Significance
Poly(ADP-ribose) polymerase-1 (PARP-1) is a DNA repair protein and part of the genome maintenance network. On the other hand, PARP-1 is involved in pathophysiological processes such as inflammation. Chronic inflammation has emerged as a key event in carcinogenesis, including the formation of colorectal cancer (CRC). Our data reveal that PARP-1 is abundantly expressed in human CRC, correlating with disease progression. Using transgenic mouse models, we show that PARP-1 fosters inflammation-driven colorectal tumor growth and stimulates the IL6-STAT3-cyclin D1 axis in tumors. In turn, PARP-1 protects against alkylation-triggered colorectal tumor induction dependent on the repair protein O6-methylguanine-DNA methyltransferase (MGMT). These findings unveil the opposing functions of PARP-1 in CRC initiation and CRC progression and its link to MGMT.
Keywords: DNA repair, PARP-1, colorectal carcinogenesis, mouse models, intestinal inflammation
Abstract
Colorectal cancer (CRC) is one of the most common tumor entities, which is causally linked to DNA repair defects and inflammatory bowel disease (IBD). Here, we studied the role of the DNA repair protein poly(ADP-ribose) polymerase-1 (PARP-1) in CRC. Tissue microarray analysis revealed PARP-1 overexpression in human CRC, correlating with disease progression. To elucidate its function in CRC, PARP-1 deficient (PARP-1−/−) and wild-type animals (WT) were subjected to azoxymethane (AOM)/ dextran sodium sulfate (DSS)-induced colorectal carcinogenesis. Miniendoscopy showed significantly more tumors in WT than in PARP-1−/− mice. Although the lack of PARP-1 moderately increased DNA damage, both genotypes exhibited comparable levels of AOM-induced autophagy and cell death. Interestingly, miniendoscopy revealed a higher AOM/DSS-triggered intestinal inflammation in WT animals, which was associated with increased levels of innate immune cells and proinflammatory cytokines. Tumors in WT animals were more aggressive, showing higher levels of STAT3 activation and cyclin D1 up-regulation. PARP-1−/− animals were then crossed with O6-methylguanine-DNA methyltransferase (MGMT)-deficient animals hypersensitive to AOM. Intriguingly, PARP-1−/−/MGMT−/− double knockout (DKO) mice developed more, but much smaller tumors than MGMT−/− animals. In contrast to MGMT-deficient mice, DKO animals showed strongly reduced AOM-dependent colonic cell death despite similar O6-methylguanine levels. Studies with PARP-1−/− cells provided evidence for increased alkylation-induced DNA strand break formation when MGMT was inhibited, suggesting a role of PARP-1 in the response to O6-methylguanine adducts. Our findings reveal PARP-1 as a double-edged sword in colorectal carcinogenesis, which suppresses tumor initiation following DNA alkylation in a MGMT-dependent manner, but promotes inflammation-driven tumor progression.
Colorectal cancer (CRC) is one of the most commonly diagnosed cancer types worldwide and is responsible for about 10% of cancer-related deaths among Western countries (1). The causes underlying the development of CRC are diverse and include genetic predisposition, inflammatory bowel disease (IBD), and lifestyle factors, such as alcohol intake and meat consumption (1). Hereditary CRC syndromes are predominantly attributable to defects in DNA repair. The prime example is Lynch syndrome, which is induced by mutations in DNA mismatch repair genes, thereby leading to microsatellite instability (2). DNA repair is also implicated in the etiology of sporadic CRC as illustrated by epigenetic inactivation of O6-methylguanine (O6-MeG)-DNA methyltransferase (MGMT), which highly predisposes to KRAS mutations induced by alkylating N-nitroso compounds (NOCs) (3). The pivotal role of MGMT in the protection against NOC-induced CRC has recently been demonstrated (4, 5).
Poly(ADP-ribose) polymerase-1 (PARP-1) is a nuclear enzyme belonging to the DNA damage surveillance network and a founding member of the PARP superfamily (6). Following activation by DNA strand breaks, PARP-1 catalyzes the synthesis of poly(ADP-ribose) (PAR) in a NAD+-dependent manner, which accounts for the vast majority of cellular PAR formation (7). The formed biopolymer is covalently linked to acceptor proteins, including PARP-1, in a process termed PARylation. This posttranslational protein modification is highly dynamic and fully reversible by enzymes responsible for PAR degradation, such as poly(ADP-ribose) glycohydrolase (PARG) (8). Furthermore, PAR interacts in a noncovalent fashion with proteins involved in the DNA damage response (DDR), DNA repair, and cell cycle regulation via conserved binding motifs (9). This process occurs with high affinity and specificity (10). By means of covalent and noncovalent PARylation, PARP-1 regulates protein–protein interactions, protein activities, and their subcellular localization (11).
PARP-1 has a fundamental role in the maintenance of genome stability and regulates chromatin structure (6). PARP-1 participates in different DNA repair pathways, including base excision repair (BER) and DNA single-strand break (SSB) repair (11). PARP-1 is also engaged in lesion sensing by recruiting ATM and MRN to sites of DNA double-strand breaks (DSBs) and of stalled replication forks (12, 13). In line with these findings, PARP-1–deficient mice exhibit hypersensitivity to DNA damaging agents, with increased genomic instability and carcinogenesis at different sites (7, 14, 15). A previous study reported an elevated number of colonic tumors in PARP-1−/− animals, which were treated repetitively with the NOC-related carcinogen azoxymethane (AOM), implicating PARP-1 in the defense against NOC-induced CRC formation (16).
On the other hand, PARP-1 is involved in pathophysiological processes such as neurodegeneration and contributes to inflammation, which occurs primarily by PARP-1–dependent coactivation of the proinflammatory transcription factor nuclear factor-kB (NF-κB) (7). This was illustrated in a study using transgenic PARP-1–deficient mice, which were resistant against the bacterial endotoxin LPS and displayed a markedly reduced expression of NF-κB–dependent proinflammatory genes (17). It has also recently been shown that PARP-1–deficient mice are protected against acute colonic mucosal injury induced by dextran sodium sulfate (DSS) (18). It is established that inflammatory processes, as observed in patients with IBD and in the inflammatory microenvironment in sporadic CRC, are tightly linked to the development and progression of malignant disease (19). In light of these apparently opposing functions of PARP-1, we set out to clarify the role of PARP-1 in colorectal cancer induction and progression using murine models and human CRC tissue. Our study revealed that PARP-1 is a double-edged sword in CRC, which suppresses NOC-induced tumor initiation in a MGMT-dependent manner at the expense of enhanced tumor growth. Consistently, PARP-1 and its product PAR were overexpressed in human colorectal carcinoma specimens and correlated with tumor progression from healthy tissue to benign lesions and to invasive carcinoma.
Results
PARP-1 Is Abundant in Human Colorectal Carcinomas and Correlates with CRC Progression.
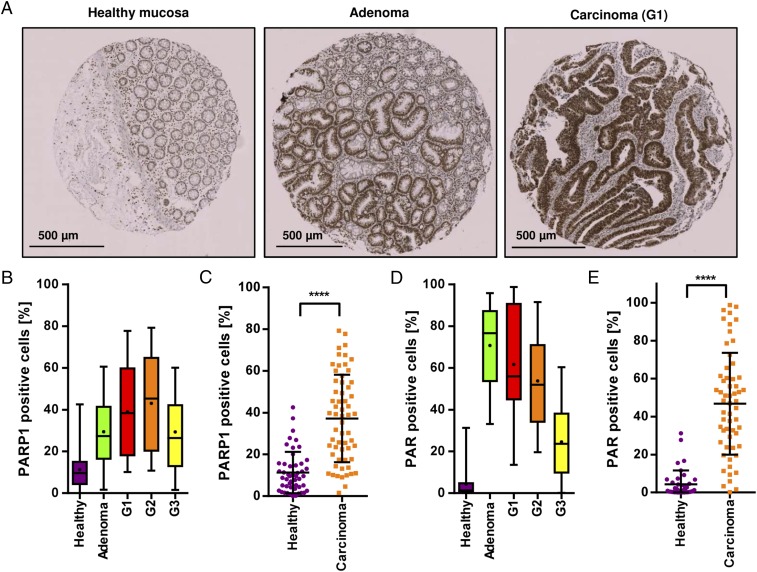
First, the protein expression of PARP-1 was assessed by immunohistochemistry (IHC) in a grading tissue microarray (TMA) comprising healthy colorectal tissue and adenoma and carcinoma tissue with different histopathological grading (G1–G3). PARP-1 levels correlated well with CRC progression, showing lowest expression in healthy tissue and highest expression in carcinomas (Fig. 1 A–C). To test whether PARP-1 expression goes along with its activity, i.e., the formation of PAR, the biopolymer was stained in the same set of tissue samples. A striking increase of PAR levels was detected in tumor tissue compared with healthy mucosa, with a gradual decline of PAR-positive cells with advanced dedifferentiation (Fig. 1 D and E and Fig. S2). In summary, both PARP-1 and its product PAR were markedly elevated in colorectal tumor tissue, suggesting a role of PARP-1 in CRC formation and progression.
