Significance
Photosystem I (PSI) is one of the most efficient nanophotochemical machines in nature. To adapt to various environments, photosynthetic organisms developed different PSI structure during evolution from prokaryotic cyanobacteria to higher plants. Red algae are one of the most primitive eukaryotic algae, and their photosynthetic apparatus represents a transitional state between cyanobacteria and eukaryotes. We determined two forms of the PSI-LHCR structure from a red alga by cryo-EM. Our results revealed unique features and energy transfer pathways in the red algal PSI supercomplex with LHCI (light-harvesting complex I), as well as its remarkable differences with those of cyanobacterial PSI and higher plant PSI-LHCI. These results provide important information for delineating the function and evolution of PSI from prokaryotic to eukaryotic photosynthetic organisms.
Keywords: cryo-EM, PSI-LHCR, red algae, PsaO, energy transfer
Abstract
Photosystem I (PSI) is one of the two photosystems present in oxygenic photosynthetic organisms and functions to harvest and convert light energy into chemical energy in photosynthesis. In eukaryotic algae and higher plants, PSI consists of a core surrounded by variable species and numbers of light-harvesting complex (LHC)I proteins, forming a PSI-LHCI supercomplex. Here, we report cryo-EM structures of PSI-LHCR from the red alga Cyanidioschyzon merolae in two forms, one with three Lhcr subunits attached to the side, similar to that of higher plants, and the other with two additional Lhcr subunits attached to the opposite side, indicating an ancient form of PSI-LHCI. Furthermore, the red algal PSI core showed features of both cyanobacterial and higher plant PSI, suggesting an intermediate type during evolution from prokaryotes to eukaryotes. The structure of PsaO, existing in eukaryotic organisms, was identified in the PSI core and binds three chlorophylls a and may be important in harvesting energy and in mediating energy transfer from LHCII to the PSI core under state-2 conditions. Individual attaching sites of LHCRs with the core subunits were identified, and each Lhcr was found to contain 11 to 13 chlorophylls a and 5 zeaxanthins, which are apparently different from those of LHCs in plant PSI-LHCI. Together, our results reveal unique energy transfer pathways different from those of higher plant PSI-LHCI, its adaptation to the changing environment, and the possible changes of PSI-LHCI during evolution from prokaryotes to eukaryotes.
Photosystem I (PSI) is one of the two huge pigment-protein complexes responsible for light-induced electron transfer reactions in photosynthesis and is located within the thylakoid membranes of cyanobacteria, various algae, and higher plants. Upon absorption of light energy, the reaction center core of PSI accepts electrons from plastocyanin or cytochrome c6 at the luminal side, and then transfers them to the soluble electron carrier ferredoxin at the stromal side (1, 2), providing the reduction power for CO2 reduction. During around 2.5 billion years of evolution from prokaryotic cyanobacteria to higher plants (3–5), the core components of PSI remained largely conserved, with slight differences such as the loss of PsaM and gain of PsaG, PsaH, PsaO, and possibly PsaN subunits in higher plant PSI in comparison with the cyanobacterial PSI (1, 3, 5). In contrast, the light-harvesting antenna system surrounding the PSI core has changed dramatically during evolution, most likely to adapt to various environments where different photosynthetic organisms inhabit. In cyanobacteria, the PSI core exists independently without directly associated antennae (6) (although in some cases, soluble phycobiliproteins serve as light-harvesting antennae, see refs. 7 and 8); on the other hand, the PSI core of various algae and higher plants is surrounded by membrane-intrinsic light-harvesting complex (LHC)I proteins, forming a PSI-LHCI supercomplex to maximize the light-energy utilization efficiency (1, 2, 9). The LHCI apoproteins, however, are remarkably different in terms of their sequences, numbers, and associated pigments among different organisms. The differences in the antenna system also resulted in different organization of the reaction center core; thus, the cyanobacterial PSI core exists predominately in trimers, whereas those of eukaryotic PSI exist in a monomeric form (1, 2).
The membrane-intrinsic LHCIs are considered to appear first in red algal PSI (10, 11), as red algae are one of the most primitive photosynthetic eukaryotic algae closely related to cyanobacteria, and their photosystems (PSI and PSII) possess unique properties. In fact, PSII of red algae uses soluble phycobiliproteins as antennae similar to that of cyanobacteria, whereas their PSI is associated with LHC proteins (termed LHCRs) encoded by lhcr genes, which are considered the origin of LHCI proteins (10, 12). Thus, the red algal photosynthetic apparatus represents a transitional state between cyanobacteria and eukaryotic organisms, and their LHCRs may possess characteristics of the earliest membrane antennae that bear implications for the early specification of the membrane-intrinsic light-harvesting antennae (10–14). The LHCI genes are then evolved and diversified in terms of both sequences and numbers in the eukaryotic algae and higher plants, yielding different numbers and probably different patterns they associate with the PSI core, as well as the different types of pigments they bind. For example, green algae LHCIs bind both chlorophyll (Chl) a and Chl b, as those of higher plant PSI, but the number of the LHCI subunits they associate with every PSI core is between 8 and 10 (15, 16), which is much larger than the 4 subunits found in higher plant PSI (17–21). Diatom and brown algae bind Chl c in addition to Chl a, as well as different types of carotenoids (22), providing their characteristic brown-yellowish color.
