Significance
Recent success of T cell-based cancer immunotherapies highlights the importance of further understanding molecular mechanisms in the regulation of T cell responsiveness. Here, we performed a genome-wide CRISPR screen to identify genes that regulate T cell activation upon anti-T cell receptor (TCR) stimulation. Our screen confirmed many of the known regulators in proximal T cell signaling. Moreover, we identified a previously uncharacterized gene named FAM49B, which acts as a negative regulator in T cell activation. Our study suggests that genome-wide CRISPR screening is a powerful means to identify key regulators of TCR signaling. The same strategy could be applied to CD28-mediated costimulatory signaling or PD-1–mediated coinhibitory signaling. The unbiased approach presented here may allow us to identify new therapeutic targets for cancer immunotherapy.
Keywords: genome-wide CRISPR screen, TCR signaling, FAM49B, Rac1, actin cytoskeleton
Abstract
Despite decades of research, mechanisms controlling T cell activation remain only partially understood, which hampers T cell-based immune cancer therapies. Here, we performed a genome-wide CRISPR screen to search for genes that regulate T cell activation. Our screen confirmed many of the known regulators in proximal T cell receptor signaling and, importantly, also uncovered a previously uncharacterized regulator, FAM49B (family with sequence similarity 49 member B). FAM49B deficiency led to hyperactivation of Jurkat T cells following T cell receptor stimulation, as indicated by enhancement of CD69 induction, PAK phosphorylation, and actin assembly. FAM49B directly interacted with the active form of the small GTPase Rac, and genetic disruption of the FAM49B–Rac interaction compromised FAM49B function. Thus, FAM49B inhibits T cell activation by repressing Rac activity and modulating cytoskeleton reorganization.
T cells play an essential role in maintaining human health against cancer, infection, and autoimmune diseases. T cell immune responses to pathogen- or tumor-antigens, as well as self-peptides, are tightly regulated (1, 2). Activation of T lymphocytes by their cognate antigens initiates a series of events. After T cell receptor (TCR) engagement, Src kinases (Lck and Fyn) phosphorylate immunoreceptor tyrosine-based activation motifs (ITAMs) in the cytoplasmic tails of CD3- and ζ-chains within the TCR:CD3 complex. These phosphorylated ITAMs recruit Zap70, which partially relieves autoinhibition of Zap70 (3). After Zap70 is further activated by Lck-mediated phosphorylation, fully activated Zap70 phosphorylates the adaptors LAT and SLP-76 on multiple tyrosines to form an effective signaling complex, which recruits a number of signaling proteins, including GADS, Grb2, SOS, Vav1, ITK, and PLC-γ1. The assembly of LAT-nucleated signalosomes subsequently leads to activation of multiple downstream events, which include intracellular calcium increases, mitogen-activated protein kinase activation, and cytoskeletal reorganization (4).
Actin cytoskeletal dynamics play an important role during T cell activation. In particular, actin remodeling is required to provide a scaffold for signaling proteins and for maintaining a stable immunological synapse between T cells and antigen-presenting cells (5). Our previous study using a chemical–genetic approach to inhibit Csk also revealed a requirement of actin remodeling in the initiation of full T cell activation (6). Perturbing the actin assembly via cytochalasin D (an inhibitor of actin polymerization) treatment has also been reported to enhance anti-CD3–induced Ca2+ flux (7). However, the details of exactly how actin dynamics influence and are controlled by TCR signaling are elusive.
Recent progress in T-cell based immunotherapies highlights the importance of further understanding the molecular mechanisms underlying T cell activation for developing effective cancer treatments. While many TCR signaling molecules have been identified and characterized by classic genetic and biochemical approaches in the past 20 y, identification of novel signaling regulators, as well as studying their contribution to T cell activation, remains a challenge. To address this, we conducted an unbiased genome-wide CRISPR-based screen in human Jurkat T cells to identify novel regulators of T cell activation.
Results
A Pooled Genome-Wide CRISPR Screen to Dissect Proximal TCR Signaling.
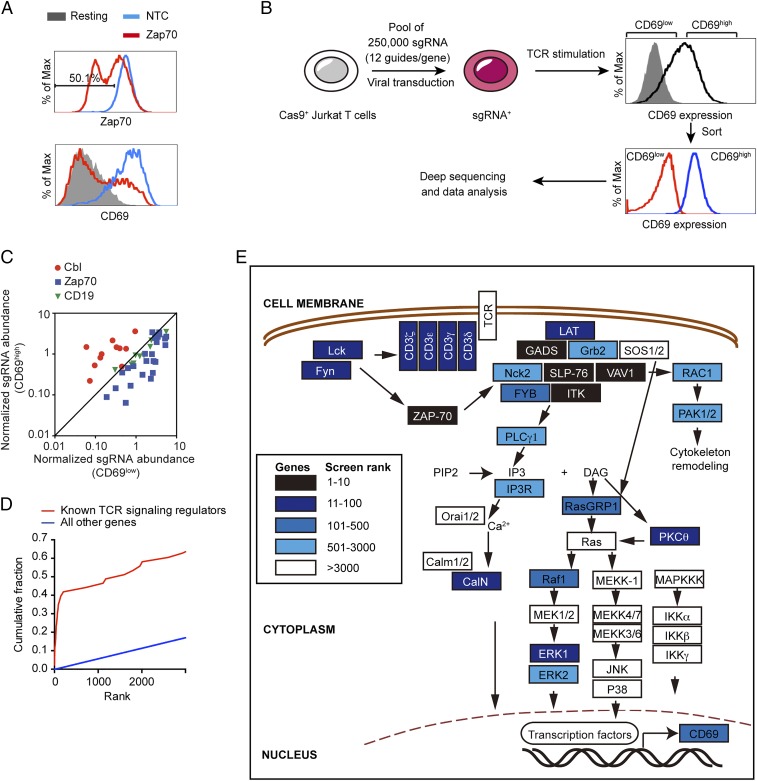
To perform a pooled genetic screen, we first developed a single-cell–based readout of T cell activation by measuring the up-regulation of CD69. We chose to assess CD69 up-regulation as a readout in our screen for the following two reasons: (i) surface up-regulation of CD69 is a well-defined early marker of T cell activation, which faithfully reflects TCR signaling strength; (ii) a genome-wide screen often requires handling of over hundreds of millions of cells, and it could be technically challenging to measure some intracellular signaling events, such as Ca2+ influx, in such a large number of cells. Our loss-of-function genetic screen was performed in our Cas9-expressing Jurkat T cells (8). We showed that transducing this cell line with a single-guide RNA (sgRNA) targeting Zap70, a critical kinase controlling T cell activation, markedly reduced Zap70 expression, and inhibited induction of CD69 (Fig. 1A), which validated our experimental system.
Fig. 1.
Genome-wide CRISPR screen correctly identifies known regulators of proximal TCR signaling. (A) CRISPR targeting Zap70 impaired T cell activation. After lentiviral transduction of sgRNA in Cas9-expressing Jurkat cells, intracellular Zap70 expression (Upper) and surface CD69 expression after TCR stimulation were assessed using flow cytometry (Lower). (B) Workflow of the CRISPR-based screen. (C) The abundance of sgRNAs against Cbl, Zap70, and NTC in both CD69high and CD69low samples. (D) Cumulative density functions of gene rank for the 43 known positive regulators of the TCR pathway (red) and all other genes (blue). (E) Components of the TCR signaling pathway derived from ref. 10 and their ranks in our screen (blue scale).