Fig. 1.
Expression of PARP-1 and its product PAR in CRC and correlation with disease progression. (A) PARP-1 staining in healthy colorectal mucosa, adenoma, and carcinoma with respect to the grade of differentiation. Representative cores of healthy tissue, adenoma tissue, and well-differentiated carcinoma (G1). (B and C) Quantitative evaluation of PARP-1 positive cells in healthy mucosa (n = 49), adenoma (n = 19), G1–G3 carcinoma (n = 19, 21, and 20, respectively). (D and E) Quantitative assessment of PAR staining, reflecting PARP-1 activity in situ, in the same set of tissue samples. Healthy mucosa (n = 39), adenoma (n = 19), G1–G3 carcinoma (n = 19, 20, and 19, respectively). Data represent median (B and D) or mean ± SD (C and E). ****P < 0.0001 as determined by Student’s t test.
PARP-1 Promotes Murine AOM/DSS-Triggered Colorectal Carcinogenesis.
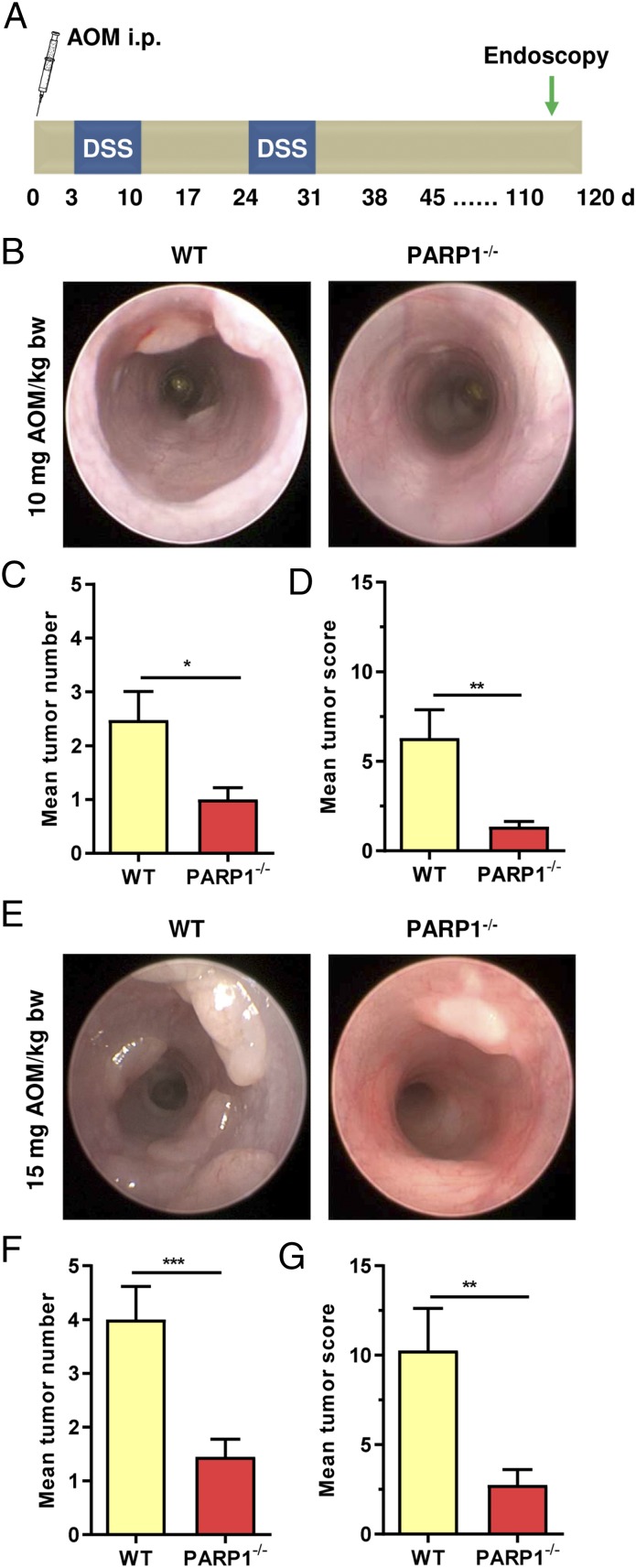
To analyze the role of PARP-1 in CRC etiology in more detail, the murine AOM/DSS model of colorectal carcinogenesis was used (Fig. 2A). AOM is a colonotropic tumor initiator causing DNA alkylation damage (e.g., N7-methylguanine, N3-methyladenine, O6-methylguanine, and others), while DSS induces tumor-promoting colitis. Tumor formation was assessed in PARP-1–proficient wild-type (WT) and PARP-1–deficient (PARP-1−/−) mice using noninvasive miniendoscopy (Fig. 2 B and E). An AOM dose of 10 mg resulted in a significantly higher tumor number in WT than in PARP-1−/− animals (Fig. 2C). This was also mirrored in the tumor score, which accounts for both tumor number and size (Fig. 2D). The difference between WT and PARP-1−/− animals was even more prominent at a high dose of 15 mg AOM (Fig. 2 F and G), revealing significantly more tumors in WT mice. In agreement with these findings, tumors formed in WT animals were also larger than those of PARP-1−/− animals (Fig. S3 C and D). Taken together, PARP-1 deficiency conferred resistance to AOM/DSS-induced CRC formation.
Fig. 2.
PARP-1 promotes AOM/DSS-triggered colorectal carcinogenesis. (A) Scheme of the used AOM/DSS model. (B–D) Tumor formation in PARP-1–proficient WT animals (n = 23) and PARP-1–deficient animals (PARP-1−/−) (n = 20) treated with 10 mg AOM per kilogram of body weight (kg/bw) followed by two cycles of 1% DSS in the drinking water. Representative images obtained during miniendoscopy (B) are shown. Tumor number (C) and tumor size were assessed after 16 wk by miniendoscopy, which was used to calculate the tumor score (D). (E–G) Tumor formation in WT (n = 15) and PARP-1−/− (n = 20) animals treated with 15 mg AOM/kg bw followed by two cycles of 1% DSS in the drinking water. Representative images obtained during miniendoscopy (E) are shown. Tumor number (F) and tumor size were assessed after 16 wk by miniendoscopy, which was used to calculate the tumor score (G). Data are shown as mean + SEM. *P < 0.05, **P < 0.01, ***P < 0.005 as determined by Student’s t test.
Lack of PARP-1 Increases DNA Strand Breaks Without Affecting AOM-Induced Autophagy and Apoptotic Cell Death in Colon Crypts.
