SUMMARY
Variants of TREM2 are associated with Alzheimer’s disease (AD). To study whether increasing TREM2 gene dosage could modify the disease pathogenesis, we developed BAC transgenic mice expressing human TREM2 (BAC-TREM2) in microglia. We found that elevated TREM2 expression reduced amyloid burden in the 5xFAD mouse model. Transcriptomic profiling demonstrated that increasing TREM2 levels conferred a rescuing effect, which includes dampening the expression of multiple disease-associated microglia genes, and augmenting downregulated neuronal genes. Interestingly, 5xFAD/BAC-TREM2 mice showed further upregulation of several reactive microglial genes linked to phagocytosis and negative regulation of immune cell activation. Moreover, these mice showed enhanced process ramification and phagocytic marker expression in plaque-associated microglia, and reduced neuritic dystrophy. Finally, elevated TREM2 gene dosage led to improved memory performance in AD models. In summary, our study shows that a genomic-transgene driven increase in TREM2 expression reprograms microglia responsivity and ameliorates neuropathological and behavioral deficits in AD mouse models.
INTRODUCTION
Alzheimer’s disease (AD) is the most common cause of dementia worldwide. AD neuropathology is characterized by amyloid plaques, neurofibrillary tangles, neuronal loss, and reactive gliosis and microgliosis. Such pathology affects brain regions critical for memory and cognition including the hippocampus, cerebral cortex, and basal forebrain. The genetic etiology of AD was revealed by studies of rare early-onset familial AD (fAD) patients as well as sporadic late-onset AD patients (LOAD; Karch et al., 2014). The pathogenic fAD-causing mutations in APP, PSEN1 and PSEN2 were found to elevate the generation of pathogenic β-amyloid (Aβ) species and amyloid deposition, hence supporting a pivotal role for amyloid in AD pathogenesis (Hardy and Selkoe, 2002). By far, the most common and potent genetic risk factor for LOAD is the Apolipoprotein E ε4 allele (APOE4) (Liu et al., 2013). Recent genome-wide association studies (GWAS) of LOAD revealed more than 20 AD-associated loci (Karch et al., 2014), implicating multiple innate immunity genes expressed in microglia and peripheral myeloid cells in AD pathogenesis (Efthymiou and Goate, 2017; Gandy and Heppner, 2013).
Microglia are resident innate immune cells in the brain derived from myeloid precursors (Graeber, 2010). In the healthy brain, resting microglia have ramified processes that constantly survey the microenvironment (Nimmerjahn et al., 2005), and contribute to synaptic plasticity and learning (Parkhurst et al., 2013). In response to injury or neurodegenerative disorders, reactive microglia mediate phagocytic uptake and secretion of inflammatory cytokines (Ransohoff, 2016). It is generally believed that short-term microglia activation may promote tissue repair. However, chronic microglia activation, such as in the case of AD, may elicit neurotoxicity and contribute to disease pathogenesis.
Rare variants in the microglia-enriched gene TREM2 confer high risk for LOAD (Guerreiro et al., 2013a; Jonsson et al., 2013; Sims et al., 2017). TREM2 is a membrane protein selectively expressed in myeloid cells, including microglia (Ulrich et al., 2017; Yeh et al., 2017). TREM2 signals through its binding partner DAP12 (TYROBP) to elicit responses including phagocytosis, suppression of proinflammatory response, and promoting cell survival (Poliani et al., 2015; Takahashi et al., 2005). Complete loss of TREM2 or DAP12 leads to Nasu-Hakola disease, a recessive disorder characterized by bone cysts and early dementia (Paloneva et al., 2002), highlighting the pivotal role of TREM2 in microglia and related myeloid cells in age-dependent disease processes. Importantly, a subset of the loss-of-function TREM2 variants are found to predispose to a frontotemporal dementia-like syndrome without apparent bone involvement (Guerreiro et al., 2013b). Together, human genetics suggests that an in-depth understanding of TREM2 biology in microglia could provide insights into the pathogenesis of AD and related neurodegenerative disorders.
Recent studies have begun to unravel the functional roles of TREM2 in molecular, cellular and animal models that are informative to AD. Substantial effort has been devoted to identify TREM2 ligands. TREM2 has been shown to bind anionic and zwitterionic lipids found on damaged neurons (Wang et al., 2015) and AD-associated proteins APOE and Clusterin (Atagi et al., 2015; Bailey et al., 2015; Yeh et al., 2016). In APP mouse models, Trem2 plays a role in clustering microglia around Aβ plaques (Jay et al., 2015; Wang et al., 2015). The impact of Trem2 deficiency on amyloid plaque formation is dynamic and complex. At an early disease stage, the plaque load is reduced in an AD model crossed to Trem2 knockout mouse (Jay et al., 2015), while in more advanced disease stages, the plaque load increased in multiple AD models (Jay et al., 2017; Wang et al., 2015). Moreover, Trem2 in plaque-associated microglia plays an important role in plaque compaction and insulation to reduce the neuritic toxicity of fibrillary Aβ (Wang et al., 2016; Yuan et al., 2016). Thus far, most Trem2 studies in disease models in vivo have used loss-of-function mutants, yet very little is known about the impact of increased Trem2 expression under its genomic regulation on normal brain function and in disease responses.
In the current study, we aimed to interrogate TREM2 function in microglia at baseline and in AD disease mice through a gain-of-function genetic approach that mimics gene dosage increase in the germline, which commonly occurs during evolution (Zarrei et al., 2015). Although in vitro studies suggest that overexpression of TREM2 in microglia promotes DAP12 signaling, phagocytosis of dead neurons and suppression of pro-inflammatory responses (Takahashi et al., 2005), currently there are no in vivo studies to assess the impact of increased TREM2 gene dosage on microglial function and AD pathogenesis. To directly test the effect of upregulating TREM2 in AD mouse models in vivo, we used Bacterial Artificial Chromosome (BAC)-mediated transgenesis to insert extra copies of the human TREM2 genomic DNA segment into the mouse genome resulting in elevated TREM2 expression selectively in microglia in the brain. We found that increase in TREM2 gene dosage, in the context of AD mice, reprogramed microglia responsivity and ameliorated disease phenotypes in multiple mouse models of AD.
RESULTS
Generation of BAC-TREM2 mice to increase TREM2 gene dosage under endogenous human regulatory elements
To increase Trem2 gene dosage, we undertook a BAC transgenic approach, which can increase the expression of genes on the BAC under genomic regulation (Yang et al., 1997). We used a human TREM2 BAC for several reasons: first, human TREM2 and murine Trem2 are highly homologous (77% protein homology) and hence should have evolutionarily conserved function. Second, human TREM2 BAC is likely to preserve regulation of the human TREM2 gene expression at baseline and in disease state (Wilson et al., 2008). Finally, the human TREM2 protein may contain residues distinct from murine Trem2 that could be relevant to the future studies of disease variants (Jordan et al., 2015).
To generate human TREM2 BAC transgenic mice, we chose a BAC (RP11-237K15) that encompasses the TREM2 coding region as well as surrounding genomic regions (>50kb on each side) with conserved gene regulatory elements (Gong et al., 2003). Since this BAC contains three other TREM-like genes, TREML1, TREML2, and TREML4, which likely serve critical innate immune functions (Colonna, 2003), their overexpression may confound the interpretation of TREM2 gene dosage studies in vivo. To ensure the BAC only overexpresses TREM2, we used sequential BAC modification steps to delete key coding exons of these TREM-like genes on the BAC (Figure 1A). The properly engineered BAC was used to generate BAC TREM2 transgenic founders in the FvB/NJ inbred background. Two independent BAC TREM2 founders (A and B) gave germline transmission of their transgenes, and genomic qPCR was used to estimate the transgene copy number to be 1–2 copies (data not shown).
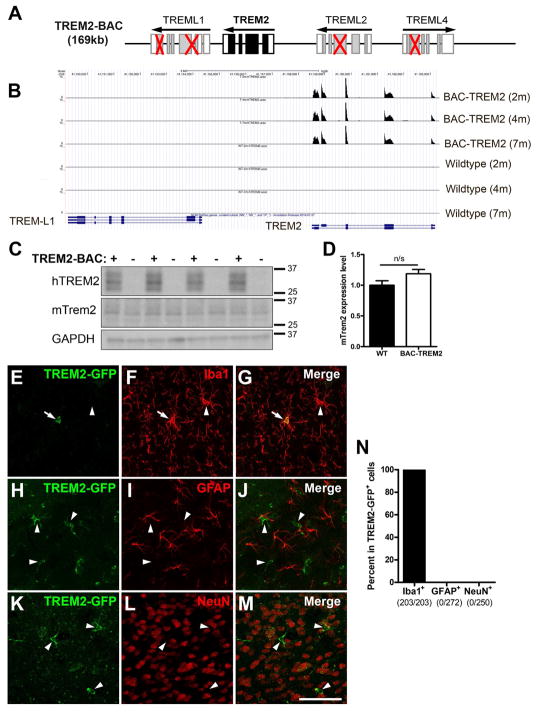
Figure 1. Generation and characterization of BAC-TREM2 mice.
(A) Schematic representation of the modification of TREM2-BAC. Red crosses indicate the deleted exons in TREM-like genes in the BAC construct.
(B) UCSC genome browser track showing read coverage at the human TREM2 locus in TREM2 transgenic and wildtype animals.
(C) Western blot was performed using the hippocampal lysates from 1.5 month-old WT and BAC-TREM2 mice with human TREM2 and mouse Trem2-specific antibodies. GAPDH served as a loading control.
(D) The band intensity of Western blots was quantified using Image J and shown as ratio of mTrem2/GAPD (n = 4).
(E–N) Brain sections from 1.5–2 month-old BAC-TREM2-GFP mice were double stained with GFP and cell-specific markers for microglia (Iba+, E–G), astrocytes (GFAP+, H–J), or neurons (NeuN+, K–M). Representative cortical images showed that BAC-TREM2-GFP colocalized with Iba1 (E–G) but not with GFAP (H–J) not NeuN (K–M). Bar = 100μm. (N) GFP+ cells were examined for the colocalization with cell-specific markers and presented as percent double labeled cells over GFP+ cells. The numbers below X-axis indicate the number of cell counted.
We next selected the BAC TREM2 line A (referred to as BAC-TREM2) to confirm the proper expression of human TREM2 RNA and protein. RNA sequencing of cortices from BAC-TREM2 and wildtype mice was used to identify unique reads of the mouse and human genomes covering murine Trem2 or human TREM2 genes (Figure 1B; see also online track links https://goo.gl/Acmcw4 and https://goo.gl/mE4b2h). The human TREM2 transcripts are only found in BAC-TREM2 mouse brains but not in wildtype brains at all ages tested. Importantly, consistent with our genetic design, we did not observe reads mapping to human TREML1, TREML2 and TREML4 among the BAC-TREM2 transcripts (Figure 1B). BAC-TREM2 mice expressed murine Trem2 transcripts at levels comparable to WT mice (Figure S1). Next, we confirmed the expression of human TREM2 protein in BAC-TREM2 mice by Western blot, and showed the expression of murine Trem2 protein is also similar between BAC-TREM2 and wildtype mice (Figure 1C).
To verify the cell-type specific expression of the TREM2 transgene, we first attempted to use available human TREM2 antibodies for immunostaining, but did not get robust signals. This might have been due to low baseline of TREM2 expression or to poor antibody specificity, as reported before (Jay et al., 2017). We thus engineered and created a BAC-TREM2-GFP reporter mouse line, which used the same TREM2 BAC as BAC-TREM2 but express TREM2 protein with a C-terminal GFP fusion. Double immunostaining of brain sections from BAC-TREM2-GFP mice demonstrated that the TREM2-GFP transgene was exclusively expressed in the Iba1+ microglia, but not in astrocytes or neurons (Figure 1E–1N). We noted that only a subset of Iba1+ cells were GFP+ (about 5.8% in the cortex and 8.7% in the hippocampus) (Figure 1E–1G; Figure S2A–S2C). This is consistent with the low levels of TREM2 expression in the homeostatic microglia in the healthy brain (Keren-Shaul et al., 2017; Krasemann et al., 2017). In summary, our novel BAC-TREM2-GFP reporter line demonstrates that the genomic regulatory elements on the human TREM2 BAC confers microglia-specific TREM2 expression in vivo.
