Abstract
Most cells experiencing heat stress reprogram their translational machinery to favor the synthesis of heat-stress proteins. Translation of other transcripts is almost completely repressed, but most untranslated messengers are not degraded. In contrast to yeast, Drosophila melanogaster, and HeLa cells, plant cells store repressed messengers in cytoplasmic nonpolysomal ribonucleoproteins (RNPs). To follow the fate of untranslated transcripts, we studied protein composition, mRNA content, and RNA-binding properties of nonpolysomal RNPs from heat-stressed tomato (Lycopersicon peruvianum) cells. Contrary to the selective interaction in vivo, RNPs isolated from tomato cells bound both stress-induced and repressed messengers, suggesting that the selection mechanism resides elsewhere. This binding was independent of a cap or a poly(A) tail. The possible role of proteasomes and heat-stress granules (HSGs) in mRNA storage is a topic of debate. We found in vitro messenger-RNA-binding activity in messenger RNP fractions free of C2-subunit-containing proteasomes and HSGs. In addition, mRNAs introduced into tobacco (Nicotiana plumbaginifolia) protoplasts were found in the cytoplasm but were not associated with HSGs.
Cells in stress alter their activities to minimize damage and facilitate recovery. The heat-stress response in eukaryotes involves remarkable reprogramming of translation: Most mRNAs are repressed, but those encoding HSPs are efficiently translated (Lindquist, 1986; Nover, 1991; Sierra and Zapata, 1994; Brostrom and Brostrom, 1998). Although translational reprogramming occurs in a wide range of cells, it is achieved in different ways (Sierra and Zapata, 1994). Yeast translates HSP and non-HSP messengers at the onset of stress (Bienz, 1982; Lindquist et al., 1982); preferential synthesis of HSPs through cytoplasmic enrichment of the corresponding messengers results from a continuous supply of HSP mRNAs, nuclear retention of other messengers (Saavedra et al., 1997), and rapid turnover of cytoplasmic transcripts. Drosophila melanogaster and HeLa cells maintain non-HSP mRNAs on polysomes, but their translation is dramatically reduced (Ballinger and Pardue, 1982; Hickey and Weber, 1982; Lindquist et al., 1982). Plants control translation during heat stress in a different way. Most repressed messengers dissociate from polysomes and are transiently stored elsewhere in the cytoplasm (Nover et al., 1989; Apuya and Zimmerman, 1992). The mechanism of this process and the proteins involved are still not known, although several models for mRNA storage have been proposed.
The appearance of large RNA-containing particles (Neumann et al., 1984) and structures called HSGs (which are rich in small HSPs; Nover et al., 1983) in heat-stressed tomato (Lycopersicon peruvianum) cells led to the hypothesis that mRNA is stored in these structures (Nover et al., 1989). This implies a novel role for small HSPs in addition to their function as molecular chaperones (Jakob et al., 1993; Lee et al., 1995; Forreiter et al., 1997). Indeed, cytoplasmic fractions enriched in small HSPs contain mRNAs that can be translated in vitro into proteins similar to those encoded by polysomal messengers from nonstressed cells, whereas in vitro translation of polysomal mRNAs from stressed cells mainly yields HSPs (Nover et al., 1989).
Translation is reprogrammed immediately upon heat stress, whereas HSGs appear only after prolonged stress. In addition, Gallie and Pitto (1996) reported that translational switch and increased transcript stability occur when small HSP synthesis is blocked. Alternatively, messengers may be stored in RNA and protease-containing particles called prosomes or proteasomes (Scherrer and Bey, 1994; Schmid et al., 1995). Their RNA content seems inversely related to the purity of the preparation, and the inhibition of translation by proteasomes in vitro that is not caused by proteolysis may be due to proteasome-associated RNase activity (Schmid et al., 1995). This activity is incompatible with RNA storage and stabilization. Other sites of storage cannot be ruled out, such as a plant homolog of a translation- regulating particle identified in animal cells that is rich in the 72-kD poly(A)-binding protein, and mRNA-binding proteins of 50 to 60 kD termed mRNP core proteins (Evdokimova et al., 1995; Spirin, 1996; Yurkova and Murray, 1997). However, plant counterparts of core mRNPs have not yet been found, so storage of repressed mRNA in plants during stress remains enigmatic.
In this study we analyzed the partitioning of stress-induced and repressed transcripts over polysomes and npRNPs from tomato cells. Messengers repressed by heat stress were found in nonpolysomal storage particles and reappeared on polysomes during recovery. However, stress-induced and repressed mRNAs could bind isolated npRNPs in vitro, suggesting that the selection mechanism resided elsewhere. Storage particles bound messengers in an RNase-resistant form in the absence of small HSPs and C2-subunit-containing proteasomes. In addition, mRNAs introduced into tobacco (Nicotiana plumbaginifolia) cells did not colocalize with small HSPs.
MATERIALS AND METHODS
Tomato Cell Culture and Heat Stress
The tomato (Lycopersicon peruvianum LpVII) cell-suspension culture was maintained as described previously (Nover et al., 1982). Heat treatments of exponentially growing cells were 25°C (control), or 15 min at 40°C followed by 3 h at 25°C (pre-induced). Stressed cells were treated for 15 min at 40°C then subsequently incubated 3 h at 25°C with an additional heat treatment of 2 h at 40°C. Recovered cells were obtained after the full stress treatment with a subsequent incubation of 2 h at 25°C.