Next, we performed a pooled genome-wide screen using a lentiviral sgRNA library consisting of over 250,000 total sgRNAs targeting every unique Refseq annotated (hg19) protein-coding isoform with up to 12 sgRNAs per gene, as well as 7,700 nontarget control sequences (NTC). In the library, the earliest possible exon of each transcript variant was targeted. We infected Cas9-expressing Jurkat T cells with the library at a multiplicity of infection (MOI) of 0.3 and selected the transduced Jurkat cells using puromycin treatment. Four-hundred million transduced Jurkat T cells were stimulated with anti-TCR Vβ8 antibody (C305) for 13 h. Based on surface CD69 expression levels, the activated T cells were sorted into two cell populations, CD69low cells and CD69high cells (Fig. 1B). After genomic DNA extraction, we PCR-amplified and sequenced sgRNAs from the two cell populations. We expected that sgRNAs targeting positive regulators of TCR signaling would be enriched in the CD69low sample, whereas sgRNAs against negative regulators would be depleted in the CD69low sample. A comparison of sgRNAs’ abundance between the two samples focusing on a known positive regulator (Zap70) and a known negative regulator (Cbl, also known as c-Cbl) was consistent with our prediction (Fig. 1C). We observed that most sgRNAs targeting Zap70 were enriched in the CD69low sample compared with CD69high. In contrast, sgRNAs against Cbl, a known negative regulator of TCR signaling, were enriched in the CD69high sample. As expected, CD19 is a B cell marker that is not expressed on Jurkat T cells and its sgRNAs were evenly distributed between two samples. We further analyzed the data using the MAGeCK program to identify both positive regulators and negative regulators of TCR signaling (9).
In the list of positive regulators (Dataset S1), the top-ranked genes were highly enriched for those annotated in an immune response-regulating cell surface receptor signaling pathway (the highest scoring categories; GOrilla, false-discovery rate q-value = 10−10). We next compared these top-ranked genes with the known TCR signaling proteins that we summarized previously (10). Many genes among the top 100-ranked positive regulators were assigned to the TCR signaling pathway (Fig. 1D). Most of the proximal TCR signaling molecules were identified by our genome-wide screen (Fig. 1E). Key positive regulators of the TCR signaling with high ranks in our screen included: the CD3 complex, comprised of CD3δ (rank 29), CD3ε (rank 26), CD3γ (rank 35), and ζ chain (rank 40); tyrosine kinases, such as Src kinases: Lck (rank 61) and Fyn (rank 55), Zap70 (rank 3), and ITK (rank 6); and the LAT signaling complex: LAT (rank 20), SLP-76 (LCP2, rank 2), GADS (rank 8), and VAV1 (rank 4). TCR stimulation triggers multiple common downstream signaling pathways, including the Ca2+, Ras/MAPK, P38, JNK, and NF-κB signaling pathways. Among these signal pathways, our results also showed that CD69 up-regulation was mainly controlled by Ca2+ signaling (Calcineurin, rank 67) and ERK (ERK1, rank 88) signaling. Overall, the top 100-ranked genes were highly enriched for central TCR signaling molecules summarized in our previous review (10) (15 of 43 previously known TCR+ regulators are in the top 100; P = 10−24, hypergeometric test).
FAM49B Is an Inhibitor of TCR Signaling.
In the list of negative regulators (Dataset S2), we identified several known regulators of TCR signaling (Table 1). For example, SLAP (rank 1) is an adaptor protein that links the E3 ligase Cbl (rank 5) to the TCR:CD3 complex and negatively regulates its expression (11, 12). The transmembrane adaptor PAG (rank 29) is able to recruit Csk (rank 268), a cytoplasmic protein tyrosine kinase that inactivates Src kinases, and plays an inhibitory role during T cell activation (13). DUSP6 (rank 76) is an ERK-specific phosphatase that dampens the ERK signal after T cell activation (14). Among the novel putative negative regulators, we focused further effort on FAM49B (family with sequence similarity 49 member B) for three reasons. First, RNA-seq analysis from the Human Protein Atlas (www.proteinatlas.org) showed that, among the 37 different human tissues and organs, FAM49B was the most highly expressed in the lymphoid organs, including the lymph node, appendix, bone marrow, spleen, and tonsil (Fig. S1A) (15). Second, the dysregulated expression of FAM49B has been previously reported to be associated with multiple sclerosis (16, 17). A genome-wide gene-expression profile of peripheral blood mononuclear cells from multiple sclerosis patients revealed that FAM49B, along with several negative regulators of inflammation, including SOCS2 and TNFAIP3, was highly expressed in multiple sclerosis patients’ peripheral blood mononuclear cells compared with healthy controls (16). Third, FAM49B is a single-domain protein containing the domain of unknown function 1394 (DUF1394), and its role in T cells has not been characterized.
Table 1.
Known negative regulators of TCR signaling
| Name | Description | Rank | Function |
| SLAP | Src like adaptor | 1 | SLAP regulates expression and trafficking of TCR:CD3 complex. |
| Cbl | Cbl proto-oncogene | 5 | E3 ligase negatively regulates TCR expression. |
| FAM49B | Family with sequence similarity 49 member B | 16 | Unknown function |
| PAG1 | Phosphoprotein membrane anchor with glycosphingolipid microdomains 1 | 29 | PAG1 binds to Csk that negatively regulates Lck activity |
| DUSP6 | Dual specificity phosphatase 6 | 76 | DUSP6 specifically dephosphorylates ERK |
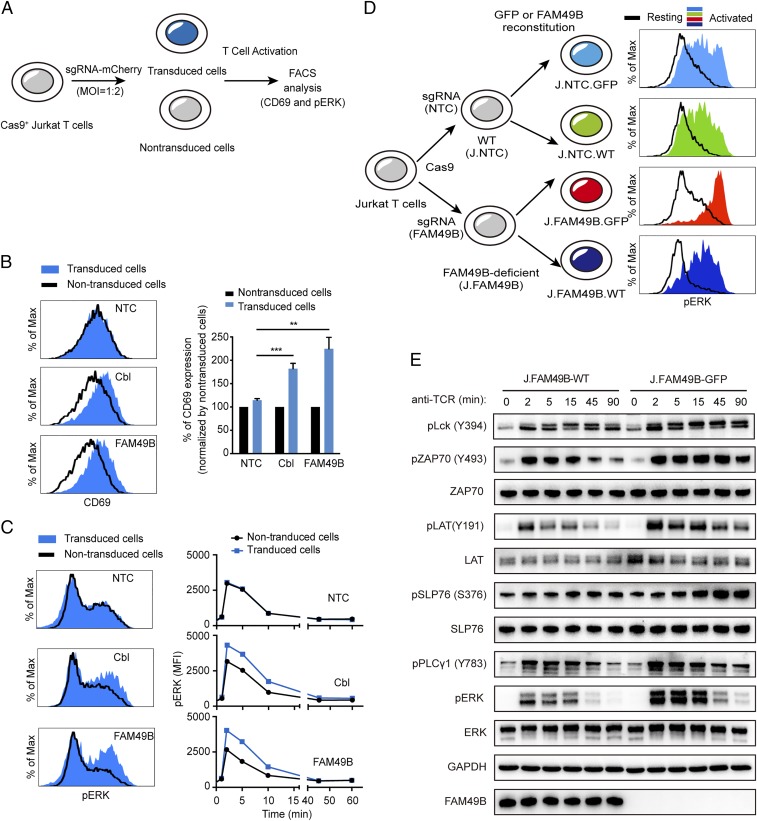
To validate the role of FAM49B during T cell activation, we chose three sgRNAs targeting FAM49B and individually introduced them into Jurkat cells using both lentivirus (Fig. 2A) and electroporation (Fig. S1B); three NTC sgRNAs and three sgRNAs against c-Cbl were used as negative and positive controls, respectively. Loss of FAM49B function enhanced CD69 induction following T cell stimulation to a similar extent as inactivation of c-Cbl (Fig. 2B and Fig. S1C). Consistent with this increased TCR induction of CD69, a larger and more prolonged increase in Erk phosphorylation appeared 2 min after T cell stimulation, peaked at 2–5 min, and lasted for at least 45 min (Fig. 2C). Taken together, these results suggested that FAM49B inhibits TCR signal transduction and negatively regulates T cell activation.
Fig. 2.