The crystal structure of cyanobacterial PSI core has been determined at 2.5 Å resolution (6), and that of higher plant PSI-LHCI was reported with increasing resolutions (17–21), which showed that the supercomplex contains 12 PSI core subunits and 4 LHCI subunits encoded by four different Lhca genes named lhca1 to lhca4. The four LHCI subunits are associated at one side of the PSI core, forming a semispherical shape for the whole supercomplex. The efficiency of excitation energy transfer (EET) from these LHCI subunits to the PSI core has been shown to be extremely high (1, 9, 23), and the structural studies with increasing resolution in plants have provided detailed organization of protein subunits and pigment–pigment interactions (2, 19–21), as well as a basis for elucidating the efficient EET process within the PSI-LHCI supercomplex. On the other hand, no high-resolution structure of PSI-LHCI from various algae has been reported so far, although some low-resolution structures have been reported (24–26).
Cyanidioschyzon merolae is a thermophilic, acidophilic red alga widely used for genetic and biochemical studies and has three Lhcr genes in contrast to the four Lhca subunits identified in the higher plant PSI-LHCI (13). Here, we report the single-particle cryogenic electron microscopy (cryo-EM) structure of PSI-LHCR from C. merolae at a resolution of 3.82 Å to 3.63 Å, which identified two forms of the PSI-LHCR supercomplex, one with three Lhcr antennae attached (designated PSI-3Lhcr) and the other one with five Lhcr antennae (designated PSI-5Lhcr). The results obtained provide important information for delineating the evolution of PSI from prokaryotic to eukaryotic photosynthetic organisms and the differences in the EET pathways among different organisms, possibly as a result of adaptation to different light environments.
Results
Overall Structure.
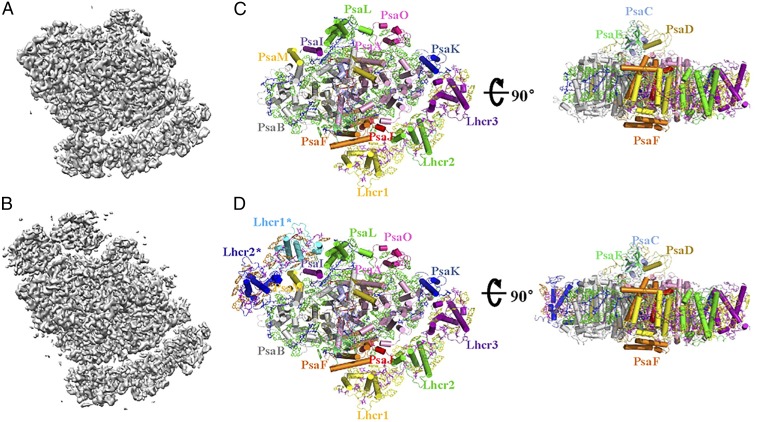
Two forms of the PSI-LHCR supercomplex purified from C. merolae cells grown under a normal light environment were classified in the cryo-EM images (Fig. 1 and SI Appendix, Table S1). The PSI-3Lhcr supercomplex binds three antenna subunits corresponding to the products of three lhcr genes (cmq142C, cmn234C, and cmn235C) identified in the C. merolae genome (13) and are named Lhcr1, Lhcr2, and Lhcr3. These three Lhcr subunits are located at positions corresponding to those of Lhca4, Lhca2, and Lcha3 in the higher plant PSI-LHCI and form an LHCR belt that interacts with PsaF, PsaJ, PsaA, and PsaK. The overall structure of PSI-3Lhcr is similar to that of plant PSI-LHCI but lacks PsaG and its neighboring Lhca1, causing a slight shift of the LHCR belt toward the PsaK side and a large marginal space in the Lhca1 position of the plant LHCI belt (Figs. 1C and 2).
Fig. 1.
Two forms of the PSI-LHCR supercomplex from C. merolae. (A) The 3D cryo-EM density map of the PSI-3Lhcr form. (B) The 3D cryo-EM density map of the PSI-5Lhcr form. (C) Structure of PSI-3Lhcr viewed along the membrane normal from the stromal side (Left) and its side view (Right). (D) Structure of PSI-5Lhcr viewed along the membrane normal from the stromal side (Left) and its side view (Right). Color codes for all panels: PSI core subunits: pink, PsaA; gray, PsaB; light blue, PsaC; olive, PsaD; mint green, PsaE; orange, PsaF; blue, PsaK; purple, PsaI; red, PsaJ; green, PsaL; yellow, PsaM; hot pink, PsaO. Lhcr proteins: yellow, Lhcr1; green, Lhcr2; purple, Lhcr3; cyan, Lhcr1*; blue, Lhcr2*. Cofactors: green, Chls a of the PSI core complex; yellow and orange, Chls a of Lhcrs; blue, carotenoids of the PSI core complex; purple, Zeas of Lhcrs; red, cofactors of the electron transfer chain (Chls a, phylloquinones, and Fe4S4 clusters).
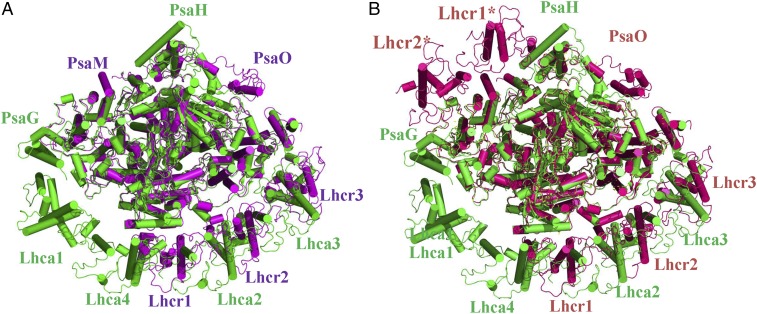
Fig. 2.