Microglia are known to have important functions in the brain, such as synaptic pruning in the hippocampus (Stephan et al., 2012). However, we found that BAC-TREM2 mice exhibited normal hippocampal long-term potentiation (LTP) compared to wild-type controls at 10 months of age (Figure S2D). Moreover, BAC-TREM2 mice did not exhibit any detectable locomotion deficits (Figure S2E). Together, these results suggested that BAC-mediated increase in TREM2 expression does not elicit overt brain functional deficits in mice.
Increased TREM2 gene dosage reduces amyloid pathology in AD mice
To address whether increased TREM2 gene dosage could alter disease-associated microglial function and other AD-related phenotypes, we bred BAC-TREM2 to 5xFAD mice, an aggressive mouse model of AD carrying 5 familial APP and PSEN1 mutations (Oakley et al., 2006). Staining of amyloid plaques with Thioflavin S (ThioS) on cortical sections from 7-month-old 5xFAD and 5xFAD/BAC-TREM2 (termed 5xFAD/TREM2 or 5xFAD/T2) mice revealed a significant reduction of amyloid plaque load in 5xFAD/TREM2 mice (Figure 2A–2C; Figure S3). We next measured the levels of soluble and insoluble Aβ40 and Aβ42 in cortical lysates using ELISA, and found both the soluble and insoluble Aβ42 were significantly decreased in 5xFAD/TREM2 mice at 4 months of age (Figure 2D and 2E). However, the difference in Aβ levels was less pronounced and no longer statistically significant at 7 months of age. We did not detect any significant differences of Aβ40 levels, the minor Aβ species in this AD mouse model, at both ages (Figure 2F and 2G). In order to examine whether the reduction of amyloid load resulted from alteration of APP expression, we examined the human-specific APP transcript reads in our RNA-sequencing and found that the transgene was expressed at comparable levels in 5xFAD and 5xFAD/TREM2 cortices (Figure S1C). Thus, the decreased plaque burden is likely due to TREM2-mediated changes in microglia-plaque interactions.
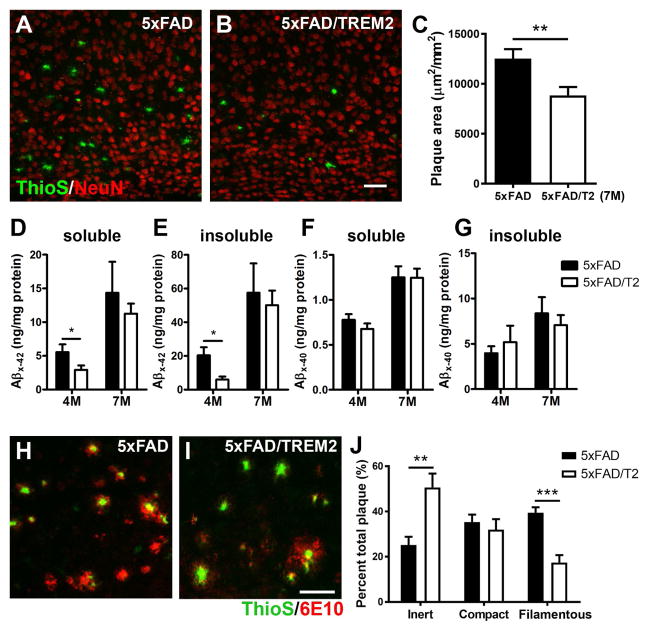
Figure 2. Increased TREM2 gene dosage ameliorates amyloid pathology and remodels amyloid plaque types.
(A–C) Matched brain sections from 7-months-old 5xFAD (A) and 5xFAD/TREM2 mice (B) were stained with ThioS (green) and NeuN (red) to visualize the amyloid plaques in the cortex. Z-stack confocal images (20X) were utilized to measure total plaque area in the field using ImageJ. The results are presented as ThioS+ plaque area (μm2) per mm2 of the cortical area (C). n = 7 per genotype, **p < 0.01. Bar = 50μm.
(D–G) The levels of soluble and insoluble Aβ42 (D and E) and Aβ40 (F and G) in the cortex of 4 and 7-month-old mice were measured by ELISA. n = 6 per genotype, *p < 0.05.
(H–J) Matched brain sections from 7-month-old 5xFAD (H) and 5xFAD/TREM2 mice (I) were stained with ThioS and anti-Aβ antibody (6E10). Z-stack confocal images (40X) were utilized to quantify 3 different forms of plaques using ImageJ (J). A total of 502 plaques were analyzed and are presented as mean ± SEM. n = 4 per genotype, **p < 0.01, ***p < 0.001. Bar = 50μm.
Recent studies have reported that Trem2 deficiency may disrupt the microglial barrier function in limiting the diffusion of fibrillary Aβ deposits (Wang et al., 2016; Yuan et al., 2016). To examine whether increased TREM2 expression alters the plaque property, we categorized and quantified distinct forms of plaques in the cortex (Yuan et al., 2016). We found that Aβ plaques in 5xFAD/TREM2 mice significantly shifted in composition towards the more inert form (strong ThioS+ with minor 6E10 staining) and less filamentous form (diffused 6E10+ staining with filamentous or missing ThioS+ labeling) compared to 5xFAD mice (Figure 2H–2J), a pattern that is opposite of that found in the Trem2-deficient mice crossed to 5xFAD (Yuan et al., 2016). Together, our study demonstrates that augmenting TREM2 expression can ameliorate Aβ pathology in 5xFAD mice.
Impact of TREM2 gene dosage increase on age-dependent transcriptional profiles in 5xFAD mice
To evaluate the impact of increasing TREM2 gene dosage on the molecular pathogenesis in 5xFAD mice, we performed transcriptional profiling of the cortical samples from WT, BAC-TREM2, 5xFAD and 5xFAD/TREM2 mice at 2, 4, and 7 months of age. We first examined the transcripts specifically mapping to murine Trem2 or human TREM2 (Figure S1A and S1B). We observed a low baseline level of murine Trem2 reads that are significantly increased in 5xFAD and 5xFAD/TREM2 cortices at an advanced (7m) but not at early disease stage (4m). Interestingly, the human TREM2 reads in 5xFAD/TREM2 mice significantly increased compared to those in BAC-TREM2 mice at 4m and 7m, but not at 2m. Together, our results suggest the BAC-TREM2 transgene drives early low level overexpression of human TREM2 in 5xFAD mouse brains, and there is a surge in the TREM2 transgene expression as disease progresses, recapitulating the disease-associated upregulation of murine Trem2 in this model.
We next performed principal component (PC) analyses including all the RNA-seq samples at the three ages (Figure S4A–S4C). We observed a more defined separation based on 5xFAD genotypes at 4m and 7m only, but did not detect a clear separation based on BAC-TREM2 genotype. These findings suggest the impact of the transgene on gene expression in WT or 5xFAD background is not robust enough to be detectable with this analysis.
We then examined genes with significant differential expression (DE) in the cortex among different genotypes across all three ages (Table S1). As shown in Figure 3A, we found that very few genes were significantly differentially expressed (FDR < 0.1) between BAC-TREM2 and WT at all ages, suggesting that the BAC-TREM2 transgene does not elicit a detectable molecular effect in the normal mouse brains. In 5xFAD mice, we observed over 1000 DE genes at 4m and 7m but not at 2m, consistent with the progression of cortical pathology in this aggressive amyloidosis model of AD (Oakley et al., 2006). Interestingly, 5xFAD/TREM2 mice had only 161 DE genes at 4m, and 916 genes at 7m compared to WT, both of which are fewer than those in 5xFAD littermates. Finally, 5xFAD/TREM2 mice had 44 and 54 DE genes when compared to 5xFAD mice at 4 and 7 months of age respectively.
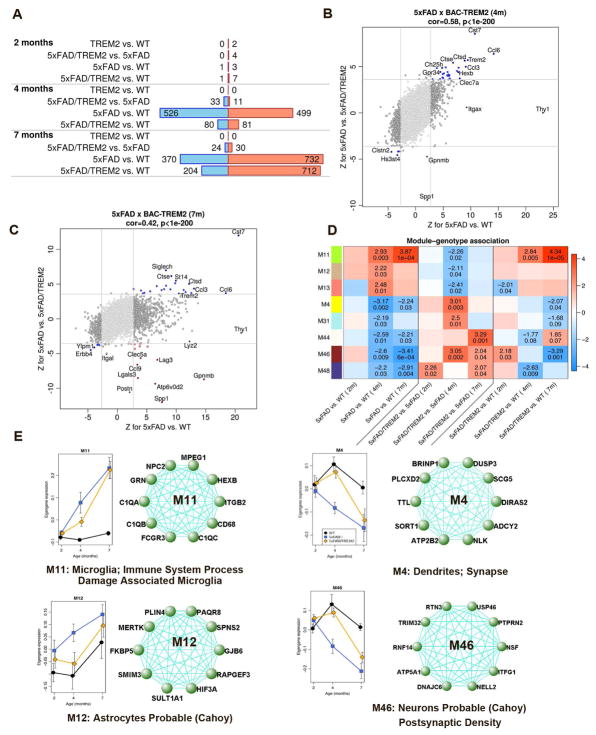
Figure 3. Transcriptomic and coexpression network analyses reveal partial rescue of transcriptomic changes in 5xFAD mice with increased TREM2 gene dosage.
(A) Numbers of DE genes (FDR < 0.1) in genotype contrasts. Blue/red bars represent significantly down–/up-regulated genes. n=6 per genotype and time point except n=5 for 5xFAD at 7 m.
(B–C) Transcriptome-wide “rescue plots” at 4 and 7 months. The plots show Z statistics for DE in 5xFAD vs. 5xFAD/TREM2 (y-axis) and 5xFAD vs. WT (x-axis) for all genes (each gene corresponds to one point). Rescued (concordant in this plot) and exacerbated (discordant) genes that pass the FDR threshold of 0.1 in both comparisons are shown in blue and red colors, respectively. Genome-wide correlations of Z statistics and the corresponding correlation p-values (for which the n=15809 genes are considered independent) are indicated on the top of each panel.
(D) DE analysis of module eigengenes. Rows correspond to modules and columns to selected genotype contrasts. Numbers in the heatmap show the Z statistics and the corresponding p-values of module eigengene association with the genotype. n=6 per genotype and time point except n=5 for 5xFAD at 7m.
(E) Expression variation of module eigengene expression with age in WT (black), 5xFAD (blue) and 5xFAD/TREM2 (Orange) samples. Points represent means of eigengene values across samples at the same age. Error bars give SEM. Network of top 10 hub genes are presented on the right.
We performed enrichment analyses of DE genes (Table S2) using both public gene sets (e.g. GO and MSigDB) and an internal gene set collection (Langfelder et al., 2016). Consistent with the disease process in 5xFAD mice and AD patients, the top enrichment terms for down-regulated genes in 5xFAD vs WT were “neurons” (p = 7.07E-55) and “synapses” (p = 6.95E-19) at both 4 and 7 months, and the top enrichment for upregulated genes in 5xFAD mice were “top microglia genes” (p = 3.19E-194) and “immune system process” (p = 9.51E-83). The few DE genes between 5xFAD/TREM2 and 5xFAD were highly enriched in microglia and immune cell activation annotations (Galatro et al., 2017).
