ABSTRACT
Among bacteria, only a single styrene-specific degradation pathway has been reported so far. It comprises the activity of styrene monooxygenase, styrene oxide isomerase, and phenylacetaldehyde dehydrogenase, yielding phenylacetic acid as the central metabolite. The alternative route comprises ring-hydroxylating enzymes and yields vinyl catechol as central metabolite, which undergoes meta-cleavage. This was reported to be unspecific and also allows the degradation of benzene derivatives. However, some bacteria had been described to degrade styrene but do not employ one of those routes or only parts of them. Here, we describe a novel “hybrid” degradation pathway for styrene located on a plasmid of foreign origin. As putatively also unspecific, it allows metabolizing chemically analogous compounds (e.g., halogenated and/or alkylated styrene derivatives). Gordonia rubripertincta CWB2 was isolated with styrene as the sole source of carbon and energy. It employs an assembled route of the styrene side-chain degradation and isoprene degradation pathways that also funnels into phenylacetic acid as the central metabolite. Metabolites, enzyme activity, genome, transcriptome, and proteome data reinforce this observation and allow us to understand this biotechnologically relevant pathway, which can be used for the production of ibuprofen.
IMPORTANCE The degradation of xenobiotics by bacteria is not only important for bioremediation but also because the involved enzymes are potential catalysts in biotechnological applications. This study reveals a novel degradation pathway for the hazardous organic compound styrene in Gordonia rubripertincta CWB2. This study provides an impressive illustration of horizontal gene transfer, which enables novel metabolic capabilities. This study presents glutathione-dependent styrene metabolization in an (actino-)bacterium. Further, the genomic background of the ability of strain CWB2 to produce ibuprofen is demonstrated.
KEYWORDS: genomic island, glutathione S-transferase, glutathione in actinobacteria, horizontal gene transfer, hybrid gene cluster, microbial ibuprofen production, proteomics, styrene monooxygenase, transcriptomics, xenobiotic compounds
INTRODUCTION
Styrene is a monoaromatic compound that naturally occurs as a component of tar and in volatile and oily substances from plants and food, but it can also be produced by microorganisms. Styrene is of high relevance in industry and produced in million tonne scale, causing substantial anthropogenic release. This is problematic, as it is hazardous for living organisms (1–3). Due to the disposability, it is corollary that organisms evolved strategies to detoxify and/or use styrene as a source of energy and carbon (3–5).
Styrene can be channeled through different unspecific degradation pathways due to relaxed substrate specificity of the respective enzymes (see the supplemental material and reference 3 for details). However, only one styrene-specific degradation pathway is known, and it seems to be favored by microorganisms under aerobic conditions (3, 5, 6). This upper degradation pathway is initiated by oxidation of the vinyl side chain. A styrene monooxygenase (SMO) produces (S)-styrene oxide, which is converted by a membrane-bound styrene oxide isomerase (SOI) to phenylacetaldehyde. A phenylacetaldehyde dehydrogenase (PAD) oxidizes the aldehyde to phenylacetic acid (PAA) (7). PAA is a central catabolite and metabolized in the so-called lower degradation pathway, which is present in about 16% of all genome-sequenced microorganisms (8, 9). That route has been described for several proteobacteria (Pseudomonas, Xanthobacter, and Sphingopyxis), actinobacteria (Rhodococcus and Corynebacterium), and fungi (Exophiala) (reviewed by Tischler [3]).
Rhodococcus sp. strain ST-10 has an incomplete degradation cluster lacking the SOI, though it is still able to use styrene as a sole source of carbon and energy (10–13). This gene cluster comprises the SMO and a putative (partial) open reading frame (ORF), designated ORF3. It was hypothesized that the SOI can be bypassed by chemical conversion of styrene oxide to phenylacetaldehyde or enzymatically (12–14). However, chemical conversion is unlikely, and no probable enzymes were identified (3); thus, the degradation pathway for strain ST-10 remains unclear.
The genus Gordonia is known to be a versatile degrader of aromatic compounds (15, 16), and Gordonia rubripertincta CWB2 in particular is able to metabolize styrene and related compounds. As previously described, strain CWB2 was obtained from a soil sample and separated via styrene-enrichment culture (17–19). Moreover, it was shown that strain CWB2 is able to produce ibuprofen from 4-isobutyl-α-methylstyrene in a cometabolic process (17, 18). Oelschlägel et al. (18) reported that other styrene degraders are not capable to catalyze this reaction and therefore proposed substantial differences in the respective enzymatic cascades. Interestingly, Gordonia rubripertincta CWB2 has a cluster that is a homolog to the partial one of strain ST-10.
In this study, we identified the complete gene cluster, which enables styrene degradation in strain CWB2. Therefore, we studied the transcriptome and the proteome under styrene exposure. Further, we measured the activity of key enzymes to clarify the root of the metabolic potential of Gordonia rubripertincta CWB2. Moreover, this gene cluster seems to be alien since it is located on a plasmid and considered a genomic island if compared to the rest of the genome. It embodies a hybrid of several homolog epoxide and aromatic compound degradation clusters from different actinobacteria.
RESULTS
Identification and annotation of gene clusters associated with styrene degradation.
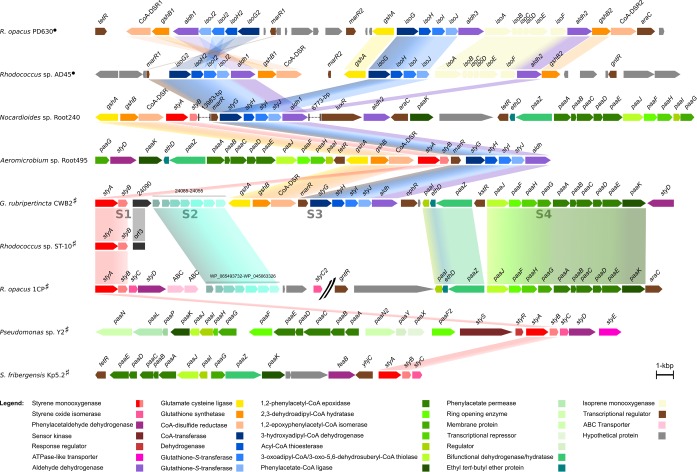
Genes that might be involved in styrene degradation in G. rubripertincta CWB2 were identified and annotated by homology search using the BLASTP algorithm (20) on the nonredundant protein database or the UniProtKB database (National Center for Biotechnology Information [NCBI]). Annotations of the putative styrene degradation clusters with respect to the closest (characterized) homologs are listed in Table 1 and Data Set S1 in the supplemental material. A 32,424-bp cluster with 36 putative ORFs (orf) was identified on plasmid pGCWB2 (∼100 kbp), which is framed by genes encoding a styrene monooxygenase (GCWB2_24100) and a phenylacetaldehyde dehydrogenase (GCWB2_23925). Interestingly, the average GC content of this cluster is 62.11% and thus 5% lower than that for the whole genome. The GC content of the whole plasmid is 3% lower than that in the chromosome. In addition to a large amount of hypothetical proteins, pGCWB2 contains four transposase-family proteins, two integrase-like proteins, one relaxase-like protein, and one type IV secretory system as an inventory for gene mobility. Genomic island analysis of the whole genome illustrates that at least parts of this styrene degradation cluster have a foreign origin (Fig. 1).
TABLE 1.
Functional categorization of proteins from G. rubripertincta CWB2 that are supposed to be involved in styrene degradation and regulation on RNA and protein levela
| Clusterc | ORF | Gene | Transcriptome |
Proteome |
Best hit on the Uniprot database at amino acid level |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | M | Cyt | Mem | Nameb | Functiond | Accession no. | % ID | Ref | |||
| S4 | 23925 | styD | 7.7 | 8.6 | 7.1 | 5.7 | styD | Phenylacetaldehyde dehydrogenase | BAL04135 | 76 | 13, 50 |
| 23930 | paaK | 8.1 | 6.0 | 8.1 | 5.9 | paaK | Phenylacetate-CoA ligase | Q9L9C1 | 68 | 94 | |
| 23935 | paaE | 8.0 | 6.2 | 11.3 | 8.2 | paaE | 1,2-Phenylacetyl-CoA epoxidase, subunit E | P76081 | 43 | 8 | |
| 23940 | paaD | 8.1 | 6.7 | NaNf | NaNf | paaD | 1,2-Phenylacetyl-CoA epoxidase, subunit D | P76080 | 42 | 8 | |
| 23945 | paaC | 8.3 | 6.7 | 10.5 | 7.4 | paaC | 1,2-Phenylacetyl-CoA epoxidase, subunit C | P76079 | 42 | 8 | |
| 23950 | paaB | 8.0 | 6.8 | 8.8 | 9.1 | paaB | 1,2-Phenylacetyl-CoA epoxidase, subunit B | P76078 | 67 | 8 | |
| 23955 | paaA | 8.5 | 7.1 | 11.4 | 9.0 | paaA | 1,2-Phenylacetyl-CoA epoxidase, subunit A | P76077 | 66 | 8 | |
| 23960 | paaG | 7.7 | 7.4 | 6.9 | 6.8 | paaG | 1,2-Epoxyphenylacetyl-CoA isomerase | P77467 | 37 | 8 | |
| 23965 | paaH | 7.6 | 7.5 | 7.1 | 6.2 | paaH | 3-Hydroxyadipyl-CoA dehydrogenase | P76083 | 36 | 8 | |
| 23970 | paaF | 7.1 | 7.4 | 7.1 | 5.8 | paaF | 2,3-Dehydroadipyl-CoA hydratase | P76082 | 36 | 8 | |
| 23975 | paaJ | 6.6 | 8.2 | 7.4 | 6.6 | paaJ | OAC/ODSCT | P0C7L2 | 55 | 8 | |
| 23980 | tetR | 7.3 | 1.8 | 2.7 | 1.8 | kstR2 | HTH-type transcriptional repressor KstR2 | A0R4Z6 | 25 | 95 | |
| 23985 | paaZ | 7.9 | 6.4 | 6.8 | 6.0 | paaZ | Bifunctional aldehyde dehydrogenase | P77455 | 51 | 8 | |
| 23990 | ethD | 8.4 | 6.3 | 5.1 | 5.7 | ethD | Uncharacterized 11.0-kDa protein | P43491 | 48 | 96 | |
| 23995 | paaI | 7.3 | 5.2 | NaNf | NaNf | paaI | Acyl-CoA thioesterase PaaI | P76084 | 45 | 8 | |
| 24000 | Partial | 7.3 | 6.4 | NaN | NaN | ||||||
| 24005 | araC | 8.0 | 6.0 | 4.1 | 8.4 | nphR | Transcriptional activator NphR | B1Q2A8 | 31 | 97 | |
| S3 | 24010 | aldH1 | 9.6 | 6.4 | 6.0 | 5.5 | styD | Phenylacetaldehyde dehydrogenase | O06837 | 36 | 98 |
