Abstract
In αI integrins, including leukocyte function–associated antigen 1 (LFA-1), ligand-binding function is delegated to the αI domain, requiring extra steps in the relay of signals that activate ligand binding and coordinate it with cytoplasmic signals. Crystal structures reveal great variation in orientation between the αI domain and the remainder of the integrin head. Here, we investigated the mechanisms involved in signal relay to the αI domain, including whether binding of the ligand intercellular adhesion molecule-1 (ICAM-1) to the αI domain is linked to headpiece opening and engenders a preferred αI domain orientation. Using small-angle X-ray scattering and negative-stain EM, we define structures of ICAM-1, LFA-1, and their complex, and the effect of activation by Mn2+. Headpiece opening was substantially stabilized by substitution of Mg2+ with Mn2+ and became complete upon ICAM-1 addition. These agents stabilized αI-headpiece orientation, resulting in a well-defined orientation of ICAM-1 such that its tandem Ig-like domains pointed in the opposite direction from the β-subunit leg of LFA-1. Mutations in the integrin βI domain α1/α1′ helix stabilizing either the open or the closed βI-domain conformation indicated that α1/α1′ helix movements are linked to ICAM-1 binding by the αI domain and to the extended-open conformation of the ectodomain. The LFA-1–ICAM-1 orientation described here with ICAM-1 pointing anti-parallel to the LFA-1 β-subunit leg is the same orientation that would be stabilized by tensile force transmitted between the ligand and the actin cytoskeleton and is consistent with the cytoskeletal force model of integrin activation.
Keywords: integrin, electron microscopy (EM), small-angle X-ray scattering (SAXS), leukocyte, cell adhesion, allostery, conformation
Introduction
Lymphocyte function–associated antigen 1 (LFA-1,3 integrin αLβ2) is important in leukocyte diapedesis, migration within tissues, and recognition processes requiring cell–cell adhesion. Two other integrins with the same β2-subunit, macrophage antigen 1 (Mac-1, αMβ2) and αXβ2, function as complement and danger receptors and are primarily expressed on myeloid cells (1, 2). Mutations in their common β2 integrin subunit in leukocyte adhesion deficiency result in life-threatening bacterial infections. LFA-1 binds intercellular adhesion molecules (ICAMs), a subfamily of cell surface molecules that contain tandem immunoglobulin-like domains. β2 integrins bind ligands through the αI domain, which is inserted in the β-propeller domain in the α-subunit (Fig. 1, A–C). The high-affinity, open conformation of the αI domain is stabilized by binding of an internal ligand to a binding site at the interface between the β-propeller and βI domains in the integrin head (3) (Fig. 1C). The αI and βI domains are structurally homologous and undergo similar conformational change between low-affinity closed and high-affinity open conformations (4, 5). Integrins that lack αI domains (αI-less integrins) bind external ligands at the same site to which αI-integrins bind the internal ligand. How signals are transmitted through integrin αLβ2 from the actin cytoskeleton to ICAM-1 on the surface of another cell and the structural characteristics of relay of activation between the αI and βI domains are major topics of this paper.
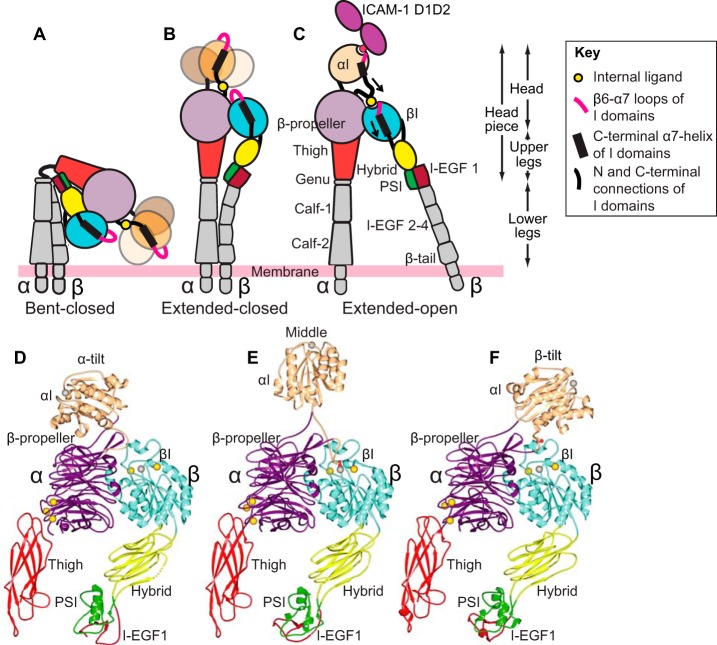
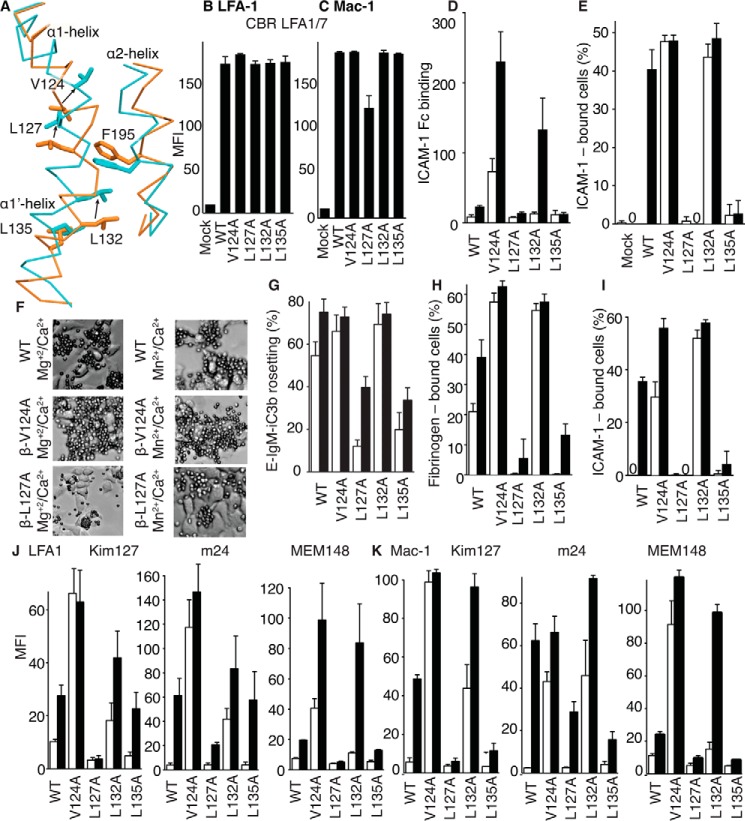
Figure 1.
Schematic of αI integrin conformational states and crystallographically determined αI-domain positions on the integrin platform. A–C, three overall conformational states of β2 integrins. Flexibility of the αI domain is schematized by making it fainter and showing three positions when its internal ligand is not engaged (A and B), consistent with findings here. D–F, ribbon diagrams of integrin headpieces with alternative positions of the αI domain seen in crystal structures with the following PDB IDs: closed αXβ2 (5ES4) (D), partially open αXβ2 (4NEH) (E), and closed αLβ2 (5E6U) (F). Spheres represent Ca2+ (gold) and Mg2+ ions (silver), with missing metal ions in D and F added as models for ease of comparison.
Integrins have three overall conformational states (Fig. 1, A–C), as shown with both αI-less integrins and the αI integrin αXβ2 (2). Visualization of αXβ2 in negative-stain EM bound to Fab fragments of allosteric, conformation-specific, activating or inhibitory antibodies, together with the effects of these Fabs on cell adhesion to the complement fragment iC3b, showed that the extended-open conformation of αXβ2 is adhesive, whereas the bent-closed and extended-closed conformations are not (6, 7). In negative-stain EM, the conformation of the headpiece can be assigned based on whether the β-subunit hybrid domain is swung away from or toward the α-subunit (Fig. 1, B and C); however, αI-domain conformation can only be inferred based on ligand-binding activity. One of the most curious features of αI integrins is how allostery can be relayed between the βI and αI domains despite evidence for marked flexibility in the αI domain (3, 8–11). One of the questions we address here is whether the αI domain adopts a defined orientation when it couples to the βI domain through its internal ligand during allostery relay. Tensile force is exerted on the cytoplasmic domain of LFA-1 when it mediates lymphocyte migration on ICAM-1 substrates (12). Thus, the specific orientation of the αI domain should be compatible with the orientation for force transmission through the integrin-ligand complex between the actin cytoskeleton and the substrate.
