According to the World Health Organization myelodysplastic/myeloproliferative neoplasms (MDS/MPN) are a heterogeneous group of myeloid neoplasms showing clinical and morphological features that overlap between MDS and MPN.1 This category includes atypical chronic myeloid leukemia (aCML), chronic myelomonocytic leukemia (CMML), MDS/MPN, unclassifiable (MDS/MPN, U) and MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T). In most cases of MDS/MPN the karyotype is normal, while targeted sequencing revealed a high frequency of mutations in MDS/MPN.1–5 Although single entities have been investigated, no comprehensive comparative analysis of MDS/MPN is available. Furthermore, MDS/MPN, U, in particular, remains a heterogeneous mixture of phenotypes and genotypes, and in some cases difficult to distinguish from aCML.5 We, therefore, aimed at discovering entity-specific mutation patterns to enable a genetic distinction of all four MDS/MPN entities for diagnostic purposes.
We investigated 177 patients at diagnosis by cytomorphology and genetic studies strictly following World Health Organization criteria: 35 patients were diagnosed as having aCML, 58 as having CMML, 39 as having MDS/MPN, U and 45 as having MDS/MPN-RS-T. The median age of the cohort was 75 years (range, 22–89 years) and comprised 72 females and 105 males. Some cases were already included in former studies.2–4 All patients were analyzed by targeted next-generation sequencing for 18 genes.6 Chromosome banding analysis was performed for 171/177 patients according to standard procedures. Further information is available in the Online Supplementary Material.
Cytogenetically, 44/171 (26%) MDS/MPN patients had an aberrant karyotype. An aberrant karyotype was most frequent in aCML (13/35, 37%), followed by MDS/MPN, U (11/38, 29%), CMML (14/55, 25%) and MDS/MPN-RS-T (6/43, 14%). The only significant difference in frequency of aberrant karyotypes occurred between aCML and MDS/MPN-RS-T (P=0.032). The only recurrent aberrations detected were trisomy 8 (n=17), monosomy 7 (n=3) and loss of the Y-chromosome (n=8), reflecting the commonly detected aberrations in MDS patients.7,8
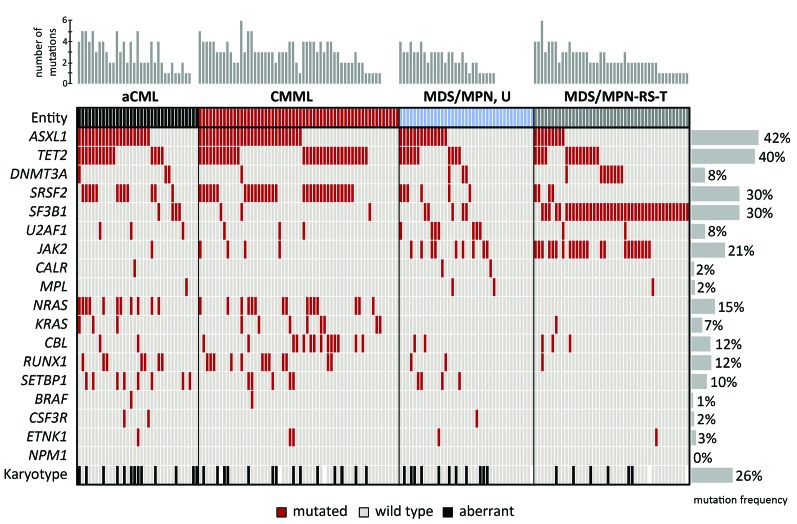
Studying the frequencies of mutations within the total cohort revealed that ASXL1 was the most frequently mutated gene (74/177, 42%), followed by TET2 (70/177, 40%), the spliceosomal genes SF3B1 and SRSF2 (both 53/177, 30%), JAK2 (37/177, 21%), NRAS (26/177, 15%), RUNX1 (22/177, 12%), CBL (21/177, 12%) and SETBP1 (17/177, 10%). All other investigated genes showed mutation frequencies <10% (Figure 1). The number of mutations per patient ranged from zero to six with a median of two mutations. This was also true for the single entities aCML, MDS/MPN, U and MDS/MPN-RS-T, while the median was three mutations per patient for cases of CMML (Figure 1).
Figure 1.
Molecular and cytogenetic characterization of patients with myelodysplastic/myeloproliferative neoplasms. Illustration of all 177 samples: each column represents one patient. All 18 analyzed genes as well as karyotype information are given for each patient. Light gray: wild type or normal karyotype, red: mutated, black: aberrant karyotype, white: no data available. The number of mutations per patient is illustrated as a bar chart above the graph, the mutation frequencies of single genes are given as a bar chart at the right.
