Abstract.
Zika virus (ZIKV) is a mosquito-borne member of the genus Flavivirus that has emerged since 2007 to cause outbreaks in Africa, Asia, Oceania, and most recently, in the Americas. Here, we used an isolate history as well as genetic and phylogenetic analyses to characterize three low-passage isolates representing African (ArD 41525) and Asian (CPC-0740, SV0127-14) lineages to investigate the potential phenotypic differences in vitro and in vivo. The African isolate displayed a large plaque phenotype (∼3–4 mm) on Vero and HEK-293 cells, whereas the Asian isolates either exhibited a small plaque phenotype (∼1–2 mm) or did not produce any plaques. In multistep replication kinetics in nine different vertebrate and insect cell lines, the African isolate consistently displayed faster replication kinetics and yielded ∼10- to 10,000-fold higher peak virus titers (infectious or RNA copies) compared with the Asian isolates. Oral exposure of Aedes aegypti mosquitoes with the African isolate yielded higher infection and dissemination rates compared with the Asian isolates. Infection of Ifnar1−/− mice with the African isolate produced a uniformly fatal disease, whereas infection with the Asian isolates produced either a delay in time-to-death or a significantly lower mortality rate. Last, the African isolate was > 10,000-fold more virulent than the Asian isolates in an interferon type I antibody blockade mouse model. These data demonstrate substantial phenotypic differences between low-passage African and Asian isolates both in vitro and in vivo and warrant further investigation. They also highlight the need for basic characterization of ZIKV isolates, as the utilization of the uncharacterized isolates could have consequences for animal model and therapeutic/vaccine development.
INTRODUCTION
Zika virus (ZIKV) is a mosquito-borne member of the Spondweni complex of the genus Flavivirus, family Flaviviridae. The genus Flavivirus comprises many important human pathogens such as Saint Louis encephalitis, yellow fever virus, dengue virus (DENV1–DENV4), West Nile virus, Japanese encephalitis virus, and tick-borne encephalitis virus.1 The ZIKV genome comprises a single-strand, positive-sense RNA of approximately 11 kb in length. The genome consists of a 5′ untranslated region (UTR) (∼107 nt), an open reading frame (ORF) ∼10.2 kb in length, and a 3′ UTR (∼428 nt). The single ORF encodes three structural proteins: capsid (C), premembrane/membrane (prM), envelope (E), and seven non–structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).1 Similar to other flaviviruses, ZIKV enters cells through receptor-mediated endocytosis, and the low endosomal pH induces fusion to release the genomic RNA into the cytoplasm. The genomic RNA is translated into a single polyprotein that is processed to facilitate replication, virion assembly, and release of mature virions. The mature ZIKV virion structure is similar to that of other flaviviruses, comprising a nucleocapsid core consisting of C protein and genomic RNA and an icosahedral shell consisting of 180 copies of E and M proteins (or prM) embedded in a host-derived lipid bilayer.2
ZIKV was first isolated from the blood of a sentinel rhesus macaque in 1947 in the Zika Forest in Uganda.3 In the following year, ZIKV was isolated from pools of Aedes africanus mosquitoes, and subsequent studies demonstrated virus transmission by Aedes aegypti to both mice and rhesus macaques.4 In the following decades (1950s–1980s), ZIKV was isolated from humans and multiple mosquito species in sub-Saharan Africa and Southeast Asia, and suggesting maintenance in both sylvatic (primarily Aedes spp. mosquitoes and nonhuman primates [NHPs]) and urban cycles (mainly A. aegypti and humans).5–19 ZIKV consists of a single serotype, with isolates comprising two geographic lineages (African and Asian) that have caused sporadic or underreported human outbreaks.20,21 However, starting in 2007, outbreaks were reported in the Yap island in the Federated States of Micronesia and in Gabon, and by 2013 ZIKV was imported from French Polynesia into northeast Brazil resulting in the largest reported epidemic to date and its subsequent expansion to many parts of Americas including mainland USA (Florida and Texas).22–27 Since the start of the 2007 epidemic, ZIKV has spread to or been detected in 84 countries and territories in the Americas, Asia, and Africa, with reported evidence of autochthonous transmission (http://apps.who.int/iris/bitstream/10665/254714/1/zikasitrep10Mar17-eng.pdf?ua=1).
Most human ZIKV infections are asymptomatic.28,29 Symptomatic infections typically present as a self-limiting acute febrile illness with symptoms ranging from fever, headache, myalgia, and rash; however, severe clinical manifestations including congenital microcephaly and other fetal defects, Guillain–Barré syndrome (GBS), and death have been reported.30,31 Although ZIKV is transmitted primarily through mosquito/human cycle, the virus can also be transmitted through sexual contact and blood transfusions.32–34
Before its arrival in the Americas, ZIKV was an obscure and understudied pathogen; as a consequence, few virus isolates were accessible for research and vaccine/therapeutic development. In addition, little or no basic in vitro and/or in vivo characterization data were available on accessible isolates. In the present study, we investigated the potential phenotypic differences between low-passage African (ArD 41525) and Asian isolates (CPC-0740 and SV0127-14) in vitro and in vivo.
MATERIALS AND METHODS
Viruses and cells.
ZIKV isolates were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch (UTMB) (SEN/1984/ArD 41525), the Armed Forces Research Institute of Medical Sciences (AFRIMS) (PHL/2012/CPC-0740, THA/2014/SV0127-14), and the Centers for Disease Control and Prevention (CDC) (PRVABC59) (Table 1). The three strains are abbreviated as ArD 41525, CPC-0740, and SV0127-14. All three viruses were titrated on Vero monolayers three separate times to ensure accuracy of virus stocks. The titers obtained from each titration were within 2-fold. Virus stocks were also deep sequenced to ensure purity.
Table 1.
Detailed histories of three ZIKV isolates
| Isolate | Source | Location | Year of collection | Passage history | Titer (PFU/mL) |
|---|---|---|---|---|---|
| CPC-0740 | Homo sapiens | Philippines | 2012 | Toxorhynchites splendens mosquito (1), C6/36 cells (1), Vero (1) | 1 × 106 |
| SV0127-14 | H. sapiens | Thailand | 2014 | Toxorhynchites splendens mosquito (1), C6/36 cells (1), Vero cells (1) | 5 × 105 |
| ArD 41525 | Aedes africanus | Senegal | 1984 | AP-61 cells (1), C6/36 cells (1), Vero cells (4) | 2 × 106 |
PFU = plaque forming unit; ZIKV = Zika virus.
Vero-WHO, RK-13, DEF, and HEK-293 cell lines were obtained from the American Type Culture Collection (Bethesda, MD). RML-12 (A. aegypti), C7/10 (Aedes albopictus), AP-61 (Aedes pseudoscutellaris), Anopheles gambiae, and PP-9 (Phlebotomus papatasi) cells were obtained from the World Reference Center for Emerging Viruses and Arboviruses at UTMB. Vero-WHO cells were obtained from internal collection. Cell lines were propagated at 37°C (vertebrate) or 28°C (insect) with 5% CO2 in Dulbecco’s Minimal Essential Medium (DMEM) containing 10% (v/v) fetal bovine serum (FBS), sodium pyruvate (1 mM), 1% (v/v) nonessential amino acids (AAs), and 50 μg/mL gentamicin. PP-9 cells were maintained in Schneider media (Sigma, St. Louis, MO) supplemented with 10% (v/v) FBS and 50 μg/mL gentamicin. Insect cell media were additionally supplemented with 1% (v/v) tryptose phosphate broth (Sigma).
Phylogenetic analysis.
Sequences of ZIKV strains were downloaded from GenBank (Table 1) and aligned in SeaView using the MUSCLE algorithm.35,36 The sequences were aligned using deduced AA sequences from ORFs and then returned to nucleotide (NT) sequences for subsequent analyses to preserve codon homology. Maximum likelihood (ML) analysis was performed using the PHYLIP package.37 Modeltest in PAUP was used to identify the best-fit NT substitution model, GTR+I+G model.38 The robustness of ML phylogeny was evaluated by bootstrap resampling with 500 replicates.
