ABSTRACT
Most RNA polymerases can initiate transcription from diverse DNA template sequences with relatively few outright sequence restraints. Recent reports have demonstrated that failure to subdue the promiscuity of RNA polymerase in vivo can severely impede cell function. This phenomenon appears common to all cell types with undesirable effects ranging from growth inhibition in prokaryotes to cancer in higher organisms. Here we discuss similarities and differences in strategies employed by cells to minimise spurious transcription across life's domains.
KEYWORDS: H-NS, RNA polymerase, cancer, E. coli, S. pombe, S. cerevisiae
Introduction
Promoters, the DNA sequences that allow RNA polymerases to initiate transcription, have few absolute DNA sequence constraints; many DNA sequences can serve as a promoter for any given RNA polymerase.1–5 As a result, promoters arise in “unexpected” genomic contexts throughout life's different domains.6–12 For example, promoters inside coding regions, or unsuitably orientated within non-coding DNA, are common.6–12 In some instances, such promoters are properly regulated and generate functional transcripts.13–19 In other cases, these promoters contribute to the phenomenon of pervasive transcription, a genome-wide background of low level RNA production, which could be beneficial in some situations.6,7 For example, spurious promoters may act as an evolutionary source of new functional RNAs.7 However, some unexpected promoters appear to occur by happenstance, and are either transcriptionally silenced, or generate RNA species that are rapidly turned over.20–22 If silencing systems fail, such transcripts can be generated at high levels.10–12,20–22 Since the synthesis of these RNAs is usually suppressed, and because the production of such transcripts can hinder correct cell function,21 we will refer to the RNAs as spurious. In this point-of-view we argue that spurious transcription is unavoidable in some circumstances given the promiscuous nature of RNA polymerases and the apparent inability of natural selection to remove all chance promoters. Consequently, all cell types have evolved mechanisms to suppress spurious transcription. We will also discuss the causes and consequences of unwanted transcription in bacteria, archaea, and eukaryotes.
Controlling spurious transcription at the level of initiation
The simplest way to prevent transcription in unwanted locations is to remove DNA sequences that can function as promoters. Natural selection appears to have been moderately successful in this regard; the occurrence of promoter-like sequences is indeed reduced within genes for many organisms.8,9,23–25 However, because the absolute sequence requirements for transcription initiation are relatively few it may not be impossible to eradicate all such sequences. For example, the housekeeping RNA polymerase in bacteria requires only a partial match to the −10 hexamer consensus (5′-TATAAT-3′) and partial matches to one of several ancillary sequences, all with an elevated A/T-content, to initiate transcription.1,3,23 Consequently, in Escherichia coli, genes with an A/T-content exceeding 60% contain many sequences capable of driving transcription both in vivo and in vitro.20,21,26 Even if natural selection could eventually remove such promoters, rampant horizontal gene transfer ensures the task is never complete.27 A similar situation may exist in archaea where there is also a close relationship between DNA A/T-content and transcription initiation.24 Indeed, it is notable that A/T-rich regions of the archaeal Methanocaldococcus jannaschii genome lacking coding potential (e.g. DNA between convergent genes) are associated with transcription.24 In eukaryotes, promoter sequences can be diverse, but A/T-rich DNA sequences disrupt nucleosome formation, and a common determinant for transcription initiation is the TATA box (consensus: 5′-TATA-3′).5,28 Hence, a TATA box alone can stimulate transcription by human RNA polymerase II.29 Consistent with this, spurious transcription initiation has been observed within many genes in Saccharomyces cerevisiae, and often coincides with the occurrence of a TATA box.11 Furthermore, many promoters in eukaryotes are bidirectional, and generate antisense transcripts in addition to the expected sense RNA.22 In fact, bidirectional transcription is likely the ground state of a newly evolved promoter, and directionality evolves over time, likely due to acquisition of binding sites for asymmetric transcriptional regulators.30
Inhibition of spurious transcription initiation