Whereas the β-propeller and βI domains have an extensive interface and highly stable orientation with respect to one another and may be termed a “platform,” crystal structures show that the αI domain can differ in orientation by up to 150° with respect to this platform (3, 8, 10) (Fig. 1, D–F). In each case, orientation is stabilized by specific contacts of the αI domain with other molecules in the crystal lattice, whereas in other cases, αI domain density is lacking, suggesting that it is flexible. Flexibility is shown schematically in Fig. 1 (A and B) as three different positions of the αI domain. In the first αXβ2 ectodomain crystal structure, all domains in the ectodomain were visualized, except most independent molecules in crystal lattices lacked density for the αI domain (8). The visualized αI domain was in the closed conformation, and crystal contacts stabilized a tilt toward the integrin β-subunit (Fig. 1F). In another αXβ2 ectodomain structure, the crystal lattice fortuitously stabilized the αI domain in the open conformation and in an orientation midway between the integrin α- and β-subunits (3) (Fig. 1E). The internal ligand at the C terminus of the αI domain was reshaped relative to the closed conformation and bound to its pocket between the β-propeller and βI domains (Fig. 1C). Furthermore, aside from internal ligand binding, there was little contact between the open αI domain and the remainder of the integrin head, suggesting that the αI domain would be capable of substantial tilting and rotation relative to the β-propeller/βI-domain platform. In a crystal structure of the αLβ2 headpiece, the αI domain was in the closed conformation, and crystal lattice contacts stabilized an extreme tilt toward the α-subunit (10) (Fig. 1D). These findings raise the question of whether the αI domain has a well-defined orientation, either in the closed conformation when not bound to the platform through the internal ligand or in the open conformation when bound through its internal ligand to the platform, and illustrate the importance of the use of techniques orthogonal to crystallography, such as EM and small angle X-ray scattering in solution (SAXS), as in the current study.
Our knowledge about how ligand binding and divalent metal ion binding influence the conformation of the headpiece in αI integrins is incomplete. Complexes thus far visualized are with isolated αI domains, with the ligand iC3b bound to the isolated αX I domain, with the ligand iC3b bound to the αMβ2 headpiece, or with the ligand iC3b bound to the αXβ2 ectodomain, which was prestabilized in the extended-open conformation with a combination of extension-stabilizing and opening-stabilizing Fabs (2, 13, 30). The headpiece conformation of αLβ2 has not previously been visualized in the presence of a biological ligand.
αI and βI domains contain a Mg2+ ion held in a metal ion–dependent adhesion site (MIDAS) that coordinates to external or internal ligands (2). Additionally, the βI domain MIDAS is flanked by an adjacent to MIDAS (ADMIDAS) that binds Ca2+. Mn2+ activates integrins by competing with Ca2+ at the ADMIDAS (14, 15). Mn2+ along with soluble ICAM-1 also enhances binding of antibodies to LFA-1 (15, 16) that stabilize extension and headpiece opening (6, 7). However, the effect of Mn2+ on β2 integrin conformation has not been addressed with structural studies.
Here, EM, SAXS, and mutational studies provide views of the headpiece of LFA-1 and its complex with ICAM-1 that are complementary to and extend previous structural studies on β2 integrins and address the open questions described above.
Results
Purified proteins and structural methods
We studied a headpiece fragment of LFA-1 containing the αI, β-propeller, and thigh domains in the αL-subunit and the βI, hybrid, PSI, and I-EGF1 domains in the β-subunit (schematized in color in Fig. 1 (A–C) and shown in ribbon representations in the same colors in Fig. 1, D–F). The LFA-1 headpiece was expressed in HEK293S GnTI−/− cells with C-terminal ACID-BASE coiled-coil peptides on the α- and β-subunits, respectively, followed by purification tags. After affinity purification using nickel-nitrilotriacetic acid–Sepharose, coiled-coil and tags were removed by 3C protease digestion, and the headpiece was purified to homogeneity by gel filtration. SDS-PAGE of gel filtration fractions showed bands corresponding only to the LFA-1 αL- and β2-subunits (Fig. 2A).
Figure 2.
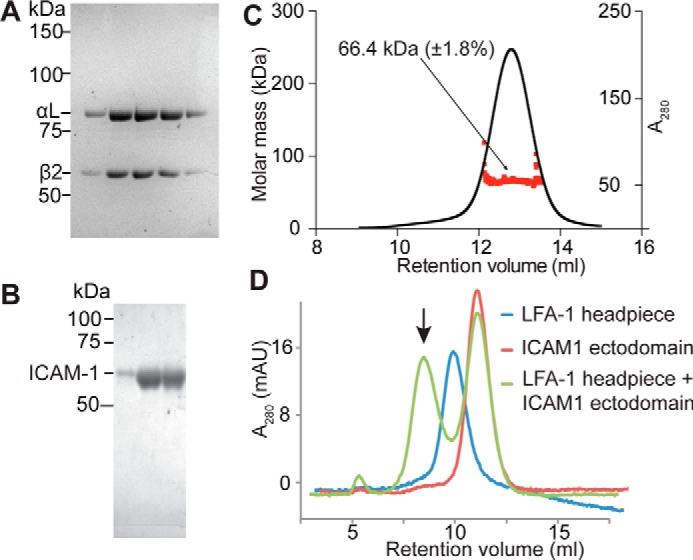
Purified protein samples. A, purified LFA-1 (αLβ2) headpiece in successive gel filtration fractions. B, purified Hi3-ICAM-1 in successive ion-exchange fractions. A and B, Coomassie Blue-stained reducing SDS-10% PAGE. C, molecular mass of Hi3-ICAM-1 D1–D5 measured by multiangle light scattering. D, size-exclusion chromatography profiles of 10 μg of LFA-1 headpiece (blue), 70 μg of ICAM-1 ectodomain (red), and their complex (green). Arrow, complex peak. mAU, milliabsorbance units.
The ICAM-1 ectodomain contains five Ig-like domains (D1–D5). To facilitate complex formation with LFA-1, we utilized D1–D5 of Hi3-ICAM-1, which contains five substitutions in the binding site for LFA-1 in D1 that increase affinity 20-fold (17, 18). Hi3-ICAM-1 with a C-terminal His tag was made in HEK293S GnTI−/− cells and purified using nickel-nitrilotriacetic acid–Sepharose, anion exchange, and gel filtration. ICAM-1 ran as a single band in SDS-PAGE (Fig. 2B). Multiangle light scattering showed a glycoprotein molecular mass of Mr 66,400 (Fig. 2C). Thus, compared with the protein mass of Mr 50,600, glycans contributed Mr 15,800. Assuming all nine potential N-linked glycosylation sites were utilized, this corresponds to Mr 1,800 per N-glycan, or 2 GlcNAc and 8.6 mannose residues per N-glycan, close to the expectation for high-mannose glycans.
Both the LFA-1 headpiece and ICAM-1 gave single, monomeric peaks in gel filtration (Fig. 2D). When the headpiece was incubated with an excess of Hi3-ICAM-1 in 1 mm Mn2+, 0.2 mm Ca2+, an early-eluting complex peak was formed, and the peak at the position of the free headpiece was depleted (Fig. 2D).
We characterized the LFA-1 headpiece, the ICAM-1 ectodomain (Ig-like domains 1–5), and their complex by both EM (Fig. 3) and SAXS (Fig. 4). We describe the results by biological unit, not by technique. Thus, we first describe the structure using both techniques of the headpiece and then ICAM-1 and then the LFA-1 headpiece–ICAM-1 complex. The LFA-1 headpiece was characterized in 5 mm Mg2+, 1 mm Ca2+ and also, to promote activation, in 1 mm Mn2+, 0.2 mm Ca2+. To maximize complex formation, we used 1 mm Mn2+, 0.2 mm Ca2+ for the LFA-1 headpiece complex with ICAM-1 and for comparison with ICAM-1 also used 1 mm Mn2+, 0.2 mm Ca2+. In EM, 6,800–10,600 particles were subjected to multireference alignment and averaging into 20 or 50 classes (Figs. S1 and S2). Representative class averages are shown in Fig. 3. SAXS data sets extended to q values ranging from 0.61 to 0.15 Å−1 (Table S1). Guinier and Kratky plots are shown in Fig. S3. Multiple ab initio molecular models constructed from the SAXS data were superimposed and averaged to calculate molecular envelopes (Fig. 4).
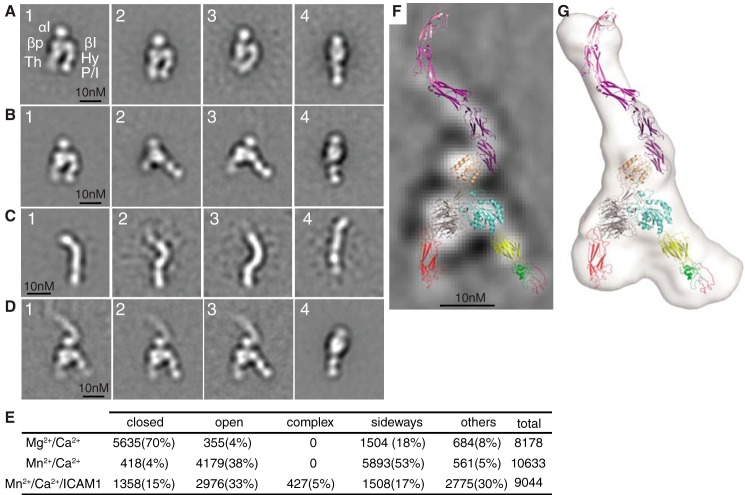
Figure 3.
