Abstract
Several macromolecular machines collaborate to produce eukaryotic messenger RNA. RNA polymerase II (Pol II) translocates along genes that are up to millions of base pairs in length and generates a flexible RNA copy of the DNA template. This nascent RNA harbours introns that are removed by the spliceosome, which is a megadalton ribonucleoprotein complex that positions the distant ends of the intron into its catalytic centre. Emerging evidence that the catalytic spliceosome is physically close to Pol II in vivo implies that transcription and splicing occur on similar timescales and that the transcription and splicing machineries may be spatially constrained. In this Review, we discuss aspects of spliceosome assembly, transcription elongation and other co-transcriptional events that allow the temporal coordination of co-transcriptional splicing.
The cellular demand for the splicing of precursor messenger RNAs (pre-mRNAs) is enormous. Typical human genes have eight introns, and each intron provokes the de novo assembly of a spliceosome1, 2. It takes only one RNA polymerase II (Pol II), one set of 5′ end capping enzymes and one 3′ end cleavage and polyadenylation complex to transcribe the pre-mRNA and to process its 5′ and 3′ ends, whereas multiple spliceosomes are necessary to remove introns from the pre-mRNA body. Nascent RNA is mostly spliced during transcription elongation3. The number of spliceosomes that simultaneously act on a given transcript is unknown. However, we can infer that RNA sequence and secondary structure continuously change with the progress of both transcription and splicing.
The recent observation that splicing is often completed as soon as the intron emerges from Pol II4, 5 invites us to consider mechanisms that can affect co-transcriptional splicing efficiency. In this Review, we discuss the minimal distances along the pre-mRNA that are required for each spliceosome assembly stage, and how transcription elongation dynamics and RNA folding may influence the identification of the introns and splicing catalysis. We evaluate how other co-transcriptional events — 5′ end capping, RNA modifications and 3′ end processing — assist in intron and exon identification. Although the spliceosome and splicing catalysis are highly conserved, higher eukaryotes have degenerate splice sites and a larger, more complex pool of splicing factors. This enables variable splicing outcomes, including frequent alternative splicing. We illustrate how the core transcription and processing machineries can be modulated to obtain different splicing outcomes. Importantly, splicing stimulates other processes — most notably, transcription itself6–13 — suggesting that the spliceosome has other direct or indirect activities beyond intron removal. We propose that the intricate coordination between DNA, RNA sequence and structure, and the transcription and processing machineries ensures highly efficient and regulated co-transcriptional mRNA processing, and that this coordination may be achieved partly by concentrating all of the involved components in subnuclear membrane-less compartments.
Gene architecture and pre-mRNA splicing
The transcription start site (TSS) marks the 5′ end of the first exon, and the poly(A) site (PAS) marks the 3′ end of the last exon (FIG. 1a). Exon–intron organization provides important additional landmarks for the alignment of signals and activities, such as Pol II density, chromatin modifications, and RNA sequence and structure elements. Mammalian genes are longer than yeast genes, primarily because they contain more and longer introns14. Nevertheless, several aspects of gene architecture are conserved from yeast to humans. For example, the last exon is almost always the longest. Moreover, intron structure is similar across evolution: the GU and AG dinucleotides, which are contained in short and conserved sequences known as splice sites, define the 5′ and 3′ intron boundaries, respectively (FIG. 1a). A third sequence, the branchpoint sequence (BPS; see below), is 18–40 nucleotides upstream of the 3′ splice site (3′SS)15–18. In metazoans, the sequence between the BPS and the 3′SS contains a polypyrimidine tract19, which helps to identify the 3′SS during spliceosome assembly2. Despite the nucleotide sequence conservation of these sites, intron ends and their differential usage are difficult to define computationally20. Therefore, most intron ends are annotated from empirical data, such as cDNA sequencing.
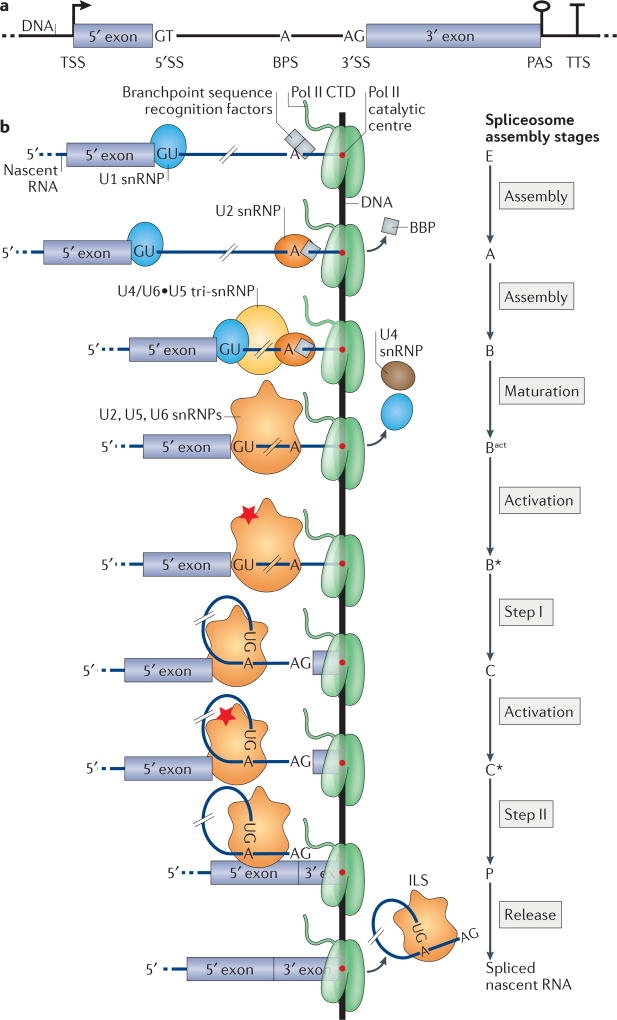
Figure 1. Yeast gene architecture and co-transcriptional spliceosome assembly.
a | Typical architecture of a budding yeast gene that contains one intron. The transcription start site (TSS), poly(A) site (PAS) and transcription termination site (TTS) are shown; for simplicity, only one of each site is represented. b | Co-transcriptional recruitment of small nuclear ribonucleoproteins (snRNPs) and splicing. The recruitment of U1 snRNP and the yeast branchpoint sequence (BPS) recognition factors (branchpoint binding protein (BBP) and Mud2) results in complex E. U2 snRNP recruitment and the concomitant displacement of BBP results in complex A. Upon recruitment of the U4/U6•U5 tri-snRNP complex B is formed. The subsequent release of the U1 and U4 snRNPs converts complex B into mature Bact, which contains the U2, U5 and U6 snRNPs. Catalytic activation (red star) yields complex B*. Step I catalysis produces complex C, which contains the 5′ exon and the branched intron lariat-3′ exon. Step II, which is catalysed by activated complex C*, produces complex P, which contains the ligated 5′ exon-3′ exon and the excised intron lariat. Spliced nascent RNA and intron lariat spliceosome (ILS) are then released. The 3′ end of the nascent RNA lies in the catalytic centre of RNA polymerase II (Pol II). The hatch marks on each nascent RNA replace the much longer sequence of the intron between the 5′SS and the BPS. CTD, carboxy-terminal domain; SS, splice site.
Pre-mRNA splicing is a two-step transesterification reaction that has been extensively characterized in vitro2. The substrates for splicing catalysis are the 5′SS, the BPS and the 3′SS21, 22 (FIG. 1b). In the first step (step I), the 2′OH of the BPS adenosine carries out a nucleophilic attack on the 5′SS guanosine, yielding a 5′ exon with a free 3′OH and a branched intron lariat that is attached to the 3′ exon. In the second step (step II), the 3′OH of the 5′ exon attacks the first nucleotide downstream of the 3′SS guanosine, releasing ligated 5′ exon-3′ exon and the excised intron lariat. How this chemistry is accomplished in the context of the growing nascent RNA polymer can be considered in light of recent data on co-transcriptional splicing and spliceosome assembly.
Spliceosome assembly on nascent RNA
The building blocks for spliceosome assembly include five U-rich small nuclear ribonucleoproteins (snRNPs), which are named after their small nuclear RNA (snRNA) component: U1, U2, U4, U5 and U6. The snRNPs coordinate dynamic base pairing between the different snRNAs and between snRNAs and the pre-mRNA to obtain secondary and tertiary RNA structures that define the spliceosome catalytic centre. Protein complexes such as the nineteen complex (NTC), nineteen-related (NTR), retention and splicing complex (RES) and other non-snRNP proteins participate in spliceosome assembly and function23. For a complete list of spliceosomal components, see REF. 24. Although splicing catalysis does not require ATP per se, essential ATPases assist the conformational transitions during assembly by establishing and rearranging RNA-RNA, RNA-protein and protein–protein interactions25. The current view is that the spliceosome positions elements within the pre-mRNA, including 5′SSs and 3′SSs, for the splicing reaction.
Co-transcriptional spliceosome assembly
In vitro splicing in cellular and nuclear extracts has revealed how snRNPs associate and are released from pre-synthesized pre-mRNAs. This has led to the formulation of a spliceosome assembly model in which the spliceosome transitions through sequential assembly stages2, 23, 25. In this model, the first stages of spliceosome assembly involve the identification of the 5′ and 3′ ends of the intron, followed by spliceosome maturation, spliceosome activation and splicing catalysis (FIG. 1b). Spliced RNA is then released, the spliceosome is disassembled and recycled, and the intron lariat is debranched and degraded2, 23. Spliceosome components and assembly are generally conserved in eukaryotes. The ~90 core proteins are conserved between the budding yeast Saccharomyces cerevisiae and human spliceosomes, although the number of human spliceosome proteins (~175) is twofold higher compared with yeast26. Because alternative splicing is extremely rare in budding yeast, this organism has been widely used as a model for identifying the basic principles of spliceosome function and assembly. Although this Review is guided by findings in budding yeast, we highlight numerous roles of metazoan proteins in these processes.