Natural selection has clearly produced organisms where promoter-like sequences within genes have been minimised. For example, in E. coli, Sulfolobus solfataricus and Schizosaccharomyces pombe, the average A/T-content of genes is often between 5% and 9% lower than that of intergenic DNA.31–33 Even so, additional mechanisms are required to suppress transcription from promoters not removed by evolutionary pressure.10,12,20,21 Prokaryotes and eukaryotes utilise analogous, but evolutionarily unrelated, repressive nucleoprotein structures to silence spurious transcription initiation (Fig. 1). In E. coli, the Histone-like nucleoid structuring (H-NS) protein specifically recognises A/T-rich DNA by virtue of an arginine side chain that interacts with the narrowed minor groove of A/T-rich DNA sequence27,34 (Fig. 1A). Interactions between DNA-bound H-NS molecules drive polymerisation of the protein and create nucleoprotein complexes capable of repressing transcription.27 Consequently, deletion of hns results in uncontrolled RNA synthesis inside genes that are A/T-rich.20,21 Although hns is not widely conserved, other bacteria express functionally related proteins that preferentially bind A/T-rich DNA, e.g. Lsr2 in mycobacteria,35 MvaT/U in pseudomonads,36 and Rok in Bacillus subtilis.37 It is likely that these H-NS analogues also prevent spurious transcription from intragenic promoters. Interestingly, the archaeal Cbp1 protein, a factor involved in chromosome packaging, is required to prevent transcription initiation within A/T-rich CRISPR loci,12 suggesting it functions analogously to H-NS.
Figure 1.
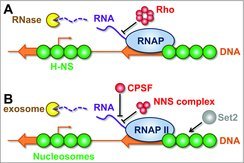
Prokaryotes and eukaryotes use analogous mechanisms to prevent spurious transcription. The DNA is shown as an orange line with genes and promoters represented by block and bent arrows respectively. All other components are individually labelled. (A) In prokaryotic cells, the Histone-like nucleoid structuring protein (H-NS) can supress the activity of spurious promoters within genes, and can impede transcription elongation. If the elongating RNA polymerase complex includes a spurious non-coding transcript, transcription is often subject to premature termination by Rho. Resulting transcripts can be degraded by RNases. (B) In eukaryotic cells, nucleosomes can impede access to spurious promoters. The repressive properties of nucleosomes can be enhanced by methyltransferase proteins such as Set2 (in yeast) or SetD2 (in metazoa) act on histone H3 residue K36. The yeast Nrd1-Nab3-Sen1 (NNS) complex recognises spurious RNAs, by virtue of their different sequence properties, and can prematurely terminate transcription elongation. The cleavage and polyadenylation specificity factor (CPSF) is a multiprotein complex and recognises poly(A) sites. Ultimately, such transcripts can be degraded by the exosome.
In eukaryotes as diverse as humans and yeast, nucleosome occupancy inhibits transcription initiation, and canonical promoters are typically nucleosome-depleted38–42 (Fig. 1B). Hence, antisense transcripts can arise near to canonical promoters or within the 3′ ends of genes.38–42 Nucleosomes also play a key role in suppressing spurious transcription by virtue of their histone modifications. For example, deletion of the gene encoding yeast Set2, which catalyzes methylation of histone H3 residue K36, allows widespread spurious transcription initiation43 (Fig. 1B). Similar effects are apparent in metazoan cells lacking the homologous SetD2 protein.44 In yeast, these phenomena appear to be mediated via differential activation of Rpd3S, a histone deacetylase complex.43,45–48 The methylation state of H3 K36 also controls recruitment of the Isw1b chromatin remodelling complex, which works with Chd1 to prevent histone exchange and maintain chromatin structure.49 Hence, yeast strains lacking both isw1 and chd1 have a prominent spurious transcription phenotype.49
Epigenetic DNA modifications occur in prokaryotic and eukaryotic cells.50,51 However, little is known about the role of such nucleic acid changes in controlling spurious transcription initiation. To date, the best characterised consequences are those identified in mouse embryonic stem cells,52 where intragenic methylation of CpG dinucleotides within the body of genes is required to prevent intragenic transcription initiation by RNA polymerase II.51 Curiously, recruitment of Dnmt3B, the enzyme responsible for this DNA modification, is mediated by the methylation state of histone H3 K36.52 Consequently, mammalian SetD2 controls histone H3 K36 methylation and co-operates with Dmnt3B to prevent spurious transcription initiation.44,52,53 Although DNA methylation is known to influence transcription initiation in bacteria,54 there is no evidence this modification controls unwanted transcription.
