Abstract
Hippo/YAP signaling plays pleiotropic roles in the regulation of cell proliferation and differentiation during organogenesis and tissue repair. Herein we demonstrate increased YAP activity in respiratory epithelial cells in lungs of patients with idiopathic pulmonary fibrosis (IPF), a common, lethal form of interstitial lung disease (ILD). Immunofluorescence staining in IPF epithelial cells demonstrated increased nuclear YAP and loss of MST1/2. Bioinformatic analyses of epithelial cell RNA profiles predicted increased activity of YAP and increased canonical mTOR/PI3K/AKT signaling in IPF. Phospho-S6 (p-S6) and p-PTEN were increased in IPF epithelial cells, consistent with activation of mTOR signaling. Expression of YAP (S127A), a constitutively active form of YAP, in human bronchial epithelial cells (HBEC3s) increased p-S6 and p-PI3K, cell proliferation and migration, processes that were inhibited by the YAP-TEAD inhibitor verteporfin. Activation of p-S6 was required for enhancing and stabilizing YAP, and the p-S6 inhibitor temsirolimus blocked nuclear YAP localization and suppressed expression of YAP target genes CTGF, AXL, and AJUBA (JUB). YAP and mTOR/p-S6 signaling pathways interact to induce cell proliferation and migration, and inhibit epithelial cell differentiation that may contribute to the pathogenesis of IPF.
Keywords: Cell Biology, Pulmonology
Keywords: Pulmonary surfactants
YAP and mTOR/p-S6 signaling pathways interact to induce cell proliferation and migration, and inhibit epithelial cell differentiation that may contribute to the pathogenesis of IPF.
Introduction
Idiopathic pulmonary fibrosis (IPF) is a common form of interstitial lung disease (ILD) resulting in alveolar remodeling and progressive loss of pulmonary function, respiratory failure, and death often within 5 years of diagnosis (1, 2). IPF pathogenesis encompasses fibrotic remodeling, inflammation, and loss of lung architecture (3). Though the underlying causes of the disease remain elusive, genetic and experimental evidence support the concept that chronic alveolar injury and failure to properly repair the respiratory epithelium are intrinsic to IPF disease pathogenesis (4). Histologically, respiratory epithelial cells in the lung parenchyma express atypical proximal airway epithelial and indeterminate cell type markers (5, 6), including goblet and basal cell characteristics that are normally restricted to conducting airways. Fibrotic lesions and honeycomb structures replace alveolar structures, the latter normally lined by AT1 and AT2 cells. Genome-wide transcriptomic analyses of lung tissue and isolated epithelial cells from IPF patients demonstrate dramatic changes in ciliated, basal, and goblet cell–associated gene expression and loss of normal alveolar epithelial cells, reflecting profound changes in epithelial cell differentiation and function in IPF (6, 7). Mutations affecting AT2 cell functions (ABCA3, SFTPA, SFTPB, and SFTPC) are among the genetic causes of chronic ILDs, further implicating abnormal alveolar epithelial cell homeostasis in the pathogenesis of these disorders (8–11). Experimental mouse models of chronically injured peripheral lung epithelial cells exhibit alveolar remodeling and fibrosis in association with activation of TGF-β (12), TGF-α, and the mTOR/PI3K/AKT pathways (13, 14). In recent single-cell RNA sequencing (RNAseq) of isolated respiratory epithelial cells, we identified distinct populations of indeterminate, basal, and goblet-like epithelial cells in IPF, identifying individual cells that shared RNA characteristics with both AT2 cells and conducting airway epithelial cells consistent with a loss of normal cell-type lineage restriction (15). Unbiased analyses of RNA profiles in IPF predicted activation of genes in the Hippo/YAP pathway, whereas inhibitors of the pathway, SAV1 and MST2, were suppressed. Likewise, RNAs associated with the activation of mTOR/PI3K/AKT signaling, including increased MLST8 and decreased phosphatase and tensin homology (PTEN) expression were predicted in IPF epithelial cells, indicating potential interactions between YAP and mTOR signaling pathways.
Major components of the Hippo signaling pathway play diverse roles in organogenesis and oncogenesis. The pathway consists of kinase-adaptor protein complexes, wherein the serine/threonine kinases Mst1 and Mst2 in concert with Salvador (Sav1) serve as YAP inhibitors by phosphorylating and activating large tumor suppressor kinases (Lats1 and Lats2), that in turn phosphorylate downstream transcriptional effectors Yap and Taz to direct their cytoplasmic localization and inhibit their transcriptional activities (16). In the absence of inhibitory phosphorylation by the Hippo kinases, Yap/Taz translocate to the nucleus where they interact with transcriptional cofactors TEAD1–4 to regulate target genes associated with cell proliferation, apoptosis, and differentiation, and induce known transcriptional targets including connective tissue growth factor (CTGF/CCN2) (17), AXL tyrosine kinase (18), and Ajuba (also known as JUB) (19). Ajuba inhibits the activity of MST1/2 and Lats1/2 and acts as a counter regulator of the pathway (20). The Hippo pathway controls organ size, cell proliferation and differentiation in stem/progenitor cells during embryogenesis and homeostasis (21–23). YAP is required for normal branching morphogenesis and epithelial differentiation in the developing lung (24). Nucleus-localized YAP is required for airway epithelial cells to respond to TGF-β and controls SOX2 expression (25). Increased YAP activity in airway basal stem cells causes epithelial hyperplasia and impairs terminal differentiation, while YAP deletion causes terminal differentiation or loss of the ability of basal cells to dedifferentiate into progenitor cells (25, 26). Genetic deletion of MST1/2 in adult and fetal mice increased nuclear YAP, causing airway hyperplasia and abnormal differentiation of airway epithelial cells (19). Herein, we demonstrate increased activity of YAP in IPF respiratory epithelial cells and identify a potentially novel pathway in which YAP interacts with mTOR/PI3K/AKT signaling to regulate abnormal cell proliferation, migration, and polarity in respiratory epithelial cells in IPF.
Results
Activation of YAP-mediated gene expression in IPF.