| 24015 | styJ | 10.1 | 6.1 | 7.7 | 5.9 | yfcG | Disulfide-bond oxidoreductase YfcG | P77526 | 47 | 99 | |
| 24020 | styI | 10.4 | 6.0 | 5.0 | 5.3 | isoI | Glutathione-S-transferase | WP_045063292 | 49 | 57 | |
| 24025 | styH | 9.5 | 6.3 | 5.0 | 4.8 | isoH | HGMBDH | WP_045063294 | 59 | 57 | |
| 24030 | styG | 10.0 | 6.3 | 6.8 | 5.5 | yfdE | Acetyl-CoA:oxalate-CoA transferase | P76518 | 33 | 100 | |
| 24035 | marR | 10.0 | 3.5 | 2.1 | 2.7 | marR | Regulatory protein | CAA52427 | 31 | 101 | |
| 24040 | dsr | 9.1 | 4.7 | 8.5 | 4.8 | dsr | CoA disulfide reductase | O58308 | 34 | 102 | |
| 24045 | gshB | 8.9 | 4.9 | 5.7 | 3.8 | gshB | Glutathione synthetase | P45480 | 50 | 103 | |
| 24050 | gshA | 9.0 | 3.4 | 5.5 | –2.2 | gshA | Glutamate-cysteine ligase EgtA | P9WPK7 | 33 | 104 | |
| S2 | 24055 | Hyp | 7.8 | 8.5 | NaNf | 8.8 | |||||
| 24060 | Hyp | 7.0 | 9.3 | 7.5 | 10.3 | ||||||
| 24065 | Hyp | 6.6 | 9.3 | 8.8 | 10.3 | ||||||
| 24070 | Hyp | 7.1 | 8.4 | 8.2 | 8.4 | ||||||
| 24075 | Hyp | 6.8 | 9.5 | 7.9 | 8.2 | ||||||
| 24080 | Hyp | 7.8 | 8.0 | 8.0 | 8.2 | ||||||
| 24085 | Hyp | 9.2 | 10.7 | 5.8 | 6.2 | ||||||
| 24090 | Hyp | 7.4 | 10.4 | 9.2 | 9.0 | ORF3 | BAL04131 | 67e | 13, 50 | ||
| S1 | 24095 | styB | 8.9 | 9.4 | 11.2 | 8.5 | styB | StyB-SMO flavin oxidoreductase | BAL04130 | 82 | 13, 50 |
| 24100 | styA | 9.0 | 9.1 | 8.1 | 6.0 | styA | StyA-SMO styrene monooxygenase | BAL04129 | 86 | 13, 50 | |
| P | 18380 | prmA | 5.3 | 3.5 | 3.4 | 3.2 | prmA | Propane monooxygenase hydroxylase LSU | BAD03956 | 96 | 25 |
| 18385 | prmB | 3.1 | 4.8 | 4.9 | 2.0 | prmB | Propane monooxygenase reductase | BAD03957 | 80 | 25 | |
| 18390 | prmC | 4.1 | 4.0 | 3.7 | 2.4 | prmC | Propane monooxygenase hydroxylase SSU | BAD03958 | 85 | 25 | |
| 18395 | prmD | 4.1 | 4.3 | 4.7 | 4.4 | prmD | Propane monooxygenase coupling protein | BAD03959 | 94 | 25 | |
| 18400 | 3.7 | 3.7 | 4.9 | 2.3 | |||||||
| 18405 | 3.3 | 3.8 | NaNf | 1.7 | |||||||
| 18410 | adh | 3.5 | 3.8 | 3.4 | 1.5 | adh1 | Alcohol dehydrogenase | BAD03962 | 88 | 25 | |
| 18415 | groL | 4.4 | 3.7 | 3.5 | 1.3 | groL | 60-kDa chaperonin | P28598 | 52 | 105 | |
| A | 14195 | ahpD | 7.1 | 3.2 | 4.7 | 4.5 | ahpD | Alkyl hydroperoxide reductase AhpD | Q50441 | 74 | 106 |
| 14190 | ahpC | 7.7 | 4.9 | 4.1 | 2.5 | ahpC | Alkyl hydroperoxide reductase subunit C | A0R1V9 | 86 | 107 | |
| 14185 | oxyR | 2.7 | 5.1 | 1.2 | 2.6 | oxyR | Probable hydrogen peroxide-inducible gene activator | Q9X5P2 | 53 | 108 | |
Transcript abundance (A) and fold change (M), as well as the protein fold change in the cytosol (Cyt) and membrane fraction (Mem), are expressed as a log2 ratio. NaNf, not detected in the fructose proteome; NaN, not detected. Ref, reference(s).
Enzymes and proteins with a reported activity or function are underlined. For further details, see the supporting information.
Clusters S1 to S4 are located on the plasmid, while clusters P and A are located on the chromosome of strain CWB2.
Abbreviations: OAC/ODSCT, 3-oxoadipyl-CoA/3-oxo-5,6-dehydrosuberyl-CoA thiolase; HGMBDH, 1-hydroxy-2-glutathionyl-2-methyl-3-butene dehydrogenase; LSU, large subunit; SSU, small subunit.
Calculated with the partial amino acid sequence of ORF3.
FIG 1.
Comparison of the styrene degradation cluster of Gordonia rubripertincta CWB2 with homologous clusters, as found in the strains Rhodococcus opacus PD630 (NZ_CP003949) (22), Rhodococcus sp. strain AD45 (NZ_CM003191) (58), Nocardioides sp. strain Root240 (NZ_LMIT01000013), Aeromicrobium sp. strain Root495 (NZ_LMFJ01000002), Rhodococcus sp. ST-10 (AB594506) (13), Rhodococcus opacus 1CP (NZ_CP009112, NZ_CP009111) (48), Pseudomonas sp. strain Y2 (AJ000330) (38, 93), and Sphingopyxis fribergensis Kp5.2 (CP009122) (17). The subclusters of strain CWB2 are indicated (S1 to S4), and gene products are indicated in the legend colored by their (predicted) function. Relevant homologous genes and clusters are emphasized by interspaced conjunctions. Clusters of marked strains are reported to be involved in isoprene (●) or styrene (#) degradation.
The cluster can be separated into four subclusters comprised as follows: cluster S1 encodes a styrene monooxygenase, which is known to initiate the styrene degradation at the vinyl side chain. The closest characterized homolog of this protein was found in Rhodococcus sp. ST-10 (StyA, 86% identity at amino acid level; StyB, 82%). A phylogenetic analysis of the amino acid sequence classifies it as an E1-type SMO (see Fig. S2 in the supplemental material). The closest match within the Gordonia species is a putative monooxygenase from Gordonia polyisoprenivorans NBRC 16320 (GAB22407, 45%; GAB22406, 39%). However, the genetic organization also refers to a close relation to the Rhodococcus cluster, as the partial sequence of ORF3 from strain ST-10 shows 67% identity to GCWB2_24090. Transmembrane domain prediction (TMHMM) of GCWB2_24090 identified four transmembrane helices that classify this protein as “hypothetical membrane associated.” Beyond that, no characterized homologs and no known domains are present in the database for this ORF. Note that no styrene oxide isomerase (styC) gene was found on the genome of strain CWB2.
The second cluster S2 embeds seven hypothetical proteins (GCWB2_24085 to GCWB2_24055). Two of these are presumably soluble, and the others are annotated as membrane proteins, while each has one transmembrane domain. Members of these clusters appear to be rare within the database and are predominantly present in rhodococci. Rhodococcus opacus 1CP contains a homologous cluster downstream of its StyABCD cluster (WP_065493732 to WP_045063326; 56 to 68%). Further, Gordonia sp. strain i37 has a homologous gene cluster in the neighborhood of a recently recorded isoprene degradation cluster (contig257, WP_079929940 to WP_079929944; contig258, OPX14963 and OPX14964 [56 and 4%, respectively]) (21).
Cluster S3 encodes proteins that might be involved in glutathione and isoprene metabolism. GCWB2_24050 and GCWB2_24045 show the highest identity to the glutamate-cysteine ligase GshA (P9WPK7, 33%) and the glutathione synthetase GshB (P45480, 50%), followed by a putative coenzyme A-disulfide reductase (CoA-DSR). The other genes of this cluster (GCWB2_24050 to GCWB2_24010) encode a putative MarR-like transcriptional regulator, a CoA-transferase, a dehydrogenase, a glutathione S-transferase (GST), a disulfide bond oxidoreductase, and an aldehyde dehydrogenase. The closest characterized homolog of the latter gene product is the phenylacetaldehyde dehydrogenase from Pseudomonas fluorescens ST (O06837, 36%). Cluster S3 can also be found in Aeromicrobium sp. strain Root495, wherein a styrene monooxygenase is located between the CoA-DSR and the transcriptional regulator. The same is true for Nocardioides sp. strain Root240, except for a 13,983-bp insertion right after the styrene monooxygenase gene. Interestingly, the closest characterized homologs of GCWB2_24025 and GCWB2_24020 can be found in Rhodococcus sp. strain AD45 (WP_045063294, 59%; WP_045063292, 49%) and are known to be a functional part of an isoprene degradation cluster that is located on a megaplasmid (300 kbp) (22). In addition, homologs of other genes from cluster S3 can be found on this plasmid, even in a similar arrangement of parts from this cluster. However, the genome of strain AD45 does not encode a styrene monooxygenase and, on the other hand, strain CWB2 lacks the isoprene monooxygenase. Homologous proteins from cluster S3 were recently found in Gordonia sp. i37 next to a homologous to cluster S2 (21). The 13,983-bp insertion of Nocardioides sp. Root240 comprises a putative mce operon, a cluster of membrane proteins whose specific function is unclear.
The putative styrene degradation cluster is completed by the fourth gene set S4 (GCWB2_24005 to GCWB2_23925) that encodes proteins required for the lower styrene degradation pathway (phenylacetic acid catabolism). (The proposed pathway of strain CWB2 is displayed in Fig. 3b below.) Besides some regulatory elements and a partial gene (GCWB2_24000), this cluster is homolog to that of Rhodococcus opacus 1CP and can also be found in Gordonia soli NBRC 108243, as well as Gordonia sp. i37 (contig69). The AraC-like transcriptional regulator (GCWB2_24005) shows homology to regulators of strain 1CP that are located before the SMO and after the PAA degradation cluster (ANS32446, 46%; WP_061046101, 41%). However, in contrast to strain CWB2, the upper and lower styrene degradation pathways are not associated in this strain. Here, the genes for the conversion of styrene to phenylacetic acid are located on plasmid pR1CP1 (NZ_CP009112), whereas the subsequent metabolization is encoded on the chromosome (NZ_CP009111). The second transcriptional regulator (GCWB2_23980) belongs to the TetR family. Cluster S4 is terminated by a phenylacetaldehyde dehydrogenase (GCWB2_23925) that is highly similar to StyD from Rhodococcus sp. strain ST-5 (BAL04135, 76%). So far, a comparable genetic environment of styrene monooxygenases can only be found in Aeromicrobium sp. strain Root495 and in five Nocardioides strains (Root79, Root190, Root240, Root614, and Root682). All of them were isolated from Arabidopsis root microbiota (23).