Comparison of the structures of red algal PSI-LHCR and pea PSI-LHCI (PDB ID code 4XK8). (A) Superposition of the structure of PSI-3Lhcr with PSI-LHCI. (B) Superposition of the structure of PSI-5Lhcr with PSI-LHCI. Color codes: green, PSI-LHCI; purple, PSI-3Lhcr; hot pink, PSI-5Lhcr.
The PSI-5Lhcr form contains two additional Lhcr subunits, Lhcr1* and Lhcr2*, which correspond to Lhcr1 and Lhcr2, respectively, and are associated with the surface of PsaL, PsaI, PsaM, and PsaB (Fig. 1D). The major difference between PSI-5Lhcr and PSI-3Lhcr lies in the additional LHCR* belt, while the core subunits and three Lhcrs of the canonical LHCR belt (including the cofactors associated with each of the Lhcr subunits) are identical. The root-mean-square deviation of the two forms, except the LHCR* belt, is 0.5 Å. Thus, we will describe the features of the red algal PSI-LHCI mainly based on the structure of PSI-5Lhcr, which contained 158 Chls a, 21 β-carotenes (BCRs), 25 zeaxanthins (Zeas), 2 phosphatidylglycerols, 1 digalactosyldiacylglycerol, 3 Fe4S4 clusters, and 2 phylloquinones (Fig. 1 and SI Appendix, Table S2), in addition to the protein components. The assignment of Chls a and BCRs was based on the cryo-EM density map (SI Appendix, Fig. S1) in combination with the results of biochemical analysis, which showed that the red alga contains only Chls a and that BCRs and Zeas are the predominant carotenoids in the PSI core and LHCR, respectively (27). Most subunits and cofactors have the same locations and orientations as those in the cyanobacterial and plant PSI; however, some subunits and cofactors show significant differences as described below.
The PSI Core.
The PSI cores of both PSI-3Lhcr and PSI-5Lhcr contain nine transmembrane (TM) subunits (PsaA, PsaB, PsaF, PsaI, PsaJ, PsaK, PsaL, PsaM, PsaO) and three extrinsic subunits (PsaC, PsaD, PsaE) located at the stromal side. Among these, 10 (PsaA through PsaF and PsaI through PsaL) are found in both cyanobacterial and higher plant PSI cores, whereas PsaM is present in the cyanobacteria PSI only, and PsaO is found in eukaryotic PSI only. In addition, the red algal PSI lacks the PsaX subunit unique to cyanobacteria and the PsaG and PsaH subunits unique to higher plants.
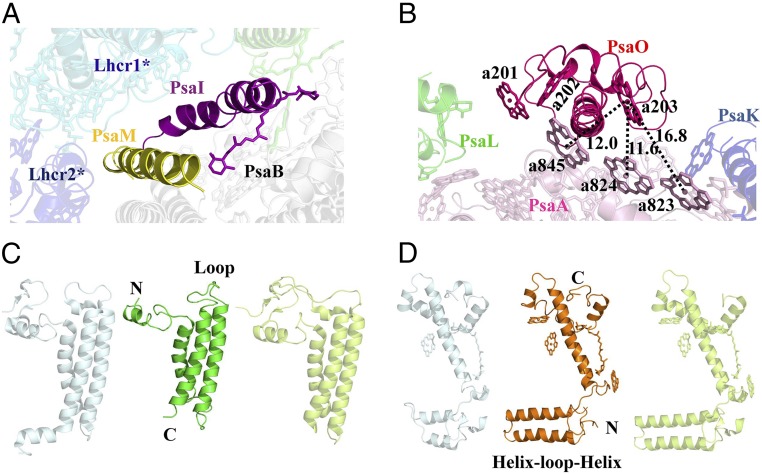
PsaM is located at the same position as that of the cyanobacterial PsaM (6) but does not bind Chl a or BCR. In PSI-3Lhcr, PsaM is located at the outlier surface in contact with PsaI and PsaB, whereas in PSI-5Lhcr, it is surrounded by Lhcr1* and Lhcr2*, suggesting that it offers an attaching site for the additional LHCR* belt (Figs. 1 C and D and 3A).
Fig. 3.
The red algal PSI core subunits. (A) Location of PsaM and PsaI in relation to Lhcr1* and Lhcr2*. (B) PsaO and its pigment–pigment interactions with PsaA. (C) Structure comparison of PsaL from cyanobacterium (Left), red alga (Middle), and higher plant (Right). (D) Structure comparison of PsaF from cyanobacterium (Left), red alga (Middle), and higher plant (Right). The color codes of PSI-LHCR subunits are the same as in Fig. 1. In C and D, light blue represents PsaL and PsaF from a cyanobacterium (PDB ID code 1JB0), and light green represents PsaL and PsaF from a higher plant (PDB ID code 4XK8).
