Abstract
Heterozygous chromosomal inversions suppress recombination. Therefore, they may potentially influence recombination-associated phenotypes of human diseases, but no studies have verified this hypothesis. Here, we describe a 35-year-old man with severe congenital ichthyosis. Mutation analysis revealed a heterozygous splice-site mutation, c.1374-2A>G (p.Ser458Argfs*120), in KRT10 on 17q21.2. This mutation was previously reported in patients with ichthyosis with confetti type I (IWC-I), a prominent skin disease characterized by the frequent occurrence of recombination-induced reversion of pathogenic mutations. Intriguingly, the number of revertant skin areas in this patient is considerably reduced compared with typical IWC-I cases. G-banded karyotyping revealed that the patient harbors a heterozygous nonpathogenic inversion, inv(17)(p13q12), whose long-arm breakpoint was subsequently refined to chromosomal positions (chr17: 36,544,407–36,639,830) via FISH. Collectively, the only chance of revertant mosaicism through somatic recombination appears to involve recombination between the KRT10 mutation and the inversion breakpoint. Indeed, in the examined revertant spot, the KRT10 mutation was diminished by somatic recombination starting from chromosomal positions (chr17: 36,915,505–37,060,285) on 17q12. This study provides the first evidence to our knowledge implicating chromosomal inversions as a potential modifier of clinical phenotypes. Furthermore, the reduced occurrence of revertant spots in the recombination-suppressed patient suggests that somatic recombination is the main mechanism of revertant mosaicism in IWC-I.
Keywords: Dermatology, Genetics
Keywords: Genetic diseases, Skin
Heterozygous chromosomal inversions have effects on not only meiotic, but also somatic recombination and are potential modifiers of human disease phenotypes.
Introduction
Inversions are a type of chromosomal rearrangement involving 2-point breakage of a given chromosome, followed by end-to-end rotation of the broken segment (1). Inversions can be pathogenic when the breakpoints disrupt a functionally important gene and/or genomic element but otherwise exhibit a minimal impact upon disease phenotypes. However, such nondamaging inversions significantly contribute to the evolution of genomes in a wide range of species, from Drosophila to humans, because heterozygous inversions protect chromosomes from recombination in the inverted regions during meiosis and allow genes in the protected regions to coevolve, eventually leading to chromosomal speciation (1). Given the recombination-suppressing nature of heterozygous inversions during meiosis, it can be hypothesized that they represent a possible phenotypic modifier of somatic recombination–associated diseases; however, human models verifying this hypothesis have not been reported.
To address the phenotype-modifying effects of inversions in human diseases, we sought to analyze patients with recombination-associated diseases who harbor heterozygous inversions. In human diseases, homologous recombination has long been associated with carcinogenesis and was more recently linked to revertant mosaicism in genetic disorders (2). Revertant mosaicism refers to the somatic reversion of a mutant phenotype resulting from the correction of a disease-causing mutation in a somatic cell, followed by the survival and expansion of the revertant cell (3). This remarkable natural gene therapy phenomenon was first reported in patients with Lesch-Nyhan disease, followed by those with genetic immunodeficiency disorders, such as Wiscott-Aldrich syndrome (4). The possible corrective mechanisms underlying the restoration of gene function include back mutation, second-site mutation, and homologous recombination (crossover or gene conversion) (3, 4). Ichthyosis with confetti (IWC; OMIM #609165), a rare autosomal dominant skin disorder caused by frameshift mutations in KRT10 (IWC-I) or KRT1 (IWC-II), represents the best example of recombination-induced revertant mosaicism (2, 5). IWC is clinically characterized by ichthyosiform erythroderma (widespread reddening of the skin) and, most remarkably, by thousands of independent small patches of mutation-free healthy skin, which increase in number and size with age (2, 6). Here, we report the effects of a heterozygous inversion on revertant phenotypes in a family with IWC-I and provide the first human evidence to our knowledge that even nondamaging inversions can modify disease phenotypes.