LFA-1 headpiece, ICAM-1, and their complex in negative-stain EM. A–D, representative negative-stain EM averages of (A) LFA-1 headpiece in Mg2+/Ca2+ (in panel 1, the domains corresponding to each EM density are abbreviated; P/I, PSI and I-EGF1); (B) LFA-1 headpiece in Mn2+/Ca2+; (C) Hi3-ICAM-1 D1–D5; and (D) LFA-1 headpiece complex with ICAM-1 D1–D5. E, quantification of LFA-1 headpiece particles based on headpiece conformation, orientation on grid, and ICAM-1 complex formation. F, the same model of the LFA-1–ICAM-1 complex as shown in Fig. 4L is superimposed on an EM class average. G, Fig. 4L for comparison.
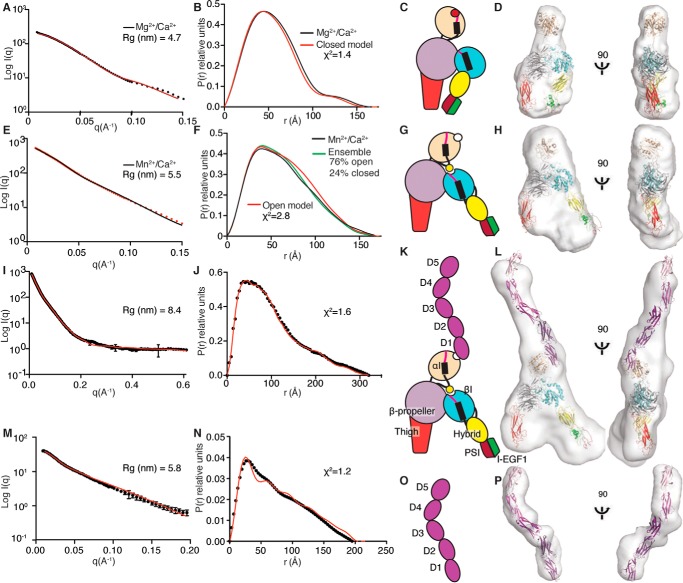
Figure 4.
Structures in solution of the LFA-1 headpiece, ICAM-1, and their complex measured by SAXS. A–P, each row corresponds to one protein sample/experimental condition, with columns from left to right of X-ray scattering (A, E, L, and M), distance distribution plot (black) with back-calculated scattering from a model (red or cyan) (B, F, J, and N), schematic diagram (C, G, K, and O), and ribbon representation of models in SAXS envelopes (D, H, L, and P). Samples were LFA-1 headpiece in Mg2+/Ca2+ (A–D) or Mn2+/Ca2+ (E–H); LFA-1 headpiece in Mn2+/Ca2+ complexed with ICAM-1 (I–L); or ICAM-1 in Mn2+/Ca2+ (M–P). The radius of gyration (Rg) was calculated using the Guinier approximation: ln(I(q)) = ln(I(0)) − Rg2 q2/3. χ2 values are with molecular models; the χ2 values for the LFA-1 headpiece in Mg2+/Ca2+ and Mn2+/Ca2+ are for the closed headpiece and open headpiece, respectively.
The LFA-1 headpiece
In Mg2+/Ca2+, EM class averages of the LFA-1 headpiece predominantly exhibited the closed-headpiece conformation (Fig. 3A, panels 1–3, representing 70% of all particles). Clear densities were present for the αI, β-propeller, and thigh domains in the αL-subunit and the βI, hybrid, and PSI/I-EGF1 unit in the β-subunit (Fig. 3A, panel 1). Only 4% of particles showed the open conformation, and 18% of particles oriented on their sides on grids, preventing classification as open or closed (Fig. 3A, panel 4). Side views contained clear density for the αI and β-propeller domains in the αL-subunit and two leg domains that appear to correspond to the hybrid and PSI/I-EGF1 unit. The presence of at least four distinct densities in side views demonstrated that side views contained both subunits, because each subunit contained only three distinct densities. LFA-1 appears to have a greater tendency than other studied integrins, such as αXβ2, to electrostatically adsorb to grids on its side, as seen previously with the complete LFA-1 ectodomain (6).
In activating conditions in Mn2+/Ca2+, 38% of particles exhibited the open-headpiece conformation (Fig. 3B, panels 2 and 3). Some particles remained closed (4%; Fig. 3B, panel 1). Furthermore, a higher proportion of particles than in Mg2+ oriented on their sides (53%; Fig. 3B, panel 4). Additionally, a large proportion of particles adopted an oblique orientation, intermediate between the view on the side and the view with the integrin legs in the plane of the grid (Fig. 3B, panel 2). Obliqueness was evidenced by the shorter length of the thigh domain and lack of separate density for the β-propeller and βI domains in the head (Fig. 3B, panel 2) compared with planar views (Fig. 3, A (panels 1–3) and B (panels 1 and 3)). We speculate that in Mn2+, the αI domain orients more out of the headpiece plane than in Mg2+, giving rise to oblique orientations and a higher proportion of side orientations on EM grids.
SAXS also showed that in Mg2+/Ca2+, the LFA-1 headpiece was closed (Fig. 4, A–D). Fits of closed LFA-1 headpiece models to the SAXS envelope in Mg2+ showed that the middle position of the αI domain (Fig. 1E) fit better (χ2 = 1.8) than α- and β-tilt positions (χ2 = 2.6–2.8), and manual fitting of the αI domain was only slightly better than the middle tilt (χ2 = 1.4).
In Mn2+/Ca2+, the headpiece was substantially more open, with an increase in the proportion of interatomic distances in a range from 75 to 140 Å and an increase in radius of gyration (Rg) to 5.5 nm (Fig. 4, E–H) compared with values in Mg2+/Ca2+ (Fig. 4, A–D). Correspondingly, the ab initio molecular envelope of the headpiece was broader in Mn2+ than in Mg2+ (Fig. 4, compare H and D). A model of the open headpiece of LFA-1, made using the swung-out orientation of the hybrid domain from the open crystal structure of integrin αIIbβ3 (4), was used to fit the SAXS data in Mn2+. The fit was reasonable, although the model showed greater separation between the upper integrin legs than implied by the distance distribution data or the ab initio molecular envelope (Fig. 4, F and H). Fits to the open headpiece with different αI domain orientations showed reasonable fits to a manually docked αI domain (Fig. 4F, χ2 = 2.8), reasonable fits to middle and β-tilts and to an orientation optimized for complexes with ICAM-1 described below (3.1–3.2), and a poorer fit to an αI α-tilt (χ2 = 3.6). As the EM data suggested both closed- and open-headpiece conformations were present in Mn2+, we used nonlinear regression analysis sampling of the back-calculated P(r) graphs of the open and closed LFA-1 headpiece models to approximate the P(r) graph generated from the experimental Mn2+ SAXS data. The best approximation, with a correlation coefficient of 0.9994, was achieved using an ensemble containing 24% closed-headpiece and 76% open-headpiece conformations (Fig. 4F). The results suggest that Mn2+ by itself stabilizes the open-headpiece relative to the closed-headpiece conformation but is not sufficient to induce a complete shift to the open conformation.
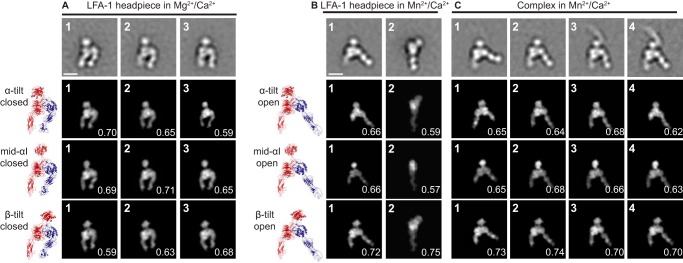
We also used EM to examine αI domain orientations. Filtered molecular envelopes at 20 Å resolution from molecular models with three alternative αI domain orientations (Fig. 1, D–F) and either the closed or open headpiece were used to calculate regularly spaced projections at 2° intervals and were cross-correlated with EM class averages using SPIDER (19) (Fig. 5). In Mg2+, differences in αI domain position were apparent in class averages, and class averages were found that cross-correlated best with αI domains in α-tilt, middle, and β-tilt positions (Fig. 5A, panels 1, 2, and 3, respectively). By contrast, in Mn2+, the best cross-correlation with all representative class averages was with the β-tilted αI domain (Fig. 5, B (panels 1 and 2) and C (panels 1 and 2)). Thus, it appears that in Mn2+, the αI domain is more constrained than in Mg2+/Ca2+ and is more proximal to the βI domain, to which it couples by binding of the internal ligand in the αI domain to the βI domain (Fig. 1C).
Figure 5.
Comparison of αI domain orientation in Mg2+/Ca2+, in Mn2+/Ca2+, and in ICAM-1 complexes in Mn2+/Ca2+. A–C, representative class averages in the indicated conditions. For ICAM-1 complexes in Mn2+/Ca2+, class averages both with and without binding to ICAM-1 are shown. The top row shows the class averages. Rows 2–4 show the best-correlating headpiece projections and their cross-correlation scores with the headpiece models in ribbon representation shown to the left. For complexes with ICAM-1, the ICAM-1 moiety was masked out before cross-correlation. Models show three different αI domain tilts defined in Fig. 1 (D–F) with the closed headpiece (A) or the open headpiece (B and C). Scale bars, 10 nm.