The structure of the eukaryotic PsaO was found (Fig. 3B) to contain two TM helices, A and C, and an amphipathic helix B at the luminal side, with both the N- and C-terminal regions located at the stromal side. It associates with PsaA mainly through its TM helix A, and binds three Chls a (a201, a202, and a203). Among these, Chl a203 is located at a close distance to the Chl a823-a824-a845 cluster of PsaA at the stromal side, with Mg-to-Mg distances of 16.8 Å, 11.6 Å, and 12.0 Å, respectively, between these adjacent Chl pairs, indicating rapid EET from PsaO to the core.
The conserved PsaL subunit fulfills different functions in cyanobacteria and higher plants (3, 18–21, 28). The red algal PsaL has the shortest structure due to the shorter N and C termini and the loop region between the second and third TM helices (Fig. 3C). Its N-terminal stromal region lacks three small β-strands that connect PsaL to the core found in cyanobacteria and higher plants (14, 29), and the C-terminal luminal region lacks a short α-helix that participates in cyanobacterial PSI trimer formation. The loop between the TM helices is involved in PsaH binding in plant PSI, but red algal PSI has no PsaH, resulting in the shortest length of the PsaL loop. The higher plant PsaH covers PsaL and hinders the formation of trimer; instead, it forms part of the docking site for LHCII (18–21, 28). Red algae have no PsaH and LHCII; thus, the monomeric PSI of red algae is caused by the unique structure of PsaL and the attachment of the additional LHCR* belt.
Red algal PsaF is a “merged” structure with its N- and C-terminal regions similar to that of plant and cyanobacterial PsaF, respectively (Fig. 3D), which is consistent with the results of sequence analysis (29). The N-terminal helix–loop–helix motif is similar to that of plant and Chlamydomonas reinhardtii PsaF where this motif was found to be necessary for the efficient electron transfer from plastocyanin (30, 31). Cyanobacterial PsaF does not have this motif, and addition of this motif to the cyanobacterial PsaF improved the binding and electron transfer between the chimeric PSI and the cyanobacterial cytochrome c6 (31). As red algae also use cytochrome c6 as the electron donor (29, 32), the helix–loop–helix motif may be beneficial for an efficient electron transfer from cytochrome c6 to PSI. On the other hand, the C terminus of the red algal PsaF is similar to that of the cyanobacterial one, which interacts with the stromal PsaE and may facilitate interactions with the membrane-extrinsic phycobiliproteins (33).
The LHC.
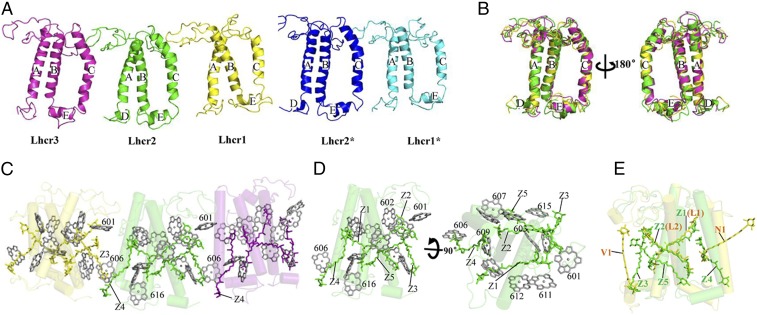
The red algal light-harvesting antennae belong to the LHC gene superfamily and have similar structure to plant LHCI (19, 20), LHCII (34), and CP29 (35) but also show distinct differences in their detailed structures and pigment binding. All Lhcr apoproteins have three TM helices (A, B, and C) and two short amphipathic helices (D and E) parallel to the luminal membrane (Fig. 4 A and B and SI Appendix, Fig. S2). Slight differences are found in the loop regions between the helices and in the lengths of helices A and C, among the different Lhcrs. The Lhcr apoproteins also differ from plant Lhcas, LHCII, and CP29 in several ways. The N-terminal stromal region of Lhcr1/Lhcr1* and Lhcr2 interacts with a loop from their neighboring Lhcr similar to that observed in plant LHCI. However, the C-terminal luminal region of the Lhcrs does not interact with their neighbors due to the lack of a plant-type short helix (SI Appendix, Fig. S3), which also shortens the distance between the two neighboring Lhcrs, resulting in a more compact LHCR belt. The length of the loop between helices C and A in Lhcrs is similar to that in Lhca1 and LHCII but shorter than that in Lhca2, Lhca3, and Lhca4. Helix E is present in all Lhcrs and in LHCII but is absent in Lhca2, Lhca3, and Lhca4.
Fig. 4.
Structures of LHCR subunits and their pigment arrangements. (A) Side view of Lhcr1, Lhcr2, Lhcr3, Lhcr1*, and Lhcr2* from the PSI core. (B) Superposition of Lhcr1, Lhcr2, and Lhcr3 subunits. (C) Chls a and Zeas in Lhcr1, Lhcr2, and Lhcr3 viewed from the PSI core. (D) Chls a and Zeas in Lhcr2 viewed from the LHCR side and rotated 90° along the membrane plane. (E) Comparison of carotenoids in Lhcr2 and LHCII (orange; PDB ID code 1RWT). The color codes of LHCR subunits are the same as in Fig. 1.