Hyperactivation of TCR signaling in FAM49B-deficient Jurkat T cells. (A) Schematic of validating screen candidate genes by transducing Cas9+ Jurkat cells with lentivirus-expressing sgRNA at an MOI of 0.5. (B) FACS analysis of CD69 levels in Jurkat cells expressing sgRNAs targeting NTC, c-Cbl, or FAM49B 13 h after anti-TCR C305 stimulation. Data are representative of three independent experiments. ***P < 0.0001, **P < 0.001 (Student’s t test). (C) FACS analysis of pERK phosphorylation in the sample described in B. Histogram shows pERK MFI from 0 to 60 min upon anti-TCR stimulation. (D) Schematic of reconstitution J.FAM49B cells and J.NTC cells with empty vector (GFP) or FAM49B vector, and FACS analysis of pERK level upon anti-TCR stimulation. (E) The FAM49B-suffcient and -deficient cells were rested and stimulated with anti-TCR antibody for times indicated. Phosphorylation of Lck, Zap70, LAT, SLP-76, PLCγ, and ERK were assessed by immunoblotting.
To exclude the off-target effect of the sgRNAs, we generated two different FAM49B-deficient Jurkat cell lines (J.FAM49B) and asked whether reexpression of FAM49B could rescue the FAM49B-deficient phenotype (Fig. 2D and Fig. S2). Upon TCR stimulation, both FAM49B-deficient clones exhibited a higher ERK phosphorylation than a control line (Fig. 2D and Fig. S2). Importantly, reconstitution with WT FAM49B largely suppressed this hyperactivation of ERK (Fig. 2D and Fig. S2). These data suggested that the T cell hyperresponsiveness resulting from sgRNA targeting FAM49B was indeed caused by the deficiency of FAM49B, rather than an off-target effect of sgRNA expression.
To explore how FAM49B deficiency increases TCR signaling, we examined other TCR signaling events. FAM49B deficiency did not affect surface CD3 or CD28 expression (Fig. S2C), but led to prolonged increases in all of the downstream phosphorylation events investigated, including pTyr394 in Lck, pTyr493 of Zap70, pTyr191 of LAT, pS376 of SLP76, pTyr783 of PLCγ1, and pERK (Fig. 2E). Taken together, these data suggested that FAM49B suppressed early T cell signal transduction.
FAM49B Directly Interacts with Active Rac1.
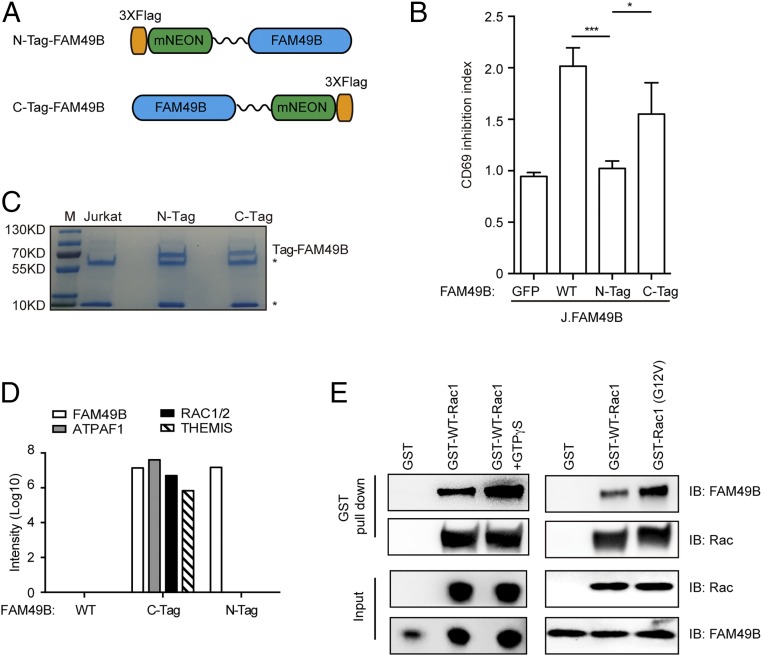
We next sought to identify the binding partners of FAM49B using immunoprecipitation in combination with mass spectrometry (IP-MS). FAM49B was tagged at either terminus with FLAG-mNeon (Fig. 3A). Interestingly, the N-terminal tag completely abolished FAM49B function, whereas the C-terminal tagged FAM49B largely maintained the suppressive function of FAM49B (Fig. 3B). Therefore, we used the N-terminal tagged FAM49B as a negative control for FAM49B-associated proteins (Fig. 3C). We reasoned that a FAM49B-associated protein mediating its suppressive function would likely interact with C-terminal tagged FAM49B, but not N-terminal tagged FAM49B. Our IP-MS result revealed that there were three proteins selectively immunoprecipitated with C-tagged FAM49B (but not the N-tagged or native FAM49B): (i) ATP synthase mitochondrial F1 complex assembly factor 1 (ATPAF1); (ii) the small GTPase Rac; and (iii) thymocyte selection associated (THEMIS) (Fig. 3D). Because ATPAF1 is a mitochondria-localized protein (18) and it is unlikely to be involved in the TCR signaling transduction, we focused on the other two candidates: Rac and THEMIS. Our biochemical experiment showed that C-terminal tagged FAM49B associated with endogenous Rac, but not THEMIS (Fig. S3). To further test whether FAM49B directly interacts with Rac, we purified the recombinant FAM49B as well as recombinant GST-WT Rac1 and active GST-Rac1 (G12V). By using a GST pull-down assay, we found that FAM49B directly interacted with Rac1 in vitro and selectively bound to activated Rac1 (GTPγS loaded WT Rac1 and the activated G12V mutant) with a higher affinity compared with WT Rac1 (Fig. 3E). Thus, our results indicated that FAM49B directly and preferentially binds to active Rac1.
Fig. 3.
IP-MS identifies FAM49B interacting protein Rac. (A) Construction FAM49B with N-terminal triple FLAG mNeonGreen or C-terminal mNeonGreen triple Flag. (B) J.FAM49B cells were reconstituted with N-tagged or C-tagged FAM49B at an MOI of 0.5. The function of N-tagged or C-tagged FAM49B function were assessed by using CD69 inhibition index, which was calculated by MFI of CD69 in the GFP− population (nontransduced J.FAM49B cells) divided by that of GFP+ population (J.FAM49B cells reconstituted with FAM49B variants). ***P < 0.0001, *P < 0.05 (Student’s t test). (C) Coomassie blue staining of immunoprecipitated samples. An asterisk (*) indicates heavy chain and light chain of IgG antibody. (D) MS data showed ATPAF1, Rac1/2, and THEMIS peptides were enriched in the C-tag, but not N-tag or nontag FAM49B sample. (E) GST pull-down assay confirming the direction of FAM49B and Rac. Recombinant purified FAM49B protein was incubated with GST-Rac1 (in the presence or absence of GTPγS) or GST-Rac (G12V) protein for 1 h and IP with GST beads. The elutates from the GST beads and input samples were analyzed by Western blot. Data are representative of three independent experiments. IB, immunoblot.
The α-Helix (Residues 150–166) of FAM49B Is Obligatory for Its Interaction with Rac1.