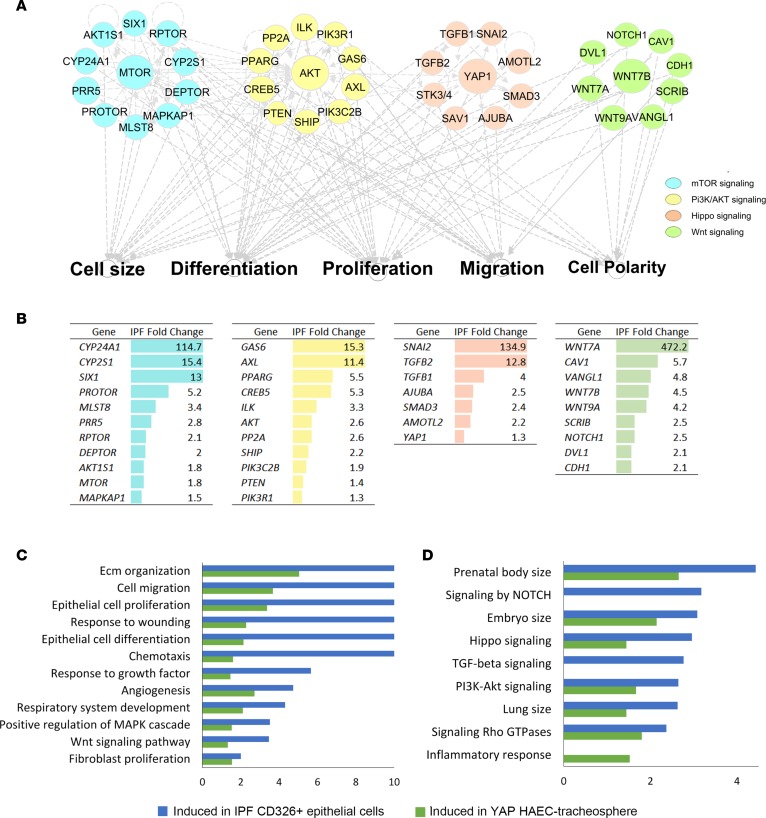
An unbiased analysis of RNAseq data from FACS isolated epithelial cells (CD326+/HTII-280+) from normal and IPF, and primary human bronchiolar epithelial cells (HBECs) expressing activated YAP (S127A) (19), was performed to predict the bioprocesses and pathways shared in these data sets. Genes encoding proteins involved in mTOR, PI3K/AKT, and Hippo/YAP and WNT signaling were predicted to be active by functional classification and network construction using ingenuity pathway analysis (IPA) (Figure 1A). Network analysis predicted extensive interactions among mTOR, PI3K/AKT, planar polarity, and Hippo/YAP signaling. These processes and pathways are involved in the regulation of epithelial cell size, migration, proliferation, differentiation, and cell polarity, supporting the hypothesis that these phenotypic features in IPF are regulated in part by activation of Hippo/YAP–associated signaling (Figure 1A). Predicted pathways and gene expression changes in IPF are shown in Figure 1B, including increased YAP1, JUB, AKT3, AMOTL2, GAS6, and CYP24A1 RNAs. Gene expression profiles of sorted IPF epithelial cells and of primary HBECs expressing activated YAP (S127A) shared enriched bioprocesses, including extracellular matrix organization, cell migration, response to wound, cell size, and epithelial proliferation/ differentiation, as well as increased expression of genes associated with canonical TGF-β, Hippo/YAP, and PI3K/AKT signaling pathways (Figure 1, C and D) essential for processes known to be regulated by the Hippo/YAP pathway (16).
Figure 1. Prediction of signaling interactions in idiopathic pulmonary fibrosis (IPF) epithelial cells.
(A) Ingenuity pathway analysis of RNA sequencing data from CD326+/HTII-280+ sorted epithelial cells from healthy donors (n = 3) and IPF (n = 3) was used to predict intensive interactions among mTOR/PI3K/AKT, Hippo/YAP, and polarity signaling pathways. (B) Genes associated with each of the pathways significantly altered in IPF are shown. Each pathway is represented by a distinct color code: mTOR (blue), PI3K/AKT (yellow), Hippo (pink), and polarity (green). (C and D) Functional enrichment analysis predicted that genes induced in CD326+/HTII-280+ IPF epithelial cells (8) and in human airway epithelial cells (HAECs) expressing YAP (20) share (C) commonly activated bioprocesses and (D) signaling pathways including those affecting epithelial cell proliferation, migration, and cell size. The x axis represents the –log10-transformed enrichment P value.
Increased YAP activity in IPF epithelial cells.
Immunofluorescence confocal microscopy and in situ hybridization RNA analyses of peripheral lung tissue demonstrated increased nuclear YAP and decreased MST1/2 in IPF epithelial cells that costained with ABCA3 or pan-cytokeratin. Consistent with increased nuclear YAP, staining for Ajuba, a known transcriptional target of YAP, was increased and primarily detected in epithelial cells in IPF lesions (Figure 2, A–C). In sharp contrast, nuclear YAP was rarely detected and MST1/2 was highly expressed in alveolar AT2 cells and bronchial epithelial cells in normal donor lung tissues. Western blot analysis of whole-lung lysates demonstrated increased total YAP in IPF and chronic obstructive pulmonary disease (COPD) tissues, while levels of phosphorylated YAP were similar in normal, IPF, and COPD lung samples (Figure 2D). The ratio of total YAP to phosphorylated YAP was significantly increased in IPF compared with normal lung tissues, consistent with increased active YAP in IPF (Figure 2E).
Figure 2. Hippo/YAP signaling in idiopathic pulmonary fibrosis (IPF).
(A–C) Representative immunofluorescence confocal microscopy of donor (n = 4) and IPF (n = 6) lung tissue was used to detect Hippo components. (A) AJUBA (white), ACTA2 (green), and ABCA3 (red). (B) YAP (green) and ABCA3 (red). (C) MST1/2 (red) and ABCA3 (green). White arrows point to cells coexpressing ABCA3 and nuclear YAP in IPF. Scale bars: 100 μm (overviews) and 10 μm (insets). (D) Representative immunoblots of YAP and phosphorylated YAP (p-YAP) from lung tissue lysates of IPF (n = 6), control (n = 4), and COPD (n = 3) patients are shown. (E) Western blots were normalized to β-actin. (F) Multiplexed proximity ligation fluorescence in situ hybridization (PLISH) staining of CTGF (red) and AXL (white) were costained using pan-cytokeratin (PanKRT) (green) in normal donor (n = 6) and IPF (n = 6) lung tissue. Scale bars: 25 μm and 2.5 μm (insets). (G) qPCR analysis of RNA from CD326+ epithelial cells isolated from peripheral lungs of healthy donor (n = 3) and IPF (n = 3) lung tissue was used to quantify SAV1, MST2, and JUB RNAs. ANOVA was used to assess Western blot quantification; Student’s t test was used for qPCR. *P < 0.05.
RNAs associated with YAP activation are increased in IPF epithelial cells.
Proximity ligation fluorescence in situ hybridization (PLISH) was used to detect YAP target genes AXL and CTGF in IPF epithelial cells (Figure 2F). AXL RNA was increased throughout the lung and particularly in epithelial cells. SAV1 and MST2, inhibitors of YAP activity, were reduced, while JUB was increased in RNA from CD326+ sorted IPF respiratory epithelial cells (Figure 2G). Loss of the YAP inhibitors, SAV1 and MST2, and increased YAP target gene expression were consistent with increased YAP transcriptional activity in IPF epithelial cells.
Epithelial cell polarity is disrupted in IPF.
Since expression of a number of genes related to planar cell polarity, including SCRIB and VANGL1, were increased in CD326+ sorted IPF epithelial cells (Figure 1, A and B), we used immunofluorescence staining of epithelial cell polarity markers scribble (SCRIB) and vang-like (VANGL) to assess epithelial cell polarity in IPF. In contrast to apical localization in normal epithelial cells, SCRIB and VANGL staining was increased and diffuse (Figure 3, A and B), indicating loss of normal apical-basal polarity in IPF epithelial cells. VANGL1, SCRIB, and CELSR1 RNAs were increased in IPF epithelial cells, consistent with disruption of normal cell polarity in IPF (Figure 3C), and consistent with RNAseq data that demonstrated increased expression of cell polarity genes in IPF (Figure 3D).
Figure 3. Vangl and Scribble in idiopathic pulmonary fibrosis (IPF) lung tissue.
(A and B) Representative immunofluorescence imaging of healthy donor (n = 4) and IPF (n = 6) lung tissue are shown. (A) Scribble (Scrib) (green) and Krt8 (red) or (B) Vangl (red) and Krt8 (green). Scale bars: 50 μm and 5 μm (insets). (C) CDH1, CELSR1, SCRIB, and VANGL1 RNAs were measured in CD326+ sorted epithelial cells from healthy donors (n = 3) and IPF (n = 3) lungs. (D) RNA sequencing analysis of polarity-associated genes of healthy donors (n = 3) and IPF (n = 3) demonstrating significant increases in epithelial cell polarity genes including WNT9A and WNT7A. *P < 0.05, calculated by Student’s t test. N.S., not significant.