Prior studies of APP transgenic models including 5xFAD showed earlier and more severe disease-related phenotypes such as amyloid plaque pathology in female compared to male mice (Oakley et al., 2006; Sadleir et al., 2015), which is reminiscent of sex differences in AD (Mazure and Swendsen, 2016). However, sex differences in the transcriptomic effects in 5xFAD mouse brains have not been reported. In our RNA-seq study, notwithstanding the limited sample size (N=3 per sex/genotype/age group; except N=2 for 5xFAD females at 7m), we observed more DE genes in female than in male 5xFAD mice at both 4m and 7m (Figure S4D–S4I; Table S3). Similarly, we detect more DE genes comparing 5xFAD/TREM2 to 5xFAD in female than male transgenic mice at these ages. These findings are consistent with the earlier and more severe cortical pathology in female compared to male 5xFAD mice (Oakley et al., 2006; Sadleir et al., 2015). We examined the correlation between DE statistics observed in male vs. female animals when comparing 1) 5xFAD vs WT; and 2) 5xFAD/TREM2 vs 5xFAD animals, at both 4m and 7m (Figure S5). This correlation between fold changes is positive and highly significant in all cases, supporting the notion that the two sexes behave similarly in all comparisons. Hence, although female 5xFAD mice have more significant DE genes when compared to WT or 5xFAD/TREM2, the transcriptional changes in the male mice are trending in the same direction. This hypothesis is further supported by formal statistical testing for interactions of genotype and sex, which showed only few sex-dependent, significant DE genes across genotypes (Figure S4D–S4F). Thus, we conclude that transcriptomic changes occur earlier and more robustly in female than male 5xFAD mice, and that the effect of TREM2 increased gene dosage is also more apparent in female than male 5xFAD mice. However, because of the strong correlation of fold changes between male and female mice in all comparisons, and the lack of strong statistical genotype/sex interaction effects, we reasoned that a combined analysis of both sexes is a reasonable approach to identify the most consistent DE genes in 5xFAD, and those that are modulated by BAC-TREM2 in 5xFAD.
Partial rescue of transcriptional dysregulation by increasing TREM2 gene dosage in 5xFAD/TREM2 mice
We next asked whether increased TREM2 gene dosage in 5xFAD/TREM2 mice leads to overall rescuing or exacerbating effects on the transcriptional dysregulation observed in 5xFAD mice (compared to WT). We compared the Z statistics for 5xFAD vs. 5xFAD/TREM2 against those for 5xFAD vs. WT in a transcriptome-wide “rescue/exacerbation plot” (Figure 3B–3C). A positive correlation can be interpreted as an overall transcriptome-wide “rescue effect”, and conversely, a negative correlation would indicate an exacerbation effect. In both 4m and 7m mice, the plots show a highly significant positive correlation (Figure 3B–3C), demonstrating an overall transcriptome-wide rescuing effect in 5xFAD/TREM2 mice.
To further identify molecular networks that may be selectively targeted by increased TREM2 gene-dosage in 5xFAD/TREM2 mouse brains, we performed consensus Weighted Gene Co-expression Network Analyses (WGCNA; Langfelder and Horvath, 2008). The WGCNA approach has been used previously to identify coherent gene modules dysregulated in AD mouse and patient brains (Matarin et al., 2015; Miller et al., 2008). Our consensus WGCNA analysis identified 28 co-expression modules (Table S4; Figure S6). By relating the eigengene (a single representative expression profile of a module, Horvath and Dong, 2008) to genotypes, we identified five modules (M11, M12, M13, M28 and M30) significantly (p < 0.05) upregulated in 5xFAD vs. WT mice at 4 or 7m, and 9 modules (M4, M15, M24, M31, M33, M37, M44, M46 and M48) down-regulated in 5xFAD mice. Importantly, three of the five upregulated 5xFAD modules (M11, M12 and M13) and five of the nine downregulated AD modules (M4, M31, M44, M46 and M48) are partially rescued in 5xFAD/BAC-TREM2 mice (Figure 3D).
Enrichment analyses of these network modules (Table S5) revealed that two of the three modules that were upregulated in 5xFAD and partially rescued in 5xFAD/TREM2 were annotated for microglia-enriched (M11) and astrocyte-enriched genes (M12). M11 is a large module (1232 genes) enriched with terms such as “immune system processes” and “damage-associated microglia” (Keren-Shaul et al., 2017) (Figure 3E; Table S5). Top hub genes in M11 module are known to be involved in microglial function (e.g. Grn and C1q). The M11 eigengene was progressively upregulated in 5xFAD compared to WT mice. This elevation was significantly reduced at 4m in the 5xFAD/TREM2 mice (Figure 3D–3E). M12, a smaller module (119 genes), was enriched with astrocyte genes. The upregulation of genes in M12 was also diminished in 5xFAD/TREM2 compared to 5xFAD mice (Figure 3D–3E). One of the M12 hub genes is Mertk, a phagocytic receptor involving in astrocyte and microglia mediated phagocytosis of synapses and neurons (Brown and Neher, 2014; Chung et al., 2015).
Among the five modules that have decreased expression in the 5xFAD mice and are partially rescued in 5xFAD/TREM2 mice, two are particularly interesting. M4 is significantly enriched in terms including “dendrite” and “synaptic genes” (e.g. Atp2b2, Grik2, Grin1, Mapt), and M46 (347 genes) is enriched in “neuronal genes” and “genes downregulated in the hippocampi of AD patients” (Figure 3D and 3E; Table S4). Increased TREM2 gene dosage in 5xFAD/TREM2 mice significantly improved these modules at 4m, and for M46 such effect appeared to be extend to 7m (Figure 3E). Thus, increased TREM2 expression in microglia may exert non-cell-autonomous effects to partially restore neuronal gene expression in the AD mice.
Reprogramming disease-associated microglia gene expression signatures in 5xFAD mice by TREM2 gene dosage increase
Systems biology has played a powerful role in the unbiased discovery of microglia function and dysfunction in the brain (Galatro et al., 2017; Gosselin et al., 2017; Zhang et al., 2013). In 5xFAD mice, Trem2 deficiency greatly impaired the overall transcriptional response of reactive microglia (Wang et al., 2015). Two recent studies examined microglia molecular signatures that are associated with disease progression in mouse models of neurodegenerative disorders, including AD and ALS (Keren-Shaul et al., 2017; Krasemann et al., 2017). These studies identified overlapping microglial gene sets termed damage-associated microglia (DAM) genes (Keren-Shaul et al., 2017) or molecular signatures of disease-associated microglia (MGnD) (Krasemann et al., 2017). Importantly, both studies showed a critical set of microglial genes that are dependent on Trem2 for their disease-associated upregulation. In this study, we asked what would be the effects of increased TREM2 gene dosage on these known disease-associated microglial molecular signatures in the AD mice.
Based on the transcriptome-wide rescue/exacerbation plots (Figure 3B and 3C), we defined three sub-groups of DE genes between 5xFAD/TREM2 and 5xFAD (termed TD1-TD3, for TREM2 Dosage-dependent genes; Figure 4A). We first selected DE genes between 5xFAD/TREM2 and 5xFAD (FDR < 0.1) at either 4m or 7m. Among these genes, TD1 genes are those with expression levels significantly upregulated in 5xFAD compared to WT (Z > 2), but are significantly downregulated in 5xFAD/TREM2 compared to 5xFAD (Z < −3) compared to 5xFAD. The TD1 genes are those located in the upper right quadrant of the plots (Figure 3B and 3C). TD2 genes are those significantly upregulated in 5xFAD mice vs WT (Z > 2), and further upregulated in 5xFAD/TREM2 vs 5xFAD (Z > 3). These are genes located in the lower right quadrant of the plots (Figure 3B and 3C). Finally, TD3 genes are in the lower left quadrant of the rescue/exacerbation plots, and are significantly downregulated in 5xFAD vs WT (Z < −2), but were significantly upregulated in 5xFAD/TREM2 vs 5xFAD (Z > 3). We did not observe any transcripts that are downregulated in 5xFAD mice and are further decreased in 5xFAD/TREM2 mice. Importantly, we are able to apply real-time reverse transcription PCR to validate a critical subset of TD1-TD3 genes for their appropriate DE profiles in 5xFAD and 5xFAD/TREM2 cortices (Figure S7).
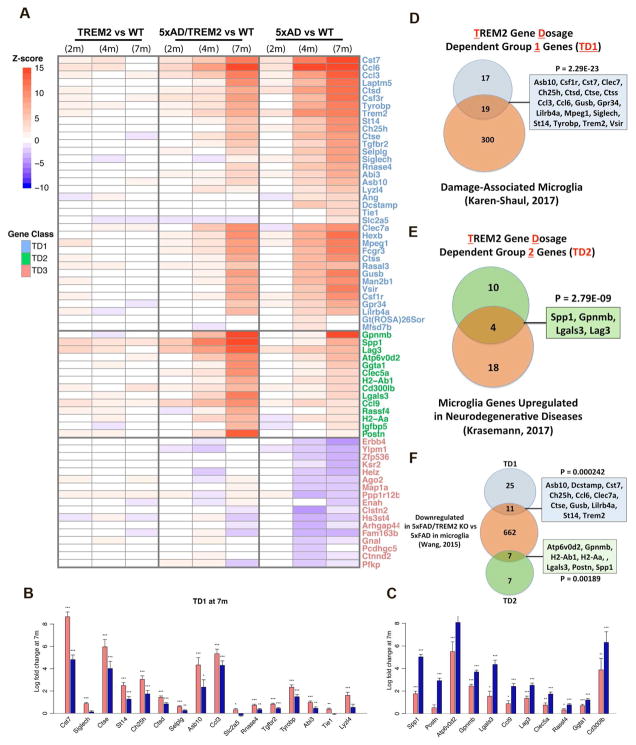
Figure 4. Increased TREM2 gene dosage reprogrammed disease-associated microglia gene expression.
(A) Heatmap representation of differential expression Z statistics for TD1-3 genes. Genes are divided into TD1 (blue), TD2 (green) and TD3 (red).
(B–C) Fold changes of individual TD1 (B) and TD2 (C) genes in 5xFAD (red) and 5xFAD/TREM2 (blue) vs. wildtype mice at 7m. Stars indicate FDR-corrected significance (***FDR<0.001, **FDR<0.01, *FDR<0.1). n=6 per genotype, except n=5 for 5xFAD.
(D–F) Venn diagrams of overlaps of TD1 (blue) and TD2 (green) genes with published genesets (orange).
We next performed enrichment analyses of TD1-3 (Table S6). The top enrichment terms for TD1 genes are “Upregulated in damage associated microglia” (p = 2.29E-23; Keren-Shaul et al., 2017), “top human microglia-specific genes” (p = 2.15E-16; Galatro et al., 2017), “Autophagy lysosome” (p = 4.90E-09), “genes upregulated with age in hippocampus” (p = 2.89E-08), and “immune system process” (p = 0.000142). These annotations suggest TD1 genes are key microglial genes normally involved in microglia activation in diseased brain, and are significantly reversed with increased TREM2 gene dosage. Despite the relatively small number of genes in TD1 group, they were among the top upregulated, disease-associated microglia genes in 5xFAD mice (based on Z statistics; Figure 3B–3C and Figure 4D). The 19 DAM genes (Keren-Shaul et al., 2017; Figure 4D) are partially, but significantly, rescued in 5xFAD/TREM2 mice include Cst7 (the top upregulated microglial gene in 5xFAD brain), several cathepsins (Ctsd, Ctse, Ctss), chemokines (Ccl3, Ccl6) and their receptors (Csf1r), and established AD-associated genes (Trem2 and Abi3) (Sims et al., 2017).
The TD2 genes constitute possibly the most interesting group, despite its relatively small number (14 genes). The top enrichment term in this group is “Microglia genes upregulated in neurodegenerative diseases” (p = 2.79E-09; Krasemann et al., 2017). Four genes belonging to this group (Spp1, Gpnmb, Lgals3 and Lag3) were significantly further upregulated in 5xFAD/TREM2 vs 5xFAD mice at 4m and 7m. Importantly, the other top enrichment terms for TD2 genes were GO terms such as “Negative regulation of T cell activation” (p = 4.96E-09) and “Innate immune response” (p = 8.87E-07). TD2 genes were well characterized for regulation of phagocytosis and microglial activation (e.g. Lgasl3; Rotshenker, 2009), microglial survival (e.g. Spp1; Rabenstein et al., 2016), alternative M2 activation of microglia (Zhou et al., 2015), and lysosomal proton pump (Atp6v0d2). These analyses suggest increasing TREM2 gene dosage selectively upregulates an interesting subset of “disease-associated microglial genes” to promote certain aspects of their function, such as phagocytosis and suppression of over-activation of the innate immune response in the AD mouse brain.