Termination of spurious transcription
Whilst all cell types take measures to block spurious transcription initiation, these inhibitory mechanisms are imperfect. Hence, bacteria and eukaryotes have each evolved mechanisms to rapidly terminate production of spurious transcripts. In both cases, the termination machinery recognises a property of spurious RNA production not associated with functional transcription. In bacteria, discrimination is based on the coupling of transcription and translation. Since appropriately positioned translation start codons and ribosome binding sites rarely occur by chance, most spurious transcripts are not translated. The Rho transcription termination factor, found in 90% of bacteria, recognises and terminates transcription of non-coding RNA55 (Fig. 1A). Hence, chemical inhibition of Rho results in increased transcription beyond gene boundaries and within A/T-rich genes.56–58 Interestingly, H-NS occupancy can enhance transcription termination by Rho56 (Fig. 1A). Thus, H-NS serves a dual purpose in suppressing pervasive transcription: silencing spurious promoters, and enhancing termination of spurious transcripts. In eukaryotes, transcription and translation are not coupled. Hence, cryptic unstable transcripts arising between genes are identified by a different mechanism. For example, in mouse embryonic stem cells, spurious antisense transcripts can arise from bidirectional promoters, with the corresponding sense transcripts being functional RNAs. Poly(A) sites are enriched in the 5′ regions of the antisense RNAs, and stimulate premature termination of antisense transcripts by cleavage and polyadenylation specificity factor (CPSF) and associated proteins.59 In contrast, binding sites for U1 snRNP are enriched in 5′ regions of the sense transcripts, and recruitment of U1 snRNP protects these RNAs from premature cleavage and polyadenylation.59 In S. cerevisiae, the Nrd1-Nab3-Sen1 (NNS) complex has a key role60 (Fig. 1B). To distinguish between spurious and functional transcripts, the yeast NNS complex also recognises specific nucleotide signatures in the RNA.61 Crucially, these sequences are depleted in mRNAs.62 In some instances, termination by the polyadenylation machinery may provide a back-up mechanism.63 Sequences recognised by the polyadenylation machinery are enriched at the 3′ ends of genes in the antisense orientation, preventing read-through of spurious transcripts into genes.63 In prokaryotes, intrinsic terminators downstream of genes can be bidirectional, but most are not.64
Degradation of spurious transcripts
In both bacteria and eukaryotes, many spurious transcripts are rapidly degraded following transcription. This process is best understood in eukaryotes, where some spurious transcripts (as well as some functional transcripts) are degraded by the exosome complex. In S. cerevisiae, Nrd1 interacts with Trf4, a member of the TRAMP polyadenylation complex.65 Thus, NNS-terminated transcripts are polyadenylated by TRAMP, which leads to degradation by the exosome66 (Fig. 1B). There is also feedback from the exosome to the NNS complex, whereby the exosome component Rrp6 stimulates NNS-mediated transcription termination of a subset of RNAs.67 The details of spurious transcript degradation are poorly understood in bacteria; the process has only been studied in the context of antisense RNAs. Thus, RNase III has been shown to degrade antisense RNAs in E. coli and Staphylococcus aureus,66–68 and may target antisense RNAs paired with their cognate mRNA.68–70 In Bacillus subtilis, RNase Y and RNase J1 play a larger role than RNase III in degradation of antisense RNAs71 (Fig. 1A).