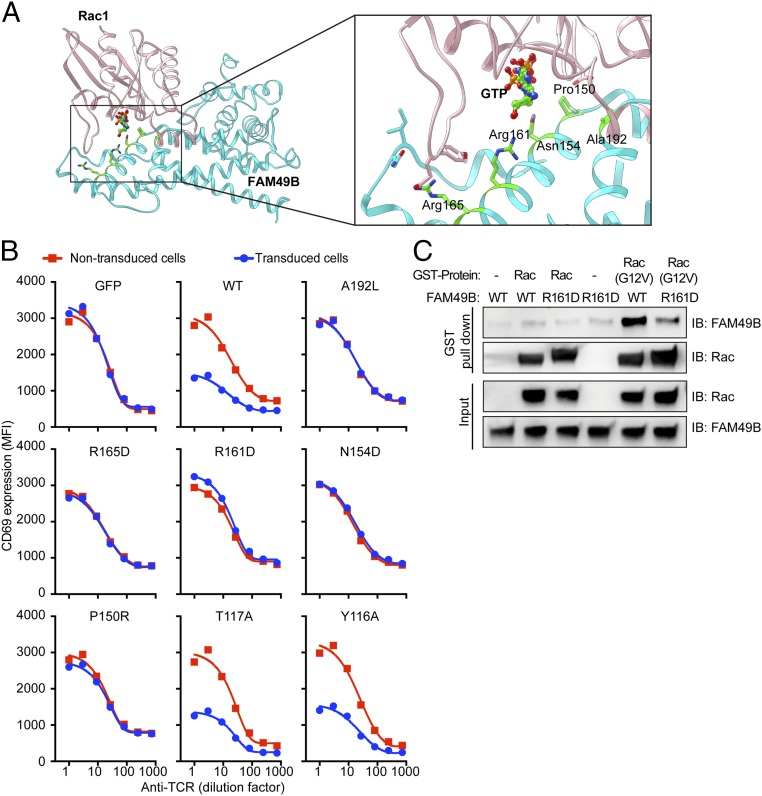
To explore how FAM49B binds Rac1, we searched for the human proteins sharing sequence similarity with FAM49B using the BLAST program. Besides its family member FAM49A, the cytoplasmic FMR1-interacting protein 1 (CYFIP1), a component of the WAVE regulatory complex that regulates actin polymerization (19), is the top hit. CYFIP1 and FAM49B share DUF1394 domains, which are 20.6% identical between the two proteins (Fig. S4A). CYFIP1 is known to interact with Rac1 through its DUF1394 domain (19). Because the structure of CYFIP1 has been solved and the key residues mediating its interaction with Rac1 have been identified (19), we used the HADDOCK server (20) to dock Rac1 with FAM49B based on three criteria: (i) the correct binding interface should cover Pro150 and Arg161 of FAM49B according to the conserved residues Cys179, Arg190 of its homolog CYFIP1 (19); (ii) if CFYIP1 is superimposed on FAM49B within the Rac1–FAM49 docking complex, Rac1 should interact with Glu434, Phe626, and Met632 of CYFIP1 (19); and (iii) the Rac1 binding interface should include the loops around GTP binding region (21–25). This analysis suggests that the α-helix at residues 150–166 of FAM49B plays a critical role in the Rac1–FAM49B complex formation (Fig. 4A). In particular, Arg161 is close to the Rac1 GTP binding region, and its positive charge forms an ionic interaction with the electronegative region of GTP. Furthermore, Pro150 forms hydrophobic interactions with Tyr64 of Rac1. Another key residue is Arg165, which forms salt bridge and cation–π interaction with Glu31 and Tyr32 of Rac1, respectively. Other residues at the interaction interface include Met147, Asn154, Glu182, Ala192, and Glu193, which may contribute to binding. To directly test the predictions from this docking model, we mutated multiple potential Rac binding sites at FAM49B—namely Ala192, Arg165, Arg161, Asn154, and Pro150—and expressed these mutants in J.FAM49B cells. Each of the five mutants completely abolished FAM49B function (Fig. 4B), which was not due to a decrease in protein stability (Fig. S4B). As a control, we showed mutations at sites outside the predicted interaction interface (T117 and Y116) had no effect on FAM49B function. Finally, among these key interaction sites, we chose and purified FAM49B (R161D) to test whether it is required for Rac1 binding. The GST pull-down assay showed that FAM49B (R161D) significantly attenuated its interaction with active Rac (G12V) (Fig. 4C). These results suggest the α-helix (150–166) of FAM49B contacts Rac1 and this interaction is important for the FAM49-mediated regulation of T cell activation.
Fig. 4.
The interaction of FAM49B and Rac. (A) Docking of FAM49B with Rac1 was performed by the HADDOCK web server, key residues in FAM49B contributing in the interaction were highlighted in green. (B) FACS analysis of CD69 level upon anti-TCR stimulation in J.FAM49B expressing indicated FAM49 variants. (C) To perform the GST pull-down assay, WT FAM49B or FAM49B (R161D) protein was incubated with GST-Rac or GST-Rac (G12V) protein in vitro in the presence of GST beads. The elutates from the GST beads and input samples were then analyzed by Western blot.
FAM49B Regulates Cytoskeletal Remodeling via Rac-PAK Axis.
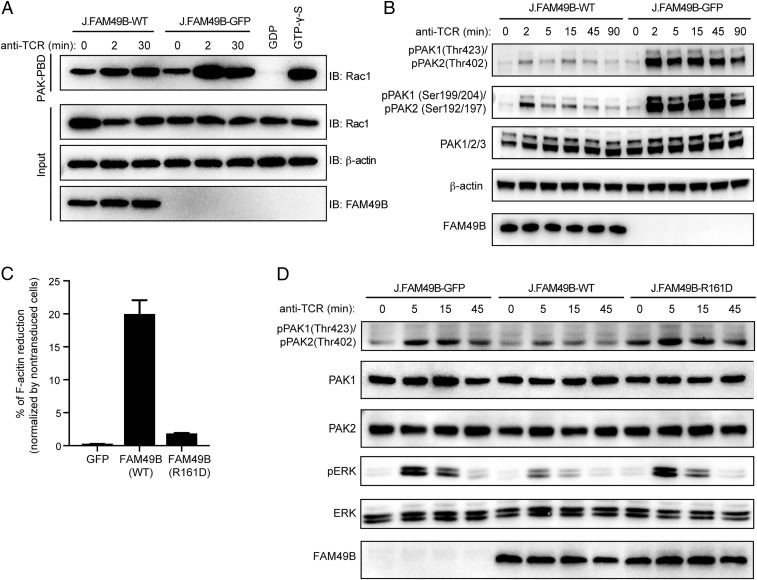
Rac exists in the active GTP-bound and inactive GDP-bound forms, and plays crucial roles in T cell signaling (26). The physical interaction of FAM49B with Rac prompted us to determine if FAM49B controls Rac function. Indeed, FAM49B deficiency enhanced the amount of Rac-GTP, especially 2 min after TCR stimulation (Fig. 5A), indicating that FAM49B inhibited Rac activation. Rac is known to act mainly via the serine/threonine p21-activated kinase (PAK) family to regulate actin dynamics (27). Therefore, we then assessed the activation of PAK and actin assembly in FAM49B KO cells. PAK phosphorylation was dramatically elevated in FAM49B-deficient cells (Fig. 5B and Fig. S5A). We found that PAK phosphorylation in J.FAM49B cells was rescued with WT-FAM49B and was comparable to WT Jurkat cells (Fig. S5A). We also measured intracellular F-actin levels using Phalloidin staining. We found a more than 20% reduction in F-actin level in FAM49B-WT reconstituted cells compared with FAM49B-deficient cells (Fig. 5C). Notably, the FAM49B mutant (R161D), which exhibited markedly impaired binding to Rac1, failed to reduce F-actin levels (Fig. 5C) or to suppress PAK activity (Fig. 5D). Taken together, these results demonstrated that formation of a FAM49B–Rac1 complex inhibits Rac1 activity and PAK phosphorylation, thereby influencing polymerization of actin.
Fig. 5.
FAM49B controls T cell activation by regulating cytoskeleton remodeling. (A) J.FAM49B cells stably expressing FAM49B or GFP were stimulated with anti-TCR. Cells were lysed at indicated times, Rac1–GTP pull-down assays were carried out using Pak-PBD beads. GTPγS and GDP-loaded lysates were used as controls. Total Rac1, β-actin, FAM49B and active Rac1 were detected by Western blot. (B) The samples in A were stimulated with C305 for times indicated. Phosphorylation of PAK was assessed by immunoblotting. (C) J.FAM49B cells were transduced with virus expressing empty vector (GFP), WT-FAM49B, or FAM49B (R161D) at an MOI of 0.5. Quantification of the F-actin level was assessed by flow cytometry. (D) J.FAM49B cells stably expressing WT-FAM49B, GFP, or FAM49B (R161D) were stimulated with anti-TCR for indicated time. Phosphorylation of PAK and ERK was assessed by immunoblotting.
FAM49B Regulated T Cell Activation by Modulating Cytoskeletal Remodeling.