We next asked what genes altered in 5xFAD/TREM2 mice overlap with those altered in 5xFAD/Trem2−/− (Wang et al., 2015). We found a total of 11 TD1 and 7 TD2 genes overlapping with the down-regulated genes in 5xFAD/Trem2−/− microglia (Figure 4F). Of these genes, the seven TD2 genes are potentially interesting, as they were down-regulated in 5xFAD mice in the absence of Trem2 but upregulated in 5xFAD mice with increased TREM2 gene dosage. They may constitute transcriptional targets of Trem2 gene dosage-dependent signaling in the context of disease-associated microglia.
Finally, the TD3 group is only modestly enriched for postsynaptic density genes (Table S6), consistent with the results of network analyses showing that a subset of neuronal and synaptic genes downregulated in 5xFAD mice were partially normalized with TREM2 overexpression.
Increased TREM2 gene dosage altered microglial interaction with amyloid plaques
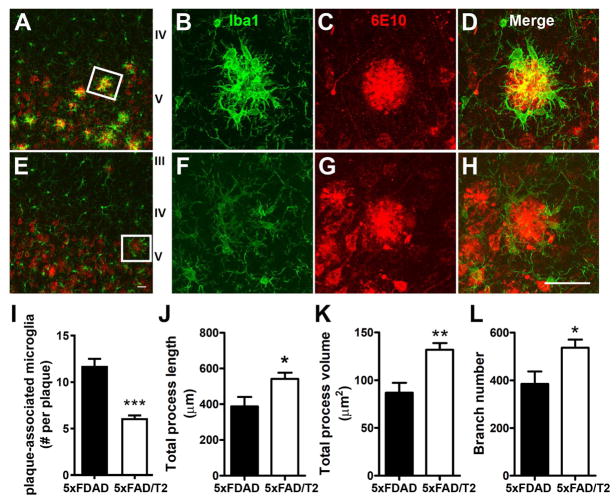
A primary feature of reactive microgliosis is morphological transformation (Stence et al., 2001). In 5xFAD mice, plaque-associated microglia exhibited shortened and thickened processes with hypertrophic amoeboid shape, representing canonical reactive microglia (Figure 5A–5D). However, these phenotypes were significantly attenuated in 5xFAD/TREM2 mice (Figure 5E–5H). We first quantified the number of plaque-associated microglia and found a significant reduction in 5xFAD/TREM2 mice compared to 5xFAD mice (Figure 5I). Unlike the Trem2-deficient microglia in AD mice (Jay et al., 2015; Wang et al., 2015), the plaque-associated microglia in 5xFAD/TREM2 mice show more elongated and ramified processes than those in the 5xAD mice, as quantified by microglial branch length, total processes volume, and branch number (Figure 5J–5L). We interpret these findings as evidence for reprogrammed microglial response in 5xFAD/BAC-TREM2 mice, so that fewer and less activated microglia per plaque are still able to function effectively in limiting the size and diffusion of the amyloid plaque in 5xFAD cortices.
Figure 5. Upregulation of TREM2 altered microglial response to the amyloid plaque.
(A–H) Representative images demonstrated the interaction between microglia (Iba1+, green) and the plaque (6E10+, red) in 7 months old 5xFAD (A–D) and 5xFAD/TREM2 mice (E–H). Bar = 25 μm.
(I) Amyloid plaques in cortex were randomly selected and Z-stack confocal images were taken for counting plaque-associated microglia (Iba1+). n = 4 per genotype. ***p < 0.001.
(J–L) The morphological properties of plaque-associated microglia were measured by Imaris using z-stack confocal images taken under a 63X objective lens. The results are presented as total process length (J), process volume (K) and branch numbers (L) per microglia. Images of more than 15 plaques with over 150 microglia per genotypes were analyzed and presented as mean ± SEM. n = 4 per genotypes, **p < 0.01, *p < 0.05.
Increased TREM2 gene dosage enhanced phagocytic microglia markers in vivo and phagocytic activity in vitro
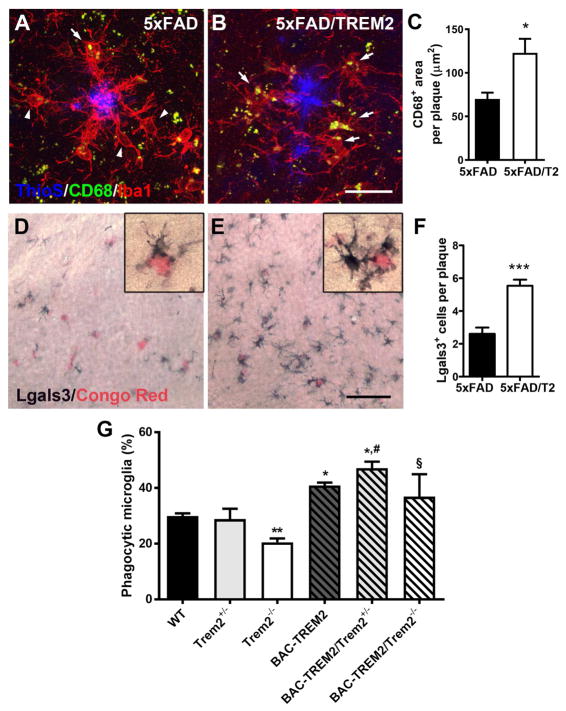
Based on the reduced amyloid burden and the upregulation of several phagocytosis-related TD2 genes in the 5xFAD/TREM2 mice, we hypothesize that phagocytic activity of microglia might be enhanced by increased TREM2 gene dosage in AD mouse brains. We first assessed CD68 protein expression, a marker for phagocytosis shown to be upregulated in plaque-associated microglia in AD brains (Yuan et al., 2016). We found a significant increase of anti-CD68 staining in the plaque-associated microglia in 5xFAD/TREM2 cortices compared to those in 5xFAD cortices at 7m of age (Figure 6A–6C and Figure S8). This result suggests that a post-transcriptional mechanism may be involved in the upregulation of CD68 in 5xFAD/TREM2 microglia surrounding the plaque, as transcript amounts are comparable between 5xFAD and 5xFAD/TREM2 at this age (Table S1). We further assessed the level of a second microglial phagocytosis related protein, Lgals3 (Rotshenker, 2009), which is one of the TD2 genes. We detected very little staining for Lgals3 in wildtype mouse brains at this age (data not shown). In the 5xFAD/TREM2 mice, we observed a significant increase in the number of Lgals3+ microglia surrounding the amyloid plaque in 5xFAD/TREM2 brains compared to the 5xFAD brains (Figure 6F, 6D), hence supporting the concept that upregulation of phagocytic markers is part of the reprogrammed microglia response in 5xFAD/TREM2 mice.
Figure 6. Increased TREM2 gene dosage upregulated expression of phagocytic markers and enhanced phagocytic activity in microglia.
(A–C) Matching cortical sections from 7 month-old 5xFAD (A) and 5xFAD/BAC-TREM2 mice (B) were stained with CD68 (green), Iba1 (red) and ThioS (blue). (C) Area of CD68+ labeling per plaque was measured on z-stack confocal images. n = 4 per genotypes, *p < 0.05. Bar = 25μm.
(D–F) Matching cortical sections from 7 month-old 5xFAD (D) and 5xFAD/BAC-TREM2 mice (E) stained with Lgals3 (dark blue) and Congo Red (pink). (F) Number of Lgal3+ cells per plaque were counted under microscope by a blinded observer. n = 3 per genotypes, ***p < 0.001. Bar = 50μm.
(G) Phagocytosis of Alexa488-conjugated microsphere by primary microglia were measured by flow cytometry. Phagocytic microglia were detected with strong fluorescent signal in the cells. The graph is pooled results from 4 independent experiments and presented as mean ± SEM. n = 3–6 per genotypes; **p < 0.01, *p < 0.05 compared to WT; #p < 0.05 compared to Trem2+/−; §p < 0.05 compared to Trem2−/−.
To functionally evaluate the phagocytic capacity of microglia in our models, at least in vitro, we performed a phagocytosis assay with primary microglia isolated from neonatal mice. Such in vitro assay provided a model to evaluate the stress-induced phagocytic activity in the microglia (Gosselin et al., 2017). Polystyrene microbeads were used to assess the general phagocytic activity, and also to ensure phagocytosis rather than pinocytosis was being measured. The results showed a significant enhancement of phagocytic activity in BAC-TREM2 microglia compared to WT microglia, while Trem2−/− microglia showed a significant decrease in phagocytic activity (Figure 6G). Importantly, such a deficit in Trem2-deficient microglia can be rescued by the BAC-TREM2 transgene. Our results suggest that Trem2 level is a critical rate-limiting factor in regulating the phagocytic activities of microglia in vitro. Furthermore, the rescue assay showed that Trem2 function in regulating microglia phagocytosis is preserved and elevated in the BAC-TREM2 transgene. In summary, our studies showed that enhanced phagocytic activity could be another key component of reprogrammed microglial function by BAC-TREM2 in AD mouse brains.
Upregulation of TREM2 in microglia reduced plaque-associated neurite dystrophy and ameliorates cognitive deficit
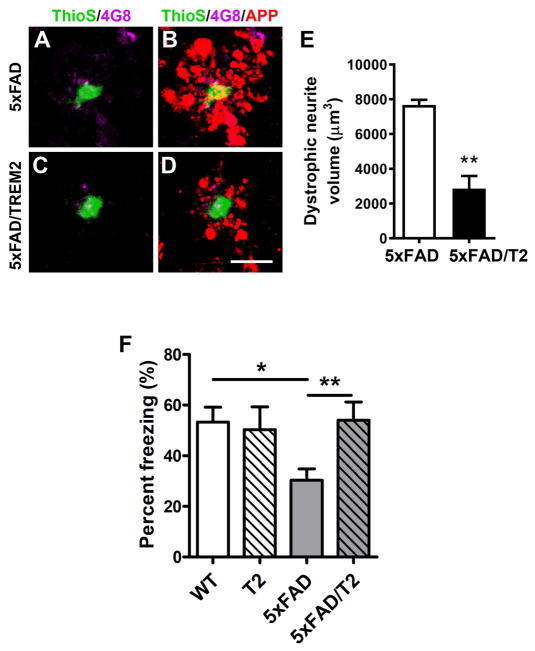
Previous studies reported the presence of neurite dystrophy in close association with Aβ deposits in AD patients and mouse models (Masliah et al., 1996; Nixon, 2007), and this phenotype is exacerbated with AD risk-associated TREM2 variants (Wang et al., 2016; Yuan et al., 2016). Our transcriptome analyses suggest that the down-regulated neuronal genes may partially be recovered in the 5xFAD/TREM2 mice. To directly assess the plaque-associated neurite dystrophy, we performed immunostaining with antibody against the N-terminus of APP, which accumulates in damaged neurites. Importantly, our RNA-seq analyses did not show any difference in the expression of APP transgene and endogenous murine App between 5xFAD and 5xFAD/TREM2 mice (Figure S1C–S1D). Our analyses showed a significant reduction of dystrophic neurites in 5xFAD/TREM2 mice (Figure 7A–7E), which provides further evidence that increased TREM2 expression in microglia confers a non-cell-autonomous, neuroprotective effect in vivo.
Figure 7. Increase TREM2 gene dosage in microglia reduced plaque-associated neuritic pathology and ameliorate cognitive deficit.
(A–D) Representative images of APP-immunolabeled dystrophic neurites (red) surrounding the plaques co-labelled with 4G8 (purple) and thioflavin S (green) in the cortex of 5xFAD and 5xFAD/TREM2 mice. Bar = 25μm.
(E) Total dystrophy neurites volume was quantified in the cortex of 5xFAD and 5xFAD/TREM2 mice. n = 3 per genotype. Data presented as mean ± SEM, **p < 0.01. (F) The contextual memory function of mice from the cohort of BAC-TREM2 × 5xFAD (n = 8–14 per genotype) was evaluated by contextual fear conditioning and is presented as percent time freezing. Power analysis was performed to ensure >80% confidence levels with the number of animals used. **p < 0.01, *p < 0.05.