Results
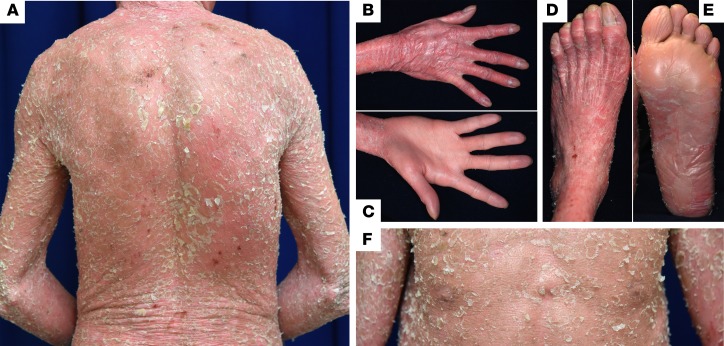
A 35-year-old Japanese man was referred to a dermatologist (T. Nomura) with severe ichthyosis since birth. Physical examination revealed erythroderma, large white or yellow adherent scales, and desquamation (skin peeling) with pruritus of varying intensity on the entire body, with the exception of the palms (Figure 1, A–F). His other clinical features included short stature (4 feet, 11 inches), low body weight (83.8 lb), ectropion, eclabion, microtia, nipple hypoplasia, hypohidrosis, and large lunulae (Figure 1F and Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97595DS1). He also exhibited thoracic kyphosis and joint contractions of the limbs that caused a gait abnormality (Supplemental Figure 1, C and D). He had no other significant medical history. He was born to unrelated healthy parents with no family history of similar skin conditions. Histologically, the lesional skin exhibited hyperkeratosis, parakeratosis, acanthosis, and perinuclear vacuolization of several spinous and granular-layer keratinocytes (Figure 2A). Keratohyalin granules were almost absent (Figure 2A). Based on these observations, the patient was initially diagnosed with congenital ichthyosiform erythroderma (CIE), an autosomal recessive genodermatosis.
Figure 1. Clinical features of the IWC-I patient.
(A–F) The patient presented with diffusely erythematous skin with scales on the whole body, except for the palms (C). Nipple hypoplasia (F) was also noted.
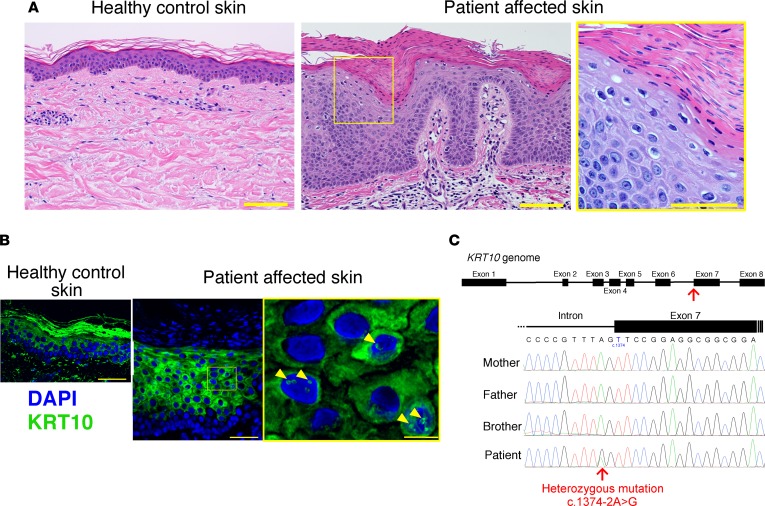
Figure 2. Histological and genetic features of the patient.
(A) Compared with healthy control skin (left panel), the patient’s affected epidermis, including the stratum corneum, was thicker (center panel). Parakeratosis, perinuclear vacuolization of keratinocytes, and marked reduction of keratohyalin granules were also noted (right panel). Scale bar: 100 μm. (B) WT KRT10 forms intermediate filaments in the cytoplasm of the suprabasal keratinocytes. In contrast, ectopic accumulation of KRT10 in the nucleus was observed in the affected epidermis (arrowheads). Scale bar: 50 μm (left and center panels), 10 μm (right panel). (C) A heterozygous splice-site mutation (c.1374-2A>G) in KRT10 was carried by the patient, whereas the unaffected family members were WT for this mutation.
To confirm the diagnosis, we performed whole-exome and Sanger sequencing using genomic DNA extracted from his peripheral blood cells. Unexpectedly, sequencing resulted in the identification of a heterozygous splice-site mutation, c.1374-2A>G, in KRT10 (Figure 2C), whereas no pathogenic homozygous or compound heterozygous mutations were identified in any of the 12 known causative genes for CIE: ABCA12, TGM1, ALOXE3, ALOX12B, CYP4F22, NIPAL4, PNPLA1, LIPN, CERS3, SDR9C7, ST14, and SULT2B1 (data not shown). His unaffected parents and brother did not carry the KRT10 mutation (Figure 2C). Notably, this de novo splice-site mutation was previously reported in patients with IWC-I (2), resulting in a single splice variant, r.1374_1381del, which results in the substitution of serine with arginine at codon 458 and in a premature termination codon 120 amino acids downstream from the mutation (p.Ser458Argfs*120). IHC revealed aberrant nuclear localization of the KRT10 protein in the affected epidermal keratinocytes (Figure 2B), which reflects the fact that the KRT10 mutant contains an arginine-rich nuclear localization signal, as previously reported (2). These results were highly suggestive of the diagnosis of IWC-I, although revertant spots, the most characteristic feature of the disease, were not noted in the initial examination.