The ICAM-1 ectodomain
Hi3-ICAM-1 appeared in EM as an asymmetric, rodlike structure, ∼20 nm in length and 3 nm in width (Fig. 3C and Fig. S2B). Discrete densities corresponding to single Ig-like domains were often present. Many class averages, including the most populous one (Fig. 3C, panel 1), displayed a bend about two-fifths along the D1–D5 ectodomain axis.
SAXS of Hi3-ICAM-1 showed an Rg of 5.8 nm and an extended molecular envelope with two kinks (Fig. 4, M–P). ICAM-1 has been crystallized as either D1-D2 or D3–D5 fragments (20–22); however, D1–D5 of ICAM-5 are 50% identical in amino acid sequence to ICAM-1, and superposition of ICAM-1 D1-D2 and D3–D5 fragments on a D1–D4 crystal structure of ICAM-5 (23) allowed us to build an ICAM D1–D5 model. The model fit into the molecular envelope well, at a low χ2 value = 1.2 for the experimental and back-calculated solution scattering data (Fig. 4, M–P). The largest kink observed in both ICAM-1 and ICAM-5 crystal structures and in an early EM study that mapped antibody binding sites is at the D3–D4 junction (21, 23, 24). The kink helps define the orientation of D1–D5 in the SAXS envelope (Fig. 4, O and P); flipping the end-to-end orientation resulted in protrusion of D1 from the SAXS envelope and a poor fit. The overall kinked, S-shaped SAXS structure is in agreement with the bent structure observed here in EM (Fig. 3C) and in early EM structures (24, 25).
The LFA-1–ICAM-1 complex
EM of the LFA-1 headpiece–Hi3-ICAM-1 complex peak from gel filtration in Mn2+/Ca2+ (Fig. 2D) showed class averages corresponding to ICAM-1–LFA-1 complexes, headpiece alone, and ICAM-1 alone (Fig. 3D). Dissociation of receptor-ligand complexes is a common problem in negative-stain EM and may relate to the low protein concentrations required by the technique. Remarkably, all three class averages of the complex showed nearly identical ICAM-1–LFA-1 orientations (Fig. 3D, panels 1–3). The rod-shaped ICAM-1 molecule bound to the face of the αI domain adjacent to the β-subunit rather than the face of the αI domain most distal from the head. Crystal structures show that the LFA-1 αI domain binds to an edge β-strand in D1 of ICAM-1 (5), as shown in the ribbon diagrams in Fig. 3 (F and G). Thus, the αI domain must be tilted toward the β-subunit. This inference is independently supported by cross-correlations of the headpiece moiety from complexes with Hi3-ICAM-1, which correlate better with the β-tilt position than the other two positions tested (Fig. 5C, panels 3 and 4). Density for individual domains of ICAM-1 was not evident in complexes, but the length of the rod of 16 nm suggests that D1–D4 were visualized, that D4 was fainter, and that D5 was averaged out. ICAM-1 D1–D4 extended along a line that was almost anti-parallel to the line made by the three densities in the integrin β-subunit that correspond to the βI domain, the hybrid domain, and the PSI plus I-EGF1 domain that attach to the same end of the hybrid domain (compare Fig. 3D (panels 1–3) with Fig. 1C). In headpiece complexes with ICAM-1, the hybrid and PSI/I-EGF1 domains were clearly swung away from the integrin α-subunit in the open conformation (Fig. 3D, panels 1–3).
For SAXS, the LFA-1–ICAM-1 complex in 1 mm MnCl2 and 0.2 mm CaCl2 was isolated by gel filtration at a ∼20-fold higher concentration than used for EM and further concentrated to 3.2 mg/ml for data collection. The Rg of the complex was 8.4 nm. Averaging of ab initio GASBOR models showed a molecular envelope with clear density for the ICAM-1 and LFA-1 headpiece moieties (Fig. 4, I–L). The open-headpiece moiety fit the envelope well with a solvent-filled gap between the knee-proximal α-subunit thigh and β-subunit PSI and I-EGF1 domains (Fig. 4L). Comparison with the molecular envelope of the headpiece alone in Mn2+ (Fig. 4H), which appeared to correspond to an ensemble of closed and open conformations, suggested that binding to ICAM-1 completely shifted the headpiece into the open conformation. A thin, elongated protrusion of the molecular envelope clearly corresponded to ICAM-1. Using the model of ICAM-1 D1–D5 described above, the structure of the LFA-1 αI domain bound to ICAM-1 D1-D2 (5), and the model of the LFA-1 headpiece in the open conformation, we constructed a model of the LFA-1–ICAM-1 complex. The model fit the molecular envelope well, and its back-calculated scattering showed excellent fit to the experimental SAXS data with a χ2 value of = 1.6 (Fig. 4J). In SAXS, the long axis of ICAM-1 was anti-parallel to the integrin β-subunit (Fig. 4L), just as seen in EM. The ICAM-1–integrin binding orientations seen in SAXS and EM are remarkably similar, as shown in the comparison in Fig. 3 (F and G). These results establish the overall structure of LFA-1 bound to its physiological ligand, ICAM-1, and that binding results in a well-defined orientation of ICAM-1 with respect to the integrin. Moreover, the particular orientation found aligns ICAM-1 anti-parallel to the LFA-1 β-subunit leg, as would occur in the presence of tensile force exerted physiologically when actin retrograde flow exerts force on the integrin that is resisted by ICAM-1 on the surface of another cell.
Mutational studies
The above experiments suggested that binding of LFA-1 to ICAM-1 is associated with opening of the integrin headpiece. We used mutations to further support this conclusion and to relate it to movement of residues in the βI domain α1 and α1′ helices that alter position upon opening of the β2 βI domain (3). During opening, axial displacement of the α1 and α1′ helices is associated with displacement of Val124 by Leu127 (Fig. 6A), as shown in a structure of partially open integrin αXβ2 (3). Val124 is more buried in the closed than partially open conformation, whereas Leu127 is more buried in the partially open than closed conformations (Fig. 6A); therefore, the mutations V124A and L127A are predicted to stabilize the open and closed conformations, respectively. We also mutated residues Leu132 and Leu135 to Ala; however, their effects were difficult to predict in advance because both residues are similarly buried in the partially open and closed conformations (Fig. 6A). Studies on integrin αIIbβ3 showed that movements in the region corresponding to residues 124 and 127 occur earlier in the opening process than movements in the region corresponding to residues 132 and 135 (26); thus, the positions of residues 132 and 135 in partially open β2 integrins are not predictive of their positions in the open state. We hypothesized that Leu132 and Leu135 would move during complete headpiece opening and that their mutation might differentially stabilize the closed and open conformations. Mutant β2-subunits were co-transfected with αL- or αM-subunits to study ligand-binding function of LFA-1 and Mac-1, respectively, in HEK293T cells. All mutants were expressed at levels comparable with those of WT (Fig. 6, B and C).
Figure 6.
Effects of mutations of putative ratchet residues in the βI-domain α1 and α1′ helices on LFA-1 and Mac-1 ligand binding and epitope exposure. A, alteration in position of the α1 and α1′ helices in partially open (orange) and closed (cyan) β2 βI domain structures shown after superposition on the βI domain. Backbone is shown as Cα trace, and key side chains are shown in stick representations. The α1 helix continues with a short break into the α1′ helix, and the neighboring α2 helix is also shown. B–K, all experiments utilized 293T cells co-transfected with the α- and β-subunits of LFA-1 or Mac-1 with WT or mutant β-subunits. Results are averages ± S.D. (error bars) of replicates from at least three independent experiments with three different replicates (each transfectant seeded in three different wells). B and C, expression of mutants measured with antibody CBR LFA-1/7 to the β2-subunit. D, binding of ICAM-1-Fcα chimera measured with fluorescent anti-IgA on LFA-1 transfectants in flow cytometry expressed as mean fluorescence intensity (MFI). E, adhesion of LFA-1 transfectants to wells coated with ICAM-1-Fcα chimera. F, representative phase-contrast photomicrographs showing rosetting of iC3b-sensitized erythrocytes (smaller cells) with Mac-1 293T transfectants (larger cells). G, quantitation of rosetting of erythrocytes sensitized with iC3b to adherent Mac-1 transfectants. H and I, adhesion of Mac-1 transfectants to wells coated with fibrinogen (H) or ICAM-1-Fcα chimera (I). J and K, Kim127, m24, and MEM148 epitope exposure on LFA-1 (J) or Mac-1 (K) transfectants measured by MFI of antibody staining normalized by CBR LFA-1/7 MFI.
Mutations of all four residues had clear effects. As measured in HEK293T LFA-1 transfectants, the V124A mutation enhanced binding of ICAM-1-Fc measured by flow cytometry (Fig. 6D) and activated cell adhesion to ICAM-1 substrates (Fig. 6E). To evaluate the commonality of the relay mechanism among β2 integrins, we also evaluated mutations in Mac-1 (αMβ2). The β2 V124A mutation in Mac-1 transfectants enhanced rosetting with iC3b opsonized erythrocytes (Fig. 6, F and G) and adhesion to fibrinogen (Fig. 6H) and activated adhesion to ICAM-1 substrates (Fig. 6I). The L127A mutation had the opposite effect on transfectants expressing LFA-1 or Mac-1. L127A eliminated manganese-stimulated LFA-1 binding and adhesion to ICAM-1 (Fig. 6, D and E) and decreased or abolished Mac-1 rosetting with iC3b opsonized erythrocytes and adhesion to fibrinogen and ICAM-1 substrates (Fig. 6, F–I). L132A strongly activated LFA-1 transfectant adhesion to ICAM-1 and had much less effect on binding to soluble ICAM-1-Fc, which reflected a trend shared with the V124A mutation (Fig. 6, D and E). L132A similarly activated Mac-1 function in all three assays (Fig. 6, F–I). In contrast, the L135A mutation inhibited manganese-stimulated binding and adhesiveness of LFA-1 and Mac-1 in all assays (Fig. 6, D–I).