To evaluate whether the neuroprotective phenotypes observed in 5xFAD/TREM2 mice may correspond to a behavioral improvement, we performed the contextual fear-conditioning test, a hippocampus-dependent memory task that is compromised in 5xFAD mice (Kimura and Ohno, 2009). Impressively, unlike the 5xFAD mice that exhibited a robust deficit in this task, the performance of 5xFAD/TREM2 mice is comparable to that of WT controls (Figure 7F). Moreover, BAC-TREM2 mice was not significantly different from wildtype mice, suggesting the BAC-TREM2 transgene alone does not affect this memory task. Thus, we conclude that the BAC-TREM2 transgene is improving both the neuritic pathology and cognitive performance in an AD mouse model.
Increased TREM2 gene dosage alters plaque-associated microglia morphology and ameliorates behavioral deficit in a second mouse model of AD
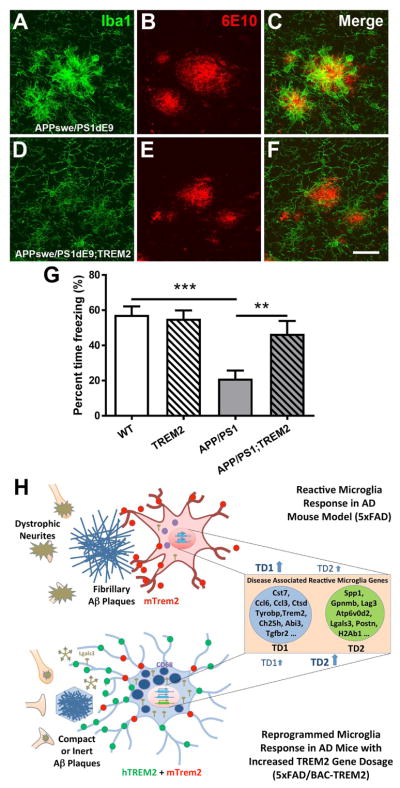
To validate and extend some of the findings in 5xFAD/TREM2 in another AD mouse model, we crossed BAC-TREM2 mice with APPswe/PS1dE9 mice (APP/PS1), another commonly used mouse model of AD (Jankowsky et al., 2004). This model has slower onset of amyloid pathology that affects hippocampus as well as the cortex. At 11 months of age, we observed a marked alteration of reactive microglia morphology surrounding the plaques (i.e. more elongated processes) in the hippocampus of the APP/PS1;BAC-TREM2 (APP/PS1;TREM2) mice (Figure 8A–8F). There is also marked reduction of Iba1 immunostaining signals in the double mice compared to APP/PS1 mice, suggesting altered microglial reactivity (Figure 8A–8F, Figure S9).
Figure 8. Increased TREM2 gene dosage alters plaque-associated microglia morphology and ameliorates behavioral deficit in a second mouse model of AD.
(A–F) Representative confocal images from 11 month-old APP/PS1 (A–C) and APP/PS1;TREM2 (D–F) mice stained with anti-Iba1 (green) and 6E10 (red) antibodies. Bar = 25μm.
(G) The contextual memory function was evaluated by contextual fear conditioning and is presented as percent time freezing. n = 14–18 per genotype, ***p < 0.001, **p < 0.01.
Similar to 5xFAD mice, APP/PS1 mice have been reported to show deficits in contextual fear conditioning (Knafo et al., 2009). Here we observed that at 11 months of age, APP/PS1 mice exhibited a robust contextual fear conditioning deficit compared to wildtype mice, and this phenotype was significantly abolished in the APP/PS1;TREM2 mice (Figure 8G). The results suggest that microglial expression of the BAC-TREM2 transgene reprograms microglial responsivity and ameliorates the disease-associated behavioral impairment in a second AD mouse model.
Discussion
Our study provides a rigorous and detailed analysis of the phenotypic impact of increased TREM2 gene dosage, through BAC-mediated transgenesis, on a variety of phenotypes in two AD mouse models. We showed that TREM2 BAC can selectively drive reporter transgene expression in microglia in the mouse brain, and elevated human TREM2 RNA and protein expression. In the context of AD mice, such elevated TREM2 expression led to reduction of the amyloid plaque load, and a shift of plaque composition from the fibrillary towards more compact and inert types. Brain transcriptomic and network analyses reveal partial rescuing effects at the transcriptome-wide level in 5xFAD/TREM2 mice. Detailed examination of known disease-associated microglial genes revealed an interesting reprogramming of the microglial transcriptomic response in the 5xFAD/TREM2 mouse brains compared to that in 5xFAD; there is a selective downregulation of a subset of reactive microglial genes (i.e. TD1) and upregulation of a second subset (TD2). Such molecular reprogramming of microglia in the diseased brain is supported by evidence of the number, morphology, and phagocytic marker expression in the plaque-associated microglia in the 5xFAD/TREM2 mice. Functional studies in vitro showed TREM2 gene dosage increase augmented microglia phagocytic activity, a phenotype opposite to that of Trem2 deficiency. We also showed evidence for reduced neuritic pathology and improved memory task in the AD models with elevated TREM2 gene dosage. Together, our study reveals that elevated TREM2 gene dosage can mediate microglia reprogramming, reduced neuropathology, and improved cognitive performance in AD mouse models.
A key strength of our study is the mouse genetic construct design, which allows us to precisely address the increase of TREM2 gene dosage on microglial function and disease-related phenotypes in vivo. BAC transgenes are known to drive more accurate, endogenous-like transgene expression (Gong et al., 2002b; Yang et al., 1997), and are suitable for studying the effects of gene dosage increase in intact animals (Yang et al., 1999). The use of human BAC transgene allows us to study TREM2 function in the intact human TREM2 genomic DNA, RNA and protein context, which could be relevant to investigating disease variants in vivo (Jordan et al., 2015). We obtained strong evidence that our BAC transgene is properly expressed in microglia cells using our novel BAC-TREM2-GFP reporter line, and showed the TREM2 is functional in complementing the Trem2 deficiency in the in vitro phagocytosis assay (Figure 6G). An important aspect of our BAC transgene design is the deletion of essential exons in three other TREM-like genes on the BAC, which are known to have important innate immunity function (Colonna, 2003; Ford and McVicar, 2009), and may play distinct roles in AD (Carrasquillo et al., 2017). Thus, in order to draw strong conclusions on the role of TREM2 gene dosage increase on the microglial and disease phenotypes in AD mouse models, one would have to genetically abolish the expression of TREM-like molecules from the BAC transgene, a strategy we have implemented in our BAC-TREM2 mice.
Another important question addressed in this study is whether increased TREM2 gene dosage, hence its expression levels under genomic regulation, is beneficial or harmful in the disease process. Trem2 deficiency induces dynamic changes but an eventual increase in plaque and neuritic pathology (Jay et al., 2017; Wang et al., 2015; Wang et al., 2016; Yuan et al., 2016). In addition to amyloid-induced pathology, two recent studies also demonstrated that Trem2-deficiency altered reactive microgliosis and proinflammantory responses of microglia in two tauopathy mouse models (Bemiller et al., 2017; Leyns et al., 2017), which are reminiscent of those observed in the APP models (Jay et al., 2017; Jay et al., 2015; Wang et al., 2015). However, its impact on tau-induced pathology remains discrepant in these models. Since deletion of Trem2 in mice blocks the proper transcriptional activation of microglia in neurodegenerative diseases (Keren-Shaul et al., 2017; Krasemann et al., 2017; Wang et al., 2015), it is difficult to predict a priori whether TREM2 gene dosage increase could broadly or subtly alter the microglial transcriptome response, and what impact it would exert on the disease process. Our study provides strong evidence for a partial, but significant, rescuing effect of TREM2 gene dosage increase on the transcriptomic phenotypes, amyloid pathology, and behavioral impairment in AD mouse models. Our transcriptome-wide rescue/exacerbation analyses and network analyses show unbiased evidence for partial normalization of the disease-associated gene expression in the 5xFAD/TREM2 mice, with reduction of expression of microglial and astrocyte module genes and upregulation of neuronal and synaptic genes. The latter notion is supported by the reduction of neuritic pathology (Figure 7) and enhancement of fear conditioning (Figure 7G and 8G) in the AD mouse models. We are cognizant of the fact that the mouse models used in the study are only partial models of AD, but they are useful to show the in vivo neuronal and microglial responses to the amyloid pathology. Thus, future studies with additional AD-relevant mouse models (e.g. Tau transgenic mice) and mouse models of other neurodegenerative disorders will allow further evaluation of the impact of TREM2 gene dosage increase on disease pathogenesis in vivo.
Our study shed some light on the kinetics of TREM2 level increase that could be effective in exerting beneficial effects in AD mice. Prior studies (e.g. Keren-Shaul et al., 2017; Krasemann et al., 2017) and our current one (Figure S1A–S1B) showed elevated Trem2 expression occurs at relatively late disease stages. Moreover, an increase in secreted extracellular fragment of TREM2 appears to be a CSF biomarker of disease progression in AD (Suárez-Calvet et al., 2016). These findings raise the question of whether elevated TREM2 during disease progression in AD is beneficial in modulating disease progression. Our study showed that BAC-TREM2 mice have elevated TREM2 levels as early as 2m of age (Figure 1B, 1C and S1). Thus, we favor a testable model that upregulation of TREM2 levels early in the AD mouse brains, prior to the upregulation of hundreds of other reactive microglial genes, may be most effective in reprogramming the microglial responsivity to ameliorate the disease.
A crucial question raised by our study is through what mechanism increased TREM2 gene dosage reprograms microglial responsivity, and what aspects of the reprogrammed microglial function is crucial to the beneficial effects observed in the BAC-TREM2 expressing AD mice. One important next question to address is whether known TREM2 downstream signaling molecules (e.g. DAP12 and SYK) are necessary in TREM2 gene- dosage dependent changes in microglial function in the AD models (Ulrich et al., 2017). Moreover, it would be interesting to explore whether other AD-associated genes, especially those showing protective effects and with myeloid cell enriched expression (Huang et al., 2017; Sims et al., 2017), could function in the same pathway as TREM2 in the diseased brain. While these questions remain unresolved, our study sheds several tantalizing clues to the TREM2-mediated microglial reprogramming. For example, we showed several dichotomous cellular and molecular phenotypes between BAC-TREM2 and Trem2-deficient microglia in mouse models of AD (Jay et al., 2017; Ulrich et al., 2017; Yuan et al., 2016). These include the compaction of amyloid plaques, the ramification of plaque-associated microglial processes, the expression of certain activated microglial markers, and phagocytosis. These microglial phenotypes may help to pinpoint the molecular function that is rate-limited by the levels of Trem2 across a broad dynamic range, hence could be more proximal to the direct molecular function of TREM2 in microglia in response to the brain disease environment.
Our molecular analyses provide tantalizing candidates that may underlie the phenotypic changes in microglia response and overall disease phenotypes in the AD mice crossed to BAC-TREM2. Although we cannot currently rule out that both down-regulated microglial genes (TD1) and upregulated microglial genes (TD2) could both play a role in the TREM2 gene dosage effect in the AD mice, we favor the latter group of genes as prime candidates to investigate first. A subset of TD2 genes show transcriptional changes that are bidirectional in Trem2 knockout and our BAC-TREM2 mice crossed to 5xFAD (Figure 4F), which could be transcriptional targets that are positively regulated by TREM2 levels (or signaling), a hypothesis that can be tested. Moreover, multiple TD2 genes have known functions that are consistent with Trem2 effects in myeloid cells. Lgals3 is a critical regulator of microglia activation (Rotshenker, 2009), and it facilitates phagocytosis (Sano et al., 2003). Interestingly, Lgals3 is shown to be an “eat-me” signal linking phagocytic receptor Mertk to its cargo (Caberoy et al., 2012). Spp1 regulates cytokine expression and promotes microglia survival (Rabenstein et al., 2016), a function similar to that of Trem2 (Yeh et al., 2017). Periostin (Postn) is shown to be required for alternative (less inflammatory) activation of microglia in brain tumors (Zhou et al., 2015). The latter finding is consistent with the enrichment in TD2 of genes for “negative regulators of T cell activation” (Table S6). These molecular changes may help to explain the overall dampened morphology of reactive microglia in BAC-TREM2 expressing AD mice. Future studies are needed to investigate whether and how the new molecular candidates as well as known TREM2 associated molecules could be tapped to reprogram microglial responsivity upon TREM2 gene dosage increase.