Cell Fractionation
Isolation of npRNPs was modified from Nover et al. (1983, 1989). Samples were kept on ice and centrifuged at 4°C. Cells were harvested by aspiration, ground in liquid nitrogen, and placed in 50 mm Tris-HCl, pH 7.8, 0.5 mm MgCl2, 25 mm KCl, 2 mm CaCl2, 1 mm EDTA, 0.1% Nonidet P-40, 0.1% β-mercaptoethanol, 25% glycerol, and 150 mm Suc at 2 mL g−1 fresh weight of cells. Debris was removed by 5 min of centrifugation at 3,400g. Polysomes were disrupted by adding 250 mm KCl, 30 mm EDTA, and 0.5% Nonidet P-40. After 20 min of centrifugation at 6,700g, the supernatant was centrifuged for 1 h at 68,000g through two Suc pads: 5 mL of 15% Suc, 1 mm EDTA, and 10 mm MgCl2 in a gradient buffer (20 mm Tris-HCl, pH 7.8, 250 mm KCl, 25% glycerol, and 0.1% β-mercaptoethanol) overlaid with 5 mL of 10% Suc and 30 mm EDTA in a gradient buffer. Supernatants (S30) were used to prepare P100 fractions (see below). Pellets were homogenized in 10 mL of 20 mm Tris-HCl, pH 7.8, 250 mm NaCl, 30 mm EDTA, 0.5% Nonidet P-40, 0.2% lauroylsarcosine, 0.1% β-mercaptoethanol, and 15% glycerol with a Teflon homogenizer and spun for 5 min at 5,000 rpm.
The resulting supernatants were centrifuged as above through 15 mL each of 15% and 10% Suc solutions. Resedimented P30 pellets and P100 fractions, prepared by spinning S30s through a 2 mL 15% Suc pad for 16 h at 220,000g, were rinsed twice with RNP buffer (20 mm Tris-HCl, pH 7.8, 50 mm NaCl, 0.1% β-mercaptoethanol, and 15% glycerol) and resuspended in the same buffer. To isolate polysomes, 25 μg mL−1 cycloheximide was added to cell cultures 1 min before harvesting. Cells were ground in liquid nitrogen and thawed in PIB buffer (100 mm Hepes-KOH, pH 8.3, 500 mm KCl, 20 mm MgCl2, 2 mm EDTA, 0.5% Triton X-100, 0.05% β-mercaptoethanol, and 250 mm Suc). Debris was removed by spinning for 10 min at 8,000g. After centrifugation for 3 h at 150,000g through pads of 20% and 60% Suc in PIB buffer, polysomes were collected from the borders of the pads.
RNA-Blot Analysis
Antisense probes were synthesized with an RNA-labeling kit (Boehringer Mannheim) from templates encoding tomato histone H4, calmodulin, and cyclophilin (Materna, 1996), and from plasmids encoding steroid dehydrogenase (Ganal et al., 1998), HsfA2 (Scharf et al., 1990), and HSP17.7 (GenBank accession no. AJ225046). RNA was isolated from cells, polysomes, and P30s using guanidinum isothiocyanate (Forreiter and Apel, 1993) and separated on 1.2% agarose gels containing 1% formaldehyde. It was then transferred to Hybond N+ membranes (Amersham) and fixed using a Stratalinker (Stratagene). Blots were prehybridized for 5 min in 5× SSC, 50% deionized formamide, 0.02% SDS, 0.1% lauroylsarcosine, and 2% Denhardt's reagent; they were hybridized in this solution with DIG-labeled probes for 16 h at 65°C and given three 10-min washes at 62°C with 0.1× SSC and 0.1% SDS. For detection we used anti- DIG-alkaline phosphatase (Boehringer Mannheim), nitroblue tetrazolium salt, and 5-bromo-4-chloro-3-indolyl phosphate, following the manufacturer's instructions.
mRNA Synthesis
Tomato histone H4 (GenBank accession no. X69179) and HSP17.7 cDNAs amplified by PCR were inserted into pBluescript SK (+) (Stratagene). The HSP17.7 construct was linearized with BamHI. A linear template containing T7 and T3 promoters was amplified from the histone clone with primers M13(−20) and M13 reverse (Stratagene). The luciferase mRNAs luc and lucA50 were produced from pT7-LUC-A50 (Gallie et al., 1991) cut with BamHI and DraI, respectively. Capped and uncapped RNAs were transcribed with the mCap kit (Stratagene) and RNase inhibitor (RNasin, Promega), [α-32P]UTP (New England Nuclear), or fluorescin-12-UTP (Boehringer Mannheim). DNA removal by DNase I (Stratagene) was followed by spinning through Sephadex G50 and ethanol precipitation as described by Sambrook et al. (1989).
RNA Protection Assay
Fluorescin-labeled mRNA (approximately 200 ng) and 40 μg of npRNPs (or 50 μg of BSA) in RNP buffer were incubated on ice for 10 min in the presence of 20 units of RNasin (Promega), which does not inhibit micrococcal nuclease. After treatment with 2 units of micrococcal nuclease (Boehringer Mannheim) and 5 mm CaCl2 for 10 min at 37°C, EDTA was added to 20 mm. RNAs were separated on 7 m urea/6% polyacrylamide gels (Sambrook et al., 1989).
Gel Retardation Assay
Radioactive mRNA (105 cpm) and 10 μg of npRNPs (or 50 μg of BSA) in RNP buffer were incubated on ice for 10 min, mixed with 5× loading buffer (50% glycerol, 5× TBE, 50 mm EDTA, and bromphenol blue), and separated on a 5% polyacrylamide/10% glycerol gel in TBE at 4°C.
Gel Filtration Chromatography
RNPs from 0.5 g fresh weight of tomato cells were incubated with 105 cpm 32P-labeled mRNA for 10 min on ice, applied to a Superdex S200 (Pharmacia) column, and eluted with 20 mm Tris-HCl, pH 7.8, at 1 mL min−1 using a Gradifrac (Pharmacia). Radioactivity in 10-mL fractions was measured in a scintillation counter, and proteins were precipitated with acetone and analyzed by SDS-PAGE.
Protein Analysis
Proteins separated on 15% SDS-PAGE gels (Laemmli, 1970) were stained with Coomassie blue or blotted onto Hybond C nitrocellulose (Amersham). Blots were blocked with 5% nonfat dry milk in TBS, probed with antibodies against HSP17 (obtained from M. Kirschner, Frankfurt, Germany), HSP70 (StressGen, Victoria, Canada), HsfA2 (Lyck et al., 1997), tubulin (Sigma), or the proteasome C2 subunit (Umeda et al., 1997), and developed with anti-rabbit horseradish peroxidase (Bio-Rad) and enhanced chemiluminescence (DuPont-New England Nuclear ) or with anti-rabbit alkaline phosphatase (Promega), nitroblue tetrazolium, and 5-bromo-4-chloro-3-indolyl phosphate, following the manufacturer's instructions.
