ABSTRACT
The integron platform allows the acquisition, expression, and dissemination of antibiotic resistance genes within gene cassettes. Wastewater treatment plants (WWTPs) contain abundant resistance genes; however, knowledge about the impacts of wastewater treatment on integrons and their gene cassettes is limited. In this study, by using clone library analysis and high-throughput sequencing, we investigated the abundance of class 1, 2, and 3 integrons and their corresponding gene cassettes in three urban WWTPs. Our results showed that class 1 integrons were most abundant in WWTPs and that wastewater treatment significantly reduced the abundance of all integrons. The WWTP influents harbored the highest diversity of class 1 integron gene cassettes, whereas class 3 integron gene cassettes exhibited highest diversity in activated sludge. Most of the gene cassette arrays detected in class 1 integrons were novel. Aminoglycoside, beta-lactam, and trimethoprim resistance genes were highly prevalent in class 1 integron gene cassettes, while class 3 integrons mainly carried beta-lactam resistance gene cassettes. A core class 1 integron resistance gene cassette pool persisted during wastewater treatment, implying that these resistance genes could have high potential to spread into environments through WWTPs. These data provide new insights into the impact of wastewater treatment on integron pools and highlight the need for surveillance of resistance genes within both class 1 and 3 integrons.
IMPORTANCE Wastewater treatment plants represent a significant sink and transport medium for antibiotic resistance bacteria and genes spreading into environments. Integrons are important genetic elements involved in the evolution of antibiotic resistance. To better understand the impact of wastewater treatment on integrons and their gene cassette contexts, we conducted clone library construction and high-throughput sequencing to analyze gene cassette contexts for class 1 and class 3 integrons during the wastewater treatment process. This study comprehensively profiled the distribution of integrons and their gene cassettes (especially class 3 integrons) in influents, activated sludge, and effluents of conventional municipal wastewater treatment plants. We further demonstrated that while wastewater treatment significantly reduced the abundance of integrons and the diversity of associated gene cassettes, a large fraction of integrons persisted in wastewater effluents and were consequentially discharged into downstream natural environments.
KEYWORDS: environmental pollution, human health, horizontal gene transfer, wastewater treatment plants
INTRODUCTION
The emergence and spread of antibiotic resistance genes (ARGs) among bacterial pathogens are threatening global public health (1). The dissemination of resistance genes in the environment could partially be attributed to horizontal gene transfer (HGT) mediated via mobile genetic elements (MGEs), which are frequently detected in human pathogens (2–4). Therefore, understanding the role of MGEs in the development of antibiotic resistance is especially important in attempting to mitigate the dissemination of resistance genes.
Integrons are genetic elements that can capture and express exogenous genes (e.g., ARGs) embedded within gene cassettes (GCs) (5, 6). All integrons contain three key elements: an intI gene encoding an integrase for catalyzing recombination between incoming genes, an attI gene for an integron-associated recombination site, and an integron-associated promoter (Pc) for driving the expression of the newly integrated genes. Gene cassettes are individually mobilizable elements, which normally couple an open reading frame (ORF) with a site-specific recombinase recognition site known as a 59-base element or attC (7). Integron gene cassettes can be integrated into bacterial chromosomes or plasmids, and their mobility allows genes to penetrate into new organisms (8, 9). Thus, integrons have access to a vast pool of gene cassettes with diverse functions. Mobile integrons are prevalent in human-dominated ecosystems with prolonged exposure to selective agents (e.g., detergents, antibiotics, and heavy metals) (10). Integrons can accumulate genes that confer advantageous phenotypes. Therefore, they are regarded as one of the major drivers contributing to the evolution and acquisition of bacterial resistance (11–13).
Integrons can be classed according to the differences in the amino acid sequence of the IntI protein (14). Three major integron classes, classes 1, 2, and 3, are the most commonly known to be associated with horizontally transferred resistance genes. Class 1 integron-integrase genes have been extensively surveyed, especially in multidrug-resistant Gram-negative clinical isolates (11, 15, 16), and they may serve as a proxy for anthropogenic pollution (17). Many known class 1 integrons contain a 3′ conserved segment (3′ CS), which is composed of qacEΔ1, conferring resistance to quaternary ammonium compounds (QACs), sul1, conferring resistance to sulfonamides, and orf5, encoding a protein of unknown function (7, 18). These integrons share a pool of gene cassettes, most of which confer resistance to a wide range of antibiotics. Currently, approximately 130 different resistance gene cassettes have been described for class 1 integrons, and these resistance gene cassettes probably have been accumulated incrementally from diverse phylogenetic backgrounds (6, 14, 19). Class 1 integrons are highly conserved and closely associated with Tn402-like transposons. Less is known about class 2 and 3 integrons, which possibly originated from a chromosomal ancestor through transposable elements (6). These two classes have been observed in several clinical isolates, and their integrase genes have been reported to be inactive or unable to carry a high diversity of gene cassettes (20, 21).
Wastewater treatment plants (WWTPs) are a critical hub for the evolution and development of anthropogenically derived antibiotic resistance genes. Studies of integrons in WWTPs have revealed variable abundances of integrons and their host bacteria (7, 22, 23). However, these studies have frequently focused on quantities of integrons using quantitative PCR (qPCR) and limited numbers of host bacteria based on culture-dependent methods. High-throughput sequencing analyses have been applied for characterizing class 1 integron gene cassettes, revealing a wide distribution of ARG-carrying class 1 integrons in different environmental samples (e.g., water environments, sediment, and feces) (24, 25). A recent study revealed that hospital effluents (EFFs) are important sources of integrons in sewage treatment plants (26). However, variations of integron abundance and their gene cassettes during the entire wastewater treatment process (influent [INF], activated sludge [AS], and EFF) is poorly understood, especially for class 3 integrons. In this study, we used the combination of clone library and high-throughput amplicon sequencing analyses to investigate the abundances of class 1, 2, and 3 integrons and to characterize class 1 and 3 integron gene cassette contents in WWTPs. Our results provide insights on the influence of wastewater treatment on the dynamics of integron abundance and their gene cassette contents, which are necessary for mitigating the dissemination of ARGs carried by integrons in WWTPs.
RESULTS
Impact of wastewater treatment on integron abundance in WWTPs.
High concentrations of integrons were detected in three WWTPs (see Table S1 in the supplemental material). The sum of the abundance for the three classes of integrons ranged from 107 copies/ml to 1010 copies/ml (Fig. 1). The concentrations of class 1 integrons were significantly higher (2 to 3 orders of magnitude; P < 0.05) than those of class 2 and 3 integrons. There was no significant difference in the abundance between class 2 integrons and class 3 integrons (P > 0.05). Integron concentrations were lower in WWTP effluents than in WWTP influents (P < 0.05) and were highest in activated sludge (Fig. 1). After wastewater treatment, approximately 96.9% of detected integrons and 87.8% of total bacteria were removed, corresponding to around a 2-log decrease in their concentrations.
FIG 1.
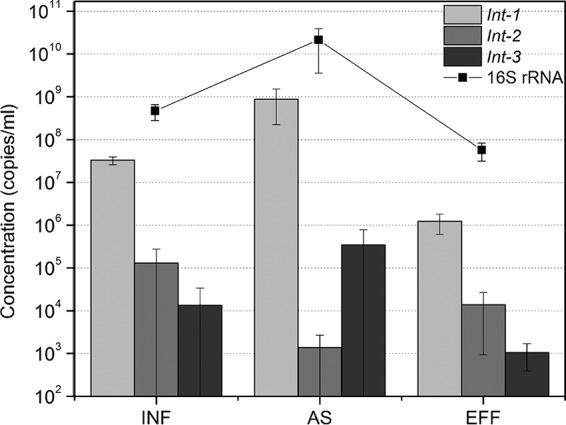
Variation in concentrations of integrons (class 1, class 2, and class 3) during wastewater treatment. The line shows the variable concentrations of the total bacterial community through wastewater treatment. INF, influent; AS, activated sludge; EFF, effluent. Means ± SDs are shown.