Increased mTOR activity in IPF respiratory epithelial cells and lung tissue.
Since the RNA network analysis predicted that YAP and mTOR/PI3K/AKT may interact to regulate gene expression in IPF epithelial cells, immunofluorescence imaging was used to assess levels of phosphorylated S6 kinase (p-S6K), a downstream component of mTOR/PI3K/AKT signaling, in IPF epithelial cells (Figure 4A). p-S6K staining was increased throughout IPF lung tissue and selectively increased in and colocalized primarily with pan-cytokeratin staining in IPF epithelial cells. Immunoblotting demonstrated increased p-PTEN, p-S6, and total S6 in IPF tissues compared with tissue from donors or COPD patients (Figure 4, B and C). Increased expression of mTOR/PI3K/AKT components and inhibition of PTEN in IPF epithelial cells were consistent with increased mTOR activity.
Figure 4. Increased mTOR signaling in idiopathic pulmonary fibrosis (IPF) respiratory epithelium.
(A) Representative immunofluorescence imaging of healthy donor (n = 3) and IPF (n = 3) lung for phosphorylated S6K (p-S6K) (red) with pan-cytokeratin (Pan-KRT) (green) is shown. Scale bars: 50 μm and 5 μm (insets). (B) Representative immunoblots were prepared from whole-lung lysates of donor (n = 4), COPD (n = 3), and IPF (n = 6) lung tissue for total S6, phosphorylated S6 (p-S6), and phosphorylated PTEN (p-PTEN). (C) Western blots were normalized to GAPDH. *P < 0.05, determined by ANOVA.
YAP activates mTOR/PI3K/AKT signaling in HBECs.
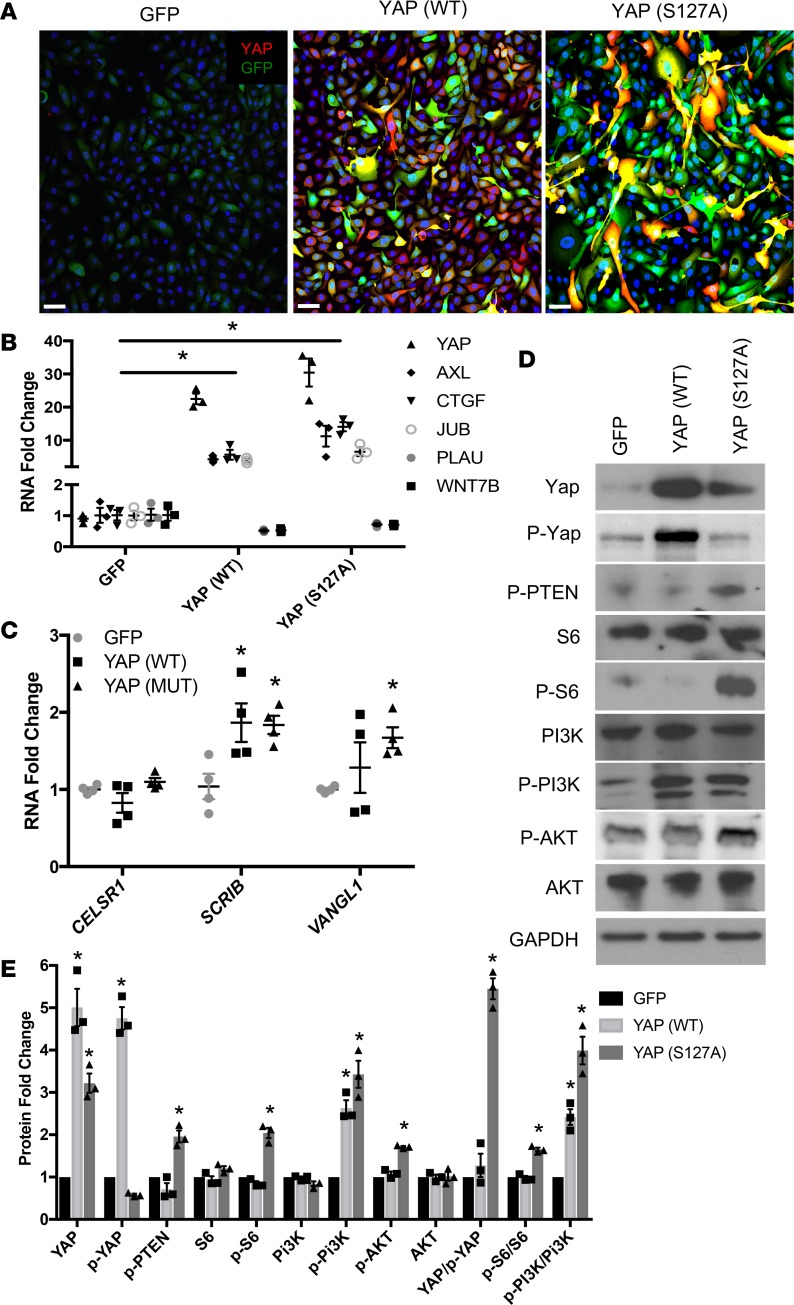
To directly test the hypothesis that YAP and mTOR interact to influence cellular functions in IPF, hTERT/CDK4–immortalized human bronchiolar epithelial cells (HBEC3KT, referred to as HBEC3) were transduced with lentivirus expressing YAP (WT) or constitutively active YAP (S127A) to test whether YAP influences mTOR/PI3K/AKT signaling. Since the characteristics of the abnormal epithelial cells in IPF are consistent with conducting airway cells, we utilized HBEC3s that maintain basal characteristics in vitro and are readily transfected with lentiviral constructs, to study the effects of YAP and mTOR interaction. Immunofluorescence confocal microscopy demonstrated increased nuclear YAP after lentiviral transduction (Figure 5A). Western blotting and quantitative PCR (qPCR) analyses demonstrated increased YAP and YAP transcriptional target RNAs, i.e., JUB, AXL, and CTGF, consistent with activation of YAP (Figure 5B). YAP induced planar polarity genes SCRIB and VANGL1 (Figure 5C). Activated YAP (S127A) increased the phosphorylation of S6, PI3K, AKT, and PTEN, indicating that YAP activates the mTOR/PI3K/AKT pathway in HBEC3s (Figure 5D). Expression of YAP (WT) increased p-PI3K but did not alter p-S6, p-AKT, or p-PTEN (Figure 5E). YAP-mediated induction of p-S6, p-PI3K, and inhibition of PTEN support the predicted interactions between YAP and mTOR/PI3K/AKT activity in IPF.
Figure 5. YAP (S127A) activates mTOR/PI3K/AKT.
(A) HBEC3s were transduced with lentiviruses expressing YAP (WT), YAP (S127A), or empty GFP vector. YAP (red) was identified by immunofluorescence. Images are representative of (n = 3) transductions. Scale bars: 10 μm. (B) qPCR analysis of cells 48 hours after transduction assessing genes regulated by YAP activity. (C) Analysis of polarity genes CELSR1, SCRIB, and VANGL1 in HBEC3s expressing YAP (WT) and YAP (MUT) for 48 hours. (D) Forty-eight hours after transduction, lysates were prepared and immunoblotted for YAP, p-YAP, p-PTEN, S6, p-S6, PI3K, p-PI3K, AKT, and p-AKT. (E) Western blot quantification normalized to GAPDH. *P < 0.05, assessed by ANOVA. HBEC3s, hTERT/CDK4–immortalized human bronchiolar epithelial cells.
Verteporfin inhibits YAP-induced targets, p-S6, cell proliferation, and migration.