Each Lhcr contains 11 to 13 Chls a and 5 Zeas; the assignment of Zeas was based on the cryo-EM density map (SI Appendix, Fig. S1) and the results of biochemical analysis, which showed that the red algal Lhcrs contains mainly Zeas as carotenoids (27). Compared with the pigments in plant LHCI and LHCII, the number of Chls a in Lhcr is 1 to 3 fewer and the carotenoids are 1 to 2 more (Fig. 4 C–E and SI Appendix, Fig. S4). Most Chls a in Lhcrs have their counterparts in both LHCI and LHCII, including the red Chl pair a603-a609 (19, 23, 34, 36) (SI Appendix, Fig. S5). However, Lhcrs lack Chls at positions 605, 608, and 614 found in plant LHCI and LHCII. The positions of 601, 606, and 607 are occupied by Chl b in LHCI (19, 21) and LHCII (34) but are Chl a in Lhcrs, with slightly different orientations and positions since there is no Chl b in the red algae. The extra Chls 616, 617, 618, and 619 in plant Lhcas are not found in Lhcrs. Instead, Lhcrs have two additional Chls a (615 and 616), which are absent in LHCI and LHCII (SI Appendix, Fig. S4). Among these two Chls, a615 is located in the gap region between Lhcrs and the PSI core and exists in all Lhcrs except Lhcr1*, indicating that it may facilitate energy transfer from LHCR to the PSI core (18–21, 36). Chl a616 is only found in Lhcr2 and Lhcr2* and is located at the outer surface, suggesting that it may easily dissociate during purification.
Among the five Zea-binding sites (designated Z1 to Z5) identified (Fig. 4 C–E), Z1 and Z2 correspond to the L1 and L2 sites in LHCI, LHCII, and CP29, but the carotenoids in these two sites are different. Lhcrs do not have the N1 site conserved in LHCII, CP29, and LHCI, or the V1 site found in LHCII. Instead, Lhcrs have three additional sites: Z3, Z4, and Z5. Z3 is located approximately parallel to helix A and surrounded by several Chls in both the stromal and luminal sides. In Lhcr1/Lhcr1* and Lhcr2, Z3 is close to helix C and Chl a606 of the neighboring Lhcr, suggesting that it may mediate interaction between adjacent Lhcrs and/or energy dissipation under strong light conditions. Z4 is located between helices B and C and surrounded by Chl a604 and Chl a606 with short distances (<5 Å). Z5 and Z2 are clustered together (with an approximate distance of 5 Å) in a similar orientation in a position facing the core and are also close to Chls a607 and a615 in the gap region between Lhcrs and the PSI core at the luminal side, suggesting that these pigments may also facilitate energy dissipation under excess light illumination.
Energy Transfer from LHCR to the PSI Core.
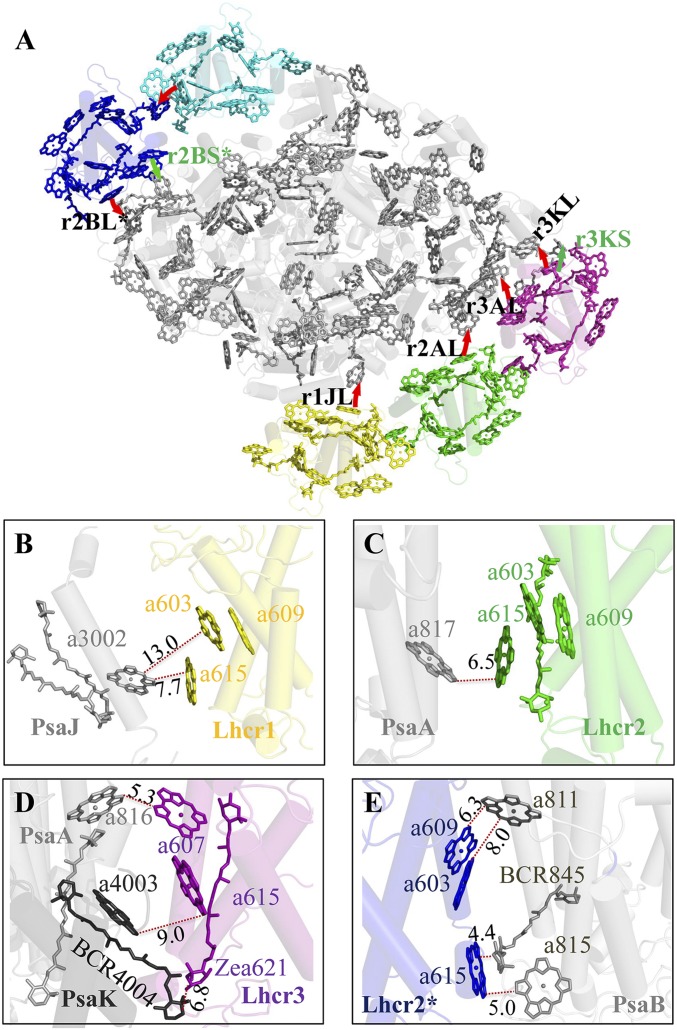
The differences in the arrangement of pigments between Lhcrs and other Lhcas, in particular the identification of pigment-binding sites and the presence of the LHCR* belt, may bring important implications for the possible EET pathways in the red algal PSI-LHCR. Based on the close distances between the pigments in Lhcrs and PSI core revealed in the present study, we propose seven possible EET pathways: r1JL, r2AL, r3AL, r3KL, r3KS, r2BL*, and r2BS* (Fig. 5 and SI Appendix, Table S3) [the nomenclature of these pathways follows those for the plant PSI-LHCI (19), with the first letter r representing Rhodophyta]. Among these pathways, five are found at the luminal side, with the r1JL, r2AL, r3KL, and r2BL* pathways employing the Chl a615 (identified in this study) bound to the helix A of Lhcrs. This Chl serves as a link between the core and Lhcr1, Lhcr2, Lhcr3, and Lhcr2*, resulting in the shortest edge-to-edge distances of 7.7 Å, 6.5 Å, 9.0 Å, and 5.0 Å, between the core and the four Lhcrs, respectively. Furthermore, Chl a615 of Lhcr2* has an edge-to-edge distance of 4.4 Å to a BCR in PsaB, which may enhance the EET efficiency or energy dissipation. The r3KL pathway is not found in the plant PSI-LHCI and arises from the shift of Lhcr1, Lhcr2, and Lhcr3 toward PsaK. The r3AL pathway occurs through Chl a607 at helix E of Lhcr3 and Chl a816 of PsaA, with a shortest edge-to-edge distance of 5.3 Å. Two pathways (r3KS and r2BS*) are found at the stromal side, in which the r2BS* pathway is formed by a red Chl pair (a603-a609) and Chl a811 at PsaA (6.3 Å).
