Abstract
Background
Several candidate genes have been identified in relation to lipid metabolism, and among these, lipoprotein lipase (LPL) and apolipoprotein E (APOE) gene polymorphisms are major sources of genetically determined variation in lipid concentrations. This study investigated the association of two single nucleotide polymorphisms (SNPs) at LPL, seven tagging SNPs at the APOE gene, and a common APOE haplotype (two SNPs) with blood lipids, and examined the interaction of these SNPs with dietary factors.
Methods
The population studied for this investigation included 660 individuals from the Prevention of Cancer by Intervention with Selenium (PRECISE) study who supplied baseline data. The findings of the PRECISE study were further replicated using 1238 individuals from the Caerphilly Prospective cohort (CaPS). Dietary intake was assessed using a validated food-frequency questionnaire (FFQ) in PRECISE and a validated semi-quantitative FFQ in the CaPS. Interaction analyses were performed by including the interaction term in the linear regression model adjusted for age, body mass index, sex and country.
Results
There was no association between dietary factors and blood lipids after Bonferroni correction and adjustment for confounding factors in either cohort. In the PRECISE study, after correction for multiple testing, there was a statistically significant association of the APOE haplotype (rs7412 and rs429358; E2, E3, and E4) and APOE tagSNP rs445925 with total cholesterol (P = 4 × 10− 4 and P = 0.003, respectively). Carriers of the E2 allele had lower total cholesterol concentration (5.54 ± 0.97 mmol/L) than those with the E3 (5.98 ± 1.05 mmol/L) (P = 0.001) and E4 (6.09 ± 1.06 mmol/L) (P = 2 × 10− 4) alleles. The association of APOE haplotype (E2, E3, and E4) and APOE SNP rs445925 with total cholesterol (P = 2 × 10− 6 and P = 3 × 10− 4, respectively) was further replicated in the CaPS. Additionally, significant association was found between APOE haplotype and APOE SNP rs445925 with low density lipoprotein cholesterol in CaPS (P = 4 × 10− 4 and P = 0.001, respectively). After Bonferroni correction, none of the cohorts showed a statistically significant SNP-diet interaction on lipid outcomes.
Conclusion
In summary, our findings from the two cohorts confirm that genetic variations at the APOE locus influence plasma total cholesterol concentrations, however, the gene-diet interactions on lipids require further investigation in larger cohorts.
Electronic supplementary material
The online version of this article (10.1186/s12944-018-0744-2) contains supplementary material, which is available to authorized users.
Keywords: APOE gene, Total cholesterol, LDL-C, PRECISE, Caerphilly prospective studies
Background
Cardiovascular diseases (CVD) are common multifactorial conditions characterized by dyslipidaemia, type 2 diabetes and hypertension [1, 2]. Elevated triacylglycerol (TAG) and reduced high density lipoprotein cholesterol (HDL-C) concentrations are associated with an increased risk of developing CVD [3–5]. Furthermore, several studies have reported that certain genetic variants influence susceptibility to altered circulating lipid concentrations, leading to an increased risk of CVD events [6–8]. Genetic variations have been shown to be associated with lipid outcomes, while dietary factors appear to modulate the effect of such genes on lipid concentrations [9, 10]. Previous studies have shown that single nucleotide polymorphisms (SNPs) of the apolipoprotein E (APOE) [6, 11] and lipoprotein lipase (LPL) [12–14] genes contribute to significant variation in lipid concentrations.
The APOE protein plays a key role in the transport and metabolism of cholesterol and TAG containing particles by serving as a receptor-binding ligand that mediates the clearance of dietary derived chylomicrons, and hepatically derived very low density lipoprotein (VLDL) and their remnants from the circulation [6]. The three most recognized alleles of the APOE gene are E2, E3 and E4, with carriage of E4 associated with CVD risk factors and increased low density lipoprotein cholesterol (LDL-C) concentrations [11, 15, 16], and hence increased CVD risk [17, 18].
Genetic variations in the LPL gene have been reported to be involved with lipid metabolism and partly explain the phenotypic variation in blood lipid levels [19]. LPL is a lipolytic enzyme that catalyses hydrolysis of TAG in all of the major classes of TAG-rich lipoproteins [20]. High enzyme activity is associated with favourable lipid levels, including relatively low TAG concentrations [21]. The two most widely studied LPL SNPs, rs328 (S447X) and rs320 (HindIII) [22, 23]. The ‘G’ minor alleles of both the SNPs, rs328 and rs320, are associated with decreased TAG concentrations and increased HDL-C concentrations, whereas the opposite association was found for the ‘C’ allele and ‘T’ allele respectively [24–26].
Data from several studies supports the role of genetic factors in lipid metabolism [27]; however, only a few studies have examined the effects of lifestyle factors such as diet on the association of polymorphisms with lipid-related outcomes [10, 28, 29]. Therefore, the present study aimed to investigate the effect of seven APOE tagSNPs (rs405509, rs769450, rs439401, rs445925, rs405697, rs1160985, and rs1064725), one APOE haplotype (rs7412 and rs429358), and two commonly studied LPL SNPs (rs328 and rs320) on blood lipid profile in 660 participants (baseline data) from the Prevention of Cancer by Intervention with Selenium (PRECISE) study. As diet type and intake is also known to modify lipid levels [30–32], the potential impact of the interaction between these SNPs and dietary factors on lipid levels was also investigated. To confirm the findings, the Caerphilly Prospective Study (CaPS; n = 1238) was used as a replication cohort.
Methods
PRECISE cohort
Participants and methods
Baseline data of 660 individuals from the PRECISE study, conducted in two populations [UK (n = 468) and Denmark (n = 192)] were used for the analysis [33, 34]. Briefly, study participants were selected from four general practices (study centres) in various areas of the UK that were affiliated with the Medical Research Council General Practice Research Framework (MRC GPRF). Between June 2000 and July 2001, research nurses recruited similar numbers of men and women from each of three age groups: 60–64, 65–69 and 70–74 years. The Danish participants were men and women recruited from the same three age groups from the County of Funen in Denmark.
The UK study obtained approval from the appropriate UK Local Research Ethics Committees [South Tees (ref: 99/69), Worcestershire Health Authority (ref: LREC 74/99), Norwich District (ref: LREC 99/ 141), Great Yarmouth and Waveney (under reciprocal arrangements with Norwich District LREC)], and the participants provided written informed consent. The regional Danish Data Protection Agency and Scientific Ethical Committees of Vejle and Funen counties approved the Danish study (Journal number. 19980186).
Dietary information
Information about each participant’s usual dietary intake was obtained using validated EPIC food frequency questionnaires (FFQ) [35]. Total energy intake and macronutrient composition were analysed using the FETA software program [36].
Anthropometric measurements and biochemical analysis
Body mass index (BMI) was calculated as body weight in kilograms divided by height in square metres (kg/m2). Participants provided non-fasting blood samples for biochemical analysis and these samples were stored at − 80 °C. Total cholesterol and HDL-C concentrations in lithium-heparin plasma were measured using an Architect c16000 analyser (Abbott) with dedicated reagents. Measurements were performed by enzymatic colorimetric analysis. Traceability for total cholesterol and HDL-C was ensured through participation in the National Reference System for Cholesterol (NRS/CHOL), as established by the Clinical and Laboratory Standards Institute, with isotope dilution-MS used as the reference method, and reference material taken from the National Institute of Standard and Technology. Evidence of equivalence in the analytical performance of the cholesterol-oxidase assays performed in the UK and Denmark from a comparison of total cholesterol on forty-four serum samples which produced a limit of variation of 2% [33].
SNP selection
The APOE gene is located on chromosome 19q13.32. It comprises four exons, which are transcribed into the APOE mRNA which is 1180 nucleotides long. The seven tagSNPs for the APOE gene were chosen based on International HapMap Phase II collected from individuals of Northern and Western European ancestry (CEU) (HapMap Data release 27 Phase 2 + 3, Feb 09, NCBI B36 assembly, dbSNP b126). The Haploview software V3.3 (http://www.broadinstitute.org/haploview/haploview-downloads) was used to assess the linkage disequilibrium between SNPs. Tagger software was used to select tagSNPs with the ‘pairwise tagging only’ option. Two criteria were used to filter the SNPs included in the analysis, minor allele frequency ≥ 5% and Hardy–Weinberg equilibrium P-value > 0.01. In total, seven tagSNPs [rs405509 (G > T), rs1160985 (C > T), rs769450 (G > A), rs439401 (C > T), rs445925 (G > A), rs405697 (G > A), and rs1064725 (T > G)] representing the entire common genetic variations across the APOE gene were selected for the study. The APOE haplotype/SNPs [6, 11, 37–44] and LPL [12, 13] SNPs were chosen based on their previous association with various lipid outcomes.