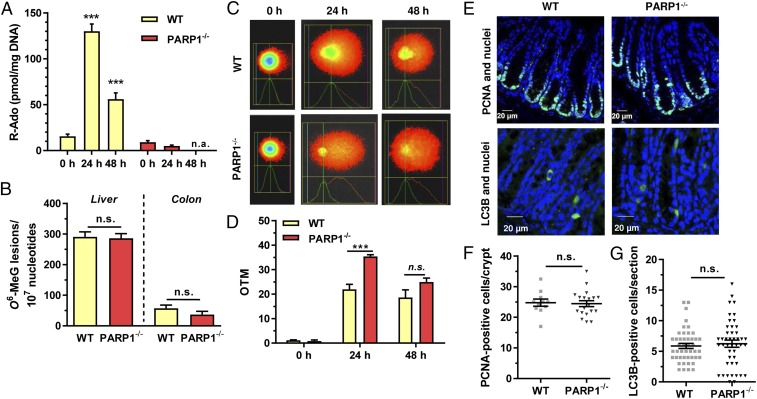
Next, we wanted to study how PARP-1 deficiency affects DNA damage induction and downstream pathways, including cell death and autophagy. PAR formation was determined by mass spectrometry in liver and colon tissue, revealing a strong AOM-dependent increase of hepatic PAR levels in WT mice, while little effect was observed in colon tissue (Fig. 3A and Fig. S4). In contrast, PARP-1−/− mice completely lacked AOM-induced PARylation and displayed only low basal PAR levels in liver and colon as expected. The formation of the DNA adduct O6-methylguanine (O6-MeG) was then studied in liver and colon tissue, since it is the main DNA lesion driving AOM-induced CRC (3). Quantitative analysis by mass spectrometry showed similar levels of O6-MeG DNA adducts in both genotypes 24 h after AOM administration (Fig. 3B). This finding was confirmed by slot blot analysis using an antibody directed against O6-MeG (Fig. S5 B–D). AOM-induced DNA strand breaks were then assessed using the alkaline Comet assay, revealing more DNA damage in liver tissue of PARP-1−/− animals after 24 h (Fig. 3 C and D) compared with WT tissue. To assess potential differences in cell proliferation, colorectal tissue of WT and PARP-1−/− animals was stained for proliferating cell nuclear antigen (PCNA) as proliferation marker. Quantitative evaluation revealed a similar level of PCNA-positive cells in the basal colon crypts of both genotypes (Fig. 3 E and F). Subsequently, we analyzed the induction of autophagy in colorectal tissue, which is an important cell survival mechanism in response to genotoxic stress. At 48 h following AOM administration, autophagic cells were labeled by the well-established autophagy marker LC3B. We observed no differences in the number of LC3B-positive cells between the genotypes, arguing against an involvement of DNA damage-triggered autophagy (Fig. 3 E and G). Moreover, AOM-triggered cell death was visualized in situ using terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining 48 h after treatment. PARP-1–deficient mice showed a low number of apoptotic cells in colon crypts, which was similar to WT animals (Fig. S5E, also see Fig. 7 D and E). In summary, although lack of PARP-1 resulted in a higher level of initial DNA strand breaks, it had no impact on cellular processes relevant to tumor initiation, including proliferation, apoptosis, and autophagy, in the colon epithelium.
Fig. 3.
Loss of PARP-1 moderately increases DNA strand break formation. (A) Time-dependent formation of PAR in WT and PARP-1−/− animals upon AOM treatment. Animals received 10 mg AOM/kg bw. PAR levels in the liver were determined as ribosyl-adenosine (R-Ado) using LC-MS/MS analysis. Data are depicted as mean + SEM (n = 3 per genotype and time point). ***P < 0.005 compared with 0 h (untreated control); n.a., not assessed. (B) Detection of hepatic and colonic O6-MeG DNA adducts in WT and PARP-1−/− animals 24 h after AOM injection using mass spectrometry. (n = 3 per genotype). (C) Assessment of AOM-induced DNA strand breaks in WT and PARP-1−/− mice. Following AOM treatment, liver tissue was isolated and subjected to alkaline Comet assay. Representative pictures are shown. (D) Quantitative evaluation of alkaline Comet assay. Data are presented as mean + SEM (n = 3 per time point and genotype). ***P < 0.005; n.s., not significant. (E) Basal proliferation (Top) and AOM-induced autophagy (Bottom) in colon crypts of WT and PARP-1−/− animals (Top). Proliferating cells were visualized by PCNA staining (green) and nuclei by TO-PRO-3 staining (blue) followed by confocal microscopy. Autophagy was assessed by LC3B staining and confocal microscopy. Representative pictures are shown. (F) Quantification of PCNA staining. Data are presented as mean + SEM (n = 3 per genotype; ≥4 sections per sample); n.s., not significant. (G) Quantification of LC3B-positive cells per section. Results are depicted as mean + SEM (n = 3 per genotype; ≥10 sections per sample); n.s. not significant. Statistical significance was determined by Student’s t test.
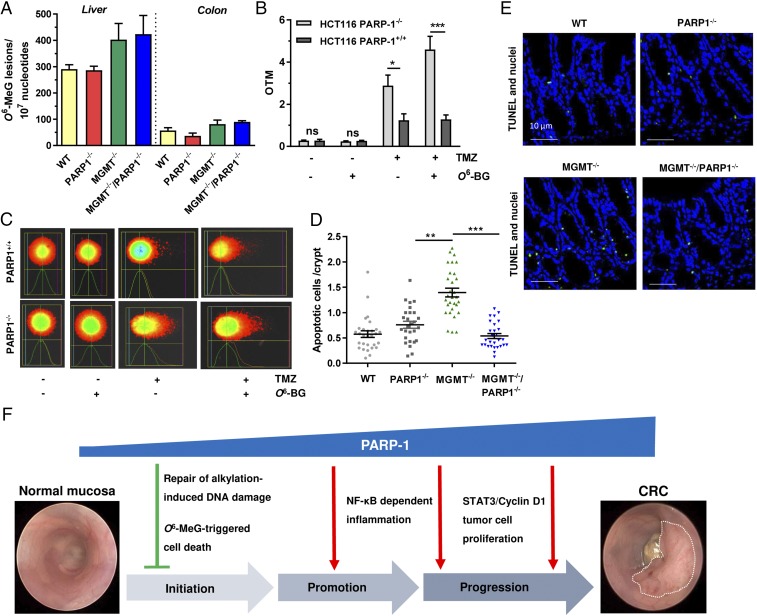
Fig. 7.
Impact of PARP-1 on alkylation-induced DNA damage and cell death depending on the MGMT status. (A) Detection of hepatic and colonic O6-MeG DNA adducts in WT, PARP-1−/−, MGMT−/−, and MGMT−/−/PARP-1−/− animals 24 h after AOM injection using mass spectrometry (n = 3 per genotype). (B and C) DNA strand break induction in PARP-1–proficient and –deficient HCT116 cells depending on the MGMT activity. Cells were exposed to the SN1-alkylating agent TMZ and subjected to the alkaline Comet assay after 24 h in the absence or presence of the MGMT inhibitor O6-BG. Representative pictures are shown. OTM, olive tail moment. Data are presented as mean + SEM (n ≥ 4 per treatment group). ***P < 0.005; *P < 0.05; n.s., not significant. (D and E) AOM-dependent cell death induction in WT, PARP-1−/−, MGMT−/−, and MGMT−/−/PARP-1−/− mice. Apoptotic cells were labeled in situ by TUNEL staining (green). Representative pictures are shown. Data are given as mean + SEM (n = 3 per genotype; ≥9 sections per sample). ***P < 0.005; **P < 0.01. (F) Involvement of PARP-1 in colorectal carcinogenesis and underlying mechanisms, which are responsible for the opposing function of PARP-1 in tumor induction vs. tumor progression.
PARP-1 Deficiency Attenuates the Innate Immune Response and Colitis Following DSS Treatment.