Chromatin immunoprecipitation (ChIP)-based experiments showed that spliceosomal components accumulate around splice sites following their transcription (Supplementary information S1 (box)). For example, U1 snRNP ChIP signals increase immediately downstream of the exon–intron boundary27–30. This in vivo observation supports the stepwise spliceosome assembly model that was derived from biochemistry (FIG. 1b). Studies in multiple laboratories and in many species indicate that splicing is mainly co-transcriptional3. Recently, single-molecule sequencing of nascent RNA from yeast showed that splicing catalysis occurs when Pol II has transcribed 26–129 nucleotides downstream of the 3′SS4, 5. These data suggest that spliceosome assembly, splicing catalysis and transcription elongation occur at similar rates. In such a model, the progress of transcription and spliceosome assembly are interwoven, raising the question of how many nucleotides downstream of the 5′SS, BPS and 3′SS need to be transcribed for spliceosome assembly and catalysis.
Commitment to splicing
The first step of spliceosome assembly is the formation of commitment complex E (FIG. 1b). The U1 snRNP binds to the 5′SS, and splicing factor 1 (SF1) and U2 auxiliary factor (U2AF) establish RNA–protein interactions with the BPS and the poly-pyrimidine tract at the 3′ end of the intron. Mammalian U2AF65 (also known as U2AF2) binds to Pol II in the early transcription stages and is delivered to nascent RNA as soon as a few nucleotides have emerged from the enzyme31. In vivo ultraviolet (UV) crosslinking experiments have shown that the binding of yeast U1 snRNP protein components is highest ~17 nucleotides downstream of the 5′SS32. This suggests that U1 snRNP contacts additional nucleotides around the 5′SS via RNA–protein interactions, in addition to base pairing its snRNA with the 5′SS23. The UV crosslinking signal of the branchpoint binding protein (BBP; the yeast orthologue of SF1) peaks on the BPS, whereas Mud2 (the yeast homologue of U2AF65) crosslinks along the entire intron32. These observations suggest that the transcription of the 5′SS, BPS and the few downstream nucleotides is necessary to trigger spliceosome assembly in yeast (FIG. 1b). Gene-specific distances between the 5′SS and the BPS, and thus the different time intervals between the emergence of the BPS from Pol II relative to the 5′SS, could influence the kinetics of commitment complex E formation to different degrees.
Assembly of a catalytically active spliceosome
Complex E is converted into the pre-spliceosome (complex A) once BBP dissociates and U2 snRNA base pairs with the BPS. The BPS adenosine bulges out of the BPS– U2 duplex and will carry out the nucleophilic attack on the 5′SS during step I. Recruitment of the tri-snRNP that contains U4, U6 and U5 results in complex B formation33–36 (FIG. 1b). Conformational rearrangements of RNA–RNA and RNA–protein interactions, the recruitment or stabilization of protein complexes, such as NTC, NTR and RES, and the release of U1 and U4 snRNPs result in spliceosome maturation into the activated complex Bact (REF. 26) (FIG. 1b). In vitro studies have shown that yeast Bact assembles on pre-mRNAs that lack the 3′SS, indicating that only the BPS is required for this assembly stage26, 37, 38. Bact contains the catalytic centre that is formed by the interactions between U2–U6 and the pre-mRNA, but the spliceosome is not catalytically active39, 40. UV crosslinking of purified yeast Bact complex revealed that the SF3a and SF3b complexes of the U2 snRNP directly contact the pre-mRNA upstream and downstream of the BPS37, 41. Recent cryo-electron microscopy (cryo-EM) structures of yeast Bact have shown that the BPS is occluded by components of SF3b and that it is kept ~50 Å away from the catalytic centre39, 40. The ATPase Prp2 triggers the conversion of Bact into the catalytically active complex B* by destabilizing SF3a and SF3b, thereby exposing the BPS adenosine37, 40. A stretch of 23–33 nucleotides downstream of the BPS is necessary and sufficient for Prp2 action37 (TABLE 1). Conversion into B* and step I catalysis do not require the 3′SS AG dinucleotide in yeast or in mammalian extracts37, 38, 42. In a co-transcriptional context, these observations suggest that step I catalysis can, in principle, take place as soon as a short stretch of ~30 nucleotides downstream of BPS emerges from Pol II. The presence of the 3′SS in this stretch of nucleotides is not necessary for step I.
Table 1.
Estimation of spatial constraints on RNA processing
| Process | Processing substep | Location on pre-mRNA | Minimal distance from processing site (nt)* |
Experimental system |
Organism | Refs |
|---|---|---|---|---|---|---|
| 5′ end capping | NA | 5′ end | ~20 | In vivo | Budding yeast | 53, 127 |
| RNA editing | NA | Gene specific | <60 | In vivo | Mammals | 109 |
| Splicing | Step I | BPS | <40 | In vitro | Budding yeast | 37 |
| Step II | 3′SS | <40 | In vivo | Budding yeast | 4 | |
| mRNA release | Splice junction | ~20 | In vitro | Budding yeast | 45 | |
| 3′ end cleavage | NA | PAS | <200 | In vivo | Budding yeast | 186, 187 |
BPS, branchpoint sequence; NA, not applicable; nt, nucleotides; PAS, poly(A) site; SS, splice site.
Distances along the pre-mRNA required to start RNA processing. In vivo distance values reflect the amount of transcribed RNA required for the listed processing event; in vitro distance values refer to minimal RNA lengths required by ATPases to trigger these reactions.
Rearrangements preceding step II
Complex C, which is formed following step I catalysis, contains the 5′ exon and intron lariat-3′ exon intermediates (FIG. 1b). The U5 snRNA base pairs with the 5′ exon, thereby retaining it in the active site. The U6 snRNA interacts with the branched 5′SS, and the U2 snRNA immobilizes the intron lariat43, 44. UV crosslinking experiments have shown that, upon step I catalysis, the major U5 snRNP protein Prp8 binds to the pre-mRNA downstream of the BPS, near the 3′SS, implying that further conformational changes are necessary for the subsequent step41. Complex C stimulates the ATPase activity of Prp16, which rearranges the catalytic site from step I to step II conformation, leading to the formation of activated complex C* (REFS 45, 46). Recent cryo-EM structures of this complex have revealed a major rearrangement of the branched adenosine and the BPS–U2 snRNA duplex, and have suggested that the vacated space caused by this reorganization in the catalytic centre contributes to 3′SS localization to the catalytic site47–49. The first AG dinucleotide encountered downstream of the BPS docks in the active site21, 50. This is known as the ‘first AG rule’, and is consistent with a co-transcriptional spliceosome assembly model, in which this AG dinucleotide is the first to be transcribed and, therefore, is favoured for recognition by the spliceosome. However, exceptions to the first AG rule have been observed. For example, if the dinucleotide is located closer than 10 nucleotides downstream of the BPS or if it is buried in secondary structures, then it is not efficiently recognized and is skipped51, 52. Alternative 3′SS selection is frequent in metazoans; how the spliceosome skips 3′SSs that are favourably positioned relative to the BPS is unclear.
Step II catalysis and post-splicing dynamics
Step II catalysis, in which the 3′OH of the 5′ exon attacks the first residue of the 3′ exon, produces the post-spliceosome (complex P), which is associated to the ligated 5′ exon–3′ exon nascent RNA and the excised intron lariat (FIG. 1b). In yeast, the earliest step II products have been observed when Pol II transcribes <40 nucleotides downstream of the 3′SS4 (TABLE 1). In light of possible crosstalk between Pol II and the spliceosome, it is interesting to consider how much RNA is protected by Pol II and the spliceosome and thus how close the two machines may be during co-transcriptional splicing. On the one hand, 15 nucleotides of nascent RNA are buried in the Pol II exit channel53. On the other hand, ~20 nucleotides downstream of the 3′SS are likely embedded in the spliceosome, because Prp22 (an ATPase with splicing fidelity and disassembly functions) requires a segment of 13–23 nucleotides downstream of the splice junction45. The sum of these nucleotides (~35) and recent measurements of Pol II position at the completion of step II (<40 nt downstream) highlight the close proximity of the spliceosome to Pol II (FIG. 1b) (TABLE 1). The observation that Prp22 can be crosslinked by UV irradiation to the 3′ exon downstream of the splice junction only after step II completion45 suggests that further transcription is necessary for mRNA release (FIG. 1b). Finally, the ATPase Prp43 disassembles the intron lariat spliceosome (ILS) into the intron lariat and the spliceosomal components, which are then recycled for further rounds of splicing54.
Transcription and splicing interactions
Spliceosome assembly and catalysis require the recognition of the splice sites and BPS on the nascent transcript. How is this achieved? Two main aspects define the co-transcriptional selection of splice sites and the BPS. First, only a subset of these sequences is present on the nascent RNA at a given time because of ongoing transcription. This restricts the choice of sites available to the spliceosome. Transcription rate determines the portion of the transcript that is available for inspection by the spliceosome, such that faster transcription rates may provide longer stretches of nascent RNA sequence. Interestingly, Pol II elongation rates are faster over introns than over exons55, 56. Second, both DNA and RNA undergo different folding states and modifications, and they exhibit dynamic protein-binding profiles that influence transcription rate, accessibility of splice sites and BPS, as well as splicing factor recruitment. Local differences in post-translational modifications (PTMs) of the Pol II carboxy-terminal domain (CTD) and chromatin environment influence transcription rates and nascent RNA processing dynamics. For example, the histone 2A variant H2A.Z promotes efficient pre-mRNA splicing of introns with non-consensus splice sites in both budding and fission yeast57, 58. The extensive crosstalk between splicing, transcription and other nuclear machineries can be appreciated by considering the multitude of reported genetic and physical interactions between them, which we summarize in FIG. 2 (see also Supplementary information S2 (table)).