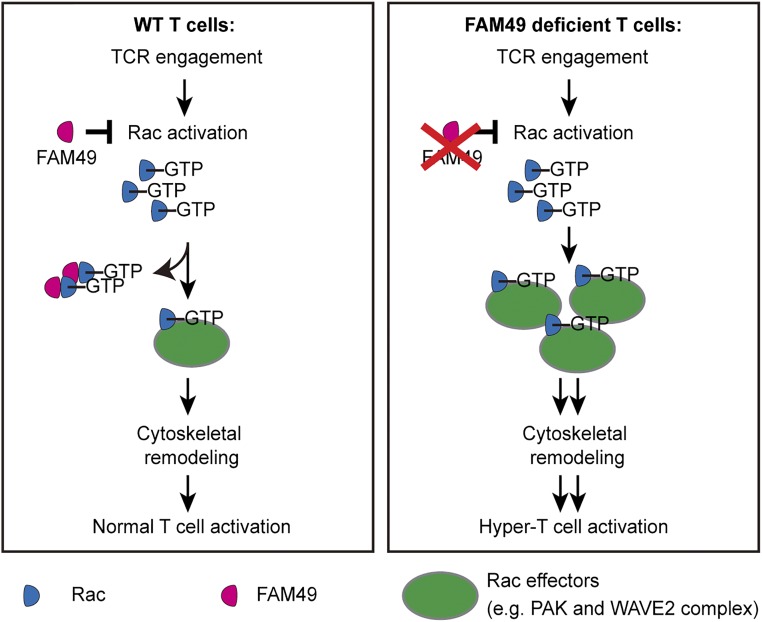
Our findings that FAM49B suppressed T cell activation and also modulated the actin cytoskeleton in T cells led us to propose a model to explain how FAM49B regulates TCR signal transduction (Fig. 6). In our model, TCR engagement activates the small GTPase Rac. FAM49B directly binds to active Rac, sequestering some active Rac-GTP, resulting in reduction of the pool of Rac-GTP available to interact with its downstream effector proteins, such as PAK, with resultant reduction Rac effectors’ activities in T cells. Consequently, these events modulate actin cytoskeletal remodeling, ensuring proper T cell activation. In the absence of FAM49B, both the available pool of Rac-GTP and activation of Rac downstream effectors are elevated, which increases actin polymerization and leads to a hyperactivation of TCR signaling. According to this model, we predicted that hyper-T cell activation in FAM49B-deficient cells would be repressed when PAK activation, a downstream event of FAM49B, is inhibited. To test this hypothesis, we utilized a small-molecule inhibitor to modulate PAK activity during T cell activation. Our results showed that treatment of PAK inhibitor markedly repressed CD69 up-regulation (Fig. S5B), which is consistent with impaired TCR signaling in PAK2-deficient T cells, as previously described (27). More importantly, this treatment significantly blocked the influence of FAM49B on T cell activation (Fig. S5C). These results support our model that PAK contributes to FAM49B-mediated regulation of the T cell activation.
Fig. 6.
Proposed model for FAM49B mediated regulation of T cell activation. Proposed model for how FAM49B regulates cytoskeleton and T cell activation.
Discussion
To our knowledge, our study is unique in presenting a genome-wide CRISPR screen focusing on the TCR signal transduction pathway in the Jurkat cell line, a frequently used model for TCR signaling function. Our screen identified most of the known proximal positive and negative T cell signaling proteins, as well as diverse novel regulators. Our strategy provides a paradigm for dissecting the TCR signaling pathway, and is obviously applicable to other immunological processes, such as CD28-mediated costimulatory signaling and PD-1–mediated coinhibitory signaling, thus facilitating the discoveries of new therapeutic targets for cancer immune therapy and autoimmune disease treatment.
Despite of the power of the CRISPR-based screening system (28), our screen missed some important regulators. We believe that the primary reason is due to gene redundancy, which obscures the effect of gene deletion. For example, Ras activation is critical for ERK activation in T cells (29). However, there are three different functional Ras genes in Jurkat cells—H-ras (rank 5498), K-ras (rank 11796), and N-ras (rank 7096)—and thus deletion of any individual Ras gene might be inconsequential. Similar examples include SOS1 (rank 15675) and SOS2 (rank 8630), Orai1 (rank 7557) and Orai2 (rank 8203), and Calmodulin1 (rank 7548) and Calmodulin2 (rank 6598). Second, some missing genes are essential for T cell survival. Inactivating these essential genes would result lack of representation from our T cell pool gradually, causing the error of omission. Third, the TCR receptor is missing in our list, because the TCR-α locus and -β locus are not included in our sgRNA library.
Our screen identified FAM49B, a previously unrecognized regulator of the actin cytoskeleton, as a negative regulator of TCR signaling. FAM49B directly interacted with active Rac via its α-helix (residues 150–166). This interaction inhibited the Rac-PAK signaling axis and reduced actin assembly in T cells. Consistent with the importance of actin dynamics in TCR signaling, our genetic screen also identifies several well-defined actin regulators in our top-ranked positive regulator list, including Vav1 (rank 4), DEF6 (also known as SLAT, rank 10), Dock2 (rank 11), and RhoH (rank 24). Vav1 is a GEF that activates Rho family GTPases Rac, RhoA and Cdc42, and plays a critical role in T cell activation (30). DEF6, another GEF for Rac and Cdc42, is required for Ca2+ signaling and NFAT activation in T cells (31, 32). Dock2 regulates the actin cytoskeleton and T cell responsiveness through controlling Rac activation (33). RhoH is a GTPase predominantly expressed in the hematopoietic system, and both RhoH-null mice and RhoH-deficient patients show defective T cell signaling (34, 35). It was recently reported that, in pancreatic ductal adenocarcinoma cells, FAM49B regulated mitochondrial fission and acted as a tumor suppressor (36). This effect is likely tissue-specific, because in Jurkat T cells, FAM49B did not affect the phosphorylation of Dynamin-related protein1 (Drp1), a hall marker of mitochondrial fission (Fig. S6A).
Several prominent questions remain about FAM49B. First, how exactly does FAM49B regulate Rac-PAK signaling? FAM49B might function as an adaptor recruiting a GAP protein or GDI protein to control Rac. Alternatively or additionally, FAM49B seems to be myristoylated at its N-terminal MG motif (37), therefore leading to membrane recruitment of FAM49B. As active Rac is also localized at the membrane, the high local concentration of membrane-associated FAM49B could sequester Rac from binding to its downstream effector kinase PAK, therefore inhibiting actin cytoskeleton reorganization. This hypothesis could also explain why adding the N-terminal tag to FAM49B completely abolished its inhibitory function in T cell activation. A second important concern about FAM49B concerns the fact that the FAM49 family has two family members, FAM49A and FAM49B, which share 79.3% sequence identity. Whereas FAM49B is selectively expressed in Jurkat T cells (Fig. S2D), the two members are coexpressed in mouse T cells (Fig. S6B). Future studies are needed to clarify why mouse T cells require two FAM49 members and how their expression is regulated during T cell development and differentiation. Third, can FAM49B regulate other Rac downstream effectors in T cells? It is known that two downstream effectors of Rac, the PAKs and the WAVE2 complex, are involved in regulating actin reorganization in T cells (38). In FAM49B-deficient T cells, Rac-GTP levels are elevated compared with WT T cells (Fig. 5A). Therefore, it is possible that these two Rac downstream pathways are affected in FAM49B-deficient T cells and they both contribute to FAM49B-mediated regulation of actin reorganization. Two PAKs, PAK1 and PAK2, are expressed in T cells (Fig. 5B) and both of them are activated after TCR engagement (27, 39, 40). Upon activation by Rac-GTP, PAK phosphorylates and activates the LIM kinase. The activated LIM kinase further phosphorylates and inactivates cofilin, a family of actin-binding proteins that disassembles actin filaments, resulting in stabilization of F-actin. In addition to cofilin, PAK1 is able to directly phosphorylate Raf-1 and Mek and activate ERK in T cells (41). The PAK inhibitor (FRAX597) we used here (Fig. S5B) inhibits both PAK1 and PAK2 kinase activity; therefore, it is not clear whether PAK1 or PAK2 (or both) mediates FAM49B’s function in these experiments. In primary mouse T cells, Pak2 seems to have the more important role (27). In addition to PAKs, it is also known that the interaction between Rac1 and CYFIP1 is able to activate the WAVE2 complex and promote actin reorganization in T cells (42). We also noticed that the inhibition of PAK kinase activity only partially blocked the effect of FAM49B on T cell activation (Fig. S5C), indicating that other Rac effector molecules, such as the WAVE2 complex, are likely to participate in the regulation of T cell activation mediated by FAM49B.