DNA isolation and genotyping
The genotyping for the selected SNPs using a KASP assay with a competitive allele-specific PCR assay® was performed on DNA samples by LGC Genomics (Hoddesdon, Herts, UK). The eleven SNPs were in Hardy Weinberg Equilibrium (HWE) (P > 0.05 for all comparisons) (Additional file 1: Table S1).
Caerphilly prospective study (CaPS)
Participants and methods
The CaPS was used to replicate the findings from the PRECISE study. The phase 1 (July 1979 to September 1983) recruitment for the CaPS included 2512 men aged 45–59 years who were living in the town of Caerphilly and five of its adjacent villages in the UK; these participants were followed up at regular intervals [45, 46]. The follow-up data collection included periods from 1984 to1988 (phase 2), from 1989 to 1993 (phase 3), from 1993 to 1997 (phase 4), and from 2002 to 2005 (phase 5). For the current study, the data analysed were taken from phase 3 (n = 1238), which had the maximum number of samples and variables appropriate to this analysis (total cholesterol and dietary information), and from phase 5 (n = 529) (HDL-C and LDL-C). Ethical approval was obtained from the South Wales Research Ethics Committee D, and each subject provided written informed consent.
Dietary information
Participants completed validated semi-quantitative FFQ in phase 3 [47, 48]. The FFQ included 50 typical food items in the British diet in order to estimate the mean daily energy intake and macronutrients and micronutrients consumption.
Anthropometric measurements and biochemical analysis
Height and weight was recorded in order to calculate the BMI. Height was measured on a stadiometer and weight was measured on a beam balance. Plasm prepared from blood samples taken after an overnight fast were transported at 4 °C to the laboratories on the day of venepuncture. Total cholesterol and HDL-C, LDL-C concentrations were measured using enzymatic procedures [49]. and the LDL-C levels were calculated using the Friedewald Formula [50].
DNA isolation and genotyping
DNA was extracted from blood samples collected during the period 1992–1994. SNP information was obtained from the Illumina Cardio Metabochip, which includes data on 200,000 SNPs from regions previously identified for associations with risk factors for cardiometabolic disease [51]. Imputation was conducted against the 1000-genomes reference panel, providing information on approximately two million typed or imputed SNPs. Duplicate samples were genotyped to compute the error rate. Quality control on genotyped samples has been previously reported [52] and the SNPs had a call rate of > 98%. The SNPs were in HWE (P > 0.05) (Additional file 1: Table S1).
Statistical analysis
Statistical analysis was performed using the SPSS software package, version 22.0. The data were presented as mean ± standard deviation (SD) in Tables 1 and 3 and beta regression coefficients and standard error (SE) were presented in Tables 2, 4, and 5. Independent t-test was used to compare means between men and women at baseline in the PRECISE cohort (Table 1). Univariate linear regression analysis was applied to test for association of the SNPs with total cholesterol and HDL-C, controlling for age, sex, BMI and country. SNP-diet interactions on total cholesterol and HDL-C were investigated using a univariate general linear model. In this model, total cholesterol and HDL-C were the dependent variables, SNPs were fixed factors, and dietary factors (fat energy %, protein energy %, carbohydrate energy %), sex, age BMI, and country were covariates. The dominant model was applied for all SNPs with minor allele frequency ≤ 0.3 and the additive model applied for SNPs with minor allele frequency ≥ 0.4. For analytical purposes, the six APOE genotype groups (E2/E2, E2/E3, E3/E3, E3/E4, E4/E4, and E2/E4) were classified into three groups. The E3/E3 genotype was classified as a group as it occurs at high frequency in the population (wild type). The E2/E2 and E2/E3 genotypes were combined and presented as E2 carriers. The E3/E4 and E4/E4 genotypes were also combined, and presented as E4 carriers [29]. Previous studies have shown that the impact of the E2 allele on serum lipids is greater than that of the E4 allele [17], therefore, the E2/E4 genotype was excluded from the analysis. The Bonferroni correction was applied separately for association and interaction analyses. For association between phenotypic and dietary factors, the Bonferroni-corrected P value was 0.008 (2 lipid outcomes* 3 dietary factors) for the PRECISE study and P value was 0.01 for CaPS (total cholesterol was the only variable available). For association between SNPs and lipids (PRECISE study), the Bonferroni corrected P value was 0.003 (10 SNPs*2 lipid outcomes = 20 tests). For interactions (PRECISE study), the Bonferroni corrected P value was 0.001 (10 SNPs*2 lipid outcomes*3 dietary factors = 60 tests). In the replication analysis (CaPS cohort), the Bonferroni corrected P value for association was 0.002 (10 SNPs*3 lipid outcomes = 30 tests), while for interactions it was 0.001 (10 SNPs*1 lipid outcome* 3 dietary factors = 30 tests).
Table 1.
Baseline characteristics of the PRECISE and Caerphilly Prospective study participants
| PRECISE study | Caerphilly Prospective study (CaPS) | |||
|---|---|---|---|---|
| Characteristics | Men (N = 248 UK, 95 Danish) |
Women (N = 220 UK, 97 Danish) |
P value | Men (N = 1238) |
| Age (years) | 67 ± 4 | 67 ± 4 | 0.12 | 62 ± 4 |
| Body mass index (kg/m2) | 27.2 ± 4.9 | 27.3 ± 4.9 | 0.82 | 26.8 ± 3.7 |
| Total Cholesterol (mmol/L) | 5.6 ± 0.9 | 6.2 ± 1.1 | 2.31 × 10−10 | 6.1 ± 1.1 |
| High density lipoprotein cholesterol (mmol/L)a | 1.5 ± 0.3 | 1.7 ± 0.4 | 2.71 × 10− 16 | 1.3 ± 0.3 |
| Protein intake (total energy %) | 17.6 ± 3.7 | 18.8 ± 3.7 | 5X10− 5 | 14.9 ± 2.7 |
| Carbohydrate intake (total energy %) | 42.8 ± 13.3 | 48.2 ± 8.7 | 1.42 × 10− 9 | 48.4 ± 7.5 |
| Fat intake (total energy %) | 35.3 ± 7.1 | 33.9 ± 6.9 | 0.01 | 36.5 ± 6.9 |
| Total energy intake (kcal) | 2256 ± 658 | 1992 ± 613 | 2.63 × 10−7 | 1964 ± 625 |
| Total energy intake (MJ) | 9.4 ± 2.7 | 8.3 ± 2.6 | 2.63 × 10−7 | 8.2 ± 2.6 |
Data shown are represented as means ± SD, wherever appropriate. P values are for the differences in the means between men and women. P values were calculated by using independent t-test
aFor CaPS, HDL-C levels were obtained from phase 5 while all other variables were obtained from phase 3
Table 3.