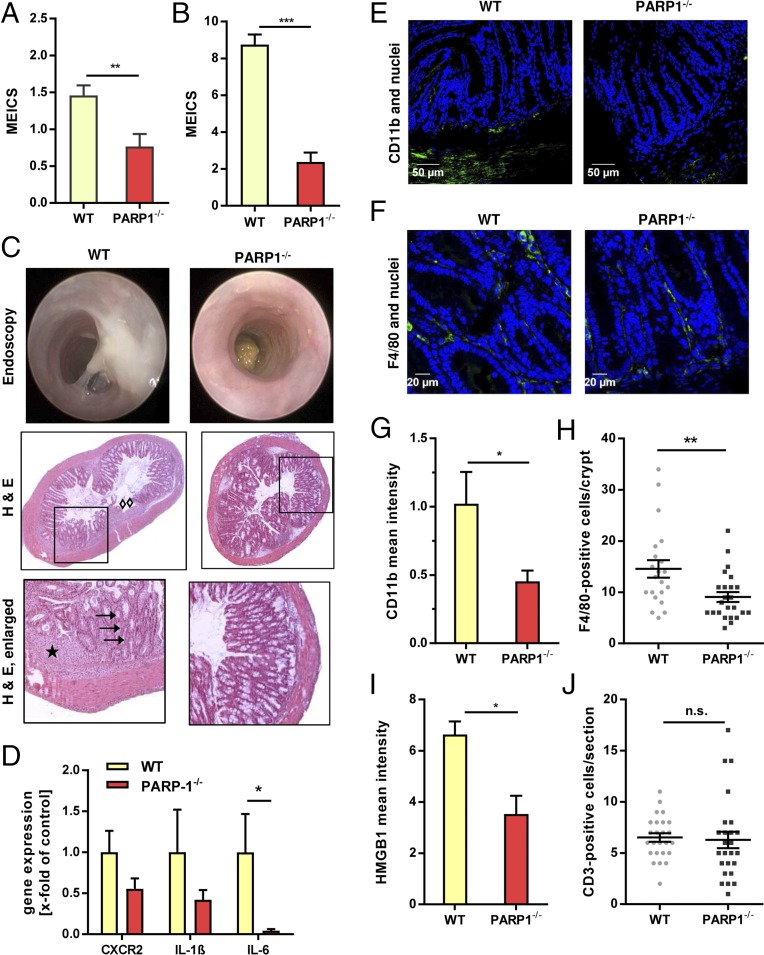
As PARP-1 is a known coregulator of the proinflammatory transcription factor NF-κB, the acute DSS-induced inflammation was assessed after the first DSS cycle using the murine endoscopic index of colitis severity (MEICS) (Fig. 4 A and B). PARP-1−/− animals displayed a significantly lower level of intestinal inflammation compared with WT animals, which was even more pronounced at higher DSS doses (2.5% instead of 1%) (Fig. 4 A and B). Endoscopy revealed massive fibrin deposition, enhanced granularity, and thickening of the colon mucosa in WT animals, whereas in PARP-1–deficient animals, only little fibrin was visible and the mucosa appeared almost healthy with normal vascular pattern, low granularity, and high translucency (Fig. 4C, Top). Microscopic evaluation of H&E-stained WT colon sections showed loss of crypt architecture (arrows), disappearance of goblet cells (arrows), regions of mucosal erosions and hyperplasia (diamonds), together with mucosal edema (star) (Fig. 4C, Bottom). In contrast, PARP-1−/− animals exhibited little histomorphological alterations (Fig. 4C). In line with these observations, reduced gene expression levels of proinflammatory cytokines (IL-1β and IL-6) and chemokine receptor (CXCR2) as well as decreased COX-2 staining were found in PARP-1–deficient mice (Fig. 4D and Fig. S6B). To detail the infiltration of different leukocyte populations, colon sections of WT and PARP-1−/− mice were stained with CD11b (monocytes), F4/80 (macrophages), and CD3 (T lymphocytes). Analysis by confocal microscopy demonstrated a reduced number of monocytes residing in the submucosa of PARP-1−/− animals (Fig. 4 E and G). Consistent with this finding, the level of macrophages localized to the lamina propria was also decreased (Fig. 4 F and H). The proinflammatory cytokine HMGB1, which is secreted by activated macrophages and natural killer cells (20), was found to be down-regulated in PARP-1−/− animals, further indicating an attenuated innate immune response (Fig. 4I and Fig. S6C). On the other hand, no differences were observed in the number of T lymphocytes present in colon tissue of both mouse strains (Fig. 4J and Fig. S6D). In conclusion, PARP-1 deficiency conferred resistance against acute DSS-induced colitis, which was primarily attributable to a reduced innate immune response.
Fig. 4.
PARP-1 deficiency confers resistance to DSS-triggered gut inflammation. (A and B) Analysis of AOM/DSS-induced mucosal inflammation in WT (n = 18 and n = 17, respectively) and PARP-1−/− (n = 15 and n = 14, respectively) animals. Mice were challenged with 15 mg AOM + 1% DSS (A) or 10 mg AOM + 2.5% DSS (B) and the MEICS was assessed by miniendoscopy after the first DSS cycle. **P < 0.005, ***P < 0.0005. (C) Representative images of B and corresponding H&E staining of colon sections. Arrows show loss of crypt architecture and disappearance of goblet cells (arrows). Diamonds indicate regions of mucosal erosions and hyperplasia. The star highlights mucosal edema. (D) Determination of proinflammatory gene expression in WT and PARP-1−/− animals. Animals were treated as described above (B), killed after the first DSS cycle, and colorectal tissue was harvested. Gene expression was normalized to the WT and is presented as mean + SEM (n ≥ 3 per genotype). *P < 0.05. (E and F) Visualization of CD11b-positive cells (monocytes) and F4/80-positive cells (macrophages) in WT and PARP-1−/− animals after AOM/DSS treatment. Representative confocal images are shown. (G and H) Quantitative assessment of CD11b staining intensity and F4/80-positive cells. Data are given as mean + SEM (n = 3 per genotype; ≥7 sections per sample). *P < 0.05, **P < 0.005. (I) Detection of the proinflammatory cytokine HMGB1 in WT and PARP-1−/− mice challenged as described in B. Results are shown as mean + SEM (n = 3 per genotype; ≥7 sections per sample). *P < 0.05. (J) CD3-positive cells (T lymphocytes) depicted as mean + SEM (n = 3 per genotype, ≥7 sections per sample); n.s., not significant. Statistical significance was determined by Student’s t test.
PARP-1 Supports Inflammation-Driven Tumor Growth and Fosters IL6-STAT3-Cyclin D1 Signaling in Colorectal Tumors.
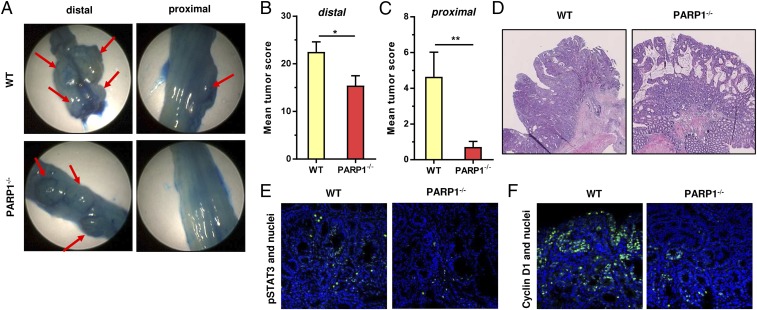
Having shown that PARP-1 knockout animals are resistant to colitis induction following a high DSS dose, we set out to determine the colitis-associated colorectal tumor formation. To this end, mice received 10 mg AOM followed by two cycles with 2.5% DSS, which resulted in strong tumor formation already detectable by miniendoscopy after 8 wk. Owing to the accelerated tumor growth, animals were killed after 12 wk and the tumors were documented by imaging with the miniendoscopy system after methylene blue staining (21), which enhances their visibility (Fig. 5A). In general, both the tumor number and tumor score in the distal part of the colon were strongly elevated in both genotypes if compared with the previous experiment with 1% DSS (Fig. 5B and Fig. S7A vs. Fig. 2 B and C). Nevertheless, PARP-1−/− animals displayed a reduced tumor burden, which was even more obvious in the proximal part of the colon (Fig. 5C and Fig. S7 B and D). Histopathological analysis of H&E-stained tumor tissue sections revealed a predominant cribriform growth pattern in both WT and PARP-1−/− animals, with a higher aggressiveness and signs of putative invasion in tumors of WT mice (Fig. 5D and Fig. S8). Furthermore, the IL6-STAT3-cyclin D1 axis, which is driven by NF-κB, was analyzed in tumor tissue sections by IHC and confocal microscopy. Interestingly, WT tumors showed a higher level of phosphorylated STAT3 (Fig. 5E) and an increased expression of its downstream target cyclin D1 (Fig. 5F). Collectively, the data show that PARP-1 promotes inflammation-driven colorectal tumor growth. The findings further indicate a PARP-1–mediated stimulation of the IL6-STAT3-cyclin D1 axis in tumors, thereby likely fostering tumor growth and progression in WT animals.
Fig. 5.
PARP-1 supports tumor growth and fosters the IL6-STAT3-cyclin D1 axis. (A) Tumor formation in WT and PARP-1−/− animals. Mice were injected with 10 mg AOM/kg bw followed by two cycles with 2.5% DSS in the drinking water. After 12 wk, isolated colon was opened, stained with methylene blue, and tumors were recorded with the miniendoscopy system. Representative distal and proximal colon sections are shown. (B and C) Tumor score in the distal colon and in the proximal colon. Data are shown as mean + SEM (n = 14 per genotype). *P < 0.05; **P < 0.01. (D) H&E staining of colorectal tumors. (E and F) p-STAT3 and cyclin D1 staining in AOM/DSS-induced colorectal tumors of WT (n = 4) and PARP-1−/− (n = 3) animals. Representative confocal images are shown.