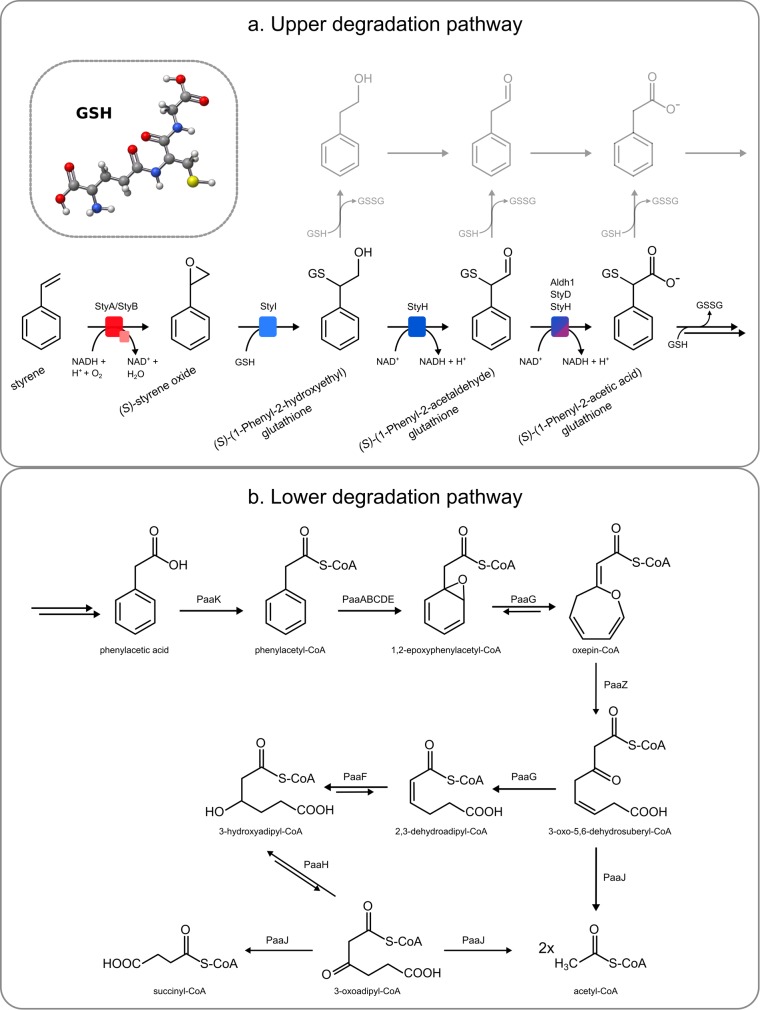
FIG 3.
(a) Proposed novel degradation pathway of styrene in Gordonia rubripertincta CWB2 (see the text for details). (b) Proposed phenylacetic acid degradation pathway of Gordonia rubripertincta CWB2. The genes of the involved enzymes are present on the genome (cluster S4) and upregulated on the transcriptome and the proteins on the proteome level, respectively (see Table 1). Starting from the product of the upper degradation pathway, phenylacetic acid, strain CWB2, is able to metabolize styrene to acetyl-CoA or succinyl-CoA. (Adapted with permission from reference 68.)
As already mentioned, the whole cluster embeds three helix-turn-helix (HTH)-type regulators, which are known to respond to aromatic compounds (24). They are not similar to the regulation machinery as described for pseudomonads (3).
In addition, the genome of strain CWB2 was examined for other genes and clusters that might enable degradation of styrene or metabolites. Since no styrene oxide isomerase gene (styC) is located on the genome of strain CWB2, this degradation step might be bypassed by a styrene oxide reductase (SOR) and a phenylacetaldehyde reductase (PAR). So far, there is no enzyme characterized that has SOR activity and thus no comparison with strain CWB2 on DNA level is possible. However, two putative ORFs (GCWB2_12345, 70%; GCWB2_18410, 35%) show similarity to the PAR of Rhodococcus sp. ST-10 (BAD51480). Interestingly, the latter one is part of a cluster with eight ORFs (GCWB2_18380 to GCWB2_18415) that is a homolog to one in Gordonia sp. strain TY-5 (BAD03956 to BAD03963 [87 to 96%]) (25). It comprises a chaperonin, a putative alcohol dehydrogenase, two hypothetical proteins, and a putative propane monooxygenase. The monooxygenase also resembles a propene monooxygenase from Mycobacterium sp. strain M156 (28 to 38%) (26) and a methane monooxygenase from Methylococcus capsulatus strain Bath (29 to 34%) (27). These binuclear iron monooxygenases are able to epoxidize styrene. Homologous clusters can be found in several actinobacteria (28–30), e.g., in R. opacus 1CP and Gordonia sp. i37 (21). The other putative PAR is not part of a cluster. In addition, several cytochrome P450 monooxygenases can be found on the CWB2 genome, which might also be able to perform the epoxidation of styrene.
Transcriptome and proteome analysis of the styrene degrader CWB2.
The transcriptome and proteome of strain CWB2 was analyzed to reveal the global profile of genes and proteins that are involved in styrene metabolism. Therefore, fructose-grown cultures served as a reference condition. The transcriptome output is summarized in Table S4 in the supplemental material.
Assuming a threshold of ≥1.5 (M value), then 2.5% of the genes of strain CWB2 are overexpressed under styrene exposure, and 30% of these are located on the plasmid (see Fig. S5 in the supplemental material). It is known that the transcriptome as the total amount of mRNA does not necessarily reflect the total amount of protein abundance in the cell. However, we were able to identify 3,691 proteins in the proteome of strain CWB2. Assuming a threshold of 1.5 (log2 ratio), then 7% of the proteins were highly abundant when strain CWB2 was grown on styrene (see Fig. S6 in the supplemental material). The gene cluster, which is framed by the styrene monooxygenase and the phenylacetaldehyde dehydrogenase, is highly upregulated on the transcriptome level (average increase, 7-fold), as well as the proteome level (average increase, 6.6-fold) (Table 1 and Data Set S1 in the supplemental material).
Validation of the enzymatic activity of selected members of the styrene degradation pathway.
After analysis of the genome, transcriptome, and proteome of strain CWB2, we screened for enzyme activities that enable styrene degradation on different pathways. For this, a crude extract from a styrene-grown biomass was prepared, and proteins were separated and enriched by different chromatography methods. The activities were measured directly or indirectly using a spectrophotometer or by quantification of the products on the reversed-phase HPLC (RP-HPLC), respectively (see Table S5 in the supplemental material). It was possible to detect SMO activity in the crude extract and to enrich the enzyme 36-fold to an activity of 6.82 mU mg−1. Due to the missing SOI, it was proposed that the conversion of styrene oxide is bypassed by the activity of a SOR, which produces 2-phenylethanol. This is supposed to be converted to phenylacetaldehyde by a PAR. Only minor activity of a PAR with 2-phenylethanol was detected in crude extract. Higher activities were determined in crude extracts, when styrene oxide was applied as the substrate. However, this might be due to activity of a GST, while residual glutathione (GSH) is present in the crude extract of strain CWB2. To further prove this assumption, a crude extract was assayed for GST activity after the supply of additional GSH. In this way, a GST activity of 44.23 U mg−1 was reached for the conversion of styrene oxide (Fig. 2).
FIG 2.
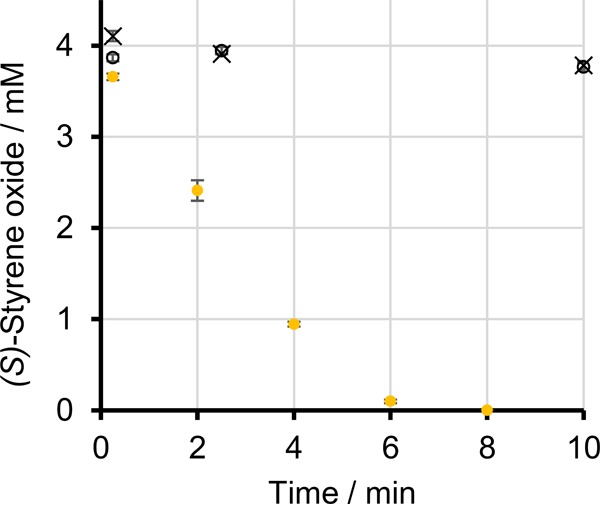
Degradation of 4 mM (S)-styrene oxide with crude extract of styrene-grown biomass of Gordonia rubripertincta CWB2 and 5 mM reduced glutathione (yellow circles). Only minor consumption was detected when excluding either reduced glutathione (open circles) or crude extract (×) from the reaction mix.
Further, crude extracts were assayed for vinylcatechol-2,3-dioxygenase and cis,cis-muconate cycloisomerase to exclude other degradation pathways. However, there was no detectable activity for one of these enzymes.
Two putative SMOs of strain CWB2 were cloned and expressed for initial characterization. Of the two putative SMOs, only one was expressed and synthesized in an active form. It is part of the styrene degradation cluster (S1; GCWB2_24100) and produces (S)-styrene oxide with a specific activity of 0.42 ± 0.02 U mg−1. The SMO can be classified as E1-type SMO (see Fig. S2 in the supplemental material). PADs are aldehyde dehydrogenases that catalyze the formation of the central intermediate phenylacetic acid. Two aldehyde dehydrogenases of strain CWB2 were recombinantly expressed in Escherichia coli. AldH1, which originates from the isoprene degradation cluster (S3), catalyzes the conversion of phenylacetaldehyde with an activity of 0.29 ± 0.01 U mg−1. StyD, which is encoded in cluster S4, is 10-fold slower and has an activity of 0.026 ± 0.001 U mg−1.
DISCUSSION
Adaption of G. rubripertincta CWB2 to styrene exposure.
Only a few reports exist for Gordonia considering the metabolization of styrene (18, 31). Further, the limited amount of SMOs that are encoded on genomes of this genus indicate that styrene degradation is not a common feature. G. rubripertincta CWB2 is able to withstand and degrade large amounts of styrene (520 g m−3 in 21 h), even compared to other efficient styrene degraders (32, 33). Some bacteria produce surfactants, when they are exposed to hydrophobic substrates, to increase their accessibility (34–36). However, we found no indication that strain CWB2 exports biosurfactants into the media, but it seems to have a hydrophobic cell surface, which improves substrate uptake. This is supported by a tendency to form agglomerates during growth in liquid media.