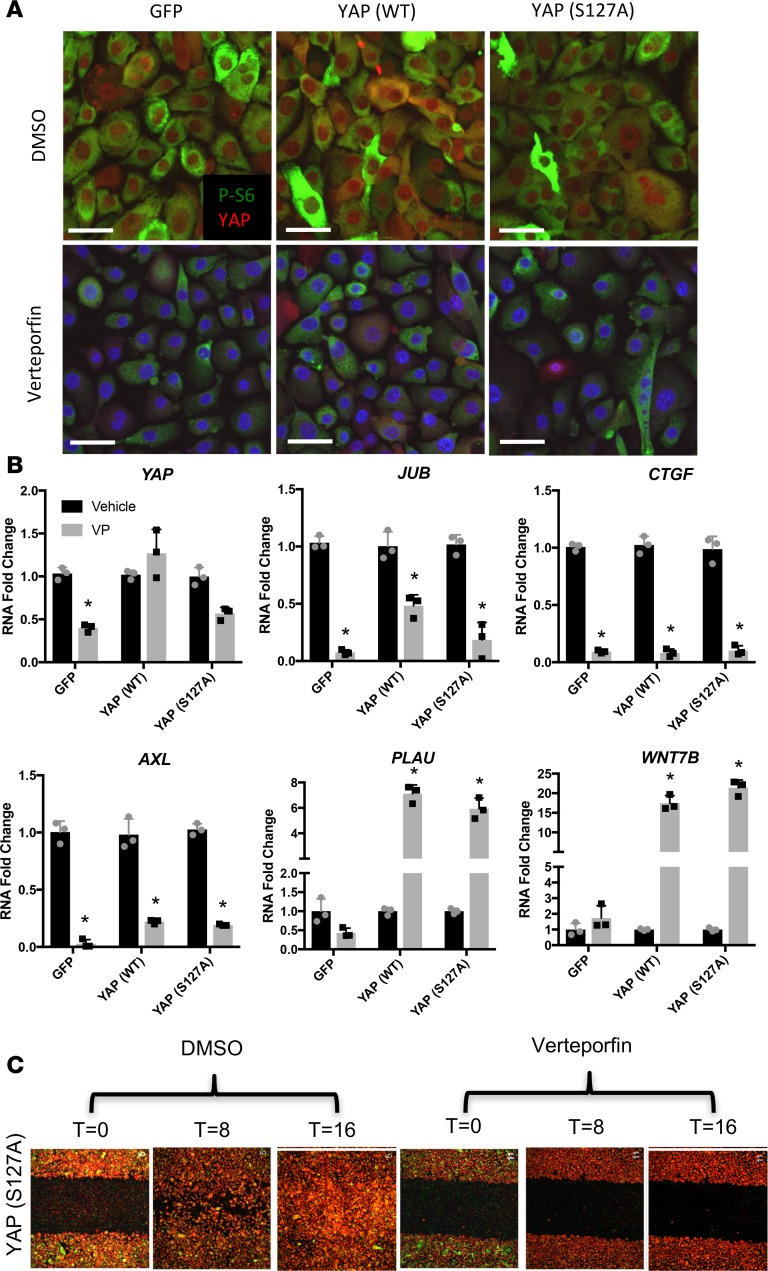
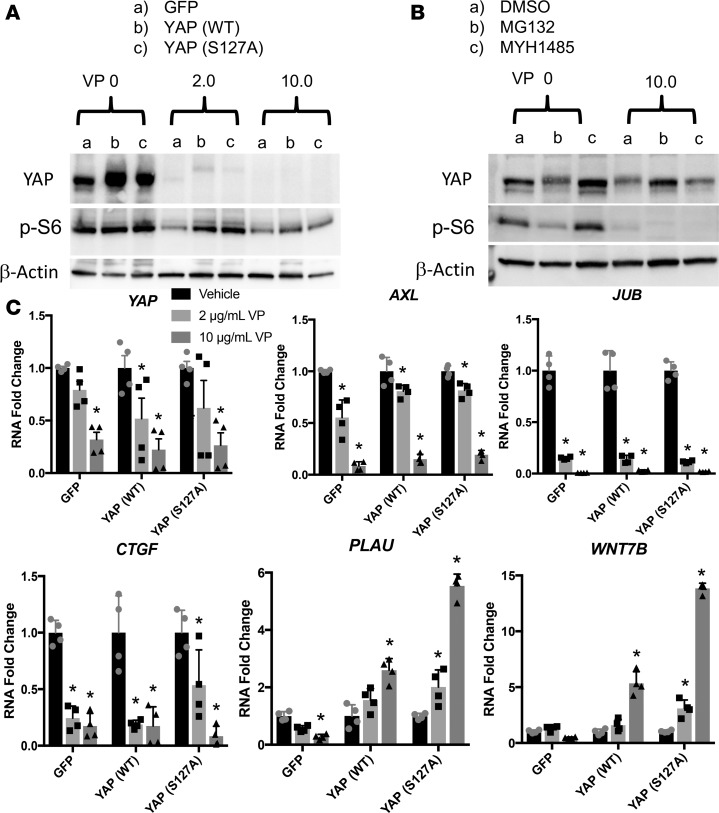
Verteporfin inhibits YAP transcriptional activity by influencing YAP-TEAD interactions (27). In HBEC3s, verteporfin inhibits YAP and YAP-induced cell proliferation, without affecting cell viability (Supplemental Figure 1, C and D; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98738DS1). Verteporfin reduced nuclear YAP staining, total YAP expression, and inhibited p-S6 in HBEC3s (Figure 6A and Supplemental Figure 1A). Since verteporfin is known to be photo activated (27), we assessed YAP and its transcriptional targets AXL, JUB, and CTGF RNAs in the presence (Figure 6) and absence of ambient light (Figure 7). Total YAP was reduced following verteporfin treatment in darkness (Figure 7A). YAP targets were significantly reduced regardless of light exposure (Figures 6B and 7B); however, in the absence of light, higher concentrations of verteporfin were required (Supplemental Figure 4). Consistent with verteporfin regulating YAP activity, PLAU and WNT7B were reduced following YAP (WT) and YAP (S127A) expression, and were significantly increased by verteporfin treatment (Figure 7B). Expression of YAP (WT) and YAP (S127A) increased cell migration, which was blocked by verteporfin in the presence (Figure 6C and Supplemental Figure 1B) or absence of ambient light (Figure 7C and Supplemental Figure 2). Verteporfin inhibited nuclear localization of YAP, total YAP mRNA, YAP target gene expression, and YAP-mediated cell migration. To test whether verteporfin regulates YAP through proteasomal degradation, YAP-expressing cells were treated with verteporfin in the presence of proteasome inhibitor MG132. MG132 protected YAP from verteporfin-induced degradation. To test if mTOR activation rescued the loss of YAP by verteporfin, cells were treated with verteporfin in the presence of mTOR activator MHY1485, whereby expression of YAP and p-S6 were increased; however, MHY1485 failed to block loss of YAP following verteporfin treatment (Figure 7B). Thus, activated mTOR was not sufficient to prevent destabilization and inhibition of YAP by verteporfin.
Figure 6. Verteporfin inhibits YAP transcriptional activity and phosphorylation of S6 in ambient light.