Association of APOE and LPL SNPs with HDL-C and total cholesterol levels in the PRECISE and Caerphilly studies
| SNP | MAF | HDL-C (mmol/L) | Total Cholesterol (mmol/L) | LDL-C a (mmol/L) |
|---|---|---|---|---|
| PRECISE | ||||
| LPL | ||||
| rs320 | 0.26 | |||
| TT | 1.6 ± 0.3 | 5.9 ± 1.1 | ||
| T/G | 1.7 ± 0.4 | 5.8 ± 1.0 | ||
| P value | 0.02 | 0.19 | ||
| rs328 | 0.10 | |||
| CC | 1.6 ± 0.3 | 5.9 ± 1.1 | ||
| C/G | 1.7 ± 0.4 | 5.7 ± 0.9 | ||
| P value | 0.04 | 0.06 | ||
| APOE | ||||
| rs405509 | 0.47 | |||
| GG | 1.7 ± 0.4 | 5.8 ± 1.1 | ||
| GT | 1.5 ± 0.3 | 5.8 ± 1.1 | ||
| TT | 1.6 ± 0.3 | 6.1 ± 1.0 | ||
| P value | 0.07 | 0.23 | ||
| rs769450 | 0.39 | |||
| GG | 1.6 ± 0.3 | 5.9 ± 1.1 | ||
| A allele | 1.6 ± 0.4 | 5.9 ± 1.1 | ||
| P value | 0.72 | 0.97 | ||
| rs439401 | 0.33 | |||
| CC | 1.6 ± 0.4 | 5.9 ± 1.1 | ||
| T allele | 1.6 ± 0.3 | 5.9 ± 1.1 | ||
| P value | 0.43 | 0.51 | ||
| rs445925 | 0.11 | |||
| GG | 1.6 ± 0.3 | 5.9 ± 1.1 | ||
| A allele | 1.7 ± 0.4 | 5.6 ± 0.9 | ||
| P value | 0.25 | 0.003 | ||
| rs405697 | 0.25 | |||
| GG | 1.6 ± 0.4 | 5.9 ± 1.1 | ||
| A allele | 1.6 ± 0.3 | 5.9 ± 1.0 | ||
| P value | 0.71 | 0.96 | ||
| rs1160985 | 0.43 | |||
| CC | 1.6 ± 0.3 | 5.9 ± 1.1 | ||
| CT | 1.6 ± 0.4 | 5.8 ± 1.0 | ||
| TT | 1.7 ± 0.4 | 5.9 ± 1.1 | ||
| P value | 0.12 | 0.44 | ||
| rs1064725 | 0.04 | |||
| TT | 1.6 ± 0.4 | 5.9 ± 1.0 | ||
| G allele | 1.7 ± 0.3 | 6.1 ± 1.2 | ||
| P value | 0.17 | 0.38 | ||
| (rs7412- rs429358) E2, E3, and E4 | ||||
| E3 | 1.6 ± 0.3 | 5.9 ± 1.1 | ||
| E4 | 1.5 ± 0.3 | 6.1 ± 1.1 | ||
| E2 | 1.7 ± 0.4 | 5.5 ± 0.9 | ||
| P value | 0.09 | 4X10−4 | ||
| Caerphilly | ||||
| LPL | ||||
| rs320 | 0.26 | |||
| TT | 1.3 ± 0.3 | 6.1 ± 1.1 | 2.7 ± 0.8 | |
| T/G | 1.4 ± 0.3 | 6.2 ± 1.2 | 2.8 ± 0.8 | |
| P value | 0.05 | 0.55 | 0.05 | |
| rs328 | 0.10 | |||
| CC | 1.3 ± 0.3 | 6.1 ± 1.1 | 2.7 ± 0.8 | |
| C/G | 1.3 ± 0.3 | 6.1 ± 1.1 | 2.9 ± 0.9 | |
| P value | 0.63 | 0.71 | 0.05 | |
| APOE | ||||
| rs405509 | 0.46 | |||
| GG | 1.4 ± 0.3 | 6.0 ± 1.1 | 2.7 ± 0.9 | |
| GT | 1.3 ± 0.3 | 6.2 ± 1.1 | 2.8 ± 0.8 | |
| TT | 1.3 ± 0.3 | 6.3 ± 1.1 | 2.9 ± 0.9 | |
| P value | 0.16 | 0.02 | 0.29 | |
| rs769450 | 0.41 | |||
| GG | 1.3 ± 0.2 | 6.1 ± 1.2 | 2.8 ± 0.9 | |
| A allele | 1.4 ± 0.3 | 6.2 ± 1.1 | 2.8 ± 0.8 | |
| P value | 0.10 | 0.41 | 0.82 | |
| rs439401 | 0.35 | |||
| CC | 1.4 ± 0.3 | 6.2 ± 1.1 | 2.8 ± 0.9 | |
| T allele | 1.3 ± 0.3 | 6.1 ± 1.1 | 2.7 ± 0.8 | |
| P value | 0.72 | 0.42 | 0.32 | |
| rs445925 | 0.11 | |||
| GG | 1.3 ± 0.3 | 6.2 ± 1.1 | 2.8 ± 0.8 | |
| A allele | 1.3 ± 0.3 | 5.9 ± 1.2 | 2.5 ± 0.9 | |
| P value | 0.99 | 3X10−4 | 0.001 | |
| rs405697 | 0.26 | |||
| GG | 1.4 ± 0.4 | 6.1 ± 1.1 | 2.8 ± 0.9 | |
| A allele | 1.3 ± 0.3 | 6.1 ± 1.1 | 2.8 ± 0.8 | |
| P value | 0.30 | 0.88 | 0.9 | |
| rs1160985 | 0.45 | |||
| CC | 1.34 ± 0.29 | 6.2 ± 1.1 | 2.8 ± 0.9 | |
| CT | 1.35 ± 0.35 | 6.2 ± 1.2 | 2.7 ± 0.8 | |
| TT | 1.37 ± 0.40 | 6.1 ± 1.0 | 2.8 ± 0.8 | |
| P value | 0.61 | 0.30 | 0.73 | |
| rs1064725 | 0.01 | |||
| TT | 1.3 ± 0.3 | 6.2 ± 1.1 | 2.8 ± 0.8 | |
| G allele | 1.4 ± 0.3 | 6.1 ± 1.1 | 2.8 ± 0.7 | |
| P value | 0.18 | 0.60 | 0.68 | |
| (rs7412- rs429358) E2, E3, and E4 | ||||
| E3 | 1.4 ± 0.4 | 6.2 ± 1.1 | 2.8 ± 0.8 | |
| E4 | 1.4 ± 0.3 | 6.4 ± 1.1 | 3.0 ± 0.9 | |
| E2 | 1.3 ± 0.3 | 5.8 ± 1.3 | 2.4 ± 0.8 | |
| P value | 0.95 | 2X10−6 | 4X10−4 | |
Values are given as mean ± SD. P values for differences between genotypes were obtained using linear regression model adjusted for age, sex, body mass index, and country
Bonferroni corrected P value < 0.003 was considered statistically significant
MAF minor allele frequency, HDL-C high density lipoprotein cholesterol, LDL-C low density lipoprotein cholesterol
a LDL-C values available only in Caerphilly prospective study
Table 2.
Association between dietary factors and lipids in PRECISE and Caerphilly Prospective studies
| PRECISE study | ||
|---|---|---|
| Association between dietary factors and total cholesterol | ||
| Fat total energy % intake Beta (± S.E), Passociation |
Protein total energy % intake Beta (± S.E), Passociation |
Carbohydrate total energy % intake Beta (± S.E), Passociation |
| 0.01 (0.01) 0.47 |
−0.01 (0.01) 0.13 |
−0.004 (0.01) 0.40 |
| Association between three dietary factors and HDL-C high density lipoprotein | ||
| Fat total energy % intake | Protein total energy % intake | Carbohydrate total energy % intake |
| −0.002 (0.002) 0.29 |
−0.002 (0.004) 0.59 |
− 0.004 (0.002) 0.02 |
| Caerphilly Prospective study | ||
| Association between three dietary factors and total cholesterol | ||
| Fat total energy % intake Beta (± S.E), Passociation |
Protein total energy % intake Beta (± S.E), Passociation |
Carbohydrate total energy % intake Beta (± S.E), Passociation |
| 0.01 (0.004) 0.06 |
−0.01 (0.01) 0.26 |
−0.01 (0.004) 0.17 |
HDL-C, high density lipoprotein cholesterol
P values were obtained using linear regression adjusted for age, sex, body mass index and country
Table 4.