Real-time reverse transcription polymerase chain reaction (RT-PCR) assay.
The ZIKV 106-nt 5′ UTR TaqMan-MGB probe assay used the six complete ZIKV genome sequences from GenBank available on December 9, 2015 (LC002520 [isolate MR766-NIID], NC_012532 [isolate MR766], AY632535 [isolate MR766], KF268949 [isolate ARB15076], KF268948 [isolate ARB13565], and KF268950 [isolate ARB7701]). The six complete ZIKV genome sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) as DNA with the format “Clustal w/numbers.” All other parameters were left as the default settings. Any nt mismatches were noted, and the reference target sequence (Accession NC_012532) was loaded into the Primer Express Version 3.0.1 for Windows 7 (Applied BioSystems®/Life Technologies, Carlsbad, CA) as a “TaqMan®-MGB Quantification” assay. Specific amplification detection was accomplished using a forward primer (5′-GARTCAGACTGCGACAGTTCGA-3′), reverse primer (5′-CCAAATCCAAATTAAACCTGTTGA-3′), and TaqMan-MGB probe (6FAM-ACTGTTGTTAGCTCTCGC–MGBNFQ). The resulting primer/probe pairs were initially tested using genomic RNA from the Zika isolate MR-766 (UCC# R4481T). All positive pairs were then screened for inclusivity with eight different ZIKV isolates available at USAMRIID: MR-766 (Uganda, 1947); ArD 41525 (Senegal, 1984); ArD 41662 (Senegal, 1984); ArD 41671 (Senegal, 1984); IBH 30656 (Nigeria, 1968); SV0127-14 (Thailand, 2010); CPC0740 (Philippines, 2010); and FSS13025 (Cambodia, 2010). The down-selected primer/probe pair was then optimized for primer concentration, tested for limit of detection (LOD) on both the LightCycler 480 (Roche Diagnostics, Indianapolis, IN) and the Bio-Rad QX200 droplet digital polymerase chain reaction (ddPCR) system (Bio-Rad, Hercules, CA). The LOD on the LightCycler 480 was 2 plaque forming units (PFUs)/mL by real-time polymerase chain reaction (PCR) and 69,900 target copies/mL by ddPCR using the ZIKV isolate MR-766. Finally, the assay was tested for specificity against genomic RNA from 65 virus isolates and genomic DNA from 11 bacterial strains and with human/mouse/mosquito genomic DNA. The cycling conditions on the LC480 were as follows: 1 cycle at 50°C for 15 minutes; 1 cycle at 95°C for 5 minutes; followed by 45 cycles at 95°C for 5 seconds and 55°C for 20 seconds; and last 1 cycle at 40°C for 30 seconds for cooling. A single read of the plate was taken at the end of each 55°C/20 second step. The QX200 ddPCR was run according to the manufacturer’s instructions.
Multistep replication kinetics.
Replication kinetics were assessed on vertebrate and insect cell lines in triplicate on 90% confluent monolayers in T-25 cm2 flasks. Virus infections were performed at a multiplicity of infection (MOI) of 0.1 PFU/cell in triplicate, and virus was adsorbed for 1 hour at 37°C (vertebrate cells) or 28°C (insect cells). After incubation, the inoculum was removed; monolayers were rinsed three to four times with room temperature (RT) DMEM to remove unbound virus; and 5 mL of growth medium was added to each flask. Aliquots of 0.5 mL were taken immediately afterward as a “time 0” sample and replaced with 0.5 mL of fresh medium. Flasks were incubated at 37°C or 28°C, and further samples were taken at 6, 24, 48, 72, 96, 120, and 144 hours post-infection (hpi), flash frozen in dry ice/ethanol bath, and stored at −80°C.
Plaque assay.
Virus titrations were performed on 100% confluent Vero or HEK-293 cell monolayers seeded overnight in six-well plates. Duplicate wells were infected with 0.1 mL aliquots of serial 10-fold diluted virus in growth medium; 0.4 mL of growth medium was also added to each well to prevent cell desiccation, and virus was adsorbed for 1 hour at 37°C. After adsorption, the virus inoculum was aspirated, and cell monolayers were overlaid with 3 mL of 0.8% w/v SeaPlaque agarose (Cambrex Bio Science, Basel, Switzerland) in DMEM containing sodium pyruvate (1 mM), 1% (v/v) nonessential AAs, and 50 μg/mL gentamicin. Cells were incubated at 37°C under 5% CO2 for 5 days to allow for plaque development. After incubation, cells were fixed with 4% formaldehyde in PBS for 24 hours; overlay was removed; and cells were stained with 2% crystal violet in 70% methanol for 5–10 minutes at RT; the excess stain was removed under running water, and plaques were counted.
Oral mosquito infections.
Strains of A. aegypti (Orlando, Florida strain) were obtained from the United States Department of Agriculture in Gainesville, FL. Cohorts of 100 adult females 5–6 days after emergence from the pupal stage were sugar starved for 24 hours. They were fed an artificial meal consisting of defibrinated sheep blood (Colorado Serum Company, Denver, CO) and ZIKV at ∼6.0 log10 PFU/mL. Mosquitoes were allowed to feed for 1 hour, and after feeding the mosquitoes, they were cold-anesthetized and sorted. Fully engorged mosquitoes at or higher than stage 3 were retained for the study.39 Mosquitoes were given 10% sucrose in cotton balls and held for an extrinsic incubation period of 7 days at 28°C. After extrinsic incubation, mosquitoes were cold-anesthetized, and bodies and legs/wings were removed. Mosquito bodies and legs/wings were triturated separately in 500 μL of 1 × DMEM containing 20% FBS (v/v), penicillin (200 U/mL), streptomycin (200 μg/mL), 100 μg/mL gentamicin, and 5 μg/mL amphotericin B, using a Mixer Mill 300 (Retsch, Haan, Germany).9 Samples were then centrifuged at 18,000 × g for 15 minutes, and supernatants from each sample were immediately analyzed by plaque assay to detect ZIKV. Monolayers were infected with supernatants, and 0.4 mL of 1 × DMEM containing penicillin (200 U/mL), streptomycin (200 μg/mL), 100 μg/mL gentamicin, and 5 μg/mL amphotericin B was also added to each well to prevent cell desiccation. After a 1 hour incubation, the monolayers were rinsed with 2 mL of 1 × DMEM containing penicillin (200 U/mL), streptomycin (200 μg/mL), 100 μg/mL gentamicin, and 5 μg/mL amphotericin B. The cell monolayers were overlaid with 3 mL of 0.8% w/v SeaPlaque agarose (Cambrex Bio Science) in 1 × DMEM containing penicillin (200 U/mL), streptomycin (200 μg/mL), 100 μg/mL gentamicin, and 5 μg/mL amphotericin B. All mosquito samples were assayed twice to confirm positive samples.
Ethics statement.
This work was approved by the IACUC. Research was in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and adheres to the principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
Mouse studies.
Wild-type and Ifnar−/− C56BL/6 mice were obtained from Jackson Laboratories and used for infection studies. Cohorts of 10 Ifnar−/− male and female mice 10 weeks of age were infected with ArD 41525, CPC-0740, or SV0127-14 isolates in 200 μL of PBS by intraperitoneal (IP) injection. Mice were observed daily for weight loss, signs of clinical illness, and were euthanized with the onset of severe clinical symptoms. Mice were assigned a clinical score (0–3) based on the following observations: normal appearance (0); lethargy, ruffled fur, hunched posture, or slow movement (1); decreased mobility or no longer socializing with other animals or 20% weight loss from initial weight (2); labored breathing or inability to move when stimulated or 25% weight loss or hind limb paralysis resulting in decreased mobility (3). Animals were assigned a score based on the most severe clinical sign observed.