There is no complete prokaryotic transcriptome under styrene exposure available yet. So far, studies focused on the transcriptional regulation of styrene degradation and a small number of target genes, solely with respect to Pseudomonas strains (37–44). A proteome of R. jostii RHA1, which uses an unspecific styrene degradation route, is available (45). So far, only one system level proteome analysis for styrene degradation in P. putida CA-3 exists (46). This was the first time where all of the respective enzymes of the upper and lower degradation pathway were detected when a strain was grown on styrene (46).
The omic analysis of strain CWB2 in this study outlines the biological background for its adaption to styrene as a source of carbon and energy. This was found to be totally different from previously characterized styrene degraders. Initially, styrene has to be imported into the cell. The only specific styrene transporter StyE was found in pseudomonads (47). However, the styE gene is not encoded in most other styrene degraders, and thus other transport mechanisms, as well as diffusion, have to be considered (3, 46). Cluster S2 of strain CWB2 contains several membrane proteins that are highly upregulated, and the same cluster is also present in the styrene degrader R. opacus 1CP (Fig. 1). Thus, it is likely that these proteins might also be involved in substrate transport or cell membrane adaption. Interestingly, Gordonia sp. i37 harbors a similar cluster in the proximity of an isoprene degradation cluster (21). However, there are no characterized homologs available in the database, and thus the specific function of these proteins remains unclear.
Strain CWB2 merged clusters to form a hybrid that enables styrene degradation.
The genetic organization of the putative styrene degradation cluster compared to other clusters with homolog proteins can be found in Fig. 1. The “classical” styrene degradation cluster of Pseudomonas sp. Y2 differs from Rhodococcus clusters, as well as the recently reported cluster of S. fribergensis Kp5.2 (17, 38, 48). Thus, it is obvious that the arrangement and regulation is variable among different organisms. The styrene degradation cluster of strain CWB2 is highly upregulated on the mRNA and protein levels, when strain CWB2 grows on styrene (Table 1 and see Data Set S1 in the supplemental material). In Pseudomonas putida CA-3, SMO and PAD were the most abundant proteins (46). In strain CWB2, they are also highly upregulated but in a range comparable to the rest of the genes and proteins of this cluster. The transcriptional regulators (GCWB2_24035 and GCWB2_23980) are less expressed and synthesized. It is also obvious that regulation of gene expression differs in strain CWB2, since no StyR/StyS homolog is associated with this cluster. However, further studies with different inducers are necessary to clarify the regulation.
There is no evidence that strain CWB2 performs direct ring cleavage of styrene or activation by an epoxide hydrolase since the respective parts of these pathways are not present on the genome or upregulated in the transcriptome or proteome when cultivated on styrene (see the supplemental material). In contrast, initial epoxidation of styrene was found to be catalyzed by a SMO. Enzyme activity was detected in crude extracts of styrene-grown cells, and the SMO was successfully enriched by ion-exchange chromatography and hydrophobic interaction chromatography (see Table S5 in the supplemental material). The SMO (GCWB2_24100; ASR05591) was recombinantly expressed and purified. The specific epoxidation activity is about 0.4 U mg−1 and thus higher than for most other characterized SMOs (48–50). However, epoxidation of styrene is usually the rate-limiting step due to the relative low activity of the SMOs (51).
The SMO is part of cluster S1, which is highly similar to the partial styrene degradation cluster of Rhodococcus sp. ST-10. Itoh and coworkers proposed the chemical conversion of styrene oxide or the cooperation of a SOR and PAR (12, 13) since no SOI is present in this strain. However, previous reports, as well as this study, indicate that this is rather unlikely, since we detected only minor SOR and PAR activity in the crude extract of strain CWB2. Both assumptions would not explain the fast degradation of styrene found in these strains (3, 52, 53). However, strain ST-10 accumulated epoxide when incubated with styrene, and thus it remains to be shown whether the rest of the genes are also homologs to the styrene degradation cluster in strain CWB2.
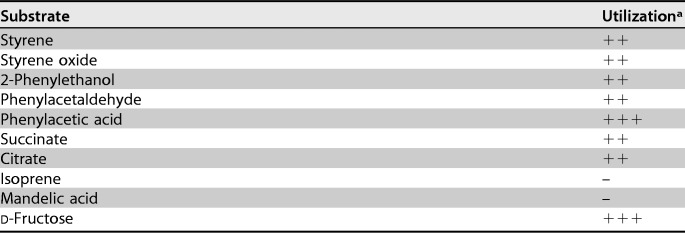
To circumvent this missing link of enzymatic styrene oxide isomerization, strain CWB2 seems to have incorporated a cluster (S3) that is very similar to those from Aeromicrobium sp. Root495 and Nocardioides sp. Root240. Interestingly, both strains were isolated at the same site (23), and both clusters are also closely located to a styrene monooxygenase in these strains. The genes of cluster S3 may originate from an isoprene degradation cluster, as found on a megaplasmid in Rhodococcus sp. AD45 (AJ249207) and also in Gordonia sp. i37 (21, 54). Actinobacteria from the genera Mycobacterium, Rhodococcus, and Gordonia were constantly detected in different environments as isoprene degraders (21, 54, 55). Rhodococcus sp. AD45 initially epoxidizes isoprene by the activity of an isoprene monooxygenase. It then uses a GST to convert the epoxide to a glutathione-alcohol adduct, which is further metabolized by a dehydrogenase to form an aldehyde and subsequently an acid (22, 54, 56–59). Remarkably, strain AD45 is also able to metabolize styrene but has no SMO (56). Based on these observations, styrene may also be channeled through the isoprene degradation pathway in G. rubripertincta CWB2. Further, strain CWB2 carries genes that are necessary for glutathione synthesis and reduction in cluster S3 (60). Interestingly, strain CWB2 does not possess an isoprene monooxygenase and cannot catabolize isoprene (Table 2).
TABLE 2.
Substrate spectra, especially those that might be related to styrene degradation in G. rubripertincta CWB2
a+++, vigorous growth; ++, good growth; +, growth; −, no growth.
Styrene oxide is channeled into a novel glutathione-dependent degradation pathway.
To prove whether glutathione-dependent metabolization occurs, we assayed crude extract from styrene-grown cells for GST activity with (S)-styrene oxide as the substrate. We found that the epoxide was degraded fast, with an activity of 44 U mg of crude extract−1 (Fig. 2). Only minor activity was detected when no reduced glutathione was added to the reaction.
Therefore, we propose a novel degradation pathway for styrene via initial epoxidation by a SMO to (S)-styrene oxide and the addition of glutathione by the GST StyI (Fig. 3a). The resulting (S)-(1-phenyl-2-hydroxyethyl) glutathione (CAS no. 64186-97-6) will be further converted by the dehydrogenase StyH to (S)-(1-phenyl-2-acetaldehyde) glutathione and (S)-(1-phenyl-2-acetic acid) glutathione. Possibly, the PAD and/or the aldehyde dehydrogenase (AldH1) is also involved in this step since both showed activity with phenylacetaldehyde (61). Since the glutathione adduct is not easily accessible, it has to be verified whether the AldH1 and the PAD can also catalyze this reaction. The aldehyde dehydrogenase of cluster S3 is induced in strain AD45, but no specific role had been ascribed (22). Subsequently, the glutathione is removed from the adduct, which may occur due to the activity of StyJ and StyG (58, 62, 63). The product of this process will be phenylacetic acid or phenylacetyl-CoA, which will be degraded via several enzymes from cluster S4 to yield acetyl-CoA and succinyl-CoA (Fig. 3b). We suppose that the 2-phenylethanol and phenylacetaldehyde, which can be detected during growth, result from side-product formation of this novel pathway due to instability of the glutathione adducts or enzymatic removal of glutathione in an earlier step (Fig. 3a).
It should be mentioned that it is unusual for actinobacteria to produce glutathione, since mycothiol is the dominant thiol in these organisms (64–66). However, it was reported that strain AD45 also produces substantial amounts of glutathione, and it was suggested that this ability was gained by the horizontal gene transfer of isoprene degradation genes (54, 56, 58, 65). It is likely that the same is true for strain CWB2 due to the plasmid uptake. This view is supported by the finding that the GC content of the plasmid and the styrene degradation cluster is much lower compared to the whole genome. The GC content of that cluster S3 is close to that of strain AD45 (61.7% [22]). Recently, cluster S3 genes were found in Gordonia sp. i37 (21). Further, strain CWB2, as well as strain 1CP, encodes several mobile elements in the direct neighborhood of the styrene degradation cluster on their plasmid, which suggests horizontal gene transfer. In addition, cluster S4 is highly similar to that of the R. opacus 1CP and strain RHA1, and the degradation of PAA likely takes place in the same way (67, 68) (Fig. 3b).
Conclusion.
Omic analyses imply that strain CWB2 incorporated a plasmid that contains an assembly of different gene clusters and forms a “hybrid” that enables it to metabolize styrene and analogous compounds. Our study illustrates the possibilities of horizontal gene transfer for Gram-positive bacteria and an ongoing adaptation to glutathione as a cofactor in actinobacteria. This adaptation is coupled with the high biotechnological potential of this organism, since G. rubripertincta CWB2 can produce ibuprofen, which is not possible through the classical styrene degradation pathway (17, 18). The involved SMO shows higher activities than have been reported for other SMOs thus far. This might be interesting since these enzymes are known to catalyze a variety of valuable reactions (3). Further, bacterial GSTs are known to be involved in the degradation of (halogenated) xenobiotics and other chemical transformations, and therefore the GST of strain CWB2 might open a new field of possible biochemical reactions to this class of enzymes (63, 69, 70).
MATERIALS AND METHODS
Isolation and cultivation of styrene-degrading strains.
Styrene-degrading bacteria were isolated from (contaminated) soil. A small amount of the soil was transferred into a 1-liter Erlenmeyer flask and suspended in 100 ml of water. Portions (10 to 40 μl) of styrene were supplied via an evaporation adaptor as the sole source of carbon and energy. The growth medium was dosed with 0.02 mg ml−1 nalidixin acid and 0.075 mg ml−1 cycloheximide to prevent the growth of Gram-negative bacteria or fungi, respectively. Then, 10 ml of the culture was plated on solid mineral medium (MM) (71) without a carbon source, followed by incubation at room temperature in a 5-liter desiccator in a styrene-containing atmosphere. The grown colonies were repeatedly transferred onto fresh solid MM, followed by incubation for 2 to 3 days in the desiccator. The isolates were stored at −80°C in 40% (vol/vol) glycerol.
Liquid cultures were kept in Erlenmeyer flasks containing MM (71). The respective carbon source was added either directly into the media or, in case of volatile compounds, via an evaporation adapter.
Characterization of G. rubripertincta CWB2.