Interaction between APOE and LPL SNPs and dietary factors on HDL-C and total cholesterol in the PRECISE study
| Interaction between rs320 at LPL*dietary factors on HDL-C | ||
|---|---|---|
| Interaction between SNP rs320* fat energy % intake | Interaction between SNP rs320* protein energy % intake | Interaction between SNP rs320* carbohydrate energy % intake |
| 0.003 (0.004) 0.46 |
0.002 (0.01) 0.76 |
−0.0004 (0.002) 0.87 |
| Interaction between rs320 at LPL *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs320* fat energy % intake | Interaction between SNP rs320* protein energy % intake | Interaction between SNP rs320* carbohydrate energy % intake |
| 0.01(0.01) 0.27 |
−0.03 (0.02) 0.13 |
−0.01 (0.01) 0.06 |
| Interaction between rs328 at LPL *dietary factors on HDL-C | ||
| Interaction between SNP rs328* fat energy % intake | Interaction between SNP rs328* protein energy % intake | Interaction between SNP rs328* carbohydrate energy % intake |
| 0.01 (0.01) 0.09 |
−0.001 (0.01) 0.89 |
0.001 (0.003) 0.63 |
| Interaction between rs328 at LPL *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs328* fat energy % intake | Interaction between SNP rs328* protein energy % intake | Interaction between SNP rs328* carbohydrate energy % intake |
| −0.002 (0.02) 0.88 |
0.003 (0.03) 0.90 |
−0.01 (0.01) 0.55 |
| Interaction between rs405509 at APOE*dietary factors on HDL-C | ||
| Interaction between SNP rs405509* fat energy % intake | Interaction between SNP rs405509* protein energy % intake | Interaction between SNP rs405509* carbohydrate energy % intake |
| 0.01 (0.01) 0.11 |
−0.001 (0.01) 0.75 |
−0.01 (0.003) 0.09 |
| Interaction between rs405509 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs405509* fat energy % intake | Interaction between SNP rs405509* protein energy % intake | Interaction between SNP rs405509* carbohydrate energy % intake |
| 0.02 (0.02) 0.39 |
−0.04 (0.03) 0.26 |
−0.01 (0.01) 0.59 |
| Interaction between rs769450 at APOE *dietary factors on HDL-C | ||
| Interaction between SNP rs769450* fat energy % intake | Interaction between SNP rs769450* protein energy % intake | Interaction between SNP rs769450* carbohydrate energy % intake |
| −0.001 (0.004) 0.88 |
0.001 (0.01) 0.88 |
0.003 (0.003) 0.19 |
| Interaction between rs769450 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs769450* fat energy % intake | Interaction between SNP rs769450* protein energy % intake | Interaction between SNP rs769450* carbohydrate energy % intake |
| −0.001 (0.01) 0.94 |
0.01 (0.02) 0.63 |
0.01 (0.01) 0.51 |
| Interaction between rs439401 at APOE *dietary factors on HDL-C | ||
| Interaction between SNP rs439401* fat energy % intake | Interaction between SNP rs439401* protein energy % intake | Interaction between SNP rs439401* carbohydrate energy % intake |
| 0.01 (0.004) 0.11 |
0.01 (0.01) 0.39 |
−0.001 (0.003) 0.64 |
| Interaction between rs439401 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs439401* fat energy % intake | Interaction between SNP rs439401* protein energy % intake | Interaction between SNP rs439401* carbohydrate energy % intake |
| 0.003 (0.01) 0.79 |
−0.02 (0.02) 0.37 |
− 0.001 (0.01) 0.89 |
| Interaction between rs445925 at APOE *dietary factors on HDL-C | ||
| Interaction between SNP rs445925* fat energy % intake | Interaction between SNP rs445925* protein energy % intake | Interaction between SNP rs445925* carbohydrate energy % intake |
| −0.003 (0.01) 0.53 |
0.01 (0.01) 0.52 |
0.0003 (0.003) 0.93 |
| Interaction between rs445925 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs445925* fat energy % intake | Interaction between SNP rs445925* protein energy % intake | Interaction between SNP rs445925* carbohydrate energy % intake |
| −0.03 (0.01) 0.05 |
0.01 (0.03) 0.66 |
0.01 (0.01) 0.36 |
| Interaction between rs405697 at APOE *dietary factors on HDL-C | ||
| Interaction between SNP rs405697* fat energy % intake | Interaction between SNP rs405697* protein energy % intake | Interaction between SNP rs405697* carbohydrate energy % intake |
| 0.01(0.004) 0.06 |
−0.002 (0.01) 0.80 |
−0.004 (0.002) 0.16 |
| Interaction between rs405697 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs405697* fat energy % intake | Interaction between SNP rs405697* protein energy % intake | Interaction between SNP rs405697* carbohydrate energy % intake |
| 0.01 (0.01) 0.22 |
−0.03 (0.02) 0.19 |
−0.003 (0.01) 0.72 |
| Interaction between rs1160985 at APOE *dietary factors on HDL-C | ||
| Interaction between SNP rs1160985* fat energy % intake | Interaction between SNP rs1160985* protein energy % intake | Interaction between SNP rs1160985* carbohydrate energy % intake |
| −0.01 (0.01) 0.08 |
−0.002 (0.01) 0.97 |
0.01 (0.004) 0.03 |
| Interaction between rs1160985 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs1160985* fat energy % intake | Interaction between SNP rs1160985* protein energy % intake | Interaction between SNP rs1160985* carbohydrate energy % intake |
| −0.01 (0.01) 0.58 |
0.05 (0.03) 0.28 |
−0.001 (0.01) 0.19 |
| Interaction between rs1064725 at APOE *dietary factors on HDL-C | ||
| Interaction between SNP rs1064725* fat energy % intake | Interaction between SNP rs1064725* protein energy % intake | Interaction between SNP rs1064725* carbohydrate energy % intake |
| −0.001 (0.01) 0.90 |
0.004 (0.02) 0.77 |
−0.002 (0.004) 0.73 |
| Interaction between rs1064725 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs1064725* fat energy % intake | Interaction between SNP rs1064725* protein energy % intake | Interaction between SNP rs1064725* carbohydrate energy % intake |
| 0.03 (0.03) 0.28 |
0.02 (0.04) 0.62 |
−0.01 (0.01) 0.48 |
| Interaction between APOE (E2, E3, and E4)*dietary factors on HDL-C | ||
| Interaction between SNP APOE (E2, E3, and E4)* fat energy % intake | Interaction between SNP APOE (E2, E3, and E4)* protein energy % intake | Interaction between SNP APOE (E2, E3, and E4)* carbohydrate energy % intake |
| −0.01 (0.01) 0.39 |
0.001 (0.01) 0.99 |
0.002 (0.003) 0.17 |
| Interaction between APOE (E2, E3, and E4)*dietary factors on Total Cholesterol | ||
| Interaction between SNP APOE (E2, E3, and E4)* fat energy % intake | Interaction between SNP APOE (E2, E3, and E4)* protein energy % intake | Interaction between SNP APOE (E2, E3, and E4)* carbohydrate energy % intake |
| −0.03 (0.02) 0.18 |
−0.02 (0.04) 0.32 |
0.01 (0.01) 0.51 |
Values represented β regression coefficients (± S.E), and Pinteraction. P values were obtained by using a general linear model adjusted for age, sex, body mass index, country and total energy intake, wherever appropriate
Bonferroni corrected P value < 0.001 was considered statistically significant
HDL-C High density lipoprotein cholesterol
Table 5.