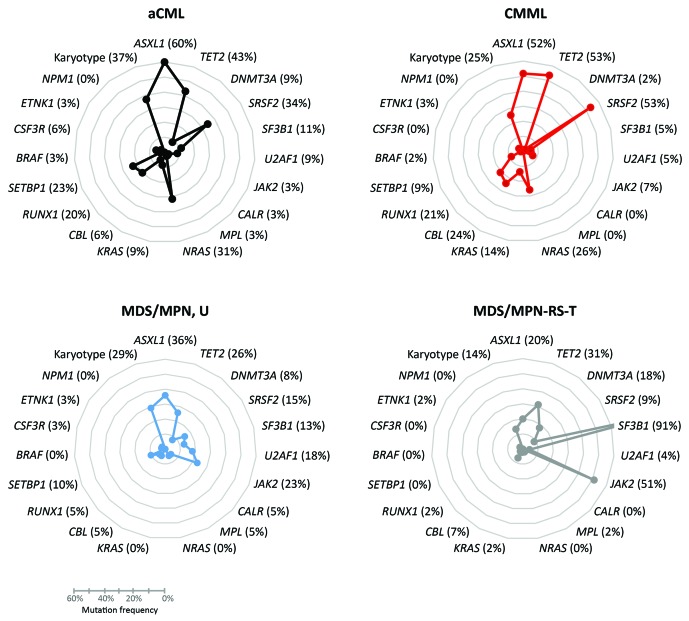
Looking at the mutational profile of the four separate entities revealed significant differences in mutation frequencies of single genes (Figure 2, Online Supplementary Table S2). ASXL1 and TET2 were frequently affected in all entities although there were significant differences: ASXL1 was less frequently mutated in MDS/MPN-RS-T (20%) than in aCML (60%; P<0.001) and CMML (52%; P=0.001). TET2 was more often mutated in CMML (53%) than in MDS/MPN, U (26%; P=0.007) or MDS/MPN-RS-T (31%; P=0.028). Spliceosomal genes are one of the most frequently mutated genes in MDS but different frequencies can be noted between the MDS subgroups.9 Within the MDS/MPN entities SRSF2 mutations appeared more often in CMML (53%) than in MDS/MPN-RS-T (9%; P<0.001) or MDS/MPN, U (15%; P<0.001). SF3B1 mutations prevailed in MDS/MPN-RS-T (91%) compared to all other entities (aCML: 11%, CMML: 5%, MDS/MPN, U: 13%; for all P<0.001). Interestingly, in MDS/MPN, U all three investigated splicing genes (SF3B1, SRSF2 and U2AF1) appeared mutated with a slight predominance in U2AF1 mutations, while in aCML and CMML SRSF2 mutations and in MDS/MPN-RS-T SF3B1 mutations were most often observed. Although the distribution was not statistically significantly different, SETBP1 mutations occurred most often in aCML (23%), followed by MDS/MPN, U (10%) and CMML (9%); this gene was not mutated in MDS/MPN-RS-T. SETBP1 mutations have been described as a marker pointing to a diagnosis of aCML, which is in line with the data presented here.2,10,11 The incidence of JAK2, the MPN entity-defining mutation, was significantly higher in MDS/MPN-RS-T (51%) than in MDS/MPN, U (23%, P=0.013), CMML (7%, P<0.001) and aCML (3%, P<0.001). Furthermore, JAK2 mutations were also significantly more frequent in MDS/MPN, U than in aCML and CMML (P=0.015 and P=0.032, respectively). Investigating the RAS signaling genes showed that NRAS is not mutated in MDS/MPN-RS-T and MDS/MPN, U but was quite frequently mutated in aCML (31%, P<0.001) and CMML (26%, P<0.001). Furthermore, CBL was more often mutated in CMML (24%) than in MDS/MPN-RS-T (7%, P=0.030), aCML (6%, P=0.025) and MDS/MPN, U (5%, P=0.013). The CBL mutation frequency therefore makes the difference between the quite similar aCML and CMML cases. Overall, these mutation frequencies are in line with published data, except for a slightly higher frequency of CBL mutations in CMML and the absence of NRAS mutations in MDS/MPN, U.2–5,10,12–14 In previous studies the frequency of the CBL mutation was found to be ~10% in CMML instead of the 24% in our study and RAS mutations were detected in 14% of MDS/MPN, U, contrasting with the 0% in the present study.14,15 These differences might be caused by overall small cohort sizes slightly over- or underestimating mutation frequencies. Taking the overall mutational landscape into account, Figure 2 shows that aCML and CMML share a similar mutation pattern, differentiated by the frequencies of SRSF2, TET2, NRAS and CBL mutations (upper panel Figure 2), reflecting the typically described mutation profiles.2,3,10,12,15 MDS/MPN-RS-T, however, showed a different pattern dominated by overall low mutation frequencies except for SF3B1 and JAK2 (lower right panel Figure 2), as described previously.4,13 MDS/MPN, U cases seem to have an intermediate mutation profile, showing features of both groups (lower left panel Figure 2): overall, there were lower mutation frequencies, but a higher frequency of JAK2 mutations, in contrast to aCML and CMML, and no NRAS mutations similar to MDS/MPN-RS-T.14 One can postulate that cases so far classified as MDS/MPN, U may be categorized according to their respective molecular patterns in the near future and that this would be a refinement compared to the criteria currently in use. It is, therefore, worth mentioning that the related entity chronic neutrophilic leukemia (CNL) shows an overlapping but different mutation pattern to the entitites addressed here. CNL can be clearly discriminated by the high occurrence of CSF3R mutations (43%).11
Figure 2.