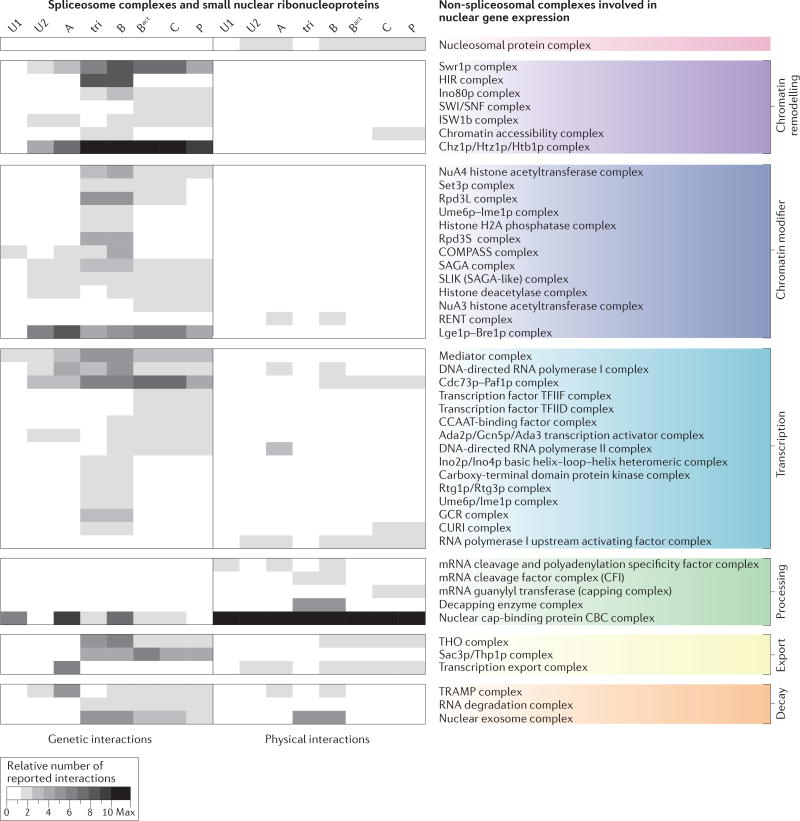
Figure 2. Crosstalk of the assembling spliceosome with nuclear gene expression machineries.
The crosstalk of the components of the small nuclear ribonucleoproteins (snRNPs) and the different spliceosome assembly stages with nuclear gene expression factors and complexes is underlined by a multitude of genetic and physical interactions. Genetic and physical interactions that involve core splicing factors of Saccharomyces cerevisiae were obtained from the Biological General Repository for Interaction Datasets (BioGRID). Protein complex annotations were derived from the CYC2008 yeast proteins catalogue182 and the Spliceosome Database24. Only predominantly nuclear complexes that are involved in chromatin biology, transcription and RNA-related nuclear processes were considered183 (Supplementary information S2 (table)). The grey scale reflects the number of reported interactions between spliceosomal and non-spliceosomal complex subunits. The number of reported interactions is adjusted to the number of reported non-spliceosomal complex subunits. A minimum of two reports for the same interaction was required. Overall, chromatin-modifying and chromatin-remodelling complexes display predominantly genetic interactions. Fewer reports exist of physical interactions, with the exception of core spliceosomal complexes (Supplementary information S2 (table)) and the 5′ and 3′ end processing machineries. In line with mechanistic studies (see the main text), specific genetic interactions have been reported between the cap-binding complex (CBC) and some spliceosomal complexes, but an extensive physical interaction network (possibly mediated through the nascent RNA) has been mapped with all spliceosomal complexes. The full non-spliceosomal complex and protein names are given in Supplementary information S3 (table). Tri, tri-snRNP; U1, U1 snRNP; U2, U2 snRNP.
Phosphorylation dynamics of the Pol II CTD
Changes in PTMs of the Pol II CTD mirror and influence the different phases of transcription and nascent RNA processing, owing to the interaction of the CTD with factors that regulate transcription, mRNA processing and downstream steps such as mRNA export11, 59, 60. CTD-modifying enzymes often have additional cellular targets, thereby integrating the CTD into a greater network of gene expression59. The CTD consists of repeats of almost the same seven amino acids Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (26 repeats in yeast and 52 in humans61, 62), which are mainly modified by phosphorylation of Ser2, Ser5, Ser7, Thr4 and Tyr1 (REFS 63–65) (FIG. 3a). Pol II is differentially modified at the start and the end of transcription units. In budding yeast, Ser5 and Ser7 phosphorylation (Ser5P and Ser7P) occurs early, during transcription initiation and early elongation along the first exon, whereas Ser2, Thr4 and Tyr1 phosphorylation occurs later, during the transcription of the second exon and transcription termination (FIG. 3a). PTM transitions have recently been mapped to transcription pause positions along yeast gene bodies and, in particular, to 3′SSs, consistent with changes in Pol II elongation rate around intron–exon boundaries66, 67. In well-spliced yeast genes, the relative abundance of ‘early’ CTD PTMs decreases over the intron, whereas ‘late’ PTMs begin to increase at the 3′SS (FIG. 3b).
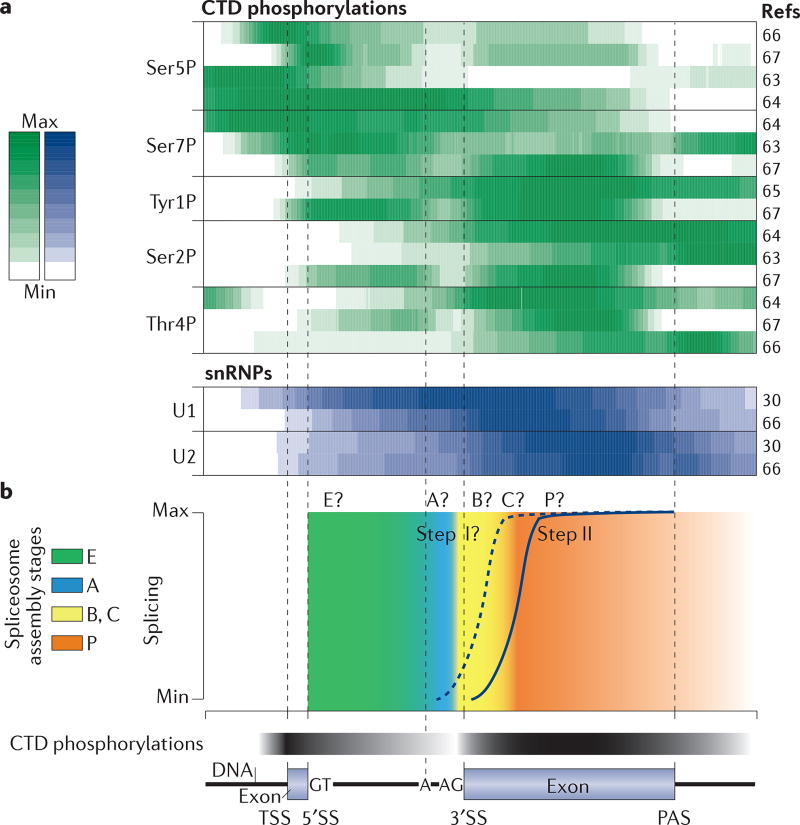
Figure 3. Patterns of RNA polymerase II C-terminal domain phosphorylation, small nuclear ribonucleoprotein binding and splicing along an average intron-containing budding yeast gene.
a | Comparison of RNA polymerase II (Pol II) carboxy-terminal domain (CTD) phosphorylation profiles and small nuclear ribonucleoprotein (snRNP) binding profiles from different studies. Heatmap of average CTD phosphorylation profiles (top) normalized to total Pol II profiles (the total Pol II profile was not available for REF. 66) for the 50% of intron-containing genes with the highest snRNP signal over terminal exons (snRNP data are from REF. 30 and Pol II CTD data are from REFS 63–67). In most data sets, Ser5 and Ser7 phosphorylation is most abundant in the first exon and in the intron (most budding yeast genes contain one intron), whereas phosphorylation of the other CTD repeat residues is high over the terminal exon and/or the poly(A) site (PAS), pointing to a transition in CTD phosphorylation profiles around the 3′ splice sites (3′SSs). Note that the data sets differ in experimental procedure (chromatin immunoprecipitation (ChIP)–chip, ChIP-Nexus, ultraviolet (UV) crosslinking and analysis of cDNAs (CRAC)) and the antibodies used. A heatmap of average U1 and U2 snRNPs binding profiles (data are from REFS 30, 66) (bottom) for the same intron-containing genes illustrates stepwise co-transcriptional spliceosome assembly. b | Schematic of step II splicing kinetics in yeast4 and hypothetical step I splicing kinetics. The kinetics of the spliceosome assembly stages — the transition from B complexes (including B, Bact and B*) to C complexes during step I, and the transition from C complexes (including C and C*) to P during step II — have not yet been determined. Hypothetical splicing assembly stage transitions and global CTD phosphorylation changes are indicated by the colour gradient. A, branch adenosine; TSS, transcription start site.
In addition to the Pol II CTD, the entire transcription elongation machinery and the nascent RNA itself interact with proteins, forming RNP complexes. Many of these proteins belong to complexes that are involved in mRNA 5′ end capping, splicing, 3′ end processing, editing, folding, nuclear export and decay, and bind to specific transcript regions, such as untranslated regions, introns and exons32, 68, 69. For example, in higher eukaryotes, tetrameric heterogeneous nuclear ribonucleo-protein C (hnRNPC) binds to introns in nascent RNA and organizes long intronic sequences for splicing70. During intron transcription, U1 snRNP is recruited to the 5′SS, and U2 snRNP levels begin to increase at the 3′SS, paralleling the transition from early to late CTD PTMs32 (FIG. 3a). Nevertheless, the relationship between specific Pol II CTD PTM profiles and nascent RNA processing events is far from being understood. Some profiles are different between species, such as that of Tyr1P between yeast and humans60, 65, 71, or between different studies in the same species (FIG. 3a). Combinations of different PTMs can occur on the same CTD, albeit rarely within the same repeat72, 73. Finally, even moderate increases in PTM levels over the gene body may be important for gene expression regulation. For example, although the Ser5P levels are highest at the beginning of transcription units, a link between this PTM and splicing has been found in both yeast and humans66, 74. Pronounced peaks of Ser5P and Pol II levels are observed at the 5′SSs of alternatively included exons compared with excluded exons74, 75.