Localization of mRNA in Tobacco Protoplasts
Tobacco (Nicotiana plumbaginifolia) leaf protoplasts were prepared according to the method of Treuter et al. (1993) and then washed five times to remove RNases from the cell wall-digesting enzyme solution. Fluorescein-labeled RNA was introduced using PEG as described for DNA by Treuter et al. (1993). After transformation, the PEG concentration was reduced by adding 10 volumes of culture medium. Control cells were kept at 25°C for 2 h following transformation. Pre-induced cells (15 min at 40°C and 2.5 h at 25°C) were transformed and stressed by gradual heating to 37°C over 15 min and subsequent incubation at this temperature for 105 min. Cells were fixed with 3% paraformaldehyde for 30 min, incubated for 15 min in 1% Nonidet P-40/PBS, and attached to polylysine-coated coverslips. After 10 min of incubation in 2% Nonidet P-40/PBS, two 10-min washes with PBS, and 30 min of blocking with 1% BSA/PBS, cells were incubated overnight at 4°C with anti-HSP17, washed three times, reblocked, and incubated for 2 h at 37°C with anti rabbit-tetramethyrhodamine isothiocyanate (Sigma). After three washes, cells were mounted in PBS (75% glycerol and 0.1% phenylenediamine) and analyzed by confocal fluorescence microscopy.
RESULTS
Protein Composition of Nonpolysomal mRNP Fractions from Tomato Cells
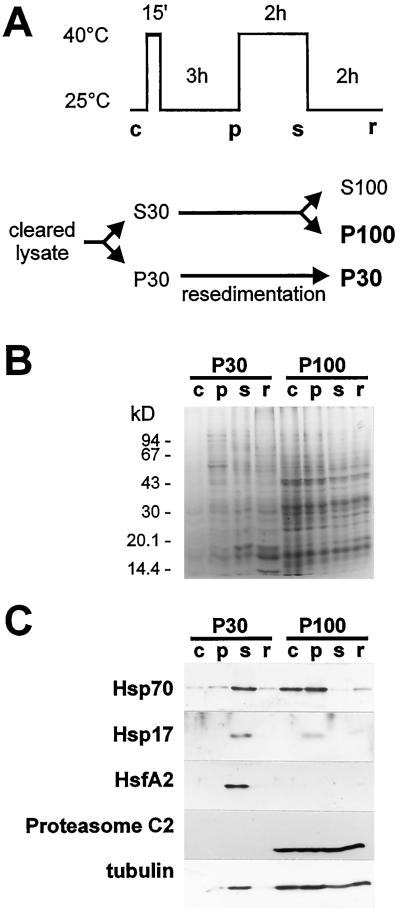
Cytoplasmic fractions enriched in RNPs were sedimented from tomato cells by ultracentrifugation, yielding P30 fractions, and by prolonged ultracentrifugation of the P30 supernatants, yielding P100s (Fig. 1A). P30s from heat- stressed cells have been reported to contain HSGs (Nover et al., 1983, 1989). P100 fractions from control and pre-induced cells contained putative HSG precursors and proteasomes (Nover et al., 1989). We prepared P30s and P100s from control, pre-induced, stressed, and recovered cells, and analyzed their proteins by SDS-PAGE, followed by Coomassie-blue staining and western blotting (Fig. 1).
Figure 1.
Proteins in npRNP sediments from control (c), pre-induced (p), stressed (s), and recovered (r) tomato cells. A, Schematic representation of heat-stress treatment and isolation of npRNPs. B, Coomassie blue staining of proteins isolated from different cytoplasmic RNP sediments after SDS-PAGE. C, Immunoblots of RNP sediments probed for HSP70, small HSPs, HsfA2, the proteasome C2 subunit, and tubulin, respectively.
Coomassie-blue staining (Fig. 1B) revealed no dramatic change in protein patterns of P30s or P100 from cells kept at different temperatures. Pellets contained HSPs and proteasome proteins, but many other proteins as well. Western blots revealed temperature-dependent changes in protein content (Fig. 1C). P30s and P100s from control cells lacked small HSPs. After pre-induction, small HSPs were present in the HSG-precursor-containing P100, which was found in P30 during stress (presumably in HSGs) and reappeared in P100s during recovery. The lack of small HSPs in P100s from stressed cells indicate that virtually all small cytoplasmic HSPs reside in granules after prolonged stress, in agreement with the cytolocalization shown in Figure 6 (see below). Stress also resulted in a large amount of HSP70 in P30. The different distribution of HSP70 in control cells, where it could be detected in all fractions, and the absence of small HSPs in sediments from control cells reflects the fact that cells synthesize constitutive and stress-induced versions of HSP70. The mRNA-rich P30 fractions lacked the proteasome C2 subunit, which was found in all P100s (Fig. 1C). HsfA2, which is thought to associate with HSGs after prolonged stress (Scharf et al., 1998), was indeed found in P30 from stressed cells. In contrast to HSP70 and the small HSPs, which were present in HSG precursors, HsfA2 associated with mature HSGs only and was not found in any of the P100s. The cytoskeleton component tubulin was abundant in all P100s and in P30s from stressed cells. A small amount was also detected in P30s from pre-induced and recovered cells. The large amount of tubulin in P30s from stressed cells coincided with a slight reduction in the corresponding P100s.
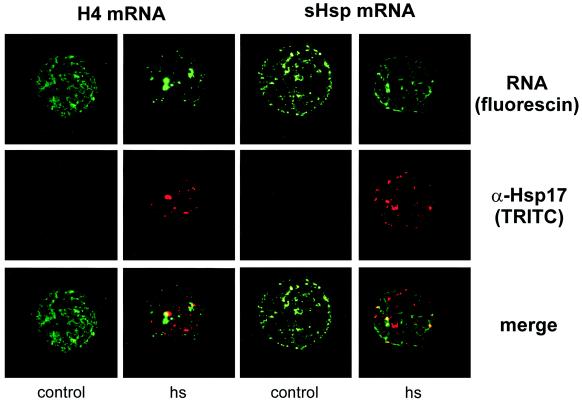
Figure 6.