Cohorts of 10 wild-type C57BL/6 female mice at 5- or 10-weeks of age were used in the interferon (IFN) antibody blockade model. Mice were injected, through an IP route, with a total of 3.0 mg (2.0 mg first dose and 0.5 mg subsequent doses) of MAb-5A3 (Leinco Technologies, St. Louis, MO)40,41 or PBS on day −1, day +1, and day 4. On day 0, mice were infected with ArD 41525, CPC-0740, or PRVABC59 through an IP route in a total volume of 200 μL 1× PBS. Mice were monitored for weight loss, signs of clinical disease, and were euthanized with the onset of severe clinical signs as per the above criteria.
Statistics.
GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla, CA, www.graphpad.com) software was used for statistical analysis. Significant differences in mean titers between the African and Asian strains were determined using two-way analysis of variance for all viruses followed by a Tukey test. A two-tailed Fisher exact test was performed to determine significant differences in mosquito infection rates, weight loss, and clinical scores. Survival curve analysis was determined by performing Mantel–Cox and Gehan–Breslow–Wilcoxon tests.
RESULTS
ZIKV isolate passage history and genetic and phylogenetic analyses.
The available ZIKV isolates and accompanying histories were obtained from UTMB and AFRIMS (Table 1). Lowest passaged isolates from both African (ArD 41525) and Asian (CPC-0740, SV0127-14) lineages were identified and amplified on Vero cells. The African isolate, ArD 41525, was isolated from a pool of A. africanus mosquitoes in 1984, Senegal. Both Asian isolates were collected from human patients in 2012 and 2014 in the Philippines and Thailand, respectively. The virus stocks were deep sequenced to determine sequences and to confirm the purity of virus stocks.40,42
The available ZIKV isolate sequences from Africa, Asia, and America were downloaded from GenBank and aligned (Supplemental Table 1). Genetic analysis was performed by comparing both NT and deduced AA sequences of the ORF (Table 2). Nucleotide and AA sequence identities of the isolates ranged from 88.3–99.8% to 96.3–99.9%, respectively. The NT and AA divergence between African and Asian/American isolates was ∼12% and ∼4%, respectively. The ArD 41525 isolate displayed ∼12% NT and ∼3% AA divergence from ZIKV isolates from the Americas. The CPC-0740 and SV0127-14 isolates displayed ∼2.3% and ∼1% NT, and ∼0.6% and ∼0.4% AA divergence compared with isolates from the Americas, respectively (Table 2).
Table 2.
Pairwise comparison of open reading frame of ZIKV isolates
| Uganda 1947 | Nigeria 1968 | Central African Republic 1976 | Senegal 1984 (41519) | Senegal 1984 (41525) | Micronesia 2007 | Malaysia 1966 | Cambodia 2010 | Philippines 2012 | Thailand 2014 | French Polynesia 2013 | Suriname 2015 | Puerto Rico 2015 | Guatemala 2015 | Mexico 2016 | Haiti 2014 | Brazil 2015 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uganda 1947 | – | 97.9 | 98.4 | 98.4 | 98.4 | 96.4 | 97 | 96.3 | 96.4 | 96.4 | 96.5 | 96.4 | 96.4 | 96.3 | 96.3 | 96.4 | 96.4 |
| Nigeria 1968 | 93.1 | – | 98.4 | 98.9 | 98.9 | 96.2 | 97 | 96.5 | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | 96.5 | 96.4 | 96.5 | 96.5 |
| Central African Republic 1976 | 94.8 | 93 | – | 98.9 | 98.9 | 96.8 | 97.6 | 97 | 97.2 | 97.1 | 97.2 | 97.1 | 97.1 | 97 | 97 | 97.1 | 97.1 |
| Senegal 1984 (41519) | 93.1 | 96.8 | 93.2 | – | 99.9 | 96.7 | 97.4 | 96.9 | 97 | 97 | 97.1 | 97 | 97 | 96.9 | 96.9 | 97 | 97 |
| Senegal 1984 (41525) | 93.2 | 96.9 | 93.3 | 99.6 | – | 96.6 | 97.4 | 96.9 | 97 | 97 | 97.1 | 97 | 97 | 96.9 | 96.9 | 97 | 97 |
| Micronesia 2007 | 88.7 | 88.4 | 88.6 | 88.6 | 88.6 | – | 98.6 | 99.2 | 99.2 | 99 | 99.2 | 99.1 | 99.1 | 99 | 98.9 | 99 | 99.1 |
| Malaysia 1966 | 89.8 | 89.7 | 90 | 90 | 90 | 95.8 | – | 98.8 | 98.9 | 98.7 | 98.9 | 98.9 | 98.9 | 98.8 | 98.7 | 98.9 | 98.8 |
| Cambodia 2010 | 88.4 | 88.3 | 88.6 | 88.5 | 88.6 | 98.3 | 95.7 | – | 99.4 | 99.4 | 99.6 | 99.5 | 99.5 | 99.4 | 99.4 | 99.5 | 99.5 |
| Philippines 2012 | 88.4 | 88.4 | 88.6 | 88.5 | 88.6 | 98.2 | 95.6 | 98 | – | 99.3 | 99.4 | 99.4 | 99.4 | 99.3 | 99.2 | 99.3 | 99.3 |
| Thailand 2014 | 88.3 | 88.3 | 88.6 | 88.4 | 88.5 | 98 | 95.4 | 98.4 | 97.7 | – | 99.7 | 99.6 | 99.6 | 99.5 | 99.5 | 99.6 | 99.6 |
| French Polynesia 2013 | 88.5 | 88.4 | 88.7 | 88.6 | 88.7 | 98.1 | 95.6 | 98.5 | 97.9 | 99.2 | – | 99.9 | 99.9 | 99.8 | 99.8 | 99.9 | 99.9 |
| Suriname 2015 | 88.4 | 88.4 | 88.7 | 88.5 | 88.6 | 97.9 | 95.4 | 98.3 | 97.6 | 99 | 99.7 | – | 99.9 | 99.8 | 99.7 | 99.8 | 99.9 |
| Puerto Rico 2015 | 88.5 | 88.4 | 88.8 | 88.6 | 88.7 | 97.9 | 95.5 | 98.3 | 97.6 | 99 | 99.7 | 99.7 | – | 99.8 | 99.7 | 99.8 | 99.9 |
| Guatemala 2015 | 88.4 | 88.3 | 88.6 | 88.5 | 88.6 | 97.9 | 95.5 | 98.3 | 97.7 | 99 | 99.7 | 99.6 | 99.6 | – | 99.6 | 99.7 | 99.8 |
| Mexico 2016 | 88.4 | 88.3 | 88.7 | 88.6 | 88.6 | 97.9 | 95.4 | 98.3 | 97.6 | 99 | 99.7 | 99.6 | 99.6 | 99.6 | – | 99.7 | 99.7 |
| Haiti 2014 | 88.4 | 88.3 | 88.7 | 88.5 | 88.6 | 98 | 95.5 | 98.4 | 97.8 | 99.1 | 99.8 | 99.7 | 99.7 | 99.7 | 99.7 | – | 99.8 |
| Brazil 2015 | 88.4 | 88.4 | 88.7 | 88.6 | 88.7 | 97.9 | 95.4 | 98.3 | 97.7 | 99 | 99.7 | 99.6 | 99.6 | 99.6 | 99.6 | 99.7 | – |
ZIKV = Zika virus. Nucleotide (NT) comparison below the diagonal in gray. Amino acid (AA) comparison above the diagonal in black.