Growth of strain CWB2 in liquid MM was assayed on various substrates (see Table S1 in the supplemental material). The production of surfactants with fructose, hexadecane, or styrene as a carbon source was examined as published earlier (36, 72). Siderophore production was determined by using a CAS-agar plate test (73). Analysis of the mycolic acid and fatty acid composition was done by the Deutsche Sammlung für Mikroorganismen und Zellkulturen (DSMZ) (see Table S2 in the supplemental material). 16S rRNA analysis and scanning electron microscopy was done as described earlier (18) (see Fig. S1 and S3 in the supplemental material). The in silico DNA-DNA hybridization was performed by using the Genome-to-Genome distance calculator 2.1 (DSMZ) (see Fig. S7 in the supplemental material). G. rubripertincta CWB2 was assayed for antibiotic resistance on chloramphenicol, ampicillin, tetracycline, nalidixic acid, gentamicin, streptomycin, and kanamycin.
DNA extraction and genome sequencing, annotation, and bioinformatic analysis.
A 50-ml culture of G. rubripertincta CWB2 was grown on fructose in MM. Cells were harvested at an optical density at 600 nm (OD600) of 0.6 by centrifugation (5,000 × g; 15 min), washed once with 100 mM phosphate buffer (pH 7.5), and centrifuged again. The cell pellet was resuspended in 2.7 ml of buffer (10 Tris-HCl, 10% sucrose, 30 mg of lysozyme; pH 7.8), followed by incubation for 1.5 h at 37°C. After centrifugation, the supernatant was discarded, and the pellet was resuspended in 2.8 ml of TE buffer 10.1 (10 mM Tris, 1 mM EDTA; pH 8) with 100 μg ml−1 proteinase K and 150 μl of 10% sodium dodecyl sulfate (SDS), followed by incubation for 2 h at 37°C. Chromosomal DNA was extracted successively with 3 to 5 ml of phenol (equilibrated two times), phenol-chloroform-isoamyl alcohol (25:24:1), and chloroform-isoamyl alcohol (24:1; two times). Each extraction was followed by a centrifugation (20,000 × g, 10 min) and separation of the aqueous phase into a new tube. RNA was then digested by adding 100 μg ml−1 RNase A at 37°C for 10 min. The DNA was precipitated on ice by adding a 1/10 volume of 3 M sodium acetate (pH 5.2) and 3 volumes of 100% ethanol. The precipitate was centrifuged at 4°C, resuspended in 70% ice-cold ethanol, and centrifuged again. The supernatant was discarded, and the pellet was dried for 10 min in a SpeedVac vacuum concentrator. The pellet was dissolved in TE buffer 10.1 for storage at 4°C, and the purity was controlled by agarose gel electrophoresis on a NanoDrop apparatus.
To obtain the complete genome sequence, two sequencing libraries were prepared: a TruSeq PCR-free whole-genome shotgun library and an 8k Nextera MatePair library (Illumina, Inc., Netherlands). Both libraries were sequenced on an Illumina MiSeq desktop sequencer with 2x 300 bp (paired end). The obtained reads were assembled using a Newbler (v2.8) de novo assembler (Roche). The initial assembly consisted of just one scaffold of 90 contigs, with 120 contigs larger than 500 bp in all. Manual inspection and assembly was performed using Consed (74, 75), which revealed a misassembly: three contigs representing a 105-kbp plasmid, henceforth called pGCWB2, were wrongly “attached” to the scaffold representing the chromosome. The sequence of both replicons could be completely established. The sequences for both replicons were annotated using PROKKA (76) (see Table S3 in the supplemental material for details). The annotated replicons were submitted to GenBank under accession numbers CP022580 (chromosome) and CP022581 (pGCWB2).
Additional functional annotation of proteins was done by using the BLASTP algorithm (20) on the nonredundant protein database or the NCBI UniProtKB database (date of search, 1/9/2017). Membrane association of proteins was verified by prediction on the TMHMM server (77). Island viewer 4 (78) was applied to detect genomic islands and foreign genes on the genome of strain CWB2 (see Fig. S4 in the supplemental material).
RNA extraction and transcriptome sequencing.
A preculture of G. rubripertincta CWB2 was grown at 30°C in minimal medium with fructose or styrene as the sole source of carbon, respectively. After 5 days of cultivation, the culture was diluted 1/10 in fresh medium, followed by incubation for 24 h with the respective substrate in a set of four Erlenmeyer flasks. Prior to harvesting the cells, a 10% ice-cold stop solution (10% buffered phenol in ethanol) was added to the culture, followed by centrifugation at 11,000 × g for 5 min at 4°C. The supernatant was discarded, and the pellet was stored until RNA isolation at −80°C. To break up the cells, 150 μl of a 5-mg ml−1 lysozyme solution was added to the pellet, mixed, and incubated at room temperature for 5 min. Next, 450 μl of buffer RLT (Qiagen) and 50 mg of 0.1-mm glass beads were added to resuspend and break the cells by repeated vortexing at 4°C. The suspension was applied to a QIAshredder column for homogenization and to remove particles from the sample. Extraction of total RNA was done by applying an RNeasy minikit, including on-column DNA digestion (Qiagen). Isolated RNA was stored at −80°C, and the quality was controlled on a 2100 Bioanalyzer using an RNA 6000 Nano kit (Agilent Technologies, Böblingen, Germany).
The RNA quality and quantity was again checked by an Agilent 2100 Bioanalyzer run (Agilent) and an Xpose system (Trinean, Gentbrugge, Belgium) before and after rRNA depletion by a Ribo-Zero rRNA removal kit (Bacteria; Illumina, San Diego, CA). A TruSeq stranded mRNA library prep kit from Illumina was used to prepare the cDNA libraries to analyze the whole transcriptome. The resulting cDNAs were sequenced on an Illumina MiSeq and HiSeq 1500 system (San Diego, CA, USA) using 2 × 75-nucleotide read length (paired end). The raw sequencing read files are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6012. Reads were mapped on the reference strain G. rubripertincta CWB2 (CP022580 and CP022581) with Bowtie2 (79) using standard settings. ReadXplorer 2.2.0 (80) was used to visualize short read alignments and for data analysis. Differential gene expression analysis was performed based on a normalized read count using TPM (transcripts per million) values of CDS calculated by ReadXplorer 2.2.0. The signal intensity value (i.e., the a-value) was calculated by 0.5 × (log2 TPM condition A + log2 TPM condition B) of each CDS, and the signal intensity ratio (i.e., the m-value) was calculated by the difference in log2 TPM. CDS determinations with m-values higher than or equal to +1.5 or lower than or equal to −1.5 were considered to be differentially transcribed.
Preparation of protein samples and identification by liquid chromatography-electrospray ionization-tandem mass spectrometry.
G. rubripertincta CWB2 was grown the same way as for RNA extraction in a set of four samples per carbon source. After cultivation, two samples were pooled, centrifuged at 5,000 × g for 30 min at 4°C, and resuspended in 2.5 ml of 50 mM phosphate buffer (PB) (pH 7.26). The cells were disrupted by sonication on ice (10 cycles, 1.5 min; power, 70%; Bandelin SONOPLUS HD2070 homogenizer) after adding 40 U of DNase I and 1 mg ml−1 lysozyme. The suspension was centrifuged at 5,000 × g for 1 h at 4°C to separate soluble from insoluble matter. The proteins were separated by size using discontinuous SDS-PAGE with 50 μg of each sample per lane. Lanes were cut into eight slices and destained, alkylated, and digested with trypsin as previously described (81, 82). Peptides were extracted from the gel pieces with acetonitrile, loaded onto STAGE tips for storage, and eluted from the tips shortly before mass spectroscopy analysis (83).
By using an EASY-nLC 1000 (Thermo Scientific) LC system, peptides were separated at a flow rate of 400 nl/min on an 18-cm self-packed column (75-μm [inner diameter], 1.9-μm Reprosil-Pur 120 C-18AQ beads; Dr Maisch HPLC GmbH, Ammerbuch, Germany) housed in a custom-built column oven (84) at 45°C. Peptides were separated using gradient of buffers A (0.1% formic acid) and B (80% acetonitrile, 0.1% formic acid) from to 60% B. The column was interfaced with a Nanospray flex ion source (Thermo Scientific) to a Q-Exactive HF mass spectrometer (Thermo Scientific). MS instrument settings were as follows: 1.5-kV spray voltage, full MS at 60K resolution, AGC target 3e6, range of 300 to 1,750 m/z, max injection time 20 ms, Top 15 MS/MS at 15K resolution, AGC target 1e5, max injection time 25 ms, isolation width 2.2 m/z, charge exclusion +1 and unassigned, peptide match preferred, exclude isotope on, and dynamic exclusion for 20 s.
Mass spectra were recorded with Xcalibur software 3.1.66.10 (Thermo Scientific). Using a custom database containing 4,831 predicted protein sequences, the proteins were identified using Andromeda and quantified with the LFQ algorithm embedded in MaxQuant v1.5.3.17 (85). The following parameters were used: main search maximum peptide mass error of 4.5 ppm, tryptic peptides of minimum 6 amino acid length with maximum of two missed cleavages, variable oxidation of methionine, protein N-terminal acetylation, fixed cysteine carbamidomethylation, LFQ minimum ratio count of 2, matching between runs enabled, PSM and (Razor) protein false-discovery rate of 0.01, and advanced ratio estimation and second peptides enabled. Proteins with a log2 ratio of higher than or equal to +1.5 or lower than or equal to −1.5 were considered to be differentially synthesized.
Cloning, expression, and purification of recombinant styA, styD, and aldH1.
The styA (GCWB2_24100, GCWB2_21620), styD (GCWB2_23925), and aldH1 (GCWB2_24010) genes were purchased from Eurofins MWG (Ebersberg) in a pEX-K2 vector system allowing for kanamycin resistance selection. The DNA sequences were optimized for the codon usage and GC content of Acinetobacter baylyi ADP1 with the Optimizer tool (48, 86). 5′-NdeI and 3′-NotI restriction sites were added and used for subcloning into pET16bP to obtain the expression constructs pSGrA1_P01, pSGrA2_P01, and pSGrD1_P01, and pSGrD2_P01, from which recombinant proteins can be obtained as His10-tagged proteins. E. coli strains DH5α and BL21(DE3)/pLysS were cultivated for cloning and expression purposes as described elsewhere (87). The plasmids are listed in Table 3.
TABLE 3.
Plasmids used in this study
| Plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| pET16bP | pET16b with additional multicloning site; allows synthesis of recombinant proteins with an N-terminal His10 tag | U. Wehmeyera |
| pEX-K2-pSGrA1 | GCWB2_24100 (1,284-bp NdeI/NotI fragment) cloned into pEX-K2 vector | MWG Eurofins |
| pEX-K2-pSGrA2 | GCWB2_21620 (1,359-bp NdeI/NotI fragment) cloned into pEX-K2 vector | MWG Eurofins |
| pEX-K2-pSGrD1 | GCWB2_23925 (1,488-bp NdeI/NotI fragment) cloned into pEX-K2 vector | MWG Eurofins |
| pEX-K2-pSGrD2 | GCWB2_24010 (1,437-bp NdeI/NotI fragment) cloned into pEX-K2 vector | MWG Eurofins |
| pSGrA1_P01 | GCWB2_24100 (1,284-bp NdeI/NotI fragment) cloned into pET16bP | This study |
| pSGrA2_P01 | GCWB2_21620 (1,359-bp NdeI/NotI fragment) cloned into pET16bP | This study |
| pSGrD1_P01 | GCWB2_23925 (1,488-bp NdeI/NotI fragment) cloned into pET16bP | This study |
| pSGrD2_P01 | GCWB2_24010 (1,437-bp NdeI/NotI fragment) cloned into pET16bP | This study |
Personal communication.