Fig. 5.
Possible energy transfer pathways from LHCR to the PSI core. (A) The energy transfer pathways (red arrows) from Lhcrs to the PSI core. (B) Luminal side pathway r1JL from Lhcr1 to the core. (C) Luminal side pathway r2AL from Lhcr2 to the core. (D) Pathways r3KL, r3AL, and r3KS from Lhcr3 to the core. (E) Pathways r2KL* and r2BS* from Lhcr2* to the core.
EET from Lhcr1* to the core is most likely conveyed by Lhcr2* at both the stromal and luminal sides, since the Chls a in Lhcr1* have longer distances to the core Chls a, and they are also blocked by PsaI, PsaL, and PsaM. The shortest edge-to-edge distance between Lhcr1* and Lhcr2* is 4.7 Å, which is found between Chl a613 at helix A of Lhcr1* and Chl a606 at helix C of Lhcr2* at the luminal side.
Among the above EET pathways, the most efficient candidates are presumably those from Lhcr3 and Lhcr2* to the core, because these two Lhcrs not only have a close distance to the core but also converge with several pathways, thus improving the EET efficiency. Together, our results show that the possible EET pathways in the red algal PSI-LHCR differ significantly from those in plant PSI-LHCI in both the locations and the pigments involved.
Discussion
The high-resolution structural analysis achieved in the present study allowed us to trace most of the amino acid residues and cofactors in each of the subunits of the red algal PSI-LHCR supercomplex, which resulted in a much more detailed structure and, therefore, to shed light on the evolutionary process of PSI during the prokaryote–eukaryote transition as well as its possible adaptation to the light environment. We found two different forms of the supercomplex: PSI-3Lhcr and PSI-5Lhcr. The major difference between the two forms is the presence of the two additional Lhcr* subunits (Lhcr1* and Lhcr2*) in the PSI-5Lhcr form attached to the opposite side of the other three Lhcrs. In fact, an enrichment of Lhcr1 over Lhcr2 and Lhcr3 has been reported from biochemical analysis (24, 27), and the presence of additional density at the position similar to that of Lhcr1* and Lhcr2* has been suggested by a low-resolution cryo-EM study on both C. merolae (24) and the microalga Nannochloropsis gaditana (26), as well as on Chlamydomonas reinhardtii (25). The previous study (24) further reported an extra density extended from PsaK, which was not assigned due to the low resolution. In the present study, this region is assigned to PsaO.
The PSI-5Lhcr form, with two Lhcr belts bound at the two opposite sides of the PSI core, may represent the form adapted to low light intensities under an aquatic environment, as the additional Lhcr subunits will facilitate light-harvesting under weak light. Thus, this form likely corresponds to a prototype of the plant PSI-LHCI when the red algae appeared in the aquatic environment. The binding of the second belt (Lhcr*) at the PsaL-PsaM-PsaI side prevented trimerization, resulting in the transition of PSI from a trimer in cyanobacteria to a monomer in eukaryotes. Upon high light illumination, the additional Lhcr* belt may have been lost, resulting in the PSI-3Lhcr form that has only one Lhcr belt attached to one side of the core. This forms the crescent shape, a common structure found in other eukaryotic algal and higher plant PSI-LHCI. In green algae and some other eukaryotic algae, additional Lhcas appear to attach to the outside of the major Lhca belt in response to the requirement for harvesting enough energy under very low light conditions under water. Furthermore, due to the loss of the second Lhcr* belt in green algae and higher plants, the PsaL-PsaH side becomes open, enabling LHCII to move and bind to this position upon state transition. Red algae do not contain LHCII, and the state transition is accomplished by the movement of phycobiliproteins; thus, the PsaL side is occupied by the additional Lhcr* belt.
The core of the red algal PSI also showed unique features that are representative of its intermediate type during evolution. In addition to the 10 subunits commonly found from cyanobacteria to higher plant PSI, the PsaM subunit (present in cyanobacteria only) and the PsaO subunit (found in the eukaryotic PSI only) were both found in the red algal PSI core. On the other hand, the red algal PSI lacks the PsaX subunit of cyanobacteria and the PsaG and PsaH subunits of higher plants. These features strongly suggest that the red algal PSI is an intermediate type during evolution from cyanobacteria to higher plants.