A detailed genetic analysis was performed comparing isolates ArD 41525, CPC-0740, SV0127-14, and PRVABC59 (Supplemental Tables 2–4). There were 119 AA differences spread throughout the entire polyprotein, and the majority (75 of 119) distinguished the Asian and African lineages of ZIKV. One of these AA differences (NS4A-L4F) is found in the NS3/NS4A polyprotein site recognized by the viral protease. A total of 7 AA differences (prM-S17N, E-V473M, E-T487M, NS3-N400H, NS3-Y584H, NS4B-M98I, and NS5-M114V) were shared by contemporary Asian lineage viruses isolated in South/Central America and the Caribbean. There was one unique AA difference each for ZIKV strains ArD 41525 (NS3-I460V) and PRVABC59 (C-I80T). The ZIKV strain SV0127-14 had four unique AA differences (E-A227T, E-S368G, NS1-A45V, and NS5-I181V). The ZIKV strain CPC-0740 had seven unique AA differences (prM-A141V, NS2B-V67I, NS3-I115T, NS3-S347T, NS4A-M7I, NS4A-I78M, and NS4B-M26T) (Supplemental Tables 2–4).
To study the evolutionary relatedness of ZIKV isolates, phylogenetic trees were generated using the ML method. Similar to previous analyses, two main clades were observed: one main branch consisting of African isolates and another consisting of Asian/American isolates (Figure 1). The ArD 41525 isolate grouped with a 1968 isolate from Nigeria. The CPC-0740 and SV0127-14 isolates grouped with a 1966 isolate from Malaysia, basal to isolates from the Americas. The phylogenetic placement of the Asian isolates suggests multiple introduction of ZIKV from Mainland Southeast Asia to islands in the Pacific Ocean. Last, the high genetic identity and phylogenetic placement of SV0127-14 with isolates from the Americas demonstrate that it is a close relative of the parental virus introduced into the Americas from French Polynesia.
Figure 1.
Midpoint rooted maximum likelihood tree based on nucleotide (NT) sequence of the open reading frame. Bootstrap values of > 75% are shown at internal nodes. The scale bar represents NT substitutions per site.
Based on low-passage history, deep sequence confirmation, and genetic and phylogenetic analysis, ArD 41525, CPC-0740, and SV0127-14 isolates were selected for in vitro and in vivo characterization to investigate potential phenotypic differences.
In vitro characterization of ZIKV isolates.
Three low-passage isolates ArD 41525, CPC-0740, and SV0127-14 were amplified on Vero cells (Table 1). The ArD 41525 isolate readily induced cytopathic effects (CPEs) by 4 days post-infection (dpi), whereas 50% CPE was observed 6–8 dpi for CPC-0740 or SV0127-14. However, all three viruses yielded similar titers at 6 dpi (Table 1).
The plaque phenotype of all isolates was also investigated on Vero and HEK-293 cells. The ArD 41525 isolate produced plaques 3–4 mm in diameter by 5 dpi on Vero cell monolayers, whereas plaque sizes were 1–2 and < 1 mm for CPC-0740 and SV0127-14, respectively (Figure 2). Similar results were obtained in HEK-293 cells, with plaque sizes of 3–4 mm and 1–2 mm produced by ArD 41525 and CPC-0740, respectively (Figure 2). In contrast to infection of Vero cell monolayers, the SV0127-14 isolate did not produce visible plaques on HEK-293 by 5 dpi (Figure 2).
Figure 2.
Plaque phenotype of African (ArD 41525) and Asian (CPC-0740 and SV0127-14) Zika virus isolates in Vero and HEK-293 cell monolayers. Plaque phenotype was assessed in Vero cell monolayers; 5 days post-infection (dpi) cells were fixed and stained with crystal violet.
Multistep replication kinetics of ArD 41525 and CPC-0740 were investigated on four cell lines representing diverse vertebrate hosts; Vero-WHO (NHP), HEK-293 (human), DEF (avian), and RK-13 (rabbit) cells (Figure 3). Monolayers were infected at an MOI of 0.1, and virus replication was determined by plaque assays on Vero cell monolayers and quantitative RT-PCR (qRT-PCR). The ArD 41525 isolate yielded peak titers and genome copies of ∼4.0–8.0 log10 PFU/mL and ∼6.0–10.0 log10 ge/mL in all four cell lines, whereas the CPC-0740 titers and genome copies ranged from ∼2.0–7.0 log10 PFU/mL to ∼4.0–8.0 log10 ge/mL (Figure 3). The difference in peak titer and genome copies of ArD 41525 and CPC-0740 ranged from 10–10,000-fold to 100–100,000-fold, respectively (Figure 3). The CPC-0740 isolate also displayed delayed replication kinetics relative to ArD 41525, by 24–72 hours in Vero-WHO and HEK-293 cells (Figure 3). In addition, only minimal replication of the CPC-0740 isolate could be detected in DEF by either plaque assay or qRT-PCR.
Figure 3.
Replication kinetics of African (ArD 41525) and Asian (CPC-0740) Zika virus isolates in Vero-WHO, HEK-293, DEF, and RK-13 cell lines. Replication kinetics of all isolates were performed at multiplicity of infection of 0.1 plaque forming units (PFU)/cell in triplicate. Average titers ± standard deviation (SD) (error bars) are shown. P values ≤ 0.01 are indicated with *.
Replication kinetics of both isolates were also investigated in five cell lines representing insect hosts; C7/10 (A. albopictus), RML-12 (A. aegypti), AP-61 (A. pseudoscutellaris), A. gambiae, and PP-9 (P. papatasi) cells (Figure 4). Similar to infection of vertebrate cell lines, the ArD 41525 isolate yielded higher peak titers and genome copies, ∼4.0–9.0 log10 PFU/mL and ∼5.0–11.0 log10 ge/mL, respectively (Figure 4). By contrast, CPC-0740 isolate yielded peak titers and genome copies, ∼2.0–8.0 log10 PFU/mL and ∼3.0–8.3 log10 ge/mL, respectively (Figure 4). CPC-0740 also displayed a delay in replication kinetics by 24 to 96 hours in C7/10, AP-61, and PP-9 cells (Figure 4). Last, minimal replication of CPC-0740 isolate was detected in A. gambiae cells by either plaque assay or qRT-PCR.
Figure 4.
Replication kinetics of African (ArD 41525) and Asian (CPC-0740) Zika virus isolates in C7/10, RML-12, AP-61, Anopheles gambiae, and sandfly (PP-9) cell lines. Replication kinetics of all isolates were performed at multiplicity of infection of 0.1 plaque forming units (PFU)/cell in triplicate. Average titers ± standard deviation (SD) (error bars) are shown. P values ≤ 0.03 are indicated with *.
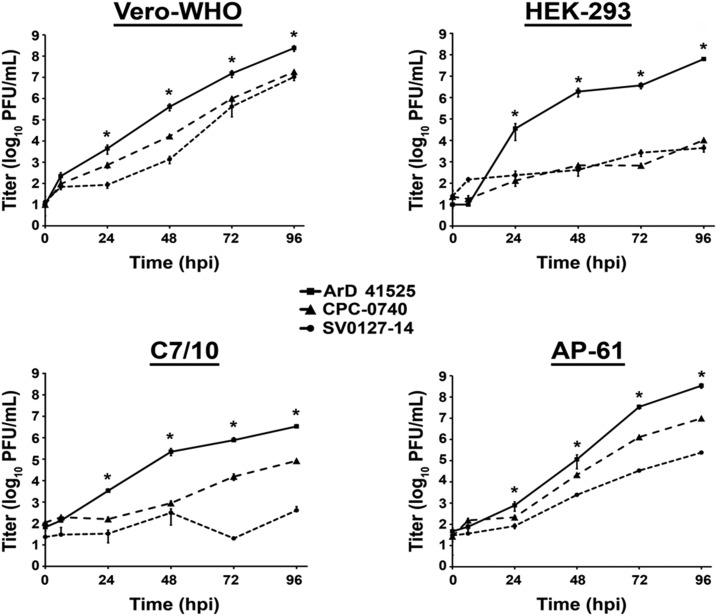
These data demonstrated that ArD 41525 isolate displayed greater fitness than CPC-0740 in vitro in the cells we tested. We next investigated whether the reduced fitness was also present in another low-passage Asian isolate, SV0127-14. Replication kinetics were performed with ArD 41525, CPC-0740, and SV0127-14 in Vero-WHO, HEK-293, C7/10, and AP-61 cells at an MOI of 0.1 (Figure 5). Similar to previous results, the peak titers of ArD 41525 isolate in Vero and HEK-293 were ∼8.0 and ∼7.0 log10 PFU/mL, respectively. By contrast, both CPC-0740 and SV0127-14 peak titers were ∼7.0 and ∼3.0 log10 PFU/mL, respectively, in Vero and HEK-293 cells (Figure 5). The difference in peak virus titer between ArD 41525 and either CPC-0740 or SV0127-14 isolates ranged from 10 to 1,000-fold throughout the course of infection (Figure 5). In addition, both CPC-0740 and SV0127-14 isolates displayed delayed replication kinetics by 24–96 hpi.