The mutation frequencies of all 18 analyzed genes as well as karyotype information for the separate subcohorts of myelodysplastic/myeloproliferative neoplasms. Each of the four subcohorts, aCML, CMML, MDS/MPN, U and MDS/MPN-RS-T, is shown separately. In the MDS/MPN-RS-T graph the mutation frequency of SF3B1 (91%) is cropped for more clarity of other gene mutation frequencies.
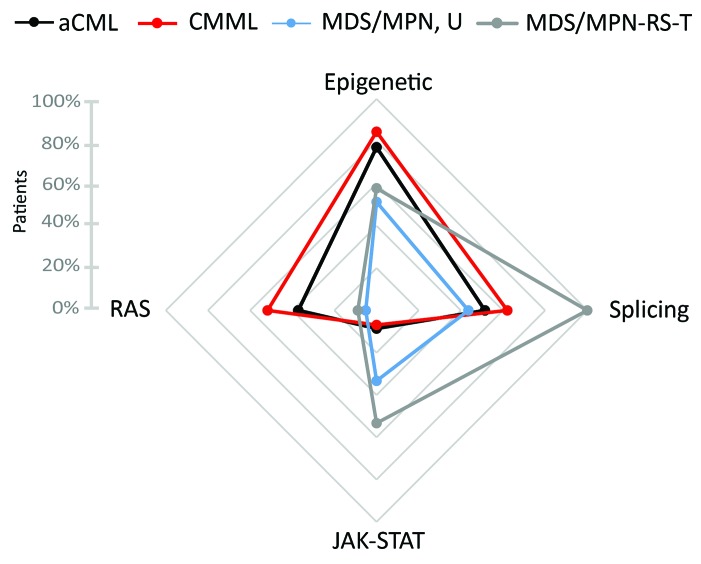
Since the most prominent differences between the four entities occurred in spliceosomal genes, JAK2 and NRAS, we grouped the genes by their respective pathways in attempt to gain further insights into the different mechanisms involved in disease manifestation. We, therefore, grouped together the genes ASXL1, TET2 and DNMT3A involved in epigenetic regulation, the spliceosomal genes SF3B1, SRSF2 and U2AF1, the genes JAK2, CALR, MPL affecting the JAK-STAT pathway and finally the genes NRAS, KRAS and CBL from the RAS signaling pathway. All four MDS/MPN entities were affected by mutations in different pathways (Figure 3). Although the most frequently mutated genes belong to the epigenetic regulators in MDS/MPN, these genes were less frequently mutated in MDS/MPN-RS-T (58%) than in CMML (84%, P<0.001) and also less frequently in MDS/MPN, U (51%) than in aCML (77%, P=0.030) or CMML (84%, P<0.001). The group of spliceosomal genes were more often mutated in MDS/MPN-RS-T (100%) than in MDS/MPN, U (44%, P<0.001), CMML (62%, P<0.001) or aCML (51%, P<0.001). Mutations in JAK2/CALR/MPL (the JAK-STAT pathway) occurred at a higher prevalence in MDS/MPN, U (33%) and MDS/MPN-RS-T (53%) than in aCML (9%) or CMML (7%; P<0.001). In contrast, NRAS/KRAS/CBL (the RAS pathway) were more often mutated in aCML (37%) and CMML (52%), as compared to MDS/MPN, U (5%) and MDS/MPN-RS-T (9%; P<0.001). Here different genetic profiles became obvious (Figure 3). While the same pathways were affected in aCML and CMML, namely the RAS pathway, epigenetic regulation and splicing with hardly any involvement of the JAK-STAT pathway, in MDS/MPN-RS-T epigenetic regulation, the JAK-STAT pathway and splicing were affected but rarely the RAS signaling pathway. Once again, MDS/MPN, U seems to be a mixture of both with affected epigenetic regulation, JAK-STAT pathway and splicing but a marginally affected RAS pathway. Considering the mean variant allele frequencies of the gene mutations in the different affected pathways revealed similar variant allele frequencies for all the pathways in all four entities pointing to driver mutations in the main cell clone (Online Supplementary Figure S1).