The characterization of isolated RNP complexes associated with specific CTD PTM profiles helps to define the dynamic nascent RNP interactome66. In yeast, all interactomes of the phosphorylated residues, except that of Thr4P, are enriched for splicing factors, which is consistent with the general trend of Thr4P increasing towards gene ends64, 66 (FIG. 3a). The combination of interactome analyses with analyses of the activity of these transient macromolecular assemblies will be crucial for the study of co-transcriptional processes, including spliceosome assembly, activity and disassembly (FIG. 3b; see Supplementary information S1 (box)). Data integration using computational modelling approaches, such as machine learning, will help to rationalize Pol II CTD phosphorylation patterns and to identify gene regions that are important for successful transcription and RNA processing, in addition to the known gene landmarks such as the splice sites. This will hopefully disentangle the causes and the consequences of different CTD profiles along genes67.
Transcription rates are affected by gene architecture and chromatin features
The transcription rate can influence splice site identification by the spliceosome. Current models suggest that different local rates of transcription elongation can influence the time frame between the synthesis of sequential splice sites, thereby possibly modulating RNA folding or the interactions with RNA-binding proteins10. The synthesis of RNA by Pol II occurs with an average elongation rate of 1–4 kb per minute55, 56, 76. However, Pol II can transiently pause, stall or terminate prematurely55, 56. Pol II elongation rate is influenced by a multitude of factors, such as the underlying DNA sequence, nucleosome position and histone modifications, which affect local chromatin structure, the activity of elongation factors, and the folding and processing of the nascent RNA55, 77. For example, the balance between histone acetylation and methylation in neuronal cells is a determinant of transcription rate and associated splicing patterns78.
The positioning of nucleosomes relative to TSSs, transcription termination sites (TTSs), exons and introns helps to define gene architecture79–82 (FIG. 4a). Conversely, nucleosome phasing seems to be facilitated by the inherent exon–intron primary structure (such as elevated GC content in exons), by the sequence elements that are required for pre-mRNA splicing79, 80, 82 and by the local activity of chromatin remodellers80, 83. Intriguingly, the median length of internal exons in the human genome is 137 bp, which is very close to the 147 bp that are wrapped around a nucleosome14 (FIG. 4a). The correlation between the length of internal exons and the nucleosomal DNA could be explained by the observation that evolutionarily recent intron generation events in the alga Micromonas pusilla occurred upon the preferential integration of DNA transposons into nucleosome spacer regions81.
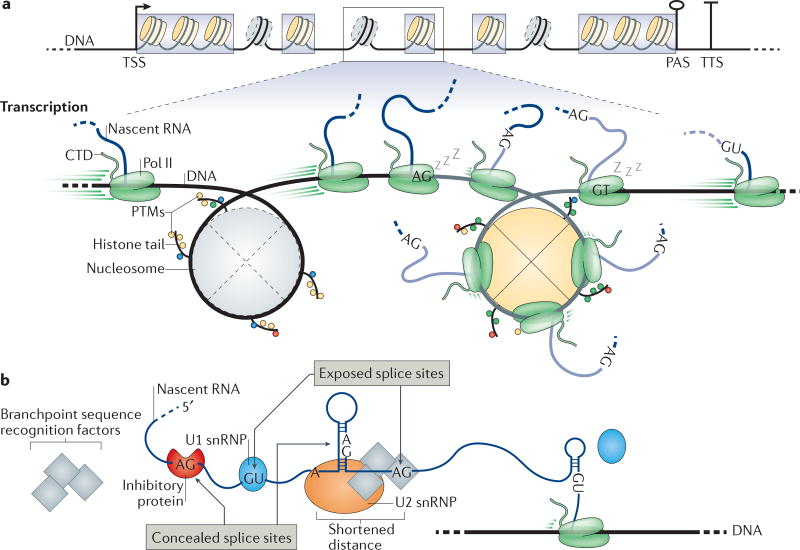
Figure 4. Gene architecture, chromatin features and nascent RNA properties influence co-transcriptional splicing.
a | The length of typical internal exons (grey boxes) is comparable to the DNA that is wrapped around a nucleosome. Nucleosome positioning relative to the transcription start site (TSS), transcription termination site (TTS) and, to a lesser extent, exons helps to define the boundaries of these elements, providing a platform for crosstalk between chromatin, transcription and splicing. Less stable nucleosomes at introns are indicated with dashed outlines. For simplicity, only one TSS, poly(A) site (PAS) and TTS are depicted. The zoomed-in section shows that RNA polymerase II (Pol II) transcription rates change along introns (black lines with grey nucleosomes) and exons (grey lines with yellow nucleosomes) from high rates to low rates. A sleeping Pol II represents pausing events at splice sites (AG and GT). Post-translational modifications (PTMs) on histone tails influence transcription and splicing. b | RNA secondary structures and RNA-binding proteins can modulate the availability of splice sites and branchpoint sequences. The splicing machinery cannot identify sites that are concealed in secondary structures or that are bound by inhibitory proteins. Pol II transcription rate and local RNA folding contribute to site accessibility. CTD, carboxy-terminal domain; snRNP, small nuclear ribonucleoprotein.
Nucleosomes can interfere with transcription progression67, 84–87. Consistent with nucleosome phasing over exons, slower transcription elongation has been measured over exonic sequences75, 86, 88. This is suggested by high Pol II density close to the nucleosome centre in all genes67, 85. The histone tails of nucleosomes that are positioned over exons can be enriched for PTMs that may facilitate exon definition by affecting the recruitment of splicing factors, as well as the transcription process itself8, 89–92 (FIG. 2). Slow passing of the transcription machinery through nucleosomes may facilitate the relocation of splicing factors and regulators from the chromatin template to Pol II or to the nascent RNA.
Splicing-dependent Pol II pausing
Recent studies have implicated the splicing process in transcriptional pausing. For example, pausing at terminal exons was detected in efficiently spliced genes in yeast93. Upon splicing inhibition, either by introducing a temperature-sensitive mutant allele of the RNA helicase Prp5, or by introducing mutations in the BPS or the U2 snRNA, the Pol II ChIP signal increases on introns, suggesting the activation of a transcription elongation checkpoint to allow spliceosome assembly94. Discrete pauses have been mapped to 5′SSs and 3′SSs66, 75, 95 (FIG. 4a). How does Pol II pausing specifically relate to intron-exon boundary identification and spliceosome assembly? The extent to which splicing-related pausing occurs and a mechanistic understanding of this process are still elusive. When Pol II pauses at the 3′SS, the intron 3′ end, including the polypyrimidine tract, is still in its catalytic centre or exit channel and is not yet available for splicing31. How does pausing at the 3′SS relate to spliceosome assembly? The distance between the BPS and the 3′SS in most introns might allow assembly to proceed during pausing. Pol II pause release can be aided by transcription elongation factors, such as transcription factor IIS (TFIIS) 85. In addition, splicing factors may stimulate transcription progress. Interestingly, the shift of U2AF65 from Pol II to the nascent RNA was suggested to stimulate transcription elongation31, 96. Explaining the prevalence and functional importance of Pol II pausing around 3′SSs and the general change of Pol II elongation rate over exons is crucial for understanding how transcription elongation and nascent RNA splicing are intertwined12. The development of new single- nucleotide-resolution single-molecule sequencing strategies that monitor splicing intermediates offers promising new avenues of investigation74, 85 (Supplementary information S1 (box)).
Nascent RNA folding and RNA modifications influence splicing
Nascent RNA folds into secondary structures that affect its subsequent processing. The propensity for RNA folding directly depends on sequence-specific folding rates, transcription elongation rates and the rate of proteins binding to the RNA97, 98 (FIG. 4b). RNA secondary structures can conceal or expose the 5′SSs, BPSs and 3′SSs, which are consequently ignored or readily recognized by the splicing machinery99, 100. By concealing or exposing alternative cis-acting elements, secondary structures may have a role in alternative splicing52 (FIG. 4b). The splicing machinery may recognize splicing targets on nascent RNA before the RNA has time to fold into a secondary structure100. Similarly, RNA-binding proteins may influence the transient folding of nascent RNA and, therefore, may modulate the timing of splice site exposure to the splicing machinery99. For example, hairpins with a small loop readily fold after transcription, thereby concealing the splice site that is contained in their stem. By contrast, the folding of hairpins with bigger loops takes longer, allowing longer splice site exposure to the splicing machinery and/or to regulatory RNA-binding proteins101.
Changes in transcription elongation rates influence nascent RNA folding and so may affect splice site selection97, 100. Intramolecular hairpins that sequester the 5′SS, BPS or 3′SS inhibit splicing both in vitro and in vivo101–103 (FIG. 4b). The inhibitory effects increase with hairpin stability 102. Conversely, secondary structures can increase the recognition efficiency of true splice sites by masking cryptic ones51, 52 (FIG. 4b). Interestingly, a systematic analysis in yeast showed that true 3′SSs are usually more accessible than cryptic ones52. Finally, secondary structures that form between the 5′SS and the BPS or between the BPS and the 3′SS without directly sequestering them may positively influence splicing by shortening the physical distance between these cis-acting elements and thus may help the spliceosome to bridge them52, 104 (FIG. 4b).