Interaction between APOE and LPL SNPs and dietary factors on total cholesterol in the CaPS
| Interaction between rs320 at LPL *dietary factors on Total Cholesterol | ||
|---|---|---|
| Interaction between SNP rs320* fat energy % intake | Interaction between SNP rs320* protein energy % intake | Interaction between SNP rs320* carbohydrate energy % intake |
| 0.01 (0.01) 0.48 |
− 0.01 (0.03) 0.57 |
− 0.004 (0.01) 0.64 |
| Interaction between rs328 at LPL *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs328* fat energy % intake | Interaction between SNP rs328* protein energy % intake | Interaction between SNP rs328* carbohydrate energy % intake |
| − 0.01 (0.01) 0.58 |
−0.04 (0.03) 0.17 |
0.01 (0.01) 0.29 |
| Interaction between rs405509 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs405509* fat energy % intake | Interaction between SNP rs405509* protein energy % intake | Interaction between SNP rs405509* carbohydrate energy % intake |
| 0.03 (0.01) 0.11 |
−0.04 (0.04) 0.52 |
−0.02 (0.01) 0.31 |
| Interaction between rs769450 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs769450* fat energy % intake | Interaction between SNP rs769450* protein energy % intake | Interaction between SNP rs769450* carbohydrate energy % intake |
| −0.01 (0.01) 0.10 |
0.05 (0.02) 0.04 |
0.01 (0.01) 0.42 |
| Interaction between rs439401 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs439401* fat energy % intake | Interaction between SNP rs439401* protein energy % intake | Interaction between SNP rs439401* carbohydrate energy % intake |
| −0.003 (0.01) 0.77 |
−0.01 (0.03) 0.68 |
0.004 (0.01) 0.65 |
| Interaction between rs445925 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs445925* fat energy % intake | Interaction between SNP rs445925* protein energy % intake | Interaction between SNP rs445925* carbohydrate energy % intake |
| −0.0003 (0.01) 0.97 |
−0.02 (0.03) 0.55 |
0.002 (0.01) 0.87 |
| Interaction between rs405697 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs405697* fat energy % intake | Interaction between SNP rs405697* protein energy % intake | Interaction between SNP rs405697* carbohydrate energy % intake |
| 0.01 (0.01) 0.51 |
−0.03 (0.03) 0.24 |
−0.002 (0.01) 0.84 |
| Interaction between rs1160985 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs1160985* fat energy % intake | Interaction between SNP rs1160985* protein energy % intake | Interaction between SNP rs1160985* carbohydrate energy % intake |
| −0.01 (0.01) 0.13 |
−0.004 (0.03) 0.19 |
0.01 (0.01) 0.43 |
| Interaction between rs1064725 at APOE *dietary factors on Total Cholesterol | ||
| Interaction between SNP rs1064725* fat energy % intake | Interaction between SNP rs1064725* protein energy % intake | Interaction between SNP rs1064725* carbohydrate energy % intake |
| −0.01 (0.03) 0.66 |
0.05 (0.11) 0.62 |
0.01 (0.03) 0.74 |
| Interaction between APOE (E2,E3, and E4)*dietary factors on Total Cholesterol | ||
| Interaction between SNP APOE (E2, E3, and E4)* fat energy % intake | Interaction between SNP APOE (E2, E3, and E4)* protein energy % intake | Interaction between SNP APOE (E2, E3, and E4)* carbohydrate energy % intake |
| −0.02 (0.02) 0.038 |
0.02 (0.04) 0.83 |
0.01 (0.01) 0.08 |
Values represented β regression coefficients (± S.E), and Pinteraction
P values were obtained by using a general linear model adjusted for age, sex, body mass index, country and total energy intake, wherever appropriate
Bonferroni corrected P value < 0.001 was considered statistically significant
Results
Participant characteristics
The general characteristics of the participants by sex are presented in Table 1. In the PRECISE study, women were found to have significantly higher total cholesterol and HDL-C concentrations than men (P = 2.31 × 10− 10 and P = 2.71 × 10− 16, respectively). The consumption of carbohydrates (P = 1.42 × 10− 9) and protein (energy %) (P = 5 × 10− 5) were higher in women than in men, whereas the consumption of fat (energy %) and total energy intake were lower in women than in men (P = 0.01). Characteristics of the individuals from CaPS are given in Table 1. Elevated total cholesterol levels were observed among men at phase 3. Dietary-pattern data showed higher consumption of energy from total fat.
Association between dietary factors and blood lipids
In both the PRECISE and CaPS, there was no association between the dietary factors and total cholesterol or high-density lipoprotein after Bonferroni correction and adjustment for confounding factors (Table 2).
Genotypes and serum lipid levels in the PRECISE study
As shown in Table 3, of the seven tagSNPs at APOE, tagSNP rs445925 was significantly associated with total cholesterol (P = 0.003) after correction for multiple testing. The ‘A’ allele carriers (5.65 ± 0.98 mmol/L) had 5% lower levels of total cholesterol than GG homozygotes (5.99 ± 1.06 mmol/L).
The levels of HDL-C were significantly different among the LPL SNP genotypes, rs328 (P = 0.04) and rs320 (P = 0.02), where the carriers of the ‘G’ minor allele of both SNPs had higher levels of HDL-C (1.68 ± 0.41 mmol/L for rs328 and 1.66 ± 0.40 mmol/L for rs320) than CC homozygotes (rs328) and TT homozygotes (rs320) (1.61 ± 0.38 and 1.60 ± 0.39 mmol/L) respectively. However, these associations were not statistically significant after Bonferroni correction.
APOE haplotype and serum lipid levels in the PRECISE study
The effects of APOE haplotypes (E2, E3, and E4) on serum lipids are shown in Table 3. These haplotypes (E2, E3, and E4) were significantly associated with total cholesterol (P = 4 × 10− 4) after correction for multiple testing. The carriers of the E2 allele (5.54 ± 0.97 mmol/L) had lower total cholesterol concentrations than the carriers of the E3 (P = 0.001) (5.98 ± 1.05 mmol/L) and E4 alleles (6.09 ± 1.06 mmol/L) (P = 2 × 10− 4) (Fig. 1).
Fig. 1.
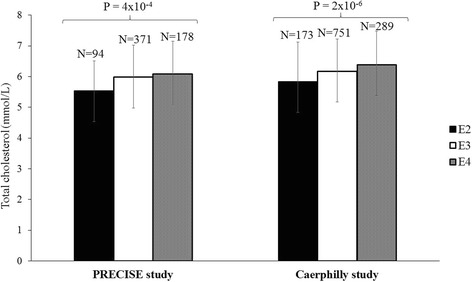
Association of APOE haplotypes (E2, E3, and E4) with total cholesterol concentrations in the Prevention of Cancer by Intervention with Selenium (PRECISE) study and Caerphilly Prospective study (CaPS). E2 allele carriers have significantly lower levels of total cholesterol than E3 (P = 0.001 and P = 4 × 10− 4 in the PRECISE and CaPS, respectively) and E4 (P = 2 × 10− 4 and P = 3 × 10− 6 in the PRECISE and CaPS, respectively) allele carriers
Interactions between genotypes and dietary factors on serum lipid in the PRECISE study
None of the dietary factors significantly interacted with the APOE SNPs, haplotypes and LPL SNPs with plasma lipids after correction for multiple testing (P > 0.001) (Table 4).
Replication analysis: Effect of SNPs at APOE and LPL on serum lipids in the CaPS
The associations of APOE and LPL SNPs with blood lipids in the CaPS are presented in Table 3. The association of APOE haplotype (E2, E3, and E4) and APOE SNP rs445925 with total cholesterol (P = 2 × 10− 6 and P = 3 × 10− 4, respectively) was replicated (Fig. 1). The ‘A’ allele carriers of APOE SNP rs445925 had lower total cholesterol (5.96 ± 1.24 mmol/l) than ‘GG’ genotypes (6.24 ± 1.08 mmol/L). In the APOE haplotype analysis, the carriers of the E2 allele had 5% and 14% lower total cholesterol than carriers of the E3 (P = 4 × 10− 4) and E4 alleles (P = 3 × 10− 6), respectively. Additionally, significant association was seen between APOE haplotypes (E2, E3, and E4) and APOE SNP rs445925and LDL-C (P = 4X10− 4, 0.001, respectively).
There was an interaction between fat (% energy) and APOE haplotype (E2, E3, and E4) on total cholesterol (P = 0.038) in CaPS. However, after correction for multiple testing, all the SNP-diet interactions were consistent with chance variation (Table 5).
Discussion
Our findings demonstrated significant associations between the APOE haplotype (E2, E3, and E4) and APOE SNP rs445925 with total plasma cholesterol and LDL-C (only CaPS) concentration, which were further replicated in an independent UK Caucasian cohort. The levels of total cholesterol were significantly lower in carriers of the APOE E2 allele and the ‘A’ allele of the SNP rs445925 than carriers of E3, E4 and ‘GG’ genotype of the APOE SNP rs445925, respectively. Given that our findings confirm that genetic polymorphisms of APOE influence the inter-individual variation in total plasma cholesterol, a marker of dyslipidemia, changes in dietary consumption to reduce disease susceptibility could be implemented for individuals at genetic risk.
The effects of APOE polymorphisms on lipid concentrations have previously been investigated in different ethnic groups [11, 53, 54] and studies have shown that the APOE gene variants contributed to 7% variability in total cholesterol [55]. The results of the current study were in line with previously reported findings that APOE haplotypes (E2, E3, and E4) are associated with serum total cholesterol and LDL-C, with E4 carriers associated with increased concentrations compared with E3/E3 wildtype and particularly E2 carriers [16, 53, 56]. One of the primary roles of APOE is binding the low density lipoprotein receptor (LDLR) and the LDLR-related protein, to facilitate cellular uptake of lipoprotein particles [57]. The three alleles, E2, E3, and E4, differ in their amino-acid sequences, resulting in functional differences in receptors-binding affinity. Amino-acid sequences of the E2 allele have lower binding affinity than those of the E3 and E4 alleles, causing decreased hepatic VLDL and chylomicron remnants clearance, thus reducing the uptake of postprandial lipoprotein particles [57]. Furthermore, it could be postulated that increase in apoE TAG-rich lipoproteins in E4 carriers could possibly increase the affinity to bind LDL-receptors resulting in decreased uptake of LDL and increased circulating plasma cholesterol [58]. E2 carriers also have an impaired conversion of the VLDL particles to LDL-C compared to E4 carriers [59], who have a higher rate of VLDL catabolism [60], which explains in part the lower total cholesterol and LDL-C in E2 allele carriers.