Genetic Ablation of MGMT Reveals the Tumor-Suppressive Function of PARP-1 at Tumor Initiation.
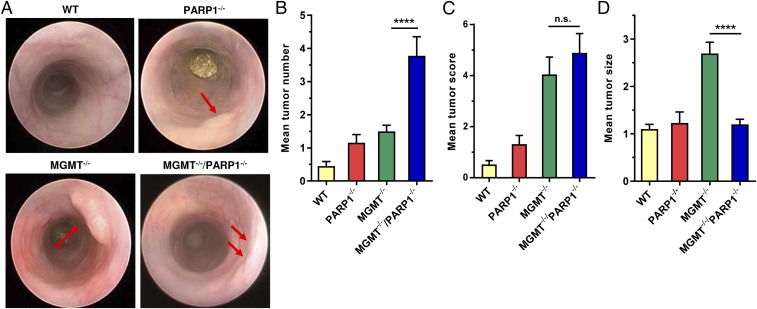
To detail the contribution of PARP-1 to CRC induction and progression, PARP-1−/− animals were crossed with MGMT−/− animals, which are highly sensitive to AOM-induced colorectal carcinogenesis due to their inability to repair O6-MeG adducts (4). The generated MGMT−/−/PARP-1−/− double knockout (DKO) animals were challenged with 3 mg AOM and two cycles of 1% DSS. Miniendoscopy revealed an increased number of tumors in DKO mice compared with MGMT single knockouts (Fig. 6 A and B), which were treated with the same AOM/DSS protocol in a previous study (4). As expected, DNA repair-competent WT animals displayed the lowest tumor number (4), while PARP-1 single knockouts showed moderately higher tumor number as the corresponding WT animals. It is important to note that PARP-1−/− animals were challenged here with 5 mg AOM and developed almost the same number of tumors as the control treated with 0 mg AOM (Fig. S9). Interestingly, DKO mice and MGMT−/− mice yielded almost comparable tumor scores (Fig. 6C). This was attributable to the tumor size, which was significantly smaller in DKO mice compared with MGMT-deficient animals (Fig. 6D), lending further support for a role of PARP-1 in tumor growth and progression.
Fig. 6.
PARP-1 supports tumor growth, but protects against tumor induction depending on MGMT. (A) Tumor formation in WT, PARP-1−/−, MGMT−/−, and MGMT−/−/PARP-1−/− double knockout animals. Animals were challenged with 3 mg AOM/kg bw followed by two cycles with 1% DSS. Please note that PARP1−/− animals received 5 mg AOM. After 16 wk, tumor number and size was determined by miniendoscopy. Representative images are depicted. Red arrows indicate tumors. (B) Tumor number, (C) tumor score, and (D) tumor size. Data of WT and MGMT−/− animals have been reported previously (4). Values are expressed as mean + SEM (n ≥ 19 for WT, PARP-1−/−, and MGMT−/− animals; n = 9 for MGMT−/−/PARP-1−/−). ****P < 0.0001. Statistical significance was determined by Student’s t test.
Lack of PARP-1 Increased Alkylation-Induced DNA Damage, but Reduced Cell Death in a MGMT-Dependent Manner.
We then wished to identify the mechanisms responsible for the increased tumor induction in MGMT−/−/PARP-1−/− animals. First, O6-MeG adduct levels were determined by mass spectrometry in liver and colon tissue of all used mouse strains 24 h after AOM administration. Importantly, both MGMT−/− and DKO animals displayed comparable AOM-induced O6-MeG levels in liver and colon, which were higher than those observed in WT and PARP-1 single knockouts (Fig. 7A). To study the interplay of PARP-1 and MGMT in response to alkylation-induced DNA damage in more detail, we switched to a cell model. PARP-1–deficient HCT116 cells were generated by means of the CRISPR/Cas9 system and validated using DNA sequencing, Western blot analysis, and immunofluorescence microscopy (Figs. S10 and S11). MGMT was inactivated in these cells or respective PARP-1+/+ cells using its highly potent pharmacological inhibitor O6-benzlyguanine (O6-BG). The cell lines were then treated for 24 h with the SN1-alkylating agent temozolomide (TMZ) that does not require metabolic activation in comparison with AOM and induces a similar DNA adduct spectrum. TMZ caused DNA strand breaks in HCT116–PARP-1−/− cells, which was further increased following MGMT inactivation as demonstrated by an alkaline Comet assay (Fig. 7 B and C). In contrast, DNA strand-break induction was lower in PARP-1–proficient HCT116 cells and was unaffected by MGMT inhibition (Fig. 7 B and C).
Since O6-MeG is known to be a cytotoxic lesion (22), apoptosis was analyzed in colorectal tissue of all genotypes 48 h after AOM treatment by means of TUNEL staining. Consistent with a previous report (4), mice lacking MGMT exhibited increased levels of apoptotic cells in colon crypts due to the accumulation of cytotoxic O6-MeG adducts (Fig. 7 D and E). Strikingly, DKO animals showed a significantly lower number of apoptotic cells per crypt compared with MGMT single knockouts (Fig. 7 D and E), although they harbor the same O6-MeG adduct levels (Fig. 7A). Altogether, these findings indicate that PARP-1 contributes to the repair of O6-MeG–induced DNA strand breaks and seems to be involved in O6-MeG–triggered cell death in colon crypts. Impairment of these mechanisms likely accounts for the increased tumor induction observed in MGMT−/−/PARP-1−/− animals.
Our findings are summarized in Fig. 7F, revealing PARP-1 as a double-edged sword in colorectal carcinogenesis. On the one hand, PARP-1 suppresses NOC-induced tumor initiation via repair of DNA alkylation damage (most likely BER and SSB repair) together with MGMT and elimination of colonocytes harboring mutagenic O6-MeG adducts. On the other hand, PARP-1 fuels NF-κB–driven intestinal inflammation and stimulates the IL6-STAT3-cyclin D1 axis, thereby promoting tumor progression. Consistent with these findings, we show that PARP-1 is overexpressed in human CRC and correlates with disease progression.
Discussion
In this study, we dissected the role of PARP-1 in colorectal carcinogenesis using transgenic mouse models and human tissue specimens. First, we performed TMA analysis of human healthy colon tissue and colorectal tumors with respect to their histological grading. Our results showed a clear association between PARP-1 and colorectal cancer progression, with highest levels found in moderately differentiated carcinomas. This extends previous studies, which indicated enhanced PARP-1 expression in adenomas versus normal mucosa using qualitative IHC and PARP-1 mRNA analysis, respectively (23, 24). Furthermore, we were able to show that increased PARP-1 expression in carcinoma tissue correlates with a higher PAR level, reflecting an enhanced PARP activity. The data indicate that high PARP-1 expression confers a growth advantage to established tumors. This notion was corroborated using the murine AOM/DSS model of colorectal carcinogenesis. Intriguingly, PARP-1–deficient mice showed strongly reduced tumor formation and tumor growth compared with WT animals.
PARP-1 is well known for participating in different DNA repair pathways, such as BER, SSB, and DSB repair, which are all relevant for protecting against DNA alkylation damage (22). Consistently, detailed analysis showed lack of AOM-induced PARylation and higher levels of DNA strand breaks in PARP-1–deficient animals, but did not translate into increased tumor formation compared with the WT. AOM-induced O6-MeG, a driver lesion in CRC, was largely unaffected by genetic ablation of PARP-1 as confirmed by mass spectrometry. To explain the observed differences in tumor formation, we analyzed cellular pathways downstream of DNA damage. These include apoptosis and autophagy, which are both tumor-suppressive mechanisms (25), in particular at early stages of carcinogenesis. However, PARP-1 deficiency affected neither AOM-induced cell death nor autophagy. It is further established that excessive PARP-1 activation is able to provoke necrosis due to NAD+ and ATP depletion as well as apoptosis-inducing factor (AIF)-mediated caspase-independent cell death (26). This PARP-1–dependent cell death was described after administration of the SN2-alkylating agent methyl methanesulfonate (MMS) at high doses, causing severe tissue toxicity (27). It is also known that a high dose of the SN1-alkylating agent streptozotocin, a glucosamine-nitrosourea conjugate, causes massive DNA alkylation damage and strong PARP activation in pancreatic β-cells, leading to their destruction in a PARP-1–dependent manner (28). However, a rather low dose of AOM was used in our study, arguing against a hyperactivation of PARP-1 and subsequent NAD+ depletion.