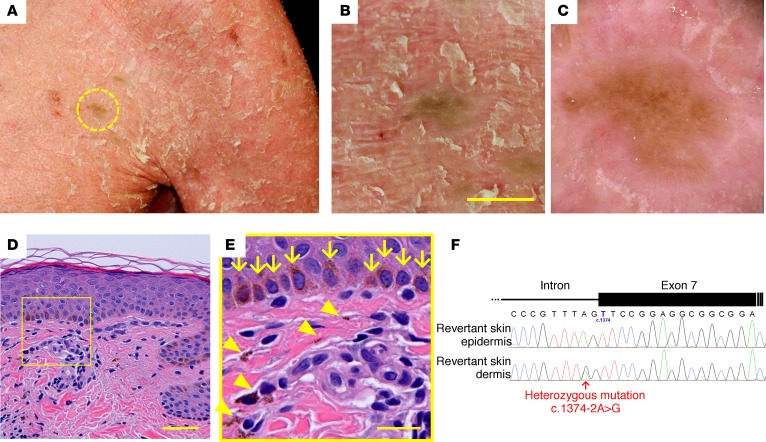
Careful reevaluation of his skin phenotypes demonstrated that his ichthyotic skin was interrupted by ~60 whitish, greenish, or brownish patches, which were up to 10 mm in size and were noted almost exclusively on the trunk and neck (Figure 3, A and B, and Supplemental Figure 2, A–E). Because the patient had not noticed these spots until we called attention to them, it was unclear when they emerged. Dermoscopic observation revealed that the patches exhibited no scaling (Figure 3C), suggesting the possibility that they represented revertant skin areas. To validate this hypothesis, we performed skin sampling from one of the greenish patches on his left chest (Figure 3, A and B), which was then subjected to histological and genetic analyses. Histologically, the stratum corneum exhibited a normal basket-weave pattern without hyperkeratosis, parakeratosis, acanthosis, or a reduced stratum granulosum (Figure 3D), all of which were observed in the lesional skin. Basal melanosis and incontinentia pigmenti histologica (accumulation of melanin pigment in the upper dermis) were also noted (Figure 3E), most likely contributing to the skin discoloration.
Figure 3. Identification of revertant spots.
(A–F) Gross (A and B), dermoscopic (C), histological (D and E), and genetic (F) features of the revertant spot on his left chest. (B) Note that no scales were observed on the revertant spot. Scale bar: 10 mm. (D) The revertant spot showed histological normalization. Scale bar: 50 μm. (E) High-power view of the square area in D. Basal melanosis (arrows) and incontinentia pigmenti histologica (arrowheads) were evident. Scale bar: 20 μm. (F) The splice-site mutation was lost in the revertant epidermis.
We next sought to determine whether this clinically and histologically verified reversal of symptoms was attributed to a correction of the mutation. We examined the KRT10 genotype in genomic DNA samples extracted separately from the epidermis and dermis of the hyperpigmented skin. Notably, the mutation was absent in the epidermis but retained in the dermis (Figure 3F). Thus, we confirmed that the somatic reversion of the mutation in the epidermis underlies the clinical and histological recovery. Taking these results together, we ultimately made a diagnosis of IWC-I. However, it was intriguing that the number of revertant skin areas in our patient was considerably reduced compared with typical IWC-I cases, in which thousands of healthy confetti-like skin spots appear in early childhood, or even at birth, and increase in number and size with time (2), although his other clinical features — such as erythroderma, adherent scales, short stature, low body weight, ectropion, eclabion, microtia, nipple hypoplasia, hypohidrosis, large lunulae, and gait abnormality — were rather typical of IWC-I. This discrepancy could not be explained by genotype-phenotype correlation or the age of the patient, as a 35-year-old IWC-I patient who is heterozygous for the same KRT10 mutation (p.Ser458Argfs*120) was reported to exhibit thousands of revertant skin spots (7). Therefore, the question arises of what determines the reduced occurrence of revertant spots in the patient.