Furthermore, our study highlights an association between APOE SNP rs445925, which is one of the selected tagSNPs within the APOE gene, and total cholesterol. The SNP rs445925 has not been extensively studied, however, a genome-wide association study showed a significant association between SNP rs445925 and LDL-C levels in 3644 black and white individuals from the US and Europe [61]. In addition, previous genome-wide linkage and association studies have shown linkage disequilibrium (LD) between APOE SNPs rs7412 and rs445925 [62] and between ‘A’ allele carriers at SNP rs445925 and E2 haplotype [63], respectively, which could explain in part a similar function in cholesterol synthesis. It is also possible that A’ allele carriers of the SNP rs445925 might exhibit lower conversion of the VLDL particles to LDL-C which could have resulted in the decreased rate of LDL formation and hence lowered the total cholesterol concentrations [63].
Besides genetic associations, our study also identified an interaction of APOE haplotypes (E2, E3, and E4) with intake from fat (%) on total cholesterol in the CaPS, where, among those who consumed a low-fat diet (%), individuals carrying the E2 allele had significantly lower total cholesterol concentrations than to E4 allele carriers. However, this interaction was not statistically significant after correction for multiple testing. A previous study has examined the response of APOE genotype to fat intake in 45 individuals using a prospective design, where after consumption of a lower-fat-cholesterol diet (34% fat, 265 mg/day) according to modified National Cholesterol Education program there was a significant reduction in total cholesterol by 14%, 9%, and 4% in E4/E4, E3/E4, and E3/E3 genotypes, respectively [64]. Another study showed that the response to a diet high in cholesterol increases total cholesterol in E3 and E4 compared to E2 allele carries in a study comprising 29 healthy men [65]. By contrast, a cross sectional study in European Caucasians (n = 996) reported that E2 allele carriers had lower total cholesterol levels, but there were no reported between interactions between saturated fatty acids and total cholesterol [66]. Given that the previous studies have given inconsistent results and have used various types of fatty acids, replication of our gene-diet interaction finding in a large well-designed randomized controlled trial is highly warranted.
Previous studies have shown that the minor allele of LPL SNP rs328 enhance lipolytic activity [12]. Increased activity of LPL results in enhance clearance of TAG from the circulation, and associated with higher HDL-C concentrations [67]. The LPL SNP rs320 (HindIII) is in LD with rs328 (S447X) and they have been shown to have similar effects on HDL-C, where minor allele was reported to increase HDL-C [24, 68]. In our study, in accordance with findings from other studies, there were associations between LPL SNPs, rs320 and rs328, and HDL-C concentrations, where common homozygotes of both SNPs had lower HDL-C [22–24, 26]. However, in our study, these associations were no longer statistically significant after Bonferroni correction. Furthermore, there were no significant LPL SNP-diet interactions with HDL-C or total cholesterol concentrations in either cohort. To date, there has only been one study that has shown an interaction between LPL rs328 and total fat intake on HDL-C in 8764 individuals from the US population, where high fat intake associated with increase HDL-C in CC homozygotes and CG heterozygotes carriers [28]. One of the main reasons we did not identify a significant interaction may be our small sample size; however, we cannot rule out an effect of differences in dietary fat sources between European and the US population.
The present study has some limitations. Importantly, some lipid-related outcomes, such as LDL-C and TAG concentrations, were not measured in the PRECISE study. The PRECISE study was also conducted in two populations, a UK cohort and a Danish cohort, which used different food frequency questionnaires and this might have introduced measurement bias, even though the current results were adjusted for country in the regression analysis to avoid confounding. Another possible limitation is the use of a cross-sectional design (in both studies) to investigate genetic effects at a single point in time, whereas a longitudinal analysis design would have captured the genetic effects on lipid outcomes over a specific time period. The effect-size of the minor allele of some of the studied SNPs was relatively small, and hence a large sample size is required to detect reliably detect any interaction between SNPs and dietary factors. Despite the fact that this study was not adequately powered to detect such an interaction, it was sufficiently powered to detect the main effects (i.e., associations). Significant gene-diet interactions were identified, however these did not reach the Bonferroni-corrected P value (P = 0.001) and hence need to be confirmed in larger cohorts. This study is strengthened by the fact that it is the first study to investigate the role of tagSNPs at the APOE gene in relation to dietary factors and lipid outcomes. The fact that genetic associations from the PRECISE study were replicated in another Caucasian cohort (CaPS) confirms the validity of our findings. Additionally, CaPS was based on a cohort with a very high response rate, and is therefore closely representative of the general population.
Conclusion
Our study, carried out in two Caucasian populations, confirmed that genetic variations at the APOE gene locus influence plasma lipid concentrations. Thus, our results suggest that APOE gene variants affect risk of dyslipidemia in individuals who carry the E4 risk allele and GG genotype at SNP rs445925. Future studies with a larger sample size examining tagSNPs at APOE, particularly prospectively genotyped dietary intervention studies are required to confirm the gene-diet interactions identified in our study.
Additional file
Table S1. Genotype distribution of SNPs at LPL and APOE genes and Hardy Weinberg Equilibrium P values. (DOCX 18 kb)
Funding
We acknowledge the support from the British Nutrition Foundation. Genotyping of the genetic variants in the PRECISE study samples were funded by the Saudi government. The PRECISE study was supported by the Danish Cancer Society; the Research Foundation of the County of Funen; Cypress Systems Inc.; the Danish Veterinary and Food Administration; the Council of Consultant Physicians, Odense University Hospital; the Clinical Experimental Research Foundation at Department of Oncology, Odense University Hospital; K.A Rohde’s Foundation; Dagmar Marshall’s Foundation. Pharma Nord ApS, Vejle, Denmark provided the selenium and placebo tablets. The Caerphilly Prospective Study was undertaken by the former MRC Epidemiology Unit (South Wales) and was funded by the Medical Research Council of the United Kingdom. The Caerphilly DNA Bank was established by an MRC Grant (G9824960). University of Bristol act as the data custodians of CaPS. The funders of the study had no influence on the study design, analysis and interpretation of the data, writing, review, approval or submission of the manuscript.
Abbreviations
- APOE
Aspolipoprotein E
- CaPS
Caerphilly Prospective cohort
- CVD
Cardiovascular disease
- FFQ
Food frequency questionnaire
- HDL-C
High density lipoprotein cholesterol
- HWE
Hardy Weinberg Equilibrium
- LD
Linkage disequilibrium
- LDL-C
Low density lipoprotein cholesterol
- LPL
Lipoprotein lipase
- PRECISE
Prevention of Cancer by Intervention with Selenium
- SNPs
Single nuclide polymorphisms
- TAG
Triacylglycerol
- VLDL
Very low density lipoprotein
Authors’ contributions
IMS performed the statistical analysis and drafted the manuscript; KSV conceived and designed the nutrigenetics study; KW and MR designed and conducted the PRECISE study; PE designed and led the conduct of the Caerphilly Prospective study and YBS was involved in the design and conduct of phase V as well as obtaining funding for genetic analysis. JAL, BE, KW, MR, YBS, PE, IG, and KSV critically reviewed the manuscript. All authors contributed to and approved the final version of the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from each study participant, and the study was approved by the regional Danish Data Protection Agency and Scientific Ethical Committees of Vejle and Funen counties approved the Danish study (PRECISE), the appropriate UK Local Research Ethics Committees [South Tees (ref: 99/69), Worcestershire Health Authority (ref: LREC 74/99), Norwich District (ref: LREC 99/ 141), Great Yarmouth and Waveney (under reciprocal arrangements with Norwich District LREC)] (PRECISE), the South Wales Research Ethics Committee D (CaPS).
Consent for publication
Written informed consent for publication was obtained from all the study participants.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12944-018-0744-2) contains supplementary material, which is available to authorized users.
Contributor Information
Israa M. Shatwan, Email: i.m.a.shatwan@pgr.reading.ac.uk
Kristian Hillert Winther, Email: Kristian.Winther@rsyd.dk.