We correlated these mutational effects on ligand binding function of the LFA-1 and Mac-1 αI domains with exposure of epitopes that measure β2 integrin extension (Kim127 epitope) and βI domain opening (m24 and MEM148 epitopes). Activating mutations V124A and L132A greatly increased LFA-1 and Mac-1 extension measured with the Kim127 epitope in both Mg2+ and Mn2+ (Fig. 6, J and K). V124A and L132A also greatly enhanced headpiece opening measured with m24 and MEM148 in both Mg2+ and Mn2+, with the MEM148 epitope showing less exposure than m124 by the L132A mutation in Mg2+ but comparable exposure in Mn2+ (Fig. 6, J and K). L127A and L135A mutations decreased exposure of all three epitopes in both Mg2+ and Mn2+ and in both LFA-1 and Mac-1 (Fig. 6, J and K). Thus, mutations demonstrate that ligand binding to the αI domain, including ICAM-1 binding to the αI domain of LFA-1, is regulated by movement of residues in the βI domain α1 and α1′ helices and is correlated with βI domain opening and integrin extension.
Discussion
Here, we have used techniques orthogonal to crystallography to investigate how the αI domain of LFA-1 is linked to the other two domains in the integrin head, the influence of the activating metal ion Mn2+ and the ligand ICAM-1 on the orientation of the αI domain and the conformation of the headpiece. Moreover, we investigated the relevance of movements of the α1 and α1′ helices in the βI domain to these linkages between the αI domain and the conformation of the headpiece and the ectodomain. Perhaps most strikingly, we have visualized how the headpiece of integrin LFA-1 binds to its physiological ligand, ICAM-1, and demonstrated a remarkably well-defined orientation in EM as well as in solution that we discuss in relation to the pathway for force transmission through the integrin between ICAM-1 and the actin cytoskeleton.
The LFA-1 headpiece and ICAM-1
Previous crystal structures of αXβ2 and αLβ2 have revealed three markedly different αI domain orientations (Fig. 1, D–F) (3, 8, 10). Among three distinct crystal lattices in one study with a total of 10 independent integrins, only two integrins showed density for the αI domain (8). Variations in lattice dimensions altered αI domain lattice contacts and orientation whether the αXβ2 ectodomain was associated with an open, internal ligand-bound αI domain (3) or a closed αI domain (8). Reconstruction of the negatively stained αMβ2 ectodomain suggested a distinct orientation for the αI domain from that seen in one αXβ2 lattice (9), which, given the marked variation in αI domain orientation in distinct lattice environments, is hardly surprising. In basal conditions in Mg2+, we found here that the αLβ2 headpiece was predominantly in the closed conformation, as shown by both EM and SAXS. In the closed conformation, we found class averages that differed in αI domain position relative to the head and that were consistent with crystal structures in which the αI domain is tilted toward the α-subunit or β-subunit side of the headpiece or in a middle position. In SAXS, we saw no evidence for broadening of the envelope for the αI domain, and it was closest to the middle position. An interpretation consistent with all of the above studies is that in the closed conformation, 1) the αI domain has a preferred orientation near the middle of the variation seen among crystal lattices, 2) there is enough variation in αI domain position among integrin ensembles to prevent resolution in crystal structures where lattice contacts are absent, and 3) dynamic motion of the αI domain relative to the remainder of the headpiece is sufficiently limited so that an average position for the center of mass of the αI domain in solution is evident in SAXS molecular envelopes. In the closed conformation of the αI domain, its flexible connection to the α-subunit β-propeller domain in which it is inserted may enable it to sample multiple orientations to bind external ligand, as well as multiple orientations in which the αI domain may reshape into the open conformation, and its internal ligand can bind to its pocket in the integrin head between the β-propeller and βI domains.
In activating conditions in Mn2+, EM showed that the LFA-1 headpiece had predominantly the open conformation, with the hybrid, PSI, and I-EGF1 domains swung away from the α-subunit. SAXS also showed opening, and the best fit to the scattering data was obtained with an ensemble containing 76% open and 24% closed conformations. This was consistent with ab initio molecular envelopes that showed greater width between the α- and β-subunit legs than in Mg2+ and less width than expected for an open-headpiece model or seen with the LFA-1 headpiece in Mn2+ when complexed with ICAM-1. In Mn2+, EM class averages of the headpiece alone or the headpiece bound to ICAM-1 showed better correlation to a crystal structure with an αI domain tilted toward the β-subunit. In Mn2+, SAXS results on the headpiece alone were consistent with either a middle αI domain position or tilt toward the β-subunit. These results support tilting toward the β-subunit of the αI domain in the open-headpiece conformation, which is consistent with binding the internal ligand at the C terminus of the αI domain to a pocket at the interface between the βI and β-propeller domains (3).
SAXS showed a relatively well-defined molecular envelope for the five Ig-like domains in the ectodomain of ICAM-1, with a clear kink. A kink was also evident in EM about two-fifths of the way along the rodlike length of the five tandem Ig-like domains (i.e. either between domains 2 and 3 or between domains 3 and 4). The bend had previously been localized to between domains 3 and 4 by antibody mapping in EM (24) and between domains 3 and 4 in crystal structures of D3–D5 of ICAM-1 and D1–D4 of ICAM-5 (21, 23, 27). A model of D1–D5 of ICAM-1 fit the SAXS molecular envelope well and confirmed the kink position. Crystal structures of ICAM-1 have shown variation in angle between D1 and D2 in lattices of up to 17° (20) and between D3 and D4 of up to 18° (21). This interdomain flexibility is far less than the ∼150° variation in orientation seen for the αI domain among integrin structures (10). The well-defined molecular envelope of ICAM-1 in SAXS suggests that in solution, there are preferred orientations between the tandem Ig-like domains in ICAM-1.
The LFA-1–ICAM-1 complex
Given the potential range of motion found in crystal lattices of the integrin αI domain relative to the remainder of the integrin head, and between tandem Ig-like domains in ICAM-1, we found remarkably well-defined orientations between the ICAM-1 moiety and the headpiece moiety in complexes. Crystal structures of the isolated LFA-1 αI domain bound to D1-D2 fragments of ICAM-1, ICAM-3, and ICAM-5 have all shown a well-defined interface of the αI domain bound to D1 of the ICAM fragment, with the MIDAS Mg2+ ion of the αI domain at the center of the interface (5, 28, 29). The Mg2+ ion coordinates a Glu side chain in an edge β-strand in D1 of ICAM-1. In agreement, our EM class averages show that the αI domain binds to one side of the rod-shaped ICAM-1 molecule and at a position on the rod consistent with binding to D1 (Fig. 3, H and I). The αI domain appears as a round density in EM, and thus its orientation is impossible to assign based on the shape of its density. However, because it is known that the MIDAS-bearing face of the αI domain binds D1 of ICAM-1, and the orientation of the αI domain complex with D1 of ICAM-1 is clearly visible in each LFA-1–ICAM-1 complex class average, the αI domain with its MIDAS-bearing face must be markedly tilted toward the β-subunit side of the integrin head. In other words, while the αI domain is inserted in the β-propeller domain, it is tilted toward the βI domain interface with the β-propeller domain, to which its internal ligand is bound in the open conformation of the αI domain (3). The tilt toward β of the αI domain in the complex with ICAM-1 and in the open LFA-1 headpiece in Mn2+ in EM class averages was independently confirmed by cross-correlation with a crystal structure with a β-tilted αI domain.
The LFA-1 headpiece and ICAM-1 moieties were also well-resolved in complexes with ICAM-1. The headpiece was fully open, with the leg of the β-subunit swung away from the α-subunit, in both EM class averages and SAXS. In contrast, the headpiece in Mn2+ alone was best represented as an ensemble with about 76% open and 24% closed conformations. The open-headpiece conformation of a β2 integrin remains to be defined with a crystal structure, but swing-out of the hybrid domain with the PSI and I-EGF1 domain matched well a model based on integrin αIIbβ3 (4). The ICAM-1–stabilized open headpiece resembled the open headpiece previously seen with a minority of integrin αXβ2 and αLβ2 ectodomain class averages in Mg2+ (6), the open αXβ2 ectodomain conformation stabilized with activating Fab (7), the open αXβ2 ectodomain simultaneously bound to activating Fabs and the ligand iC3b (13), and the open headpiece visualized in complexes of the ligand-bound αXβ2 headpiece (30). The orientation of the rodlike density for ICAM-1 relative to the integrin headpiece was remarkably similar among three independent EM class averages and was uniform enough in solution to give a rodlike shape, and the orientation in solution obtained in ab initio SAXS models was essentially identical to that seen in EM, as shown by superposition of the model that fit the SAXS envelope with an EM class average (Fig. 3, F and G). The uniform orientation seen in three EM class averages confirms that the five Ig-like domains of ICAM-1 have a well-defined average position, as independently shown with SAXS of ICAM-1 alone.