In conclusion, our study provides strong genetic evidence that increased TREM2 gene dosage can modify microglial transcriptional programs and morphological and functional responses in the brain of AD mouse models. Such microglia molecular reprogramming led to reduced plaque load and enhanced plaque compaction, reduced dystrophic neurites, and improved behavioral outcomes. Our study supports the future pursuit of early boosting of TREM2 levels or signaling to prevent the onset or reduce the severity of pathological microglial response and overall disease phenotypes in neurodegenerative diseases including AD.
STAR Methods
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Aβ (clone 6E10) | BioLegend | Cat# 803015; RRID: AB_2565328 |
| Mouse monoclonal anti-Aβ (4G8) | BioLegend | Cat# 800709; RRID: AB_2565325 |
| Mouse monoclonal anti-Aβ40 | BioLegend | Cat# 805401; RRID: AB_2564680 |
| Mouse monoclonal anti-Aβ42 | BioLegend | Cat# 805501; RRID: AB_2564683 |
| Rabbit polyclonal anti-Amyloid Precursor Protein C-Terminal (751-770) | Millipore | Cat# 171610; RRID: AB_211444 |
| Mouse monoclonal anti-CD68 | Bio-Rad | Cat# MCA1815; RRID: AB_322866 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling Technology | Cat# 2118; RRID: AB_561053 |
| Rabbit polyclonal anti-GFP | Thermo Fisher | Cat# A-11122; RRID: AB_221569 |
| Chicken polyclonal anti-GFP | Millipore | Cat# AB5541; RRID: AB_177521 |
| Rabbit polyclonal anti-Iba1 | Wako | Cat# 019-19741; RRID: AB_839504 |
| Mouse monoclonal anti-Iba1 (clone NCNP24) | Wako | Cat# 016-26721 |
| Rabbit polyclonal anti-Lgals3 | Cell Signaling Technology | Cat# 12733S |
| Sheep polyclonal anti-mouse Trem2 | R&D Systems | Cat# AF1729 |
| Goat polyclonal anti-human TREM2 | R&D Systems | Cat# AF1828 |
| Recombinant DNA | ||
| RP11-237K15 BAC | BACPAC Resources Center | clone: RP11-237K15 |
| Oligonucleotides | ||
| Primers for BAC modification (see Supplementary Table S7) | This paper | N/A |
| Primers for real-time PCR (see Supplementary Table S7) | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Thioflavin S | Sigma-Aldrich | T1892 |
| Congo Red | Sigma-Aldrich | 60910 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw data files for RNA-seq | This paper | NCBI Gene Expression Omnibus record: GSE104775 |
| Track for human TREM2-specific reads | This paper | https://goo.gl/dJ3MmK |
| Track for mouse Trem2-specific reads | This paper | https://goo.gl/JY6ne2 |
| Experimental Models: Organisms/Strains | ||
| BAC-TREM2 mice | This paper | N/A |
| BAC-TREM2-GFP mice | This paper | N/A |
| 5xFAD mice | The Jackson Laboratory | Cat# 034840-JAX, RRID:MMRRC_034840-JAX |
| APPswe/PS1dE9 mice | The Jackson Laboratory | Cat# 034829-JAX; RRID: MMRRC_034829-JAX |
| Software and Algorithms | ||
| ZEN Imaging Software | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Imaris | Bitplane | http://www.bitplane.com/imaris |
| ImageJ | NIH | https://imagej.nih.gov/ij/index.html |
| Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| STAR aligner | Dobin et al, 2013 | https://github.com/alexdobin/STAR |
| HTSeq | Anders et al, 2014 | http://htseq.readthedocs.io/en/release_0.9.1/ |
| R, version 3.4.1 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| DESeq2 | Love et al, 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| WGCNA | Langfelder and Horvath, 2008 | https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA |
| anRichment | Peter Langfelder, Jeremy A. Miller and Steve Horvath | https://labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/GeneAnnotation/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by Lead Contact X. William Yang (xwyang@mednet.ucla.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Generation of BAC Transgenic Mice
RP11-237K15 BAC contains the human TREM2 gene, as well as surrounding TREML1, TREML2, & TREML4 genes. The fidelity of the TREM2 gene was confirmed with Sanger sequencing of PCR products covering the entirety of the gene. TREML1, TREML2, and TREML4 genes were deleted with 4 sequential modification steps using RecA-based shuttle vector plasmids described previously (Yang et al., 1997; Gong et al., 2002; Gong and Yang, 2005; see Table S6). Exons 1–3 with proximal promoter region were deleted from TREML4, excising a majority of the protein coding sequence. Due to concern for a downstream in-frame ATG site in TREML1, Exons 5–6 were deleted along with exons 1–2 and the proximal promoter region, abolishing 80% of TREML1’s protein-coding sequences. For TREML2, exon 2–3 were deleted, resulting in a frame-shift and early stop site in exon 4. All the BAC modification products were confirmed using established methods (e.g. PCR, restriction mapping, etc; Gong and Yang, 2005). TREM2-GFP BAC was modified from TREM2 BAC by introducing the EGFP sequence to the 3′ end of TREM2 before the stop codon with the methods described above. The modified BAC DNA was prepared according to our published protocols and microinjected into FvB fertilized oocytes. BAC-TREM2 and BAC-TREM2-GFP mice were maintained in the FvB/NJ background.
Animal breeding and husbandry
5xFAD and APPswe/PS1dE9 mice were purchased from the Jackson Laboratory (MMRRC) and crossed to BAC-TREM2 mice in FvB/NJ inbred background. Thus, all genotypes of mice used in the current study were generated and analyzed in the F1 hybrid background (C56BL6J;FvB/NJ F1), which is suitable for phenotypic study of genetically engineered mutant mice (Silva et al., 1997). Animals were housed in standard mouse cages under conventional laboratory conditions, with constant temperature and humidity, 12h/12h light/dark cycle and food and water ad libitum. All animal studies were carried out in strict accordance with National Institutes of Health guidelines and approved by the UCLA Institutional Animal Care and Use Committees. Matched number of mice in both genders were used in the study. Age and the number (n) of mice used are as indicated in the individual experiments and figures.
METHOD DETAILS
Tissue collection and sample preparation
Mice were anesthetized with pentobarbital and perfused with ice-cold PBS. Brains were bisected. The right hemispheres were immediately submerged in ice-cold DEPC/PBS and cortices and hippocampi were carefully dissected out under a dissection microscope. Dissected tissues were snap frozen in dry ice and stored in −80ºC before further processing. The left hemispheres were fixed in 4% PFA/PBS overnight followed by submergence in 30% sucrose before freezing. Coronal sections (40 μm) were obtained using a cryostat and stored in cryopreserve solution at −20ºC.
For preparing the samples for RNA sequencing and biochemistry, dissected brain tissues were homogenized and aliquoted as described previously (Cramer et al., 2012). In brief, cortical and hippocampal tissues from one hemisphere were homogenized in tissue homogenization buffer (250 mM sucrose, 20 mM Tris at pH 7.4, 1 mM EDTA, and 1 mM EGTA in DEPC-treated water) and centrifuged at 5000 × g for 10 min at 4ºC. Supernatants were aliquoted and stored at −80ºC.
Aβ ELISA
Homogenates of cortical samples were subjected to sequential extraction using DEA (0.4% diethylamine in 100 mM NaCl) and FA (formic acid, >95%) solutions as described previously (Cramer et al., 2012). Concentration of soluble (DEA) and insoluble (FA) Aβ fractions were measured by ELISA using anti-Aβ1-16 (6E10) as a capturing antibody. Specific Aβ species were detected by anti-Aβ40-HRP and anti-Aβ42-HRP antibodies with chromogenic substrate TMB (ThermoFisher). Absorbance at 650 nm was read on a Spectramax colorimetric plate reader (Molecular Devices).
Immunohistochemistry and image analysis
Coronal sections were blocked in the blocking buffer (3% BSA, 2% normal goat serum and 0.3% Triton X-100 in PBS) for 1 hour at room temperature and then incubated with primary antibodies at 4ºC overnight. Incubation in secondary antibodies was performed for 2h at room temperature before mounting on slides with Prolong Diamond anti-fade mountant (ThermoFisher). Aβ plaques were visualized by ThioS and Congo Red staining or by immunostaining using anti-Aβ antibodies 6E10 and 4G8. For plaque number and categorization, 3 matched coronal sections/mouse spacing out across 1 mm (0.5 mm apart) were stained with 6E10 and ThioS. Z-stack 20× images covering 30 μm thickness were taken on Zeiss LSM510 confocal microscope and analyzed using ImageJ. Pixels with < 1% of max intensity were discarded as background and were not counted as a part of the plaque. All images were preprocessed using the same threshold setting prior to analysis. For microglial morphology, Z-stack 63× images of 50–55 overlapping optical slices aligned along the center of the plaques were collected from matched cortical regions. More than 12 plaques per genotype from 3 gender matched animals were taken. Morphology of all plaque-associated microglia in the images was analyzed using the FilamentTracer feature in Imaris 9.0 (Bitplane). For analyzing dystrophic neurites, Z-stack images were acquired from each section using a Zeiss confocal microscope (LSM 710) with 40× oil lens. The volume of dystrophic neurites in each section were quantified by Imaris and normalized to the number of plaques in the field. The image acquisition and quantification described above were performed in a blinded manner.
RNA purification and mRNA sequencing
Total RNA was extracted using RNeasy kit (Qiagen). Library preparation and RNA sequencing were performed by the UCLA Neuroscience Genomics Core (UNGC). Libraries were prepared using the Illumina TruSeq RNA Library Prep Kit v2 and sequenced on an Illumina HiSeq4000 sequencer using strand-specific, paired-end, 69-mer sequencing protocol to a minimum read depth of 30 million reads per sample. Reads were aligned to mouse genome mm10 using the STAR aligner (Dobin et al., 2013) with default settings. Read counts for individual genes were obtained using HTSeq.
Human-specific TREM2 reads were obtained by aligning to the human reference genome (build GRCh38) reads that failed to align to the mouse genome (build mm10). Mouse-specific Trem2 reads were obtained in similar way. Mapped reads were quantified by the htseq-count tool (Anders et al., 2015). TREM2 counts were divided by the library size per million to determine the counts per million (CPM) TREM2 level. Homer (Heinz et al., 2010) makeTagDirectory (parameters: -format sam -flip -sspe) and makeUCSCfile (parameters: -fragLength given -o auto -raw) functions, bedtools (Quinlan and Hall, 2010) and bedGraphToBigWig tools were used to create CPM bigwig tracks for visualization onto the UCSC genome browser. Read coverage at the TREM2 locus are available in a custom UCSC genome browser track: https://goo.gl/mE4b2h. Read coverage at the murine Trem2 locus is available at https://goo.gl/Acmcw4.
Differential expression analysis
For outlier removal and for network analysis using WGCNA, we retained mRNA profiles whose observed counts are 5 or more in at least one-quarter of the samples and transformed the raw counts using variance stabilization. Outlier samples were removed as described (Oldham et al., 2012), using the Euclidean distance-based sample connectivity Z.k threshold of −6. This procedure resulted in the removal of a single sample (7 month old, female 5xFAD).
For DE testing and network analysis, we used individual observation weights constructed as follows. Tukey bi-square-like weights (Wilcox 2012) are calculated for each (variance-stabilized) observation x, as
where u = min(1,|x – m|/(9MAD)), and m and MAD are median and median absolute deviation of the observations of the gene.
For each gene, MAD is adjusted such that (1) 10th percentile of the weights λ is at least 0.1 (that is, the proportion of observations with coefficients <0.1 is less than 10%) (Langfelder and Horvath 2012) and (2) for each individual time point and genotype, 40th percentile of the weights λ is at least 0.9 (that is, at least 40% of the observation have a high coefficient of at least 0.9).
