Abstract
Following decades of ecologic and economic impacts from a growing list of nonindigenous and invasive species, government and management entities are committing to systematic early- detection monitoring (EDM). This has reinvigorated investment in the science underpinning such monitoring, as well as the need to convey that science in practical terms to those tasked with EDM implementation. Using the context of nonindigenous species in the North American Great Lakes, this article summarizes the current scientific tools and knowledge – including limitations, research needs, and likely future developments – relevant to various aspects of planning and conducting comprehensive EDM. We begin with the scope of the effort, contrasting target-species with broad-spectrum monitoring, reviewing information to support prioritization based on species and locations, and exploring the challenge of moving beyond individual surveys towards a coordinated monitoring network. Next, we discuss survey design, including effort to expend and its allocation over space and time. A section on sample collection and analysis overviews the merits of collecting actual organisms versus shed DNA, reviews the capabilities and limitations of identification by morphology, DNA target markers, or DNA barcoding, and examines best practices for sample handling and data verification. We end with a section addressing the analysis of monitoring data, including methods to evaluate survey performance and characterize and communicate uncertainty. Although the body of science supporting EDM implementation is already substantial, research and information needs (many already actively being addressed) include: better data to support risk assessments that guide choice of taxa and locations to monitor; improved understanding of spatiotemporal scales for sample collection; further development of DNA target markers, reference barcodes, genomic workflows, and synergies between DNA-based and morphology-based taxonomy; and tools and information management systems for better evaluating and communicating survey outcomes and uncertainty.
Keywords: early-detection monitoring, survey design, morphological taxonomy, DNA-based taxonomy, uncertainty characterization
1. Introduction
Nonindigenous species (NIS) threaten biodiversity and the functioning of ecosystems and economies worldwide (Pejchar and Mooney 2009). In the USA, invasive species cause damage exceeding $120 billion annually (Pimentel et al. 2005), and in the North American Great Lakes, the most heavily invaded freshwater system in the world (Pagnucco et al. 2015), the loss in ecosystem services caused by NIS introduced by the ship-borne invasion pathway alone is an estimated $138 million annually (Rothlisberger et al. 2012). Even localized invasions can result in dramatic and costly losses in ecosystem services (Walsh et al. 2016). With ever-increasing global commerce, and in the absence of effective surveillance and management, the danger posed by NIS introduced to ecosystems worldwide will also increase (Keller et al. 2011; Lodge et al. 2006; Pagnucco et al. 2015). Early detection monitoring (EDM) is a critical component of efforts to mitigate threats posed by NIS, allowing for new invaders to be responded to (e.g., by eradication or containment – Lodge et al. 2006; Mehta et al. 2007; Vander Zanden et al. 2010) and the effectiveness of prevention measures to be evaluated. Accordingly, EDM is now being called for in national and international policies and initiatives. For example, the 2012 Great Lakes Water Quality Agreement between Canada and the USA includes a commitment to establishing an aquatic NIS early detection and rapid response network.
Early detection monitoring falls under the umbrella of biological assessment, and shares the general aims and concerns regarding survey designs, collection approaches, and taxonomic challenges. However, EDM differs from other biological assessment in ways that have significance for sampling design and resources. Because detection is only “early” if organisms are found while still few and localized (i.e., rare) and rare organisms are inherently difficult to find, EDM programs are particularly challenged by the need to reconcile limited resources with comprehensive sampling. EDM programs may need to search a broad suite of habitats with a broad range of methods to avoid missing elusive NIS, which runs counter to the repeatability and standardization that other biological surveys typically seek. Early detection monitoring is particularly demanding regarding taxonomic processing because of the need for thorough sample searches and highly-resolved organism identification, whereas assessments of biologic condition via indicator taxa can be robust with coarser searches and taxonomy (Carter and Resh 2001). Finally, EDM programs particularly need to quantify survey effectiveness and uncertainty, as a firm basis for communicating outcomes, choosing management responses, and adaptively refining survey designs.
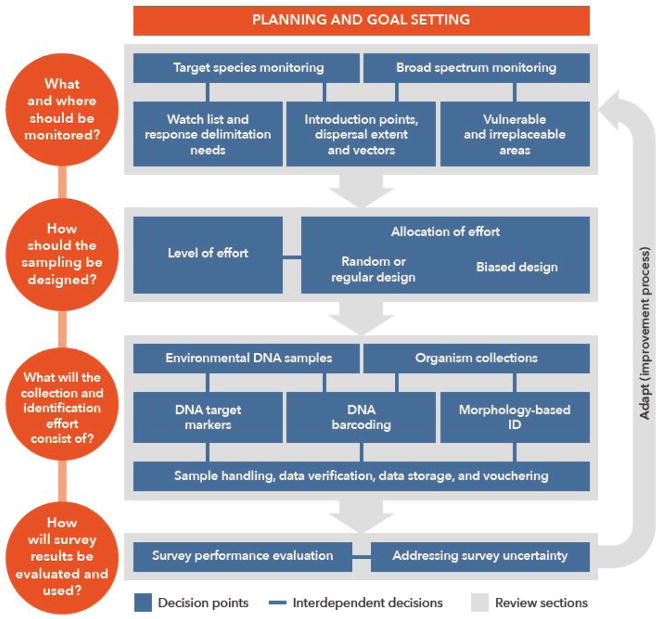
In this article, we review current capabilities and scientific understanding for the design and implementation of EDM and discuss areas where further development and testing is needed. Our presentation is structured around major decision points confronting those charged with implementing EDM (Figure 1) namely: 1) What and where should be monitored? 2) How should the survey be designed with respect to effort and its allocation? 3) What will the collection and identification effort consist of? And 4) How will the survey outcome be evaluated and communicated? We frame our review in the context of aquatic NIS in the North American Great Lakes, but the challenges, decision points, and science tools discussed are applicable to EDM broadly.
Figure 1.
Diagram of decision points involved in implementing NIS early detection monitoring (blue boxes), and the sections of our review that addresses them (grey shaded areas). Narrow blue arrows connect boxes where decisions are interdependent, wider grey arrows show the progression among EDM implementation steps which, when iterated, constitute an adaptive learning cycle.
2. What and where should be monitored?
Two contrasting approaches to the question of “what to monitor for” have been the subject of EDM development. One approach searches for preidentified species of concern (target-species monitoring, hereafter), while the second conducts broad-spectrum monitoring aimed at finding any new NIS within broad taxonomic groups. While target-species monitoring most directly and efficiently incorporates knowledge of imminent NIS, broad-spectrum monitoring enables the discovery of unexpected NIS while also yielding biodiversity information relevant to evaluating NIS impacts and other ecological questions (Simmons et al. 2015). Both target-species monitoring and broad-spectrum monitoring (also sometimes referred to as active vs. passive surveillance – Simmons et al. 2015) should be considered valuable components to a surveillance toolbox.
Target-species monitoring choices are generally informed by taxonomic prioritizations (watch lists) constructed from a combination of information on ecological suitability, previous invasion history and impacts, and ability to block or eradicate (McGeoch et al. 2016). Monitoring choices can also be driven by recently detected NIS incursions or range expansions (e.g., Bowen and Keppner 2015; Jerde et al. 2013). For example, substantial effort has been invested in conducting risk assessments for species with the potential to invade and impact the Great Lakes (Gantz et al. 2015; Grippo et al. 2017; Snyder et al. 2014; U.S. Fish and Wildlife Service 2015). Such assessments not only identify likely target species, but also provide an obvious basis for deciding where and how to monitor by identifying relevant habitats, entry points, and collection devices (e.g., Herborg et al. 2007; Howeth et al. 2016; Muirhead et al. 2011). Species watch lists may also be filtered by vulnerability to specific introduction pathways -- e.g., ballast water (Ricciardi 2006; Wonham et al. 2013), live-organism trade (Gertzen et al. 2008), or recreational boating (Schneider et al. 1998) -- which provides additional information to support EDM design. Currently, the effort surrounding monitoring for Asian Carps near at-risk entry points is the best-developed example of target-species EDM in the Great Lakes (Jerde et al. 2013; Wilson et al. 2014).
Broad-spectrum monitoring is also backed by an extensive scientific literature aimed at understanding the geographic distribution of invasion risk. Although this type of monitoring precludes the specificity provided by species-based risk assessment, options for prioritizing locations include compiling introduction vectors and estimating their propagule pressure, extracting spatial patterns from NIS incidence databases, and identifying existing NIS hotspots (e.g., Grigorovich et al. 2003; O’Malia 2015). Site prioritization also may consider ecological characteristics of recipient habitats (McGeoch et al. 2016), potential to serve as epicenter for further NIS spread (Rothlisberger et al. 2012), value for ecosystem services or biodiversity reservoirs (Margules and Pressey 2000; Walsh et al. 2016), and place- specific societal tolerance for NIS (e.g., nature park vs. commercial port). In the Great Lakes, fish community monitoring implemented at various shipping ports currently constitutes the most mature example of broad-spectrum monitoring (Hoffman et al. 2016).
Regardless of the approach, there is reason to believe that the scientific basis for monitoring targets can continue to be improved. Invasion risk assessments are limited by the quality of available data, which can leave existing watch lists taxonomically incomplete. For example, multiple non-indigenous mollusks and oligochaetes are already established in the Great Lakes (Ricciardi 2006) yet these taxa are underrepresented in watch lists, because risk assessments have focused on higher profile species (e.g., fish and zooplankton) and poor distribution information limits the ability to assess ecological tolerances and spread vectors. Likewise, the availability and quality of data on anthropogenic vectors varies greatly. Quantitative data are simply lacking for some vectors (e.g., bait bucket transfers), and others are difficult to synthesize due to disparate data collection methods and scales (e.g., commercial and recreational vessel traffic). These limitations highlight opportunities for adaptive EDM, wherein both taxonomic and geographic targets are modified as additional information is acquired.