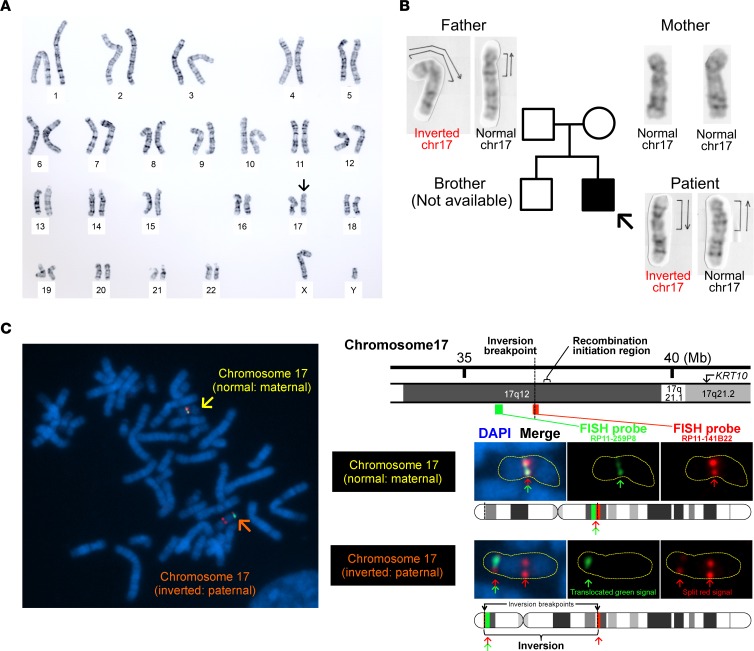
We hypothesized that chromosomal structural abnormality might underlie this intriguing phenomenon because heterozygous inversions mediate inhibition of somatic recombination, which is the only reported mechanism underlying the reversion of pathogenic mutations in IWC-I patients (2, 8). To address this hypothesis, we performed G-banded karyotyping using peripheral blood lymphocytes. Notably, we identified a heterozygous pericentric inversion (an inversion that includes the centromere) of chr17, inv(17)(p13q12), in the proband (Figure 4A). This previously unreported inversion itself does not confer pathogenicity upon the skin phenotype, as the patient’s unaffected father is also heterozygous with respect to the inversion, whereas his mother is WT (Figure 4B). Cloning and sequencing of each allele subsequently revealed that the de novo KRT10 mutation is located on the paternal inverted chr17 (data not shown).
Figure 4. Detection of inv(17)(p13q12) and its breakpoints.
(A) G-banded karyotype analysis using peripheral blood lymphocytes from the proband revealed that the patient was heterozygous for a pericentric inversion (arrow), inv(17)(p13q12). (B) His unaffected father is also heterozygous with respect to inv(17)(p13q12), suggesting that the inversion itself is nonpathogenic. His mother is WT for the inversion. (C) Two-color FISH analysis using 2 BAC clone probes, RP11-141B22 (red) and RP11-259P8 (green), revealed a split red signal, suggesting that the breakpoint of the inversion is located in the RP11-141B22 locus.
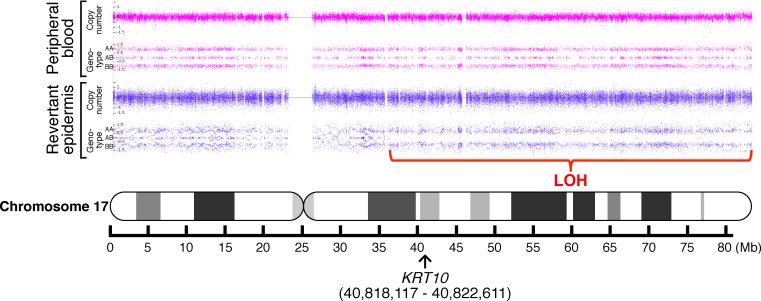
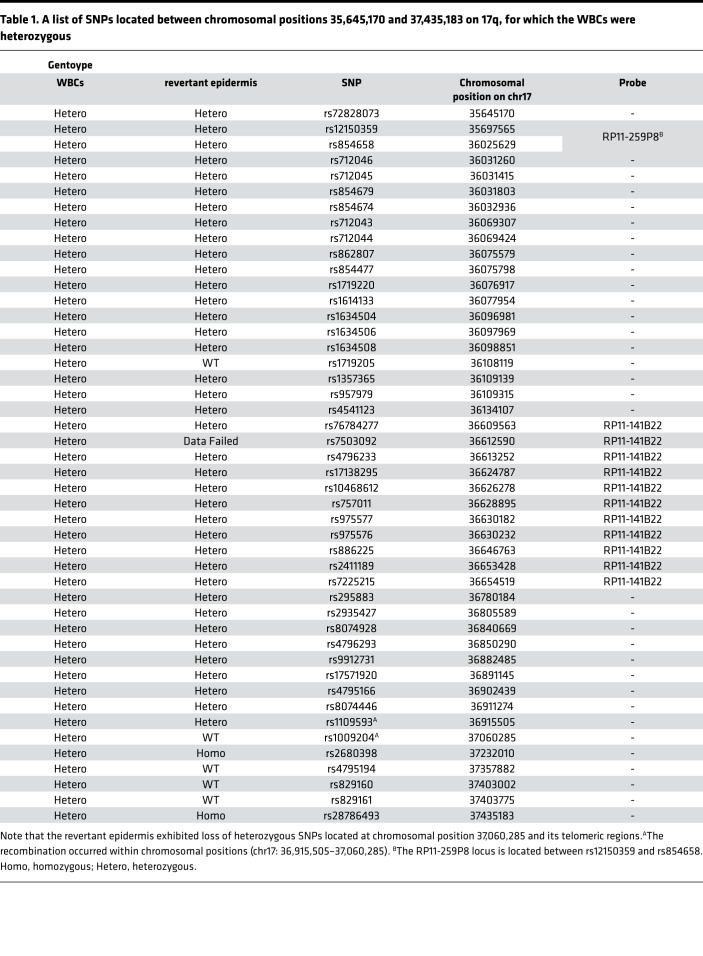
Notably, heterozygous pericentric inversions act as a recombination inhibitor for the following reasons: in the inverted region, (i) they result in a reduction of chiasma (the point of chromosomal crossover that is a prerequisite for recombination) formation, and (ii) recombination produces an unbalanced chromosome, with duplication or deletion of some portion of the chromosome, which typically makes cells inviable (9). Under this scenario, the only chance of mutation reversion via somatic recombination is for recombination to occur between the KRT10 mutation site and the breakpoint of the pericentric inversion on the long arm of chr17 (17q). Thus, we first performed genome-wide