The binding of PsaM is relevant to the association of the two additional Lhcr* belts, since PsaM provides a binding site for these two Lhcrs. Furthermore, since PsaM is located in a position close to the adjacent monomer in the cyanobacterial PSI trimer, the binding of the additional Lhcr* belt covering PsaM may have hampered the trimerization, making PSI a monomer in the red algae PSI. The loss of PsaM in green algae and higher plant PSI may be one of the reasons that these eukaryotic PSI cores have only one belt of LHCI bound to the PsaF-PsaJ-PsaK side. However, the green algal and higher plant PSI is also a monomer; this may be caused by the acquisition of the PsaH subunit that is located close to PsaL and, therefore, hampers the trimer formation.
The absence of PsaG in the red algal PSI also implies its premature nature as a eukaryotic PSI, since PsaG has some sequence and structural similarities with that of PsaK and has been considered to be a product of gene duplication from PsaK. The red algal PSI contains PsaK, suggesting that it is closer to the cyanobacterial PSI in this respect and that PsaG appeared after the evolution of red algae. As a consequence of the absence of PsaG, the red algal PSI was not able to bind Lhcr in the PsaG side, resulting in the major Lhcr belt containing only three Lhcr subunits, which is one less than found in higher plant PSI-LHCI.
Materials and Methods
Cells of C. merolae were cultivated under a normal light condition (50 μmol photons per meter squared per second), and the PSI-LHCR supercomplexes were isolated and purified following the protocol described by Tian et al. (27) (summarized in SI Appendix, Fig. S6). The crude PSI-LHCR supercomplexes were first isolated from the thylakoid membranes by a strong anion-exchange column (Q Sepharose High-Performance; GE Healthcare) (SI Appendix, Fig. S7A), which were further purified by sucrose density gradient centrifugation (SI Appendix, Fig. S7B). The dark green band (SI Appendix, Fig. S7C) after sucrose density centrifugation was collected for cryo-EM studies.
The structures of PSI-5Lhcr and PSI-3Lhcr were solved at 3.63 Å and 3.82 Å resolution, respectively (SI Appendix, Fig. S8). Detailed procedures for cryo-EM image collection and structural analysis are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Lei and the staff at the Tsinghua University Branch of National Center for Protein Sciences Beijing for providing facility support, and the “Explorer 100” cluster system of the Tsinghua National Laboratory for Information Science and Technology for providing the computation resources. This work was supported by grants from the National Key R&D Program of China (2017YFA0503700), the National Basic Research Program of China (2016YFA0501100) (to S.-F.S. and L.C.), the National Natural Science Foundation of China [31230016 and 31370717 (to S.-F.S.) and 31622007 and 31670237 (to X.Q.)], the Strategic Priority Research Program of Chinese Academy of Sciences (XDB17000000), the Chinese Academy of Sciences Key Research Project for Frontier Science (QYZDY-SSW-SMC003), the National Basic Research Program of China (2015CB150100) (to T.K.), and the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (JP17H0643419) (to J.-R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM density maps and atomic models have been deposited in the Electron Microscopy Data Bank and the Protein Data Bank for the PSI-5Lhcr structure at 3.63 Å (EMD ID code 6929 and PDB ID code 5ZGB) and for the PSI-3Lhcr structure at 3.82 Å resolution (EMD ID code 6930 and PDB ID code 5ZGH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722482115/-/DCSupplemental.
References
- 1.Nelson N, Junge W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu Rev Biochem. 2015;84:659–683. doi: 10.1146/annurev-biochem-092914-041942. [DOI] [PubMed] [Google Scholar]
- 2.Suga M, Qin X, Kuang T, Shen JR. Structure and energy transfer pathways of the plant photosystem I-LHCI supercomplex. Curr Opin Struct Biol. 2016;39:46–53. doi: 10.1016/j.sbi.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Nelson N, Ben-Shem A. The structure of photosystem I and evolution of photosynthesis. BioEssays. 2005;27:914–922. doi: 10.1002/bies.20278. [DOI] [PubMed] [Google Scholar]
- 4.Blankenship RE. Origin and early evolution of photosynthesis. Photosynth Res. 1992;33:91–111. [PubMed] [Google Scholar]
- 5.Nelson N. Evolution of photosystem I and the control of global enthalpy in an oxidizing world. Photosynth Res. 2013;116:145–151. doi: 10.1007/s11120-013-9902-6. [DOI] [PubMed] [Google Scholar]
- 6.Jordan P, et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 7.Kondo K, Ochiai Y, Katayama M, Ikeuchi M. The membrane-associated CpcG2-phycobilisome in Synechocystis: A new photosystem I antenna. Plant Physiol. 2007;144:1200–1210. doi: 10.1104/pp.107.099267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong C, et al. ApcD is necessary for efficient energy transfer from phycobilisomes to photosystem I and helps to prevent photoinhibition in the cyanobacterium Synechococcus sp. PCC 7002. Biochim Biophys Acta. 2009;1787:1122–1128. doi: 10.1016/j.bbabio.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Croce R, van Amerongen H. Light-harvesting in photosystem I. Photosynth Res. 2013;116:153–166. doi: 10.1007/s11120-013-9838-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe GR, Cunningham FX, Durnfordt D, Green BR, Gantt E. Evidence for a common origin of chloroplasts with light-harvesting complexes of different pigmentation. Nature. 1994;367:566–568. [Google Scholar]