Localization of fluorescin-labeled histone H4 and small Hsp mRNAs (green) and HSGs in control and stressed tobacco protoplasts. HSGs were labeled with anti-Hsp17 and TRITC-conjugated secondary antibodies (red). Overlays of the top and middle panels are displayed at the bottom.
Repressed Messengers Shuttle between Polysomes and Nonpolysomal RNPs
In vitro translation of mRNAs from polysomes and npRNPs from tomato cells suggests that most translatable housekeeping messengers leave the polysomes upon heat stress and are replaced by HSP transcripts (Nover et al., 1989). However, the poly(A)-tail length of HSP mRNAs varied with temperature (Osteryoung et al., 1993; Dellavalle et al., 1994), and repressed mRNAs on polysomes and heat-stress messengers in npRNPs could escape detection if their translatability was affected by poly(A)-tail shortening or other modifications. To detect mRNAs independent of their translatability, we analyzed mRNA distribution by RNA blotting instead of indirectly assaying them by in vitro translation; this way, we could study defined messengers as opposed to proteins produced from a large pool of unidentified transcripts.
Total, polysomal, and nonpolysomal RNA were isolated from cells prior to stress and after pre-induction, heat stress, and recovery. Following the method of Nover et al. (1989), we isolated nonpolysomal RNAs from P30 fractions after the polysomes were disrupted with high amounts of KCl, EDTA, and Nonidet P-40 to prevent their cosedimentation with npRNPs. RNA from different RNP fractions was separated on a gel and stained with ethidium bromide (Fig. 2A). The lanes with RNA from npRNPs were deliberately overloaded. Despite the large amount of RNA applied, rRNAs are not visible in these lanes, indicating that these fractions are indeed nonpolysomal and the messengers therein are therefore not translated.
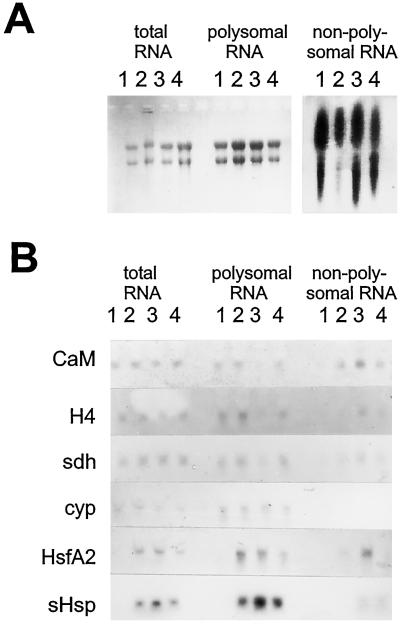
Figure 2.
RNA distribution during heat stress and recovery. Total, polysomal, and nonpolysomal RNA was isolated from control (lanes 1), pre-induced (lanes 2), stressed (lanes 3), and recovered (lanes 4) cells separated by electrophoresis and stained with ethidium bromide (A) or blotted and probed for the transcripts indicated (B). RNA was normalized to the number of cells isolated, except the last four lanes in A were deliberately overloaded to display the presence of mRNAs and the absence of rRNAs. CaM, calmodulin; H4, histone H4; sdh, steroid dehydrogenase; cyp, cyclophilin; sHsp, small HSP.
For northern analysis (Fig. 2B), however, RNPs were normalized for the number of cells to avoid loading artifacts, because the amount of mRNA in npRNPs varied with temperature and was markedly increased by stress (not shown). Additionally, normalization for RNA content would result in overrepresentation of messengers from control npRNPs because RNA is almost absent in these particles. We propose that mRNAs detected in nonpolysomal populations indeed reflect inactive transcripts, a conclusion supported by the fact that P30 fractions from stressed cells contained much more RNA than other P30s, in accord with the effects of translational repression during heat stress.
Stress and recovery had no detectable influence on the total amount of mRNAs for calmodulin, histone H4, steroid dehydrogenase, and cyclophilin (Fig. 2B). HsfA2 and small HSP (HSP17.7) transcripts were not found in control cells, but appeared upon pre-induction. Cyclophilin, small HSP, and HsfA2 messengers were translated during heat stress. Figure 2B shows that these transcripts were indeed present mainly in polysomes. However, HsfA2 mRNA appeared in npRNPs after prolonged stress. Together with the appearance of the corresponding protein in cytoplasmic aggregates (Scharf et al., 1998; Fig. 1C), this result suggests that HsfA2 was active in the early heat-stress response but was repressed when stress continued. Moreover, appearance of this stress-induced transcript proves that the mRNAs in our npRNPs from stressed cells represented messengers inactivated by stress and did not merely reflect aggregation of pre-existing mRNPs.
Some small HSP RNA was detected in npRNPs after large numbers of small HSPs were produced, but most small HSP messengers remained on polysomes and the synthesis of small HSPs continued during the entire stress phase and far into the recovery period. Histone H4, calmodulin, and steroid dehydrogenase were not stress induced, because their messengers were found in npRNPs during heat stress and reappeared in polysomes during recovery. In agreement with the observation that translation of non-HSP proteins resumed upon recovery from stress when de novo synthesis of mRNA was blocked by actinomycin D (Nover and Scharf, 1984), our data show that a set of selected heat-stress-repressed messengers is stored in npRNPs in tomato cells during stress.
Isolated Storage RNPs Bound mRNAs in a Nuclease-Resistant Form Independent of Transcript Type
Selective mRNA distribution over polysomes and npRNPs may be caused by a combination of the translational apparatus, the storage machinery, and/or other factors. To study the possible role of storage RNPs in this process, we tested their capacity to bind mRNAs in vitro. NpRNPs from control, pre-induced, stressed, and recovered cells were incubated with transcripts encoding tomato histone H4 and HSP17.7, the former being repressed and the latter induced upon stress. We also tested firefly luciferase messengers with or without the poly(A) tail for interaction with tomato RNPs. These luciferase constructs were translated when introduced into plant cells, but were repressed and stabilized by heat stress (Gallie et al., 1995).