As inflammation is an important driver in CRC and PARP-1 a known coactivator of NF-κB, we analyzed the effects of PARP-1 deficiency on DSS-induced colitis. Lack of PARP-1 strongly reduced inflammation in the large intestine, which was primarily attributable to an attenuated innate immune response. This was evidenced in PARP-1−/− animals by reduced infiltration of monocytes and macrophages, lower levels of proinflammatory cytokines such as IL-6 and IL-1β, and enhanced levels of HMGB1 and COX-2. In contrast, no differences were observed with regard to the infiltration of CD3-positive lymphocytes. These findings extend a previous study, in which acute colitis was induced by rectal instillation of trinitrobenzene sulfonic acid (TNBS), revealing impaired neutrophil activation and decreased mucosal injury in PARP-1–deficient mice (29). Our data are further supported by a recent study, which revealed a transcriptional reprogramming of the colon mucosa and reduced levels of the proinflammatory cytokines TNFα and IL-17 following high dose DSS administration, leading to colitis resistance in PARP-1−/− animals (18). In line with our observation, PARP-1 knockout was reported to attenuate the proinflammatory gene expression and intestinal inflammation following infection with Salmonella enterica (30), highlighting the driving role of PARP-1 in intestinal inflammation. Interestingly, mice with a deficiency for PARG, the enzyme responsible for PAR degradation, are also protected against intestinal inflammation induced by splanchnic ischemia/reperfusion or TNBS instillation (31, 32), indicating that both PARG and PARP-1 are vital for the intestinal inflammatory response by regulating cellular PAR levels and NAD+ consumption.
The observed protection of PARP-1–deficient animals against DSS-induced colitis was associated with a significantly reduced colonic tumor formation, even at high DSS doses. The AOM/DSS-induced tumors from WT animals exhibited a higher degree of aggressiveness and signs of invasion. Further IHC analysis of the isolated tumors revealed higher levels of STAT3 activation and up-regulation of cyclin D1 in WT tumors, thereby likely fostering tumor progression and growth. This finding is consistent with the strong down-regulation of the NF-κB target IL-6 shown herein. IL-6 is a key molecule in chronic inflammatory bowel disease, which links inflammation to CRC development via STAT3 activation (33). STAT3, in turn, plays an important role in the maintenance of a procarcinogenic microenvironment in tumorigenesis, stimulates tumor cell proliferation, and inhibits antitumor immunity (34). STAT3 has also been implicated in the expression of cyclin D1, which promotes cell proliferation and is overexpressed in human CRC (35).
The role of PARP-1 in tumor initiation and progression was dissected using MGMT/PARP-1 DKO animals. These mice developed even more tumors than MGMT single knockouts, which are already highly sensitive to AOM-induced colon carcinogenesis (4). This was not attributable to differences in AOM-induced colonic O6-MeG adducts, which were very similar in both genotypes (DKO and MGMT−/−). Using a CRISPR/Cas9-engineered cell model, we provide evidence that PARP-1–deficient cells display more DNA strand breaks after DNA methylation, which further increased if MGMT was inactivated. This finding implicates PARP-1 in the response to persistent O6-MeG adducts, e.g., by resolving replication stress triggered by O6-MeG adducts or by promoting the repair of secondary DNA strand breaks. In line with these observations, PARP-1 deficiency was shown to increase genomic instability in vivo following exposure to the alkylating agent N-nitrosobis(2-hydroxypropyl)amine (36). Moreover, our data revealed that AOM-induced cell death in colon crypts is reduced in DKO animals compared with MGMT−/− animals, although the level of cytotoxic O6-MeG lesions is similar in both genotypes, suggesting a hitherto unknown role of PARP-1 in O6-MeG–triggered colonic cell death. The increased survival of cells with persistent O6-MeG will likely enhance the mutation rates in colon, finally resulting in higher tumor induction as observed in DKO mice.
Intriguingly, the tumors in DKO mice were much smaller compared with those of MGMT−/− mice, corroborating the importance of PARP-1 for tumor progression. It should be mentioned that PARP-1 was previously identified as a component and coactivator of the oncogenic T-cell factor-4 (TCF-4)/β-catenin complex (23), whose deregulation is an important step in early colorectal carcinogenesis (2), and may thereby also promote tumor growth. In agreement with this notion, it was shown that PARP-1 promotes hepatocellular carcinoma growth and is critical for proinflammatory and angiogenic gene expression, which is significantly reduced by PARP inhibition (37).
In view of our findings, pharmacological inhibition of PARP-1 may be an interesting therapeutic strategy to prevent or reduce the risk of CRC development in patients with IBD, who are notoriously difficult to treat and have limited therapeutic options. This idea is supported by previous studies with PARP inhibitors (PARPi), demonstrating a reduced inflammation in rodents upon TNBS-induced colitis and a normalized colon permeability with reduced expression of proinflammatory cytokines in IL-10–deficient mice (38, 39). Considering our results in MGMT−/−/PARP-1−/− DKO mice, it would however be important to assess the MGMT status in patients with IBD before starting the treatment with PARPi. Inactivation of MGMT by promoter hypermethylation predisposes to alkylation-induced CRC and is frequently found in sporadic colorectal carcinogenesis (3), but also occurs in ulcerative colitis-associated dysplasia (40). Thus, PARPi treatment could increase the risk of these patients to develop CRC. Finally, the high levels of PARP-1 and its product PAR in human colorectal carcinoma and its significant role in stimulating tumor growth in mice shown herein suggest that colorectal tumors may rely on PARP-1 expression and/or its activity. This might offer novel treatment options in CRC therapy by combination of PARPi with chemotherapeutics or biologicals in advanced stages of CRC. Interestingly, a few clinical trials are already ongoing or have been launched in patients with CRC, in which PARPi are used as monotherapy or in combination with DNA-damaging anticancer drugs (41).
In conclusion, our study revealed that PARP-1 is a double-edged sword in colorectal carcinogenesis. On the one hand, PARP-1 counteracts NOC-induced tumor initiation by repair of DNA alkylation damage and elimination of colonocytes harboring mutagenic O6-MeG adducts. On the other hand, PARP-1 drives intestinal inflammation via the innate immune response and promotes colorectal tumor growth through activation of the IL6-STAT3-cyclin D1 axis.
Materials and Methods
Mouse Models, Induction of Colitis-Associated Colorectal Cancer, and Miniendoscopy.
Parp-1-null (PARP-1−/−) and Mgmt-null (MGMT−/−) mice on a C57BL/6 background were previously described (5, 15). Parp-1/Mgmt DKO animals on a C57BL/6 background were generated in the context of this work. To this end, PARP-1−/− and MGMT−/− animals were crossed and the offspring’s genotype was determined using the REDExtract-N-Amp Tissue PCR kit (Sigma-Aldrich) as previously reported (42). In vivo experiments were performed with 8- to 14-wk-old sex-matched PARP-1−/−, MGMT−/−, DKO, and C57BL/6 WT animals. All mouse strains were obtained from the in-house animal breeding facility at the University Medical Center, Mainz. The AOM/DSS model was applied to induce DNA damage-initiated, inflammation-driven colorectal carcinogenesis (43). AOM (Sigma-Aldrich) was dissolved in distilled water and diluted with PBS to the desired concentration, ranging from 0 to 15 mg/kg body weight (BW). A single initial i.p. injection of AOM was followed by two subsequent cycles of 1% or 2.5% DSS (MP Biomedicals) in the drinking water as described previously (4). A high-resolution miniendoscopy system (Karl Storz) was utilized to monitor intestinal inflammation after the first DSS cycle and the formation of colorectal tumors after 16 wk. Inflammation was determined as MEICS (44). The MEICS is based upon five different parameters: thickening of the colon wall, changes of the normal vascular pattern, presence of fibrin, granularity of the mucosal surface, and stool consistency (44). Colorectal tumors were scored with regard to their number and size (4). In the experimental setup with 2.5% DSS, miniendoscopy could not be performed due to a high number of large tumors localized to the rectum. Instead, mice were killed and the isolated colon was longitudinally opened to evaluate tumor number and size, which was documented with the camera of the miniendoscopy system.
Cell Culture, Creation, and Validation of HCT116–PARP-1−/− Cells.