We can infer that the LFA-1 αI domain in the LFA-1 complex with ICAM-1 is activated. There is no direct evidence at the resolution of the EM and SAXS studies here that the LFA-1 αI domain is in the open conformation or that its internal ligand is bound to its pocket at the β-propeller–βI domain interface. However, it is known that the open αI domain binds with ∼1,000-fold higher affinity than the closed αI domain to ICAM-1 (18, 31); that stabilizing the αLβ2 headpiece in the open conformation with activating Fabs to the β-subunit increases αI domain affinity for ICAM-1 by ∼1,000-fold (18); and that mutation of LFA-1 αI domain internal ligand residue Glu-310 or antagonists directed to the pocket that binds the internal ligand abolish activation of LFA-1 adhesiveness, including by activating Fabs to the β-subunit (32–34). This previous evidence, together with the evidence here that the LFA-1 headpiece is open with its β-leg swung out when it binds ICAM-1, suggests with great confidence that in the complex with ICAM-1 visualized here, the ICAM-1–bound αI domain is open and its internal ligand is bound to its pocket at the interface between the β-propeller and βI domains.
αI domain orientation and linkage to the headpiece
We may further deduce that binding of the internal ligand of the open αI domain to its pocket enforces a highly preferred orientation of the αI domain in which it is tilted toward its internal ligand binding pocket at the β-subunit side of the head (Figs. 1C and 3 (F and G)). This contrasts with the middle orientation of the αI domain (Fig. 1B) in a structure of the αXβ2 ectodomain in which the crystal lattice enforced the open αI domain conformation and in which the αI domain internal ligand bound to its pocket and the βI domain was in an intermediate, partially open conformation (3). The αI domain was held away from the remainder of the head by lattice contacts, and its α7 helix was largely unwound to span the distance to the internal ligand-binding pocket. It was therefore predicted that the open αI domain should be able to substantially rotate while remaining allosterically engaged (3). This prediction is consistent with the middle αI domain orientation observed in the latter structure and the β-tilt of the αI domain in the ICAM-1–engaged αLβ2 headpiece structure observed here. The unwinding of the αI domain α7 helix must have been energetically unfavorable, and imposed by the crystal lattice, which held the αI domain 15 Å farther away from the binding pocket for the internal ligand than would have been possible without α7 helix unwinding (3). It is reasonable to assume that in the LFA-1 complex with ICAM-1, the αI domain α7 helix remains helical, which would bring it ∼15 Å closer to the internal ligand binding pocket and tilt the αI domain toward the β-subunit, exactly as observed here in the LFA-1–ICAM-1 complex.
We previously visualized in EM a complex between the ectodomain of integrin αXβ2, three activating Fabs, and fragments of complement component C3 that are ligands of αXβ2, iC3b, and C3c (13). The αXβ2 αI domain bound to a specific site on the “key ring” moiety of C3c, which lies predominantly in one plane. Both the αXβ2 ectodomain and iC3b/C3c had planar orientations on the EM grid. In some class averages, the orientation of C3c was flipped, whereas interaction of the αI domain was maintained. We speculated that the orientation of the iC3b/C3c key ring and extended-open integrin planes might have been more perpendicular than co-planar in solution and that adsorption on the grid forced them to be co-planar and resulted in occasional rotation of the αI domain C3c moiety relative to the remainder of the integrin ectodomain (13). This result did not imply lack of a preferred orientation of the ligand-bound αI domain in solution. In the case of binding of ICAM-1 to the LFA-1 headpiece, our SAXS data suggest that in solution, the ICAM-1 rod is in the same plane as the LFA-1 headpiece. Thus, there should be no tendency for ICAM-1 to flip during adsorption to the substrate, consistent with the excellent agreement among distinct EM class averages of ICAM-1–LFA-1 complexes and the SAXS molecular envelope.
Our mutational data demonstrate that movements in the α1 and α1′ helices of the βI domain are coupled to adhesiveness, ligand binding, headpiece opening, and extension of integrins LFA-1 and Mac-1 on the cell surface. Shifts in these helices were noted in a crystal structure of an intermediate conformation of the αXβ2 βI domain (3). As predicted by the structure and confirmed by mutation of αXβ2, β2 V124A and L127A mutations activated and inhibited, respectively, ligand binding and opening and extension of αXβ2. Here, we extended these results to LFA-1 and Mac-1. Furthermore, we tested two residues that are more C-terminal in the α1′ helix, Leu132 and Leu135. This region underwent less movement in the intermediate conformation but is expected to be involved in more extensive, ratchet-like exchanges in position in an open βI domain conformation. We found that mutations L132A and L135A activated and inhibited, respectively, binding to soluble ICAM-1 and adhesiveness to ICAM-1 of LFA-1, adhesiveness to ligands including rosetting with iC3b-sensitized erythrocytes of Mac-1, and exposure of epitopes associated with headpiece opening and ectodomain extension on cell surfaces of both LFA-1 and Mac-1. These results suggest that movements may occur throughout the length of the α1 and α1′ helices in the open conformation of the β2 I domain and predict that burial of Leu132 is important to stabilize the open conformation, whereas burial of Leu135 (perhaps in the same ratchet pocket) is important to stabilize the closed conformation of the β2 I domain. Overall, the mutational results extend our observations on purified fragments to LFA-1 on cell surfaces and show that high-affinity binding to ICAM-1, headpiece opening, and extension are linked and that this linkage requires specific conformational changes in the LFA-1 βI domain.
The anti-parallel orientation of ICAM-1 and the integrin β-leg in complexes visualized here is relevant to force-dependent activation by the actin cytoskeleton (35). Actin polymerization and myosin-dependent filament contractility apply force through adaptor proteins to integrin cytoplasmic domains, which is then transmitted through integrins to ligands, which resist the applied force as a consequence of anchorage on the cell surface or in the extracellular matrix (36). Tensile force measurements in the β-subunit cytoplasmic domain of LFA-1 show that tension generated by the actin cytoskeleton is required to induce and stabilize adhesion to ICAM-1 (12). Tensile force stabilizes more extended and open integrin conformations because of their increased length in the direction of force transmission (37, 38). Our study shows that the ICAM-1–LFA-1 headpiece complex has a preferred orientation in which the pathways for force transmission through ICAM-1 and the upper β-subunit leg are parallel to one another. As force is transmitted through receptors and their ligands, force balance requires that the pathway for force transmission become aligned with the direction of force application. Flexible domains and flexible linkages between ectodomains and the plasma membrane will straighten out, like links in a tow chain, to align the receptor-ligand complex with the points where force is applied by the cytoskeleton to the integrin and resisted by the ligand. Fluorescence polarization microscopy shows that LFA-1 is aligned in the same direction as, and is tilted by, the force applied by actin retrograde flow (39). The structure of the ICAM-1—LFA-1 complex shows that ICAM-1 and the integrin β-leg are pre-aligned in an orientation that requires little readjustment after force application, which makes the complex well-suited for stabilization by cytoskeletal force of the high-affinity, extended-open integrin conformation.
Experimental procedures
Integrin αLβ2 headpiece protein and high-affinity ICAM-1 ectodomain
αL- and β2-subunit cDNAs encoding mature residues Tyr1–Asn745 and Gln1–Glu460, respectively, were expressed in HEK293S N-acetylglucosaminyltransferase I-negative (GnTI−/−) cells and purified by His tag affinity and gel filtration as described (10). ICAM-1 ectodomain was expressed in stably transfected HEK293S GnTI−/− cells and purified by His tag affinity, anion-exchange, and Superdex S200 gel chromatography as described (18). The Mg2+/Ca2+ buffer condition was 20 mm Hepes, pH 7.4, 150 mm NaCl, 5 mm MgCl2, and 1 mm CaCl2. The Mn2+ buffer condition was 20 mm Hepes, pH 7.4, 150 mm NaCl, 1 mm MnCl2, and 0.2 mm CaCl2.
Synchrotron SAXS measurements
Purified LFA-1 headpiece in 5 mm Mg2+, 1 mm Ca2+ at 1 mg/ml and purified LFA-1 headpiece, ICAM-1 ectodomain, and their complex in 1 mm Mn2+, 0.2 mm Ca2+ at concentrations of 1.4, 0.85, and 3.2 mg/ml, respectively, were shipped on ice to the synchrotron and incubated at ambient temperature for 15–20 min before X-ray solution scattering measurements were performed. Small and wide angle X-ray scattering measurements were collected at beam line X9 at the Brookhaven National Synchrotron Light Source (Upton, NY). The detector was a high sensitivity 300K Pilatus, at a sample-to-detector distance of 3.5 m. Samples were passed through a flow capillary, collecting data at 20-s exposures in triplicate at the specified concentrations. Guinier analysis showed no signs of radiation damage or aggregation. I(0) and the pair distance distribution function P(r) were calculated by circular averaging of the scattering intensities I(q) and scaling using the software GNOM (40). LFA-1 and ICAM-1 alone scattering data were processed to a q(A−1) of 0.15 and 0.2, respectively, and 12 ab initio models were generated using DAMMIF (41). The LFA-1–ICAM-1 complex scattering data were processed to a q(A−1) of 0.61. GASBOR (42), which can utilize wide-angle scattering, was used to generate 12 ab initio models. Models were superimposed and averaged using the program DAMAVER (43). The theoretical solution scattering of the crystal structures of LFA-1 headpiece in the closed and open states, ICAM-1, and the open LFA-1 headpiece–ICAM-1 complex were calculated using CRYSOL (44).