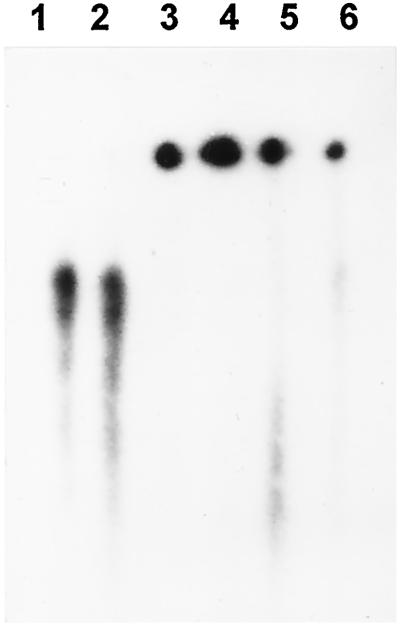
The npRNPs in the P30s and P100s protected messengers from degradation by micrococcal nuclease (shown for histone H4 mRNA in Fig. 3A), regardless of the transcript (shown for the P30 from stressed cells in Fig. 3B) . All other combinations of npRNPs and mRNAs yielded the same result (not shown). Messengers incubated with BSA but without nuclease remained intact (Fig. 3). BSA itself did not protect the transcript, and neither did the P30 from stressed cells when it was heated to 95°C for 5 min before the addition of RNA (Fig. 3A), indicating that the mRNAs were protected from the nuclease by proteins in the npRNP preparations. The poly(A) tail was not involved in this protection, because messengers with and without the tail survived the nuclease treatment (Fig. 3B).
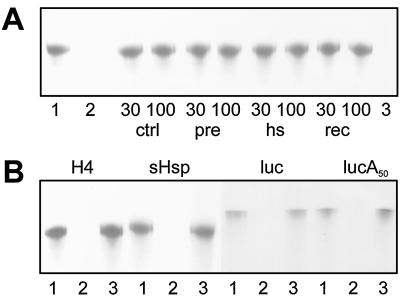
Figure 3.
Protection of mRNAs from nuclease digestion by npRNPs. RNA survival was monitored after electrophoresis. A, H4 mRNA incubated with BSA (lanes 1 and 2), P30s (lanes 30) and P100s (lanes 100) from control (ctrl), pre-induced (pre), heat-stressed (hs), and recovered (rec) cells, and with heat-denatured P30 from stressed cells (lane 3). No nuclease was added to the samples in lane 1. B, Messengers for histone H4 , HSP17.7 (sHsp), and luciferase with (lucA50) and without Luc the poly(A+) tail incubated with BSA (lanes 1 and 2) and P30s from stressed cells (lanes 3).
Transcripts with or without a methylated GpppG cap were protected equally efficiently (not shown). The same RNAs were retarded on a nondenaturing gel after incubation with npRNPs but not when incubated with BSA, as shown in Figure 4 for histone H4 mRNA with P30 from stressed cells and P100s from control, pre-induced, and stressed cells. We also observed insensitivity to nuclease and a shift in the elution peak upon gel filtration (see below) when mRNAs were incubated with npRNPs in a buffer containing high amounts of salt and detergent (250 mm NaCl, 0.5% Nonidet P-40, and 0.2% sodium-lauroyl-sarcosine), which suggests a specific interaction. In the nuclease and gel-retardation assays and in the gel-filtration experiment, the RNPs did not distinguish stress-induced from repressed messengers, suggesting that selection in vivo requires other factors.
Figure 4.
Gel retardation of mRNA by npRNPs. Radiolabeled histone H4 mRNA (lane 1) and the same RNA incubated with BSA (lane 2), P100s from control (lane 3), pre-induced (lane 4), and stressed cells (lane 5), and P30 from stressed cells (lane 6) was separated on a nondenaturing gel.
mRNA Binding by Fractions of RNP Sediments
The idea that HSGs or proteasomes are associated with mRNPs is based on cosedimentation after ultracentrifugation. However, sediments did not only contain proteasomes, HSGs, and their precursors, but many other proteins as well. Unfortunately, the large aggregates in short-spin sediments (P30s) resisted chromatographic separation. Proteasomes and HSG precursors in P100s were separated by chromatography, but the RNA content of the resulting fractions was not determined (Nover et al., 1989). Detection of endogenous messengers in the P100s was not feasible as the lengthy isolation procedure yielded RNA that was not suitable for northern analysis. Therefore, we again fractionated the sedimented RNPs by gel filtration after mRNA binding in vitro. For this purpose, RNP fractions were incubated with radiolabeled mRNA and separated on a Superdex S200 column.
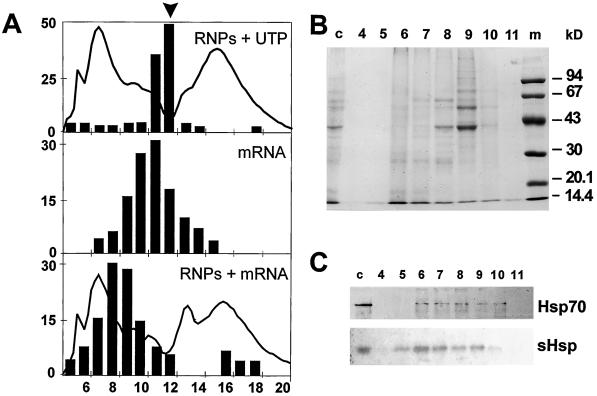
P100s from control and pre-induced cells eluted as complexes of distinct protein composition, as shown in Figure 5 for P100s from pre-induced cells incubated with histone H4 mRNA. Elution of labeled messenger was shifted (Fig. 5A) to fractions with major protein bands at approximately 40, 55, 60, 70, and >100 kD and minor bands in the 25- to 35-kD range (Fig. 5B). This shift was also observed using mRNAs coding for small HSPs and luciferase (not shown), but elution of UTP remained unaltered (Fig. 5A). HSP70 and small HSPs eluted in a broad range of fractions, reflecting the size heterogeneity of small HSP oligomers and (pre)HSGs. The peak fractions did not coincide with those of the reporter RNA (Fig. 5C). This result is in agreement with observations that these HSPs were not required for mRNA binding (Gallie and Pitto, 1996; Figs. 3 and 4) and with our finding that elution of all messengers was shifted when incubated with npRNPs from control cells, which do not contain small HSPs (not shown).
Figure 5.