HBEC3s were transduced with GFP, YAP (WT), or YAP (S127A) lentiviral vectors and treated with either vehicle (DMSO) or 0.25 μg/ml verteporfin for 48 hours (n = 3). (A) Immunofluorescence of p-S6 (green) and YAP (red) was assessed following verteporfin (0.25 μg/ml) or DMSO. Scale bars: 10 μm. (B) YAP, JUB, AXL, CTGF, WNT7B, and PLAU RNAs were quantified following 0.25 μg/ml verteporfin (VP) or vehicle exposed to ambient light (n = 3). Expression is normalized to DMSO controls. (C) Scratch assay of YAP (S127A)–transduced HBEC3s following DMSO or verteporfin treatment (0.25 μg/ml) in ambient light at T = 0, 8, and 16 hours of assay (n = 3). *P < 0.05, determined by ANOVA. HBEC3s, hTERT/CDK4–immortalized human bronchiolar epithelial cells.
Figure 7. Verteporfin reduces YAP transcriptional activity and phosphorylation of S6 in absence of light.
HBEC3s transduced with GFP, YAP (WT), or YAP (S127A) lentiviral vectors were treated with either vehicle (DMSO) or verteporfin (2.0 or 10.0 μg/ml) for 48 hours (n = 3). (A) Western blot for YAP and p-S6 following a verteporfin dose curve in complete darkness show reduced total YAP and p-S6. (B) Western blot analysis of YAP and p-S6 following verteporfin treatment for 48 hours with MHY1485 treatment for the final 24 hours shows increased YAP after mTOR activation. Addition of MG132 for the final 24 hours of verteporfin treatment protects YAP from degradation caused by verteporfin. (C) Yap, JUB, CTGF, AXL, PLAU, and WNT7B RNAs were assessed in experiments performed in complete darkness (n = 4 transfections). RNA expression is normalized to DMSO-treated cells. *P < 0.05, determined by ANOVA. HBEC3s, hTERT/CDK4–immortalized human bronchiolar epithelial cells.
Temsirolimus blocks nuclear YAP, YAP-induced gene targets, cell proliferation and migration.
To test if mTOR influences YAP activity, HBEC3s expressing YAP (WT) or YAP (S127A) were treated with temsirolimus, an inhibitor of the mTOR pathway. Temsirolimus reduced YAP-mediated cell proliferation but did not affect cell viability (Supplemental Figure 3, B and C). Temsirolimus inhibited phosphorylation of S6 and remarkably, reduced nuclear YAP staining (Figure 8, A and B). Proteasome blocking with MG132 or activation of mTOR with MHY1485 did not prevent the loss of YAP by temsirolimus (Supplemental Figure 3D). Consistent with the inhibitory effect of temsirolimus on YAP activity, CTGF, JUB, and AXL were reduced (Figure 8C and Supplemental Figure 5). To further assess the interaction of YAP/mTOR in regulating cell behavior, migration scratch assays were performed in HBEC3s expressing YAP (WT) and YAP (S127A). Temsirolimus prevented YAP-induced cell migration in scratch assays (Figure 8D and Supplemental Figure 3A), supporting a network in which mTOR signaling cooperates with YAP to regulate epithelial cell migration and proliferation.
Figure 8. Temsirolimus inhibits phosphorylation of S6, YAP transcriptional activity, and YAP-induced migration.
(A) Representative immunofluorescence microscopy was performed in HBEC3s transfected with GFP, YAP (WT), and YAP (S127A) and stained for p-S6 (green) and YAP (red) in the presence of temsirolimus (25 μg/ml) or DMSO (n = 5). Scale bars: 10 μm. (B) Western blot analysis of YAP following temsirolimus treatments shows reduced total YAP expression. (C) YAP, JUB, AXL, and CTGF RNAs were assessed after 48 hours (n = 3). RNA expression is normalized to DMSO-treated cells. (D) Time-lapse imaging of HBEC3s transduced with YAP (S127A) following treatment with temsirolimus at T = 0, 8, and 16 hours of a scratch assay (n = 3). *P < 0.05, assessed by ANOVA. HBEC3s, hTERT/CDK4–immortalized human bronchiolar epithelial cells.
Discussion
IPF involves extensive remodeling of the peripheral lung parenchyma resulting in loss of alveolar gas exchange regions of the lung. Normal alveoli, lined by AT1 and AT2 epithelial cells, are replaced by diverse pathological lesions lined by epithelial cells with abnormal conducting airway epithelial cell characteristics, including goblet, ciliated, basal, and indeterminate cells in IPF (5, 6, 15, 28). These pathological findings include profound changes in epithelial cell differentiation, shape, migration, and/or proliferation in IPF. The present unbiased pathway analysis derived from RNAseq from CD326+/HT280+ sorted epithelial cells from IPF lungs and from primary HBECs expressing constitutively active YAP, indicated activation of canonical signaling pathways regulated by Hippo/YAP and mTOR in both data sets. YAP regulates respiratory epithelial cell differentiation during both development and lung remodeling correlating to the abnormal differentiation seen in IPF epithelial cells (15, 19, 25). The present immunofluorescence and in vitro studies demonstrated increased nuclear YAP, YAP target gene expression, and S6 kinase activity in IPF epithelial cells. In vitro and in vivo data support the concept that YAP and mTOR signaling interact to influence epithelial cell shape, proliferation, and migration, processes that are likely to contribute to the pathogenesis of IPF (Figure 9).
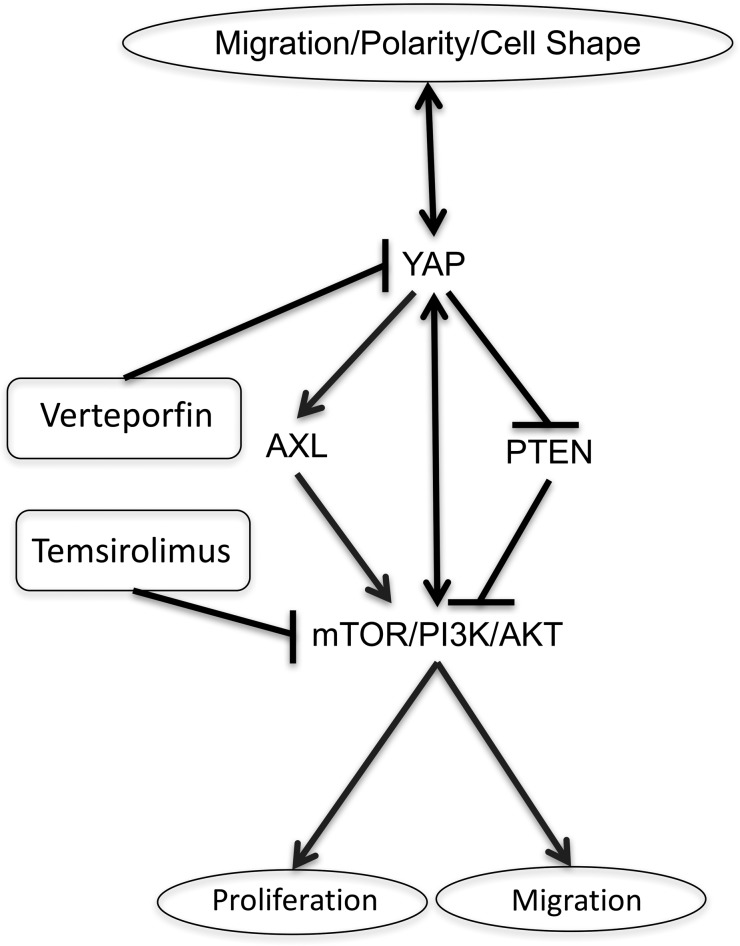
Figure 9. Proposed polarity/YAP/mTOR network regulating epithelial cell processes.