Molecular models
LFA-1 headpiece models utilized PDB entry 5E6U (10). Because the LFA-1 headpiece crystal structure lacks a thigh domain, that of αXβ2 was added from PDB entry 5ES4. Three different orientations of the αI domain were utilized: PDB entries 5E6U for the βI-tilt, 4NEH for the mid-αI tilt, and 5ES4 for the αI-tilt. To model the ICAM-1 D1–D5 complex with the αL I domain, crystal structures of ICAM-1 D3–D5 (27) and the αL I domain–ICAM-1 D1-D2 complex (5) were superimposed onto the ICAM-5 D1–D4 structure (23). The ICAM-1 D1–D5/αL I domain model was used to model the LFA-1 headpiece–ICAM-1 D1–D5 structure. To obtain the model of the ICAM-1 D1–D5 ectodomain alone, the αL I domain was removed from the ICAM-1 D1–D5/αL I domain structure.
Swing-out motion of the hybrid domain relative to the β3 I domain was modeled into the β2-subunit (4) as follows. Comparison of the closed β2- and β3-subunits after superimposing only the βI domains shows that the closed β2 and β3 hybrid domain positions differ by an in-plane angle of ∼20º, with the β2 hybrid domain more swung in. Therefore, the same amount of rotation as seen between the closed and open conformations of β3 was applied to the β2 hybrid domain to swing it out.
Two models, 1) the ICAM-1 D1–D5 model that contains the αL I domain and 2) the open LFA-1 headpiece lacking the αL I domain, were simultaneously docked to the SAXS profile using the FOXSDOC server. The position of the LFA-1 headpiece was then manually adjusted, which decreased the χ value from 1.7 to 1.6.
Negative-stain EM of LFA-1 headpiece, ICAM-1 D1–D5, and their complex
Within 2 h after Superdex 200 10/30 column purification of ∼30 μg of the LFA-1 headpiece in 20 mm Hepes, pH 7.0, 150 mm NaCl, 5 mm MgCl2, and 1 mm CaCl2; the ICAM-1 ectodomain; or the LFA-1 headpiece alone or its complex with the ICAM-1 ectodomain in 20 mm Hepes, pH 7.0, 150 mm NaCl, 1 mm MnCl2, and 0.2 mm CaCl2, peak fractions were adsorbed to glow-discharged carbon-coated copper grids, stained with uranyl formate, and inspected with an FEI Tecnai 12 electron microscope (Hillsboro, OR) operated at 120 kV. Low-dose images were acquired with an FEI Tecnai 12 electron microscope at 120 kV and a nominal magnification of ×67,000 or ×52,000 using a defocus of −1.5 μm. About 10,000 particles were interactively picked, windowed into individual images using the BOXER module of EMAN (45), and subjected to 10 cycles of multireference alignment and K-means classification into 50 or 20 classes using SPIDER (19) as described (6).
Rosetting assay
Sheep erythrocytes were sensitized with IgM anti-Forssman and human C5-deficient serum as described previously (46). E-IgM-iC3b and E-IgM as control were assayed for binding to αXβ2 HEK293T transfectants (105 cells/well in 12-well cluster plates) (47). Briefly, E-IgM-iC3b or E-IgM in HBS was incubated for 1.5 h at 37 °C in the presence of 1 mm Mg2+/Ca2+ or Mn2+. After three washes, the percentage of cells with rosette formation (>10 erythrocytes/HEK293T cell, over 100 HEK293T cells examined) was assessed by microscopy.
V-bottom cell adhesion assays
Cell adhesion to a V-bottom-well plate was assayed as described previously (48). Briefly, V-bottom 96-well plates (Corning) were coated with a chimera containing the five Ig domains of ICAM-1 fused to the Fc portion of IgA (ICAM-1-Fcα) or human fibrinogen (1 μg/ml) at 4 °C overnight and then blocked with Hepes-buffered saline, pH 7.4, and 2% BSA for 1 h at ambient temperature. LFA-1 and Mac-1 HEK293T transfectants were labeled for 30 min at 37 °C with 2′,7′-bis-(carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester (Molecular Probes), washed, resuspended in HBS (5 × 104 cells/50 μl) with 2 mm Mg2+/Ca2+ or Mn2+ or 10 mm EDTA, and incubated for 30 min at ambient temperature. 50 μl of transfectants were added to each well and incubated for 30 min and then centrifuged at 200 × g for 5 min at ambient temperature.
Soluble ICAM-1 binding
HEK293T transient transfectants were washed with HBS (20 mm HEPES, 150 mm NaCl, pH 7.3) containing 5 mm EDTA and resuspended in HBS buffer containing Mg2+/Ca2+ or Mn2+ or EDTA. Binding of dimeric soluble ICAM-1 was assayed as follows. ICAM-1-Fcα was added to the cells at 10 μg/ml and incubated at 37 °C for 30 min in the presence of 1 mm Mg2+/Ca2+ or Mn2+. The cells were washed and incubated with a 1:100 dilution of goat anti-human IgA-FITC (Zymed Laboratories Inc.) for 30 min at room temperature, washed, and analyzed by flow cytometry.
Epitope exposure
For immunofluorescent flow cytometry (49), HEK293T transfectants were stained with FITC-labeled CBR LFA-1/7 and biotinylated KIM127, MEM148, or m24, referenced elsewhere (7), in the presence of 1 mm MgCl2/CaCl2 or 1 mm MnCl2 and then with phycoerythrin (PE)-streptavidin to recognize the activation-dependent epitopes. An FITC gate was set to define transfected cells and used to collect PE fluorescence. Mutant PE mean fluorescence intensity (MFI) was normalized by multiplying it by (FITC MFI of WT)/(FITC MFI of mutant).
Author contributions
M. S. and T. A. S. conceived the work and wrote the manuscript; A. C. K. carried out and wrote sections on SAXS data collection and analysis; M. S. expressed and purified αLβ2 and high-affinity ICAM-1, prepared samples, analyzed SAXS and EM data, fit molecular models, tested β2 mutations for functional analysis, and wrote the paper; K. Y. made and tested β2 mutations in the binding assays; J. W. acquired EM data.
Supplementary Material
Acknowledgments
We thank Wei Xia for help with EM data acquisition, the staff at beam line X9 of the National Synchrotron Light Source (Upton, NY), and Structural Biology Grid (SBGRID) and the University of Houston Center for Advanced Computing and DataScience for assistance with the calculations carried out in this work.
This work was supported by National Institutes of Health NCI Grant CA031798 (to T. A. S.), by a GlaxoSmithKline fellowship (to M. S.), and by National Institutes of Health Grant NIGMS R01GM118277 (to K. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
- LFA-1
- leukocyte function–associated antigen 1
- Mac-1
- macrophage antigen 1
- ICAM-1
- intercellular adhesion molecule-1
- SAXS
- small-angle X-ray scattering
- MIDAS
- metal ion–dependent adhesion site
- ADMIDAS
- adjacent to MIDAS
- PE
- phycoerythrin
- MFI
- mean fluorescence intensity
- PDB
- Protein Data Bank.