Gel-filtration fractionation of npRNP sediments. A, Elution of [α-32P]UTP and npRNPs (P100) from pre-induced tomato cells (top) and radiolabeled histone H4 mRNA in the absence (middle) and presence (bottom) of npRNPs. Bars, eluted radioactivity as percentage of total counts applied. Arrowhead, Elution of UTP in the absence of npRNPs. Solid line, A280 in arbitrary units. Numbers below the graphs mark fractions of 10 mL. B, Fractions separated by SDS-PAGE and stained with Coomassie blue. Lane m, Marker. C, Immunoblots of fractions using antisera for Hsp70 and small Hsp (sHsp). Numbers in B and C correspond to the fraction numbers in A. Lanes c, Control (nonpolysomal sediment before gel filtration).
Localization of Fluorescein-Labeled mRNAs in Tobacco Protoplasts
In our in vitro RNA-binding experiments, mRNPs did not distinguish stress-induced from repressed messengers. To study mRNA during stress within cells, we introduced fluorescent transcripts into protoplasts and studied their localization by fluorescence microscopy. Since our tomato cell culture resisted transformation (not shown) we used tobacco leaf protoplasts, which are able to translate synthetic mRNAs (Gallie et al., 1991). UV crosslinking of radioactive messengers to proteins in intact tobacco protoplasts and lysed tomato cells labeled three major bands at approximately 30, 55, and >200 kD (R. Stuger, unpublished data), which is similar to the labeling pattern seen after the cross-linking of transcripts injected into Xenopus laevis oocytes by Meric et al. (1997).
After insertion of RNAs encoding histone H4 and HSP17.7, protoplasts were heat stressed or kept at 25°C for 2 h. This amount of time was necessary to form HSGs. Analysis of radiolabeled mRNAs re-isolated after introduction into tobacco protoplasts indicated that approximately one-half of the RNA remained intact after 2 h (not shown), comparable to the half-life of approximately 100 min for capped luciferase mRNA in tobacco protoplasts (Gallie et al., 1991). Such luciferase mRNA could still be translated into active luciferase after this time. The HSGs were detected with an antibody against small HSPs.
HSGs were visible in most of the heat-stressed cells. In some cells the amount of RNA was below the detection limit, and in others the amount was too high to draw conclusions on the subcellular localization. The majority of cells contained intermediate amounts of RNA. Figure 6 shows RNAs and small HSPs as they appeared in most of the cells. The messengers for histone H4 and HSP17.7 resided in the cytoplasm but did not colocalize with HSGs, although a little overlap was sometimes visible. We obtained the same result with luciferase transcripts with and without a poly(A) tail: Neither of the tested reporter mRNAs colocalized with the small HSPs (not shown).
DISCUSSION
Transfer of heat-stress-repressed mRNAs from polysomes to cytoplasmic nonpolysomal storage particles in tomato cells and relocation to polysomes during recovery illustrates the diversity of selective mRNA repression during stress in different organisms. Whereas the investigated non-heat-stress transcripts were driven to npRNPs upon heat stress, stress-induced messengers were associated mainly with polysomes. Messengers for HsfA2, although present in polysomes early in the stress response, entered npRNPs after prolonged stress. This relocation coincided with the clustering of HsfA2 in cytoplasmic HSGs, where it was obviously not active as a transcriptional activator. The increased amount of mRNA and aggregated protein in sediments from stressed cells and the fact that cells also keep transcripts in npRNPs under nonstress conditions raise the possibility that the mRNAs in the P30s from stressed cells do not represent messengers silenced during stress but instead reflect the aggregation of npRNPs present before stress. However, the presence of the stress-induced HsfA2 mRNA on polysomes after pre-induction and its appearance in npRNPs during stress shows that the messengers in the P30s did indeed represent mRNAs dissociated from polysomes as a consequence of stress.
Our data and previous findings (Nover and Scharf, 1984) suggest that non-heat-stress-induced transcripts shuttle from polysomes to npRNPs and back. Similar shuttling was proposed for somatic carrot embryos, whereas small HSP RNA was not found on polysomes in heat-stressed carrot suspension-cultured cells (Apuja and Zimmerman, 1992). However, small HSP mRNA was abundant in polysomes from tomato suspension-cultured cells. Discrimination between different transcripts was not seen when mRNAs were added to isolated npRNPs. All npRNPs bound non-heat-stress mRNA and heat-stress mRNA, although the latter were mainly polysome-associated in vivo. We propose that the selection mechanism must reside elsewhere.
The binding of transcripts to small HSP-free RNPs together with different cytoplasmic localization of HSGs and mRNAs are in agreement with the finding that repression and stabilization of non-heat-stress transcripts does not require HSPs or heat-stress mRNAs (Gallie and Pitto, 1996). We cannot rule out the possibility that RNA-binding proteins (partially) unfolded by stress are captured by HSGs, an interaction that may be expected because small HSPs and other molecular chaperones are abundant in HSGs (Nover et al., 1983; Arrigo, 1987; Helm et al., 1997; Jinn et al., 1997). Although we found no interaction between messengers and small HSPs in vitro or in tobacco protoplasts, it remains possible that small HSPs are involved in mRNA handling during recovery from stress.
The binding of mRNAs to C2 proteasome subunit-free RNP preparations is in contradiction to the proposed role of proteasomes in mRNA repression, although proteasomes from sea urchin (Akhayat et al., 1987), duck, mouse, and HeLa cells (Schmid et al., 1984) were found to cosediment with mRNPs. The observation that X. laevis proteasomes form granular clusters that colocalize with actin and myosin (Ryabova et al., 1994) and that proteasomes from mammalian cells also interact with the cytoskeleton (Olink-Coux et al., 1994) may explain cosedimentation, because nonpolysomal mRNPs (Scherrer and Bey, 1994), polysomes (Hesketh, 1994), and possibly mRNA itself (Muench et al., 1998) also associate with the cytoskeleton.