Cell polarity is regulated by and regulates YAP activity. YAP expression alters cell shape. YAP (S127A) expression inhibits PTEN, increases AXL, and activates mTOR with increased p-S6, p-S6K, and p-PI3K. Blocking mTOR activity with temsirolimus inhibits YAP-induced cell proliferation and migration, and also reduces nuclear YAP staining. Verteporfin inhibits YAP nuclear localization and transcriptional activity, while reduced p-S6 staining indicates reduced mTOR.
While the genetic and cellular processes underlying the pathophysiological changes in IPF remain poorly understood, genetic causes of ILDs are closely associated with chronic epithelial cell injury and failure of normal alveolar epithelial cell repair (2, 7, 8, 12, 29). Present and published studies demonstrate the loss of normal alveolar structures, and the presence of epithelial cells with more proximal airway epithelial cell characteristics in the alveolar regions in IPF (15). Recent studies of a murine model of influenza infection support the concept that conducting airway basal cells migrate and proliferate in the alveolar regions, but fail to differentiate and regenerate functional alveoli (30, 31).
Immunofluorescence staining of IPF lung tissue demonstrated the loss of normal epithelial cell shape, mislocalization of Scribble and Vangl, and qPCR analyses of purified IPF epithelial cells demonstrated increased expression of SCRIB, VANGL1, and CELSR1 mRNAs. These findings are consistent with previous findings that YAP caused the mislocalization of Scrib (32) and altered cell shape in conducting airways of MST1/2 deleted mice (19). YAP-mediated changes in planar polarity–associated genes may in turn influence cell shape, cell-cell contacts, and migration (33). Likewise, disruption of cell-cell contacts and cell shape may influence YAP activity via the planar cell polarity pathway. For example, Crumbs increased YAP-LATS1/2 interactions enhancing YAP phosphorylation in mouse airway epithelial cells (34). Planar cell polarity influences epithelial cell proliferation and migration through YAP (35, 36), consistent with the currently proposed role for YAP in regulating epithelial cell activities in IPF epithelial cells.
The present study demonstrates that YAP and mTOR/PI3K/AKT interact to influence cell proliferation and migration in HBEC3s. Phosphorylated PTEN was increased in IPF lung lysates, and expression of YAP (S127A) increased p-PTEN in HBEC3s, supporting a potential mechanism by which YAP activates mTOR signaling. p-S6K staining and p-S6 protein were increased in IPF epithelial cells and expression of YAP (S127A)–induced p-S6 in HBEC3s. Verteporfin inhibited YAP and reduced p-S6 staining in HBEC3s, demonstrating the direct interactions between the pathways. The present data are consistent with findings in MCF10A mammary epithelial cells and in intestinal cells in which YAP induced mTOR (37). The present data showing that temsirolimus inhibited YAP, YAP target genes, and blocked YAP-induced increases in cell migration demonstrated a feed-forward signaling loop between YAP and mTOR. Taken together, the present findings that pharmacological activation of mTOR with MHY1485 induced YAP, temsirolimus decreased YAP, and expression of YAP (S127A) increases p-S6 in HBEC3s, provide evidence of a mechanism by which mTOR is both regulated by and regulates YAP activity in respiratory epithelial cells. In the bleomycin model of lung injury, inhibitors of the mTOR pathway were not found to be effective in preventing lung injury or enhancing repair (38–40). The present data demonstrate complex interactions among a diversity of signaling pathways that may contribute to the pathogenesis of IPF, findings that support the need to understand and target multiple signaling pathways in this complex disorder and to develop IPF models reflecting the complexity of IPF.
YAP induced airway epithelial cell migration, proliferation, and activated known targets including AXL, CTGF, and JUB. YAP inhibited WNT7B and PLAU RNAs, consistent with our previous findings in the mouse (19). We found that verteporfin blocked YAP-induced cell proliferation, migration, and reduced YAP transcriptional targets, while PLAU and WNT7B RNAs were increased in HBEC3s. Importantly, verteporfin is photo activated and inhibits YAP in the presence and absence of light (27, 41–44). In the presence of light, verteporfin caused cross-linking of proteins in vitro and in vivo, and may therefore cause pleiotropic effects on cellular functions (45). We therefore tested verteporfin activity in YAP-transduced HBEC3s in the presence and absence of light, and found verteporfin to be more active in light without affecting cell viability. We found that YAP activity in HBEC3s was readily inhibited by verteporfin. We repeatedly tested primary HBECs at air-liquid interface cultures under identical light or dark conditions, and found that verteporfin was not effective under these conditions, thereby failing to inhibit YAP, and prioritized HBEC3s for our studies. The precise mechanisms by which verteporfin inhibits YAP remain unclear. Verteporfin has been shown to block YAP-TEAD interactions without altering total YAP levels in retinoblastoma cells (27). In another study, verteporfin induced 14-3-3α–mediated cytoplasmic sequestering of YAP, resulting in degradation of YAP (44). In the present study, verteporfin reduced YAP RNA, total YAP, nuclear YAP, and YAP targets, and treatment with MG132 protected YAP from verteporfin-mediated loss, supporting a potential role for a proteasomal degradation mechanism of inhibition in HBEC3s.
Pathway analysis of RNA expression data predicted interactions between YAP and mTOR/PI3K/AKT in IPF epithelial cells supported by immunofluorescence, in situ hybridization and expression studies in IPF tissue and in vitro. The present data are consistent with a model in which activation of YAP regulated epithelial cell gene expression, cell shape, migration, and proliferation, consistent with diverse effects of YAP on cellular processes (16, 46). Although initial clinical studies using mTOR inhibitors for treatment in IPF were unsuccessful, the present studies demonstrated extensive crosstalk between YAP and mTOR/PI3K/AKT (40, 47). The data predict extensive interactions among multiple signaling pathways that may influence the pathogenesis of IPF. Herein, we identified direct intersections between mTOR, YAP, and planar polarity pathways that together may influence cellular behaviors active in IPF. The present work identifies YAP/mTOR signaling components as potential prognostic and therapeutic targets for the treatment of IPF.
Methods
RNA analysis.
Comparative analysis of RNAs from sorted epithelial cells (CD326+/HTII-280+) from control (n = 3) and IPF donors (n = 3) (GSE94555) (15), and RNA sequences from primary HBECs expressing YAP (S127A) (19) were carried out to reveal commonly regulated pathways. Briefly, differentially expressed genes in IPF epithelial cells and in HBEC-YAP (S127A) were identified by Student’s t test with FDR-adjusted P value < 0.05. Differentially expressed genes from the 2 data sets were subjected to gene set enrichment analysis using ToppGene Suite (https://toppgene.cchmc.org/), and commonly enriched bioprocesses and pathways were compared. Biological cross-talk and related networks among mTOR, PI3K/AKT, and Hippo/YAP signaling pathways were constructed using IPA (http://www.ingenuity.com). Regulatory relationships connecting gene nodes and cellular functions were identified through literature mining using the Ingenuity knowledge base. qPCR was used for validation, and RNA was extracted using the RNeasy Micro kit (Qiagen). cDNA was produced using an iScript cDNA synthesis kit (Bio-Rad). PCR was performed using a StepOne Plus Real-Time PCR system (ABI 7500) utilizing TaqMan gene expression assays (Supplemental Table 2) (Applied Biosystems). All samples were assayed in duplicate for each target gene and quantification was performed using ΔΔCT to determine gene fold changes.
Western blot.