Figure 3.
The frequencies of patients showing at least one mutation within genes grouped according to their functional properties. The gene groups were: epigenetic regulation (ASXL1, TET2, DNMT3A), splicing (SF3B1, SRSF2, U2AF1), the JAK-STAT pathway (JAK2, CALR, MPL) and the RAS pathway (NRAS, KRAS, CBL).
Signaling mutations (the JAK-STAT and RAS signaling pathways) result in aberrant activation of proliferation as well as anti-apoptotic pathways and therefore correlate with a myeloproliferative phenotype. Overall our data reflect the described incidences of signaling mutations (80% JAK-STAT mutations in MDS/MPN-RS-T and 50% signaling mutations in CMML): the here presented slightly lower mutation frequencies of 53% in the JAK-STAT pathway in MDS/MPN-RS-T and 53% signaling mutations in CMML might be caused by different grouping of analyzed genes to more subdivided pathways.5
In conclusion, genes involved in epigenetic regulation are overall the most frequently mutated genes in MDS/MPN. For discrimination, SRSF2 is most frequently mutated in CMML, but also in aCML, while SF3B1 is predominantly mutated in MDS/MPN-RS-T. The JAK-STAT pathway is more often affected in MDS/MPN, U and MDS/MPN-RS-T in contrast to the RAS pathway, which is more often affected in aCML and CMML. The two entities aCML and CMML show very similar mutation profiles, which are clearly different from the profile of MDS/MPN-RS-T. MDS/MPN, U seems to be a mixture of both groups with a specific mutation profile and would be better classified according to gene patterns than by phenotype.
Supplementary Material
Acknowledgments
We thank all clinicians for sending samples to our laboratory for diagnostic purposes and for providing clinical information and follow-up data. In addition, we would like to thank all the co-workers at the MLL Munich Leukemia Laboratory for approaching together many aspects in the field of leukemia diagnostics and research.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 2.Meggendorfer M, Bacher U, Alpermann T, et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia. 2013;27(9):1852–1860. [DOI] [PubMed] [Google Scholar]
- 3.Meggendorfer M, Roller A, Haferlach T, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood. 2012;120(15):3080–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeromin S, Haferlach T, Weissmann S, et al. Refractory anemia with ring sideroblasts and marked thrombocytosis (RARS-T) cases harbor mutations in SF3B1 or other spliceosome genes accompanied by JAK2V617F and ASXL1 mutations. Haematologica. 2014; 100(4):e125–e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mughal TI, Cross NC, Padron E, et al. An International MDS/MPN Working Group’s perspective and recommendations on molecular pathogenesis, diagnosis and clinical characterization of myelodysplastic/myeloproliferative neoplasms. Haematologica. 2015; 100(9):1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delic S, Rose D, Kern W, et al. Application of an NGS-based 28-gene panel in myeloproliferative neoplasms reveals distinct mutation patterns in essential thrombocythaemia, primary myelofibrosis and polycythaemia vera. Br J Haematol. 2016;175(3):419–426. [DOI] [PubMed] [Google Scholar]
- 7.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. [DOI] [PubMed] [Google Scholar]
- 10.Piazza R, Valletta S, Winkelmann N, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013; 45(1):18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meggendorfer M, Haferlach T, Alpermann T, et al. Specific molecular mutation patterns delineate chronic neutrophilic leukemia, atypical chronic myeloid leukemia, and chronic myelomonocytic leukemia. Haematologica. 2014;99(12):e244–e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cazzola M, la Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122(25):4021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiter A, Invernizzi R, Cross NC, Cazzola M. Molecular basis of myelodysplastic/myeloproliferative neoplasms. Haematologica. 2009;94(12):1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang SA, Hasserjian RP, Fox PS, et al. Atypical chronic myeloid leukemia is clinically distinct from unclassifiable myelodysplastic/myeloproliferative neoplasms. Blood. 2014;123(17):2645–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itzykson R, Kosmider O, Renneville A, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428–2436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.