Figure 5.
Replication kinetics of African (ArD 41525) and Asian (CPC-0740, SV0127-14) Zika virus isolates in Vero-WHO, HEK-293, C7/10, and AP-61 cell lines. Replication kinetics of all isolates were performed at an multiplicity of infection of 0.1 plaque forming units (PFUs)/cell in triplicate. Average titers ± standard deviation (SD) (error bars) are shown. P values ≤ 0.02 are indicated with *.
ArD 41525 also achieved higher peak titers ∼6.0 and ∼8.0 log10 PFU/mL in both C7/10 and AP-61 cells, respectively (Figure 5). Whereas peak titers in C7/10 cells of CPC-0740 and SV0127-14 were ∼4.7 and ∼2.7 log10 PFU/mL, respectively. Similarly, CPC-0740 and SV0127-14 yielded titers of ∼6.7 and ∼4.7 log10 PFU/mL, respectively, in AP-61 cells (Figure 5). The differences in virus titer between isolates ranged from 10 to 10,000-fold. Last, the replication kinetics of CPC-0740 and SV0127-14 isolates were delayed relative to ArD 41525 by 24 to 96 hpi (Figure 5). Taken together, these data suggest that both low-passage human isolates from Asia display a fitness reduction in vitro.
Oral infection studies in A. aegypti mosquitoes.
We next investigated whether the phenotypic differences observed in vitro extend to in vivo models. Cohorts of female A. aegypti mosquitoes were fed 6.0 log10 PFU/mL of ArD 41525, CPC-0740, and SV0127-14 isolates. Kinetics of virus replication was measured by detecting virus using the plaque assay at 5 and 7 dpi (Table 3). The body infection rates of mosquitoes exposed to ArD 41525 were 47% and 44% at 5 and 7 dpi, respectively. Virus titer in infected bodies ranged from ∼1.0 to 5.0 log10 PFU, and geometric mean virus titers of 2.7 and 3.0 log10 PFU at 5 and 7 dpi, respectively (Table 3). By contrast, infection rates with CPC-0740 were 15% and 5% at 5 and 7 dpi, respectively. In addition, the mean body titer was ∼10-fold lower in CPC-0740–infected mosquitoes (Table 3). Surprisingly, the SV0127-14 isolate-exposed mosquitoes had comparable body infection rates and viral load at both 5 and 7 dpi. The infection rates at 5 and 7 dpi were 35% and 45% versus 47% and 44%, respectively, for SV0127-14 and ArD 41525. However, disseminated infection was not observed in mosquitoes infected with either of the Asian isolates. Disseminated infection was observed only in mosquitoes infected with ArD 41525 isolate at 5 dpi with a rate of 12% and an average virus titer of 102 PFU/mosquito.
Table 3.
Oral infections of African (ArD 41525) and Asian (CPC-0740 and SV0127-14) ZIKV isolates in Aedes aegypti
| ZIKV Isolates | Blood meal titers | 5 DPI | 7 DPI | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Body | Legs and wings | Body | Legs and wings | ||||||
| log10 PFU/mL | % Infected (infected/total) | Average titer (log10 PFU/mosquito) (± SD) | % Infected (infected/total) | Average titer (log10 PFU/mosquito) (± SD) | % Infected (infected/total) | Average titer (log10 PFU/mosquito) (± SD) | % Infected (infected/total) | Average titer (log10 PFU/mosquito) (± SD) | |
| ArD 41525 | 6.32 | 47 (9/19) | 2.9 (± 1.8) | 12 (2/19) | 2.0 (± 1.2) | 44 (7/16) | 2.5 (± 1.2) | 0 | B.L.D. |
| CPC-0740 | 6.33 | 15 (3/20) | 2.0 (± 0.5) | 0 | B.L.D. | 5 (1/20) | 4.2 | 0 | B.L.D. |
| SV0127-14 | 6.36 | 35 (7/20) | 3.0 (± 0.4) | 0 | B.L.D. | 45 (9/20) | 2.8 (± 0.7) | 0 | B.L.D. |
B.L.D. = below the limit of detection (10 PFU/mL); DPI = days post-infection; PFU = plaque forming unit; SD = standard deviation; ZIKV = Zika virus. Mosquito bodies and legs/wings homogenates at 5 and 7 dpi were analyzed for infectious virus via plaque assays on Vero cell monolayers. Bold indicates P value ≤ 0.01.
Murine infections.
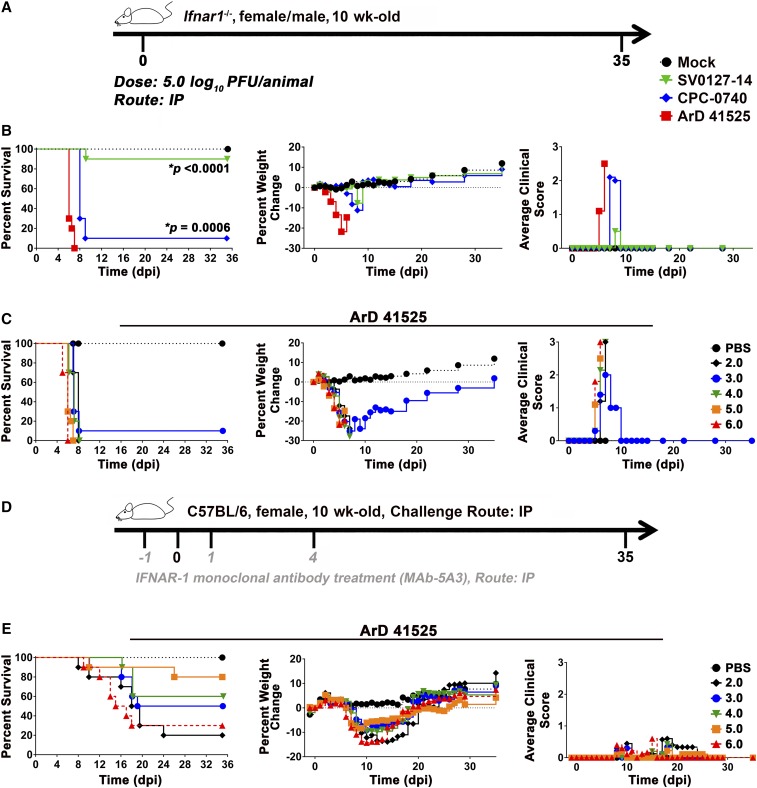
The potential phenotypic differences of ArD 41525, CPC-0740, and SV0127-14 were also investigated in 10-week-old C56BL/6 Ifnar1−/− mice and 10- and 5-week-old wild-type C56BL/6 mice in the IFN type I (IFN-I) antibody blockade model recently described by Smith et al.43 Cohorts of 10 Ifnar1−/− mice were inoculated at 5.0 log10 PFU/animal IP and followed for clinical disease (Figure 6A and B). All mice infected with isolate ArD 41525 displayed clinical signs, weight loss, and met euthanasia criteria by 7 dpi. Similar results were obtained for mice infected with CPC-0740 isolate, and 90% of mice were euthanized by 9 dpi. By contrast, mice infected with the SV0127-14 isolate displayed minimal clinical signs and weight loss, and only 10% of the mice met euthanasia criteria by 9 dpi (Figure 6B).