genotyping to compare the genotypes of the revertant epidermal cells and peripheral blood cells. We identified a copy-neutral loss of heterozygosity (LOH) on 17q only in the revertant epidermis (Figure 5), whereas no large LOHs were detected in other chromosomes (data not shown). The LOH started from a chromosomal position (chr17: 37,060,285 in hg38 coordinates) centromeric to KRT10 and extended to the telomere (Figure 5 and Table 1), indicating that somatic recombination had occurred within chromosomal positions (chr17: 36,915,505–37,060,285). To refine the breakpoint of inv(17)(p13q12), we next performed FISH analysis using paired BAC clone probes, RP11-141B22 (chr17: 36,544,407–36,735,254) and RP11-259P8 (chr17: 35,748,902–35,920,353), which correspond to the immediate centromeric region of the recombination initiation site and the more centromeric region of 17q12, respectively (Figure 4C). This finding revealed that the breakpoint of the inversion resides on the centromeric side of the RP11-141B22 locus (chr17: 36,544,407–36,639,830, Figure 4C). Taken together, these results indicate that homologous recombination in the examined revertant epidermis occurred between the inversion breakpoint and the KRT10 mutation, which is consistent with our hypothesis (Figure 6). Thus, the heterozygous pericentric inversion, inv(17)(p13q12), is highly likely to suppress the occurrence of revertant mosaicism in the IWC-I patient via inhibition of somatic recombination within the inverted region. However, we cannot exclude the possibility that some SNPs on 17q reduced the growth advantage of the revertant keratinocytes by becoming homozygous as a consequence of homologous recombination.
Figure 5. A copy-neutral LOH on 17q.
Whole-genome SNP microarray analysis identified a copy-neutral LOH on 17q in the revertant epidermis, whereas no LOH was detected in peripheral blood cells. The LOH began in regions centromeric to KRT10 (arrow) and extended to the telomere of the chromosome. No LOH was present on other chromosomes (data not shown).
Table 1. A list of SNPs located between chromosomal positions 35,645,170 and 37,435,183 on 17q, for which the WBCs were heterozygous.
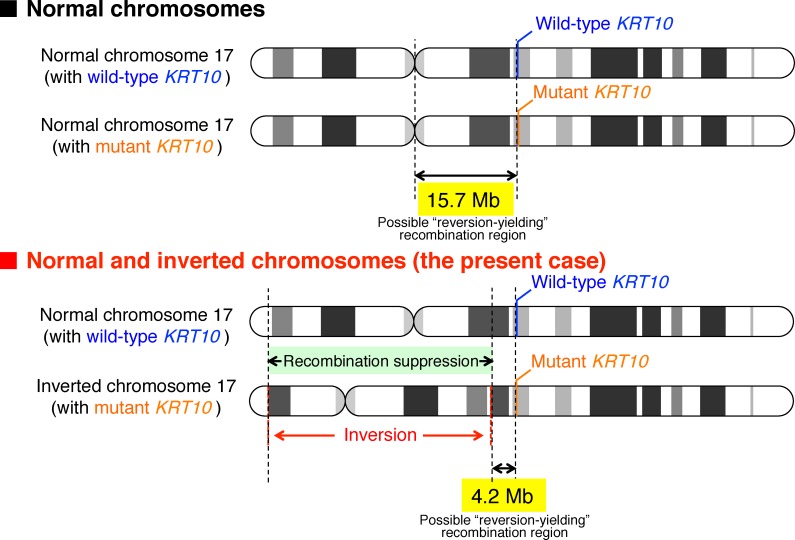
Figure 6. Schematic diagram explaining the possible mechanism of fewer revertant skin spots in the present case.