There is also more work to be done concerning how EDM efforts are best coordinated and synthesized. Although the rationale for monitoring high-risk species and locations is obvious, such monitoring fails to constitute a comprehensive EDM program for a large and ecologically diverse region such as the Great Lakes. Monitoring only a few taxa at a limited number of locations can leave large areas unsearched, leads to incomplete understanding about spread and impacts of NIS, and foregoes the opportunity to assess the generality of monitoring strategies. Language in policy documents such as the 2012 Great Lakes Water Quality Agreement makes clear the intent for developing an EDM network and not just EDM of individual places. Such a network arguably should include background as well as hot-spot sampling, needs to incorporate searches capable of uncovering taxa not suspected to be invasive or imminent, and might entail combinations of approaches (e.g., broad-spectrum and target-species sampling, opportunistic sampling and pathway monitoring) which would increase the complexity of the design and outcome evaluation. There is also potential for incorporating EDM into biological monitoring being conducted for other purposes, but exactly how that is best accomplished remains to be determined.
3. How should the survey be designed with respect to effort and its allocation?
Survey design and execution arguably are the most crucial elements of early detection monitoring. Where, in what manner, and how intensively monitoring is conducted governs the adequacy and efficiency of the effort, sets the context for interpreting technical details, and underpins the ability to make informed policy and management decisions. In this section, we discuss two major dimensions of survey design, namely overall level of effort to expend and how to allocate that effort in space and time. Information relevant to probing these questions derives in part from evaluating the outcome of previous surveys (section 5). Consequently, it is fruitful to approach EDM as an adaptive management process of “implement – evaluate – refine” (Hoffman et al. 2016).
Whether the plan is to conduct target-species or broad-spectrum monitoring has substantial influence on survey design. Target-species EDM planning has the benefit of considerable up-front information; questions of where and how to sample can be approached by building on known habitat preferences and collection vulnerabilities and expert opinion concerning detectability (e.g., Jarrad et al. 2011). Broad-spectrum EDM, in contrast, needs to be approached from a cover-all-bases perspective: sampling across all relevant environmental dimensions to avoid presupposing where new species might establish and employing multiple collection devices because of the varying organisms and habitats for which they are effective. Combining multiple sampling methods and habitats might also be necessary for target-species EDM, if no one collection device is effective for all locations or a suite of species with disparate habitat associations is being targeted (e.g., Gladman et al. 2010; Jarrad et al. 2011).
3.1 Level of effort
Deciding on the overall level of effort is probably of foremost concern, because of the close connection to available personnel and funding. Because the primary goal of EDM is detecting new species while they are still rare (low abundance, limited distribution), detection sensitivity -- the probability of finding species with a given sampling method and intensity – and the converse non-detection risk are the obvious basis for setting EDM performance goals. Searches for rare species have a high probability of failure by definition and no survey can prove something absent; rather the goal should be reasonable certainty that the effort was sufficient to detect ‘rare’ species, however that is defined.
While the desired detection probability is ultimately a matter of policy and resources, EDM science provides the datasets and tools to estimate detection probabilities for a given level of effort (see section 5) and has demonstrated that early detection is a resource-intensive proposition. For example, a study examining the role of timing in monitoring for an NIS zooplankter with greatly varying seasonal abundance revealed a need for upwards of 100 samples to achieve 95% detection probability (Harvey et al. 2009a). Analyses of broad-spectrum monitoring data from a Great Lakes port found that detecting 95% of the total species pool takes on the order of 100, 200, and >500 samples for fish, benthic macroinvertebrates, and zooplankton respectively (Hoffman et al. 2011). A study monitoring planktonic NIS in reservoirs suggested a need for a tenfold increase in net-tow volume to confidently detect a large proportion of the taxa present (Counihan and Bollens 2017). Furthermore, important endpoints related to survey performance evaluation (e.g., asymptotic richness, rare taxa occurrence) are adequately estimated only if the survey pushes well out on the species-effort curve. Accordingly, there is value to “oversampling” to amass data with which to conduct numerical experiments aimed at refining future sampling protocols (e.g., Trebitz et al. 2009) – especially when a new system or taxonomic group is undertaken.
The EDM level-of-effort question can also be approached from the perspective of balancing initial surveillance versus eventual control (eradication, containment, etc.). Detection becomes easier while control becomes harder as NIS become more abundant and widespread (Hulme 2006, Vander Zanden et al. 2010), yet the resources for accomplishing both often come from a single pool. Optimizing this trade-off has been approached via decision-support models that are parameterized with data on population size and dynamics (e.g., initial number, rates of survival and spread), costs of surveillance and control (personnel hours or monetary units), and costs of damages that NIS might inflict (e.g., Epanchin-Niell et al. 2012; Hauser and McCarthy 2009, Mehta et al. 2006; Yemshanov et al. 2017). Generally, these studies find that greater initial surveillance effort is justified for NIS that are more likely to establish and expand, more difficult to control, and more likely to cause damages – in other words NIS that are typically high on watch-list priorities. Because of the information needs for these models, this approach to the surveillance effort question is primarily applicable to target-species monitoring.
3.2 Allocation of effort
EDM effort should be allocated with a formal sample design, so that key parameters such as NIS distribution patterns can be statistically quantified. The effort allocation should also consider whether the EDM is well-established and tested or still in a learning mode regarding how best to sample. Efficiencies in EDM arise from identifying the habitats and collection protocols that most likely yield the target species or the combinations of habitats and collection devices that most rapidly accumulate taxa across the biological groups of interest. If this is not yet known, the monitoring also needs to produce the data to determine these efficiencies, for example by embedding various possible comparisons of space, time, and sampling devices in the survey designs and by collecting supporting environmental data.
In the absence of specific knowledge concerning patterns in the taxonomic group and system of interest, random probability or uniform grid designs – potentially stratified across key habitat dimensions such as water depth or vegetation structure (Smokorowski and Pratt 2007) -- are an effective starting point. Their benefits include enforcing spatial coverage, eliminating subjective choices, providing an explicit framework for allocation of effort, supporting statistical extrapolation from samples to populations, and providing a “null” design against which to test the performance of later alternative designs (Hirzel and Guisan 2002; Hui et al. 2011; Rew et al. 2006; Stevens and Olsen 2004). With greater knowledge about the species being sought or system being monitored, monitoring can be optimized by deliberately biasing the sampling design towards the most informative habitats and collection devices, while continuing to dedicate some effort to broader spatial coverage to avoid the risk of false negatives (Carvalho et al. 2016; Hoffman et al. 2016). Broad-spectrum monitoring is likely to find certain habitats or collection devices contribute most strongly to the total species pool, while others yield comparatively few unique species (Trebitz et al. 2009; Vermonden et al. 2010). Target-species monitoring may emphasize invasion fronts or the presumed preferred habitats, although examples of NIS exploiting novel habitats (Gutsch and Hoffman 2016) support the need for distributed sampling.
The spacing among sampling locations also deserves some attention. Collection efforts should document the volume sampled, so that organism spatial coverage can be evaluated and effectiveness of alternate surveys can be properly compared, and should set a minimum among-sample distance that avoids sweeping the same space twice. Which spacing most effectively detects a given organism distribution pattern can be explicitly addressed by comparing alternate survey designs. For example, a study exploring strategies for detecting a NIS zooplankter concluded it better to sample a larger area with greater spacing than to sample a small area with closer-spaced samples (Harvey et al. 2009a).
Once regular EDM has been implemented, the temporal distribution of the sampling effort also becomes relevant. This is an aspect of EDM sampling design that is not yet well explored. Concern about not letting too much time pass before a new NIS is detected (to maximize response success) is likely to favor expending a smaller amount of effort annually rather than conducting a more intensive effort every few years; however, reductions in within-year effort will be associated with an increased risk of missed detection in any given year. In large, open systems such as the Great Lakes, rare-species encounters tend to accumulate over years (due to population fluctuations, in- and out-migrations, etc.) so understanding of sampling sufficiency will become deeper over multi-year efforts (Hoffman et al. 2016).
The burgeoning interest in biological monitoring via DNA that organisms have shed into the environment presents additional sampling design questions. Some studies have assumed that shed DNA accumulates at the water surface and therefore focused collection there (e.g., US Geological Survey 2013), while others have assumed that shed DNA is bound to particles that settle to the bottom (e.g., Turner et al. 2014), but understanding of the appropriate sampling grain is still limited (Schultz and Lance 2015). A concerted effort to better understand the distribution and fate of shed DNA is underway and necessary to inform sampling design recommendations (Barnes and Turner 2016, Goldberg et al. 2016; Yoccoz 2012). For example, recent studies have explored the size distribution of DNA-bearing particulates (Turner et al 2014), the transport and retention of shed DNA in lotic systems (Deiner and Altermatt 2014, Jerde et al. 2016), and the persistence of shed DNA in relationship to various physiochemical aspects of the environment (Barnes et al 2014; Dejean et al. 2011; Strickland et al. 2015).
4. What will the collection and identification effort consist of?
Until quite recently, biological monitoring was almost entirely based on collecting organisms and identifying them from morphological traits. Now, organisms can also be identified from their DNA and that DNA can be recovered from environmental media (e.g., water or sediment) as well as extracted from collected tissues. Two different DNA-based identification approaches are relevant to NIS monitoring. One is the DNA target marker approach, wherein samples are screened for preselected species by addition of an analytically detectable molecular probe designed to bind only to unique segments of their DNA. The other is the DNA barcoding approach, wherein species present are determined by analyzing the base-pair sequences of DNA in a sample and matching them to reference sequences (known as “barcodes”) from sequence databases (Hebert et al. 2003). DNA barcoding becomes DNA metabarcoding when applied to genetic material from mixed-organism samples rather than to material from a single organism at a time, which is made possible through recent advances in high-throughput sequencing and bioinformatics (Halanych and Mahon 2014).