Modifications that alter the chemical properties of RNA, such as base modifications and substitutions, may also affect splicing by altering local RNA secondary structures or the binding sites for the splicing machinery105. Nucleotide substitution through RNA editing is widespread in metazoa106, 107 and occurs co-transcriptionally108, 109. Nucleotides can be edited as close as ~55 nucleotides upstream of the 3′ end of nascent RNAs, in the Pol II catalytic centre, suggesting that the introduction of these modifications may be very rapid109 (TABLE 1). All 12 types of nucleotide editing have been identified on endogenous transcripts107, 109. A-to-I (adenosine to inosine, which is recognized as a guanosine by cellular machineries) is the most abundant modification107 and is catalysed by adenosine deaminase acting on RNA (ADAR). ADAR can act on double-stranded RNA substrates that form through base pairing between exonic and intronic sequences, pointing towards coupling between editing and splicing110. It is possible that the splicing of these introns is delayed owing to occupancy of the 5′SS by ADAR. ADAR2 directly alters the splicing of its own pre-mRNA by introducing an alternative 3′SS. Moreover, the Pol II CTD is required for efficient editing111. The glutamate receptor subunit B (GluRB; also known as GluR2) pre-mRNA contains edited sites that are close to the 5′SSs112. In vitro, the editing and splicing of these sites on pre-synthesized GluRB pre-mRNA are mutually exclusive. However, such mutual exclusion is not observed in vivo, suggesting that transcription coordinates the editing and splicing machineries that act on these sites to ensure that both can take place112. Transient local RNA folding is likely to have a role in this coordination. Thus, changes in secondary structures that are induced upon RNA editing may conceal or expose the splice sites, resulting in alternative splicing113.
Strategies for splice site identification
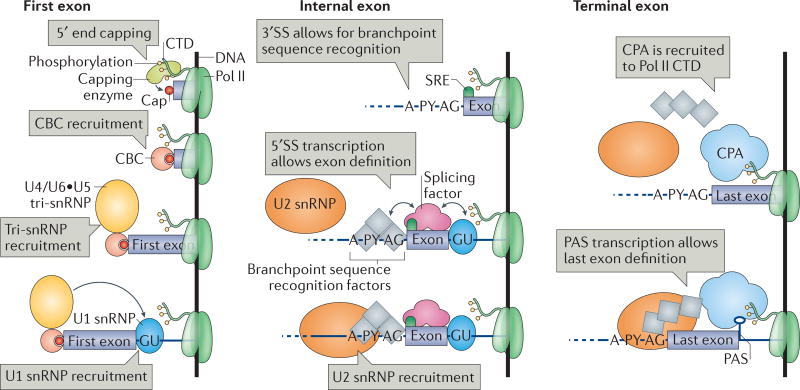
The complexity of gene architecture varies between phyla114, 115, requiring different mechanisms to identify splice sites. Metazoan splice sites are short and poorly conserved, in contrast to budding yeast splice sites23. This implies that the spliceosome might encounter frequent ‘incorrect’ splice sites along the transcripts, or at least a multiplicity of splice sites from which to choose116. The high complexity of metazoan gene architecture requires strategies to identify bonafide splice sites and lends itself to various means of pre-mRNA splicing regulation, including alternative splicing116. In metazoa, accurate splice site selection depends on short conserved sequences, known as splicing regulatory elements (SREs), that reside in introns or exons117, 118. Regulatory proteins, such as SR proteins and hnRNPs, specifically bind to SREs and influence splice site recognition and/or spliceosome assembly116. Productive or unproductive interactions between SRE-binding proteins and the spliceosomal machinery generally depend on the location of the SREs relative to the splice sites, underscoring a large regulatory potential116 (FIG. 5).
Figure 5. First, internal and terminal exon definition.
As a prerequisite to first exon definition, the capping enzyme that is bound to the phosphorylated RNA polymerase II (Pol II) carboxy-terminal domain (CTD) adds a cap to the 5′ end of the nascent RNA. The cap-binding complex (CBC) recruits the U4/U6•U5 tri-small nuclear ribonucleoprotein (snRNP) and mediates the association of the U1 snRNP with the first 5′ splice site (SS). In internal exon definition, the transcription of an internal 3′SS and the downstream 5′SS triggers the recruitment of the U1 snRNP, the branchpoint sequence recognition factors (splicing factor 1 (SF1), U2AF65 and U2AF35) and the U2 snRNP. Splicing factors facilitate or inhibit exon definition by binding to splicing regulatory elements (SREs), leading to alternative splicing. As a prerequisite to terminal exon definition, the cleavage and polyadenylation complex (CPA) interacts with the poly(A) site (PAS), phosphorylated Pol II CTD and the splicing machinery, aiding 3′SS identification. In addition, the splicing machinery helps CPA recruitment onto the PAS. A, branch adenosine; P Y, polypyrimidine tract.
Intron and exon definition
How are introns and exons recognized by the spliceosome? In vertebrates, intron length varies from a few hundred nucleotides to several thousand nucleotides, and the median length of internal exons is approximately 137 nucleotides114. Surrounding the internal exon, the 3′SS of the upstream intron and the 5′SS of the downstream intron pair across the exon, thereby committing the upstream intron to splicing through an ‘exon definition’ mechanism119, 120 (FIG. 5). By contrast, transcripts in lower eukaryotes usually contain introns that are shorter than 250 nucleotides (REF. 114). In this case, a 5′SS pairs with the downstream 3′SS of the same intron, and splicing is triggered through an ‘intron definition’ mechanism121 (FIG. 1). In exon definition, a splice site mutation causes exon skipping, whereas in intron definition it causes intron retention121, 122. Thus, exon and intron definition mechanisms explain the different phenotypes of single splice site inactivation in vertebrates and in lower eukaryotes119.
Intron definition fits with the stepwise co-transcriptional spliceosome assembly model discussed above. In vivo, yeast complexes E and A assemble onto the 5′SS and BPS32, suggesting that the BPS rather than the 3′SS is required for splicing commitment (FIG. 1b). Indeed, budding yeast lack an obvious homologue of U2AF35, which binds to the 3′SS AG dinucleotide in the mammalian complex E. In vitro, transcripts with one short intron are also spliced upon intron definition119, and the AG dinucleotide is not necessary for step I catalysis38, 42. These observations suggest that transcription of the 5′SS and the BPS may be sufficient for splicing commitment following intron definition (FIG. 1b). In higher eukaryotes, in which splicing is triggered by exon definition, U2AF35 bound to the 3′SS participates in splicing commitment together with the U1 snRNP bound to the downstream 5′SS23, 119 (FIG. 5). This inverted order of identification of intronic elements may partly account for the slower splicing rates that are found in higher eukaryotes5.
Nascent RNA 5′ end capping and the definition of the first exon
Splicing of the first intron depends on first exon definition119. First exon boundaries consist of the 7-methylguanosine (m7G) cap structure at the 5′ end of the transcript and the 5′SS of the first intron (FIG. 5). The capping enzyme adds the m7G cap to the 5′ end of all Pol II-transcribed RNAs when the nascent RNA is less than 20 nucleotides in length53, 123–127 (TABLE 1). The nuclear cap-binding complex (CBC) serves as a platform for interacting with factors that are involved in RNA processing128. In vitro, CBC enhances the splicing of the first intron, suggesting that it contributes to first exon definition119, 123, 129. In vivo, CBC directly interacts with tri-snRNP protein components and its depletion impairs both U1 snRNP and tri-snRNP recruitment to the pre-mRNA28, 130, 131. Genetic interactions were observed between the yeast CBC, U1 snRNP proteins and BBP, suggesting that CBC functions in the formation of complex E132 (FIG. 2). Tri-snRNP recruitment by CBC was proposed to mediate U1 snRNP association and the consequent identification of the first 5′SS130. Nevertheless, all spliceosomal complexes physically interact to some degree with the CBC, either directly or indirectly through the nascent RNA (FIG. 2).
Nascent RNA 3′ end processing and the definition of the last exon
Splicing of the last intron depends on terminal exon definition119. The 3′SS of the last intron and the PAS set the terminal exon boundaries133 (FIG. 5). The PAS, the nearby AU-rich sequences and other cis-elements on the nascent RNA are bound by the cleavage and polyadenyl ation complex (CPA). It is not currently known how far transcription proceeds past the PAS before 3′ end cleavage takes place (TABLE 1). However, the PAS is required for termination, and its elimination leads to transcription readthrough134–136. PAS elimination also results in the specific inhibition of last intron splicing, indicating that 3′ end processing contributes to terminal exon definition137, 138. The U2 snRNP, U2AF65 and cleavage and poly adenylation specificity factor (CPSF; a component of the CPA) functionally and physically interact (FIG. 2); this supports a model in which the PAS triggers the removal of the last intron by facilitating spliceosome assembly139–142. Splicing and the regulation of 3′ end processing is reciprocal, as the inactivation of the terminal 3′SS inhibits 3′ end processing and transcription termination137–139, 143, 144. The splicing and 3′ end processing machineries seem to serve as recruitment platforms, as the physical presence of the two machineries is sufficient for coupling between splicing and 3′ end processing, and neither catalytic activity is required for the regulation of the complementary process138, 139, 144, 145. Indeed, artificially induced cleavage of the nascent RNA impairs splicing and 3′ end processing in vitro138. In vivo, the Pol II CTD stimulates coupling between splicing and 3′ end processing146. Taken together, these observations suggest that coupling between splicing and 3′ end processing can determine whether splicing occurs before transcription termination142.
The molecular mechanisms of 3′ end processing involve components of the splicing machinery. The mammal ian U1 snRNP component U1 70k directly interacts with poly(A) polymerase, a component of the CPA, and inhibits polyadenylation147. In vivo, functional inhibition of U1 snRNP inhibits splicing and causes premature 3′ end processing148. Interestingly, U1 snRNP is much more abundant than all other spliceosomal snRNPs149, and this may reflect its role in protecting nascent RNA from premature 3′ end processing. In such a model, the U1 snRNP binds to nascent RNA at frequent cryptic 5′SSs and suppresses the activity of adjacent cryptic PASs148. The relative abundance of PASs and U1 binding sites on transcripts may modulate premature transcription termination. Indeed, upstream antisense transcripts are enriched in PASs but depleted of U1 recognition sites, and their expression is efficiently suppressed150. Changes in transcript and U1 snRNP abundance ratios result in changes in alternative PAS selection, and consequently modulate transcript length151. Overall, the PAS and CPA have a major role in defining the last exon and thus the 3′SS for last intron removal. In addition, splicing aids 3′ end processing by preventing the recruitment of the CPA to cryptic PASs.