Expression of StyA took place in a 3-liter biofermenter. E. coli BL21 strains with the respective plasmids were cultivated in Luria-Bertani (LB) medium (100 μg ml−1 ampicillin and 50 μg ml−1 chloramphenicol) at 30°C until an OD600 of 0.4 was reached. The batch was subsequently cooled to 20°C. Expression was induced at an OD600 of 0.6 by adding 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to the culture, followed by growth for 20 h at 20°C (120 rpm). The cells were harvested by centrifugation (5,000 × g, 30 min, 4°C), resuspended in 10 mM Tris-HCl buffer (pH 7.5), and stored at −80°C. The formation of the blue dye indigo is observable if active SMOs are produced during expression in LB media (88). Since this was not the case for the expression of GCWB2_21620, we assumed that the protein is not synthesized or active.
For the purification of StyA, crude extracts were prepared from freshly thawed biomass by disruption in a precooled French pressure cell, followed by centrifugation to remove cell debris (50,000 × g, 2 h, 4°C). The supernatants were applied to a 1-ml HisTrap FF column. The column was washed with 10 column volumes (CV) of binding buffer (10 mM Tris-HCl, 0.5 M NaCl, 25 mM imidazole [pH 7.5]) to remove nonspecific bound proteins. Enzymes were eluted with a linear imidazole gradient up to 500 mM over 30 CV. Fractions with respective enzyme activities were pooled and concentrated using Sartorius Vivaspin 20 filters (5,000-molecular-weight cutoff) at 4°C. The concentrates were passed through a 10-ml Econo-Pac 10DG desalting gravity-flow column (Bio-Rad) to remove remaining imidazole and sodium chloride. The protein obtained was kept in storage buffer (10 mM Tris-HCl, 50% [vol/vol] glycerol [pH 7.5]) at −20°C. Expression of GCWB2_21620 did not yield active protein, as already mentioned. Preparation of StyD and AldH1 was done according to the method of Zimmerling et al. (61).
Purification of wild-type proteins.
All of the following purification steps were performed on an ÄKTA fast-performance liquid chromatograph (GE Healthcare). Selected wild-type enzymes were enriched from crude extracts by ion-exchange chromatography. Strain CWB2 was cultivated on styrene, and soluble crude extract was prepared as described above. The supernatant was loaded with buffer A (20 mM Tris-HCl [pH 7.5]) on a MonoQ HR 5/5 column (GE Healthcare) at a flow rate of 1 ml min−1. Nonspecific bound proteins were removed by washing with 5 CV of buffer A. The elution of proteins was done over 25 CV with a linear gradient of buffer B (20 mM Tris-HCl, 1 M NaCl [pH 7.5]). Fractions of 1 ml were collected and tested for the respective enzyme activities. A second purification step was applied to some enzymes using hydrophobic interaction chromatography. The fractions that showed the respective enzyme activities were pooled, and (NH4)2SO4 was added to a final concentration of 460 mM. The sample was loaded with buffer C [20 mM Tris-HCl, 0.8 M (NH4)2SO4; pH 7.5] on a 1-ml Phenyl HP HiTrap column (GE Healthcare) at a flow rate of 1 ml min−1. Nonspecific bound proteins were removed by washing with 5 CV of buffer A. Elution of proteins was performed over 25 CV with a linear gradient of buffer A (20 mM Tris-HCl; pH 7.5). Fractions of 1 ml were tested for the enzyme activity.
For VC12DO, gel filtration was done after hydrophobic interaction chromatography. The fractions containing VC12DO activity were pooled and applied with buffer D (25 mM Tris-HCl, 0.5 M NaCl; pH 7.5) to a Superdex 200 HR 10/30 column at a flow rate of 0.4 ml min−1. Fractions of 1 ml were tested for VC12DO activity. Recombinant and wild-type proteins were subjected to discontinuous SDS-PAGE (87) in order to determine purity and subunit molecular size.
Enzyme assays.
Crude extracts and enriched or purified protein preparations from G. rubripertincta CWB2 were assayed for enzyme activities that are representative for known degradation pathways of styrene. Wild-type catechol 1,2-dioxygenase, catechol 2,3-dioxygenase, and cis,cis-muconate cycloisomerase activities were measured spectrophotometrically (Cary 50; Varian) by following the product formation or substrate depletion according to the method of Warhurst et al. (89) using catechol, protocatechuate, and cis,cis-muconate, respectively, as the substrates.
Styrene monooxygenase (SMO) activity of wild-type and recombinant enzyme preparations with styrene were measured by quantification of the reaction product styrene oxide on an RP-HPLC system as described previously (90).
SOR and PAR wild-type activity with styrene oxide and 2-phenylethanol was determined by quantification of the reaction products phenylacetaldehyde or 2-phenylethanol on an RP-HPLC system according to the protocol described previously for the SOI (17, 18).
Wild-type PAD and wild-type PAR activity were assayed indirectly on a spectrophotometer (Cary 50) by monitoring the reduction of NAD+ to NADH at 340 nm (ε340 nm = 6.22 mM−1 cm−1) (91). The 1-ml assay mixture contained 0.5 mM phenylacetaldehyde or 2-phenylethanol in 10 mM Tris-HCl (pH 7.5), 1 mM NAD+, and 50 μl of protein-containing sample, respectively. Recombinant PAD activity was assayed as previously described (61).
GST wild-type activity was assayed in soluble crude extract by monitoring (S)-styrene oxide consumption over time. Thus, G. rubripertincta CWB2 was grown on MM with styrene as the sole source of carbon. A 100-ml preculture was prepared and used to inoculate the main culture 1:50 in 500 ml of fresh MM. The main culture was incubated at 30°C for 5 days by adding 20- to 80-μl portions of styrene via the gas phase. Cells were harvested by centrifugation at 5,000 × g for 20 min at 4°C. The supernatant was discarded, and the pellet was resuspended and washed two times in 10 ml of 20 mM PB (pH 8). The cells were disrupted by sonication on ice (10 cycles, 1 min; power, 70%; Bandelin SONOPLUS HD2070 homogenizer) after adding 40 U of DNase I and 1 mg ml−1 lysozyme. Soluble crude extracts were obtained by centrifugation at 50,000 × g at 4°C for 1 h and separation from the insoluble matter. The reaction mix (600 μl) contained 20 mM PB (pH 8), 4 mM (S)-styrene oxide, 5 mM GSH, and an appropriate amount of soluble crude extract. Blank measurements were carried out by omitting either GSH or enzyme preparation. Samples were tempered for 10 min at 30°C, and the reaction was initiated by the addition of the substrate (S)-styrene oxide. Then, 25-μl samples were quenched at certain time points in 50 μl of ice-cold acetonitrile-methanol (1:1) and centrifuged at 16,000 × g for 10 min at 4°C to remove precipitates. Supernatants were applied to RP-HPLC by the injection of 10-μl samples. All measurements were done in triplicates. Enzyme activities are given in 1 U mg−1 representing the conversion μmol of substrate per min per mg of protein.
All RP-HPLC measurements were carried out with a Eurospher C18 column (125-mm length by 4-mm inner diameter, 5-μm particle size, 100-Å pore size; Knauer, Germany). The protein content was determined by the Bradford method (92), using BradfordUltra reagent (Expedeon) and bovine serum albumin (Sigma) as a reference protein.
Accession number(s).
The genome and assembly of G. rubripertincta CWB2 are deposited at the NCBI (BioProject accession no. PRJNA394617) with the chromosome (CP022580) and plasmid (CP022581) sequences. The raw sequencing read files are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6012. The newly characterized recombinant proteins in this study include StyA (ASR05591), StyD (ASR05556), AldH1 (ASR05573), and the monooxygenase (ASR05096).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge funding of this project by the Deutsche Bundesstiftung Umwelt (DBU), the European Social Fund, and the Saxonian Government (GETGEOWEB 100101363).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00154-18.