The relationship between spurious transcription and impaired cell function
As described above, all cell types appear to permit low levels of pervasive transcription, but multiple systems exist to avoid high level production of spurious transcripts. When these control measures fail, cell function is impaired. For example, in many bacteria, deletion of hns results in a slow growth phenotype, and such strains rapidly acquire compensatory mutations to alleviate these effects.21,72,73 The underlying mechanism involves titration of the limited RNA polymerase pool and a consequent down-regulation of housekeeping genes.21 Formation of R-loops following inhibition of Rho is also likely to be deleterious.74 Adverse consequences of spurious transcription initiation or read-through in eukaryotes have also been reported.75,76 Of particular note are observations identifying SetD2 as a tumour suppressor.77–80 For example, loss of SetD2 activity in renal carcinoma cells causes inefficient transcription termination. As a result, transcription elongation complexes for spurious RNAs invade oncogenes and increase their expression.81 Similarly, in some melanomas, aberrant chromatin modifications are associated with intron derived RNAs and expression of a novel anaplastic lymphoma kinase isoform.82 Chromatin alterations, and the activation of otherwise cryptic promoters, are also common in gastric adenocarcinoma.83 More anecdotally, there are many accounts of A/T-rich DNA sequences being associated with chromosome instability and the synthesis of poorly defined microRNAs.84,85 This is significant, given the likelihood of such DNA sequences being enriched for spurious promoter elements.
Concluding remarks
The structure and function of housekeeping RNA polymerases is conserved throughout life.2 In particular, RNA polymerase has a conserved propensity to initiate transcription with relatively low sequence specificity. Consequently, most organisms have evolved mechanisms to minimise the occurrence of spurious transcription (Fig. 1). In both bacteria and eukaryotes, derepression of spurious transcription leads to impaired cell function. In metazoans, this can manifest as disease. We argue that such spurious transcriptional events are an unavoidable consequence of DNA-based life where the flow of genetic information via an RNA intermediate requires a transcriptional apparatus that is unable to differentiate between promoters of functional RNAs and promoters that occur spuriously. Importantly, spurious transcription may also play a positive role, serving as a rich source for the evolution of functional transcripts.86
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Fred Winston for support and critical reading of the manuscript.
References
- 1.Decker KB, Hinton DM. Transcription regulation at the core: Similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu Rev Microbiol. 2013;67:113–39. doi: 10.1146/annurev-micro-092412-155756. PMID:23768203 [DOI] [PubMed] [Google Scholar]
- 2.Hantsche M, Cramer P. Conserved RNA polymerase II initiation complex structure. Curr Opin Struct Biol. 2017;47:17–22. doi: 10.1016/j.sbi.2017.03.013. PMID:28437704 [DOI] [PubMed] [Google Scholar]
- 3.Browning DF, Busby SJ. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol. 2016;14:638–50. doi: 10.1038/nrmicro.2016.103. PMID:27498839 [DOI] [PubMed] [Google Scholar]
- 4.Bartlett MS. Determinants of transcription initiation by archaeal RNA polymerase. Curr Opin Microbiol. 2005;8:677–84. doi: 10.1016/j.mib.2005.10.016. PMID:16249119 [DOI] [PubMed] [Google Scholar]
- 5.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–92. doi: 10.1101/gad.1026202. PMID:12381658 [DOI] [PubMed] [Google Scholar]
- 6.Grainger DC. The unexpected complexity of bacterial genomes. Microbiology. 2016;162:1167–72. doi: 10.1099/mic.0.000309. PMID:27663516 [DOI] [PubMed] [Google Scholar]
- 7.Wade JT, Grainger DC. Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol. 2014;12:647–53. doi: 10.1038/nrmicro3316. PMID:25069631 [DOI] [PubMed] [Google Scholar]
- 8.Dornenburg JE, Devita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. mBio. 2010;1:pii: e00024–10. doi: 10.1128/mBio.00024-10. PMID:20689751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, Stadler PF, Vogel J. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. PMID:20164839 [DOI] [PubMed] [Google Scholar]
- 10.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–9. doi: 10.1126/science.1087374. PMID:12934008 [DOI] [PubMed] [Google Scholar]