HCT116–PARP-1−/− cells were generated by CRISPR-based targeting as outlined in SI Materials and Methods. Generated clones were screened for PARP-1 expression via immunofluorescence and Western blot using a rabbit monoclonal anti-PARP1 antibody (9532, Cell Signaling) (Fig. S9A). Genomic DNA from PARP-1 knockout clones was extracted and the sequence surrounding the CRISPR-target sequence was amplified by PCR using primers TTCTAAAGTGTGGGAGGGGC and AGAACTGGTGGGAAAGCCTG. Sequencing of the obtained PCR product revealed a one-nucleotide insertion right after the CRISPR-targeted sequence in clones 9 and 10, resulting in an early stop codon (Fig. S9B). Clone 9 was used for the experiments.
PARP-1 and PAR formation were assessed in HCT116–PARP-1 KO and parental cells exposed to 1 mM H2O2 for 5 min. After fixation, cells were stained with a PARP-1 antibody (GTX112864, 1:250; Genetex) or a PAR antibody (clone 10H; 1:250) and appropriate secondary antibodies. Slides were then analyzed by confocal microscopy as described below.
All HCT116-derived cell lines were cultured in RPMI containing 10% FCS and 1% penicillin/streptomycin (p/s). All cell lines were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Cell culture medium and supplements were obtained from Gibco Life Technologies unless otherwise stated. Cell lines were mycoplasma negative as revealed by PCR detection and immunofluorescence microscopy with nuclear staining.
Tissue Collection, Processing, and Histopathology.
Mice were killed at the respective time points after AOM injection (24 and 48 h), after the first DSS cycle (10 d) or at the end of the experiment (12 or 16 wk). Collected tissue was processed as described in SI Materials and Methods. For histopathological evaluation, H&E staining was performed. Samples were analyzed with a Zeiss Axiovert 35 microscope, which was equipped with an Olympus Colorview I camera. Images were acquired with Cell-A 5.1 software (Olympus).
Immunohistochemistry and Confocal Microscopy.
Paraffin sections of formaldehyde-fixed colon tissue were processed for immunohistochemistry as previously described (4). The following primary antibodies were used for the immunofluorescence staining: COX-2 (610204, 1:400; BD Transduction Laboratories), CD11b (NB110-89474, 1:400; Novus Biologicals), CD3 (MCA500A488, 1:400; AbD Serotec), cyclin D1 (2978, 1:50; Cell Signaling Technology), F4/80 (BM4007, 1:400; Acris Antibodies), HMGB1 (GTX101277, 1:400; GeneTex), LC3B (3868, 1:250; Cell Signaling Technology), PCNA (sc-56, 1:400; Santa Cruz Biotechnology), and phosphorylated STAT3 (9145, 1:200; Cell Signaling Technology). The following secondary antibodies were used: goat anti-mouse Alexa Fluor 488 (A11001, 1:500; Life Technologies), goat anti-rabbit Alexa Fluor 488 (A11008, 1:500; Life Technologies), and goat anti-rat Cy3 (112–165-167, 1:500; Jackson ImmunoResearch). IHC staining and confocal microscopy were performed as recently described (4). To detect apoptotic cells in colorectal tissue, the TUNEL assay was used as reported previously (4).
mRNA Extraction, cDNA Synthesis, and Quantitative PCR.
Snap-frozen colon tissue was pestled and mRNA was isolated using the NucleoSpin RNA kit (Macherey-Nagel) according to the manufacturer’s protocol. Subsequent cDNA synthesis and Real-Time PCR were performed as described in SI Materials and Methods.
Isolation of Genomic DNA and Detection of O6-MeG Adducts by Immuno Slot Blot Analysis.
Snap-frozen colon and liver tissue was homogenized and samples were digested with RNase A (Sigma) followed by proteinase K (Sigma) overnight. After phenol-chloroform extraction, isolated genomic DNA was precipitated with ethanol and dissolved in Tris-ethyldiethylamine tetraacetic acid (TE) buffer (pH 7.4). O6-MeG DNA adducts were then analyzed via an immuno slot blot assay (45).
Quantification of O6-MeG Adducts by UPLC-MS/MS.
DNA samples were spiked with the internal standard (IS), d3-O6-methylguanine (Toronto Research Chemicals). Thermal acid hydrolysis was performed for 2 h at 70 °C. After cooling and neutralization, an aliquot was removed for HPLC analysis to verify complete release of guanine, as described previously (46). Sample analysis by LC-MS/MS and quantification is detailed in SI Materials and Methods.
LC-MS/MS Analysis of PAR Levels.
Snap-frozen mouse tissues (30–50 mg) were cut into pieces and homogenized with a tissue disruptor (QIAGEN) in 1 mL of 20% TCA (wt/vol). Afterward, PAR was purified and analyzed by mass spectrometry as described previously (47).
Alkaline Comet Assay.
AOM-induced DNA strand breaks in liver tissue were analyzed via single-cell gel electrophoresis. Briefly, frozen mouse liver tissue was homogenized in Merchant’s medium and mechanically separated into a single-cell suspension with a 70-µm cell strainer (48). To this end, 1 × 103 cells per milliliter were mixed with 5% low melting point agarose, transferred onto agarose-coated slides, and allowed to settle. The alkaline Comet assay was then performed as reported (49, 50). In each experiment, at least 50 cells were scored per sample using the software Comet Assay IV (Perceptive Instruments).
HCT116–PARP-1+/+ and HCT116–PARP-1−/− cells were grown overnight and incubated for 24 h with the SN1-alkylating agent TMZ (500 µM; Schering-Plough), which induces the same spectrum of alkylated DNA adducts as AOM used in the animal experiments. To inactivate cellular MGMT, cells were incubated with the potent MGMT inhibitor O6-benzylguanine (20 µM; Sigma Deisenhofen) for 2 h before TMZ addition. Cells were then harvested and processed for the alkaline Comet assay as described (49, 50).
Human Tissue Samples.
Two different TMAs with a total of n = 128 patients with colorectal adenoma and carcinoma were analyzed. Patients were stratified according to the histopathologic grading of their respective tumor (grading TMA). Additionally, normal colonic mucosa as well as colorectal adenoma were evaluated. In short, for TMA generation, tumorous regions were identified by a pathology expert and two representative tissue cores of 1.0-mm diameter were transferred to the final TMA acceptor block using the TMArrayer (Pathology Devices, Inc.). Formalin-fixed paraffin-embedded (FFPE) sections were generated and immunohistochemistry was performed on the Dako Autostainer (Dako Agilent Pathology Solutions) according to the manufacturer’s guidelines. The staining procedure and image analysis are described in SI Materials and Methods.
Tissue samples were provided by the tissue bank of the University Medical Center, Mainz, and procedures were in accordance with the regulations of the tissue bank. Sample acquisition was approved by the appropriate ethics committee in accordance with all relevant guidelines.
Statistics.
Experiments were performed independently three times, except where otherwise stated. Figures show representative images. Values are depicted as mean ± SEs (SEM) using GraphPad Prism 6.0 Software. Statistical analysis was performed using two-sided Student’s t test and statistical significance was defined as P < 0.05.
Study Approval.
All animal experiments were approved by the government of Rhineland-Palatinate and the Animal Care and Use Committee of the University Medical Center, Mainz, and performed according to German federal law and the guidelines for the protection of animals.
The use of human tissue for tissue microarray generation was approved by the ethical committee of the medical association of the State of Rhineland-Palatinate (responsible institutional review board) and the committee waived the need for specific written informed consent [ref. no. 837.075.16 (10394)].
Supplementary Material
Acknowledgments
We thank Dr. Alexander Bürkle (University of Konstanz) for the kind gift of PARP-1 and PAR antibodies, Dr. Leona D. Samson (Massachusetts Institute of Technology) for providing MGMT knockout animals, and Dr. Shana J Sturla (ETH Zürich) for providing support with mass spectrometry-based O6-MeG analysis. This work was supported by the University Medical Center, Mainz (MAIFOR), and the German Research Foundation (DFG-FA1034/3-1, DFG-KA724/29-1, and INST 38/537-1). S.F. was supported by a TransMed Fellowship of the UCT (Universitaeres Centrum fuer Tumorerkrankungen) Mainz. Tissue samples were provided by the tissue bank of the University Medical Center, Mainz, in accordance with the regulations of the tissue bank.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712345115/-/DCSupplemental.