3D organization of gene expression
The 3D chromatin conformation can enable crosstalk between different mRNA processing machineries. Similar to promoters that associate with distant enhancers152, gene ends may loop over long distances. Gene looping occurs when the transcription initiation and termination machineries juxtapose TSSs and TTSs153, 154. Gene looping may regulate transcriptional output and may provide promoter directionality by depositing termination factors close to the promoter, which can lead to the termination of upstream antisense transcripts155 (FIG. 6a). In yeast, introns can further enhance looping by bridging the 5′SS with the TSS and the 3′SS with the PAS156. It is tempting to speculate that looping may facilitate splice site usage in subsequent rounds of transcription by promoting the efficient recycling of splicing factors. It remains to be seen how widespread this phenomenon is and how spliceosome assembly and catalysis contribute to gene looping.
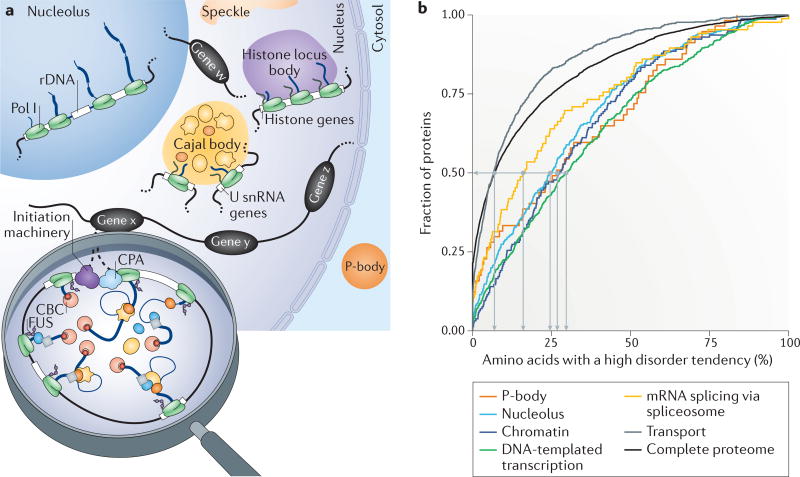
Figure 6. Higher-order organization of the gene expression machineries.
a | The nucleus and cytoplasm contain membrane-less compartments, known as bodies, such as the nucleolus, Cajal bodies, histon locus bodies, speckles and P-bodies. Such bodies form through liquid–liquid phase separation (LLPS) and are often linked to the transcription of specific genes, for example, ribosomal DNA (rDNA) in the nucleolus, small nuclear RNA (snRNA) genes in Cajal bodies and histone genes in histone locus bodies. We propose that looped and actively transcribed genes (genes w, x, y and z) are also likely to form nuclear bodies. b | Spliceosome proteins, particularly chromatin and transcription-associated proteins, are predicted to have a similar proportion of unstructured protein regions to those of other groups of proteins that are known to be involved in LLPS, as shown in the cumulative representation of the complete proteome and the protein groups associated with specific Gene Ontology terms (cellular component: P-body, nucleolus, chromatin (binding); biological process: DNA-templated transcription, mRNA splicing via spliceosome and transport). The data were downloaded from the Saccharomyces Genome Database (Supplementary information S3 (table)). The cumulative fraction of proteins (y axis) is given in association with the percentage of amino acids per protein that have a high probability of being in a disordered region (x axis), according to predictions by IUPred184,185. Whereas 50% of transport proteins and the entire proteome contain 7% or fewer amino acids with a high tendency to form disordered regions, 50% of the P-body, nucleolar or chromatin and transcription-associated proteins contain 25–30% of such amino acids. The median fraction of amino acid disorder tendency for spliceosomal proteins is 16%. Medians are represented by light grey lines. CBC, cap-binding complex; CPA, cleavage and polyadenylation complex; Pol I, RNA polymerase I.
Co-transcriptional splicing presupposes that spliceosome components are in the vicinity of the transcribed regions and are readily available for spliceosome assembly and splicing catalysis. Live cell- imaging experiments have measured diffusion constants in the order of 0.2–0.8 µm2 s−1 for snRNPs, as well as other RNPs157. This slow rate of diffusion, which is ~100 times slower than free diffusion in water158, has been attributed to transient interactions with binding partners such as pre-mRNA157. An alternative explanation is that splicing factors dwell at sites of transcription owing to liquid–liquid phase separation (LLPS). Such phase transitions are favoured by proteins with intrinsically disordered regions (IDRs) and RNA-binding domains159–163. Proteins with IDRs are found to be enriched in membrane-less compartments that are formed by LLPS, for example, SR proteins in nuclear speckles and coilin in Cajal bodies159. Approximately 18 polypeptides in the B complex harbour IDRs that are similar to SR proteins164. Interestingly, IDRs of several mammalian hnRNP isoforms are encoded by alternative exons; depending on exon inclusion, the resulting hnRNP isoform can promote multi- hnRNP assemblies that affect alternative splicing of target transcripts165. Members of the Fox1 family of RNA-binding proteins (Rbfox) can undergo higher-order interactions with the large assembly of splicing regulators (LASR) complex via a Tyr-rich low-complexity region, which is required for splicing regulation by Rbfox166. We suggest that spliceosomal components could be greatly concentrated at transcription sites as a consequence of LLPS (FIG. 6a). There are four reasons to suspect that this might be the case. First, snRNPs can reside in nuclear compartments that are formed by LLPS; they assemble from precursors and are highly concentrated in Cajal bodies167. Second, the Pol II CTD has been shown to undergo LLPS, owing to its low complexity and intrinsically disordered structure60, 168, and it was recently proposed that CTD LLPS may be important for transcription initiation60. Third, the low-complexity protein FUS mediates interactions between Pol II and the U1 snRNP169, 170, potentially promoting the recruitment of U1 to the first 5′SS. Fourth, yeast and human chromatin, transcription and spliceo-some proteins have a strong tendency to disorder, similar to proteins of P-bodies and nucleolar proteins that are known to undergo LLPS171 (FIG. 6b; see Supplementary information S3 (table)). Intriguingly, the visualization of lampbrush chromosomes reveals numerous active gene loops that take on droplet-like morphologies, suggesting the local occurrence of LLPS during gene expression172. Thus, LLPS may facilitate local high concentrations of spliceosomal components at transcription sites, thereby creating miniature nuclear bodies at each active gene (FIG. 6a); similar phase transition phenomena have been suggested to explain transcription regulation by super-enhancers163. This scenario may explain how unprocessed transcripts are retained at sites of transcription173–175 and how particular splicing isoforms predominate in a given cell or for a given gene176, 177. For example, the same pool of splicing regulators could be reused in multiple rounds of splicing. Spliceosome disassembly could also take place in this sequestered environment, while the mature messenger ribonucleoprotein (mRNP) is prepared for nuclear export.
Conclusions and perspectives
Coupling 5′ end capping, splicing and 3′ end processing to transcription ensures the formation of a fully processed mRNA that is ready for export and translation. In this Review, we rationalize how these individual steps are coordinated at the transcription unit. Delaying, preventing or enhancing splice site recognition may modulate splicing outcome, leading to mature mRNA isoforms with diverse cellular fates178–181. We highlight links between splicing outcome and transcription dynamics, nucleosome positioning and RNA folding. Spliceosome proteins interact with proteins that are involved in transcription and nuclear RNA processing, and extensive crosstalk and regulation are promoted by protein– protein interactions between different machineries (FIG. 2).
Recent cryo-EM structures have visualized spliceosome assembly complexes and have revealed new conformational details34–36, 39, 40, 43, 44, 47–49. The joint structural analysis of interfaces between splicing complexes and the transcription and processing machineries, and the use of high-resolution nascent RNA sequencing for the analysis of individual processing steps, have the potential to reveal new aspects of the coordination between these large nuclear machines. Finally, the precise quantification of local protein and RNA abundance and RNA– protein interactions, as well as the visualization of the dynamics along transcription units, will help in formulating new models of gene expression regulation, such as the recently invoked LLPS mechanism.
Supplementary Material
Acknowledgments
The authors are grateful to H. Herzel and K. Reimer for comments on the manuscript. This work was supported in part by NIH R01GM112766 from the National Institute of General Medical Sciences (to K.M.N.) and by T32GM007223 (to T.A.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the US National Institutes of Health.
Glossary
- 5′ end capping
The addition of an untemplated guanosine to the 5’ end of an RNA polymerase I transcript followed by its methylation at the N7 position Capping protects the mRNA 5′ end from endonucleases.