Our genome-wide CRISPR screen allowed us to identify FAM49B as a regular T cell activation through its ability to modulate cytoskeletal remodeling. We believe that validating and investigating other promising hits revealed by our screen will shed more light on the molecular mechanisms that influence TCR signaling transduction.
Materials and Methods
Reagents and Antibodies.
All Western blot antibodies [pLck(Y394)#2101, pZap70(Y493)#2704, pLAT(Y191)#3584, LAT#9166, pSLP76(S376)#92711, SLP76#4958, pPLCγ1(Y783)#14008, pERK#4377, ERK#9102, pPAK1(Thr423)/pPAK2(Thr402), pPAK1(Ser199/204)/pPAK2 (Ser192/197), and PAK1/2/3#4750] were purchased from Cell Signaling Technology except for FAM49B (sc-390478; Santa Cruz), pan-RAC1(sc-514583; Santa Cruz), GAPDH (Ab105428; Abcam). Anti-human CD69 (BioLegend 310910) FACS antibodies were purchased from BioLegend. The Rac1 activation Assay Biochem Kit (BK035) was purchased from Cytoskeleton.
T Cell Activation Assay.
Jurkat cells were cultured at a concentration of less than 1 million cells/mL 3 d after viral transduction; transduced cells were stimulated by a titrated anti-TCR antibody (C305). Thirteen hours after stimulation, the surface CD69 level was assessed using flow cytometry. To quantitatively assess the suppressive ability of FAM49B variant, we defined the CD69 inhibition index, which was calculated by dividing the mean fluorescent intensity (MFI) of surface CD69 of the nontransduced cell population by that of transduced cell population.
sgRNA Library Design and Cloning.
The genome-scale sgRNA library consists of over 250,000 total sgRNAs targeting every unique Refseq annotated (hg19) protein coding isoform with up to 12 sgRNAs, plus 7,700 NTCs. The earliest possible exon of each transcript variant was targeted. All sgRNAs were designed against target sites that are of the format (N)20NGG, and selected sgRNAs must pass the following off-targeting criteria: (i) the 11-bp seed must not have an exact match in any other promoter region, and (ii) if there is an exact off-target seed match, then the rest of the sgRNA must have at least seven mismatches with the potential off-target site. After all sgRNAs that pass off-targeting criteria were generated, up to 12 sgRNAs per transcript were selected. All sgRNA sequences are shown in Dataset S3. The 20-nt target-specific sgRNA sequences were synthesized as a pool on microarray surfaces (CustomArray), flanked by overhangs compatible with Gibson Assembly into the pSico-based barcoded sgLenti sgRNA library vector (Addgene #105996). The synthesized sgRNA template sequences were of the format: 5′-GGAGAACCACCTTGTTGG-(N)20-GTTTAAGAGCTATGCTGGAAAC-3′. Template pools were PCR-amplified using Phusion Flash High-Fidelity PCR Master Mix (ThermoFisher Scientific) according to the manufacturers protocol, with 1 ng/µL sgRNA template DNA, 1 µM forward primer (5′-GGAGAACCACCTTGTTGG-3′), 1 µM reverse primer (5′- GTTTCCAGCATAGCTCTTAAAC-3′), and the following cycle numbers: 1× (98 °C for 3 min), 15× (98 °C for 1 s, 55 °C for 15 s, 72 °C for 20 s), and 1× (72 °C for 5 min). PCR products were purified using Minelute columns (Qiagen). The library vector sgLenti was prepared by restriction digest with AarI (Thermo-Fischer) at 37 °C overnight, followed by 1% agarose gel excision of the digested band and purification via NucleoSpin columns (Macherey-Nagel). Using a Gibson Assmbly Master Mix (New England Biolabs), 1,000-ng digested sgLenti and 100-ng amplified sgRNA library insert were assembled in a total 200-µL reaction volume. The reaction was purified using P-30 buffer exchange columns (Bio-Rad) that were equilibrated 5× with H2O and the total eluted volume was transformed into three vials of Electromax DH5α (ThermoFisher). Escherichia coli were recovered, cultured overnight in 500 mL LB (100 μg/mL ampicillin), and used for Maxiprep (Qiagen). In parallel, a fraction of the transformation reaction was plated and used to determine the total number of transformed clones. The library cloning coverage (number of E. coli colonies per sgRNA plasmid) was determined to be >100×.
Pooled Genome-Wide CRISPR Screens.
Cas9-expressing Jurkat T cells, described previously (8), were cultured in RPMI with 10% FBS and PSG. Cells were infected with the pooled lentiviral library at an MOI of 0.3 to ensure that only one gene was targeted for Cas9-mediated editing in each cell. To ensure full representation of the library sgRNA sequences in the transduced cell population, the library coverage at transduction was determined to be ∼100 transduced cells for each sgRNA. The transduced cells were selected for 5 d with puromycin (2 μg/mL). Following puromycin selection, the cell populations were maintained at a low cell concentration (less than 1 million cell/mL) and a library coverage of >1,000× was maintained through the screen. For the genome-wide screen, 400 million transduced Jurkat cells were stimulated with anti-TCR antibody (C305, 6.8 ng/mL). After 13 h of C305 stimulation, cells were harvested. CD69high and CD69low cell population were sorted using MACS according to the manufacturer’s instruction.
Genomic DNA Extraction from Cells and Deep Sequencing.
Cells were resuspended in 20 mL P1 buffer (Qiagen) with 100 μg/mL RNaseA and 0.5% SDS. After incubating at 37 °C for 30 min, the lysate was heated at 55 °C for 30 min in the presence of Proteinase K (100 μg/mL). After digestion, samples were passed through a needle for multiple times. Next, 20 mL Phenol:Chloroform:Isoamyl Alcohol (Invitrogen #15593–031) was added into homogenized samples. After mixing, the samples were transferred into 50 mL MaXtract tubes (Qiagen) and then centrifuged at 1,500 × g for 5 min at room temperature. The aqueous phase was transferred into ultracentrifuge tubes and thoroughly mixed with 2 mL 3 M sodium acetate plus 16 mL isopropanol at room temperature before centrifugation at 15,000 × g for 15 min. The gDNA pellets were carefully washed with 10 mL 70% ethanol and dried at 37 °C. Dry pellets were resuspended in water and gDNA concentration was adjusted to 1 μg/uL.
PCR Recovery of sgRNA Sequences from gDNA and Analysis of the CRISPR Screen.
Multiple PCR reactions were prepared to allow amplification of the total harvested gDNA from a 1,000× cell coverage for each sample. For the first round of two nested PCRs, the total volume was 100 μL containing 50 μg sheared gDNA, 0.3 μM forward (5′-ggcttggatttctataacttcgtatagca-3′) and reverse (5′-cggggactgtgggcgatgtg-3′) primer, 200 μM each dNTP, 1× Titanium Taq buffer and 1 μL Titanium Taq (Clontech). PCR cycles were: 1× (94 °C 3 min), 16× (94 °C 30 s, 65 °C 10 s, 72 °C 20 s), 1× (68 °C 2 min). All first round PCRs were pooled and a fraction was used as template for the second round PCR. The total volume of the second round PCR was 100 μL containing 2-μL pooled first round PCR, 0.5 μM forward (5′-AATGATACGGCGACCACCGAGATCCACAAAAGGAAACTCACCCTAAC-3′) and reverse (5′-CAAGCAGAAGACGGCATACGAGAT-(N)6-GTGACTGGAGTTCAGACGTG- 3′) primer, where (N)6 is a 6-nt index for sequencing on the Illumina HiSeq platform, 200 μM each dNTP, 1× Titanium Taq buffer, and 1 μL Titanium Taq (Clontech). PCR cycles were: 1× (94 °C 3 min), 16× (94 °C 30 s, 55 °C 10 s, 72 °C 20 s), 1× (68 °C 2 min). The resulting PCR product (344 bp) was extracted from a 1% agarose gel. Gel extracted bands were submitted for sequencing on an Illumina HiSEq. 2500 platform using paired-end 50 kits with the custom sequencing primer 5′- GAGACTATAAGTATCCCTTGGAGAACCACCTTGTTGG-3′ and the standard Truseq Illumina reverse primer. The sequencing results were analyzed using MAGeCK program, with default parameters to detect sgRNAs that were positively enriched in the CD69low sample (positive regulators) and CD69high sample (negative sample).