Basma Ellahi, Email: b.ellahi@chester.ac.uk.
Peter Elwood, Email: peter.c.elwood@gmail.com.
Yoav Ben-Shlomo, Email: Y.Ben-Shlomo@bristol.ac.uk.
Ian Givens, Email: d.i.givens@reading.ac.uk.
Margaret P. Rayman, Email: m.rayman@surrey.ac.uk
Julie A. Lovegrove, Email: j.a.lovegrove@reading.ac.uk
Karani S. Vimaleswaran, Phone: +44 118 378 8702, Email: v.karani@reading.ac.uk
References
- 1.Wang J, et al. The metabolic syndrome predicts cardiovascular mortality: a 13-year follow-up study in elderly non-diabetic Finns. Eur Heart J. 2007;28(7):857–864. doi: 10.1093/eurheartj/ehl524. [DOI] [PubMed] [Google Scholar]
- 2.McNeill AM, et al. Metabolic syndrome and cardiovascular disease in older people: the cardiovascular health study. J Am Geriatr Soc. 2006;54(9):1317–1324. doi: 10.1111/j.1532-5415.2006.00862.x. [DOI] [PubMed] [Google Scholar]
- 3.Barter P, et al. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N Engl J Med. 2007;357(13):1301–1310. doi: 10.1056/NEJMoa064278. [DOI] [PubMed] [Google Scholar]
- 4.Gotto AM., Jr High-density lipoprotein cholesterol and triglycerides as therapeutic targets for preventing and treating coronary artery disease. Am Heart J. 2002;144(6 Suppl):S33–S42. doi: 10.1067/mhj.2002.130301. [DOI] [PubMed] [Google Scholar]
- 5.Forrester JS. Triglycerides: risk factor or fellow traveler? Curr Opin Cardiol. 2001;16(4):261–4. doi: 10.1097/00001573-200107000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Song Y, Stampfer MJ, Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann Intern Med. 2004;141(2):137–147. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]
- 7.Ahmadzadeh A, Azizi F. Genes associated with low serum high-density lipoprotein cholesterol. Arch Iran Med. 2014;17(6):444–450. [PubMed] [Google Scholar]
- 8.Nettleton JA. Associations between HDL-cholesterol and polymorphisms in hepatic lipase and lipoprotein lipase genes are modified by dietary fat intake in African American and white adults. Atherosclerosis. 2007;194:e131–e140. doi: 10.1016/j.atherosclerosis.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho-Wells AL, et al. APOE genotype influences triglyceride and C-reactive protein responses to altered dietary fat intake in UK adults. Am J Clin Nutr. 2012;96(6):1447–1453. doi: 10.3945/ajcn.112.043240. [DOI] [PubMed] [Google Scholar]
- 10.Couture P, et al. Influences of apolipoprotein E polymorphism on the response of plasma lipids to the ad libitum consumption of a high-carbohydrate diet compared with a high-monounsaturated fatty acid diet. Metabolism. 2003;52(11):1454–1459. doi: 10.1016/S0026-0495(03)00275-0. [DOI] [PubMed] [Google Scholar]
- 11.Bennet AM, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298(11):1300–11. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 12.Radha V, et al. Association of lipoprotein lipase hind III and Ser 447 Ter polymorphisms with dyslipidemia in Asian Indians. Am J Cardiol. 2006;97(9):1337–1342. doi: 10.1016/j.amjcard.2005.11.056. [DOI] [PubMed] [Google Scholar]
- 13.Shatwan IM, et al. Impact of lipoprotein lipase gene polymorphism, S447X, on postprandial triacylglycerol and glucose response to sequential meal ingestion. Int J Mol Sci. 2016;17(3):397. doi: 10.3390/ijms17030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munshi A, et al. Association of LPL gene variant and LDL, HDL, VLDL cholesterol and triglyceride levels with ischemic stroke and its subtypes. J Neurol Sci. 2012;318(1–2):51–54. doi: 10.1016/j.jns.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Calabuig-Navarro MV, et al. Apolipoprotein E genotype has a modest impact on the postprandial plasma response to meals of varying fat composition in healthy men in a randomized controlled trial. J Nutr. 2014;144(11):1775–1780. doi: 10.3945/jn.114.197244. [DOI] [PubMed] [Google Scholar]
- 16.Shahid SU, et al. Effect of SORT1, APOB and APOE polymorphisms on LDL-C and coronary heart disease in Pakistani subjects and their comparison with Northwick Park heart study II. Lipids Health Dis. 2016;15:83. doi: 10.1186/s12944-016-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson PW, et al. Apolipoprotein E alleles, dyslipidemia, and coronary heart disease The Framingham Offspring Study. JAMA. 1994;272(21):1666–1671. doi: 10.1001/jama.1994.03520210050031. [DOI] [PubMed] [Google Scholar]
- 18.Giger JN, et al. Genetic predictors of coronary heart disease risk factors in premenopausal African-American women. Ethn Dis. 2005;15(2):221–232. [PubMed] [Google Scholar]
- 19.Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297(2):E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- 20.Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43(12):1997–2006. doi: 10.1194/jlr.R200015-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Friday KE, et al. Black-white differences in postprandial triglyceride response and postheparin lipoprotein lipase and hepatic triglyceride lipase among young men. Metabolism. 1999;48(6):749–754. doi: 10.1016/S0026-0495(99)90175-0. [DOI] [PubMed] [Google Scholar]
- 22.Nierman MC, et al. Enhanced conversion of triglyceride-rich lipoproteins and increased low-density lipoprotein removal in LPLS447X carriers. Arterioscler Thromb Vasc Biol. 2005;25(11):2410–2415. doi: 10.1161/01.ATV.0000188506.79946.ce. [DOI] [PubMed] [Google Scholar]
- 23.Rip J, et al. Lipoprotein lipase S447X: a naturally occurring gain-of-function mutation. Arterioscler Thromb Vasc Biol. 2006;26(6):1236–1245. doi: 10.1161/01.ATV.0000219283.10832.43. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Miranda J, et al. The influence of lipoprotein lipase gene variation on postprandial lipoprotein metabolism. J Clin Endocrinol Metab. 2004;89(9):4721–4728. doi: 10.1210/jc.2003-031642. [DOI] [PubMed] [Google Scholar]
- 25.Sagoo GS, et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am J Epidemiol. 2008;168(11):1233–1246. doi: 10.1093/aje/kwn235. [DOI] [PubMed] [Google Scholar]
- 26.Ukkola O, et al. Genetic variation at the lipoprotein lipase locus and plasma lipoprotein and insulin levels in the Quebec family study. Atherosclerosis. 2001;158(1):199–206. doi: 10.1016/S0021-9150(01)00413-0. [DOI] [PubMed] [Google Scholar]
- 27.Ordovas JM. Genetic influences on blood lipids and cardiovascular disease risk: tools for primary prevention. Am J Clin Nutr. 2009;89(5):1509s–1517s. doi: 10.3945/ajcn.2009.27113E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nettleton JA, et al. Associations between HDL-cholesterol and polymorphisms in hepatic lipase and lipoprotein lipase genes are modified by dietary fat intake in African American and white adults. Atherosclerosis. 2007;194(2):e131–e140. doi: 10.1016/j.atherosclerosis.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K, et al. Apolipoprotein E polymorphisms, dietary fat and fibre, and serum lipids: the EPIC Norfolk study. Eur Heart J. 2007;28(23):2930–2936. doi: 10.1093/eurheartj/ehm482. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, et al. Interactions between the -514C->T polymorphism of the hepatic lipase gene and lifestyle factors in relation to HDL concentrations among US diabetic men. Am J Clin Nutr. 2005;81(6):1429–1435. doi: 10.1093/ajcn/81.6.1429. [DOI] [PubMed] [Google Scholar]
- 31.Mensink RP, Katan MB. Effect of dietary fatty acids on serum lipids and lipoproteins. A meta-analysis of 27 trials. Arterioscler Thromb. 1992;12(8):911–919. doi: 10.1161/01.ATV.12.8.911. [DOI] [PubMed] [Google Scholar]
- 32.Hwang JY, et al. Carbohydrate intake interacts with SNP276G>T polymorphism in the adiponectin gene to affect fasting blood glucose, HbA1C, and HDL cholesterol in Korean patients with type 2 diabetes. J Am Coll Nutr. 2013;32(3):143–150. doi: 10.1080/07315724.2013.791795. [DOI] [PubMed] [Google Scholar]
- 33.Cold F, et al. Randomised controlled trial of the effect of long-term selenium supplementation on plasma cholesterol in an elderly Danish population. Br J Nutr. 2015;114(11):1807–1818. doi: 10.1017/S0007114515003499. [DOI] [PubMed] [Google Scholar]