Except for class 3 integrons, the relative abundance of integrons (number of copies/copy of the 16S rRNA gene) was significantly reduced across the wastewater treatment process (P < 0.05), with the highest normalized copy numbers observed in the influents (Fig. 2). The relative abundance of intI1 had a higher range, with normalized copy numbers of 1.1 × 10−2 to 9.3 × 10−2 copies/copy of 16S rRNA. The normalized abundances of intI2 and intI3 were lower and showed wider ranges, 2.5 × 10−7 to 1.2 × 10−3 and 4.5 × 10−7 to 1.8 × 10−4 copies/copy of 16S rRNA, respectively. Based on the average copy number (4.1) of the 16S rRNA gene in one bacterial cell, the copies of integrons per bacterial cell were estimated at 4.5 × 10−2 to 3.8 × 10−1 copies per cell for class 1 integrons, 1.0 × 10−6 to 5.0 × 10−3 copies per cell for class 2 integrons, and 7.3 × 10−4 to 1.9 × 10−6 copies per cell for class 3 integrons.
FIG 2.
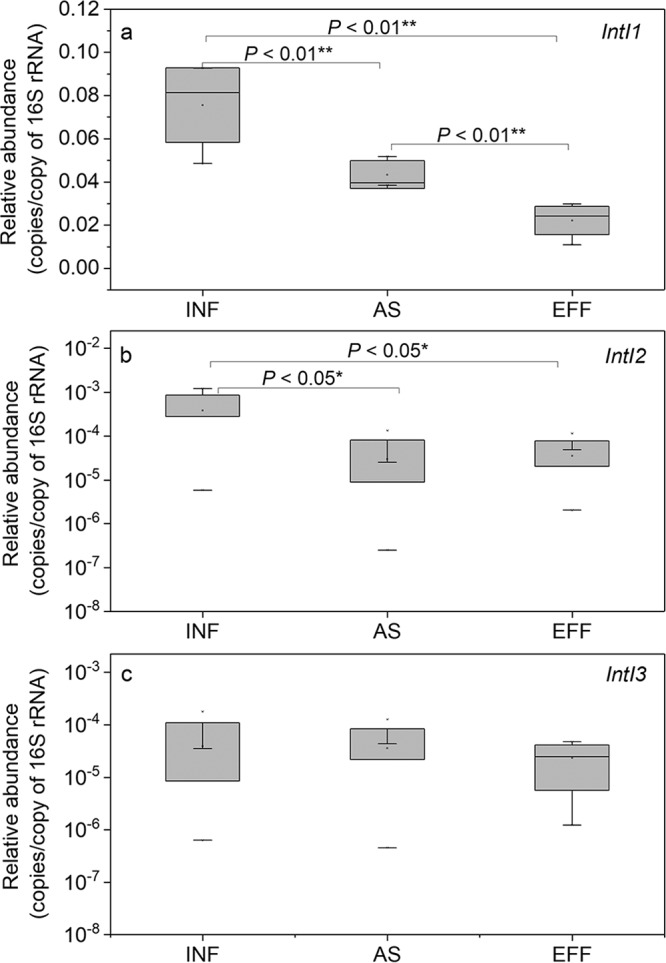
Normalized relative abundance (copies per copy of 16S rRNA gene) of integrons, including class 1 (a), class 2 (b), and class 3 (c), at different wastewater treatment processes. Means ± SDs are shown. ANOVA was performed. *, P < 0.05; **, P < 0.01.
Characterization of class 1 integron gene cassette and gene cassette array.
To further assess the impact of wastewater treatment on integrons and their associated ARG cassettes, we investigated the gene cassette contents of class 1 and class 3 integrons by clone library analysis. For each sample, we randomly picked at least 138 class 1 integron gene cassette clones, and we observed that a high proportion of class 1 integron clones (77.2%) were empty, especially those in the effluents (see Table S2). About 26.4%, 29.1%, and 12.9% of clones carried ≥1 gene cassette in the influents, activated sludge, and effluents, respectively (Table S2). A total of 79 unique gene cassettes were found in WWTPs, in which 50 different gene cassettes were detected in influents, 36 gene cassettes in activated sludge, and 27 gene cassettes in effluents (Fig. 3). Eight shared gene cassettes were found across all samples, most of which were the gene cassettes conferring resistance to aminoglycosides (e.g., aadA1, aadA2, and aadA5). The prevalence of these shared ARGs was further verified by qPCR, with the highest abundance in influents (P < 0.05) (Fig. S1). In addition, 16 unique gene cassette arrays were observed, of which 9 contained 2 gene cassettes, 6 carried 3 gene cassettes, and only 1 array carried 4 gene cassettes (Table 1). The majority of the detected arrays were absent from the INTEGRALL database, some of which (e.g., blaOXA-21-aadA2, aadA2-linF, and catB8-blaOXA-1-aadA1) were frequently found in influents and effluents (Table 1; see also Table S3).
FIG 3.
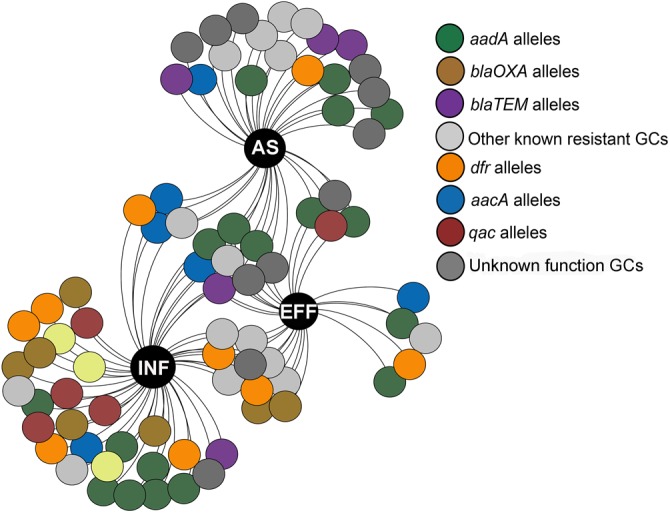
Diversity and persistence of gene cassettes (GCs) across wastewater treatment. The large black dots represent the wastewater treatment processes. Each small dot stands for a different gene cassette encoding a functional protein (based on ≥99% nucleic acid identity). Colors in dots represent different categories of gene cassettes in class 1 integrons.
TABLE 1.