IWC patients carry a heterozygous mutation in KRT10. The mutation can be reverted if recombination occurs at the chromosomal regions centromeric to KRT10. Given that a heterozygous inversion inhibits recombination in the inverted chromosomal regions, the physical size of a possible reversion-yielding recombination region in IWC-I patients with or without the heterozygous inversion, inv(17)(p13q12), is approximately 4.2 or 15.7 Mb, respectively, according to the UCSC Genome Browser Human Dec. 2013 (GRCh38/hg38) (https://genome.ucsc.edu/).
There are other possible reasons for the considerably reduced number of revertant spots presented by our patient. For example, he might carry mutations in recombination-associated genes that reduce the recombination rate. To test this, we reanalyzed the whole-exome sequencing data derived from the patient’s peripheral blood cells. This analysis revealed that he harbors no pathogenic mutations in any of the following 15 recombination-associated genes: BARD1, FAM175A, XRCC2, NBN, ATM, CHEK1, MRE11A, BRCA2, XRCC3, RAD51, PALB2, BRCA1, BRIP1, RAD51C, and CHEK2 (10–13) (Supplemental Table 1).
Discussion
This study gives rise to 2 clinically important ideas. First, when inversions are heterozygous, they have effects on not only meiotic, but also somatic recombination and are potential modifiers of human disease phenotypes. To the best of our knowledge, our patient represents the first human example of this situation. Furthermore, a Robertsonian translocation has been reported to exert karyotypic suppression of somatic recombination and to protect against LOH and cancer development in familial adenomatous polyposis–model mice (14). Thus, chromosomal abnormalities, even if nonpathogenic, can affect the phenotypes of genetic diseases associated with somatic recombination, such as IWC (2, 5), Bloom syndrome (OMIM #210900) (15), Fanconi anemia (OMIM #227650) (16), and dyskeratosis congenita (OMIM #127550) (17), which further emphasizes the importance of karyotype tests not only in genomic disorders, but also in genetic disorders.
Second, somatic recombination serves as a key process in the initiation of revertant mosaicism in IWC-I. To date, a total of 31 revertant skin spots from 7 patients with IWC-I have been analyzed with respect to LOH, all of which exhibited reversion of pathogenic KRT10 mutations via somatic recombination (2, 8). However, the major limitation of these studies lies in the limited number of spots examined, despite the fact that thousands of revertant spots exist in each patient. The reduced occurrence of revertant spots in our recombination-suppressed patient strongly suggests that somatic recombination is the major, and likely the exclusive, mechanism of revertant mosaicism in IWC-I.
Interestingly, inversions on a given chromosome affect recombination rates even in the uninverted regions (9). Given that approximately 20% of individuals of European ancestry carry a 900-kb paracentric inversion polymorphism on 17q21.31, which is next to the KRT10 locus (17q21.2), and that this inversion is associated with an increased recombination rate genome-wide (in addition to having effects of its own during meiosis; ref. 18), one would predict that IWC-I patients of such ancestry might exhibit phenotypic variations with regard to revertant skin spots. Indeed, the age at detection for revertant skin spots in IWC-I has been reported to vary greatly among patients aged 0–57 years (2, 7). Although we have no access to chromosomal data from these patients and, therefore, cannot draw any conclusion regarding whether inversion polymorphisms (including the 900-kb inversion) have an effect on the occurrence of revertant phenotypes in IWC-I, it is worth addressing this point in future studies.
Methods
IHC.
Punch-biopsied skin samples were immediately immersed in Cryo Mount (Muto Pure Chemicals) and frozen at –80°C. Briefly, 4-μm–thick frozen tissue sections were obtained using a Leica CM1900 cryostat (Leica) and dried for 30 minutes at room temperature. After rinsing in PBS and blocking with 10% normal goat serum, the sections were incubated with the primary anti–human KRT10 antibody (LH2, Santa Cruz Biotechnology Inc.) diluted in PBS, followed by incubation with the Alexa Fluor 680–conjugated goat anti–mouse IgG (H + L) secondary antibody (A21057, Thermo Fisher Scientific). Nuclei were counterstained with DAPI (Dojindo), and fluorescence images were obtained using an Olympus FV1000 confocal laser scanning microscope (Olympus).
Genomic DNA extraction.
For mutational analysis, genomic DNA from all participants was extracted from peripheral blood or saliva using a QIAamp DNA Blood Maxi Kit (Qiagen) or an Oragene DNA Self-Collection Kit (DNA Genotek), respectively. Genomic DNA was also extracted from the proband’s skin using a QIAamp DNA Mini kit (Qiagen) after separation of the epidermis and dermis from punch-biopsied skin samples using ammonium thiocyanate solution (MilliporeSigma), as described previously (19).