References
- 1.Kuipers EJ, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahrer J, Kaina B. O6-methylguanine-DNA methyltransferase in the defense against N-nitroso compounds and colorectal cancer. Carcinogenesis. 2013;34:2435–2442. doi: 10.1093/carcin/bgt275. [DOI] [PubMed] [Google Scholar]
- 4.Fahrer J, et al. DNA repair by MGMT, but not AAG, causes a threshold in alkylation-induced colorectal carcinogenesis. Carcinogenesis. 2015;36:1235–1244. doi: 10.1093/carcin/bgv114. [DOI] [PubMed] [Google Scholar]
- 5.Bugni JM, Meira LB, Samson LD. Alkylation-induced colon tumorigenesis in mice deficient in the Mgmt and Msh6 proteins. Oncogene. 2009;28:734–741. doi: 10.1038/onc.2008.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31:101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangerich A, Bürkle A. Pleiotropic cellular functions of PARP1 in longevity and aging: Genome maintenance meets inflammation. Oxid Med Cell Longev. 2012;2012:321653. doi: 10.1155/2012/321653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pascal JM, Ellenberger T. The rise and fall of poly(ADP-ribose): An enzymatic perspective. DNA Repair (Amst) 2015;32:10–16. doi: 10.1016/j.dnarep.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krietsch J, et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med. 2013;34:1066–1087. doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahrer J, Kranaster R, Altmeyer M, Marx A, Bürkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Haince JF, et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 13.Bryant HE, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: What have we learned from the deficient mouse model? Mutat Res. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 15.de Murcia JM, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nozaki T, et al. Parp-1 deficiency implicated in colon and liver tumorigenesis induced by azoxymethane. Cancer Sci. 2003;94:497–500. doi: 10.1111/j.1349-7006.2003.tb01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver FJ, et al. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larmonier CB, et al. Transcriptional reprogramming and resistance to colonic mucosal injury in poly(ADP-ribose) polymerase 1 (PARP1)-deficient mice. J Biol Chem. 2016;291:8918–8930. doi: 10.1074/jbc.M116.714386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat Immunol. 2016;17:230–240. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- 20.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 21.Becker C, et al. In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut. 2005;54:950–954. doi: 10.1136/gut.2004.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idogawa M, et al. Poly(ADP-ribose) polymerase-1 is a component of the oncogenic T-cell factor-4/beta-catenin complex. Gastroenterology. 2005;128:1919–1936. doi: 10.1053/j.gastro.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Nosho K, et al. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur J Cancer. 2006;42:2374–2381. doi: 10.1016/j.ejca.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 25.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 26.Bürkle A, Virág L. Poly(ADP-ribose): PARadigms and PARadoxes. Mol Aspects Med. 2013;34:1046–1065. doi: 10.1016/j.mam.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Calvo JA, et al. Aag DNA glycosylase promotes alkylation-induced tissue damage mediated by Parp1. PLoS Genet. 2013;9:e1003413. doi: 10.1371/journal.pgen.1003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pieper AA, et al. Poly(ADP-ribose) polymerase-deficient mice are protected from streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96:3059–3064. doi: 10.1073/pnas.96.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zingarelli B, Szabó C, Salzman AL. Blockade of Poly(ADP-ribose) synthetase inhibits neutrophil recruitment, oxidant generation, and mucosal injury in murine colitis. Gastroenterology. 1999;116:335–345. doi: 10.1016/s0016-5085(99)70130-7. [DOI] [PubMed] [Google Scholar]
- 30.Altmeyer M, et al. Absence of poly(ADP-ribose) polymerase 1 delays the onset of Salmonella enterica serovar Typhimurium-induced gut inflammation. Infect Immun. 2010;78:3420–3431. doi: 10.1128/IAI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuzzocrea S, et al. Role of poly(ADP-ribose) glycohydrolase in the development of inflammatory bowel disease in mice. Free Radic Biol Med. 2007;42:90–105. doi: 10.1016/j.freeradbiomed.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 32.Cuzzocrea S, et al. PARG activity mediates intestinal injury induced by splanchnic artery occlusion and reperfusion. FASEB J. 2005;19:558–566. doi: 10.1096/fj.04-3117com. [DOI] [PubMed] [Google Scholar]
- 33.Waldner MJ, Neurath MF. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin Immunol. 2014;26:75–79. doi: 10.1016/j.smim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 36.Shibata A, et al. Parp-1 deficiency causes an increase of deletion mutations and insertions/rearrangements in vivo after treatment with an alkylating agent. Oncogene. 2005;24:1328–1337. doi: 10.1038/sj.onc.1208289. [DOI] [PubMed] [Google Scholar]
- 37.Quiles-Perez R, et al. Inhibition of poly adenosine diphosphate-ribose polymerase decreases hepatocellular carcinoma growth by modulation of tumor-related gene expression. Hepatology. 2010;51:255–266. doi: 10.1002/hep.23249. [DOI] [PubMed] [Google Scholar]
- 38.Jijon HB, et al. Inhibition of poly(ADP-ribose) polymerase attenuates inflammation in a model of chronic colitis. Am J Physiol Gastrointest Liver Physiol. 2000;279:G641–G651. doi: 10.1152/ajpgi.2000.279.3.G641. [DOI] [PubMed] [Google Scholar]
- 39.Sánchez-Fidalgo S, Villegas I, Martín A, Sánchez-Hidalgo M, Alarcón de la Lastra C. PARP inhibition reduces acute colonic inflammation in rats. Eur J Pharmacol. 2007;563:216–223. doi: 10.1016/j.ejphar.2007.01.070. [DOI] [PubMed] [Google Scholar]
- 40.Konishi K, et al. Rare CpG island methylator phenotype in ulcerative colitis-associated neoplasias. Gastroenterology. 2007;132:1254–1260. doi: 10.1053/j.gastro.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Guerreroa SM, et al. Expression and single nucleotide polymorphism of poly (ADP-ribose) polymerase-1 in gastrointestinal tumours: Clinical involvement. Curr Med Chem. 2017;24:2156–2173. doi: 10.2174/0929867324666170316115039. [DOI] [PubMed] [Google Scholar]
- 42.Boehler C, et al. Phenotypic characterization of Parp-1 and Parp-2 deficient mice and cells. Methods Mol Biol. 2011;780:313–336. doi: 10.1007/978-1-61779-270-0_19. [DOI] [PubMed] [Google Scholar]
- 43.Wirtz S, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 44.Becker C, Fantini MC, Neurath MF. High resolution colonoscopy in live mice. Nat Protoc. 2006;1:2900–2904. doi: 10.1038/nprot.2006.446. [DOI] [PubMed] [Google Scholar]
- 45.Göder A, et al. Lipoic acid inhibits the DNA repair protein O 6-methylguanine-DNA methyltransferase (MGMT) and triggers its depletion in colorectal cancer cells with concomitant autophagy induction. Carcinogenesis. 2015;36:817–831. doi: 10.1093/carcin/bgv070. [DOI] [PubMed] [Google Scholar]
- 46.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Analysis of pyridyloxobutyl DNA adducts in F344 rats chronically treated with (R)- and (S)-N′-nitrosonornicotine. Chem Res Toxicol. 2007;20:246–256. doi: 10.1021/tx060208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zubel T, Martello R, Bürkle A, Mangerich A. Quantitation of poly(ADP-ribose) by isotope dilution mass spectrometry. Methods Mol Biol. 2017;1608:3–18. doi: 10.1007/978-1-4939-6993-7_1. [DOI] [PubMed] [Google Scholar]
- 48.Jackson P, et al. Validation of freezing tissues and cells for analysis of DNA strand break levels by comet assay. Mutagenesis. 2013;28:699–707. doi: 10.1093/mutage/get049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dörsam B, Göder A, Seiwert N, Kaina B, Fahrer J. Lipoic acid induces p53-independent cell death in colorectal cancer cells and potentiates the cytotoxicity of 5-fluorouracil. Arch Toxicol. 2015;89:1829–1846. doi: 10.1007/s00204-014-1434-0. [DOI] [PubMed] [Google Scholar]
- 50.Mimmler M, et al. DNA damage response curtails detrimental replication stress and chromosomal instability induced by the dietary carcinogen PhIP. Nucleic Acids Res. 2016;44:10259–10276. doi: 10.1093/nar/gkw791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.