Figure 6.
Infection of African and Asian isolated in ifnar−/− and IFNAR1-blocking MAb murine models. Outline of the study design in mice (A and D). African (ArD 41525) and Asian (CPC-0740 and SV0127-14) Zika virus isolates in 5-week-old C57BL/6 ifnar−/− mice (B). Cohorts of 10 males and females were infected with 5.0 log10 plaque forming units (PFUs) through intraperitoneal (IP) route. Fifty percent lethal dose (LD50) studies of the African (ArD 41525) isolate in ifnar−/− and IFNAR1-blocking MAb murine models (C and E). Cohorts of 10 males and females were infected with doses ranging from 6.0 to 2.0 log10 PFU through IP route. After infection, animals were monitored for survival, weight loss, and clinical scores (C and E). P values ≤ 0.0006 are indicated with *.
In an attempt to determine the 50% lethal dose (LD50) in both the Ifnar1−/− and IFN-I antibody blockade mouse models, cohorts of 10 mice were inoculated at doses ranging from 6.0 to 2.0 log10 PFU/animal IP and followed for clinical disease (Figure 6C–E). The infection of Ifnar1−/− mice with ArD 41525 produced clinical scores of 2–3, 20–30% weight loss, and almost all animals met the euthanasia criteria by 8 dpi regardless of the dose (Figure 6D). By contrast, the infection in the IFN-I antibody blockade model was less severe with delay in onset of clinical signs, weight loss, and mortality. The clinical scores ranged from 0 to 1, 5–15% weight loss, mortality rates of 20–80% (Figure 6E). The 6.0 and 2.0 log10 PFU/animal doses produced 80% and 70% mortality rates, respectively, whereas, mortality rates ranged from 20% to 50% at the intermediate doses (3.0–5.0 log10 PFU/animal) (Figure 6E). Although the sensitivity to ArD 41525 infection differed in both Ifnar1−/− and IFN-I antibody blockade models, we were unable to obtain an LD50 in either model.
We next investigated the LD50 of multiple ZIKV strains in younger 5-week-old mice using the IFN-I antibody blockade model at doses ranging from 6.0 to 2.0 log10 PFU/animal (Figure 7). Mice infected with ArD 41525 isolate displayed significant clinical signs, weight loss, and 80–100% of the mice were euthanized at all doses (Figure 7B). By contrast, mice infected with CPC-0740 isolate did not display any clinical signs or weight loss, and all animals survived (Figure 7C). We also evaluated the LD50 of another more recent Asian/American isolate, PRVABC59 isolated from Puerto Rico, by IP exposure of 5-week-old wild-type C56BL/6 mice in the IFN-I antibody blockade murine model. Similar to CPC-0740, none of the mice succumbed or showed signs of disease (Figure 7D). Collectively, the infection with ArD 41525 isolate of both 5-week-old Ifnar1−/− and wild-type C56BL/6 mice in the IFN-I antibody blockade model demonstrated that the isolate was more virulent than the Asian/American isolates.
Figure 7.
Infection of African (ArD 41525) (B) and Asian (CPC-0740 [C], PRVABC59 [D]) Zika virus isolates in 5-week old C57BL/6 mice treated with IFNAR1-blocking MAb. Outline of the study design in mice (A). Cohorts of 10 females were infected with 6.0–2.0 log10 plaque forming units (PFUs) through intraperitoneal (IP) route. After infection, animals were monitored for survival, weight loss, and clinical scores (B–D). P values < 0.0001 are indicated with *.
DISCUSSION
The sudden and unexpected arrival of ZIKV in the Americas and the severe complication of infection including GBS and microcephaly have prompted intense research on many aspects of the ZIKV life cycle, therapeutics, and vaccines. However, there are extensive gaps in many aspects of ZIKV biology. Before the outbreak in the Americas, limited studies were available on characterization of ZIKV isolates, and these almost exclusively used African lineage isolates. The aim of this study was to systematically characterize and investigate the potential phenotypic differences among Asian and African isolates. The isolates were selected based on several criteria: complete isolation and passage histories, genomic sequence data determined by deep sequencing, and genetic and phylogenetic analyses to determine the relationship between isolates. Based on all these criteria, three low-passage and uncontaminated isolates representing African (ArD 41525) and Asian (CPC-0740 and SV0127-14) lineages were identified for in vitro and in vivo studies.
All three isolates were able to infect cell lines from representative vertebrate and insect hosts at both 28°C and 37°C, demonstrating broad host and temperature range. However, substantial differences were observed between African and Asian isolates in mammalian and vertebrate cell lines. ArD 41525 consistently reached higher titers at many time points in multistep growth kinetics in all cell lines. In addition, the isolate displayed severe clinical disease and mortality rates in both Ifnar1−/− and IFN-I antibody blockade murine models. These data suggested greater virulence of ArD 41525 than either of the two Asian isolates. This was further supported by the inability to achieve an LD50 with the Asian isolates in both Ifnar1−/− and IFN-I antibody blockade murine models. In contrast to the in vitro and murine studies, infection of A. aegypti with the isolates yielded varied results. Both SV0127-14 and CPC-0740 displayed reduced fitness compared with ArD 41525; however, the infection in mosquitoes yielded comparable or lower body infection rates and virus titers, respectively. Although body infection rates varied among the isolates, only ArD 41525-infected mosquitoes exhibited disseminated infection. These data demonstrate substantial phenotypic differences between ArD 41525 and either of the Asian isolates.
There were also phenotypic differences observed between Asian isolates. SV0127-14 virus titers were ∼10- to 100-fold lower than CPC-0740 in both vertebrate and mosquito cells. It produced minimal clinical disease and only 10% mortality in Ifnar1−/− mice even with a high virus titer exposure. The isolate was not further characterized in the IFN-I antibody blockade model because of the attenuated phenotype in Ifnar1−/− mice, which is the more susceptible mouse model. The differing susceptibilities of the Ifnar1−/− mice versus mice treated with the IFN-I antibody are highlighted by the results for CPC-0740 in the two mouse models. When Ifnar1−/− mice were infected with CPC-0740, 90% mortality was observed, which is in contrast to no mortality in the IFN-1 antibody blockade mouse model. Ifnar1−/− mice are likely more susceptible because the IFN-I response is completely eliminated compared with a temporal blockade with the IFN-I blocking antibody. Last, infection with CPC-0740 in mosquitoes produced 2- to 9-fold lower infection rates than with SV0127-14. These data show that closely related spatiotemporal isolates can exhibit considerable phenotypic differences that warrant further investigation.
Limited studies have compared phenotypic differences among ZIKV isolates. Previous studies with Asian isolates showed reduced replication kinetics and peak virus titers in vitro, mosquito infection and dissemination rates, reduced virulence in various murine knockout models, tissue distribution, virus replication, weight loss, clinical disease when compared with African isolates.43–51 To further examine these differences, we examined the virulence of the PRVABC59 isolate from Puerto Rico in the IFN-I antibody blockade mouse model. Similar to the results obtained with the CPC-0740 isolate, infection produced no clinical disease or lethality. One potential complication of comparing of ZIKV isolates is passage histories. Isolates passaged in vitro and/or in vivo may have fitness trade-offs in either vertebrate or insect hosts. However, a recent study using a cDNA clone derived from RNA isolated from tissues of a microcephaly case (ZIKVNatal) demonstrated an attenuated phenotype both in vitro and in vivo relative to the African MR-766 isolate.50 Taken together, the data support the hypothesis that there may be lineage-specific differences of ZIKV isolates.