Whole-exome sequencing.
Following the enrichment of coding regions and intron/exon boundaries using the SureSelect Human All Exon V5 Kit (Agilent Technologies), whole-exome sequencing was performed on the HiSeq 2000 platform (Illumina) with 100-bp paired-end reads to a median of 100× coverage. The resulting data were analyzed and annotated using DNAnexus software (DNAnexus Inc., https://www.dnanexus.com/, accessed May 2013).
Sanger sequencing.
A genomic region spanning the c.1374-2A>G mutation in KRT10 was amplified via PCR using AmpliTaq Gold PCR Master Mix (Thermo Fisher Scientific). The primer sequences were as follows: forward, 5′- CCTGGCAGGGTTTTATTTTT-3′, and reverse, 5′- TTCTTGGTTTCTGATTCAACCAT-3′. The amplification conditions were as follows: 1 cycle at 96°C for 10 minutes; 35 cycles at 96°C for 30 seconds, 56°C for 30 seconds, and 72°C for 1 minute; and 1 cycle at 72°C for 7 minutes. The amplicon was treated with the ExoSAP-IT reagent (Thermo Fisher Scientific), followed by the sequencing reaction using BigDye Terminator version 3.1 (Thermo Fisher Scientific). An ABI 3130xl genetic analyzer (Thermo Fisher Scientific) was used with standard running conditions for electrophoresis and data collection to obtain the sequence data.
Whole-genome oligo-SNP array.
An Affymetrix CytoScan HD array (Affymetrix) was used to identify copy number variations and LOH using genomic DNA prepared from the peripheral blood and epidermis as described previously (6).
G-banded karyotyping.
G-banding was performed using peripheral blood lymphocytes as described previously (20).
Metaphase chromosome preparation.
Heparinized whole blood was incubated for 48 hours in RPMI 1640 medium (MilliporeSigma) supplemented with FBS (MilliporeSigma), fibronectin human protein (Thermo Fisher Scientific), and an antibiotic antimycotic solution (MilliporeSigma) under standard cell culture conditions, at 37°C with 5% CO2, followed by the addition of 300 μg/ml thymidine (Tokyo Kasei). The thymidine block was released after 16 hours of incubation by replacement of the medium with RPMI 1640 medium supplemented with FBS (MilliporeSigma), antibiotic antimycotic solution (MilliporeSigma), and 25 μg/ml bromodeoxyuridine (MilliporeSigma). Five hours after the release of the block, colcemid (MilliporeSigma) was added to a final concentration of 0.02 μg/ml. One hour later, the cells were collected and treated with 0.075 M KCl for 20 minutes at 37°C, followed by suspension in fixative (3:1 methanol/acetic acid) 3 times.
Probe preparation.
BAC clones, RP11-141B22 (chr17: 36,544,407–36,735,254) and RP11-259P8 (chr17: 35,748,902–35,920,353) (BACPAC Resources Center), were used to generate FISH probes. The BAC DNAs were labeled via nick translation with Cy3-dUTP (GE Healthcare) and Green-dUTP (Abbott Laboratories). The labeled probes were mixed with sonicated salmon sperm DNA and dog cot-1 DNA in hybridization solution.
FISH analysis.
Metaphase chromosome samples in fixative were seeded onto slide glass, followed by a 2-hour hardening treatment at 70°C. Then, probe solutions were applied to the pretreated sections, which were subsequently covered with cover slips and simultaneously denatured at 70°C for 5 minutes, followed by hybridization for 16 hours at 37°C. Next, the hybridized slides were washed with standard saline citrate and 50% formamide and counterstained with DAPI. Separate fluorochrome images were captured using a Leica DC 350FX cooled CCD camera (Leica Microsystems) mounted on a Leica DMRA2 microscope employing Leica CW4000 FISH software. The images were analyzed using Leica CW4000 karyo software (Leica).
Study approval.
The patient and his family members provided written informed consent to participate in this study, in compliance with the Declaration of Helsinki. The IRB at the Hokkaido University Graduate School of Medicine approved this study (project No. 14-063).
Author contributions
TN and S. Suzuki conceived and designed this study. TN, TM, S. Shinkuma, YF, WN, MA, and HS obtained clinical materials. S. Suzuki and MT conducted the genetic analysis. S. Suzuki conducted the immunohistochemical analysis. TN and S. Suzuki analyzed the data. TN and S. Suzuki wrote the manuscript. All authors approved the final manuscript.