Kraemer and Blobel (1997) found mRNPs appearing as coarse aggregates in HeLa cells treated with cytoskeleton-disrupting agents. RNPs and proteasomes, separated because of their association with different sections of the cytoskeleton, may come together when this structure collapses. We found that the HSG- and mRNA-rich P30s from stressed cells contained tubulin, whereas P100s from stressed cells contained less tubulin than the other P100s. Although P30s from stressed cells carried a large amount of tubulin, the proteasome C2 subunit was not present. However, the possibility remains that proteasomes without the C2 subunit interact with RNPs in a biologically significant way.
Translation-initiation factors and the 72-kD poly(A)- binding protein represent the only identified and characterized plant cytoplasmic mRNA-binding proteins (for review, see Albá and Pagés, 1998). The UV-crosslinking of radiolabeled mRNA to proteins in tomato cell lysate and tobacco protoplasts yielded a labeling pattern similar to the one found using X. laevis oocytes, with an intensely labeled band at about 55 kD. The X. laevis protein was identified as core mRNP FRGY2, which is abundant in repressed mRNPs (Meric et al., 1997). Whether core mRNPs and poly(A)-binding proteins are present in npRNPs from tomato awaits further investigation. The type of mechanism that targets mRNAs to storage particles remains unknown. The competition between the translation machinery and npRNPs may separate translation-competent transcripts from inactive mRNAs. Targeting of messengers to storage RNPs and the subsequent recruitment of HSP mRNAs for translation is also possible. It will be easier to investigate these options when we know more about cytoplasmic mRNA-binding proteins in plants.
ACKNOWLEDGMENTS
We thank Daniel Gallie for the generous gift of pT7-LUC-A50; Martin Ganal for the steroid dehydrogenase clone; Masaaki Umeda for sharing the proteasome antibody; Michael Hoff and Daniela Löw for subcloning the small HSP cDNAs; Lutz Nover, Rob Benne, Dave Speijer, and Daniela Löw for helpful comments on the manuscript; and all members of the Molecular Cell Biology Department of Goethe University for sharing clones, antibodies, and ideas.
Abbreviations:
- Hsf
heat-stress transcription factor
- HSG
heat-stress granule
- HSP
heat-stress protein
- npRNP
nonpolysomal RNP
- RNP
ribonucleoprotein
Footnotes
This work was funded by the Deutsche Forschungsgemeinschaft (grant no. No 249/1).
LITERATURE CITED
- Akhayat O, Grossi de Sa F, Infante AA. Sea urchin prosome: characterization and changes during development. Proc Natl Acad Sci USA. 1987;84:1595–1599. doi: 10.1073/pnas.84.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albá MM, Pagés M. Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 1998;3:15–21. [Google Scholar]
- Apuya NR, Zimmerman JL. Heat shock gene expression is controlled primarily at the translational level in carrot cells and somatic embryos. Plant Cell. 1992;4:657–665. doi: 10.1105/tpc.4.6.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo A-P. Cellular localization of HSP23 during Drosophila development and following subsequent heat shock. Dev Biol. 1987;122:39–48. doi: 10.1016/0012-1606(87)90330-7. [DOI] [PubMed] [Google Scholar]
- Ballinger DG, Pardue ML. The subcellular compartmentalization of mRNAs in heat-shocked Drosophila cells. In: Schlesinger MJ, Ashburner M, Tissières A, editors. Heat Shock from Bacteria to Man. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. pp. 183–190. [Google Scholar]
- Bienz M. The heat-shock response in Xenopus oocytes and somatic cells: differences in phenomena and control. In: Schlesinger MJ, Ashburner M, Tissières A, editors. Heat Shock from Bacteria to Man. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. pp. 177–181. [Google Scholar]
- Brostrom CO, Brostrom MA. Regulation of translational initiation during cellular responses to stress. Prog Nucleic Acid Res Mol Biol. 1998;58:79–125. doi: 10.1016/s0079-6603(08)60034-3. [DOI] [PubMed] [Google Scholar]
- Dellavalle RP, Petersen R, Lindquist S. Preferential deadenylation of HSP70 mRNA plays a key role in regulating HSP70 expression in Drosophila melanogaster. Mol Cell Biol. 1994;14:3646–3659. doi: 10.1128/mcb.14.6.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova VM, Wei C-L, Sitikov AS, Simonenko PN, Lazarev OA, Vasilenko KS, Ustinov VA, Hershey JWB, Ovchinnikov LP. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J Biol Chem. 1995;270:3186–3192. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- Forreiter C, Apel K. Light-independent and light-dependent protochlorophyllide-reducing activities and two distinct NADPH-protochlorophyllide oxidoreductase polypeptides in mountain pine (Pinus mugo) Planta. 1993;190:536–545. doi: 10.1007/BF00224793. [DOI] [PubMed] [Google Scholar]
- Forreiter C, Kirschner M, Nover L. Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell. 1997;9:2171–2181. doi: 10.1105/tpc.9.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Caldwell C, Pitto L. Heat shock disrupts cap and poly(A+) tail function during translation and increases mRNA stability of introduced reporter mRNA. Plant Physiol. 1995;108:1703–1713. doi: 10.1104/pp.108.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Feder JN, Schimke RT, Walbot V. Posttranscriptional regulation in higher eukaryotes: the role of the reporter gene in controlling expression. Mol Gen Genet. 1991;228:258–264. doi: 10.1007/BF00282474. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Pitto L. Translational control during recovery from heat shock in the absence of heat shock proteins. Biochem Biophys Res Commun. 1996;227:462–467. doi: 10.1006/bbrc.1996.1529. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Czihal R, Hannappel U, Kloos DU, Polley A, Ling HQ. Sequencing of cDNA clones from the genetic map of tomato (Lycopersicon esculentum) Genome Res. 1998;8:842–847. doi: 10.1101/gr.8.8.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm KW, Lee GJ, Vierling E. Expression and native structure of cytosolic class II small heat-shock proteins. Plant Physiol. 1997;114:1477–1485. doi: 10.1104/pp.114.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh J. Translation and the cytoskeleton: a mechanism for targeted protein synthesis. Mol Biol Rep. 1994;19:233–243. doi: 10.1007/BF00986965. [DOI] [PubMed] [Google Scholar]