DE testing was carried out in R using package DESeq2 (Love et al., 2014) version 1.16.1.
DESeq2 models observed counts using Negative Binomial General Linear Models with dispersion estimated from data. Wald test was used to for significance calculations, and independent filtering was disabled. For differential expression testing between genotypes, sex was used as a covariate. Genotype-sex interactions were tested using models with genotype × sex terms (with genotype and sex turned into binary indicator variables).
For each genotype contrast or interaction, DE tests result in gene-wise Z statistics (fold changes divided by their standard errors). A “rescue/exacerbation” plot assesses overall similarity of genome-wide effects of two genotype contrasts using a scatter plot of their gene-wise DE Z statistics. A positive linear trend (correlation) indicates that the effects of the two genotype contrasts are broadly similar, whereas a negative correlation indicates broadly opposing effects. Although other measures are possible, the correlation value can be used as a measure of similarity.
Consensus Weighted Gene Co-expression Network Analysis
We carried out Consensus Weighted Gene Co-expression Network Analysis (WGCNA) essentially as described previously (Langfelder and Horvath, 2007, 2008). Since the experimental design contains two variables of interest (age and genotype) with strong effects on expression, we carried out a consensus network analysis (Langfelder and Horvath, 2007) of two data sets: data from WT, 5xFAD and 5xFAD/TREM2 genotypes at 2, 4, and 7 months, and data from the same genotypes at 4 and 7 months adjusted for age. The rationale is that a consensus analysis identifies modules that group together genes correlated in both data sets, i.e., both with respect to time point as well as genotype. We left the TREM2 genotype out of the network analysis since it is overall not different from WT and we wanted to focus the network analysis on the effects of BAC-TREM2 in the 5xFAD background (5xFAD vs. 5xFAD/TREM2). We used weighted correlation with individual sample weights determined as described above and the “signed hybrid” network in which negatively correlated genes are considered unconnected.
This analysis identified 28 co-expression modules ranging from 52 to 1767 genes per module (Table S4; Figure S4). Since genes in each module are co-expressed, it is advantageous to represent each module by a single representative expression profile (i.e. the module eigengene, which explains most of the variance of the module genes) (Horvath and Dong, 2008). Eigengenes were tested for DE between genotypes using standard linear models with sex as a covariate. Module eigengenes allow one to define a continuous (“fuzzy”) measure of membership of all genes in all modules (Horvath and Dong, 2008, Langfelder et al, 2016). Genes with high fuzzy module memberhsip in a module are called intramodular hub genes for the module.
Gene Set Enrichment Calculations
We used the freely available, open source R package anRichment (https://labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/GeneAnnotation/) to calculate the enrichment of DE genes and WGCNA modules in a large collection of reference gene sets that includes Gene Ontology (GO) terms, KEGG pathways, literature gene sets collected in the userListEnrichment R function (Miller et al., 2011), Molecular Signatures Database gene sets (Subramanian et al., 2005), aging gene sets from Enrichr (Chen et al., 2013) and other gene sets. In particular, we collected microglia-relevant gene sets from several recent articles (Butovsky et al, 2014, Wang el al, 2015, Gokce et al, 2016, Galatro et al, 2017, Kraserman et al, 2017, Keren-Shaul et al, 2017). Fisher exact test was used to evaluate overlap significance.
Primary Microglial Culture and phagocytosis assay
Primary microglia were isolated from the brains of neonatal mice at postnatal days 2–3 using a mild trypsinization protocol as previously described (Lee et al., 2012). For phagocytosis assay, purified microglia were maintained in DMEM/F-12 (ThermoFisher) containing 2% heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin plated at a density of 250,000 cells/well in 24-well plates for 3–5 days before further experiments. The culture media was replenished with serum-free DMEM/F12 overnight. Cells were incubated with BSA (0.5 mg/ml in PBS)-preblocked microsphere (1 μm, Alexa 488-conjugated; ThermoFisher) for 30 min, followed by extensive washing with PBS and fixation with 4% paraformaldedyde. After fixation, cells were washed with PBS and collected for analysis using an LSR II flow cytometer (BD Biosciences).
Behavioral tests
Open-field exploration tests were performed for WT and BAC-TREM2 mice at 10 months of age (n = 11 per genotype, with matched gender ratio) using our established protocols (Wang et al., 2014). Open-field testing was performed during the dark phase of the 12h/12h light-dark cycle.
Contextual fear conditioning test was performed with minor adjustment as describe previously (Curzon et al., 2009) and conducted in the Behavioral Testing Core (BTC) at UCLA. In brief, mice were handled daily for a week prior to the behavior test. In the training phase, mice were placed individually in the conditioning chamber to explore the environment freely for 2 min before the first unconditioned stimulus (US: 0.75 mA, 2 s) was delivered. The animals were exposed to 2 US’s with an intertrial interval of 3 min. After the last shock, the mice were left in the chamber for another 1 min and then placed back in their home cages. Retention tests were performed 24 hours later. Each mouse was returned to the same chamber for measuring the percent of time frozen and number of freezes. No shocks are given during the test session. Both training and testing procedures were videotaped and the freezing behavior was measured by an automated tracking system (Med Associates).
Slice preparation and electrophysiology
Hippocampal slices were prepared as previously reported (Fernado et al., 2016). Briefly, mice were anesthetized with isoflurane and decapitated following UCLA Chancellor’s Animal Research Committee protocol. Coronal slices 350 μm thick were cut on a Leica VT1000S vibratome in N-methyl-d-glucamine (NMDG)-based HEPES-buffered solution, containing, in mM: 135 NMDG, 10 D-glucose, 4 MgCl2, 0.5 CaCl2, 1 KCl, 1.2 KH2PO4, 20 HEPES, 27 sucrose, 3 kynurenic acid. Slices were incubated at 34°C in an interface chamber in a reduced sodium artificial CSF (aCSF), containing, in mM: 85 NaCl, 25 D-glucose, 55 sucrose, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2, 4 MgCl2, 26 NaHCO3. Low-sodium aCSF was substituted for normal aCSF (naCSF) at room temperature, containing, in mM: 126 NaCl, 10 D-glucose, 2 MgCl2, 2 CaCl2, 2.5 KCl, 1.25 NaH2PO4, 1.5 sodium pyruvate, 1 L-glutamine, 26 NaHCO3. pH for all solutions = 7.3 – 7.4. Recordings were done in an interface chamber at 35 °C perfused with naCSF. The Schaffer collateral pathway was stimulated every 30 s with two pulses, 50 ms apart. LTP was induced by stimulating the Schaffer collaterals with twice the duration of baseline stimuli with a theta burst stimulation (TBS) repeated twice, 30 s apart (4 pulses, 100 s−1 repeated 20 times every 350 ms). Evoked fEPSPs were recorded in CA1 stratum radiatum (SR) with the use of a patch pipette (3 – 5 M3 resistance) filled with naCSF connected to the headstage of an amplifier (A-M Systems Inc., model 3000) where it was bandpass filtered between 0.1 and 1,000 Hz. The signal was fed through an instrumentation amplifier (Brownlee BP Precision, model 210A) and sampled at 10,000 100 s−1 with a National Instruments A/D board. Field potentials were recorded using EVAN (custom-designed LabView-based software from Thotec) and analyzed with a custom written procedure (Wavemetrics, IGOR Pro 6.22A). The slope of the fEPSPs was measured during a 0.5 – 1.0 ms window of their steepest rising phase. Presence or absence of an LTP at 40 minutes was determined as previously described (Fernado et al., 2016) and recordings in which LTP was not induced were not included in the overall LTP averages. The fraction of slices exhibiting no LTP was not different among groups (Chi-square with Yates’ correction = 0.6994; p = 0.4030).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Statistics for transcriptomic analyses were described as above. Other quantitative results, unless otherwise specified, were analyzed using one-way ANOVA with Turkey post-hoc analysis or unpaired t-test to determine the p value. Morphological and behavioral studies were subjected to Power analysis to determine the biological replicates (n) needed to reach >80% confidence level. n for individual experiments could be found in the results and figure legends.
DATA AND SOFTWARE AVAILABILITY
RNA-seq data has been deposited within the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo), accession number GSE104775. The links to the tracks for human and mouse TREM2-specific reads are listed in the Key Resource Table.
Supplementary Material
Table S1: DE genes for genotype contrasts at 2m, 4m and 7m, related to Figure 3
Table S2: Enrichment analyses of DE genes, related to Figure 3
Table S3: DE genes with significant genotype-gender interaction at 2m, 4m and 7m, related to Figure 3
Table S4: WGCNA analyses of RNA-seq data, related to Figure 3
Table S5: Enrichment analyses of WGCNA modules, related to Figure 3
Table S6: Enrichment analyses of TD1, TD2 and TD3 Genes, related to Figure 4
Table S7: Primers for BAC modification and real-time PCR study, related to Key Resource Table
Highlights.
Elevating TREM2 gene dosage altered microglial morphology and interaction with Aβ
Increasing TREM2 gene dosage reprograms microglial responsivity in AD mouse brains
Transcriptomic profiling identified 3 groups of TREM2 gene-dosage-dependent genes
Extra TREM2 gene dosage ameliorates neuropathology and memory deficits in AD mice
Acknowledgments
The research was supported by NIA/NIH (AG056114 to X.W.Y. and H.X.). Yang lab is also supported by NIH grants (NS074312, NS084298, MH106008), David Weill fund from the Semel Institute at UCLA, a grant from UCLA Neurology Department, CHDI Foundation, Inc., and Leslie Gehry Brenner Award from Hereditary Disease Foundation. The H.X. lab is supported by NIH grants (AG048519, AG021173, AG038710, AG044420, NS046673, and AG056130 to H.X.) as well as by the Tanz Family Fund and Cure Alzheimer’s Fund. A.D. was supported by NIH predoctoral training grant (T32 MH073526). We would like to thank the UCLA Neuroscience Genomics Core, Fuying Gao and Yue Qin for assistance in RNA-seq data acquisition and analyses. We acknowledge the help from the NINDS Informatics Center grant (P30 NS062691), and UCLA Behavioral Testing Core. We thank Dr. Matthew Veldman for editing the manuscript.