REFERENCES
- 1.Bond JA. 1989. Review of the toxicology of styrene. Crit Rev Toxicol 19:227–249. doi: 10.3109/10408448909037472. [DOI] [PubMed] [Google Scholar]
- 2.Gibbs BF, Mulligan CN. 1997. Styrene toxicity: an ecotoxicological assessment. Ecotoxicol Environ Saf 38:181–194. doi: 10.1006/eesa.1997.1526. [DOI] [PubMed] [Google Scholar]
- 3.Tischler D. 2015. Microbial styrene degradation. Springer Verlag, Berlin, Germany. [Google Scholar]
- 4.van Agteren MH, Keuning S, Janssen DB, Janssen JP, Oosterhaven J. 2010. Handbook on biodegradation and biological treatment of hazardous organic compounds. Environment and chemistry, vol 2 Springer, Dordrecht, The Netherlands. [Google Scholar]
- 5.Tischler D, Kaschabek SR. 2012. Microbial styrene degradation: from basics to biotechnology, p 67–99. In Singh SN. (ed), Microbial degradation of xenobiotics. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 6.Mooney A, Ward PG, O'Connor KE. 2006. Microbial degradation of styrene: biochemistry, molecular genetics, and perspectives for biotechnological applications. Appl Microbiol Biotechnol 72:1–10. doi: 10.1007/s00253-006-0443-1. [DOI] [PubMed] [Google Scholar]
- 7.Crabo AG, Singh B, Nguyen T, Emami S, Gassner GT, Sazinsky MH. 2017. Structure and biochemistry of phenylacetaldehyde dehydrogenase from the Pseudomonas putida S12 styrene catabolic pathway. Arch Biochem Biophys 616:47–58. doi: 10.1016/j.abb.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A 107:14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grishin AM, Cygler M. 2015. Structural organization of enzymes of the phenylacetate catabolic hybrid pathway. Biology (Basel) 4:424–442. doi: 10.3390/biology4020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh N, Yoshida K, Okada K. 1996. Isolation and identification of styrene-degrading Corynebacterium strains, and their styrene metabolism. Biosci Biotechnol Biochem 60:1826–1830. doi: 10.1271/bbb.60.1826. [DOI] [PubMed] [Google Scholar]
- 11.Itoh N, Hayashi K, Okada K, Ito T, Mizuguchi N. 1997. Characterization of styrene oxide isomerase, a key enzyme of styrene and styrene oxide metabolism in Corynebacterium sp. Biosci Biotechnol Biochem 61:2058–2062. doi: 10.1271/bbb.61.2058. [DOI] [PubMed] [Google Scholar]
- 12.Itoh N, Morihama R, Wang J, Okada K, Mizuguchi N. 1997. Purification and characterization of phenylacetaldehyde reductase from a styrene-assimilating Corynebacterium strain, ST-10 Appl Environ Microbiol 63:3783–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toda H, Itoh N. 2012. Isolation and characterization of styrene metabolism genes from styrene-assimilating soil bacteria Rhodococcus sp. ST-5 and ST-10. J Biosci Bioeng 113:12–19. doi: 10.1016/j.jbiosc.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Marconi AM, Beltrametti F, Bestetti G, Solinas F, Ruzzi M, Galli E, Zennaro E. 1996. Cloning and characterization of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol 62:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arenskötter M, Bröker D, Steinbüchel A. 2004. Biology of the metabolically diverse genus Gordonia. Appl Environ Microbiol 70:3195–3204. doi: 10.1128/AEM.70.6.3195-3204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drzyzga O. 2012. The strengths and weaknesses of Gordonia: a review of an emerging genus with increasing biotechnological potential. Crit Rev Microbiol 38:300–316. doi: 10.3109/1040841X.2012.668134. [DOI] [PubMed] [Google Scholar]
- 17.Oelschlägel M, Zimmerling J, Schlömann M, Tischler D. 2014. Styrene oxide isomerase of Sphingopyxis sp. Kp5.2. Microbiology (Reading, Engl) 160:2481–2491. doi: 10.1099/mic.0.080259-0. [DOI] [PubMed] [Google Scholar]
- 18.Oelschlägel M, Kaschabek SR, Zimmerling J, Schlömann M, Tischler D. 2015. Co-metabolic formation of substituted phenylacetic acids by styrene-degrading bacteria. Biotechnol Rep 6:20–26. doi: 10.1016/j.btre.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esuola CO, Babalola OO, Heine T, Schwabe R, Schlömann M, Tischler D. 2016. Identification and characterization of a FAD-dependent putrescine N-hydroxylase (GorA) from Gordonia rubripertincta CWB2. J Mol Catal B Enzym 134:378–389. doi: 10.1016/j.molcatb.2016.08.003. [DOI] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Johnston A, Crombie AT, El Khawand M, Sims L, Whited GM, McGenity TJ, Colin Murrell J. 2017. Identification and characterisation of isoprene-degrading bacteria in an estuarine environment. Environ Microbiol doi: 10.1111/1462-2920.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crombie AT, Khawand ME, Rhodius VA, Fengler KA, Miller MC, Whited GM, McGenity TJ, Murrell JC. 2015. Regulation of plasmid-encoded isoprene metabolism in Rhodococcus, a representative of an important link in the global isoprene cycle. Environ Microbiol 17:3314–3329. doi: 10.1111/1462-2920.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Munch PC, Spaepen S, Remus-Emsermann M, Huttel B, McHardy AC, Vorholt JA, Schulze-Lefert P. 2015. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 24.Tropel D, van der Meer JR. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev 68:474–500. doi: 10.1128/MMBR.68.3.474-500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotani T, Yamamoto T, Yurimoto H, Sakai Y, Kato N. 2003. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J Bacteriol 185:7120–7128. doi: 10.1128/JB.185.24.7120-7128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan Kwo Chion CK, Askew SE, Leak DJ. 2005. Cloning, expression, and site-directed mutagenesis of the propene monooxygenase genes from Mycobacterium sp. strain M156. Appl Environ Microbiol 71:1909–1914. doi: 10.1128/AEM.71.4.1909-1914.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colby J, Stirling DI, Dalton H. 1977. The soluble methane monooxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic, and heterocyclic compounds. Biochem J 165:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuya T, Hirose S, Osanai H, Semba H, Kino K. 2011. Identification of the monooxygenase gene clusters responsible for the regioselective oxidation of phenol to hydroquinone in mycobacteria. Appl Environ Microbiol 77:1214–1220. doi: 10.1128/AEM.02316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuya T, Hayashi M, Kino K. 2013. Reconstitution of active mycobacterial binuclear iron monooxygenase complex in Escherichia coli. Appl Environ Microbiol 79:6033–6039. doi: 10.1128/AEM.01856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuya T, Hayashi M, Semba H, Kino K. 2013. The mycobacterial binuclear iron monooxygenases require a specific chaperonin-like protein for functional expression in a heterologous host. FEBS J 280:817–826. [DOI] [PubMed] [Google Scholar]
- 31.Narancic T, Djokic L, Kenny ST, O'Connor KE, Radulovic V, Nikodinovic-Runic J, Vasiljevic B. 2012. Metabolic versatility of Gram-positive microbial isolates from contaminated river sediments. J Hazard Mater 215–216:243–251. doi: 10.1016/j.jhazmat.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 32.Braun-Lüllemann A, Majcherczyk A, Hüttermann A. 1997. Degradation of styrene by white-rot fungi. Appl Microbiol Biotechnol 47:150–155. doi: 10.1007/s002530050904. [DOI] [PubMed] [Google Scholar]
- 33.Gaszczak A, Bartelmus G, Gren I. 2012. Kinetics of styrene biodegradation by Pseudomonas sp. E-93486. Appl Microbiol Biotechnol 93:565–573. doi: 10.1007/s00253-011-3518-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bredholt H, Josefsen K, Vatland A, Bruheim P, Eimhjellen K. 1998. Emulsification of crude oil by an alkane-oxidizing Rhodococcus species isolated from seawater. Can J Microbiol 44:330–340. doi: 10.1139/cjm-44-4-330. [DOI] [Google Scholar]
- 35.Lang S, Philp JC. 1998. Surface-active lipids in rhodococci. Antonie Van Leeuwenhoek 74:59–70. doi: 10.1023/A:1001799711799. [DOI] [PubMed] [Google Scholar]
- 36.Franzetti A, Bestetti G, Caredda P, La Colla P, Tamburini E. 2008. Surface-active compounds and their role in the access to hydrocarbons in Gordonia strains. FEMS Microbiol Ecol 63:238–248. doi: 10.1111/j.1574-6941.2007.00406.x. [DOI] [PubMed] [Google Scholar]
- 37.O'Connor K, Buckley CM, Hartmans S, Dobson AD. 1995. Possible regulatory role for nonaromatic carbon sources in styrene degradation by Pseudomonas putida CA-3. Appl Environ Microbiol 61:544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velasco A, Alonso S, Garcia JL, Perera J, Diaz E. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol 180:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos PM, Blatny JM, Di Bartolo I, Valla S, Zennaro E. 2000. Physiological analysis of the expression of the styrene degradation gene cluster in Pseudomonas fluorescens ST. Appl Environ Microbiol 66:1305–1310. doi: 10.1128/AEM.66.4.1305-1310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Leary ND, O'Connor KE, Duetz W, Dobson AD. 2001. Transcriptional regulation of styrene degradation in Pseudomonas putida CA-3. Microbiology 147:973–979. doi: 10.1099/00221287-147-4-973. [DOI] [PubMed] [Google Scholar]
- 41.Leoni L, Ascenzi P, Bocedi A, Rampioni G, Castellini L, Zennaro E. 2003. Styrene-catabolism regulation in Pseudomonas fluorescens ST: phosphorylation of StyR induces dimerization and cooperative DNA-binding. Biochem Biophys Res Commun 303:926–931. doi: 10.1016/S0006-291X(03)00450-9. [DOI] [PubMed] [Google Scholar]
- 42.Leoni L, Rampioni G, Di Stefano V, Zennaro E. 2005. Dual role of response regulator StyR in styrene catabolism regulation. Appl Environ Microbiol 71:5411–5419. doi: 10.1128/AEM.71.9.5411-5419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milani M, Leoni L, Rampioni G, Zennaro E, Ascenzi P, Bolognesi M. 2005. An active-like structure in the unphosphorylated StyR response regulator suggests a phosphorylation-dependent allosteric activation mechanism. Structure 13:1289–1297. doi: 10.1016/j.str.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 44.del Peso-Santos T, Shingler V, Perera J. 2008. The styrene-responsive StyS/StyR regulation system controls expression of an auxiliary phenylacetyl-coenzyme A ligase: implications for rapid metabolic coupling of the styrene upper- and lower-degradative pathways. Mol Microbiol 69:317–330. doi: 10.1111/j.1365-2958.2008.06259.x. [DOI] [PubMed] [Google Scholar]
- 45.Patrauchan MA, Florizone C, Eapen S, Gomez-Gil L, Sethuraman B, Fukuda M, Davies J, Mohn WW, Eltis LD. 2008. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J Bacteriol 190:37–47. doi: 10.1128/JB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikodinovic-Runic J, Flanagan M, Hume AR, Cagney G, O'Connor KE. 2009. Analysis of the Pseudomonas putida CA-3 proteome during growth on styrene under nitrogen-limiting and non-limiting conditions. Microbiology 155:3348–3361. doi: 10.1099/mic.0.031153-0. [DOI] [PubMed] [Google Scholar]
- 47.Mooney A, O'Leary ND, Dobson ADW. 2006. Cloning and functional characterization of the styE gene, involved in styrene transport in Pseudomonas putida CA-3. Appl Environ Microbiol 72:1302–1309. doi: 10.1128/AEM.72.2.1302-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riedel A, Heine T, Westphal AH, Conrad C, Rathsack P, van Berkel WJH, Tischler D. 2015. Catalytic and hydrodynamic properties of styrene monooxygenases from Rhodococcus opacus 1CP are modulated by cofactor binding. AMB Express 5:112. doi: 10.1186/s13568-015-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otto K, Hofstetter K, Röthlisberger M, Witholt B, Schmid A. 2004. Biochemical characterization of StyAB from Pseudomonas sp. strain VLB120 as a two-component flavin-diffusible monooxygenase. J Bacteriol 186:5292–5302. doi: 10.1128/JB.186.16.5292-5302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toda H, Imae R, Komio T, Itoh N. 2012. Expression and characterization of styrene monooxygenases of Rhodococcus sp. ST-5 and ST-10 for synthesizing enantiopure (S)-epoxides. Appl Microbiol Biotechnol 96:407–418. doi: 10.1007/s00253-011-3849-3. [DOI] [PubMed] [Google Scholar]