4.1 Collecting Organisms versus shed DNA
Collected organisms are amenable to identification by both their morphological traits and their DNA. Physical collections remain the gold standard for verifying presence of suspected new NIS, can serve as reference specimens for future morphological or genetic analyses, and can yield information relevant to NIS monitoring that shed DNA alone cannot such as life-stage, condition, and breeding status. The major drawbacks to organism collection in the context of NIS monitoring involve resource needs and sampling efficiency, namely: 1) limited effectiveness of collection devices due to low capture probabilities and environmental conditions hindering their use; and 2) need for specialized equipment and trained personnel to accomplish the collection. These make mounting an organism collection effort of sufficient intensity to find rare or elusive NIS a resource-intensive endeavor. Possible impacts of intensive sampling on desirable native species might also be a concern.
Monitoring for NIS via shed DNA has the potential for increasing EDM efficiency and reducing resource demands, because shed DNA can be more widely distributed and readily detectable than the organisms it came from, and environmental samples (e.g., water or sediment) can be collected relatively quickly and with little training or equipment. Numerous studies have demonstrated that shed DNA (also ‘ environmental DNA ’ or ‘eDNA ’ in the literature) can reveal presence of difficult-to-find organisms as well as recover biodiversity information relevant to broad-spectrum monitoring (e.g., reviews by Bohmann et al. 2014; Cristescu 2014; Lodge et al. 2012; Rees et al. 2014; Thomsen and Willerslev 2015). In the context of EDM, the major drawbacks to collecting shed DNA rather than organisms are: 1) uncertainty concerning whether finding DNA translates into live NIS presence, 2) difficulty in inferring organism abundance from the DNA quantity, and 3) failure to provide ancillary information such as size and life-stage that is relevant to gaging NIS establishment and impacts (Diaz-Ferguson and Moyer 2014; Goldberg et al. 2015; Shaw et al. 2016).
4.2 Identification via morphology, DNA target markers, or DNA (meta)barcoding
Identifying organisms by means of distinguishing morphological traits is the historical underpinning of taxonomic science, and the resulting composition data are well established as the basis for ecological assessment. For taxa that are large and fully developed, morphological identification is readily accomplished in the field (e.g., adult fishes, amphibians), but morphological identification of other taxa or life-stages (e.g., eggs, larval fishes, zooplankton, benthic invertebrates) requires a laboratory effort. This laboratory effort can make substantial demands on resources and expertise – especially for EDM because searching for rare or unknown NIS forestalls the use of shortcuts such as coarser-than-species level identification, counting only a fraction of the sample, or focusing on indicator taxa only (Carter and Resh 2001; Mack et al. 2012). Furthermore, immature or damaged specimens may defy full identification and available taxonomic keys may fail to cover potential invaders. Consequently, completion of laboratory morphological identification typically lags sample collection by many months, and NIS can be missed entirely due to inability to identify them.
In contrast to morphological identification, DNA-based identification is not dependent on life stage, taxonomic expertise, or even securing a physical specimen if shed DNA is available. In practice, however, the capacity for DNA-based identification remains limited by 1) the narrow suite of species for which target markers have been developed; 2) insufficiently resolved genetic loci to differentiate some species; 3) incomplete barcode reference databases; and 4) in the case of metabarcoding, the technical complexity of the analysis (Bourlat et al. 2013; Darling and Mahon 2011). The DNA target marker approach is analytically rapid and sensitive: the markers can be detected by routine analyses and can be highly specific given adequate genetic distinctness of the target organisms. DNA barcoding of individual taxa is also analytically straightforward, and while requiring the same preparatory picking and sorting as morphological identification, can provide better taxonomic resolution for damaged or immature specimens (Bourlat et al. 2013). Extension of DNA barcoding to mixed-organism samples (i.e., metabarcoding) can bypass much of this sample picking and sorting, but involves complex analytical processes (see below) that to date, makes metabarcoding dependent on specialized equipment and highly trained personnel, and typically no faster than laboratory morphological identification (Stein et al. 2014).
Importantly, organism identification methods need not be an either/or choice. Samples can both be screened for high-profile NIS via DNA target-markers and subject to morphological identification or DNA metabarcoding of a broader suite of taxa. Comparisons between morphology-based and DNA-based data allows for assessment of taxa each method may miss and the development of environmental indicators robust to their differences in quantification and taxonomic resolution (e.g., Aylagas et al. 2016; Pilgrim et al. 2011; Shaw et al. 2016; Sweeny et al. 2011). In fact, morphological identification and DNA barcoding benefit from a close and reciprocal interaction: DNA-based identification helps to improve taxonomic keys and expand biodiversity knowledge, while trained morphological taxonomists are needed to corroborate new species and construct the organism collections needed to expand barcode databases (Hajibabaei et al. 2007; Kim and Byrne 2006; Kvist 2013).
4.3 Challenges and progress in identification technology
The limitations of morphological identification are well known to practitioners and unlikely to be substantially overcome, but some improvements are possible. For instance, the accuracy of NIS detection can be enhanced by development of invasion-appropriate keys that recognize morphological distinctions that were not considered when regional taxonomic compendiums were constructed (Peterson and Lietz 2017). Whether such distinctions can be identified a priori (e.g., for watch-list species) remains an open question. Laboratory procedures could also institute systematic scans for rare and unusual taxa – which both increases the chances of detecting otherwise unrecognized NIS and improves estimates of biodiversity indices such as richness that are relevant to evaluating survey outcome (Bellier et al. 2012). Because knowledge gaps concerning phylogeny and biogeography can preclude correct identification for both NIS and uncommon native species (e.g., Bishop and Hutchings 2011; Stepien and Neilson 2013), development of curated information systems that bring together regionally appropriate taxonomic keys, sequences, voucher specimens, and natural history information would support both morphological identification and DNA barcoding (Blanco-Bercial 2014; Bourlat et al. 2013; Cristescu 2014).
Various research efforts, some already ongoing, stand to improve the utility of DNA-based organism identification. With target-species monitoring, development of additional markers – guided by lists of the most likely and worrisome invaders – will expand the suite of taxa amenable to DNA-based detection. To date, DNA target markers relevant to the Great Lakes have been published for just a few high-profile NIS (e.g., Asian carp, Eurasian ruffe, rusty crayfish, New Zealand mudsnails – Dougherty et al. 2016; Goldberg et al. 2013; Mahon et al. 2013; Tucker et al. 2016). The procedure for developing target markers for additional species is straightforward (Rees et al. 2014), but the iterative process of designing and testing the sensitivity and specificity of potential markers can be lengthy (Harvey et al. 2009b) and genetic polymorphism can pose a barrier to finding unique markers for species (Funk and Omland 2003). Clearinghouses where target markers are catalogued would be very useful, as would creation of tissue banks to support their development and testing.
Further investigation of genetic markers is one research priority for DNA barcoding. Although much work has been directed at the mitochondrial CO1 gene for metazoans (Hebert et al. 2003), suitable barcodes have yet to be identified for some taxa (e.g., Hollingsworth et al. 2011; Pawlowski et al. 2012), and other taxa have insufficient genetic divergence (April et al. 2011; Brown et al. 2016), particularly for sequencing platforms having limited read lengths (e.g., Leray et al. 2014). Rather than focus on one ‘universal’ primer, the application of multiple barcode regions simultaneously is also being pursued (e.g., Chesters et al. 2015). A second research priority for DNA barcoding is expansion of reference databases. Despite the large number of DNA sequences already deposited in public databases such as BOLD (Ratnasingham and Hebert 2007) and GenBank (Benson et al. 2013), these databases are still substantially incomplete for important taxonomic groups (e.g., invertebrates), and can be insufficiently taxonomically resolved or lacking in supporting biogeographic information to confidently identify some organisms (Briski et al. 2016; Kvist 2013; Trebitz et al. 2015). Research teams are responding to this information deficiency by generating their own barcode databases (e.g., Failla et al. 2015; Valentini et al. 2016), but incentives for sharing this knowledge across the broader research community are needed.
Also needed are refinements to the pre- and post-sequencing processing that is required for DNA metabarcoding. For a variety of reasons (e.g., varying PCR primer affinities and genetic locus copy number, interference from other biological material), DNA is not extracted and amplified equally from all organisms in a mixed sample, which in extreme cases can lead to presence of some taxa being entirely masked by others (Evans et al. 2016; Hatzenbuhler et al. 2017; Pochon et al. 2013). Ongoing research seeks to understand, correct for, and minimize these biases (e.g., Bronnenhuber et al. 2013; Geller et al. 2013; Zhou et al. 2013). Aspects of the bioinformatics post-processing that are significant to the metabarcoding outcome include sequence filtering (which must balance removing erroneous sequences against eliminating legitimate rare sequences), classification of sequences into operational taxonomic units (e.g., clustering techniques, genetic similarity criteria), and methods for matching sequences to reference databases (Brown et al. 2015; Evans et al. 2017; Gaspar and Thomas 2013; Huse et al. 2010; Nguyen et al. 2015; Schloss et al, 2011). Research is needed to assess the implications of all these not-yet-standardized choices and to develop supporting workflow procedures (Comtet 2015; Cristescu 2014; Thomsen and Willerslev 2015). During this research phase, close attention to sample processing to help interpret DNA metabarcoding outcomes is important. For example, the ability to decipher anomalous sequencing outcomes may be greater when running taxonomically sorted subsets than running an entire sample at once.