The genetic determinants of ZIKV that enable phenotypic differences among isolates remain unknown. The African and Asian isolates differ at ∼75–100 AA residues spread throughout an ORF consisting of 3,424 AAs. The Asian and American isolates differ by ∼10–30 AAs. The impact of small AA divergence in context of protection against heterologous ZIKV infections is likely to be minimal; however, the mutation/s could have considerable impact on isolate phenotype. Studies with flaviviruses have shown that substantial fitness gains or losses can be achieved by single or a few point mutation/s.52–73 The SV0127-14 and CPC-0740 isolates differ by five unique AAs and yet produce substantially different phenotypes in vitro and in vivo. Two recent ZIKV studies have shown similar results. Two cDNA clones of isolates from the Americas (BeH819015 and SPH2015) that differed by 6 NTs yielded phenotypic differences in mice. The BeH819015 isolate produced lower clinical disease, weight loss, and mortality in a murine model than SPH2015.41 Another study demonstrated that an alanine-to-valine AA substitution in NS1 enhanced infection in A. aegypti.74
Mutagenesis studies of flaviviruses have shown the importance of the E protein in regard to virus fitness. Mutation/s in all three domains of the E protein can drastically alter phenotype both in vitro and/or in vivo. The ZIKV E protein of the four isolates that we studied differs by seven mutations that are spread over domains I, II, and III (Supplemental Figure 1).53–56,71–73 Two mutations in the SV0127-14 isolate are of note, A227T (domain II) and S368G (domain III). Both A227 and S368 are highly conserved in both African and Asian lineages and may contribute to the attenuated phenotype of SV0127-14. These mutation/s altering fitness require further investigation.
In summary, we have demonstrated substantial phenotypic differences both in vitro and in vivo between ZIKV isolates representing African and Asian lineages. These data highlight the need for basic characterization of ZIKV isolates, as the utilization of uncharacterized isolates could have potential consequences for animal model and therapeutic/vaccine development when wild-type virulence and mosquito infectivity are desired.
Supplementary Material
Supplemental Table and Figure.
Note: Supplemental tables and figures appear at www.ajtmh.org.
REFERENCES
- 1.Fields BN, Knipe DM, Howley PM, 2013. Flaviviridae in Fields Virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins. [Google Scholar]
- 2.Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ, 2016. The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352: 467–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick GW, Kitchen SF, Haddow AJ, 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46: 509–520. [DOI] [PubMed] [Google Scholar]
- 4.Boorman JP, Porterfield JS, 1956. A simple technique for infection of mosquitoes with viruses; transmission of Zika virus. Trans R Soc Trop Med Hyg 50: 238–242. [DOI] [PubMed] [Google Scholar]
- 5.Dick GW, 1952. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg 46: 521–534. [DOI] [PubMed] [Google Scholar]
- 6.Moore DL, Causey OR, Carey DE, Reddy S, Cooke AR, Akinkugbe FM, David-West TS, Kemp GE, 1975. Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann Trop Med Parasitol 69: 49–64. [DOI] [PubMed] [Google Scholar]
- 7.Fagbami A, 1977. Epidemiological investigations on arbovirus infections at Igbo-Ora, Nigeria. Trop Geogr Med 29: 187–191. [PubMed] [Google Scholar]
- 8.Fagbami AH, 1979. Zika virus infections in Nigeria: virological and seroepidemiological investigations in Oyo State. J Hyg (Lond) 83: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robin Y, Mouchet J, 1975. Serological and entomological study on yellow fever in Sierra Leone [in French]. Bull Soc Pathol Exot Filiales 68: 249–258. [PubMed] [Google Scholar]
- 10.Olson JG, Ksiazek TG, Suhandiman, Triwibowo, 1981. Zika virus, a cause of fever in Central Java, Indonesia. Trans R Soc Trop Med Hyg 75: 389–393. [DOI] [PubMed] [Google Scholar]
- 11.Jan C, Languillat G, Renaudet J, Robin Y, 1978. A serological survey of arboviruses in Gabon [in French]. Bull Soc Pathol Exot Filiales 71: 140–146. [PubMed] [Google Scholar]
- 12.Saluzzo JF, Gonzalez JP, Herve JP, Georges AJ, 1981. Serological survey for the prevalence of certain arboviruses in the human population of the south-east area of Central African Republic (author’s transl) [in French]. Bull Soc Pathol Exot Filiales 74: 490–499. [PubMed] [Google Scholar]
- 13.Saluzzo JF, Ivanoff B, Languillat G, Georges AJ, 1982. Serological survey for arbovirus antibodies in the human and simian populations of the south-east of Gabon (author’s transl) [in French]. Bull Soc Pathol Exot Filiales 75: 262–266. [PubMed] [Google Scholar]
- 14.Marchette NJ, Garcia R, Rudnick A, 1969. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg 18: 411–415. [DOI] [PubMed] [Google Scholar]
- 15.Monlun E, et al. 1993. Surveillance of the circulation of arbovirus of medical interest in the region of eastern Senegal [in French]. Bull Soc Pathol Exot Filiales 86: 21–28. [PubMed] [Google Scholar]
- 16.Akoua-Koffi C, Diarrassouba S, Benie VB, Ngbichi JM, Bozoua T, Bosson A, Akran V, Carnevale P, Ehouman A, 2001. Investigation surrounding a fatal case of yellow fever in Cote d’Ivoire in 1999 [in French]. Bull Soc Pathol Exot Filiales 94: 227–230. [PubMed] [Google Scholar]
- 17.Olson JG, Ksiazek TG, Gubler DJ, Lubis SI, Simanjuntak G, Lee VH, Nalim S, Juslis K, See R, 1983. A survey for arboviral antibodies in sera of humans and animals in Lombok, Republic of Indonesia. Ann Trop Med Parasitol 77: 131–137. [DOI] [PubMed] [Google Scholar]
- 18.McCrae AW, Kirya BG, 1982. Yellow fever and Zika virus epizootics and enzootics in Uganda. Trans R Soc Trop Med Hyg 76: 552–562. [DOI] [PubMed] [Google Scholar]
- 19.Darwish MA, Hoogstraal H, Roberts TJ, Ahmed IP, Omar F, 1983. A sero-epidemiological survey for certain arboviruses (Togaviridae) in Pakistan. Trans R Soc Trop Med Hyg 77: 442–445. [DOI] [PubMed] [Google Scholar]
- 20.Dowd KA, et al. 2016. Broadly neutralizing activity of Zika virus-immune sera identifies a single viral serotype. Cell Reports 16: 1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC, 2012. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis 6: e1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy MR, et al. 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360: 2536–2543. [DOI] [PubMed] [Google Scholar]
- 23.Grard G, Caron M, Mombo IM, Nkoghe D, Mboui Ondo S, Jiolle D, Fontenille D, Paupy C, Leroy EM, 2014. Zika virus in Gabon (Central Africa)–2007: a new threat from Aedes albopictus? PLoS Negl Trop Dis 8: e2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musso D, Nilles EJ, Cao-Lormeau VM, 2014. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infect 20: O595–O596. [DOI] [PubMed] [Google Scholar]
- 25.Campos GS, Bandeira AC, Sardi SI, 2015. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis 21: 1885–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanluca C, Melo VC, Mosimann AL, Santos GI, Santos CN, Luz K, 2015. First report of autochthonous transmission of Zika virus in Brazil. Mem Inst Oswaldo Cruz 110: 569–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musso D, 2015. Zika virus transmission from French Polynesia to Brazil. Emerg Infect Dis 21: 1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musso D, Gubler DJ, 2016. Zika virus. Clin Microbiol Rev 29: 487–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen LR, Jamieson DJ, Powers AM, Honein MA, 2016. Zika virus. N Engl J Med 374: 1552–1563. [DOI] [PubMed] [Google Scholar]
- 30.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F, 2014. Zika virus infection complicated by Guillain-Barre syndrome–case report, French Polynesia, December 2013. Euro Surveill 19: 20720. [DOI] [PubMed] [Google Scholar]
- 31.Krauer F, Riesen M, Reveiz L, Oladapo OT, Martinez-Vega R, Porgo TV, Haefliger A, Broutet NJ, Low N; WHO Zika Causality Working Group , 2017. Zika virus infection as a cause of congenital brain abnormalities and Guillain-Barre syndrome: systematic review. PLoS Med 14: e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB, 2011. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis 17: 880–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Ortenzio E, Matheron S, Yazdanpanah Y, de Lamballerie X, Hubert B, Piorkowski G, Maquart M, Descamps D, Damond F, Leparc-Goffart I, 2016. Evidence of sexual transmission of Zika virus. N Engl J Med 374: 2195–2198. [DOI] [PubMed] [Google Scholar]