- 51.Cheng L, Yin S, Chen M, Sun B, Hao S, Wang C. 2016. Enhancing indigo production by overexpression of the styrene monooxygenase in Pseudomonas putida. Curr Microbiol 73:248–254. doi: 10.1007/s00284-016-1055-3. [DOI] [PubMed] [Google Scholar]
- 52.Han JH, Park MS, Bae JW, Lee EY, Yoon YJ, Lee S-G, Park S. 2006. Production of (S)-styrene oxide using styrene oxide isomerase negative mutant of Pseudomonas putida SN1. Enzyme Microb Technol 39:1264–1269. doi: 10.1016/j.enzmictec.2006.03.002. [DOI] [Google Scholar]
- 53.Oelschlägel M, Gröning JAD, Tischler D, Kaschabek SR, Schlömann M. 2012. Styrene oxide isomerase of Rhodococcus opacus 1CP, a highly stable and considerably active enzyme. Appl Environ Microbiol 78:4330–4337. doi: 10.1128/AEM.07641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Khawand M, Crombie AT, Johnston A, Vavlline DV, McAuliffe JC, Latone JA, Primak YA, Lee S-K, Whited GM, McGenity TJ, Murrell JC. 2016. Isolation of isoprene degrading bacteria from soils, development of isoA gene probes and identification of the active isoprene-degrading soil community using DNA-stable isotope probing. Environ Microbiol 18:2743–2753. doi: 10.1111/1462-2920.13345. [DOI] [PubMed] [Google Scholar]
- 55.Alvarez LA, Exton DA, Timmis KN, Suggett DJ, McGenity TJ. 2009. Characterization of marine isoprene-degrading communities. Environ Microbiol 11:3280–3291. doi: 10.1111/j.1462-2920.2009.02069.x. [DOI] [PubMed] [Google Scholar]
- 56.van Hylckama VJ, Kingma J, van den Wijngaard AJ, Janssen DB. 1998. A glutathione S-transferase with activity towards cis-1,2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl Environ Microbiol 64:2800–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Hylckama VJ, Kingma J, Kruizinga W, Janssen DB. 1999. Purification of a glutathione S-transferase and a glutathione conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol 181:2094–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Hylckama VJ, Leemhuis H, Spelberg JHL, Janssen DB, Spelberg JH, van Hylckama Vlieg JE. 2000. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol 182:1956–1963. doi: 10.1128/JB.182.7.1956-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El Khawand M. 2014. Bacterial degradation of isoprene in the terrestrial environment. PhD dissertation. University of East Anglia, Norwich, United Kingdom. [Google Scholar]
- 60.Smirnova GV, Oktyabrsky ON. 2005. Glutathione in bacteria. Biochemistry 70:1199–1211. doi: 10.1007/s10541-005-0248-3. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerling J, Tischler D, Grossmann C, Schlömann M, Oelschlägel M. 2017. Characterization of aldehyde dehydrogenases applying an enzyme assay with in situ formation of phenylacetaldehydes. Appl Biochem Biotechnol 182:1095–1107. doi: 10.1007/s12010-016-2384-1. [DOI] [PubMed] [Google Scholar]
- 62.Fall R, Copley SD. 2000. Bacterial sources and sinks of isoprene, a reactive atmospheric hydrocarbon. Environ Microbiol 2:123–130. doi: 10.1046/j.1462-2920.2000.00095.x. [DOI] [PubMed] [Google Scholar]
- 63.Allocati N, Federici L, Masulli M, Di Ilio C. 2009. Glutathione transferases in bacteria. FEBS J 276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 64.Fahey RC, Brown WC, Adams WB, Worsham MB. 1978. Occurrence of glutathione in bacteria. J Bacteriol 133:1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson T, Newton GL, Fahey RC, Rawat M. 2009. Unusual production of glutathione in actinobacteria. Arch Microbiol 191:89–93. doi: 10.1007/s00203-008-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allocati N, Federici L, Masulli M, Di Ilio C. 2012. Distribution of glutathione transferases in Gram-positive bacteria and Archaea. Biochimie 94:588–596. doi: 10.1016/j.biochi.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 67.Navarro-Llorens JM, Patrauchan MA, Stewart GR, Davies JE, Eltis LD, Mohn WW. 2005. Phenylacetate catabolism in Rhodococcus sp. strain RHA1: a central pathway for degradation of aromatic compounds. J Bacteriol 187:4497–4504. doi: 10.1128/JB.187.13.4497-4504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X, Kohl TA, Rückert C, Rodionov DA, Li L-H, Ding J-Y, Kalinowski J, Liu S-J. 2012. Phenylacetic acid catabolism and its transcriptional regulation in Corynebacterium glutamicum. Appl Environ Microbiol 78:5796–5804. doi: 10.1128/AEM.01588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Hylckama VJ, Janssen DB. 2001. Formation and detoxification of reactive intermediates in the metabolism of chlorinated ethenes. J Biotechnol 85:81–102. doi: 10.1016/S0168-1656(00)00364-3. [DOI] [PubMed] [Google Scholar]
- 70.Rui L, Kwon YM, Reardon KF, Wood TK. 2004. Metabolic pathway engineering to enhance aerobic degradation of chlorinated ethenes and to reduce their toxicity by cloning a novel glutathione S-transferase, an evolved toluene o-monooxygenase, and gamma-glutamylcysteine synthetase. Environ Microbiol 6:491–500. doi: 10.1111/j.1462-2920.2004.00586.x. [DOI] [PubMed] [Google Scholar]
- 71.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol 99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 72.Cooper DG, Goldenberg BG. 1987. Surface-active agents from two bacillus species. Appl Environ Microbiol 53:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ames-Gottfred NP, Christie BR, Jordan DC. 1989. Use of the Chrome Azurol S agar plate technique to differentiate strains and field isolates of Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol 55:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res 8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 75.Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29:2936–2937. doi: 10.1093/bioinformatics/btt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 77.Krogh A, Larsson B, Heijne G von, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 78.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FS. 2017. IslandViewer 4. Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hilker R, Stadermann KB, Schwengers O, Anisiforov E, Jaenicke S, Weisshaar B, Zimmermann T, Goesmann A. 2016. ReadXplorer 2-detailed read mapping analysis and visualization from one single source. Bioinformatics 32:3702–3708. doi: 10.1093/bioinformatics/btw541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 82.Al-Furoukh N, Ianni A, Nolte H, Hölper S, Krüger M, Wanrooij S, Braun T. 2015. ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells. Biochim Biophys Acta 1853:2580–2591. doi: 10.1016/j.bbamcr.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 83.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 84.Thakur SS, Geiger T, Chatterjee B, Bandilla P, Fröhlich F, Cox J, Mann M. 2011. Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol Cell Proteomics 10:M110.003699. doi: 10.1074/mcp.M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. 2007. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res 35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sambrook J, Russell DW (ed). 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 88.O'Connor KE, Dobson AD, Hartmans S. 1997. Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl Environ Microbiol 63:4287–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Warhurst AM, Clarke KF, Hill RA, Holt RA, Fewson CA. 1994. Metabolism of styrene by Rhodococcus rhodochrous NCIMB 13259. Appl Environ Microbiol 60:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tischler D, Eulberg D, Lakner S, Kaschabek SR, van Berkel WJH, Schlömann M. 2009. Identification of a novel self-sufficient styrene monooxygenase from Rhodococcus opacus 1CP. J Bacteriol 191:4996–5009. doi: 10.1128/JB.00307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dawson RMC. 1986. Data for biochemical research, 3rd ed Clarendon Press, Oxford, United Kingdom. [Google Scholar]
- 92.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 93.Alonso S, Bartolomé-Martín D, del Álamo M, Díaz E, García JL, Perera J. 2003. Genetic characterization of the styrene lower catabolic pathway of Pseudomonas sp. strain Y2. Gene 319:71–83. doi: 10.1016/S0378-1119(03)00794-7. [DOI] [PubMed] [Google Scholar]
- 94.El-Said Mohamed M. 2000. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J Bacteriol 182:286–294. doi: 10.1128/JB.182.2.286-294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kendall SL, Burgess P, Balhana R, Withers M, Bokum A ten, Lott JS, Gao C, Uhia-Castro I, Stoker NG. 2010. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiology 156:1362–1371. doi: 10.1099/mic.0.034538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagy I, Schoofs G, Compernolle F, Proost P, Vanderleyden J, Mot R de. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J Bacteriol 177:676–687. doi: 10.1128/jb.177.3.676-687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takeo M, Murakami M, Niihara S, Yamamoto K, Nishimura M, Kato D-i, Negoro S. 2008. Mechanism of 4-nitrophenol oxidation in Rhodococcus sp. strain PN1: characterization of the two-component 4-nitrophenol hydroxylase and regulation of its expression. J Bacteriol 190:7367–7374. doi: 10.1128/JB.00742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beltrametti F, Marconi AM, Bestetti G, Colombo C, Galli E, Ruzzi M, Zennaro E. 1997. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol 63:2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wadington MC, Ladner JE, Stourman NV, Harp JM, Armstrong RN. 2009. Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase. Biochem 48:6559–6561. doi: 10.1021/bi9008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mullins EA, Sullivan KL, Kappock TJ. 2013. Function and X-ray crystal structure of Escherichia coli YfdE. PLoS One 8:e67901. doi: 10.1371/journal.pone.0067901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reverchon S, Nasser W, Robert-Baudouy J. 1994. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol Microbiol 11:1127–1139. doi: 10.1111/j.1365-2958.1994.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 102.Harris DR, Ward DE, Feasel JM, Lancaster KM, Murphy RD, Mallet TC, Crane EJ3. 2005. Discovery and characterization of a coenzyme A disulfide reductase from Pyrococcus horikoshii: implications for this disulfide metabolism of anaerobic hyperthermophiles. FEBS J 272:1189–1200. doi: 10.1111/j.1742-4658.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 103.Zhang CC, Huguenin S, Friry A. 1995. Analysis of genes encoding the cell division protein FtsZ and a glutathione synthetase homologue in the cyanobacterium Anabaena sp. PCC. 7120 Res Microbiol 146:445–455. [DOI] [PubMed] [Google Scholar]
- 104.Harth G, Maslesa-Galic S, Tullius MV, Horwitz MA. 2005. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol Microbiol 58:1157–1172. doi: 10.1111/j.1365-2958.2005.04899.x. [DOI] [PubMed] [Google Scholar]
- 105.Gupta RS. 1994. Identification of a GroES (CPN10)-related sequence motif in the GroEL (CPN60) chaperonins. Biochem Mol Biol Int 33:591–595. [PubMed] [Google Scholar]
- 106.Hahn J-S, Oh S-Y, Roe J-H. 2002. Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J Bacteriol 184:5214–5222. doi: 10.1128/JB.184.19.5214-5222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dhandayuthapani S, Zhang Y, Mudd MH, Deretic V. 1996. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J Bacteriol 178:3641–3649. doi: 10.1128/jb.178.12.3641-3649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ramachandran S, Magnuson TS, Crawford DL. 2000. Isolation and analysis of three peroxide sensor regulatory gene homologs ahpC, ahpX, and oxyR in Streptomyces viridosporus T7A: a lignocellulose-degrading actinomycete. DNA Seq 11:51–60. doi: 10.3109/10425170009033969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.