commun. 2015;6:7245. doi: 10.1038/ncomms8245. [DOI] [PubMed] [Google Scholar]
- 42.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 43.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 44.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 46.Samach A, et al. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 1999;20:433–445. doi: 10.1046/j.1365-313x.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhao D, Yang M, Solava J, Ma H. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev Genet. 1999;25:209–223. doi: 10.1002/(SICI)1520-6408(1999)25:3<209::AID-DVG4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 48.Gray WM, Estelle I. Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem Sci. 2000;25:133–138. doi: 10.1016/s0968-0004(00)01544-9. [DOI] [PubMed] [Google Scholar]
- 49.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 50.Ariizumi T, Lawrence PK, Steber CM. The role of two f-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 2011;155:765–775. doi: 10.1104/pp.110.166272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA. 2004;101:12771–12776. doi: 10.1073/pnas.0404287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majee M, et al. A misannotated locus positively influencing Arabidopsis seed germination is deconvoluted using multiple methods, including surrogate splicing. Plant Gene. 2017;10:74–85. [Google Scholar]
- 53.Salaita L, Kar RK, Majee M, Downie AB. Identification and characterization of mutants capable of rapid seed germination at 10 degrees C from activation-tagged lines of Arabidopsis thaliana. J Exp Bot. 2005;56:2059–2069. doi: 10.1093/jxb/eri204. [DOI] [PubMed] [Google Scholar]
- 54.Hua Z, et al. Epigenomic programming contributes to the genomic drift evolution of the F-Box protein superfamily in Arabidopsis. Proc Natl Acad Sci USA. 2013;110:16927–16932. doi: 10.1073/pnas.1316009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 56.Kim J, et al. PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell. 2016;28:1388–1405. doi: 10.1105/tpc.16.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.la Cour T, et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 58.Deng M, et al. Role of the sonchus yellow net virus N protein in formation of nuclear viroplasms. J Virol. 2007;81:5362–5374. doi: 10.1128/JVI.02349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moon J, Zhu L, Shen H, Huq E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:9433–9438. doi: 10.1073/pnas.0803611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi H, et al. Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci USA. 2015;112:3817–3822. doi: 10.1073/pnas.1502405112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen H, et al. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008;20:1586–1602. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sliwinska E, Bassel GW, Bewley JD. Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J Exp Bot. 2009;60:3587–3594. doi: 10.1093/jxb/erp203. [DOI] [PubMed] [Google Scholar]
- 64.Inoue Y, Nagashima H. Photoperceptive site in phytochrome-mediated Lettuce (Lactuca sativa L. cv. Grand Rapids) seed germination. J Plant Physiol. 1991;137:669–673. [Google Scholar]
- 65.Penfield S, et al. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol. 2005;15:1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 66.Kim HJ, Kim YK, Park JY, Kim J. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 2002;29:693–704. doi: 10.1046/j.1365-313x.2002.01249.x. [DOI] [PubMed] [Google Scholar]
- 67.Kurepin LV, et al. Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. Int J Mol Sci. 2013;14:12729–12763. doi: 10.3390/ijms140612729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sysoeva MI, Markovskaya EF, Sherudilo EG. Role of phytochrome B in the development of cold tolerance in cucumber plants under light and in darkness. Russ J Plant Physiol. 2013;60:383–387. [Google Scholar]
- 69.Toole EH, Toole VK, Borthwick HA, Hendricks SB. Interaction of temperature and light in germination of seeds. Plant Physiol. 1955;30:473–478. doi: 10.1104/pp.30.5.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heschel MS, et al. A new role for phytochromes in temperature-dependent germination. New Phytol. 2007;174:735–741. doi: 10.1111/j.1469-8137.2007.02044.x. [DOI] [PubMed] [Google Scholar]
- 71.Ni W, et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 73.Finch-Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2007;51:60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- 74.Preston J, et al. Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: A comparative study on dormant and non-dormant accessions. Plant Cell Physiol. 2009;50:1786–1800. doi: 10.1093/pcp/pcp121. [DOI] [PubMed] [Google Scholar]
- 75.Shinomura T, et al. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fleet CM, Sun TP. A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr Opin Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 77.Ariizumi T, Steber CM. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell. 2007;19:791–804. doi: 10.1105/tpc.106.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piskurewicz U, et al. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 2008;20:2729–2745. doi: 10.1105/tpc.108.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bu Q, Zhu L, Huq E. Multiple kinases promote light-induced degradation of PIF1. Plant Signal Behav. 2011;6:1119–1121. doi: 10.4161/psb.6.8.16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bu Q, Castillon A, Chen F, Zhu L, Huq E. Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol Biol. 2011;77:501–511. doi: 10.1007/s11103-011-9827-4. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, et al. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9:e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y, Li X, Li K, Liu H, Lin C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013;9:e1003861. doi: 10.1371/journal.pgen.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Denay G, et al. Endosperm breakdown in Arabidopsis requires heterodimers of the basic helix-loop-helix proteins ZHOUPI and INDUCER OF CBP EXPRESSION 1. Development. 2014;141:1222–1227. doi: 10.1242/dev.103531. [DOI] [PubMed] [Google Scholar]
- 84.Huq E, et al. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 85.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller JH. Experiments in Molecular Genetics. 3rd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 87.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- 88.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 89.Martin K, et al. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009;59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- 90.Dinkins RD, et al. Changing transcriptional initiation sites and alternative 5′- and 3′-splice site selection of the first intron deploys Arabidopsis protein isoaspartyl methyltransferase2 variants to different subcellular compartments. Plant J. 2008;55:1–13. doi: 10.1111/j.1365-313X.2008.03471.x. [DOI] [PubMed] [Google Scholar]
- 91.Dinkins RD, Conn HM, Dirk LMA, Williams MA, Houtz RL. The Arabidopsis thaliana peptide deformylase 1 protein is localized to both mitochondria and chloroplasts. Plant Sci. 2003;165:751–758. [Google Scholar]
- 92.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 93.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 94.Untergasser A, et al. Primer3–New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anonymous . Statistical Analysis Systems. SAS Institute Inc.; Cary, NC: 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.