- 11.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. PMID:18998772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–80. doi: 10.1093/nar/gkr1111. PMID:22139923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–19. doi: 10.1038/emboj.2012.229. PMID:22922465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol. 2010;75:1215–31. doi: 10.1111/j.1365-2958.2010.07044.x. PMID:20070527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. PMID:19148191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. PMID:14744438 [DOI] [PubMed] [Google Scholar]
- 17.Haycocks JRJ, Grainger DC. Unusually situated binding sites for bacterial transcription factors can have hidden functionality. PloS one 2016;11:e0157016. doi: 10.1371/journal.pone.0157016. PMID:27258043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahuja R, Kumar V. Stimulation of Pol III-dependent 5S rRNA and U6 snRNA gene expression by AP-1 transcription factors. FEBS J 2017;284:2066–77. doi: 10.1111/febs.14104. PMID:28488757 [DOI] [PubMed] [Google Scholar]
- 19.McKnight K, Liu H, Wang Y. Replicative stress induces intragenic transcription of the ASE1 gene that negatively regulates Ase1 activity. Curr Biol. 2014;24:1101–6. doi: 10.1016/j.cub.2014.03.040. PMID:24768052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014;28:214–9. doi: 10.1101/gad.234336.113. PMID:24449106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamberte LE, Baniulyte G, Singh SS, Stringer AM, Bonocora RP, Stracy M, Kapanidis AN, Wade JT, Grainger DC. Horizontally acquired AT-rich genes in Escherichia coli cause toxicity by sequestering RNA polymerase. Nat Microbiol. 2017;2:16249. doi: 10.1038/nmicrobiol.2016.249. PMID:28067866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Mol Cell. 2013;52:473–84. doi: 10.1016/j.molcel.2013.10.032. PMID:24267449 [DOI] [PubMed] [Google Scholar]
- 23.Singh SS, Typas A, Hengge R, Grainger DC. Escherichia coli σωΠ senses sequence and conformation of the promoter spacer region. Nucleic Acids Res. 2011; 39:5109–18. doi: 10.1093/nar/gkr080. PMID:21398630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smollett K, Blombach F, Reichelt R, Thomm M, Werner F. A global analysis of transcription reveals two modes of Spt4/5 recruitment to archaeal RNA polymerase. Nat Microbiol. 2017;2:17021. doi: 10.1038/nmicrobiol.2017.21. PMID:28248297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Hou J, Bai L, Hu C, Tong P, Kang Y, Zhao X, Shao Z. Genome-wide analysis of core promoter structures in Schizosaccharomyces pombe with DeepCAGE. RNA Biol. 2015;12:525–37. doi: 10.1080/15476286.2015.1022704. PMID:25747261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chintakayala K, Singh SS, Rossiter AE, Shahapure R, Dame RT, Grainger DC. E. coli Fis protein insulates the cbpA gene from uncontrolled transcription. PLoS Genet. 2013;9:e1003152. doi: 10.1371/journal.pgen.1003152. PMID:23341772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grainger DC. Structure and function of bacterial H-NS protein. Biochem Soc Trans. 2016;44:1561–9. doi: 10.1042/BST20160190. PMID:27913665 [DOI] [PubMed] [Google Scholar]
- 28.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–35. doi: 10.1038/ng1789. PMID:16645617 [DOI] [PubMed] [Google Scholar]
- 29.Malecová B, Gross P, Boyer-Guittaut M, Yavuz S, Oelgeschläger T. The initiator core promoter element antagonizes repression of TATA-directed transcription by negative cofactor NC2. J Biol Chem. 2007;282:24767–76. doi: 10.1074/jbc.M702776200. PMID:17584739 [DOI] [PubMed] [Google Scholar]
- 30.Jin Y, Eser U, Struhl K, Churchman LS. The ground state and evolution of promoter region directionality. Cell. 2017;170:889–98. doi: 10.1016/j.cell.2017.07.006. PMID:28803729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–74. doi: 10.1126/science.277.5331.1453. PMID:9278503 [DOI] [PubMed] [Google Scholar]
- 32.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC-Y, Clausen IG, et al.. The complete genome of the crenarchaeon Sulfolobus solfataricus P2 2001;98:7835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–80. doi: 10.1038/nature724. PMID:11859360 [DOI] [PubMed] [Google Scholar]