Class 1 integron GC arrays and their structures detected in WWTPs, based on a clone librarya
| GC array | GC structure | Source description | Sample(s) in which GC was detected | No. of GCs |
|---|---|---|---|---|
| aacA4-ereA1 | 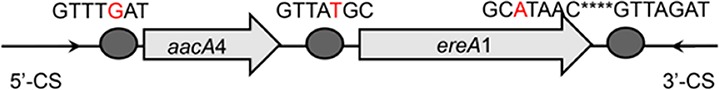 |
This study | QPE2 | 1 |
| blaOXA-101-orf-aadA2 | 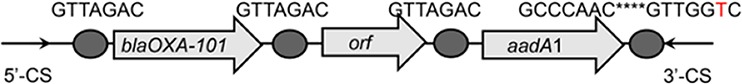 |
This study | LYE1 | 1 |
| trm-aacA4-ereA1 | 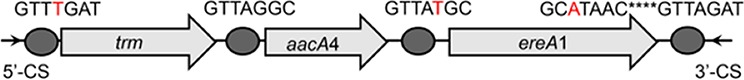 |
This study | QPI1 | 1 |
| blaOXA-101-orf-aadA2 | 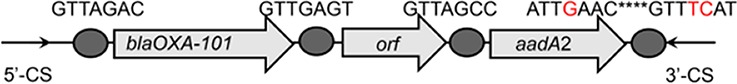 |
This study | LYE1 | 1 |
| orf-aadA2-blaOXA-129-linF | 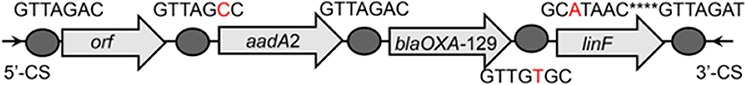 |
This study | LYI1 | 1 |
| blaOXA-21-aadA2 | 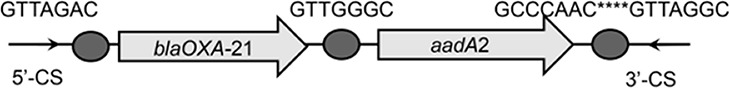 |
This study | LYI1 | 1 |
| aacA4-ereA-aadA2 | 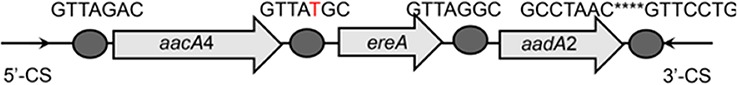 |
This study | LYA2 | 1 |
| aadA2-linF | 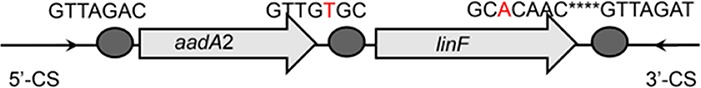 |
This study | QPI1, JME1 | 2 |
| arr2-aacA4 | 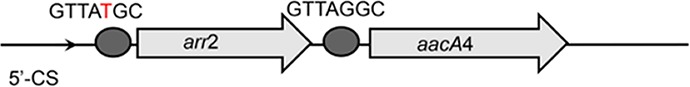 |
EU340416; 63 | LYI1 | 1 |
| blaOXA-10-aadA1 | 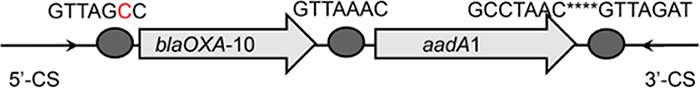 |
This study | QPI1, JME2 | 2 |
| catB8-blaOXA-1-aadA1 | 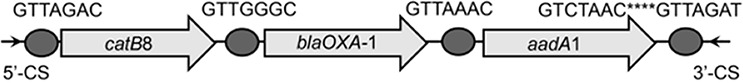 |
This study | QPI1 | 1 |
| blaVEB-1-aadB | 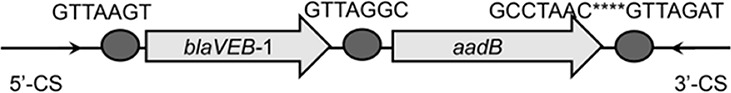 |
This study | LYI1 | 1 |
| dfrA14-nit1-nit2 | 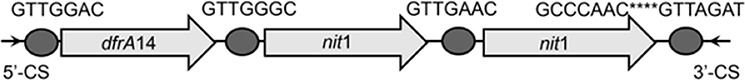 |
This study | QPA1 | 1 |
| arr2-dfrA27 | 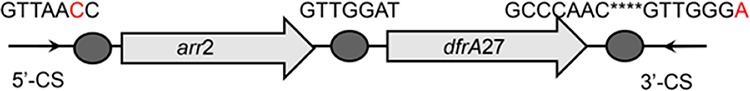 |
This study | JME1 | 1 |
| aadA16-orfD | 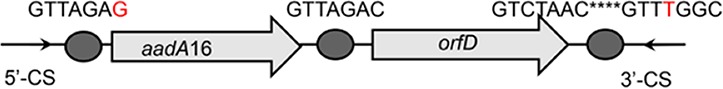 |
This study | JME1 | 1 |
| qacE2-orfD | 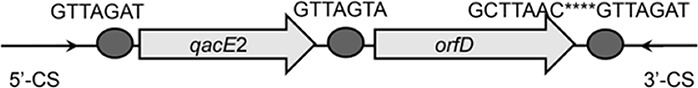 |
DQ462520; 64 | LYA1, LYE1 | 3 |
The circle in the gene cassette (GC) structure represents the attC site, where the base in red indicates a variant in the conserved region. Nucleotide sequences with asterisks indicate the inverted repeats. Samples are named on the basis of the abbreviation for the WWTPs and sample types. For example, “QPE” refers to the effluent sample (E) from of Qianpu (QP) wastewater treatment plant. LY, Longyan; JM, Jimei.
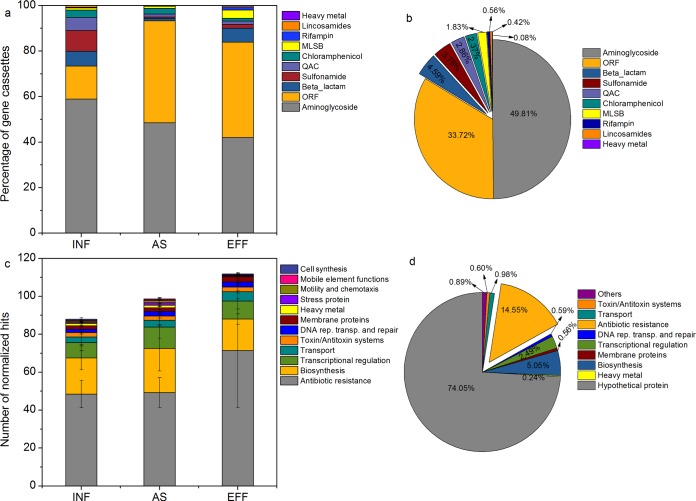
A majority of the gene cassettes identified in this study consisted of aminoglycoside genes (average, 49.8%) and beta-lactam resistance genes (average, 4.6%) (Fig. 4a and b). In addition, heavy metal resistance gene chrA, encoding chromate ion transport protein, was also detected in an activated sludge sample (Table S3). A total of 39 novel gene cassettes were found in WWTPs, and most of them were predicted to encode conserved hypothetical proteins or contained open reading frames for which no homology could be found in databases (e.g., orf, orfA, and orfD). For example, the orfD gene cassette was detected in almost all samples, accounting for 29.9% of total gene cassettes. orfD shared a high protein sequence identity (99%) with the partial sequence corresponding to GenBank accession number DQ091179 as well as structural homology (100%) with hypothetical 11.6K protein (GenBank accession number JQ1757). The sequence with GenBank accession number DQ091179 was annotated as the aadA6 gene cassette, and the hypothetical 11.6K protein was from plasmid R46. The other new gene cassette, orfA, potentially encoded a protein sharing 73% identity with a deazaflavin-dependent nitroreductase (GenBank accession number CFE35199.1). Additionally, some different resistance gene alleles were found to be likely new resistance gene variants (90 to 99% identity with known resistance gene cassettes), e.g., aadA alleles and blaTEM-1 alleles.
FIG 4.
Percentage of class 1 integron-associated gene cassettes during wastewater treatment based on a clone library (a and b) and high-throughput sequencing data (c and d). The bar chart shows the variation in abundance of gene cassettes during wastewater treatment, and the pie chart reveals the percentage of all gene cassettes from 18 WWTP samples. The number of normalized hits was calculated by normalization to the average number of total reads for each sample. MLSB, macrolide-lincosamide-streptogramin B; QAC, quaternary ammonium compound; ORF, open reading frame; DNA rep. transp. and repair, DNA replication, transport, and repair. Means ± SD are shown.
High-throughput analysis of class 1 integron gene cassettes.