Western blot was performed on human whole-tissue lysates obtained from resected normal donors, IPF, or COPD patient lungs. Diagnoses were made using criteria established by the American Thoracic Society and the European Respiratory Society to identify IPF and COPD samples postmortem. Control tissue was acquired from explanted lung from normal individuals with lungs rejected for transplant. Tissue lysates were prepared using RIPA buffer with protease and phosphatase inhibiters (MilliporeSigma) (19). Protein was measured using a Direct Detect infrared spectrometer (MilliporeSigma). Samples were run in a NOVEX 10%–20% Tris-glycine gel (Invitrogen). iBlot2 (Invitrogen) was used for dry transfer onto PVDF membranes (MilliporeSigma). Membranes were blocked using 5% bovine serum albumin (BSA) in Tris-buffered saline, 0.1% Tween 20 (TBST). Primary antibodies were diluted in 0.5% BSA/TBST. Western blots were quantified using Image Lab software (Bio Rad V5.2.1) and normalized to GAPDH or β-actin.
Immunofluorescence.
Immunofluorescence confocal microscopy was performed using paraffin-embedded samples with 5-μm sections. Antigen retrieval in 10 mM citrate buffer (pH 6.0) or Tris-EDTA (pH 9.0) was used for some antibodies. Samples were blocked in 4% normal donkey serum in PBS, 0.1% Triton X-100 (PBST) for 1 hour. Slides were incubated in primary antibodies that were diluted in blocking buffer for 24–48 hours at 4°C. Antibody dilutions are listed in Supplemental Table 1. Samples were washed in PBST (3 times). Secondary antibody (1:200) and DAPI (1 μg/ml) were added for 2 hours at room temperature. Samples were then washed in PBST, rinsed in phosphate buffer, and No. 1.5 cover slips were mounted with Prolong Gold (Thermo Fisher Scientific). Images were captured on an inverted Nikon A1R confocal microscope (×10, ×20, or ×60 magnification). Maximum intensity projections of Z-stack images were generated using NIS-Elements software (Nikon).
PLISH.
The PLISH protocol was provided by Tushar Desai, Stanford University, Stanford, California, USA (48). Paraffin sections were processed as described for immunofluorescence using antigen retrieval. RNA hybridization probes (100 nM) (Supplemental Table 3) were incubated in hybridization buffer (1 M sodium trichloroacetate, 5 mM EDTA, 50 mM Tris pH 7.4, 0.2 mg/ml heparin in DEPC water) for 2 hours at 37°C and 100% humidity. Sections were incubated with T4 ligase buffer (NEB) and phosphorylated common bridge and connector circle oligos (10 nM) for 60 minutes and followed with T4 ligase buffer and T4 ligase for 2 hours. NxGen phi29 DNA polymerase (Lucigen, 30221) was used for DNA amplification overnight. Slides were washed with label buffer (2× SCC/20% formamide in DEPC water) and incubated with 100 nM Tye 705– or Alexa 568–labeled probe for 1 hour. Immunofluorescence costaining was performed and images obtained and analyzed as above. Reactions performed with probes for the Bacillus subtilis gene mgsA or mock reactions lacking hybridization probes were used for controls. Hybridization probes were ordered from Integrated DNA Technologies with standard desalting and stored as 100 μM stocks.
Cell culture and lentiviral transduction.
HBEC3s were cultured and transduced as previously reported (19) using lentivirus (MOI = 1) containing SFFV-FLAG-hYAP-ires-eGFP (YAP), SFFV-FLAG-hYAPS127A-ires-eGFP (YAP S127A), or SFFV-ires-eGFP (control) (19). Verteporfin and temsirolimus stocks were resuspended in DMSO. For experiments conducted without ambient light, verteporfin stocks were prepared in the dark and all stages of cell culturing and sample collection were performed in the dark. Temsirolimus experiments were performed following standard culture protocols in ambient light. Cells were treated with verteporfin or temsirolimus for 48 hours. For immunofluorescence staining experiments, cells were cultured on ibiTreat chambered μ-Slide coverslips (ibidi, 80826). For proteasome blocking studies, MG132 (2 μM) (MilliporeSigma, M8699) or the mTOR activator MHY1485 (1 μM) (MilliporeSigma, 500554), were added after 24 hours of exposure to verteporfin or temsirolimus. After 48 hours of verteporfin or temsirolimus and 24 hours of MG132 or MHY1485, protein lysates were collected for immunoblotting.
Cell count and viability.
A Muse Cell Analyzer and the Muse Cell Count and Viability assay were used (MilliporeSigma) to determine cell counts and viability, which were confirmed manually using trypan blue and a hemocytometer. For each experiment (n = 3) treatments were assessed in duplicate.
Cell migration assay.
HBEC3s transduced with control, YAP (WT), and YAP (S127A) cells were cultured on 48-well plates. At 100% confluence, cells were treated with DMSO, verteporfin, or temsirolimus. Scratches were performed using a P200 pipette tip. Time-lapse imaging of cell migration was recorded over 16 hours, with images captured every 10 minutes using a ×10 objective. Temsirolimus and control treatments were imaged on an inverted Nikon A1R LUNV confocal microscope. A SpectraX wide-field microscope with a long-pass filter (>680 nm) was used to image verteporfin experiments to minimize photo activation of the drug. For each experimental replicate (n = 3), n = 4 for each treatment.
Statistics.
Error bars are represent SEM. Statistical significance was defined as P < 0.05, as determined by a paired Student’s t test for single comparison or ANOVA for multiple comparisons.
Study approval.
Deidentified patient samples were acquired in accordance with approval from the IRBs of Cincinnati Children’s Hospital Medical Center, Cedar Sinai Medical Center, and Vanderbilt University Medical Center.
Author contributions
JJG designed and performed experiments, analyzed data, and co-wrote the manuscript. AS designed, carried out, and analyzed experiments and edited the manuscript. JG and GC performed experiments. BRS designed experiments and edited the manuscript. YX analyzed and interpreted RNAseq data. AKTP designed experiments and edited the manuscript. JAW designed experiments, interpreted data, and co-wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank Jason Woods (Cincinnati Children’s Hospital Medical Center) for COPD and IPF patient samples. We thank Yina Du for data analysis. We thank the patients who generously donated tissue. Funding for this project was from NIH grants T32 HL007752 (to J.J. Gokey and J.A. Whitsett), U01 HL122642 (to J.A. Whitsett and Y. Xu), U01 HL134745 (to J.A. Whitsett and Y. Xu), U01 HL122638 (to J.A. Whitsett), and R01 HL131661 (to J. Green and A.K.T. Perl).
Version 1. 03/22/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(6):e98738. https://doi.org/10.1172/jci.insight.98738.
Contributor Information
Anusha Sridharan, Email: anusha.sridharan@cchmc.org.
Yan Xu, Email: yan.xu@cchmc.org.
Jenna Green, Email: jenna.green@cchmc.org.
Gianni Carraro, Email: Gianni.Carraro@cshs.org.
Barry R. Stripp, Email: barry.stripp@cshs.org.