- 34.Gordon BR, Li Y, Cote A, Weirauch MT, Ding P, Hughes TR, Navarre WW, Xia B, Liu J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci USA. 2011;108:10690–5. doi: 10.1073/pnas.1102544108. PMID:21673140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon BR, Imperial R, Wang L, Navarre WW, Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J Bacteriol. 2008;190:7052–99. doi: 10.1128/JB.00733-08. PMID:18776007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castang S, Dove SL. Basis for the essentiality of H-NS family members in Pseudomonas aeruginosa. J Bacteriol. 2012;194:5101–9. doi: 10.1128/JB.00932-12. PMID:22821971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smits WK, Grossman AD. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 2010;6:e1001207. doi: 10.1371/journal.pgen.1001207. PMID:21085634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mai X, Chou S, Struhl K. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol Cell Biol. 2000;20;6668–76. doi: 10.1128/MCB.20.18.6668-6676.2000. PMID:10958664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–9. PMID:7781610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. PMID:19056940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. PMID:19056938 [DOI] [PubMed] [Google Scholar]
- 42.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–7. doi: 10.1038/nature07728. PMID:19169243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. PMID:16286007 [DOI] [PubMed] [Google Scholar]
- 44.Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013;41:2881–93. doi: 10.1093/nar/gks1472. PMID:23325844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. PMID:16286008 [DOI] [PubMed] [Google Scholar]
- 46.Drouin S, Laramée L, Jacques PÉ, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. PMID:21060864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010; 39:234–46. doi: 10.1016/j.molcel.2010.07.003. PMID:20670892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–8. doi: 10.1016/j.molcel.2005.11.021. PMID:16364921 [DOI] [PubMed] [Google Scholar]
- 49.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 2012;19:884–92. doi: 10.1038/nsmb.2312. PMID:22922743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez-Romero MA, Busby SJ, Dyer NP, Ott S, Millard AD, Grainger DC. Dynamic distribution of seqa protein across the chromosome of Escherichia coli K-12. mBio. 2010;1:e00012–10. doi: 10.1128/mBio.00012-10. PMID:20689753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang B, Zhou Y, Wang CM, Huang TH, Jin VX. Integration of DNA methylation and gene transcription across nineteen cell types reveals cell type-specific and genomic region-dependent regulatory patterns. Sci Rep. 2017;7:3626. doi: 10.1038/s41598-017-03837-z. PMID:28620196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–7. doi: 10.1038/nature21373. PMID:28225755 [DOI] [PubMed] [Google Scholar]
- 53.Teissandier A, Bourc'his D. Gene body DNA methylation conspires with H3K36me3 to preclude aberrant transcription. EMBO J. 2017;36:1471–3. doi: 10.15252/embj.201796812. PMID:28442531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sánchez-Romero MA, Cota I, Casadesús J. DNA methylation in bacteria: From the methyl group to the methylome. Curr Opin Microbiol. 2015;25:9–16. doi: 10.1016/j.mib.2015.03.004. PMID:25818841 [DOI] [PubMed] [Google Scholar]
- 55.D'Heygère F, Rabhi M, Boudvillain M. Phyletic distribution and conservation of the bacterial transcription termination factor Rho. Microbiology. 2013;159:1423–36. doi: 10.1099/mic.0.067462-0. PMID:23704790 [DOI] [PubMed] [Google Scholar]
- 56.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–33. doi: 10.1101/gad.196741.112. PMID:23207917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106:15406–11. doi: 10.1073/pnas.0903846106. PMID:19706412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–8. doi: 10.1126/science.1152763. PMID:18487194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–3. doi: 10.1038/nature12349. PMID:23792564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–51. doi: 10.1016/j.molcel.2006.07.024. PMID:16973436 [DOI] [PubMed] [Google Scholar]
- 61.Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/MCB.16.12.6993. PMID:8943355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uwimana N, Collin P, Jeronimo C, Haibe-Kains B, Robert F. Bidirectional terminators in Saccharomyces cerevisiae prevent cryptic transcription from invading neighboring genes. Nucleic Acids Res. 2017;45:6417–26. doi: 10.1093/nar/gkx242. PMID:28383698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen YJ, Liu P, Nielsen AA, Brophy JA, Clancy K, Peterson T, Voigt CA. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10:659–64. doi: 10.1038/nmeth.2515. PMID:23727987 [DOI] [PubMed] [Google Scholar]
