Abstract
Summary
The Genome Context Viewer is a visual data-mining tool that allows users to search across multiple providers of genome data for regions with similarly annotated content that may be aligned and visualized at the level of their shared functional elements. By handling ordered sequences of gene family memberships as a unit of search and comparison, the user interface enables quick and intuitive assessment of the degree of gene content divergence and the presence of various types of structural events within syntenic contexts. Insights into functionally significant differences seen at this level of abstraction can then serve to direct the user to more detailed explorations of the underlying data in other interconnected, provider-specific tools.
Availability and implementation
GCV is provided under the GNU General Public License version 3 (GPL-3.0). Source code is available at https://github.com/legumeinfo/lis_context_viewer.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Advances in sequencing technology and algorithms for genome assembly and gene annotation have led to widespread availability of annotated whole genome assemblies. These vary widely in terms of the technologies and algorithms used, the complexity of the genomes targeted, the quality of their representation and the magnitude of the phylogenetic distances among them (Bradnam et al., 2013). For many comparative applications, users of these resources are less concerned with low-level details of sequence alignment than with basic questions surrounding functional content and the genomic contexts in which it occurs. Consequently, there is a need for tools that can facilitate the use of contextualized functional annotation as a unit of search and comparison among sets of genomes spanning diverse taxonomic groups that may reside in a distributed set of databases.
Here, we present the Genome Context Viewer (GCV), a web-based visual data-mining tool for dynamically identifying syntenic genomic segments and enabling interactive exploration of gene content similarities and differences among distributed collections of annotated genomes.
2 Motivation
Consider the situation faced by a breeder assessing trait-linked regions across multiple genomes within a single species (pan-genomics) or among wild species that are candidates for introgression with a congeneric domesticated species. In the first place, given the relatively small phylogenetic distances involved, one could expect fairly high levels of collinear gene content, though possibly subject to lineage-specific losses and expansions. On the other hand, it is likely that relatively few of the assemblies would exceed draft genome quality standards with significant fragmentation of regional content across multiple scaffolds, or that only targeted sequencing and assembly of the regions of interest would have been performed. Finally, in addition to the interest in determining candidate elements in each region that are most likely to be causal for variation in the trait of interest, it is also important to be able to assess conservation of content in the surrounding regions, since the effects of linkage drag will depend on an extended neighborhood around the causal locus. It is also worth noting that a user with this background may be relatively unfamiliar with the complexities inherent in performing whole-genome sequence alignments, though they are likely to have good familiarity with the classes of genes responsible for the traits of interest to their breeding programs.
Using GCV, a simple request using a gene known to reside in the region of interest is sufficient to retrieve all genomic segments with similar gene content (regardless of whether a match is present to the query gene itself), align the genes in the returned segments to account for presence/absence, copy number and structural variation, and present the result in an intuitive interactive view for in depth exploration.
3 GCV application
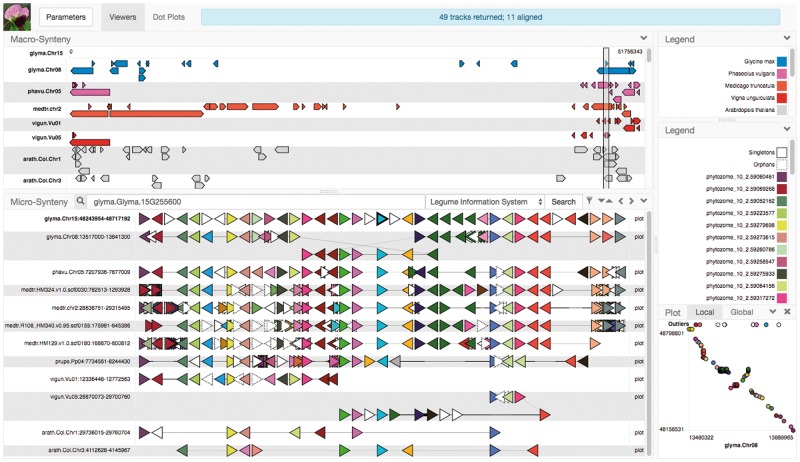
A genome context is a region considered primarily with respect to the ordering and orientation of its functionally significant elements. The primary visualization of a genome context in GCV is a horizontal track in which triangular glyphs represent genes ordered according to their occurrence in the segment, with directionality indicating orientation and intergenic distances represented by the thickness of connecting lines; see Figure 1. Colors are assigned to reflect membership in families, providing a visual overview of homologies within and between tracks. The association of each color with a gene family is presented in an interactive legend which can be used to highlight all members of a family present in the view, or to access more information about a family.