- 3′ end cleavage and polyadenylation
Endonucleolytic cleavage that defines the 3′ ends of RNA polymerase II transcripts by cleavage and polyadenylation specificity factor (CPSF) and other factors followed by the addition of non-templated poly(A) tails by poly(A) polymerase
- Nascent RNA
RNA that is tethered to DNA by any elongating RNA polymerase
- Gene architecture
The ensemble of cis-regulatory, coding and non-coding elements of a gene, including length, position and sequence
- Ultraviolet (UV) crosslinking
UV irradiation-induced covalent bonds that link amino acids with nucleic acids
- SR proteins
RNA-binding proteins with long repeats of arginine (Arg) and serine (Ser) residues that are involved in the regulation of alternative splicing and other steps of gene expression
- Intrinsically disordered regions
Protein regions that contain little amino acid diversity and appear to lack well-defined secondary and tertiary structures
- Speckles
Membrane-less subnuclear granules that are enriched in splicing factors, particularly the SR proteins
- Cajal bodies
Membrane-less subnuclear compartments (2–4 per cell) that are the sites of small nuclear RNA modification and small nuclear ribonucleoprotein assembly. Cajal bodies are not the sites of splicing
- P-bodies
Membrane-less cytoplasmic compartments that are involved in mRNA turnover
- Lampbrush chromosomes
Giant meiotic chromosomes that are formed in oocyte nuclei owing to the looping of chromosomal regions that are highly transcribed and coated with nascent RNA
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
CYC2008: http://wodaklab.org/cyc2008/
Saccharomyces Genome Database: http://www.yeastgenome.org
Spliceosome Database: http://spliceosomedb.ucsc.edu/
Biological General Repository for Interaction Datasets (BioGRID): https://thebiogrid.org
FURTHER INFORMATION
IUPred: http://iupred.enzim.hu/
References
- 1.Sakharkar MK, Perumal BS, Sakharkar KR, Kangueane P. An analysis on gene architecture in human and mouse genomes. In Silico Biol. 2005;5:347–365. [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000 Prime Reports. 2013;5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrillo Oesterreich F, et al. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell. 2016;165:372–381. doi: 10.1016/j.cell.2016.02.045. Shows a tight match between progress in transcription and splicing completion in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alpert T, Herzel L, Neugebauer KM. Perfect timing: splicing and transcription rates in living cells. Wiley Interdiscip. Rev. RNA. 2016;8:e1401. doi: 10.1002/wrna.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2:62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moehle EA, Braberg H, Krogan NJ, Guthrie C. Adventures in time and space: splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014;11:313–319. doi: 10.4161/rna.28646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 11.Custódio N, Carmo-Fonseca M. Co-transcriptional splicing and the CTD code. Crit. Rev. Biochem. Mol. Biol. 2016;51:395–411. doi: 10.1080/10409238.2016.1230086. [DOI] [PubMed] [Google Scholar]
- 12.Saldi T, Cortazar MA, Sheridan RM, Bentley DL. Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 2016;428:2623–2635. doi: 10.1016/j.jmb.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace E, Beggs J. Extremely fast and incredibly close: co-transcriptional splicing in budding yeast. RNA. 2017;23:601–610. doi: 10.1261/rna.060830.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander ES, et al. International human genome sequencing consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Gould GM, et al. Identification of new branch points and unconventional introns in Saccharomyces cerevisiae. RNA. 2016;22:1522–1534. doi: 10.1261/rna.057216.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin D, Huang L, Wlodaver A, Andrade J, Staley JP. Sequencing of lariat termini in S cerevisiae reveals 5’ splice sites, branch points, and novel splicing events. RNA. 2016;22:237–253. doi: 10.1261/rna.052829.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercer TR, et al. Genome-wide discovery of human splicing branchpoints. Genome Res. 2015;25:290–303. doi: 10.1101/gr.182899.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taggart AJ, et al. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 2017;27:639–649. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim L, Burge C. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl Acad. Sci. USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 22.Moore M, Query C, Sharp P. In: The RNA World. Gesteland RF, Atkins JF, editors. Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 23.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cvitkovic I, Jurica MS. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res. 2013;41:D132–D141. doi: 10.1093/nar/gks999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordin O, Beggs JD. RNA helicases in splicing. RNA Biol. 2013;10:83–95. doi: 10.4161/rna.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabrizio P, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 27.Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5’ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol. Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. Analyses co-transcriptional recruitment of U snRNPs onto nascent RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Újvári A, Luse DS. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J. Biol. Chem. 2004;279:49773–49779. doi: 10.1074/jbc.M409087200. [DOI] [PubMed] [Google Scholar]
- 32.Baejen C, et al. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol. Cell. 2014;55:745–757. doi: 10.1016/j.molcel.2014.08.005. Systematic mapping of the binding sites of several factors involved in mRNP biogenesis on yeast transcripts. [DOI] [PubMed] [Google Scholar]
- 33.Rigo N, Sun C, Fabrizio P, Kastner B, Lührmann R. Protein localisation by electron microscopy reveals the architecture of the yeast spliceosomal B complex. EMBO J. 2015;34:3059–3073. doi: 10.15252/embj.201592022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen TH, et al. Cryo-EM structure of the yeast U4/U6. U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen TH, et al. The architecture of the spliceosomal U4/U6. U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan R, et al. The 3.8 Å structure of the U4/U6. U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 37.Liu HL, Cheng SC. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol. Cell. Biol. 2012;32:5056–5066. doi: 10.1128/MCB.01109-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rymond BC, Rosbash M. Cleavage of 5’ splice site and lariat formation are independent of 3’ splice site in yeast splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 39.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016;353:905–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 40.Rauhut R, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353:1399–1405. doi: 10.1126/science.aag1906. Cryo-EM structure of the Bact complex, revealing details of spliceosome active site conformation before step I catalysis and suggesting that conformation rearrangement is needed for catalysis. [DOI] [PubMed] [Google Scholar]
- 41.Schneider C, et al. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3’ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet. 2015;11:e1005539. doi: 10.1371/journal.pgen.1005539. Evaluates direct interactions between splicing proteins and pre-mRNA at different stages of spliceosome assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruskin B, Green M. Role of the 3’ splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985;317:732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- 43.Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science. 2016;353:895–904. doi: 10.1126/science.aag2235. [DOI] [PubMed] [Google Scholar]
- 44.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohrt T, et al. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in purified system. RNA. 2013;19:902–915. doi: 10.1261/rna.039024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast step II catalytically activated spliceosome. Science. 2017;355:149–155. doi: 10.1126/science.aak9979. Cryo-EM structure of C* complex, revealing details of spliceosome active site conformation before step II catalysis and suggesting that conformation rearrangement is needed for catalysis. [DOI] [PubMed] [Google Scholar]
- 48.Fica SM, et al. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542:377–380. doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertram K, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 50.Smith C, Chu T, Nadal-Ginard B. Scanning and competition between AGs are involved in 3’ splice site selection in mammalian introns. Mol. Cell. Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gahura O, Hammann C, Valentová A, Puta F, Folk P. Secondary structure is required for 3’ splice site recognition in yeast. Nucleic Acids Res. 2011;39:9759–9767. doi: 10.1093/nar/gkr662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer M, Plass M, Peréz-Valle J, Eyras E, Vilardell J. Deciphering 3’ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol. Cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. Analyses the role of RNA secondary structure in 3′SS selection. [DOI] [PubMed] [Google Scholar]
- 53.Martinez-Rucobo FW, et al. Molecular basis of transcription-coupled pre-mRNA capping. Mol. Cell. 2015;58:1079–1089. doi: 10.1016/j.molcel.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Fourmann JB, et al. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013;27:413–428. doi: 10.1101/gad.207779.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwak H, Lis JT. Control of transcriptional elongation. Annu. Rev. Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neves LT, et al. The histone variant H2A.Z promotes efficient cotranscriptional splicing in S. cerevisiae. Genes Dev. 2017;31:702–717. doi: 10.1101/gad.295188.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nissen KE, et al. The histone variant H2A.Z promotes splicing of weak introns. Genes Dev. 2017;31:688–701. doi: 10.1101/gad.295287.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaborowska J, Egloff S, Murphy S. The pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol. 2016;23:771–777. doi: 10.1038/nsmb.3285. [DOI] [PubMed] [Google Scholar]
- 60.Harlen KM, Churchman LS. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017;18:263–273. doi: 10.1038/nrm.2017.10. [DOI] [PubMed] [Google Scholar]
- 61.Allison L, Moyle M, Shales M, Ingles C. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 62.Corden J, Cadena D, Ahearn J, Dahmus M. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. Natl Acad. Sci. USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim H, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mayer A, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 65.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 66.Harlen KM, et al. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Rep. 2016;15:2147–2158. doi: 10.1016/j.celrep.2016.05.010. Analyses phospho-specific Pol II C-terminal domain interactomes by mass spectrometry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milligan L, et al. Strand-specific, high-resolution mapping of modified RNA polymerase II. Mol. Syst. Biol. 2016;12:874. doi: 10.15252/msb.20166869. Identifies distinct Pol II C-terminal domain PTMs associated with transcription initiation, and early and late elongation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh G, Pratt G, Yeo GW, Moore MJ. The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Müller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 70.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Descostes N, et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. eLife. 2014;3:e02105. doi: 10.7554/eLife.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schüller R, et al. Heptad-specific phosphorylation of RNA polymerase II CTD. Mol. Cell. 2016;61:305–314. doi: 10.1016/j.molcel.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 73.Suh H, et al. Direct analysis of phosphorylation sites on the Rpb1 C-terminal domain of RNA polymerase II. Mol. Cell. 2016;61:297–304. doi: 10.1016/j.molcel.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nojima T, et al. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mayer A, et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veloso A, et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nedelcheva-Veleva MN, et al. The thermodynamic patterns of eukaryotic genes suggest a mechanism for intron-exon recognition. Nat. Commun. 2013;4:2101. doi: 10.1038/ncomms3101. [DOI] [PubMed] [Google Scholar]
- 78.Schor IE, Fiszbein A, Petrillo E, Kornblihtt AR. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J. 2013;32:2264–2274. doi: 10.1038/emboj.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 80.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat. Struct Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 81.Huff JT, Zilberman D, Roy SW. Mechanism for DNA transposons to generate introns on genomic scales. Nature. 2016;538:533–536. doi: 10.1038/nature20110. Explains the correlation between internal exon length and nucleosome DNA length. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beckmann J, Trifonov E. Splice junctions follow a 205-base ladder. Proc. Natl Acad. Sci. USA. 1991;88:2380–2383. doi: 10.1073/pnas.88.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 84.Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. Describes the effects of nucleosomes on transcription elongation dynamics in vivo. [DOI] [PubMed] [Google Scholar]