Class 1 integron gene cassettes were further analyzed using high-throughput sequencing to obtain a comprehensive overview of gene cassette composition in WWTPs. A total of 11,664,734 clean reads were generated from 18 samples, ranging from 179,488 to 974,652 per sample (mean = 648,040). A total of 23,370 contigs were assembled, and 6,559 coding sequences (CDSs) (ranging from 164 to 763 for each sample) were inferred with diverse putative functions (Table S1). A total of 74.1% of these CDSs in all the samples were predicted to encode hypothetical proteins (Fig. 4d). A total of 25.7% of the CDSs (528 unique gene cassettes) encoded functionally characterized proteins, which were involved in biosynthesis, transcriptional regulation, heavy metal resistance, antibiotic resistance, transport, toxin/antitoxin systems, and DNA replication, transport, and repair or were membrane proteins (Fig. S2). Among the CDSs encoding functional proteins, genes related to antibiotic resistance (14.6%) and biosynthesis (5.1%) were the predominant gene cassette-associated genes in WWTPs (Fig. 4c; see also Table S4). Genes (yoeB and yefM) encoding toxin-antitoxin (TA) systems were found in almost all samples.
A total of 37 unique ARG cassettes were detected during the wastewater treatment, including genes encoding resistance to aminoglycosides, beta-lactams, bicyclomycin, chloramphenicol, fosfomycin, gentamicin, macrolides, multiple drugs, trimethoprim, and QACs (Table S5). Among them, trimethoprim resistance genes were most frequently observed on gene cassettes, accounting for 44.5% of all ARG cassettes, followed by aminoglycoside resistance genes (21.5%), beta-lactamase genes (18.2%), and multidrug resistance genes (5.9%). aacA4 and ant1 (aminoglycoside resistance), dfrA and dhfrI (trimethoprim resistance), blaOXA-2 (beta-lactamase), cat_1 (chloramphenicol resistance), and emrE (multidrug resistance) gene cassettes were found in all 18 samples. In addition, heavy metal resistance gene cassettes were also detected, including genes conferring resistance to As (arsA, arsC, and arsD), Cu (copA), Hg (merR and merC), and Co-Zn-Cd (czcA); the arsD gene cassette (encoding arsenic resistance operon trans-acting repressor) was the most frequently observed in WWTPs (Table S5).
Characterization of class 3 integron gene cassettes.
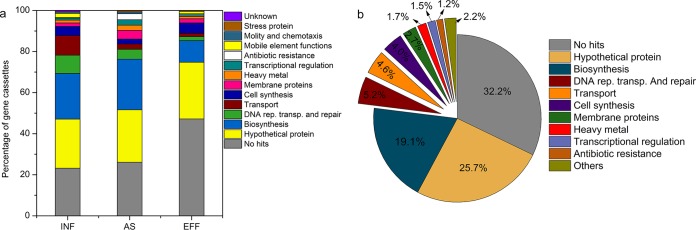
Class 3 integron gene cassette contents were characterized by clone library analysis, and 100 clones for each sample were randomly picked for sequencing analysis. We found that 44% of clones carried ≥1 gene cassette in WWTPs (55.2% in influents, 48.3% in activated sludge, and 28.5% in effluents). A total of 162 different gene cassettes were found, and most of them were first reported for class 3 integrons (Table S6). Among them, the majority of gene cassettes encoded biosynthesis-associated proteins (19.1%), followed by DNA replication, transport, and repair proteins (5.2%) and transport proteins (4.6%) (Fig. 5a and b). These gene cassettes, including genes encoding ABC transporter ATP-binding protein, PilZ domain-containing protein, zinc ribbon domain-containing protein, saccharopine dehydrogenase, and potassium transporter TrkA, were most frequently found in WWTPs. Antibiotic and heavy metal resistance gene cassettes were also detected, accounting for 1.2% and 1.7% of all gene cassettes, respectively. A total of 8 ARG cassettes were detected, and most of them were found in activated sludge, including the genes encoding resistance to beta-lactams (2 GcuF/OXA-28 fusion protein and OXA-10 family class D beta-lactamase), acriflavin (acriflavin resistance protein), aminoglycoside (aminoglycoside adenyltransferase), and multiple drugs (cation/multidrug efflux pump). Heavy metal gene cassettes mainly contained Ca-, Co-, and Ni/Fe-associated functional genes (Table S6).
FIG 5.
Percentage of class 3 gene cassettes during wastewater treatment. The bar chart (a) shows the variations in abundance of gene cassettes during wastewater treatment, and the pie chart (b) reveals the percentages of all gene cassettes from 18 WWTP samples.
Diversity of gene cassettes.
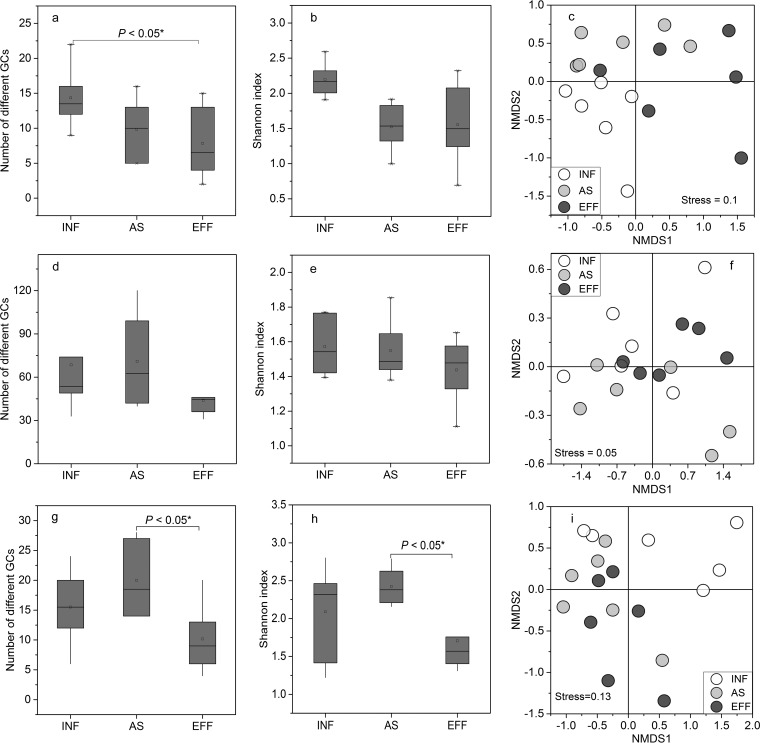
Significant differences in the detected numbers of integron gene cassettes were observed across the wastewater treatment process. The rarefaction curves based on the clone library were unsaturated, indicating that identified gene cassettes represented only a fraction of the total diversity of the gene cassette arrays at the sampling sites (Fig. S3). Wastewater treatment significantly reduced the diversity of class 1 integron gene cassettes (P < 0.05), and WWTP influents harbored the most diverse gene cassettes. Based on high-throughput analysis of class 1 integrons, there was no significant difference in the alpha-diversities across the wastewater treatment process (Fig. 6a, b, d, and e). For class 3 integrons, wastewater treatment had no impact on gene cassette diversity (P > 0.05), while the highest gene cassette number was observed in activated sludge (P < 0.05) (Fig. 6g and h). Nonmetric multidimensional scaling (NMDS) analysis revealed that the gene cassette structure of neither class 1 nor class 3 showed significant clustering with different wastewater treatment processes (Fig. 6c, f, and i).
FIG 6.
Alpha-diversity and beta-diversity of class 1 integron gene cassette (a to c, based on clone library data of class 1 integron gene cassettes; d to f, based on high-throughput sequencing data of class 1 integron gene cassettes) and class 3 integron gene cassette (g to i, based on class 3 integron gene cassettes derived from a clone library) profiles across wastewater treatment plants. Means ± SDs are shown. ANOVA was performed. *, P < 0.05; **, P < 0.01.