- 11.Green BR, Durnford DG. The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- 12.Durnford DG, et al. A phylogenetic assessment of the eukaryotic light-harvesting antenna proteins, with implications for plastid evolution. J Mol Evol. 1999;48:59–68. doi: 10.1007/pl00006445. [DOI] [PubMed] [Google Scholar]
- 13.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 14.Neilson JAD, Durnford DG. Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth Res. 2010;106:57–71. doi: 10.1007/s11120-010-9576-2. [DOI] [PubMed] [Google Scholar]
- 15.Drop B, et al. Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J Biol Chem. 2011;286:44878–44887. doi: 10.1074/jbc.M111.301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi H, Okamuro A, Minagawa J, Takahashi Y. Biochemical characterization of photosystem I-associated light-harvesting complexes I and II isolated from state 2 cells of Chlamydomonas reinhardtii. Plant Cell Physiol. 2014;55:1437–1449. doi: 10.1093/pcp/pcu071. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Shem A, Frolow F, Nelson N. Crystal structure of plant photosystem I. Nature. 2003;426:630–635. doi: 10.1038/nature02200. [DOI] [PubMed] [Google Scholar]
- 18.Amunts A, Drory O, Nelson N. The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature. 2007;447:58–63. doi: 10.1038/nature05687. [DOI] [PubMed] [Google Scholar]
- 19.Qin X, Suga M, Kuang T, Shen JR. Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science. 2015;348:989–995. doi: 10.1126/science.aab0214. [DOI] [PubMed] [Google Scholar]
- 20.Mazor Y, Borovikova A, Nelson N. The structure of plant photosystem I super-complex at 2.8 Å resolution. eLife. 2015;4:e07433. doi: 10.7554/eLife.07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazor Y, Borovikova A, Caspy I, Nelson N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat Plants. 2017;3:17014. doi: 10.1038/nplants.2017.14. [DOI] [PubMed] [Google Scholar]
- 22.Kuczynska P, Jemiola-Rzeminska M, Strzalka K. Photosynthetic pigments in diatoms. Mar Drugs. 2015;13:5847–5881. doi: 10.3390/md13095847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wientjes E, van Stokkum IH, van Amerongen H, Croce R. The role of the individual Lhcas in photosystem I excitation energy trapping. Biophys J. 2011;101:745–754. doi: 10.1016/j.bpj.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busch A, Nield J, Hippler M. The composition and structure of photosystem I-associated antenna from Cyanidioschyzon merolae. Plant J. 2010;62:886–897. doi: 10.1111/j.1365-313X.2010.04202.x. [DOI] [PubMed] [Google Scholar]
- 25.Drop B, Yadav KNS, Boekema EJ, Croce R. Consequences of state transitions on the structural and functional organization of photosystem I in the green alga Chlamydomonas reinhardtii. Plant J. 2014;78:181–191. doi: 10.1111/tpj.12459. [DOI] [PubMed] [Google Scholar]
- 26.Alboresi A, et al. Conservation of core complex subunits shaped the structure and function of photosystem I in the secondary endosymbiont alga Nannochloropsis gaditana. New Phytol. 2017;213:714–726. doi: 10.1111/nph.14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L, et al. Isolation and characterization of PSI-LHCI super-complex and their sub-complexes from a red alga Cyanidioschyzon merolae. Photosynth Res. 2017;133:201–214. doi: 10.1007/s11120-017-0384-9. [DOI] [PubMed] [Google Scholar]
- 28.Busch A, Hippler M. The structure and function of eukaryotic photosystem I. Biochim Biophys Acta. 2011;1807:864–877. doi: 10.1016/j.bbabio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Vanselow C, Weber APM, Krause K, Fromme P. Genetic analysis of the Photosystem I subunits from the red alga, Galdieria sulphuraria. Biochim Biophys Acta. 2009;1787:46–59. doi: 10.1016/j.bbabio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Hippler M, Drepper F, Haehnel W, Rochaix JD. The N-terminal domain of PsaF: Precise recognition site for binding and fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1998;95:7339–7344. doi: 10.1073/pnas.95.13.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hippler M, Drepper F, Rochaix JD, Mühlenhoff U. Insertion of the N-terminal part of PsaF from Chlamydomonas reinhardtii into photosystem I from Synechococcus elongatus enables efficient binding of algal plastocyanin and cytochrome c6. J Biol Chem. 1999;274:4180–4188. doi: 10.1074/jbc.274.7.4180. [DOI] [PubMed] [Google Scholar]
- 32.Allen JF, de Paula WBM, Puthiyaveetil S, Nield J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011;16:645–655. doi: 10.1016/j.tplants.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Fromme P, Melkozernov A, Jordan P, Krauss N. Structure and function of photosystem I: Interaction with its soluble electron carriers and external antenna systems. FEBS Lett. 2003;555:40–44. doi: 10.1016/s0014-5793(03)01124-4. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z, et al. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 35.Pan X, et al. Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat Struct Mol Biol. 2011;18:309–315. doi: 10.1038/nsmb.2008. [DOI] [PubMed] [Google Scholar]
- 36.Morosinotto T, Breton J, Bassi R, Croce R. The nature of a chlorophyll ligand in Lhca proteins determines the far red fluorescence emission typical of photosystem I. J Biol Chem. 2003;278:49223–49229. doi: 10.1074/jbc.M309203200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.