References
- 1.King TE, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378(9807):1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 2.Steele MP, Schwartz DA. Molecular mechanisms in progressive idiopathic pulmonary fibrosis. Annu Rev Med. 2013;64:265–276. doi: 10.1146/annurev-med-042711-142004. [DOI] [PubMed] [Google Scholar]
- 3.Barkauskas CE, Noble PW. Cellular mechanisms of tissue fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. Am J Physiol, Cell Physiol. 2014;306(11):C987–C996. doi: 10.1152/ajpcell.00321.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camelo A, Dunmore R, Sleeman MA, Clarke DL. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front Pharmacol. 2014;4:173. doi: 10.3389/fphar.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plantier L, et al. Ectopic respiratory epithelial cell differentiation in bronchiolised distal airspaces in idiopathic pulmonary fibrosis. Thorax. 2011;66(8):651–657. doi: 10.1136/thx.2010.151555. [DOI] [PubMed] [Google Scholar]
- 6.Seibold MA, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS ONE. 2013;8(3):e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kropski JA, Blackwell TS, Loyd JE. The genetic basis of idiopathic pulmonary fibrosis. Eur Respir J. 2015;45(6):1717–1727. doi: 10.1183/09031936.00163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009;84(1):52–59. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogee LM, Dunbar AE, Wert S, Askin F, Hamvas A, Whitsett JA. Mutations in the surfactant protein C gene associated with interstitial lung disease. Chest. 2002;121(3 Suppl):20S–21S. doi: 10.1378/chest.121.3_suppl.20s. [DOI] [PubMed] [Google Scholar]
- 10.Nogee LM, Dunbar AE, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med. 2001;344(8):573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 11.Whitsett JA, Wert SE, Weaver TE. Diseases of pulmonary surfactant homeostasis. Annu Rev Pathol. 2015;10:371–393. doi: 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, et al. Epithelium-specific deletion of TGF-β receptor type II protects mice from bleomycin-induced pulmonary fibrosis. J Clin Invest. 2011;121(1):277–287. doi: 10.1172/JCI42090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonniaud P, et al. Smad3 null mice develop airspace enlargement and are resistant to TGF-beta-mediated pulmonary fibrosis. J Immunol. 2004;173(3):2099–2108. doi: 10.4049/jimmunol.173.3.2099. [DOI] [PubMed] [Google Scholar]
- 14.Chang W, et al. A critical role for the mTORC2 pathway in lung fibrosis. PLoS One. 2014;9(8):e106155. doi: 10.1371/journal.pone.0106155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1(20):e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfleger CM. The Hippo pathway: A master regulatory network important in development and dysregulated in disease. Curr Top Dev Biol. 2017;123:181–228. doi: 10.1016/bs.ctdb.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Shimomura T, Miyamura N, Hata S, Miura R, Hirayama J, Nishina H. The PDZ-binding motif of Yes-associated protein is required for its co-activation of TEAD-mediated CTGF transcription and oncogenic cell transforming activity. Biochem Biophys Res Commun. 2014;443(3):917–923. doi: 10.1016/j.bbrc.2013.12.100. [DOI] [PubMed] [Google Scholar]
- 18.Cui ZL, et al. YES-associated protein 1 promotes adenocarcinoma growth and metastasis through activation of the receptor tyrosine kinase Axl. Int J Immunopathol Pharmacol. 2012;25(4):989–1001. doi: 10.1177/039463201202500416. [DOI] [PubMed] [Google Scholar]
- 19.Lange AW, Sridharan A, Xu Y, Stripp BR, Perl AK, Whitsett JA. Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J Mol Cell Biol. 2015;7(1):35–47. doi: 10.1093/jmcb/mju046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20(7):657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13(8):877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141(8):1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- 23.Volckaert T, et al. Fgf10-Hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev Cell. 2017;43(1):48–59.e5. doi: 10.1016/j.devcel.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C, et al. YAP is essential for mechanical force production and epithelial cell proliferation during lung branching morphogenesis. Elife. 2017 doi: 10.7554/eLife.21130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahoney JE, Mori M, Szymaniak AD, Varelas X, Cardoso WV. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev Cell. 2014;30(2):137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao R, et al. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev Cell. 2014;30(2):151–165. doi: 10.1016/j.devcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brodowska K, et al. The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Exp Eye Res. 2014;124:67–73. doi: 10.1016/j.exer.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chilosi M, et al. Abnormal re-epithelialization and lung remodeling in idiopathic pulmonary fibrosis: the role of deltaN-p63. Lab Invest. 2002;82(10):1335–1345. doi: 10.1097/01.LAB.0000032380.82232.67. [DOI] [PubMed] [Google Scholar]
- 29.Yan Z, Kui Z, Ping Z. Reviews and prospectives of signaling pathway analysis in idiopathic pulmonary fibrosis. Autoimmun Rev. 2014;13(10):1020–1025. doi: 10.1016/j.autrev.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 30.Vaughan AE, Chapman HA. Regenerative activity of the lung after epithelial injury. Biochim Biophys Acta. 2013;1832(7):922–930. doi: 10.1016/j.bbadis.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech. 2010;3(9–10):545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohseni M, et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol. 2014;16(1):108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song JY, et al. Dual function of Yap in the regulation of lens progenitor cells and cellular polarity. Dev Biol. 2014;386(2):281–290. doi: 10.1016/j.ydbio.2013.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szymaniak AD, Mahoney JE, Cardoso WV, Varelas X. Crumbs3-mediated polarity directs airway epithelial cell fate through the Hippo pathway effector Yap. Dev Cell. 2015;34(3):283–296. doi: 10.1016/j.devcel.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gumbiner BM, Kim NG. The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci. 2014;127(Pt 4):709–717. doi: 10.1242/jcs.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence PA, Casal J. The mechanisms of planar cell polarity, growth and the Hippo pathway: some known unknowns. Dev Biol. 2013;377(1):1–8. doi: 10.1016/j.ydbio.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tumaneng K, et al. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14(12):1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Washino S, et al. Temsirolimus induces surfactant lipid accumulation and lung inflammation in mice. Am J Physiol Lung Cell Mol Physiol. 2014;306(12):L1117–L1128. doi: 10.1152/ajplung.00251.2013. [DOI] [PubMed] [Google Scholar]
- 39.Madala SK, et al. Rapamycin regulates bleomycin-induced lung damage in SP-C-deficient mice. Pulm Med. 2011;2011:653524. doi: 10.1155/2011/653524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buschhausen L, Kamm M, Arns W, Schulze-Lohoff E, Weber M. [Successful treatment of a severe case of idiopathic pulmonary fibrosis with rapamycin] Med Klin (Munich) 2005;100(3):161–164. doi: 10.1007/s00063-005-1015-3. [DOI] [PubMed] [Google Scholar]
- 41.Al-Moujahed A, et al. Verteporfin inhibits growth of human glioma in vitro without light activation. Sci Rep. 2017;7(1):7602. doi: 10.1038/s41598-017-07632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng J, Gou J, Jia J, Yi T, Cui T, Li Z. Verteporfin, a suppressor of YAP-TEAD complex, presents promising antitumor properties on ovarian cancer. Onco Targets Ther. 2016;9:5371–5381. doi: 10.2147/OTT.S109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma YW, Liu YZ, Pan JX. Verteporfin induces apoptosis and eliminates cancer stem-like cells in uveal melanoma in the absence of light activation. Am J Cancer Res. 2016;6(12):2816–2830. [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, et al. Verteporfin inhibits YAP function through up-regulating 14-3-3σ sequestering YAP in the cytoplasm. Am J Cancer Res. 2016;6(1):27–37. [PMC free article] [PubMed] [Google Scholar]
- 45.Konstantinou EK, et al. Verteporfin-induced formation of protein cross-linked oligomers and high molecular weight complexes is mediated by light and leads to cell toxicity. Sci Rep. 2017;7:46581. doi: 10.1038/srep46581. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Hansen CG, Moroishi T, Guan KL. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25(9):499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malouf MA, Hopkins P, Snell G, Glanville AR, Everolimus in IPF Study Investigators An investigator-driven study of everolimus in surgical lung biopsy confirmed idiopathic pulmonary fibrosis. Respirology. 2011;16(5):776–783. doi: 10.1111/j.1440-1843.2011.01955.x. [DOI] [PubMed] [Google Scholar]
- 48.Nagendran M, Riordan DP, Harbury PB, Desai TJ. Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife. 2018:e30510. doi: 10.7554/eLife.30510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.