References
- 1. Luo B.-H., Carman C. V., and Springer T. A. (2007) Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Springer T. A., and Dustin M. L. (2012) Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24, 107–115 10.1016/j.ceb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sen M., Yuki K., and Springer T. A. (2013) An internal ligand-bound, metastable state of a leukocyte integrin, αXβ2. J. Cell Biol. 203, 629–642 10.1083/jcb.201308083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiao T., Takagi J., Wang J.-H., Coller B. S., and Springer T. A. (2004) Structural basis for allostery in integrins and binding of fibrinogen-mimetic therapeutics. Nature 432, 59–67 10.1038/nature02976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shimaoka M., Xiao T., Liu J.-H., Yang Y., Dong Y., Jun C.-D., McCormack A., Zhang R., Joachimiak A., Takagi J., Wang J.-H., and Springer T. A. (2003) Structures of the αL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell 112, 99–111 10.1016/S0092-8674(02)01257-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishida N., Xie C., Shimaoka M., Cheng Y., Walz T., and Springer T. A. (2006) Activation of leukocyte β2 integrins by conversion from bent to extended conformations. Immunity 25, 583–594 10.1016/j.immuni.2006.07.016 [DOI] [PubMed] [Google Scholar]
- 7. Chen X., Xie C., Nishida N., Li Z., Walz T., and Springer T. A. (2010) Requirement of open headpiece conformation for activation of leukocyte integrin αXβ2. Proc. Natl. Acad. Sci. U.S.A. 107, 14727–14732 10.1073/pnas.1008663107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xie C., Zhu J., Chen X., Mi L., Nishida N., and Springer T. A. (2010) Structure of an integrin with an αI domain, complement receptor type 4. EMBO J. 29, 666–679 10.1038/emboj.2009.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adair B. D., Xiong J. P., Alonso J. L., Hyman B. T., and Arnaout M. A. (2013) EM structure of the ectodomain of integrin CD11b/CD18 and localization of its ligand-binding site relative to the plasma membrane. PLoS One 8, e57951 10.1371/journal.pone.0057951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sen M., and Springer T. A. (2016) Leukocyte integrin αLβ2 headpiece structures: the αI domain, the pocket for the internal ligand, and concerted movements of its loops. Proc. Natl. Acad. Sci. U.S.A. 113, 2940–2945 10.1073/pnas.1601379113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weitz-Schmidt G., Schürpf T., and Springer T. A. (2011) The C-terminal αI domain linker as a critical structural element in the conformational activation of αI integrins. J. Biol. Chem. 286, 42115–42122 10.1074/jbc.M111.282830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nordenfelt P., Elliott H. L., and Springer T. A. (2016) Coordinated integrin activation by actin-dependent force during T-cell migration. Nat. Commun. 7, 13119 10.1038/ncomms13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen X., Yu Y., Mi L. Z., Walz T., and Springer T. A. (2012) Molecular basis for complement recognition by integrin αXβ2. Proc. Natl. Acad. Sci. U.S.A. 109, 4586–4591 10.1073/pnas.1202051109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J., Salas A., and Springer T. A. (2003) Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat. Struct. Biol. 10, 995–1001 10.1038/nsb1011 [DOI] [PubMed] [Google Scholar]
- 15. Chen J., Yang W., Kim M., Carman C. V., and Springer T. A. (2006) Regulation of outside-in signaling by the β2 I domain of integrin αLβ2. Proc. Natl. Acad. Sci. U.S.A. 103, 13062–13067 10.1073/pnas.0605666103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dransfield I., Cabañas C., Craig A., and Hogg N. (1992) Divalent cation regulation of the function of the leukocyte integrin LFA-1. J. Cell Biol. 116, 219–226 10.1083/jcb.116.1.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song G., Lazar G. A., Kortemme T., Shimaoka M., Desjarlais J. R., Baker D., and Springer T. A. (2006) Rational design of ICAM-1 variants for antagonizing integrin LFA-1-dependent adhesion. J. Biol. Chem. 281, 5042–5049 10.1074/jbc.M510454200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schürpf T., and Springer T. A. (2011) Regulation of integrin affinity on cell surfaces. EMBO J. 30, 4712–4727 10.1038/emboj.2011.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., and Leith A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- 20. Casasnovas J. M., Stehle T., Liu J.-H., Wang J.-H., and Springer T. A. (1998) A dimeric crystal structure for the N-terminal two domains of ICAM-1. Proc. Natl. Acad. Sci. U.S.A. 95, 4134–4139 10.1073/pnas.95.8.4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen X., Kim T. D., Carman C. V., Mi L. Z., Song G., and Springer T. A. (2007) Structural plasticity in IgSF domain 4 of ICAM-1 mediates cell surface dimerization. Proc. Natl. Acad. Sci. U.S.A. 104, 15358–15363 10.1073/pnas.0707406104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bella J., Kolatkar P. R., Marlor C. W., Greve J. M., and Rossmann M. G. (1998) The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc. Natl. Acad. Sci. U.S.A. 95, 4140–4145 10.1073/pnas.95.8.4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Recacha R., Jiménez D., Tian L., Barredo R., Gahmberg C. G., and Casasnovas J. M. (2014) Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions. Acta Crystallogr. D Biol. Crystallogr. 70, 1934–1943 10.1107/S1399004714009468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kirchhausen T., Staunton D. E., and Springer T. A. (1993) Location of the domains of ICAM-1 by immunolabeling and single-molecule electron microscopy. J. Leukoc. Biol. 53, 342–346 10.1002/jlb.53.3.342 [DOI] [PubMed] [Google Scholar]
- 25. Staunton D. E., Dustin M. L., Erickson H. P., and Springer T. A. (1990) The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell 61, 243–254 10.1016/0092-8674(90)90805-O [DOI] [PubMed] [Google Scholar]
- 26. Zhu J., Zhu J., and Springer T. A. (2013) Complete integrin headpiece opening in eight steps. J. Cell Biol. 201, 1053–1068 10.1083/jcb.201212037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Y., Jun C.-D., Liu J.-H., Zhang R., Jochimiak A., Springer T. A., and Wang J.-H. (2004) Structural basis for dimerization of ICAM-1 on the cell surface. Mol. Cell 14, 269–276 10.1016/S1097-2765(04)00204-7 [DOI] [PubMed] [Google Scholar]
- 28. Song G., Yang Y., Liu J.-H., Casasnovas J. M., Shimaoka M., Springer T. A., and Wang J.-H. (2005) An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin αLβ2. Proc. Natl. Acad. Sci. U.S.A. 102, 3366–3371 10.1073/pnas.0500200102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H., Casasnovas J. M., Jin M., Liu J.-H., Gahmberg C. G., Springer T. A., and Wang J.-H. (2008) An unusual allosteric mobility of the C-terminal helix of a high-affinity αL integrin I domain variant bound to ICAM-5. Mol. Cell 31, 432–437 10.1016/j.molcel.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu S., Wang J., Wang J. H., and Springer T. A. (2017) Distinct recognition of complement iC3b by integrins αXβ2 and αMβ2. Proc. Natl. Acad. Sci. U.S.A. 114, 3403–3408 10.1073/pnas.1620881114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin M., Song G., Carman C. V., Kim Y.-S., Astrof N. S., Shimaoka M., Wittrup D. K., and Springer T. A. (2006) Directed evolution to probe protein allostery and integrin I domains of 200,000-fold higher affinity. Proc. Natl. Acad. Sci. U.S.A. 103, 5758–5763 10.1073/pnas.0601164103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huth J. R., Olejniczak E. T., Mendoza R., Liang H., Harris E. A., Lupher M. L. Jr, Wilson A. E., Fesik S. W., and Staunton D. E. (2000) NMR and mutagenesis evidence for an I domain allosteric site that regulates lymphocyte function-associated antigen 1 ligand binding. Proc. Natl. Acad. Sci. U.S.A. 97, 5231–5236 10.1073/pnas.97.10.5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alonso J. L., Essafi M., Xiong J. P., Stehle T., and Arnaout M. A. (2002) Does the integrin αA domain act as a ligand for its βA domain? Curr. Biol. 12, R340–R342 10.1016/S0960-9822(02)00852-7 [DOI] [PubMed] [Google Scholar]
- 34. Salas A., Shimaoka M., Kogan A. N., Harwood C., von Andrian U. H., and Springer T. A. (2004) Rolling adhesion through an extended conformation of integrin αLβ2 and relation to αI and βI-like domain interaction. Immunity 20, 393–406 10.1016/S1074-7613(04)00082-2 [DOI] [PubMed] [Google Scholar]
- 35. Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., and Springer T. A. (2008) Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 32, 849–861 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Z., Guo S. S., and Fässler R. (2016) Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 10.1083/jcb.201609037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Astrof N. S., Salas A., Shimaoka M., Chen J., and Springer T. A. (2006) Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry 45, 15020–15028 10.1021/bi061566o [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J., and Springer T. A. (2017) Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc. Natl. Acad. Sci. U.S.A. 114, 4685–4690 10.1073/pnas.1704171114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nordenfelt P., Moore T. I., Mehta S. B., Kalappurakkal J. M., Swaminathan V., Koga N., Lambert T. J., Baker D., Waters J. C., Oldenbourg R., Tani T., Mayor S., Waterman C. M., and Springer T. A. (2017) Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat. Commun. 8, 2047 10.1038/s41467-017-01848-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 10.1107/S0021889892001663 [DOI] [Google Scholar]
- 41. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 10.1107/S0021889809000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Svergun D. I., Petoukhov M. V., and Koch M. H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 10.1016/S0006-3495(01)76260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 44. Svergun D. I., Barberato C., and Koch M. H. (1995) CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 10.1107/S0021889895007047 [DOI] [Google Scholar]
- 45. Ludtke S. J., Baldwin P. R., and Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 46. Bilsland C. A. G., Diamond M. S., and Springer T. A. (1994) The leukocyte integrin p150,95 (CD11c/CD18) as a receptor for iC3b: activation by a heterologous β subunit and localization of a ligand recognition site to the I domain. J. Immunol. 152, 4582–4589 [PubMed] [Google Scholar]
- 47. Zang Q., and Springer T. A. (2001) Amino acid residues in the PSI domain and cysteine-rich repeats of the integrin β2 subunit that restrain activation of the integrin αXβ2. J. Biol. Chem. 276, 6922–6929 10.1074/jbc.M005868200 [DOI] [PubMed] [Google Scholar]
- 48. Kim M., Carman C. V., and Springer T. A. (2003) Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301, 1720–1725 10.1126/science.1084174 [DOI] [PubMed] [Google Scholar]
- 49. Lu C., Ferzly M., Takagi J., and Springer T. A. (2001) Epitope mapping of antibodies to the C-terminal region of the integrin β2 subunit reveals regions that become exposed upon receptor activation. J. Immunol. 166, 5629–5637 10.4049/jimmunol.166.9.5629 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.