On the technology front, we expect continued reductions in analytical turnaround times and physical footprint of DNA detection platforms, driven by advances in DNA processing methods and sensor technology. Two examples of approaches under development are 1) using laser transmission spectroscopy to detect increases in the diameter of nanoparticles that occur when DNA from target species binds to the coating of oligonucleotide tags (Egan et al. 2015); and 2) using microfluidic chips to directly amplify DNA without the need for extraction and purification (Stedtfeld et al. 2014). DNA detection platforms are also being integrated with electronic applications and dashboards for easier transfer of information to researchers (e.g., Stedtfeld et al. 2016). We can envision future NIS monitoring using real-time field-operable DNA-based detection tools, with benefits including the ability to follow new finds with immediate management response, and eliminating the need for sample shipment and curation. Development of such tools will require active collaboration between practitioners of molecular biology and engineers capable of designing useful EDM detection platforms.
4.4 Sample handling and data verification
Sample handling and data verification are extremely important aspects of organism collection and identification, because they determine the quality of the taxonomic data, the degree to which sources of error are bounded and controlled for (Table 1), the comparability among efforts, and the extent to which collections are amenable to follow-up investigations. Organisms that are released after enumeration cannot later be re-examined, so field crews need to be instructed to retain representative specimens, photograph diagnostic features, or collect body parts (e.g., fin clips, scales) that allow for DNA analysis for any taxa that look at all unusual. Collection and processing procedures that damage tissues should be avoided, and organisms should be preserved in 95% ethanol rather than formalin if DNA analyses are envisioned (Pilgrim et al. 2011). Some aquatic taxa require specialized handling to allow preservatives to reach internal tissues (e.g., mollusks). Quality-assurance checks should be a routine part of laboratory morphological identification procedures, with outside experts asked to verify specimens of unfamiliar species or atypical morphology. Samples intended for DNA analysis must be subject to great care to avoid cross-contamination, and the workflow should include control samples known to be negative (blanks) or positive (spikes) at each step of the process (Goldberg et al. 2016; Nguyen et al. 2015; Rees et al. 2014) to help identify issues such as contamination or PCR inhibition (Table 1). Standardization is crucial for comparing surveys based on shed DNA, whose quantities vary not only with sample volume but also the methods used to concentrate and preserve the DNA (e.g., Deiner et al. 2015; Goldberg et al. 2015; Minamoto et al. 2016).
Table 1.
Summary of factors contributing to uncertainty in survey outcome. Remedies for addressing them are classified as procedural (changes in protocol can reasonably address), design (changes to sampling design can potentially address), and research (additional research required to address).
| Source of uncertainty | Primary concern | Remedy |
|---|---|---|
| sample contamination | false positive | procedural: run DNA blanks, decontaminate equipment |
| organism or DNA from outside system (e.g., hydraulic transport) | false positive | design: spatial controls, follow-up investigations for unexpected taxa |
| organism misidentified, not fully identifiable, or missed by morphological ID | false negative (false positive also possible) |
procedural: independent verification, confirmatory DNA, deeper search of samples research: improved taxonomic keys |
| DNA misidentified, not fully identifiable, or missed by genetic sequencing | false negative, false positive |
procedural: sequencing error checks, bioinformatics choice checks research: expanded barcode databases, alternative barcode loci, more specific target markers |
| organism or DNA too degraded to identify | false negative |
procedural: attention to preservation, avoid damaging field devices research: methods for shorter DNA fragments |
| PCR inhibition masks organism DNA | false negative |
procedural: DNA positive controls, comparison to morphological ID research: better primers, different PCR protocols |
| Insufficient extent or spatial coverage to find organism | false negative, uncertain distribution | design: increase survey effort, diversify habitats sampled |
| species not vulnerable to collection devices | false negative, uncertain distribution | design: diversify sampling methods |
| species not “watched” for | false negative | research: better watch lists, additional target markers, shift to broad-spectrum monitoring |
| organism number or density poorly characterized | uncertain distribution | research: better understand appropriate sampling grain, factors affecting DNA copy number |
5. How will survey results be evaluated and used?
As with any assessment endeavor, EDM-related data do not become information until they are evaluated and communicated to the research and management community (Darling 2015; Lehtiniemi et al. 2015). EDM surveys are resource intensive, so periodic performance evaluations are imperative and comparison to different surveys are often of interest. Aspects of performance to be considered include detection probability attained for a given effort (i.e., sensitivity), efficiency with which detection is achieved, and uncertainty in the survey outcome (Darling and Mahon 2011; Darling 2015). Sensitivity and efficiency can be evaluated for a survey overall or incrementally for various field and laboratory components. Performance metrics can be based on specified NIS (e.g., percentage of samples in which detected), but also on community-level information such as total number of NIS, species richness, and number of rare species (whether native or NIS). Incorporating and subsequently evaluating design or methodology changes in an adaptive manner can be used to improve survey performance over time (Hoffman et al. 2016; Jones et al. 2015). Various approaches and tools to evaluating survey effort, sensitivity, efficiency and uncertainty are available, but typically require some statistical or numerical expertise to apply.
5.1 Survey performance evaluation
One approach to evaluating survey performance involves settings where the species composition is already known. For example, serial dilutions or mesocosm experiments can be used to examine appropriate sample volumes and extraction protocols for DNA analyses (Evans et al. 2016; Kelly et al. 2014; Wilcox et al. 2013), deliberately constructed organism mixes can be used to examine PCR amplification bias and ability to detect rare DNA signals among common ones (Brown et al. 2016; Hatzenbuhler et al. 2017; Zhan et al. 2013), and the efficacy of alternate sampling designs and collection devices can be evaluated at locations where the species of interest are known to be present (Dejean et al. 2012; Harvey et al. 2009a). Survey performance can also be evaluated by comparison to a different, more thorough survey that serves as the “truth” (Goldberg et al. 2013; Mächler et al. 2014; Treguier et al. 2014). Virtual experiments are also an option, where hypothetical species distributions constructed across computer-based landscapes are used to generate find-rate statistics for various effort allocation schemes (Hirzel and Guisan 2002; Rew et al. 2006).
However, EDM survey evaluation is often of most interest where the true state is not known, which requires approaches that do not hinge on the presence or identity of any particular NIS. With data from repeat sampling events, species occupancy models (MacKenzie et al. 2006) can be used to estimate detection probability for any species encountered at least once, which then establishes the level of sampling needed to confidently infer that a species of given rarity and detectability was absent rather than simply missed (Diaz-Ferguson and Moyer 2014). Occupancy models are relevant both to assessing effort expenditure and allocation in the field (e.g., Dougherty et al. 2016; Sliwinski et al. 2016; Valentini et al. 2016) and the sensitivity of results to various analytical decision points (e.g., Ficetola et al. 2016; Schmidt et al. 2013). Broad-spectrum monitoring data are also amenable to a community rarefaction approach to evaluating survey performance (Gotelli and Colwell 2001), which uses information on the rate at which additional samples acquire additional species to estimate the total species pool (asymptotic richness), and from that the portion of the species pool detected for a given level of effort (e.g., Hoffman et al. 2011; Ram et al. 2014).
Analysis of survey outcomes also can identify means for making them more efficient, typically by finding sampling design that concentrate effort in those habitats that yield the most ‘finds’. The degree of overlap among possible sample groupings (e.g., collection devices, habitat types) can be examined via summary statistics such as the number of unique taxa for each, and via ordination plots that show similarities and differences in species composition. Numerical experiments on the survey data can also be informative; for example, subsets representing various potential effort allocations can be randomly drawn from the bigger dataset and compared with respect to richness and species detection rates (Trebitz et al. 2009). Rarefaction analysis is useful here too, as designs that more efficiently find rare taxa will approach the asymptote of the species-effort curve more quickly (e.g., Hoffman et al. 2011).
Because many aspects of survey design and methodology can influence performance, it can be helpful to have statistical models that integrate this information to assist with evaluating the alternatives. As already discussed (Section 3.1), various models examining the tradeoffs between resources dedicated to detection versus later control of target species are available in the literature. Other models enable users to explore how alternate sampling protocols (e.g., water volume, number of replicates) and physical factors (e.g., stream flow, temperature) influence detection probability for shed DNA (Furlan et al. 2016; Schultz and Lance 2015).
5.2 Addressing survey uncertainty
Monitoring for potential new NIS – without knowing whether introductions have occurred and what species are involved -- is inherently uncertain. Some of these sources of uncertainty – including false positives and false negatives (Darling and Mahon 2011) -- can be procedurally controlled for, and others can be addressed through sampling design and additional research (Table 1). Nevertheless, uncertainty will never be eliminated entirely, and should be quantified and clearly communicated, so that EDM practitioners understand the quality of the information on which they are basing decisions and potential for misinterpretation in the public sphere is minimized.
With EDM that collects actual organisms, the most worrisome component of uncertainty is missing NIS that are in fact present (false negatives). As discussed above, organism collection devices typically have limited capture probability, aquatic habitats are difficult to sample comprehensively, and specimens in hand can defy full identification by either morphology or DNA. False positives are possible with physical organism surveys (specimen mistakenly identified as an NIS) but can be procedurally controlled for, and true positives (actual NIS captures) are readily verified and therefore informative. Thus, evaluations of EDM surveys based on physical organism collections typically emphasize detection probability achieved, and securing sufficient resources to keep detection probability high is a primary concern.