DISCUSSION
WWTPs serve as a rich repository of ARGs and a hot spot for the release of antibiotic resistance determinants into the environment (27, 28). However, the impact of wastewater treatment on integrons and gene cassettes is not well addressed. Hence, this study focused on the dynamics of integrons and diversity of class 1 and class 3 integron gene cassettes during wastewater treatment.
As stated above, WWTPs contain massive amounts of integrons with total concentrations of 107 to 1010 copies/ml, similar to those reported from other wastewater environments (26, 29, 30). The highest concentrations of integrons were found in activated sludge, whereas the highest normalized copy number of integrons (except for class 3 integrons) was detected in influents, highlighting that anthropogenic activities are a major source of integrons in WWTPs since most urban discharges possess an abundance of antibiotic-resistant bacteria and genes (26, 31, 32). The differences in microbial biomass might be mainly responsible for the higher abundance of integrons in sludge. Samples from different treatment stages harbored distinct microbes with varied profiles of integron gene cassettes (24), and the bacteria that proliferate in activated sludge do not potentially harbor integrons to the same extent as influent microbiota (30, 33). The concentration of integrons in effluents was 1 to 2 orders of magnitude lower than that in influents, suggesting that the wastewater treatment process could effectively reduce the concentration of integrons. However, integrons persisted with substantial copy numbers (105 to 106 copies/ml) and considerable diversity in effluents, manifesting the potential risks caused by integron-mediated dissemination of ARGs into downstream environments (25).
Class 1 integrons were the most prevalent among the three classes of integrons in this study, and this observation was consistent with previously reported studies (26, 29). The class 1 integron-integrase gene was considered a proxy for anthropogenic pollution (17). Thus, the high concentrations of class 1 integrons detected in this study further support the view that human activities contribute significantly to the prevalence of class 1 integrons (26, 31). Compared with those of class 1 integrons, the concentrations of class 2 and 3 integrons were fairly low. Class 1 integron gene cassettes were found to carry more antibiotic resistance genes than class 3 integron gene cassettes, suggesting that class 1 integrons might be the main mobile elements responsible for spread of integron-mediated antibiotic resistance, probably due to the active characteristic of its integrase gene (6).
Class 1 integron gene cassettes and gene cassette arrays were investigated by clone library analysis and high-throughput sequencing. Both approaches have their advantages and limitations. High-throughput sequencing can provide a high-throughput pipeline for tracking gene cassettes in integrons (24) and cover the whole picture of gene cassette distribution owing to greater sequencing depth while avoiding the bias caused by cloning. Clone library analysis may overcome the drawbacks of high-throughput sequencing caused by assembling bias of short reads derived from amplified sequences with high similarity. Despite this, the sampling depth of integron gene cassette clones by Sanger sequencing is likely unable to provide comprehensive profiles of integron gene cassette contents. Additionally, the integron-related database INTEGRALL was applied to annotate integron gene cassettes from a clone library, whereas it was not feasible to analyze short reads from data set in high-throughput sequencing (25). The sequencing depth, cloning bias, and different reference databases could explain the discrepancy between the clone library and the high-throughput sequencing data for the integron gene cassettes. For instance, our clone library analysis showed that aminoglycoside and beta-lactam resistance genes were the most frequently observed resistance gene cassettes. High-throughput sequencing data revealed that the trimethoprim resistance gene cassettes were most prevalent in class 1 integrons, but aminoglycoside and beta-lactam resistance gene cassettes also accounted for a considerable proportion of gene cassettes in class 1 integrons, a finding consistent with a previous metagenomic study (25). Based on the clone library analysis, we could estimate the abundance of these ARGs harboring integrons by multiplying the concentration of class 1 integrons by the corresponding percentage of the resistance gene cassettes. We estimated that the abundance of aminoglycoside resistance gene cassettes harboring integrons ranged from 5.1 × 105 copies/ml to 1.9 × 107 copies/ml and the abundance of beta-lactam resistance gene cassettes harboring integrons was from 7.5 × 104 copies/ml to 2.1 × 106 copies/ml. However, it was difficult to estimate the abundance of these resistance gene cassettes from our high-throughput sequencing data, since class 1 integrons can carry more than one type of antibiotic resistance gene cassette.
Several shared antibiotic resistance gene cassettes were persistent during wastewater treatment (Fig. 4). These abundant and persistent ARG-harboring gene cassettes should have priority over other gene cassettes when evaluating the human health risk of ARGs, because they are more likely to be transferred from environmental bacteria to human pathogens via integron-associated HGT events (2, 34). Anthropogenic antibiotic usage (e.g., regular use of therapeutic combination of trimethoprim and sulfamethoxazole) could trigger a bacterial SOS response and exert selection pressure on bacteria carrying integrons with cassettes, which might partially explain the high frequency of these genes in integron gene cassettes (26, 35–37). In addition, it was recently observed that the low fitness cost of these resistance genes on class 1 integrons could favor their maintenance and prevalence in cassette networks (38).
High-throughput sequencing analysis has reinforced the fact that up to 74% of cassettes and their encoded polypeptides had no known homologues in protein databases or exhibited homology to conserved hypothetical proteins. Only about 30% of gene cassettes could be functionally characterized, and most of these functional proteins were associated with bacterial adaptation to stressful environments (34, 39, 40). For example, toxin-antitoxin (TA) systems (e.g., yoeB-yefM), which are a common feature of cassette arrays and responsible for maintaining array stability (6), were frequently found across wastewater treatment plants in this study. Previous studies showed that TA systems had an important role in bacterial stress physiology and might form the basis of multidrug resistance (41, 42). Only a few qac gene cassettes (qacE2, qacG, and qacH) were found (mainly in influents) based on a clone library, which was consistent with a previously reported study (26), since in class 1 integrons with a 3′ CS, this qacE gene has undergone a deletion (5, 43).
Class 3 integron gene cassettes were scarcely involved in previous studies. Our results provide the exploratory investigation of class 3 gene cassettes in WWTPs. Although class 3 integrons were significantly less abundant than class 1 integrons, a great diversity and a high percentage of detected gene cassettes carried by class 3 integrons were obviously different from the case with class 1 integrons, indicating that class 3 integrons are continuously evolving, colonizing new species, and acquiring novel gene cassettes (44, 45). These findings increased awareness of class 3 gene cassettes, which were previously reported to be less active than those of the other classes of integron (46). Class 3 integrons could harbor gene cassettes identical to those in bacteria carrying class 1 integrons (47); for example, gene cassettes encoding ABC transporter ATP-binding protein, saccharopine dehydrogenase, and antibiotic resistance (e.g., blaOXA-10 and blaOXA-28) were found in both class 1 and 3 integrons. It is likely that gene cassette rearrangements may have occurred between the two classes of integron (48), considering that class 3 integrons have an evolutionary history (Tn402 transposon) similar to that of the class 1 integrons (21). However, the proportion of identified ARGs in class 3 integrons was significantly lower than that in class 1 integrons. The genetic and evolutionary mechanisms and the ecological significance of this phenomenon remain unknown. Class 3 integron gene cassettes had higher diversity in activated sludge, in which most ARG cassettes were observed. These data suggested that activated sludge could provide a suitable condition for enhanced exchange and shuffling of antibiotic resistance gene cassettes in class 3 integrons.