Generation of FAM49B-Deficient Jurkat Cell Lines (J.FAM49B).
FAM49B-deficient cell lines were generated by transiently expressing both sgRNA against FAM49B and Cas9 into Jurkat cells. sgRNA oligos targeting FAM49B were cloned into the pX330 vector (Addgene). Jurkat cells were electroporated with the sgRNA-pX330 plasmid as well as a GFP reporter plasmid according to the protocol in Genepulser (Bio-Rad). To obtain single clones, GFP+ cells were single-cell sorted using a BD FACSAria III into a 96-well plate. After several weeks expansion, the expression of FAM49B of each clone was screened by Western blot. Reconstituted FAM49B-deficient (J.FAM49B) lines were generated by lentiviral transduction of J.FAM49B cells with WT FAM49B or FAM49B mutant. Expanded clones were assessed of surface expression of CD3 and CD28 via FACS.
Immunoprecipitation and Mass Spectrometry Sample Preparation.
Jurkat cells stably expressing triple FLAG-mNeonGreen-FAM49B or FAM49B-mNeonGreen-triple FLAG were rested in RPMI medium for 1 h followed by multiple washings with PBS. The cells were lysed in Nonidet P-40 lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Nonidet P-40, phosphatase and protease inhibitor) (Invitrogen). The soluble supernatant was incubated with prewashed anti-FLAG(R) M2 beads (A2220; Sigma) at 4 °C overnight. After incubation, the beads were washed with lysis buffer four times, boiled, and the supernatant was loaded to a SDS/PAGE gel. The gel was fixed with buffer containing 50% methanol and 7% acetic acid solution and then washed with deionized water three times. The gel was then stained with gelcode reagent solution (Invitrogen) according to the manufacturer’s instruction. The gels were cut and digested as previously described (43).
Protein Expression and Purification.
WT Human Rac and human Rac-G12V were cloned into pGEX vector with a N-terminal GST tag. WT Human FAM49b and FAM49b (R161D) mutant were cloned into pET23b vector with a C-terminal hexahistidine tag. All vectors were transformed into Transetta (DE3) E. coli cells (CD801; TransGen). Followed by induction with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when OD600 reached 0.6, proteins were expressed for 16 h at 18 °C. The cells were harvested by centrifugation and resuspended in cold His-tag lysis buffer (50 mM Tris pH 8.0, 200 mM NaCl, 10 mM imidazole, 2.5 mM PMSF) or GST-tag lysis buffer (50 mM Tris pH 8.0, 200 mM NaCl, 2.5 mM PMSF, 5 mM DTT). Then cells were lysed with cell disruptor (Union) at 800 MPa for 2–4 min and spun at 13,000 rpm for 30 min at 4 °C. The supernatant was collected and mixed with 1mL Ni-NTA beads (his-tag protein) or 1 mL Glutathione-Sepharose beads (GST-tag protein). The mixture was rotated at 4 °C for 1 h and then loaded into a column. His-tag proteins were washed by ice cold washing buffer (50 mM Tris pH 8.0, 200 mM NaCl, 20 mM imidazole) and GST-tag proteins were washed by GST-tag lysis buffer. His-tagged WT FAM49b and FAM49b mutant proteins were eluted with 5 mL His-tag elution buffer containing 50 mM Tris, 200 mM NaCl, and 250 mM imidazole. GST-tagged Rac protein and its active-form mutant protein were eluted with buffer containing 10 mM reduced Glutathione, 50 mM Tris pH 8.0 and 200 mM NaCl. Proteins were concentrated to 1 mL using Amicon (R4BA26623; Millipore) and centrifuged at maximal speed for 10 min at 4 °C. This fraction was loaded to size-exclusion chromatography with a Superdex 200 column (17517501; GE Healthcare) with a sample pump. Fractions were collected and analyzed by SDS/PAGE. Fractions with the right-size protein were combined and concentrated. Purified proteins were stored in the storage buffer (20 mM Hepes, 150 mM NaCl, 1 mM DTT) at −80 °C until use.
Intracellular pERK Staining.
Cells were rinsed with RPMI twice, resuspended at the cell concentration of 5 million cells/mL and rested for 1 h at 37 °C. Cells were activated by anti-TCR antibody (C305) at desired time and fixed with 4% PFA in PBS. Next, cells were permeabilized in 90% MeOH and incubated on ice for 40 min. Samples were barcoded as previously described (44). Then cells were pooled and stained with pERK antibody (Cell signaling) for 1 h at room temperature followed by staining with allophycocyanin-conjugated anti-Rabbit IgG for 40 min. Samples were analyzed on Flow cytometer (BD LSRFortessa).
Molecular Docking.
The homology model of FAM49B was constructed by MODELER (45) from the HHpred server of MPI bioinformatics Toolkit (46) with template structure CYFIP1 from 3P8C (19). Rac1 structure was obtained from PDB structure 2NZ8 (24). Docking of FAM49B with Rac1 was performed by the HADDOCK web server (20). Residues Pro150, Arg161 were selected as active residues of FAM49B; passive residues are automatically defined around the active residues in HADDOCK. Residue 64 was selected as an active residue of Rac1, residues 12–14, 25–39, 57–67, and 123–132 are defined as passive residues in HADDOCK. The GTP was docked into Rac1 by superimposing a Rac1–GTP complex PDB structure 1MH1 (47) on the Rac1–FAM49B docking complex.
In Vitro Protein Interaction Assay.
GST-Rac, GST-Rac-G12V, and GST-tag were incubated with his-FAM49B and his-FAM49B (R161D) protein, and immobilized at 4 °C for 1 h with prewashed Glutathione-Sepharose beads in GST binding buffer [50 mM Hepes pH 8, 100 mM (NH4)2SO4, 150 mM MgSO4, 1 mM EDTA, 0.7% Nonidet P-40). In some experiments, WT GST-Rac was loaded with GTPγS at the final concentration of 100 μM. The mixture was rinsed with GST binding buffer three times and boiled with sample buffer at 95 °C for 10 min. The samples were centrifuged at maximum speed for 3 min at 4 °C and the supernatants were transferred to clean tubes. The samples were analyzed by Western blot.
Supplementary Material
Acknowledgments
The authors thank T. Chi, G. Fan, Z. Lin (ShanghaiTech University), and J. M. Shen (The Semiconductor Manufacturing International Corporation Private School) for their critical reading of the manuscript. This work was supported by National Basic Research Program of China Grant 2015CB964601 (to L.W.). H.W. is funded by National Natural Science Foundation of China Grant 31670919 as well as the 1,000-Youth Elite Program of China. This work was supported, in part, by a grant to A.W. from the NIH (R37AI114575).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801340115/-/DCSupplemental.