Surveys based on collection of shed DNA bring a more complex set of uncertainties. The ability of shed DNA to disperse widely in the environment is part of its appeal as a sampling target but complicates inferences about species distributions, especially in systems that are large, open, or hydrologically complex (Darling and Mahon 2011; Denier and Altermatt 2014; Merkes et al. 2014). False positives are much more likely with shed DNA than with physical specimens, with possibilities including sample contamination and accidental transport of DNA from outside the system (Table 1). Organism abundance and biomass are correlated with shed DNA amounts in controlled experiments, but different DNA shedding rates among species and life stages and physiochemical factors influencing persistence in the environment complicate this relationship in the field (Evans et al. 2016; Klymus et al. 2015; Takahara et al. 2012). Inferring organism distributions from the pattern of positive and negative shed-DNA detections remains a major challenge (Dougherty et al. 2016; Jerde et al. 2013), and shed-DNA surveys are difficult to compare quantitatively (Diaz-Ferguson and Moyer 2014; Goldberg et al. 2015; Rees et al. 2014).
Regardless of whether EDM surveys collect physical organisms or shed DNA, the greatest challenge may lie in generating appropriate models for translating patterns of detections into information that is usable to managers and understandable to the public (Darling 2015). There are alternative ways of expressing the reliability of a survey. For example, a change in sampling protocol can be evaluated either in terms of increased detection probability for a given concentration of organisms and DNA, or a decreased minimum concentration that can be detected with a specified probability. The latter is preferred because it allows EDM practitioners to assess survey effectiveness even in the absence of NIS finds (Schultz and Lance 2015). A single positive detection may be of limited information value compared to models that estimate target population distributions based on observed detection patterns and knowledge of error rates associated with the surveillance methodology (Ficetola et al. 2016, Lahoz-Monfort et al. 2016, Schmidt et al. 2013). Further developing the decision-support and information-sharing aspects of EDM surveys remains not only a research priority but an area that EDM policy needs to address (Darling 2015).
6. Conclusions
The key decision points in implementing NIS early-detection monitoring merit substantial attention. These include whether to target a few species considered to be high-risk or to conduct broad-spectrum monitoring to detect any new species; how to decide the monitoring effort and allocate it over space and time; whether to identify organisms using morphological characteristics, DNA or both; and how to implement data management practices and survey performance evaluations to support an adaptive monitoring cycle. As we have shown, a considerable body of scientific literature already exists to inform NIS monitoring at each of these decision points. We anticipate continued scientific development, motivated by increasing pressure to formalize and expand EDM and supported by continuing technical and informational advances. With more practiced and efficient workflows at all steps, we also anticipate the EDM timeline will continue to shorten such that detection truly is early and can be followed by rapid response. This does not mean EDM will be easy – early detection is likely to remain a resource-intensive proposition even with such advances – but EDM is clearly scientifically tractable given sufficient policy and management support.
A substantial factor overarching more systematic EDM monitoring is a need for data integration and communication tools to share new species finds and convey risks to managers and the public (Katsanevakis and Roy 2015). Information management is necessary for spanning the gap between a collection of disjunct local monitoring programs and a broadly coordinated and cost-effective EDM network, and provides the crucial link between early detection of incipient or expanding NIS and subsequent management responses. Significant questions remain regarding the value of different types of information to different end-users, the factors that determine whether management action will be taken in the face of uncertainty, and the social, political and economic pressures that shape these decisions (e.g., Finnoff et al. 2007). Such decision-science research is outside the scope of this paper, but would support more effective translation of NIS knowledge into management action.
We also strongly encourage efforts towards integration of EDM with other biological assessment programs. For example, the objectives and challenges of EDM have commonalities with monitoring for rare and endangered species (Field et al. 2005; Manley et al. 2004), such that communication and coordination may benefit both fields. New DNA-based identification technologies present opportunities to improve the sensitivity and efficiency of EDM, but implementation into existing assessment programs must proceed cautiously in regards to interpreting and communicating survey outcomes. We recommend greater integration of morphological-based and DNA-based identification, both to address their respective uncertainties and to evaluate how synergies can improve NIS early detection. The research directions we have identified here (Box 1) would substantially improve the robustness and reliability of surveillance, increase transparency, and facilitate the application of EDM results in decision-making contexts.
Box 1. Summary of recommendations for future research efforts.
What and where should be monitored?
Expand taxonomic coverage of watch lists
Improve information basis for species-specific and vector-specific risk assessments
How should the survey be designed?
Improve understanding of persistence and distribution of DNA shed into the environment
Improve understanding of how target taxa distribution dictates spatiotemporal sampling design
What will the collection and identification effort consist of?
Develop invasion-appropriate morphological keys
Develop additional and improved markers for targeted DNA-based detection
Assemble a suite of effective barcode loci with known efficacy
Better populate DNA barcode databases
Further develop and standardize molecular and bioinformatics workflows for metabarcoding
Explore in situ sensor platforms that enable real-time DNA-based monitoring
How will survey results be evaluated and used?
More accurate models to infer species distributions from positive and negative detection patterns
Develop resources that bring together numerical tools and supporting data for managers to use
Improve methods for assessing and communicating survey uncertainty
Develop performance metrics that adequately assess detection probabilities, frequency of false positives and negatives, and cost-efficiency
Overall EDM network development and information management
Improve optimization of resource allocation to support both breadth and depth of monitoring
Create and maintain repositories for tissues and resources relevant to DNA-based monitoring
Improve methods for data curation, synthesis, and communication
Improve understanding of factors affecting how end-users value and apply scientific information
Pursue integration of EDM monitoring with other bioassessment efforts
Acknowledgments
This paper was developed from a workshop on the state of EDM science held at the U.S. EPA’s Mid-Continent Ecology division in October of 2014. Great Lakes Restoration Initiative support to several of the authors contributed to research that informed the content of this manuscript. We thank Erika Jensen of the Great Lakes Commission and two anonymous reviewers for their comments. The views expressed in this paper are those of the authors and do not necessary reflect the views or policies of their respective agencies or universities. This is contribution 4611 from the NOAA Pacific Marine Environmental Lab.
References
- April J, Mayden RL, Hanner RH, Bernatchez L. Genetic calibration of species diversity among North America’s freshwater fishes. PNAS. 2011;1088:10602–10607. doi: 10.1073/pnas.1016437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylagas E, Borja A, Irigoien X, Rodríguez-Ezpeleta N. Benchmarking DNA metabarcoding for biodiversity-based monitoring and assessment. Front Mar Sci. 2015;3:96. [Google Scholar]
- Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM. Environmental conditions influence eDNA persistence in aquatic systems. Environ Sci Tech. 2014;48:1819–1827. doi: 10.1021/es404734p. [DOI] [PubMed] [Google Scholar]
- Barnes MA, Turner CR. The ecology of environmental DNA and implications for conservation genetics. Conserv Genetics. 2016;17:1–17. [Google Scholar]
- Bellier E, Grøtan V, Engen S, Schartau AK, Diserud OH, Finstad AG. Combining counts and incidence data: an efficient approach for estimating the log-normal species abundance distribution and diversity indices. Oecologia. 2012;170:477–488. doi: 10.1007/s00442-012-2311-2. [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop MJ, Hutchings PA. How useful are port surveys focused on target pest identification for exotic species management? Mar Poll Bull. 2011;62:36–42. doi: 10.1016/j.marpolbul.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Blanco-Bercial L, Cornils A, Copley N, Bucklin A. DNA barcoding of marine copepods: assessment of analytical approaches to species identification. PLoS Currents. 2014:6. doi: 10.1371/currents.tol.cdf8b74881f87e3b01d56b43791626d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Evans A, Gilbert MT, Carvalho GR, Creer S, Knapp M, Douglas WY, De Bruyn M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol. 2014;29:358–67. doi: 10.1016/j.tree.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Borja A, Gilbert J, Taylor MI, Davies N, Weisberg SB, Griffith JF, Lettieri T, Field D, Benzie J, Glöckner FO. Genomics in marine monitoring: new opportunities for assessing marine health status. Mar Poll Bull. 2013;74:19–31. doi: 10.1016/j.marpolbul.2013.05.042. [DOI] [PubMed] [Google Scholar]