- 85.Churchman LS, Weissman JS. Nascent transcrip sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. Describes the NET-seq method, which enables the analysis of transcription elongation dynamics at high resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwak H, Fuda N, Core LJ, Lis JT. Precise map of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. Describes the PRO-seq assay, which enables the analysis of transcription elongation dynamics at high resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Voong LN, et al. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016;167:1555–1570.e15. doi: 10.1016/j.cell.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014;24:1572–1583. doi: 10.1101/gr.176487.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-codin RNA in alternative splicing regulation. Curr. Opin. Genet. Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kfir N, et al. SF3B1 association with chromatin determines splicing outcomes. Cell Rep. 2015;11:618–629. doi: 10.1016/j.celrep.2015.03.048. [DOI] [PubMed] [Google Scholar]
- 91.Hnilicová J, et al. Histone deacetylase activity modulates alternative splicing. PLoS ONE. 2011;6:e16727. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herissant L, et al. H2B ubiquitylation modulates spliceosome assembly and function in budding yeast. Biol. Cell. 2014;106:126–138. doi: 10.1111/boc.201400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 94.Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol. Cell. 2014;53:779–790. doi: 10.1016/j.molcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Újvári A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- 97.Lai D, Proctor JR, Meyer IM. On the importance of cotranscriptional RNA structure formation. RNA. 2013;19:1461–1473. doi: 10.1261/rna.037390.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu SR, Hu CG, Zhang JZ. Regulatory effects of cotranscriptional RNA structure formation and transitions. Wiley Interdiscip. Rev. RNA. 2016;7:562–574. doi: 10.1002/wrna.1350. [DOI] [PubMed] [Google Scholar]
- 99.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eperon L, Graham I, Griffiths A, Eperon I. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 102.Goguel V, Wang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol. Cell. Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deshler J, Rossi J. Unexpected point mutations activate cryptic 3’ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991;5:1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- 104.Charpentier B, Rosbash M. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA. 1996;2:509–522. [PMC free article] [PubMed] [Google Scholar]
- 105.Nilsen T. Internal mRNA methylation finally finds functions. Science. 2014;343:1207–1208. doi: 10.1126/science.1249340. [DOI] [PubMed] [Google Scholar]
- 106.Reenan R. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434:409–413. doi: 10.1038/nature03364. [DOI] [PubMed] [Google Scholar]
- 107.Peng Z, et al. Comprehensive analysis of RNA-seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 108.Rodriguez J, Menet JS, Rosbash M. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol. Cell. 2012;47:27–37. doi: 10.1016/j.molcel.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang IX, et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 2014;6:906–915. doi: 10.1016/j.celrep.2014.01.037. Shows that RNA nucleotides can be edited as soon as they emerge from Pol II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rieder LE, Reenan RA. The intricate relationship between RNA structure, editing, and splicing. Semin. Cell Dev. Biol. 2012;23:281–288. doi: 10.1016/j.semcdb.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 111.Laurencikiene J, Källman AM, Fong N, Bentley DL, Öhman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bratt E, Öhman M. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA. 2003;9:309–318. doi: 10.1261/rna.2750803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Solomon O, et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hawkins J. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Deutsch M, Long M. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang XH-F, Leslie C, Chasin L. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 119.Berget S. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 120.Robberson B, Cote G, Berget S. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Talerico M, Berget S. Intron definition in splicing of small Drosophila introns. Mol. Cell. Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sterner D, Carlo T, Berget S. Architectural limits on plit genes. Proc. Natl Acad. Sci. USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Izaurralde E, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 124.Izaurralde E, et al. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 125.Rottman F, Shatkin A, Perry R. Sequences containing methylated nucleotides at the 5’ termini of messenger RNAs: possible implications in processing. Cell. 1974;3:197–199. doi: 10.1016/0092-8674(74)90131-7. [DOI] [PubMed] [Google Scholar]
- 126.Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rasmussen E, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl Acad. Sci. USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gonatopoulos-Pournatzis T, Cowling VH. Cap-binding complex (CBC) Biochem. J. 2014;457:231–242. doi: 10.1042/BJ20131214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Konarska MM, Padgett R, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 130.Pabis M, et al. The nuclear cap-binding complex interacts with the U4/U6.U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA. 2013;19:1054–1063. doi: 10.1261/rna.037069.112. Shows an association between the cap-binding complex and snRNPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lewis J, Izaurralde E, Jarmolozski A, McGuigan C, Mattaj I. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5’ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 132.Qiu ZR, Chico L, Chang J, Shuman S, Schwer B. Genetic interactions of hypomorphic mutations in the m7G cap-binding pocket of yeast nuclear cap binding complex: an essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA. RNA. 2012;18:1996–2011. doi: 10.1261/rna.033746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Proudfoot NJ. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science. 2016;352:1291–1300. doi: 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 135.Logan J, Falck-Pedersen E, Darnell J, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse βmaj globin gene. Proc. Natl Acad. Sci. USA. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Whitelaw E, Proudfoot N. α-Thalassemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cooke C, Hans H, Alwine J. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 1999;19:4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3′ end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol. Cell. Biol. 2008;28:849–862. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 140.Millevoi S, et al. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′ end processing. EMBO Rep. 2002;3:869–874. doi: 10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vagner S, Vagner C, Mattaj I. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- 142.Kaida D. The reciprocal regulation between splicing and 3′-end processing. Wiley Interdiscip. Rev. RNA. 2016;7:499–511. doi: 10.1002/wrna.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dye M, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 144.Davidson L, West S. Splicing-coupled 3′ end formation requires a terminal splice acceptor site, but not intron excision. Nucleic Acids Res. 2013;41:7101–7114. doi: 10.1093/nar/gkt446. Shows that early stages of spliceosome assembly are sufficient to couple splicing to 3’ end processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bento Martins S, et al. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat. Struct. Mol. Biol. 2011;18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- 146.Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gunderson S, Polycarpou-Schwarz M, Mattaj I. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 148.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. Shows that U1 snRNP protects the transcriptome by suppressing premature cleavage and polyadenylation from cryptic poly(A) sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baserga S, Steitz J. In: The RNA World. Gesteland RF, Atkins JF, editors. Cold Spring Harbor Laboratory Press; 1993. pp. 359–381. [Google Scholar]
- 150.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 153.El Kaderi B, Medler S, Raghunayakula S, Ansari A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J. Biol. Chem. 2009;284:25015–25025. doi: 10.1074/jbc.M109.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mukundan B, Ansari A. Srb5/Med18-mediated termination of transcription is dependent on gene looping. J. Biol. Chem. 2013;288:11384–11394. doi: 10.1074/jbc.M112.446773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Agarwal N, Ansari A. Enhancement of transcription by a splicing-competent intron is dependent on promoter directionality. PLoS Genet. 2016;12:e1006047. doi: 10.1371/journal.pgen.1006047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Moabbi AM, Agarwal N, El Kaderi B, Ansari A. Role for gene looping in intron-mediated enhancement of transcription. Proc. Natl Acad. Sci. USA. 2012;109:8505–8510. doi: 10.1073/pnas.1112400109. Shows that introns help gene looping and hence transcription in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huranová M, et al. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. J. Cell Biol. 2010;191:75–86. doi: 10.1083/jcb.201004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Phair R, Misteli T. High mobility of proteins in the mammalian nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 159.Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J. 2016;35:1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kwon I, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.He C, et al. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell. 2016;64:416–430. doi: 10.1016/j.molcel.2016.09.034. Identifies and characterizes new protein domains, and unannotated and/or disordered regions that interact with RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Castello A, et al. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell. 2016;63:696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PAA. Phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Neugebauer KM, Stolk JA, Roth MB. A conserved epitope on a subset of SR proteins defines a larger family of pre-mRNA splicing factors. J. Cell Biol. 1995;129:899–908. doi: 10.1083/jcb.129.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gueroussov S, et al. Regulatory expansion in mammals of multivalent hnRNP assemplies that globally control alternative splicing. Cell. 2017;170:324–339. doi: 10.1016/j.cell.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 166.Ying Y, et al. Splicing activation by RBfox requires self-aggregation through its tyrosine-rich domain. Cell. 2017;170:312–323. doi: 10.1016/j.cell.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Machyna M, Neugebauer KM, Staněk D. Coilin: the first 25 years. RNA Biol. 2015;12:590–596. doi: 10.1080/15476286.2015.1034923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kwon I, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yu Y, Reed R. FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc. Natl Acad. Sci. USA. 2015;112:8608–8613. doi: 10.1073/pnas.1506282112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sun S, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 2015;6:6171. doi: 10.1038/ncomms7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Radó-Trilla N, Albà M. Dissecting the role of low-complexity regions in the evolution of vertebrate proteins. BMC Evol. Biol. 2012;12:155. doi: 10.1186/1471-2148-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Callan H. Lampbrush Chromosomes. Vol. 36. Springer-Verlag Berlin Heidelberg: 1986. [Google Scholar]
- 173.Custódio N, Vivo M, Antoniou M, Carmo-Fonseca M. Splicing- and cleavage-independent requirement of RNA polymerase II CTD for mRNA release from the transcription site. J. Cell Biol. 2007;179:199–207. doi: 10.1083/jcb.200612109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Custódio N, et al. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–2866. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tress ML, Abascal F, Valencia A. Alternative splicing may not be the key to proteome complexity. Trends Biochem. Sci. 2017;42:98–110. doi: 10.1016/j.tibs.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tilgner H, et al. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat. Biotechnol. 2015;33:736–742. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Taliaferro JM, et al. Distal alternative last exons localize mRNAs to neural projections. Mol. Cell. 2016;61:821–833. doi: 10.1016/j.molcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. eLife. 2016;5:e10921. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Berkovits BD, Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gowda N, et al. Cytosolic splice isoform of Hsp70 nucleotide exchange factor Fes1 is required for the degradation of misfolded proteins in yeast. Mol. Biol. Cell. 2016;27:1210–1219. doi: 10.1091/mbc.E15-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pu S, Wong J, Turner B, Cho E, Wodak SJ. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Huh W, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 184.Dosztányi Z, Csizmók V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 185.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 186.Birse C, Minvielle-Sebastia L, Lee B, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 187.Baejen C, et al. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol. Cell. 2017;66:38–49. doi: 10.1016/j.molcel.2017.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.