Distinct clustering for profiles of either class 1 or class 3 integron gene cassettes was not observed among influent, activated sludge, and effluent samples, indicating that horizontal gene transfer could frequently occur between bacterial species residing in similar environments. Since horizontal gene transfers are the exchanges of genes, frequent HGT events could result in a high similarity of integron gene cassette contents among various samples. Thus, we could not observe distinct clustering of gene cassettes in WWTPs. Our observation further supports the finding that integrons could be clustered by environmental compartments, rather than by the identity of their host cells (6, 17). In addition, the sampling depth of the clone library might be insufficient to capture the variation of gene cassette contents in WWTPs, and thus, further studies of integron gene cassettes with deep sequencing are necessary for comprehensive understanding of HGT events. Wastewater treatment could significantly decrease the number and the diversity of class 1 and class 3 integron gene cassettes, highlighting the efficiency of wastewater treatment for the integron pool in WWTPs. The variation of bacterial communities could be one factor affecting the diversity of integron gene cassettes. This is because wastewater treatment could significantly influence the structures of microbial communities and reduce the microbial biomass (49), thus affecting the diversity of integron-carrying bacteria. Additionally, the removal of organic matters, solids, antibiotics, or heavy metals in effluents (50) may limit the proliferation of bacteria and their access to the vast genetic diversity of gene cassettes. A higher abundance and diversity of class 1 gene cassettes were observed in influents, which was unsurprising since influents contained a mixture of the class 1 integrons from various sources, such as human feces, hospital effluents, and livestock wastewater (51). Several novel gene cassette arrays were detected in this study (e.g., aacA4-ereA1, orf-aadA2-blaOXA-129-linF, and catB8-blaOXA-1-aadA1), implying that gene cassettes and gene cassette arrays of integron pools remained largely unexplored and that the acquisition and exchange of class 1 integron gene cassettes stay fairly active in wastewater treatment systems (34). These results highlight the need to comprehensively investigate and constantly monitor integron gene cassette contents for assessment of integrin-mediated ARG transfer in the environment.
MATERIALS AND METHODS
Sampling and DNA extraction.
Samples, including influent (INF), activated sludge (AS), and effluent (EFF) samples, were collected from three urban WWTPs located in the cities of Xiamen (XM) and Longyan (LY), China (see Table S1), in August 2014. Two replicate samples of each treatment stage were collected on two successive days without recent rainfall, and the effects of hydraulic retention time were considered when collecting the effluent samples. All samples were stored at 4°C within less than 6 h before DNA extraction. To collect the bacterial pellets, 200 ml of each influent was centrifuged at 10,000 × g and 4°C for 20 min and 400 ml of each effluent was filtered through a 0.22-μm cellulose nitrate membrane. Sludge (2 ml) was pelleted by centrifugation at 10,000 × g and 4°C for 20 min. Genomic DNA was extracted using a FastDNA spin kit for soil (MP Biomedicals, USA) according to the manufacturer's instructions. DNA quality and quantity were analyzed using a NanoDrop spectrophotometer (ND-1000).
qPCR.
Quantitative PCR (qPCR) analyses (copies) of 16S rRNA gene, class 1, class 2, and class 3 integron-integrase genes, aadA1, aadA2, and aadA5, were performed using a SYBR green-based approach (TaKaRa, Japan) on a Roche 480 (Roche Inc., USA) (Table 2). The amplification conditions were as reported previously (28, 52). To generate standard curves for qPCR, PCR amplicons were purified with a Wizard SV gel and PCR cleanup system (Promega, USA), ligated into pMD19-T vector (TaKaRa, Japan), and transferred into Escherichia coli DH5α (TaKaRa) by following the manufacturer's protocol. To confirm the identities of genes contained in each plasmid, ligated PCR products were sequenced and subjected to BLAST searching against the GenBank database using BLASTX. Serial 10-fold dilutions of plasmids harboring these gene fragments were used as templates to construct standard curves. qPCR assays of each sample were run in triplicate. Melting-curve analysis was used to check the specificity of PCR products. Based on the slope of the standard curve, the amplification efficiency, expressed as a percentage, was calculated using the formula E = (10−1/slope − 1) × 100. The relative abundance of integrase genes (copies/copy of the 16S rRNA gene) was calculated by normalization to bacterial 16S rRNA gene copy numbers. The average 16S rRNA gene copy numbers of bacteria were estimated at 4.1 based on the rRNA operon copy number database (rrnDB version 4.4.4) (53). Thus, the bacterial cell numbers were calculated by dividing the copy number of the 16S rRNA gene by 4.1, and the average copies of integrons/cell could be estimated.
TABLE 2.
Primers used in PCR and qPCR of class 1, 2, and 3 integrons
| Target gene | Primer name | Primer sequence (5′–3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | 515 F | GTGCCAGCMGCCGCGG | 410 | 59 |
| 907 R | CCGTCAATTCMTTTRAGTTT | |||
| intI1 | intI1-LC1 | GCCTTGATGTTACCCGAGAG | 196 | 45 |
| intI1-LC5 | GATCGGTCGAATGCGTGT | |||
| intI2 | intI2-LC2 | TGCTTTTCCCACCCTTACC | 195 | 45 |
| intI2-LC3 | GACGGCTACCCTCTGTTATCTC | |||
| intI3 | intI3-LC1 | GCCACCACTTGTTTGAGGA | 138 | 45 |
| intI3-LC2 | GGATGTCTGTGCCTGCTTG | |||
| 5′ conserved segment of class 2 integrons | int2S | ACCTTTTTGTCGCATATCCGTG | 60 | |
| 3′ conserved segment of class 2 integrons | intCS2 | TACCTGTTCTGCCCGTATCT | ||
| intI3 gene cassette | attI3L | GGTATCCGGTGTTTGGTCAG | 45 | |
| intI3 gene cassette | class3R | CGTCAAACGGGTAAGCAGT | ||
| 5′ conserved segment of class 1 integrons | 5′CS | GGCATCCAAGCAGCAAG | 61 | |
| 3′ conserved segment of class 1 integrons | 3′CS | AAGCAGACTTGACCTGA | ||
| aadA2 | aadA2 R | ACGGCTCCGCAGTGGAT | 62 | |
| aadA2 F | GGCCACAGTAACCAACAAATCA | |||
| aadA5 | aadA5 R | ATCACGATCTTGCGATTTTGCT | 62 | |
| aadA5 F | CTGCGGATGGGCCTAGAAG | |||
| aadA1 | aadA1 R | AGCTAAGCGCGAACTGCAAT | 62 | |
| aadA1 F | TGGCTCGAAGATACCTGCAA |
Construction of integron gene cassette libraries.
For class 1, class 2, and class 3 integrons, the variable gene cassette-containing regions were amplified with three reported primer sets complementary to conserved segments, respectively (Table 2). The PCR products of the variable regions of class 2 integrons could not be found using the current primer pairs for further clone library analysis in this study. Amplification of the variable regions of class 1 and class 3 integrons was performed with a 25-μl reaction mixture consisting of 1 U of premix Ex Taq polymerase (TaKaRa, Japan), a 10 nM (nmol/m3) concentration of each primer, 0.5 U of bovine serum albumin (BSA), and 20 ng of DNA under the following thermal conditions: 95°C for 10 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and then 72°C for 2 min 30 s, with a final extension at 72°C for 10 min. To create clone libraries, PCR fragments were cleaned up, ligated to the vector, and then transferred to component cells as previously described by Stalder et al. in 2014 (26). For each sample, at least 138 clones and 100 clones were randomly picked for class 1 and class 3 integrons, respectively (Tables S2 and S6). Random colonies were picked into a 25-μl reaction mixture for PCR amplification using the same PCR programs to validate the clone with at least one gene cassette (>153 bp for class 1 integrons and >318 bp for class 3 integrons). Validated clones with gene cassettes were sequenced with M13 primers.
Analysis of clone libraries.