- Bowen A, Keppner S. Surveillance for ruffe in the Great Lakes, 2015. U.S. Fish and Wildlife Service, Alpena Fish and Wildlife Conservation Office; Alpena, MI: 2015. Available https://www.fws.gov/midwest/alpena/documents/2015-GL-Ruffe-Surveillance.pdf. [Google Scholar]
- Briski E, Ghabooli S, Bailey SA, MacIsaac HJ. Are genetic databases sufficiently populated to detect non-indigenous species? Biol Invas. 2016;18:1911–1922. doi: 10.1007/s10530-016-1134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronnenhuber JE, Wilson CC. Combining species-specific COI primers with environmental DNA analysis for targeted detection of rare freshwater species. Conserv Genetics Res. 2013;5:971–975. [Google Scholar]
- Brown EA, Chain FJJ, Crease TJ, MacIsaac HJ, Cristescu ME. Divergence thresholds and divergent biodiversity estimates: can metabarcoding reliably describe zooplankton communities? Ecol Evol. 2015;5:2234–2251. doi: 10.1002/ece3.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EA, Chain FJJ, Zhan A, MacIsaac HJ, Cristescu ME. Early detection of aquatic invaders using metabarcoding reveals a high number of non-indigenous species in Canadian ports. Diversity Distrib. 2016;22:1045–1059. [Google Scholar]
- Carter JL, Resh VH. After site selection and before data analysis: sampling, sorting, and laboratory procedures used in stream benthic macroinvertebrate monitoring programs by USA state agencies. J N Am Benthol Soc. 2001;200:658–682. [Google Scholar]
- Carvalho SB, Goncalves J, Guisan A, Honrado JP. Systematic site selection for multispecies monitoring networks. J Applied Ecol. 2016;53:1305–1316. [Google Scholar]
- Chesters D, Zheng WM, Zhu CD. A DNA Barcoding system integrating multigene sequence data. Meth Ecol Evol. 2015;6:930–937. [Google Scholar]
- Comtet T, Sandionigi A, Viard F, Casiraghi M. DNA (meta)barcoding of biological invasions: a powerful tool to elucidate invasion processes and help manage aliens. Biol Invas. 2015;17:905–922. [Google Scholar]
- Cristescu ME. From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity. Trends Ecol Evol. 2014;29:566–571. doi: 10.1016/j.tree.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Counihan TD, Bollens SM. Early detection monitoring for larval dreissenid mussels: how much plankton sampling is enough? Envi Monit Assess. 2017 doi: 10.1007/s10661-016-5737-x. doi:10:1007/s10661-016-5737-x. [DOI] [PubMed] [Google Scholar]
- Darling JA. Genetic studies of aquatic biological invasions: closing the gap between research and management. Biol Invas. 2015;17:951–971. [Google Scholar]
- Darling JA, Mahon AR. From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ Res. 2011;111:978–988. doi: 10.1016/j.envres.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Deiner K, Altermatt F. Transport distance of invertebrate environmental DNA in a natural river. PLoS One. 2014;9:e88786. doi: 10.1371/journal.pone.0088786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiner K, Walser JC, Mächler E, Altermatt F. Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol Conserv. 2015;183:53–63. [Google Scholar]
- Dejean T, Valentini A, Duparc A, Pellier-Cuit S, Pompanon F, Taberlet P, Miaud C. Persistence of environmental DNA in freshwater ecosystems. PloS One. 2011;6:e23398. doi: 10.1371/journal.pone.0023398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean T, Valentini A, Miquel C, Taberlet P, Bellemain E, Miaud C. Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog (Lithobates cathesbeianus) J Appl Ecol. 2012;49:953–959. [Google Scholar]
- Diaz-Ferguson EE, Moyer GR. History, applications, methodological issues, and perspectives for the use of environmental DNA (eDNA) in marine and freshwater environments. Revista Biologia Tropical. 2014;62:1273–1284. doi: 10.15517/rbt.v62i4.13231. [DOI] [PubMed] [Google Scholar]
- Dougherty MM, Larson ER, Renshaw MA, Gantz CA, Egan SP, Erickson DM, Lodge DM. Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. J Appl Ecol. 2016;53:722–732. doi: 10.1111/1365-2664.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan SP, Olds B, Grey E, Feder JL, Lodge DL. Rapid molecular detection of invasive species in ballast and harbor water by integrating environmental DNA and light transmission spectroscopy. Environ Sci Tech. 2015;7:4113–4121. doi: 10.1021/es5058659. [DOI] [PubMed] [Google Scholar]
- Epanchin-Niell RS, Haight RG, Berec L, Kean JM, Liebhold AM. Optimal surveillance and eradication of invasive species in heterogeneous landscapes. Ecol Lett. 2012;15:803–812. doi: 10.1111/j.1461-0248.2012.01800.x. [DOI] [PubMed] [Google Scholar]
- Evans NT, Olds BP, Renshaw MA, Turner CR, Li Y, Jerde CL, Mahon AR, Pfrender ME, Lamberti GA, Lodge DM. Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Molec Ecol Res. 2016;16:29–41. doi: 10.1111/1755-0998.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NT, Li Y, Renshaw MA, Olds BP, Deiner K, Turner CR, Jerde CL, Lodge DM, Lamberti GA, Pfrender ME. Fish community assessment with eDNA metabarcoding: effects of sampling design and bioinformatic filtering. Can J Fish Aquati Sci. 2017 in press. [Google Scholar]
- Failla AJ, Vasquez AA, Hudson P, Fujimoto M, Ram JL. Morphological identification and COI barcodes of adult flies help determine species identities of chironomid larvae (Diptera, Chironomidae) Bull Entom Res. 2015;106:34–46. doi: 10.1017/S0007485315000486. [DOI] [PubMed] [Google Scholar]
- Ficetola GF, Taberlet P, Coissac E. How to limit false positives in environmental DNA and metabarcoding? Molec Ecology Res. 2016;16:604–607. doi: 10.1111/1755-0998.12508. [DOI] [PubMed] [Google Scholar]
- Field SA, Tyre AJ, Possingham HP. Optimizing allocation of monitoring effort under economic and observational constraints. J Wildlife Manage. 2005;69:473–482. [Google Scholar]
- Finnoff D, Shogren JF, Leung B, Lodge D. Take a risk: Preferring prevention over control of biological invaders. Ecol Econ. 2007;62:216–222. [Google Scholar]
- Funk DJ, Omland KE. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Sys. 2003;34:397–423. [Google Scholar]
- Furlan EM, Gleeson D, Hardy CM, Duncan RP. A framework for estimating the sensitivity of eDNA surveys. Molec Ecol Res. 2016;16:641–654. doi: 10.1111/1755-0998.12483. [DOI] [PubMed] [Google Scholar]
- Gantz CA, Gordon DR, Jerde CL, Keller RP, Chadderton WL, Champion PD, Lodge DM. Managing the introduction and spread of non-native aquatic plants in the Laurentian Great Lakes: a regional risk assessment approach. Manage Biol Invas. 2015;6:45–55. [Google Scholar]
- Gaspar JM, Thomas WK. Assessing the consequences of denoising marker-based metagenomic data. PLoS One. 2013;8:e60458. doi: 10.1371/journal.pone.0060458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller J, Meyer C, Parker M, Hawk H. Redesign of PCR primers for mitochondrial cytochrome C oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Molec Ecol Res. 2013;13:851–861. doi: 10.1111/1755-0998.12138. [DOI] [PubMed] [Google Scholar]
- Gertzen E, Familiar O, Leung B. Quantifying invasion pathways: fish introductions from the aquarium trade. Can J Fish Aquatic Sci. 2008;65:1265–1273. [Google Scholar]
- Gladman ZF, Yeomans WE, Adams CE, Bean CW, McColl D, Olszewska JP, McGillivray CW, McCluskey R. Detecting North American signal crayfish (Pacifastacus leniusculus) in riffles. Aquatic Conserv Marine Freshw Ecosys. 2010;20:588–594. [Google Scholar]
- Goldberg CS, Sepulveda A, Ray A, Baumgardt J, Waits LP. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum) Freshw Sci. 2013;32:792–800. [Google Scholar]
- Goldberg CS, Strickler KM, Pilliod DS. Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol Conserv. 2015;183:1–3. [Google Scholar]
- Goldberg CS, Turner CR, Deiner K, Klymus KE, Thomsen PF, Murphy MA, Spear SF, McKee A, Oyler-McCance S, Cornman RS, Laramie MB, Mahon AR, Lance RF, Pilliod DS, Strickler KM, Waits LP, Fremier AK, Takahara T, Herder JE, Taberlet MB. Critical considerations for the application of environmental DNA methods to detect aquatic species. Meth Ecol Evol. 2016;7:1299–1307. [Google Scholar]
- Gotelli NJ, Colwell RK. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett. 2001;4:379–391. [Google Scholar]
- Grigorovich IA, Korniushin AV, Gray DK, Duggan IC, Colautti RI, MacIsaac HJ. Lake Superior: an invasion coldspot? Hydrobiologia. 2003;499:191–210. [Google Scholar]
- Grippo MS, Hlohowskyi I, Fox L, Herman B, Pothoff J, Yoe C, Hayse J. Aquatic Nuisance Species in the Great Lakes and Mississippi River Basin—A Risk Assessment in Support of GLMRIS. Envi Manager. 2017;59:154–173. doi: 10.1007/s00267-016-0770-7. [DOI] [PubMed] [Google Scholar]
- Gutsch M, Hoffman JC. A review of ruffe (Gymnocephalus cernuus) life history in its native versus non-native ranges. Rev Fish Biol Fisheries. 2016;26:213–233. [Google Scholar]
- Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA. DNA barcoding: how it complements taxonomy, molecular phylogenetics, and population genetics. Trends Genetics. 2007;23:167–172. doi: 10.1016/j.tig.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Halanych KM, Mahon AR. Discovering diversity with high-throughput approaches: introduction to a virtual symposium in the Biological Bulletin. Biol Bull. 2014;227:91–92. doi: 10.1086/BBLv227n2p91. [DOI] [PubMed] [Google Scholar]