Footnotes
Author Contributions
X.W.Y. provided the conceptual framework and provide funds for the study. C.D.L., A.D. and X.W.Y. designed the experiments and interpreted the results. C.D.L. and X.W.Y. wrote the manuscript. A.D. and X.G. generated the BAC-TREM2 and BAC-TREM2-GFP mice (Fig. 1). C.D.L. performed experiments and analyzed data shown in Fig. 1E–1N, Fig. 2, Fig. 6A–6C, 6G, Fig. 7F, Fig. 8, and Figures S3, S7–S9. A.D. and C.D.L. contributed Fig. 5. X.G. contributed to Fig. 1E–M, Figure S2. RNA-seq was performed by C.D.L., N.W. and C.S.P., and data analyses were performed by P.L., G.C., Y.C. (Fig. 3, Fig. 4, Fig. S1, S4–S6). L.-L.J., Y.Z., X.L., and H.X. contributed to Fig. 1C–1D, Fig. 5I–5L and Fig. 7A–7E. I.F. and I.M. contributed Fig. S2D–S2E.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atagi Y, Liu CC, Painter MM, Chen XF, Verbeeck C, Zheng H, Li X, Rademakers R, Kang SS, Xu H, et al. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2) J Biol Chem. 2015;290:26043–26050. doi: 10.1074/jbc.M115.679043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CC, DeVaux LB, Farzan M. The Triggering Receptor Expressed on Myeloid Cells 2 Binds Apolipoprotein E. J Biol Chem. 2015;290:26033–26042. doi: 10.1074/jbc.M115.677286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemiller SM, McCray TJ, Allan K, Formica SV, Xu G, Wilson G, Kokiko-Cochran ON, Crish SD, Lasagna-Reeves CA, Ransohoff RM, et al. TREM2 deficiency exacerbates tau pathology through dysregulated kinase signaling in a mouse model of tauopathy. Mol Neurodegener. 2017;12:74. doi: 10.1186/s13024-017-0216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC, Neher JJ. Microglial phagocytosis of live neurons. Nat Rev Neurosci. 2014;15:209–216. doi: 10.1038/nrn3710. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Alvarado G, Bigcas JL, Li W. Galectin-3 is a new MerTK-specific eat-me signal. J Cell Physiol. 2012;227:401–407. doi: 10.1002/jcp.22955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Allen M, Burgess JD, Wang X, Strickland SL, Aryal S, Siuda J, Kachadoorian ML, Medway C, Younkin CS, et al. A candidate regulatory variant at the TREM gene cluster associates with decreased Alzheimer’s disease risk and increased TREML1 and TREM2 brain gene expression. Alzheimers Dement. 2017;13:663–673. doi: 10.1016/j.jalz.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Welsh CA, Barres BA, Stevens B. Do glia drive synaptic and cognitive impairment in disease? Nat Neurosci. 2015;18:1539–1545. doi: 10.1038/nn.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3:445–453. doi: 10.1038/nri1106. [DOI] [PubMed] [Google Scholar]
- Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, et al. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curzon P, Rustay NR, Browman KE. Cued and Contextual Fear Conditioning for Rodents. In: Buccafusco JJ, editor. Methods of Behavior Analysis in Neuroscience. Boca Raton (FL): 2009. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiou AG, Goate AM. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol Neurodegener. 2017;12:43. doi: 10.1186/s13024-017-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferando I, Faas GC, Mody I. Diminished KCC2 confounds synapse specificity of LTP during senescence. Nat Neurosci. 2016;19:1197–1200. doi: 10.1038/nn.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, Veras MM, Pereira TF, Leite REP, Möller T, et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat Neurosci. 2017;20:1162–1171. doi: 10.1038/nn.4597. [DOI] [PubMed] [Google Scholar]
- Gandy S, Heppner FL. Microglia as dynamic and essential components of the amyloid hypothesis. Neuron. 2013;78:575–577. doi: 10.1016/j.neuron.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Sudhof TC, Quake SR. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 2016;16:1126–1137. doi: 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JS, Kobayashi M, Hayashi H, Zou K, Sawamura N, Fujita SC, Yanagisawa K, Michikawa M. Apolipoprotein E (ApoE) isoform-dependent lipid release from astrocytes prepared from human ApoE3 and ApoE4 knock-in mice. J Biol Chem. 2002a;277:29919–29926. doi: 10.1074/jbc.M203934200. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang XW. Modification of bacterial artificial chromosomes (BACs) and preparation of intact BAC DNA for generation of transgenic mice. Curr Protoc Neurosci. 2005;Chapter 5(Unit 5):21. doi: 10.1002/0471142301.ns0521s31. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002b;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356 doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber MB. Changing face of microglia. Science. 2010;330:783–788. doi: 10.1126/science.1190929. [DOI] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013a;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Lohmann E, Bras JM, Gibbs JR, Rohrer JD, Gurunlian N, Dursun B, Bilgic B, Hanagasi H, Gurvit H, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 2013b;70:78–84. doi: 10.1001/jamaneurol.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, Harari O, Bertelsen S, Fairfax BP, Czajkowski J, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci. 2017;20:1052–1061. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- Jay TR, Hirsch AM, Broihier ML, Miller CM, Neilson LE, Ransohoff RM, Lamb BT, Landreth GE. Disease Progression-Dependent Effects of TREM2 Deficiency in a Mouse Model of Alzheimer’s Disease. J Neurosci. 2017;37:637–647. doi: 10.1523/JNEUROSCI.2110-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay TR, Miller CM, Cheng PJ, Graham LC, Bemiller S, Broihier ML, Xu G, Margevicius D, Karlo JC, Sousa GL, et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J Exp Med. 2015;212:287–295. doi: 10.1084/jem.20142322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DM, Frangakis SG, Golzio C, Cassa CA, Kurtzberg J, Davis EE, Sunyaev SR, Katsanis N Genomics T.F.f.N. Identification of cis-suppression of human disease mutations by comparative genomics. Nature. 2015;524:225–229. doi: 10.1038/nature14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Cruchaga C, Goate AM. Alzheimer’s disease genetics: from the bench to the clinic. Neuron. 2014;83:11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169:1276–1290. e1217. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Kimura R, Ohno M. Impairments in remote memory stabilization precede hippocampal synaptic and cognitive failures in 5XFAD Alzheimer mouse model. Neurobiol Dis. 2009;33:229–235. doi: 10.1016/j.nbd.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knafo S, Venero C, Merino-Serrais P, Fernaud-Espinosa I, Gonzalez-Soriano J, Ferrer I, Santpere G, DeFelipe J. Morphological alterations to neurons of the amygdala and impaired fear conditioning in a transgenic mouse model of Alzheimer’s disease. J Pathol. 2009;219:41–51. doi: 10.1002/path.2565. [DOI] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 2017;47:566–581. e569. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Cantle JP, Chatzopoulou D, Wang N, Gao F, Al-Ramahi I, Lu XH, Ramos EM, El-Zein K, Zhao Y, et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19:623–633. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Tse W, Smith JD, Landreth GE. Apolipoprotein E promotes beta-amyloid trafficking and degradation by modulating microglial cholesterol levels. J Biol Chem. 2012;287:2032–2044. doi: 10.1074/jbc.M111.295451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyns CEG, Ulrich JD, Finn MB, Stewart FR, Koscal LJ, Remolina Serrano J, Robinson GO, Anderson E, Colonna M, Holtzman DM. TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc Natl Acad Sci U S A. 2017;114:11524–11529. doi: 10.1073/pnas.1710311114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, Sisk A, Mallory M, Mucke L, Schenk D, Games D. Comparison of neurodegenerative pathology in transgenic mice overexpressing V717F beta-amyloid precursor protein and Alzheimer’s disease. J Neurosci. 1996;16:5795–5811. doi: 10.1523/JNEUROSCI.16-18-05795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarin M, Salih DA, Yasvoina M, Cummings DM, Guelfi S, Liu W, Nahaboo Solim MA, Moens TG, Paublete RM, Ali SS, et al. A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or tau pathology. Cell Rep. 2015;10:633–644. doi: 10.1016/j.celrep.2014.12.041. [DOI] [PubMed] [Google Scholar]
- Mazure CM, Swendsen J. Sex differences in Alzheimer’s disease and other dementias. Lancet Neurol. 2016;15:451–452. doi: 10.1016/S1474-4422(16)00067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL, Bose HS. Early steps in steroidogenesis: intracellular cholesterol trafficking. J Lipid Res. 2011;52:2111–2135. doi: 10.1194/jlr.R016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Langfelder P, Horvath S. Network methods for describing sample relationships in genomic datasets: application to Huntington’s disease. BMC Syst Biol. 2012;6:63. doi: 10.1186/1752-0509-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, 3rd, Lafaille JJ, Hempstead BL, Littman DR, Gan WB. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliani PL, Wang Y, Fontana E, Robinette ML, Yamanishi Y, Gilfillan S, Colonna M. TREM2 sustains microglial expansion during aging and response to demyelination. J Clin Invest. 2015;125:2161–2170. doi: 10.1172/JCI77983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein M, Vay SU, Flitsch LJ, Fink GR, Schroeter M, Rueger MA. Osteopontin directly modulates cytokine expression of primary microglia and increases their survival. J Neuroimmunol. 2016;299:130–138. doi: 10.1016/j.jneuroim.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science. 2016;353:777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- Rotshenker S. The role of Galectin-3/MAC-2 in the activation of the innate-immune function of phagocytosis in microglia in injury and disease. J Mol Neurosci. 2009;39:99–103. doi: 10.1007/s12031-009-9186-7. [DOI] [PubMed] [Google Scholar]
- Sadleir KR, Eimer WA, Cole SL, Vassar R. Abeta reduction in BACE1 heterozygous null 5XFAD mice is associated with transgenic APP level. Mol Neurodegener. 2015;10:1. doi: 10.1186/1750-1326-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Hsu DK, Apgar JR, Yu L, Sharma BB, Kuwabara I, Izui S, Liu FT. Critical role of galectin-3 in phagocytosis by macrophages. J Clin Invest. 2003;112:389–397. doi: 10.1172/JCI17592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Simpson EM, Takahashi JS, Lipp HP, Nakanishi S, Wehner JM, Giese KP, Tully T, Abel T, Chapman PF, et al. Mutant mice and neuroscience: recommendations concerning genetic background. Banbury Conference on genetic background in mice. Neuron. 1997;19:755–759. doi: 10.1016/s0896-6273(00)80958-7. [DOI] [PubMed] [Google Scholar]
- Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerrett R, Pellegrino MP, Casali BT, Taraboanta L, Landreth GE. Combined Liver X Receptor/Peroxisome Proliferator-activated Receptor Agonist Treatment Reduces Amyloid β Levels and Improves Behavior in Amyloid Precursor Protein/Presenilin 1 Mice. J Biol Chem. 2015;290:21591–21602. doi: 10.1074/jbc.M115.652008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AH, Barres BA, Stevens B. The Complement System: An Unexpected Role in Synaptic Pruning During Development and Disease. Annual Review of Neuroscience. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- Suárez-Calvet M, Kleinberger G, Araque Caballero MÁ, Brendel M, Rominger A, Alcolea D, Fortea J, Lleó A, Blesa R, Gispert JD, et al. sTREM2 cerebrospinal fluid levels are a potential biomarker for microglia activity in early-stage Alzheimer’s disease and associate with neuronal injury markers. EMBO Mol Med. 2016;8:466–476. doi: 10.15252/emmm.201506123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Rochford CDP, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med. 2005;201:647–657. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich JD, Ulland TK, Colonna M, Holtzman DM. Elucidating the Role of TREM2 in Alzheimer’s Disease. Neuron. 2017;94:237–248. doi: 10.1016/j.neuron.2017.02.042. [DOI] [PubMed] [Google Scholar]
- Wang N, Gray M, Lu XH, Cantle JP, Holley SM, Greiner E, Gu X, Shirasaki D, Cepeda C, Li Y, et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat Med. 2014;20:536–541. doi: 10.1038/nm.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ulland TK, Ulrich JD, Song W, Tzaferis JA, Hole JT, Yuan P, Mahan TE, Shi Y, Gilfillan S, et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J Exp Med. 2016;213:667–675. doi: 10.1084/jem.20151948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox RR. Introduction to robust estimation and hypothesis testing. 3. Amsterdam ; Boston: Academic Press; 2012. [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VLJ, Fisher EMC, Tavaré S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Yang XW, Wynder C, Doughty ML, Heintz N. BAC-mediated gene-dosage analysis reveals a role for Zipro1 (Ru49/Zfp38) in progenitor cell proliferation in cerebellum and skin. Nat Genet. 1999;22:327–335. doi: 10.1038/11896. [DOI] [PubMed] [Google Scholar]
- Yeh FL, Hansen DV, Sheng M. TREM2, Microglia, and Neurodegenerative Diseases. Trends Mol Med. 2017;23:512–533. doi: 10.1016/j.molmed.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Yuan P, Condello C, Keene CD, Wang Y, Bird TD, Paul SM, Luo W, Colonna M, Baddeley D, Grutzendler J. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron. 2016;90:724–739. doi: 10.1016/j.neuron.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ke SQ, Huang Z, Flavahan W, Fang X, Paul J, Wu L, Sloan AE, McLendon RE, Li X, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol. 2015;17:170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: DE genes for genotype contrasts at 2m, 4m and 7m, related to Figure 3
Table S2: Enrichment analyses of DE genes, related to Figure 3
Table S3: DE genes with significant genotype-gender interaction at 2m, 4m and 7m, related to Figure 3
Table S4: WGCNA analyses of RNA-seq data, related to Figure 3
Table S5: Enrichment analyses of WGCNA modules, related to Figure 3
Table S6: Enrichment analyses of TD1, TD2 and TD3 Genes, related to Figure 4
Table S7: Primers for BAC modification and real-time PCR study, related to Key Resource Table