Potential class 1 integrons attC sites were manually searched after trimming and assembly of the sequences. Assembled sequences were interrogated by searching against the INTEGRALL integron database (http://integrall.bio.ua.pt/?search#) using the BLAST algorithm (54), and sequence homology was checked with previously reported gene cassettes. Putative ORFs were identified using NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) when homologies of the sequence were not detected in the integron database. If a potential ORF was found between two putative attC sites, the ORF was considered to be a potential gene cassette. A “variant” of a known gene cassette was defined when the sequence showed an identity of 90 to 99% with a reference gene.
Class 3 integron sequences were assembled and primers were removed, and then the resulting sequences were subjected to automated annotation by searching against the NCBI nonredundant protein database using BLASTX with a threshold E value of 1e−5. A sequence was annotated as a potential gene with a loose cutoff value of sequence identity of ≥35% and an alignment length corresponding to ≥30 amino acids (12).
High-throughput sequencing of gene cassettes.
Amplification and purification of class 1 integron gene cassettes were performed as mentioned before. PCR products were assessed by electrophoresis on 1.2% (mass/vol) agarose gels. For each sample, 4 μg of purified PCR amplicons was used for DNA 500-bp shotgun library construction, followed by Miseq PE300 sequencing (Anoroad Genome, China). After filtering and removal of ambiguities, amplicon reads were assembled using IDBA (version 1.1.1) with default parameters (55). Contigs were subjected to CDS prediction using PROKKA (56) at an E value of ≤1e−5. CDSs were searched by using BLASTX, classified into gene subtypes with an E value of 1e−5, and then imported into best-hit tables to assess relative abundance and diversity. Gene subtype abundance was normalized by multiplying BLASTX hit numbers by a coefficient which was calculated by dividing the total number of reads for each sample by the average number of reads for all samples (24).
Statistical analysis.
Averages and standard deviations were calculated using Excel 2010 (Microsoft, USA). Nonmetric multidimensional scaling (NMDS), Adonis test, and cluster analysis were performed in R 3.2.3 (R Foundation for Statistical Computing, Vienna, Austria) with vegan 2.0-10 (57). One-way analysis of variance (ANOVA) and significance testing were performed using SPSS v20.0 (IBM, USA), with differences considered significant at a P value of <0.05. Gephi 0.9.1 software was applied for network visualization (58).
Accession number(s).
All clean reads retrieved from high-throughput sequencing analysis and all clone assembly sequences were deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject accession numbers PRJNA420645 and SRP136070.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the Natural Science Foundation of China (31722004), the National Key Research and Development Program of China-International collaborative project from Ministry of Science and Technology (2017YFE0107300), the Knowledge Innovation Program of the Chinese Academy of Sciences (IUEQN201504), K. C. Wong Education Foundation, and Youth Innovation Promotion Association, CAS.
We declare that there are no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02766-17.
REFERENCES
- 1.Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10(Suppl 1):S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 2.Rossolini GM, D'Andrea MM, Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect 14(Suppl 1):33–41. doi: 10.1111/j.1469-0691.2007.01867.x. [DOI] [PubMed] [Google Scholar]
- 3.Cambray G, Guerout AM, Mazel D. 2010. Integrons. Annu Rev Genet 44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 4.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian Gb Dong BL, Huang XH, Yu LF, Gu DX, Ren HW, Chen XJ, Lv LC, He DD, Zhou HW, Liang ZS, Shen JZ. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 5.Gaze WH, Zhang LH, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, Brown H, Davis S, Kay P, Boxall ABA, Wellington EMH. 2011. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 5:1253–1261. doi: 10.1038/ismej.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillings MR. 2014. Integrons: past, present, and future. Microbiol Mol Biol Rev 78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stalder T, Barraud O, Casellas M, Dagot C, Ploy MC. 2012. Integron involvement in environmental spread of antibiotic resistance. Front Microbiol 3:119. doi: 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Recchia GD, Hall RM. 1995. Gene cassettes, a new mobile element. Microbiology 141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 9.Stokes HW, Nesbø CL, Holley M, Bahl MI, Gillings MR, Boucher Y. 2006. Class 1 integrons predating the association with Tn402-like transposition genes are present in a sediment microbial community. J Bacteriol 188:5722–5730. doi: 10.1128/JB.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu YG, Gillings M, Simonet P, Stekel D, Banwart S, Penuelas J. 20 December 2017. Human dissemination of genes and microorganisms in Earth's Critical Zone. Glob Chang Biol doi: 10.1111/gcb.14003. [DOI] [PubMed] [Google Scholar]
- 11.Rosewarne CP, Pettigrove V, Stokes HW, Parsons YM. 2010. Class 1 integrons in benthic bacterial communities: abundance, association with Tn402-like transposition modules and evidence for coselection with heavy-metal resistance. FEMS Microbiol Ecol 72:35–46. doi: 10.1111/j.1574-6941.2009.00823.x. [DOI] [PubMed] [Google Scholar]
- 12.Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegård B, Söderström H, Larsson DGJ. 2011. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One 6(2):e17038. doi: 10.1371/journal.pone.0017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillings MR. 2013. Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Front Microbiol 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev 33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Byelashov OA, Geornaras I, Goodridge LD, Nightingale KK, Belk KE, Smith GC, Sofos JN. 2010. Characterization and transferability of class 1 integrons in commensal bacteria isolated from farm and nonfarm environments. Foodborne Pathog Dis 7:1441–1451. doi: 10.1089/fpd.2010.0555. [DOI] [PubMed] [Google Scholar]
- 16.Zhu YG, Gillings M, Simonet P, Stekel D, Banwart S, Penuelas J. 2017. Microbial mass movements. Science 357:1099–1100. doi: 10.1126/science.aao3007. [DOI] [PubMed] [Google Scholar]
- 17.Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu YG. 2015. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J 9:1269–1279. doi: 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillings MR, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, Stokes HW. 2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol 190:5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazel D. 2006. Integrons: agents of bacterial evolution. Nat Rev Microbiol 4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 20.White PA, McIver CJ, Rawlinson WD. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother 45:2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collis CM, Kim MJ, Stokes HW, Hall RM. 2002. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol Microbiol 46:1415–1427. doi: 10.1046/j.1365-2958.2002.03260.x. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh S, Ramsden SJ, LaPara TM. 2009. The role of anaerobic digestion in controlling the release of tetracycline resistance genes and class 1 integrons from municipal wastewater treatment plants. Appl Microbiol Biotechnol 84:791–796. doi: 10.1007/s00253-009-2125-2. [DOI] [PubMed] [Google Scholar]
- 23.Ma LP, Zhang XX, Zhao FZ, Wu B, Cheng SP, Yang LY. 2013. Sewage treatment plant serves as a hot-spot reservoir of integrons and gene cassettes. J Environ Biol 34:391–399. [PubMed] [Google Scholar]
- 24.Gatica J, Tripathi V, Green S, Manaia CM, Berendonk T, Cacace D, Merlin C, Kreuzinger N, Schwartz T, Fatta-Kassinos D, Rizzo L, Schwermer CU, Garelick H, Jurkevitch E, Cytryn E. 2016. High throughput analysis of integron gene cassettes in wastewater environments. Environ Sci Technol 50:11825–11836. doi: 10.1021/acs.est.6b03188. [DOI] [PubMed] [Google Scholar]