- Hickey E, Weber LA. Preferential translation of heat-shock mRNAs in HeLa cells. In: Schlesinger MJ, Ashburner M, Tissières A, editors. Heat Shock From Bacteria to Man. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. pp. 199–206. [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- Jinn T-S, Chang P-FL, Chen Y-M, Key JL, Lin C-Y. Tissue-type-specific heat-shock response and immunolocalization of class I low-molecular-weight heat-shock proteins in soybean. Plant Physiol. 1997;114:429–438. doi: 10.1104/pp.114.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee GH, Pokala N, Vierling E. Structure and in vitro chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat stock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lindquist S, DiDomenico B, Bugaisky G, Kurtz S, Petko L, Sonoda S. Regulation of the heat-shock response in Drosophila and yeast. In: Schlesinger MJ, Ashburner M, Tissières A, editors. Heat Shock from Bacteria to Man. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. pp. 167–175. [Google Scholar]
- Lyck R, Harmening U, Höhfeld I, Scharf K-D, Nover L. Intracellular distribution and identification of the nuclear localization signals of two tomato heat stress transcription factors. Planta. 1997;202:117–125. doi: 10.1007/s004250050110. [DOI] [PubMed] [Google Scholar]
- Materna T (1996) Das Gen für den Hitzestreβtranscriptionsfaktor A2 der Tomate-Strukturelle und funktionelle Charakterisierung seiner multiplen RNA-Spezies. PhD thesis. Goethe University, Frankfurt am Main, Germany
- Meric F, Matsumoto K, Wolffe AP. Regulated unmasking of in vivo synthesized maternal mRNA at oocyte maturation. J Biol Chem. 1997;272:12840–12846. doi: 10.1074/jbc.272.19.12840. [DOI] [PubMed] [Google Scholar]
- Muench DG, Wu Y, Coughlan SJ, Okita TW. Evidence for a cytoskeleton-associated binding site involved in prolamine mRNA localization to the protein bodies in rice endosperm tissue. Plant Physiol. 1998;116:559–569. doi: 10.1104/pp.116.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann D, Scharf K-D, Nover L. Heat shock-induced changes of plant cell ultrastructure and autoradiographic localization of heat shock proteins. Eur J Cell Biol. 1984;34:254–264. [PubMed] [Google Scholar]
- Nover L. Translational control. In: Nover L, editor. Heat Shock Response. Boca Raton, FL: CRC Press; 1991. pp. 299–324. [Google Scholar]
- Nover L, Kranz E, Scharf K-D. Growth cycle of suspension cultures of Lycopersicon esculentum and L. peruvianum. Biochem Physiol Pflanz. 1982;177:483–499. [Google Scholar]
- Nover L, Scharf K-D. Synthesis, modification and structural binding of heat-shock proteins in tomato cell cultures. Eur J Biochem. 1984;139:303–313. doi: 10.1111/j.1432-1033.1984.tb08008.x. [DOI] [PubMed] [Google Scholar]
- Nover L, Scharf K-D, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf K-D, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olink-Coux M, Arcangeletti C, Pinardi F, Minisini R, Huesca M, Chezzi C, Scherrer K. Cytolocation of prosome antigens on intermediate filament subnetworks of cytokeratin, vimentin and desmin type. J Cell Sci. 1994;107:353–366. [PubMed] [Google Scholar]
- Osteryoung KW, Sundberg H, Vierling E. Poly(A) tail length of a heat shock protein RNA is increased by severe heat stress, but intron splicing is unaffected. Mol Gen Genet. 1993;239:323–333. doi: 10.1007/BF00276930. [DOI] [PubMed] [Google Scholar]
- Ryabova LV, Virtanen I, Olink-Coux M, Scherrer K, Vassetzky SG. Distribution of prosome proteins and their relationship with the cytoskeleton in oogenesis of Xenopus laevis. Mol Reprod Dev. 1994;37:195–203. doi: 10.1002/mrd.1080370210. [DOI] [PubMed] [Google Scholar]
- Saavedra CA, Hammell CM, Heath CV, Cole CN. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scharf K-D, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system:HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol. 1998;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K-D, Rose S, Zott W, Schöffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer K, Bey F. The prosomes (multicatalytic proteinases; proteasomes) and their relationship to the untranslated messenger ribonucleoproteins, the cytoskeleton, and cell differentiation. Prog Nucleic Acid Res Mol Biol. 1994;49:1–64. doi: 10.1016/s0079-6603(08)60047-1. [DOI] [PubMed] [Google Scholar]
- Schmid H-P, Akhayat O, Martins De Sa C, Puvion F, Koehler K, Scherrer K. The prosome: an ubiquitous morphologically distinct RNP particle associated with repressed mRNPs and containing specific ScRNA and a characteristic set of proteins. EMBO J. 1984;3:29–34. doi: 10.1002/j.1460-2075.1984.tb01757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid H-P, Pouch M-N, Petit F, Dadet M-H, Badaoui S, Boissonnet G, Buri J, Norris V, Briand Y. Relationships between proteasomes and RNA. Mol Biol Rep. 1995;21:43–47. doi: 10.1007/BF00990969. [DOI] [PubMed] [Google Scholar]
- Sierra JM, Zapata JM. Translational regulation of the heat shock response. Mol Biol Rep. 1994;19:211–220. doi: 10.1007/BF00986963. [DOI] [PubMed] [Google Scholar]
- Spirin AS. Masked and translatable messenger ribonucleoproteins in higher eukaryotes. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 319–334. [Google Scholar]
- Treuter E, Nover L, Ohme K, Scharf K-D. Promoter specificity and deletion analysis of three tomato heat stress transcription factors. Mol Gen Genet. 1993;240:113–125. doi: 10.1007/BF00276890. [DOI] [PubMed] [Google Scholar]
- Umeda M, Fujii N, Manabe Y, Uchimiya H. Molecular and biochemical characterization of a proteasome subunit from rice and carrot cells. Mol Gen Genet. 1997;255:19–27. doi: 10.1007/s004380050470. [DOI] [PubMed] [Google Scholar]
- Yurkova MS, Murray MT. A translation regulatory particle containing the Xenopus oocyte Y box protein mRNP3+4. J Biol Chem. 1997;272:10870–10876. doi: 10.1074/jbc.272.16.10870. [DOI] [PubMed] [Google Scholar]