- Harvey CT, Quershi SA, MacIsaac HJ. Detection of a colonizing, aquatic, non-indigenous species. Diversity Distrib. 2009a;15:429–437. [Google Scholar]
- Harvey JB, Hoy MS, Rodriguez RJ. Molecular detection of native and invasive marine invertebrate larvae present in ballast and open water environmental samples collected in Puget Sound. J Exp Marine Biol Ecol. 2009b;369:93–99. [Google Scholar]
- Hatzenbuhler CL, Kelly JR, Martinson J, Okum S, Pilgrim E. Sensitivity and accuracy of high-throughput metabarcoding methods for early detection of invasive fish species. Sci Rep. 2017;7:46393. doi: 10.1038/srep46393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser C, McCarthy M. Streamlining ‘search and destroy’: cost-effective surveillance for invasive species management. Ecol Lett. 2009;12:683–692. doi: 10.1111/j.1461-0248.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, Dewaard JR. Biological identification through DNA barcodes. Proc Royal Soc Lond B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herborg L, Jerde C, Lodge D, Ruiz G, MacIsaac H. Predicting invasion risk using measures of introduction effort and environmental niche model. Ecol Applic. 2007;17:663–674. doi: 10.1890/06-0239. [DOI] [PubMed] [Google Scholar]
- Hirzel A, Guisan A. Which is the optimal sampling strategy for habitat suitability modelling. Ecol Model. 2002;157:331–341. [Google Scholar]
- Hoffman JC, Kelly JR, Trebitz AS, Peterson GS, West CW. Effort and potential efficiencies for aquatic non-native species early detection. Can J Fish Aquat Sci. 2011;68:2064–2079. [Google Scholar]
- Hoffman J, Schloesser J, Trebitz A, Peterson G, Gutsch M, Quinlan H, Kelly JR. Sampling design for early detection of aquatic invasive species in Great Lakes ports. Fisheries. 2016;41:26–37. [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howeth JG, Gantz CA, Angermeier PL, Frimpong EA, Hoff MH, Keller RP, Mandrak NE, Marchetti MP, Olden JD, Romagosa CM, Lodge DM. Predicting invasiveness of species in trade: climate match, trophic guild and fecundity influence establishment and impact of non-native freshwater fishes. Diversity Distrib. 2016;22:148–160. [Google Scholar]
- Hui C, Foxcroft LC, Richardson DM, MacFadyen S. Defining optimal sampling effort for large-scale monitoring of invasive alien plants: a Bayesian method for estimating abundance and distribution. J Appl Ecol. 2011;48:768–776. [Google Scholar]
- Hulme PE. Beyond control: wider implications for the management of biological invasions. J Appl ecol. 2006;43:835–847. [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Envi Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrad FC, Barrett S, Murray J, Stoklosa R, Whittle P, Mengersen K. Ecological aspects of biosecurity surveillance design for the detection of multiple invasive animal species. Biol Invas. 2011;13:803–818. [Google Scholar]
- Jerde CL, Chadderton WL, Mahon AR, Renshaw MA, Corush J, Budny ML, Mysorekar S, Lodge DM. Detection of Asian carp DNA as a part of a Great Lakes basin-wide surveillance program. Can J Fish Aquat Sci. 2013;70:522–526. [Google Scholar]
- Jerde CL, Olds BP, Shogren AJ, Andruszkiewicz EA, Mahon AR, Bolster D, Tank JL. Influence of stream bottom substrate on retention and transport of vertebrate environmental DNA. Environ Sci Tech. 2016;50:8770–8779. doi: 10.1021/acs.est.6b01761. [DOI] [PubMed] [Google Scholar]
- Jones ML, Brenden TO, Irwin BJ. Re-examination of sea lamprey control policies for the St. Marys River: completion of an adaptive management cycle. Can J Fish Aquat Sci. 2015;72:1538–1551. [Google Scholar]
- Katsanevakis S, Roy HE. Alien species related information systems and information management. Manage Biol Invas. 2015;6:115–117. [Google Scholar]
- Keller RP, Drake JM, Drew MB, Lodge DM. Linking environmental conditions and ship movements to estimate invasive species transport across the global shipping network. Divers Distrib. 2011;17:93–102. [Google Scholar]
- Kelly RP, Port JA, Yamahara KM, Crowder LB. Using Environmental DNA to Census Marine Fishes in a Large Mesocosm. PLoS One. 2014;9:e86175. doi: 10.1371/journal.pone.0086175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Byrne LB. Biodiversity loss and the taxonomic bottleneck: Emerging biodiversity science. Ecol Res. 2006;21:794–810. [Google Scholar]
- Klymus KE, Richter CA, Chapman DC, Paukert C. Quantification of eDNA shedding rates from invasive bighead carp (Hypophthalmichthys nobilis) and silver carp (Hypophthalmichthys molitrix) Biol Conserv. 2015;183:77–84. [Google Scholar]
- Kvist S. Barcoding in the dark? A critical view of the sufficiency of zoological DNA barcoding databases and a plea for broader integration of taxonomic knowledge. Molec Phylogen Evol. 2013;69:39–45. doi: 10.1016/j.ympev.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Lahoz-Monfort JJ, Guillera-Arroita G, Tingley R. Statistical approaches to account for false-positive errors in environmental DNA samples. Molec Ecol Res. 2016;16:673–685. doi: 10.1111/1755-0998.12486. [DOI] [PubMed] [Google Scholar]
- Lehtiniemi M, Ojaveer H, David M, Galil B, Gollasch S, McKenzie C, Minchin D, Occhipinti-Ambrogi A, Olenin S, Pederson J. Dose of truth—monitoring marine non-indigenous species to serve legislative requirements. Marine Policy. 2015;54:26–35. [Google Scholar]
- Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V, Boehm JT, Machida RJ. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Frontiers Zool. 2013;10:34. doi: 10.1186/1742-9994-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge DM, Williams S, MacIsaac HJ, Hayes KR, Leung B, Reichard S, Mack RN, Moyle PB, Smith M, Andow DA, Carlton JT, McMichael A. Biological invasions: recommendations for U.S. policy and management. Ecol Applic. 2006;16:2035–2054. doi: 10.1890/1051-0761(2006)016[2035:birfup]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lodge DM, Turner CR, Jerde CL, Barnes MA, Chadderton L, Egan SP, Feder JL, Mahon AR, Pfrender ME. Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA. Molec Ecol. 2012;21:2555–2558. doi: 10.1111/j.1365-294X.2012.05600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mächler E, Deiner K, Steinmann P, Altermatt F. Utility of environmental DNA for monitoring rare and indicator macroinvertebrate species. Freshw Sci. 2013;33:1174–1183. [Google Scholar]
- Mack HR, Conroy JD, Blocksom KA, Stein RA, Ludsin SA. A comparative analysis of zooplankton field collection and sample enumeration methods. Limnol Oceangr Meth. 2012;10:41–53. [Google Scholar]
- MacKenzie DI, Nichols D, Royle JA, Pollock KH, Hines JE, Bailey LL. Occupancy estimation and modeling: Inferring patterns and dynamics of species occurrence. Elsevier; San Diego: 2006. [Google Scholar]
- Mahon AR, Jerde CL, Galaska M, Bergner JL, Chadderton WL, Lodge DM, Hunter ME, Nico LG. Validation of eDNA surveillance sensitivity for detection of Asian carps in controlled and field experiments. PLoS One. 2013;8:e58316. doi: 10.1371/journal.pone.0058316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley PN, Zielinski WJ, Schlesinger MD, Mori SR. Evaluation of a multiple-species approach to monitoring species at the ecoregional scale. Ecol Applic. 2004;14:296–310. [Google Scholar]
- Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405:243–253. doi: 10.1038/35012251. [DOI] [PubMed] [Google Scholar]
- McGeoch MA, Genovensi P, Bellingham PJ, Costello MJ, McGrannachan C, Sheppard A. Prioritizing species, pathways, and sites to achieve conservation targets for biological invasion. Biol Invas. 2016;18:299–314. [Google Scholar]
- Mehta SV, Haight RG, Homans FR, Polasky S, Venette RC. Optimal detection and control strategies for invasive species management. Ecol Econ. 2007;61:237–245. [Google Scholar]
- Merkes CM, McCalla SG, Jensen NR, Gaikowski MP, Amberg JJ. Persistence of DNA in carcasses, slime, and avian feces may affect interpretation of environmental DNA data. PLoS One. 2014;9:e113346. doi: 10.1371/journal.pone.0113346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamoto T, Naka T, Moji K, Maruyama A. Techniques for the practical collection of environmental DNA: filter selection, preservation, and extraction. Limnology. 2016;17:23–32. [Google Scholar]
- Muirhead JR, Lewis MA, MacIsaac HJ. Prediction and error in multi-stage models for spread of aquatic non-indigenous species. Diversity Distrib. 2011;17:323–337. [Google Scholar]
- Nguyen NH, Smith D, Peay K, Kennedy P. Parsing ecological signal from noise in next generation amplicon sequencing. New Phyto. 2015;205:1389–1393. doi: 10.1111/nph.12923. [DOI] [PubMed] [Google Scholar]
- O’Malia EM. Thesis. University of Minnesota; 2015. Land-Use Proxies for Aquatic Species Introductions in the Laurentian Great Lakes. [Google Scholar]
- Pagnucco KS, Maynard GA, Fera SA, Yan ND, Nalepa TF, Ricciardi A. The future of species invasions in the Great Lakes – St. Lawrence River basin. J Great Lakes Res. 2015;41s1:96–107. [Google Scholar]
- Pawlowski J, Audic S, Adl S, Bass D, Berney C, Bowser S, Cepicka I, Declle J, Dunthorn M, Fiore-Donno AM, Gile GH, Holzmann M, Jahn R, JIrkû M, Keeling PJ, Kostka M, Kudryavtsev Am Kara Em Lukeŝ J, Mann DG, Mitchell EAD, Nitsche F, Romeralo M, Saunders GW, Simpson AGB, Smirnov AV, Spouge JL, Stern RF, Stoeck T, Zimmermann J, Schindel D, de Vargas C. CBOL Protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 2012;10:e1001419. doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchar L, Mooney H. Invasive species, ecosystem services and human well-being. Trends Ecol Evol. 2009;24:497–504. doi: 10.1016/j.tree.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Peterson GS, Lietz JE. Identification of Ruffe larvae (Gymnocephalus cernuus) in the St. Louis River, Lake Superior: Clarification and guidance regarding morphological descriptions. J Great Lakes Res. 2017;43:205–210. doi: 10.1016/j.jglr.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim EM, Jackson SA, Swenson S, Turcsanyi I, Friedman E, Weigt L, Bagley MJ. Incorporation of DNA barcoding into a large-scale biomonitoring program: opportunities and pitfalls. J N Am Benth Soc. 2011;30:217–231. [Google Scholar]
- Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ. 2005;52:273–288. [Google Scholar]
- Pochon X, Bott NJ, Smith KF, Wood SA. Evaluating detection limits of next-generation sequencing for the surveillance and monitoring of international marine pests. PLoS One. 2013;8:e73935. doi: 10.1371/journal.pone.0073935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram JL, Banno F, Gala RR, Gizicki JP, Kashian DR. Estimating sampling effort for early detection of non-indigenous benthic species in the Toledo Harbor region of Lake Erie. Manage Biol Invas. 2014;5:209–216. [Google Scholar]
- Ratnasingham S, Hebert PDN. BOLD: The barcode of life data system ( www.barcodinglife.org) Molec Ecol Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HC, Maddison BC, Middleditch DJ, Patmore JRM, Gough KC. The detection of aquatic animal species using environmental DNA – a review of eDNA as a survey tool in ecology. J Appl Ecol. 2014;51:1450–1459. [Google Scholar]
- Rew LJ, Maxwell BD, Dougher FL, Aspinall R. Searching for a needle in a haystack: evaluating survey methods for non-indigenous plant species. Biol Invas. 2006;8:523–539. [Google Scholar]
- Ricciardi A. Patterns of invasion in the Laurentian Great Lakes in relation to changes in vector activity. Divers Distrib. 2006;12:425–433. [Google Scholar]
- Rothlisberger JD, Finnoff DC, Cooke RM, Lodge DM. Ship-borne nonindigenous species diminish Great Lakes ecosystem services. Ecosystems. 2012;15:462–476. [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt BR, Kéry M, Ursenbacher S, Hyman OJ, Collins JP. Site occupancy models in the analysis of environmental DNA presence/absence surveys: A case study of an emerging amphibian pathogen. Meth Ecol Evol. 2013;4:646–653. [Google Scholar]
- Schneider D, Ellis C, Cummings K. A transportation model assessment of the risk to native mussel communities from zebra mussel spread. Conserv Biol. 1998;12:788–800. [Google Scholar]
- Schultz MT, Lance RF. Modeling the sensitivity of field surveys for detection of environmental DNA (eDNA) PLoS One. 2015;10:e0141503. doi: 10.1371/journal.pone.0141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JLA, Clarke LJ, Wedderburn SD, Barnes TC, Weyrich LA, Cooper A. Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol Conserv. 2016;197:131–138. [Google Scholar]
- Simmons M, Tucker A, Chadderton WL, Jerde CL, Mahon AR. Active and passive environmental DNA surveillance of aquatic invasive species. Can J Fish Aquatic Sci. 2015;73:76–83. [Google Scholar]
- Sliwinski M, Powell L, Koper N, Giovanni M, Schacht W. Research design considerations to ensure detection of all species in an avian community. Meth Ecol Evol. 2015;7:456–462. [Google Scholar]
- Smokorowski KE, Pratt TC. Effect of a change in physical structure and cover on fish and fish habitat in freshwater ecosystems - a review and meta-analysis. Envi Rev. 2007;15:15–41. [Google Scholar]
- Snyder RJ, Burlakova LE, Karatayev AY, MacNeill DB. Updated invasion risk assessment for Ponto-Caspian fishes to the Great Lakes. J Great Lakes Res. 2014;40:360–36. [Google Scholar]
- Stedtfeld R, Stedtfeld TM, Kronlein M, Seyrig G, Steffan RJ, Cupples A, Hashsham SA. DNA extraction-free quantification of Dehalococcoides spp. in groundwater using a hand-held device. Env Sci Tech. 2014;48:13855–13863. doi: 10.1021/es503472h. [DOI] [PubMed] [Google Scholar]
- Stedtfeld RD, Williams MR, Fakher U, Johnson TA, Stedtfeld TM, Wang F, Khalife WT, Hughes M, Etchebarne BE, Tiedje JM, Hashsham SA. Antimicrobial resistance dashboard application for mapping environmental occurrence and resistant pathogens. FEMS Microbiol Ecol. 2016;92:fiw020. doi: 10.1093/femsec/fiw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein ED, Martinez MC, Stiles S, Miller PE, Zakharov EV. Is DNA barcoding actually cheaper and faster than traditional morphological methods? Results from a survey of freshwater bioassessment efforts in the United States. PLoS One. 2014;9:e95525. doi: 10.1371/journal.pone.0095525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien CA, Neilson ME. What’s in a Name? Taxonomy and nomenclature of invasive gobies in the Great Lakes and beyond. J Great Lakes Res. 2013;39:555–559. [Google Scholar]
- Stevens DL, Olsen AR. Spatially-balanced sampling of natural resources. J Am Stat Assoc. 2004;99:262–278. [Google Scholar]
- Strickland KM, Fremier AF, Goldberg CS. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol Conserv. 2015;183:85–92. [Google Scholar]
- Sweeney BW, Battle JM, Jackson JK, Dapkey T. Can DNA barcodes of stream macroinvertebrates improve descriptions of community structure and water quality? J N Am Benth Soc. 2011;30:195–216. [Google Scholar]
- Takahara T, Toshifumi M, Yamanaka H, Doi H, Kawabata Z. Estimation of fish biomass using environmental DNA. PLoS One. 2012;7:e35868. doi: 10.1371/journal.pone.0035868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen PF, Willerslev E. Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv. 2015;183:4–18. [Google Scholar]
- Trebitz AS, Hoffman JC, Grant G, Billehus T, Pilgrim E. Potential for DNA-based identification of Great Lakes fauna: match and mismatch between species inventories and DNA barcode libraries. Sci Reports. 2015;5:12162. doi: 10.1038/srep12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitz AS, Kelly JR, Hoffman JC, Peterson GS, West CW. Exploiting habitat and gear patterns for efficient detection of rare and non-native benthos and fish in Great Lakes coastal ecosystems. Aquat Invas. 2009;4:651–667. [Google Scholar]
- Treguier A, Paillisson JM, Dejean T, Valentini A, Schlaepfer MA, Roussel JM. Environmental DNA surveillance for invertebrate species: advantages and technical limitations to detect invasive crayfish Procambrus clarkia in freshwater ponds. J Appl Ecol. 2014;51:871–879. [Google Scholar]
- Tucker AJ, Chadderton WL, Jerde CL, Renshaw MA, Uy K, Gantz C, Mahon AR, Bowen A, Strakosh T, Bossenbroek JM, Sieracki JL. A sensitive environmental DNA (eDNA) assay leads to new insights on Ruffe (Gymnocephalus cernua) spread in North America. Biol Invasions. 2016;2016:1–8. [Google Scholar]
- Turner CR, Barnes MA, Xu CC, Jones SE, Jerde CL, Lodge DM. Particle size distribution and optimal capture of aqueous macrobial eDNA. Meth Ecol Evol. 2014;5:676–684. [Google Scholar]
- U.S. Fish and Wildlife Service. Species ecological risk screening summaries. 2015 Available https://www.fws.gov/injuriouswildlife/species_erss_reports.html.
- U.S Geological Survey. Detection of environmental DNA of bigheaded carps in samples collected from selected locations in the St. Croix River and in the Mississippi River, Open File report 2013-1080 2013 [Google Scholar]
- Valentini A, Taberlet P, Miaud C, Civade R, Herder J, Thomsen PF, Bellemain E, Besnard A, Coissac E, Boyer F, Gaboriaud C, Jean P, Poulet N, Roset N, Copp GH, Geniez P, Pont D, Argillier C, Baudoin JM, Peroux T, Crivelli AJ, Olivier A, Acqueberge M, Le Brun M, Møller PR, Willerslev E, Dejean T. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Molec Ecol. 2016;25:929–942. doi: 10.1111/mec.13428. [DOI] [PubMed] [Google Scholar]
- Vander Zanden MJ, Hansen GJ, Higgins SN, Kornis MS. A pound of prevention, plus a pound of cure: early detection and eradication of invasive species in the Laurentian Great Lakes. J Great Lakes Res. 2010;36:199–205. [Google Scholar]
- Vermonden K, Leuven RS, Van Der Velde G. Environmental factors determining invasibility of urban waters for exotic macroinvertebrates. Divers Distrib. 2010;16:1009–1021. [Google Scholar]
- Yoccoz NG. The future of environmental DNA in ecology. Molec Ecol. 2012;21:2031–2038. doi: 10.1111/j.1365-294X.2012.05505.x. [DOI] [PubMed] [Google Scholar]
- Walsh JR, Carpenter SR, Vander Zanden MJ. Invasive species triggers a massive loss of ecosystem services through a trophic cascade. Proc Nat Academy Sci. 2016;113:4081–4085. doi: 10.1073/pnas.1600366113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox TM, McKelvey KS, Young MK, Jane SF, Lowe WH, Whiteley AR, Schwartz MK. Robust detection of rare species using environmental DNA: the importance of primer specificity. PLoS One. 2013;8:e59520. doi: 10.1371/journal.pone.0059520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Wright E, Bronnenhuber J, MacDonald F, Belore M, Locke B. Tracking ghosts: combined electrofishing and environmental DNA surveillance efforts for Asian carps in Ontario waters of Lake Erie. Manage Biol Invas. 2014;5:225–231. [Google Scholar]
- Wonham M, Byers J, Grosholz E, Leung B. Modeling the relationship between propagule pressure and invasion risk to inform policy and management. Ecol Applic. 2013;23:1691–1706. doi: 10.1890/12-1985.1. [DOI] [PubMed] [Google Scholar]
- Yemshanov D, Haight RG, Koch FH, Lu B, Venette R, Fournier RE, Turgeon JJ. Robust surveillance and control of invasive species using a scenario optimization approach. Ecol Econ. 2017;133:86–98. [Google Scholar]
- Zhan A, Hulak M, Sylvester F, Huang X, Abebayo AA, Abbott CL, Adamowicz SJ, Heath DD, Cristescu ME, MacIsaac HJ. High sensitivity of 454 pyrosequencing for detection of rare species in aquatic communities. Meth Ecol Evol. 2013;4:558–565. [Google Scholar]
- Zhou X, Li Y, Liu S, Yang Q, Su X, Zhou L, Tang M, Fu R, Li J, Huang Q. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. GigaScience. 2013;2:1. doi: 10.1186/2047-217X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]