Supplementary Material
Acknowledgments
We are most indebted to the patient and his family members for their participation in this study. This work was supported by Rohto Dermatology Research Award (to TN), the JSPS KAKENHI (grant no. JP15K09738) (to TN), the Terumo Foundation for Life Sciences and Arts (grant no. 16-II 330) (to TN), and the Northern Advancement Center for Science & Technology (NOASTEC) Foundation (grant no. H28 T-1-42) (to TN).
Version 1. 03/22/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(6):e97595. https://doi.org/10.1172/jci.insight.97595.
Contributor Information
Toshifumi Nomura, Email: nomura@huhp.hokudai.ac.jp.
Shotaro Suzuki, Email: s_suzuki@albion.co.jp.
Toshinari Miyauchi, Email: billlauren64@gmail.com.
Masae Takeda, Email: ma-takeda@med.hokudai.ac.jp.
Satoru Shinkuma, Email: qxfjc346@yahoo.co.jp.
Yasuyuki Fujita, Email: yfujita@med.hokudai.ac.jp.
Wataru Nishie, Email: nishie@med.hokudai.ac.jp.
Masashi Akiyama, Email: makiyama@med.nagoya-u.ac.jp.
Hiroshi Shimizu, Email: shimizu@med.hokudai.ac.jp.
References
- 1.Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol. 2010;8(9):e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choate KA, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330(6000):94–97. doi: 10.1126/science.1192280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonkman MF, Pasmooij AM. Revertant mosaicism--patchwork in the skin. N Engl J Med. 2009;360(16):1680–1682. doi: 10.1056/NEJMc0809896. [DOI] [PubMed] [Google Scholar]
- 4.Lai-Cheong JE, McGrath JA, Uitto J. Revertant mosaicism in skin: natural gene therapy. Trends Mol Med. 2011;17(3):140–148. doi: 10.1016/j.molmed.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choate KA, et al. Frequent somatic reversion of KRT1 mutations in ichthyosis with confetti. J Clin Invest. 2015;125(4):1703–1707. doi: 10.1172/JCI64415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki S, et al. Revertant Mosaicism in Ichthyosis with Confetti Caused by a Frameshift Mutation in KRT1. J Invest Dermatol. 2016;136(10):2093–2095. doi: 10.1016/j.jid.2016.05.109. [DOI] [PubMed] [Google Scholar]
- 7.Hotz A, et al. Expanding the Clinical and Genetic Spectrum of KRT1, KRT2 and KRT10 Mutations in Keratinopathic Ichthyosis. Acta Derm Venereol. 2016;96(4):473–478. doi: 10.2340/00015555-2299. [DOI] [PubMed] [Google Scholar]
- 8.Lim YH, Qiu J, Saraceni C, Burrall BA, Choate KA. Genetic Reversion via Mitotic Recombination in Ichthyosis with Confetti due to a KRT10 Polyalanine Frameshift Mutation. J Invest Dermatol. 2016;136(8):1725–1728. doi: 10.1016/j.jid.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navarro A, Betrán E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics. 1997;146(2):695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George E, et al. A patient-derived-xenograft platform to study BRCA-deficient ovarian cancers. JCI Insight. 2017;2(1):e89760. doi: 10.1172/jci.insight.89760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruno PM, et al. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat Med. 2017;23(4):461–471. doi: 10.1038/nm.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennington KP, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20(3):764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues DN, Boysen G, Sumanasuriya S, Seed G, Marzo AM, de Bono J. The molecular underpinnings of prostate cancer: impacts on management and pathology practice. J Pathol. 2017;241(2):173–182. doi: 10.1002/path.4826. [DOI] [PubMed] [Google Scholar]
- 14.Haigis KM, Dove WF. A Robertsonian translocation suppresses a somatic recombination pathway to loss of heterozygosity. Nat Genet. 2003;33(1):33–39. doi: 10.1038/ng1055. [DOI] [PubMed] [Google Scholar]
- 15.Ellis NA, Lennon DJ, Proytcheva M, Alhadeff B, Henderson EE, German J. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am J Hum Genet. 1995;57(5):1019–1027. [PMC free article] [PubMed] [Google Scholar]
- 16.Lo Ten Foe JR, et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet. 1997;5(3):137–148. [PubMed] [Google Scholar]
- 17.Jongmans MC, et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am J Hum Genet. 2012;90(3):426–433. doi: 10.1016/j.ajhg.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37(2):129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 19.Clemmensen A, et al. Extraction of high-quality epidermal RNA after ammonium thiocyanate-induced dermo-epidermal separation of 4 mm human skin biopsies. Exp Dermatol. 2009;18(11):979–984. doi: 10.1111/j.1600-0625.2009.00921.x. [DOI] [PubMed] [Google Scholar]
- 20.Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;2(7731):971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.