- 65.Tudek A, Porrua O, Kabzinski T, Lidschreiber M, Kubicek K, Fortova A, Lacroute F, Vanacova S, Cramer P, Stefl R, Libri D. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol Cell. 2014;55:467–81. PMID:25066235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuparevic I, Mosrin-Huaman C, Hervouet-Coste N, Remenaric M, Rahmouni AR. Cotranscriptional recruitment of RNA exosome cofactors Rrp47p and Mpp6p and two distinct Trf-Air-Mtr4 polyadenylation (TRAMP) complexes assists the exonuclease Rrp6p in the targeting and degradation of an aberrant messenger ribonucleoprotein particle (mRNP) in yeast. J Biol Chem. 2013;288:31816–29. PMID:24047896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fox MJ, Gao H, Smith-Kinnaman WR, Liu Y, Mosley AL. The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway. PLoS Genet. 2015;11:e1004999. PMID:25680078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lybecker M, Zimmermann B, Bilusic I, Tukhtubaeva N, Schroeder R. The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci USA. 2014;111;3134–9. PMID:24453212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, Solano C, Gingeras TR. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA. 2011;108:20172–7. PMID:22123973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lioliou E, Sharma CM, Caldelari I, Helfer AC, Fechter P, Vandenesch F, Vogel J, Romby P. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012;8:e1002782. PMID:22761586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Durand S, Gilet L, Bessières P, Nicolas P, Condon C. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 2012;8:e1002520. PMID:22412379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ali SS, Soo J, Rao C, Leung AS, Ngai DH, Ensminger AW, Navarre WW. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog. 2014;10:e1004500. PMID:25375226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–8. PMID:16763111 [DOI] [PubMed] [Google Scholar]
- 74.Leela JK, Syeda AH, Anupama K, Gowrishankar J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci USA. 2013;110:258–63. doi: 10.1073/pnas.1213123110. PMID:23251031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muratani M, Deng N, Ooi WF, Lin SJ, Xing M, Xu C, Qamra A, Tay ST, Malik S, Wu J. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat Commun. 2014;5:4361. doi: 10.1038/ncomms5361. PMID:25008978 [DOI] [PubMed] [Google Scholar]
- 76.McDaniel SL, Strahl BD. Shaping the cellular landscape with Set2/SETD2 methylation. Cell Mol Life Sci. 2017;74:3317–34. doi: 10.1007/s00018-017-2517-x. PMID:28386724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. PMID:22237106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–91. doi: 10.1158/0008-5472.CAN-10-0120. PMID:20501857 [DOI] [PubMed] [Google Scholar]
- 79.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. PMID:20054297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Al Sarakbi W, Sasi W, Jiang WG, Roberts T, Newbold RF, Mokbel K. The mRNA expression of SETD2 in human breast cancer: correlation with clinico-pathological parameters. BMC Cancer. 2009;9:290. doi: 10.1186/1471-2407-9-290. PMID:19698110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vítor AC, Desterro JM, Carmo-Fonseca M, de Almeida SF. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife. 2015;4:e09214. doi: 10.7554/eLife.09214. PMID:26575290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiesner T, Lee W, Obenauf AC, Ran L, Murali R, Zhang QF, Wong EW, Hu W, Scott SN, Shah RH. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015;526:453–7. doi: 10.1038/nature15258. PMID:26444240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muratani M, Deng N, Ooi WF, Lin SJ, Xing M, Xu C, Qamra A, Tay ST, Malik S, Wu J. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat Commun. 2014;5:4361. doi: 10.1038/ncomms5361. PMID:25008978 [DOI] [PubMed] [Google Scholar]
- 84.Galindo CL, McIver LJ, Tae H, McCormick JF, Skinner MA, Hoeschele I, Lewis CM, Minna JD, Boothman DA, Garner HR. Sporadic breast cancer patients' germline DNA exhibit an AT-rich microsatellite signature. Genes Chromosomes Cancer. 2011;50:275–83. PMID:21319262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre HJ, Lapsys N, Le Paslier D. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–74. doi: 10.1016/S0092-8674(00)81875-9. PMID:9039263 [DOI] [PubMed] [Google Scholar]
- 86.Wu X, Sharp PA. Divergent transcription: A driving force for new gene origination? Cell. 2013;155:990–6. doi: 10.1016/j.cell.2013.10.048. PMID:24267885 [DOI] [PMC free article] [PubMed] [Google Scholar]