- 25.Ma LP, Li AD, Yin XL, Zhang T. 2017. The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environ Sci Technol 51:5721–5728. doi: 10.1021/acs.est.6b05887. [DOI] [PubMed] [Google Scholar]
- 26.Stalder T, Barraud O, Jove T, Casellas M, Gaschet M, Dagot C, Ploy MC. 2014. Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. ISME J 8:768–777. doi: 10.1038/ismej.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaPara TM, Burch TR, McNamar PJ, Tan DT, Yan M, Eichmiller JJ. 2011. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ Sci Technol 45:9543–9549. doi: 10.1021/es202775r. [DOI] [PubMed] [Google Scholar]
- 28.Chen QL, An XL, Li H, Su JQ, Ma YB, Zhu YG. 2016. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ Int 92:1–10. doi: 10.1016/j.envint.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XX, Zhang T, Zhang M, Fang HHP, Cheng SP. 2009. Characterization and quantification of class 1 integrons and associated gene cassettes in sewage treatment plants. Appl Microbiol Biotechnol 82:1169–1177. doi: 10.1007/s00253-009-1886-y. [DOI] [PubMed] [Google Scholar]
- 30.Ma LP, Zhang XX, Cheng SP, Zhang ZY, Shi P, Liu B, Wu B, Zhang Y. 2011. Occurrence, abundance and elimination of class 1 integrons in one municipal sewage treatment plant. Ecotoxicology 20:968–973. doi: 10.1007/s10646-011-0652-y. [DOI] [PubMed] [Google Scholar]
- 31.Wright MS, Baker-Austin C, Lindell AH, Stepanauskas R, Stokes HW, McArthur JV. 2008. Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J 2:417–428. doi: 10.1038/ismej.2008.8. [DOI] [PubMed] [Google Scholar]
- 32.Su JQ, An XL, Li B, Chen QL, Gillings MR, Chen H, Zhang T, Zhu YG. 2017. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 5:84. doi: 10.1186/s40168-017-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Li B, Zou SC, Fang HHP, Zhang T. 2014. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res 62:97–106. doi: 10.1016/j.watres.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Gillings MR. 2017. Lateral gene transfer, bacterial genome evolution, and the Anthropocene: lateral gene transfer in the Anthropocene. Ann N Y Acad Sci 1389:20–36. doi: 10.1111/nyas.13213. [DOI] [PubMed] [Google Scholar]
- 35.Guerin É, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbé J, Ploy MC, Mazel D. 2009. The SOS response controls integron recombination. Science 324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 36.Hocquet D, Llanes C, Thouverez M, Kulasekara HD, Bertrand X, Plésiat P, Mazel D, Miller SI. 2012. Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog 8(6):e1002778. doi: 10.1371/journal.ppat.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barraud O, Ploy MC. 2015. Diversity of class 1 integron gene cassette rearrangements selected under antibiotic pressure. J Bacteriol 197:2171–2178. doi: 10.1128/JB.02455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacotte Y, Ploy MC, Raherison S. 2017. Class 1 integrons are low-cost structures in Escherichia coli. ISME J 11:1535–1544. doi: 10.1038/ismej.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koenig JE, Boucher Y, Charlebois RL, Nesbø C, Zhaxybayeva O, Bapteste E, Spencer M, Joss MJ, Stokes HW, Doolittle WF. 2008. Integron-associated gene cassettes in Halifax Harbour: assessment of a mobile gene pool in marine sediments. Environ Microbiol 10:1024–1038. doi: 10.1111/j.1462-2920.2007.01524.x. [DOI] [PubMed] [Google Scholar]
- 40.Rapa RA, Labbate M. 2013. The function of integron-associated gene cassettes in Vibrio species: the tip of the iceberg. Front Microbiol 4:385. doi: 10.3389/fmicb.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buts L, Lah J, Minh-Hoa DT, Lode W, Remy L. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci 30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Lewis K. 2005. Persister cells and the riddle of biofilm survival. Biochemistry 70:267–274. doi: 10.1007/s10541-005-0111-6. [DOI] [PubMed] [Google Scholar]
- 43.Stokes HW, Gillings MR. 2011. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev 35:790–819. doi: 10.1111/j.1574-6976.2011.00273.x. [DOI] [PubMed] [Google Scholar]
- 44.Correia M, Boavida F, Grosso F, Salgado M, Lito L, Cristino JM, Mendo S, Duarte A. 2003. Molecular characterization of a new class 3 integron in Klebsiella pneumoniae. Antimicrob Agents Chemother 47:2838–2843. doi: 10.1128/AAC.47.9.2838-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barraud O, Casellas M, Dagot C, Ploy MC. 2013. An antibiotic-resistant class 3 integron in an Enterobacter cloacae isolate from hospital effluent. Clin Microbiol Infect 19:E306–E308. doi: 10.1111/1469-0691.12186. [DOI] [PubMed] [Google Scholar]
- 46.Collis CM, Kim MJ, Stokes HW, Hall RM. 1998. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol Microbiol 29:477–490. doi: 10.1046/j.1365-2958.1998.00936.x. [DOI] [PubMed] [Google Scholar]
- 47.Sajjad A, Holley MP, Labbate M, Stokes HW, Gillings MR. 2011. Preclinical class 1 integron with a complete Tn402-like transposition module. Appl Environ Microbiol 77:335–337. doi: 10.1128/AEM.02142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simo Tchuinte PL, Stalder T, Venditti S, Ngandjio A, Dagot C, Ploy MC, Barraud O. 2016. Characterisation of class 3 integrons with oxacillinase gene cassettes in hospital sewage and sludge samples from France and Luxembourg. Int J Antimicrob Agents 48:431–434. doi: 10.1016/j.ijantimicag.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 49.Hu M, Wang X, Wen X, Xia Y. 2012. Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour Technol 117:72–79. doi: 10.1016/j.biortech.2012.04.061. [DOI] [PubMed] [Google Scholar]
- 50.Guardabassi L, Wong DMLF, Dalsgaard A. 2002. The effects of tertiary wastewater treatment on the prevalence of antimicrobial resistant bacteria. Water Res 36:1955–1964. doi: 10.1016/S0043-1354(01)00429-8. [DOI] [PubMed] [Google Scholar]
- 51.Kang HY, Jeong YS, Oh JY, Tae SH, Choi CH, Moon DC, Lee WK, Lee YC, Seol SY, Cho DT, Lee JC. 2005. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother 55:639–644. doi: 10.1093/jac/dki076. [DOI] [PubMed] [Google Scholar]
- 52.Uyaguari MI, Scott GI, Norman RS. 2013. Abundance of class 1-3 integrons in South Carolina estuarine ecosystems under high and low levels of anthropogenic influence. Mar Pollut Bull 76:77–84. doi: 10.1016/j.marpolbul.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 53.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res 29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moura A, Soares M, Pereira C, Leitão N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 55.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2010. IDBA—a practical iterative de Bruijn graph De Novo assembler. Res Comput Mol Biol 6044:426–440. [Google Scholar]
- 56.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 57.Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens MHH. 2007. The vegan package. Community ecology package. http://ftp.uni-bayreuth.de/math/statlib/R/CRAN/doc/packages/vegan.pdf.
- 58.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. In Proceedings of the 3rd International ICWSM Conference; AAAI Press, Menlo Park, CA. [Google Scholar]
- 59.Stubner S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen™ detection. J Microbiol Methods 50:155–164. doi: 10.1016/S0167-7012(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 60.Ploy MC, Denis F, Courvalin P, Lambert T. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob Agents Chemother 44:2684–2688. doi: 10.1128/AAC.44.10.2684-2688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lévesque C, Piche L, Larose C, Roy PH. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 39:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A 110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu X, Kong F, Cheng X, Yan B, Du X, Gai J, Ai H, Shi L, Iredell J. 2008. Integron gene cassettes in Acinetobacter spp. strains from South China. Int J Antimicrob Agents 32:441–445. doi: 10.1016/j.ijantimicag.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 64.Chang Y-C, Shih DY-C, Wang J-Y, Yang S-S. 2007. Molecular characterization of class 1 integrons and antimicrobial resistance in Aeromonas strains from foodborne outbreak-suspect samples and environmental sources in Taiwan. Diagn Microbiol Infect Dis 59:191–197. doi: 10.1016/j.diagmicrobio.2007.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.