References
- 1.Courtney AH, Lo WL, Weiss A. TCR signaling: Mechanisms of initiation and propagation. Trends Biochem Sci. 2017;43:108–123. doi: 10.1016/j.tibs.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15:798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, et al. ZAP-70: An essential kinase in T-cell signaling. Cold Spring Harb Perspect Biol. 2010;2:a002279. doi: 10.1101/cshperspect.a002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: A tale of cooperativity, coordination, and choreography. Cold Spring Harb Perspect Biol. 2010;2:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkhardt JK, Carrizosa E, Shaffer MH. The actin cytoskeleton in T cell activation. Annu Rev Immunol. 2008;26:233–259. doi: 10.1146/annurev.immunol.26.021607.090347. [DOI] [PubMed] [Google Scholar]
- 6.Tan YX, et al. Inhibition of the kinase Csk in thymocytes reveals a requirement for actin remodeling in the initiation of full TCR signaling. Nat Immunol. 2014;15:186–194. doi: 10.1038/ni.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valitutti S, Dessing M, Aktories K, Gallati H, Lanzavecchia A. Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. J Exp Med. 1995;181:577–584. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi S, Weiss A, Wang H. A CRISPR-based toolbox for studying T cell signal transduction. BioMed Res Int. 2016;2016:5052369. doi: 10.1155/2016/5052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Weiss A. T cell receptor signalling. J Cell Sci. 2001;114:243–244. doi: 10.1242/jcs.114.2.243. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, et al. Tonic ubiquitylation controls T-cell receptor:CD3 complex expression during T-cell development. EMBO J. 2010;29:1285–1298. doi: 10.1038/emboj.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers MD, et al. Src-like adaptor protein regulates TCR expression on thymocytes by linking the ubiquitin ligase c-Cbl to the TCR complex. Nat Immunol. 2006;7:57–66. doi: 10.1038/ni1291. [DOI] [PubMed] [Google Scholar]
- 13.Davidson D, Bakinowski M, Thomas ML, Horejsi V, Veillette A. Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol Cell Biol. 2003;23:2017–2028. doi: 10.1128/MCB.23.6.2017-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, et al. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med. 2012;18:1518–1524. doi: 10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 16.Gilli F, et al. Loss of braking signals during inflammation: A factor affecting the development and disease course of multiple sclerosis. Arch Neurol. 2011;68:879–888. doi: 10.1001/archneurol.2011.32. [DOI] [PubMed] [Google Scholar]
- 17.Gilli F, et al. Learning from nature: Pregnancy changes the expression of inflammation-related genes in patients with multiple sclerosis. PLoS One. 2010;5:e8962. doi: 10.1371/journal.pone.0008962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang ZG, White PS, Ackerman SH. Atp11p and Atp12p are assembly factors for the F(1)-ATPase in human mitochondria. J Biol Chem. 2001;276:30773–30778. doi: 10.1074/jbc.M104133200. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Zundert GCP, et al. The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Stebbins CE, Galán JE. Modulation of host signaling by a bacterial mimic: Structure of the Salmonella effector SptP bound to Rac1. Mol Cell. 2000;6:1449–1460. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- 22.Lucato CM, et al. The phosphatidylinositol (3,4,5)-trisphosphate-dependent Rac exchanger 1·Ras-related C3 botulinum toxin substrate 1 (P-Rex1·Rac1) complex reveals the basis of Rac1 activation in breast cancer cells. J Biol Chem. 2015;290:20827–20840. doi: 10.1074/jbc.M115.660456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell CH, Aricescu AR, Jones EY, Siebold C. A dual binding mode for RhoGTPases in plexin signalling. PLoS Biol. 2011;9:e1001134. doi: 10.1371/journal.pbio.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chhatriwala MK, Betts L, Worthylake DK, Sondek J. The DH and PH domains of Trio coordinately engage Rho GTPases for their efficient activation. J Mol Biol. 2007;368:1307–1320. doi: 10.1016/j.jmb.2007.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prehna G, Ivanov MI, Bliska JB, Stebbins CE. Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell. 2006;126:869–880. doi: 10.1016/j.cell.2006.06.056. [DOI] [PubMed] [Google Scholar]
- 26.Arrieumerlou C, Randriamampita C, Bismuth G, Trautmann A. Rac is involved in early TCR signaling. J Immunol. 2000;165:3182–3189. doi: 10.4049/jimmunol.165.6.3182. [DOI] [PubMed] [Google Scholar]
- 27.Phee H, et al. Pak2 is required for actin cytoskeleton remodeling, TCR signaling, and normal thymocyte development and maturation. eLife. 2014;3:e02270. doi: 10.7554/eLife.02270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang W, Wang F, Fan G, Wang H. Key elements for designing and performing a CRISPR/Cas9-based genetic screen. J Genet Genomics. 2017;44:439–449. doi: 10.1016/j.jgg.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Perez de Castro I, Bivona TG, Philips MR, Pellicer A. Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol Cell Biol. 2004;24:3485–3496. doi: 10.1128/MCB.24.8.3485-3496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tybulewicz VL. Vav-family proteins in T-cell signalling. Curr Opin Immunol. 2005;17:267–274. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Bécart S, et al. Tyrosine-phosphorylation-dependent translocation of the SLAT protein to the immunological synapse is required for NFAT transcription factor activation. Immunity. 2008;29:704–719. doi: 10.1016/j.immuni.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bécart S, Altman A. SWAP-70-like adapter of T cells: A novel Lck-regulated guanine nucleotide exchange factor coordinating actin cytoskeleton reorganization and Ca2+ signaling in T cells. Immunol Rev. 2009;232:319–333. doi: 10.1111/j.1600-065X.2009.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanui T, et al. DOCK2 is essential for antigen-induced translocation of TCR and lipid rafts, but not PKC-theta and LFA-1, in T cells. Immunity. 2003;19:119–129. doi: 10.1016/s1074-7613(03)00169-9. [DOI] [PubMed] [Google Scholar]
- 34.Crequer A, et al. Human RHOH deficiency causes T cell defects and susceptibility to EV-HPV infections. J Clin Invest. 2012;122:3239–3247. doi: 10.1172/JCI62949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu Y, et al. RhoH GTPase recruits and activates Zap70 required for T cell receptor signaling and thymocyte development. Nat Immunol. 2006;7:1182–1190. doi: 10.1038/ni1396. [DOI] [PubMed] [Google Scholar]
- 36.Chattaragada MS, et al. FAM49B, a novel regulator of mitochondrial function and integrity that suppresses tumor metastasis. Oncogene. 2017;37:697–709. doi: 10.1038/onc.2017.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thinon E, et al. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Commun. 2014;5:4919. doi: 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari S, Curado S, Mayya V, Dustin ML. T cell antigen receptor activation and actin cytoskeleton remodeling. Biochim Biophys Acta. 2014;1838:546–556. doi: 10.1016/j.bbamem.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu PC, et al. A novel role for p21-activated protein kinase 2 in T cell activation. J Immunol. 2004;172:7324–7334. doi: 10.4049/jimmunol.172.12.7324. [DOI] [PubMed] [Google Scholar]
- 40.Yablonski D, Kane LP, Qian D, Weiss A. A Nck-Pak1 signaling module is required for T-cell receptor-mediated activation of NFAT, but not of JNK. EMBO J. 1998;17:5647–5657. doi: 10.1093/emboj/17.19.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouquette-Jazdanian AK, Sommers CL, Kortum RL, Morrison DK, Samelson LE. LAT-independent Erk activation via Bam32-PLC-γ1-Pak1 complexes: GTPase-independent Pak1 activation. Mol Cell. 2012;48:298–312. doi: 10.1016/j.molcel.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nolz JC, et al. The WAVE2 complex regulates actin cytoskeletal reorganization and CRAC-mediated calcium entry during T cell activation. Curr Biol. 2006;16:24–34. doi: 10.1016/j.cub.2005.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gundry RL, et al. Preparation of proteins and peptides for mass spectrometry analysis in a bottom-up proteomics workflow. Curr Protoc Mol Biol. 2009;Chapter 10:Unit10.25. doi: 10.1002/0471142727.mb1025s88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krutzik PO, Clutter MR, Trejo A, Nolan GP. Fluorescent cell barcoding for multiplex flow cytometry. Curr Protoc Cytom. 2011;Chapter 6:Unit 6.31. doi: 10.1002/0471142956.cy0631s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 46.Alva V, Nam SZ, Söding J, Lupas AN. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016;44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirshberg M, Stockley RW, Dodson G, Webb MR. The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nat Struct Biol. 1997;4:147–152. doi: 10.1038/nsb0297-147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.