Fig. 1.
A GCV search view exhibiting copy number presence/absence/variation and structural rearrangements. (lower left) Micro-synteny relationships generated from search results aligned with the Repeat algorithm to capture inversion. (upper left) Precomputed macro-synteny blocks indicating the chromosome-scale context of the micro-synteny tracks below. (lower right) A local dot plot of the query track and a selected result track, giving a complementary view of microsynteny features. (upper right) Gene family legend with focus family highlighted (Color version of this figure is available at Bioinformatics online.)
GCV uses a service-oriented design to achieve a separation of server-side functions of content match and retrieval from client-side functions of segment alignment and display. This enables federation of data from multiple providers into a single comparative context, depending only on their adoption of a consistent classification into families. See Supplementary Material for information regarding service APIs.
The main view of GCV (Fig. 1) is built to represent a set of genome contexts in terms of their functional content. The most simple form (not pictured) is the basic view built from a user-specified set of genes, each of which serves as the focus gene of a genome context, flanked by a user-specified number of genes. A more powerful variant on this theme is presented in the search view, in which a single gene is specified as the focus of a query track and a user-specified number of flanking genes determines the track extent. Provider services are invoked to locate segments similar in content to the query, and matched segments are aligned to it based on gene family membership and ordering using modified sequence alignment algorithms. The algorithms operate on the gene family alphabet, greatly reducing the computational requirements and allowing them to compute optimal alignments within the context of the responsive user interface. All parameters for defining acceptable gene content similarity and alignment scoring may be altered by the user to suit their specific application. In addition, further algorithmic modifications make it possible to correctly align inversions and segmental tandem duplications, events occurring frequently at the scale of multi-gene segments in genomes but which are outside the scope of traditional sequence alignment algorithms (Durbin et al., 1998). See the Supplementary Material for information about alignment algorithms and track similarity services, and comparison to related tools.
Pairwise dot plots are used to represent spatial distributions of corresponding annotations and aid in identifying elements lost, gained or subjected to copy-number alterations. Local plots are composed of the gene family content of the query and result track segments, whereas global plots display all instances of genes from the families of the query track and result track across the chromosome from which the matched syntenic segment was taken. This gives a better sense for the frequency with which members of these families occur outside the matched context and can reveal wider syntenic properties, such as the existence of multiple collinear matches to a region in disparate locations on a single chromosome.
GCV can also display pre-computed macro-synteny blocks in a viewer similar to genome browser feature tracks, with the region corresponding to the current genome context highlighted. Dragging this to a different region triggers a new search, allowing the user to quickly move to regions that may be of interest for higher-level structural properties, such as breakpoints. Ordering and filtering of macro-synteny tracks is coordinated with the corresponding micro-synteny tracks below.
All GCV visualizations provide a context-menu allowing users to download high-quality images of the visualizations as well as the underlying data for further analysis. Individual elements such as genes, genomic regions and gene families also provide context-menus whose specific content can be customized to interlink with other resources providing complementary functionality.
GCV can be used as a standalone application or integrated into an existing website. It currently has instances at the Legume Information System (https://legumeinfo.org/lis_context_viewer) (Dash et al., 2016) and the Legume Federation where it is used to compare across genomes hosted at multiple member sites (https://legumefederation.org/lis_context_viewer). See the Supplementary Material for details regarding website integration.
Supplementary Material
Acknowledgements
We would like to thank our collaborators on the Legume Information System and the Legume Federation for their feedback and support; special thanks to Steven Cannon for providing the data backing the macro-synteny block view. We would also like to thank the Phytozome group for producing the genes families we have been using to enable this work.
Funding
This study was supported by U.S. Department of Agriculture Agricultural Research Service project funding for the Legume Information System and the National Science Foundation [grant number IOS-1444806 to A.D.F.].
Conflict of Interest: none declared.
References
- Bradnam K.R. et al. (2013) Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience, 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S. et al. (2016) Legume information system (legumeinfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Research, 44, D1181–D1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. et al. (1998) Biological Sequence Analysis: probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.