- 34.Rayman MP, et al. A randomized trial of selenium supplementation and risk of type-2 diabetes, as assessed by plasma adiponectin. PLoS One. 2012;7(9):e45269. doi: 10.1371/journal.pone.0045269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKeown NM, et al. Use of biological markers to validate self-reported dietary intake in a random sample of the European prospective investigation into Cancer United Kingdom Norfolk cohort. Am J Clin Nutr. 2001;74(2):188–196. doi: 10.1093/ajcn/74.2.188. [DOI] [PubMed] [Google Scholar]
- 36.Mulligan AA, et al. A new tool for converting food frequency questionnaire data into nutrient and food group values: FETA research methods and availability. BMJ Open. 2014;4(3):e004503. doi: 10.1136/bmjopen-2013-004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komurcu-Bayrak E, et al. The APOE -219G/T and +113G/C polymorphisms affect insulin resistance among Turks. Metabolism. 2011;60(5):655–663. doi: 10.1016/j.metabol.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Viiri LE, et al. Interactions of functional apolipoprotein E gene promoter polymorphisms with smoking on aortic atherosclerosis. Circ Cardiovasc Genet. 2008;1(2):107–116. doi: 10.1161/CIRCGENETICS.108.791764. [DOI] [PubMed] [Google Scholar]
- 39.Son KY, et al. Genetic association of APOA5 and APOE with metabolic syndrome and their interaction with health-related behavior in Korean men. Lipids Health Dis. 2015;14:105. doi: 10.1186/s12944-015-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kring SI, et al. Impact of psychological stress on the associations between apolipoprotein E variants and metabolic traits: findings in an American sample of caregivers and controls. Psychosom Med. 2010;72(5):427–433. doi: 10.1097/PSY.0b013e3181de30ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trompet S, et al. Replication of LDL GWAs hits in PROSPER/PHASE as validation for future (pharmaco)genetic analyses. BMC Med Genet. 2011;12:131. doi: 10.1186/1471-2350-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, et al. Association of genetic loci with blood lipids in the Chinese population. PLoS One. 2011;6(11):e27305. doi: 10.1371/journal.pone.0027305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L, et al. A genome wide association study identifies common variants associated with lipid levels in the Chinese population. PLoS One. 2013;8(12):e82420. doi: 10.1371/journal.pone.0082420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seripa D, et al. TOMM40, APOE, and APOC1 in primary progressive aphasia and frontotemporal dementia. J Alzheimers Dis. 2012;31(4):731–740. doi: 10.3233/JAD-2012-120403. [DOI] [PubMed] [Google Scholar]
- 45.The Caerphilly and speedwell collaborative Group Caerphilly and Speedwell collaborative heart disease studies. J Epidemiol Community Health. 1984;38(3):259–262. doi: 10.1136/jech.38.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mertens E, et al. Dietary patterns in relation to cardiovascular disease incidence and risk markers in a middle-aged British male population: data from the Caerphilly prospective study. Nutrients. 2017;9(1):75. doi: 10.3390/nu9010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fehily AM, Yarnell JW, Butland BK. Diet and ischaemic heart disease in the Caerphilly study. Hum Nutr Appl Nutr. 1987;41(5):319–326. [PubMed] [Google Scholar]
- 48.Yarnell JW, et al. A short dietary questionnaire for use in an epidemiological survey: comparison with weighed dietary records. Hum Nutr Appl Nutr. 1983;37(2):103–112. [PubMed] [Google Scholar]
- 49.Yarnell JW, et al. Do total and high density lipoprotein cholesterol and triglycerides act independently in the prediction of ischemic heart disease? Ten-year follow-up of Caerphilly and speedwell cohorts. Arterioscler Thromb Vasc Biol. 2001;21(8):1340–1345. doi: 10.1161/hq0801.093505. [DOI] [PubMed] [Google Scholar]
- 50.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 51.Voight BF, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah T, et al. Population genomics of cardiometabolic traits: design of the University College London-London School of Hygiene and Tropical Medicine-Edinburgh-Bristol (UCLEB) consortium. PLoS One. 2013;8(8):e71345. doi: 10.1371/journal.pone.0071345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Lebedy D, Raslan HM, Mohammed AM, Apolipoprotein E. Gene polymorphism and risk of type 2 diabetes and cardiovascular disease. Cardiovasc Diabetol. 2016;15:12. doi: 10.1186/s12933-016-0329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ken-Dror G, et al. APOE/C1/C4/C2 gene cluster genotypes, haplotypes and lipid levels in prospective coronary heart disease risk among UK healthy men. Mol Med. 2010;16(9–10):389–399. doi: 10.2119/molmed.2010.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mozas P, et al. Apolipoprotein E genotype is not associated with cardiovascular disease in heterozygous subjects with familial hypercholesterolemia. Am Heart J. 2003;145(6):999–1005. doi: 10.1016/S0002-8703(02)94788-5. [DOI] [PubMed] [Google Scholar]
- 56.Suwalak T, et al. Polymorphisms of the ApoE (apolipoprotein E) gene and their influence on dyslipidemia in HIV-1-infected individuals. Jpn J Infect Dis. 2015;68(1):5–12. doi: 10.7883/yoken.JJID.2013.190. [DOI] [PubMed] [Google Scholar]
- 57.Eichner JE, et al. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol. 2002;155(6):487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 58.Jackson KG, et al. Saturated fat-induced changes in sf 60-400 particle composition reduces uptake of LDL by HepG2 cells. J Lipid Res. 2006;47(2):393–403. doi: 10.1194/jlr.M500382-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Ehnholm C, et al. Role of apolipoprotein E in the lipolytic conversion of beta-very low density lipoproteins to low density lipoproteins in type III hyperlipoproteinemia. Proc Natl Acad Sci U S A. 1984;81(17):5566–5570. doi: 10.1073/pnas.81.17.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregg RE, et al. Abnormal in vivo metabolism of apolipoprotein E4 in humans. J Clin Invest. 1986;78(3):815–821. doi: 10.1172/JCI112645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith EN, et al. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet. 2010;6(9):e1001094. doi: 10.1371/journal.pgen.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hellwege JN, et al. Genome-wide family-based linkage analysis of exome chip variants and cardiometabolic risk. Genet Epidemiol. 2014;38(4):345–352. doi: 10.1002/gepi.21801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deshmukh HA, et al. Genome-wide association study of genetic determinants of LDL-c response to atorvastatin therapy: importance of Lp(a) J Lipid Res. 2012;53(5):1000–1011. doi: 10.1194/jlr.P021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarkkinen E, et al. Effect of apolipoprotein E polymorphism on serum lipid response to the separate modification of dietary fat and dietary cholesterol. Am J Clin Nutr. 1998;68(6):1215–1222. doi: 10.1093/ajcn/68.6.1215. [DOI] [PubMed] [Google Scholar]
- 65.Gylling H, Miettinen TA. Cholesterol absorption and synthesis related to low density lipoprotein metabolism during varying cholesterol intake in men with different apoE phenotypes. J Lipid Res. 1992;33(9):1361–1371. [PubMed] [Google Scholar]
- 66.Petkeviciene J, et al. Associations between apolipoprotein E genotype, diet, body mass index, and serum lipids in Lithuanian adult population. PLoS One. 2012;7(7):e41525. doi: 10.1371/journal.pone.0041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaser S, et al. Phospholipid and cholesteryl ester transfer are increased in lipoprotein lipase deficiency. J Intern Med. 2003;253(2):208–216. doi: 10.1046/j.1365-2796.2003.01091.x. [DOI] [PubMed] [Google Scholar]
- 68.Humphries SE, et al. Lipoprotein lipase gene variation is associated with a paternal history of premature coronary artery disease and fasting and postprandial plasma triglycerides: the European atherosclerosis research study (EARS) Arterioscler Thromb Vasc Biol. 1998;18(4):526–534. doi: 10.1161/01.ATV.18.4.526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genotype distribution of SNPs at LPL and APOE genes and Hardy Weinberg Equilibrium P values. (DOCX 18 kb)