- 34.Haddow AD, et al. 2017. High infection rates for adult macaques after intravaginal or intrarectal inoculation with Zika virus. Emerg Infect Dis 23: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouy M, Guindon S, Gascuel O, 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- 37.Felsenstein J, 1989. PHYLIP: phylogeny inference package (Version 3.2). Cladistics 5: 164–166. [Google Scholar]
- 38.Posada D, Crandall KA, 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 39.Pilitt DR, Jones JC, 1972. A qualitative method for estimating the degree of engorgement of Aedes aegypti adults. J Med Entomol 9: 334–337. [DOI] [PubMed] [Google Scholar]
- 40.Ellison DW, et al. 2016. Complete genome sequences of Zika virus strains isolated from the blood of patients in Thailand in 2014 and the Philippines in 2012. Genome Announc 4: pii: e00359–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widman DG, et al. 2017. A reverse genetics platform that spans the Zika virus family tree. MBio 8: pii: e02014–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ladner JT, et al. 2016. Complete genome sequences of five Zika virus isolates. Genome Announc 4: pii: e00377–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith DR, et al. 2017. Neuropathogenesis of Zika virus in a highly susceptible immunocompetent mouse model after antibody blockade of type I interferon. PLoS Negl Trop Dis 11: e0005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS, 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe 19: 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weger-Lucarelli J, et al. 2016. Vector competence of American mosquitoes for three strains of Zika virus. PLoS Negl Trop Dis 10: e0005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simonin Y, et al. 2016. Zika virus strains potentially display different infectious profiles in human neural cells. EBioMedicine 12: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC, 2016. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg 94: 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan JF, et al. 2016. Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg Microbes Infect 5: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R, 2016. A susceptible mouse model for Zika virus infection. PLoS Negl Trop Dis 10: e0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Setoh YX, Prow NA, Peng N, Hugo LE, Devine G, Hazlewood JE, Suhrbier A, Khromykh AA, 2017. De novo generation and characterization of New Zika virus isolate using sequence data from a microcephaly case. mSphere 2: pii: e00190–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duggal NK, Ritter JM, McDonald EM, Romo H, Guirakhoo F, Davis BS, Chang GJ, Brault AC, 2017. Differential neurovirulence of African and Asian genotype Zika virus isolates in outbred immunocompetent mice. Am J Trop Med Hyg 97: 1410–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muylaert IR, Galler R, Rice CM, 1997. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol 71: 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arroyo J, Guirakhoo F, Fenner S, Zhang ZX, Monath TP, Chambers TJ, 2001. Molecular basis for attenuation of neurovirulence of a yellow fever virus/Japanese encephalitis virus chimera vaccine (ChimeriVax-JE). J Virol 75: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arroyo J, Miller C, Catalan J, Myers GA, Ratterree MS, Trent DW, Monath TP, 2004. ChimeriVax-West Nile virus live-attenuated vaccine: preclinical evaluation of safety, immunogenicity, and efficacy. J Virol 78: 12497–12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandl CW, Allison SL, Holzmann H, Meixner T, Heinz FX, 2000. Attenuation of tick-borne encephalitis virus by structure-based site-specific mutagenesis of a putative flavivirus receptor binding site. J Virol 74: 9601–9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monath TP, Arroyo J, Levenbook I, Zhang ZX, Catalan J, Draper K, Guirakhoo F, 2002. Single mutation in the flavivirus envelope protein hinge region increases neurovirulence for mice and monkeys but decreases viscerotropism for monkeys: relevance to development and safety testing of live, attenuated vaccines. J Virol 76: 1932–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Z, Date T, Li Y, Kato T, Miyamoto M, Yasui K, Wakita T, 2005. Characterization of the E-138 (Glu/Lys) mutation in Japanese encephalitis virus by using a stable, full-length, infectious cDNA clone. J Gen Virol 86: 2209–2220. [DOI] [PubMed] [Google Scholar]
- 58.Hurrelbrink RJ, McMinn PC, 2001. Attenuation of Murray Valley encephalitis virus by site-directed mutagenesis of the hinge and putative receptor-binding regions of the envelope protein. J Virol 75: 7692–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu WJ, Chen HB, Khromykh AA, 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J Virol 77: 7804–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Wispelaere M, Yang PL, 2012. Mutagenesis of the DI/DIII linker in dengue virus envelope protein impairs viral particle assembly. J Virol 86: 7072–7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bray M, Men R, Tokimatsu I, Lai CJ, 1998. Genetic determinants responsible for acquisition of dengue type 2 virus mouse neurovirulence. J Virol 72: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gualano RC, Pryor MJ, Cauchi MR, Wright PJ, Davidson AD, 1998. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J Gen Virol 79: 437–446. [DOI] [PubMed] [Google Scholar]
- 63.Prestwood TR, Prigozhin DM, Sharar KL, Zellweger RM, Shresta S, 2008. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J Virol 82: 8411–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tajima S, Nerome R, Nukui Y, Kato F, Takasaki T, Kurane I, 2010. A single mutation in the Japanese encephalitis virus E protein (S123R) increases its growth rate in mouse neuroblastoma cells and its pathogenicity in mice. Virology 396: 298–304. [DOI] [PubMed] [Google Scholar]
- 65.Rumyantsev AA, Murphy BR, Pletnev AG, 2006. A tick-borne Langat virus mutant that is temperature sensitive and host range restricted in neuroblastoma cells and lacks neuroinvasiveness for immunodeficient mice. J Virol 80: 1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jerzak GV, Brown I, Shi PY, Kramer LD, Ebel GD, 2008. Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology 374: 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grubaugh ND, et al. 2015. Experimental evolution of an RNA virus in wild birds: evidence for host-dependent impacts on population structure and competitive fitness. PLoS Pathog 11: e1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deardorff ER, Fitzpatrick KA, Jerzak GV, Shi PY, Kramer LD, Ebel GD, 2011. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog 7: e1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vasilakis N, Deardorff ER, Kenney JL, Rossi SL, Hanley KA, Weaver SC, 2009. Mosquitoes put the brake on arbovirus evolution: experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog 5: e1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goo L, VanBlargan LA, Dowd KA, Diamond MS, Pierson TC, 2017. A single mutation in the envelope protein modulates flavivirus antigenicity, stability, and pathogenesis. PLoS Pathog 13: e1006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Erb SM, Butrapet S, Moss KJ, Luy BE, Childers T, Calvert AE, Silengo SJ, Roehrig JT, Huang CY, Blair CD, 2010. Domain-III FG loop of the dengue virus type 2 envelope protein is important for infection of mammalian cells and Aedes aegypti mosquitoes. Virology 406: 328–335. [DOI] [PubMed] [Google Scholar]
- 72.Beasley DW, Whiteman MC, Zhang S, Huang CY, Schneider BS, Smith DR, Gromowski GD, Higgs S, Kinney RM, Barrett AD, 2005. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J Virol 79: 8339–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gromowski GD, Firestone CY, Whitehead SS, 2015. Genetic determinants of Japanese encephalitis virus vaccine strain SA14-14-2 that govern attenuation of virulence in mice. J Virol 89: 6328–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, et al. 2017. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature 545: 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table and Figure.