ABSTRACT
Despite the benefits to the global food supply and agricultural economies, pesticides are believed to pose a threat to the health of both humans and wildlife. Chlorpyrifos (CP), a commonly used organophosphate insecticide, has poor target specificity and causes acute neurotoxicity in a wide range of species via the suppression of acetylcholinesterase. This effect is exacerbated 10- to 100-fold by chlorpyrifos oxon (CPO), a principal metabolite of CP. Since many animal-associated symbiont microorganisms are known to hydrolyze CP into CPO, we used a Drosophila melanogaster insect model to investigate the hypothesis that indigenous and probiotic bacteria could affect CP metabolism and toxicity. Antibiotic-treated and germfree D. melanogaster insects lived significantly longer than their conventionally reared counterparts when exposed to 10 μM CP. Drosophila melanogaster gut-derived Lactobacillus plantarum, but not Acetobacter indonesiensis, was shown to metabolize CP. Liquid chromatography tandem-mass spectrometry confirmed that the L. plantarum isolate preferentially metabolized CP into CPO when grown in CP-spiked culture medium. Further experiments showed that monoassociating germfree D. melanogaster with the L. plantarum isolate could reestablish a conventional-like sensitivity to CP. Interestingly, supplementation with the human probiotic Lactobacillus rhamnosus GG (a strain that binds but does not metabolize CP) significantly increased the survival of the CP-exposed germfree D. melanogaster. This suggests strain-specific differences in CP metabolism may exist among lactobacilli and emphasizes the need for further investigation. In summary, these results suggest that (i) CPO formation by the gut microbiota can have biologically relevant consequences for the host, and (ii) probiotic lactobacilli may be beneficial in reducing in vivo CP toxicity.
IMPORTANCE An understudied area of research is how the microbiota (microorganisms living in/on an animal) affects the metabolism and toxic outcomes of environmental pollutants such as pesticides. This study focused specifically on how the microbial biotransformation of chlorpyrifos (CP; a common organophosphate insecticide) affected host exposure and toxicity parameters in a Drosophila melanogaster insect model. Our results demonstrate that the biotransformation of CP by the gut microbiota had biologically relevant and toxic consequences on host health and that certain probiotic lactobacilli may be beneficial in reducing CP toxicity. Since inadvertent pesticide exposure is suspected to negatively impact the health of off-target species, these findings may provide useful information for wildlife conservation and environmental sustainability planning. Furthermore, the results highlight the need to consider microbiota composition differences between beneficial and pest insects in future insecticide designs. More broadly, this study supports the use of beneficial microorganisms to modulate the microbiota-mediated biotransformation of xenobiotics.
KEYWORDS: biopesticides, colony collapse disorder, detoxification, environmental toxins, honey bees, lactobacillus, microbiota, pesticides, probiotics, xenobiotics
INTRODUCTION
The agricultural industry relies on pesticides to maintain a high crop yield and economic feasibility. Consequently, persistent pesticide usage has led to the widespread contamination of the global food supply and natural environment. Synthetic organophosphates (OPs) account for ∼34% of worldwide insecticide sales and exhibit broad-spectrum activities toward a variety of insects (1). In particular, chlorpyrifos (O,O-diethyl O-3,5,6-trichloro-2-pyridyl [CP]) is an extensively used OP (2). Though banned from residential usage due to pervasive environmental toxicity, CP remains widely used commercially (3). Consequently, nontarget wildlife experience CP exposure through contaminated aquatic and terrestrial ecosystems (4–6). CP is structurally similar to other OPs and consists of three phosphoester linkages (often called phosphotriesters) that induce neurotoxicity through the inhibition of acetylcholinesterase (AchE) (7).
The major metabolites produced during CP metabolism are chlorpyrifos oxon (CPO) and 3,5,6-trichloro-2-pyridinol (TCP). CPO is the more toxic/potent metabolite, with a 10- to 100-fold greater inhibition of AchE than its parent compound (8). In contrast, the less toxic metabolite, TCP, is environmentally persistent and often refractory to microbial degradation (9, 10). TCP is the predominant metabolite formed in animals via cytochrome P450-mediated hydrolysis of CP (11). Microbial hydrolases appear more variable with regard to the end by-product formation, with a preference toward CPO production observed in many microorganisms (12–14). Numerous studies have explored the role of microbes for environmental bioremediation of CP (15), and some have looked at how CP alters the microbiotas (communities of microorganisms residing on/in multicellular organisms) of insects, rodents, and human models (16–19). However, there has been substantially less investigation into how the microbiota affects CP toxicity in vivo.
OP exposure is known to dysregulate insect immunity (a major regulator of the microbiota) (20) and alter the microbiota composition in rodents (18, 21, 22). Honey bees (Apis mellifera), which are integral to agricultural pollination (23), are experiencing drastic population declines in North America, Europe, and Asia, most likely due to the combination of habitat loss (24), infection (25), and pesticide exposure (26, 27). The effect of environmentally relevant OP exposure on acute honey bee mortality has been debated (28–31). However, there appears to be agreement that environmentally relevant OP exposure has the potential to chronically modulate honey bee immunity (30), impair learning (31), and reduce their life span (28, 29). There is also a major concern and lack of knowledge regarding the potential synergistic toxicity of OPs to honey bees in combination with other environmental toxins, such as neonicotinoid pesticides, fungicides, and pollutants (30, 32, 33). The microbiota composition of pest insects is variable but often dominated by Proteobacteria (34), which is in stark contrast to the Lactobacillus-dominant microbiota of honey bees (35). Interestingly, bacterial symbionts of the pest insects Bactrocera dorsalis (36) and Riptortus pedestris (37) have been shown to confer resistance to OP-induced toxicity, though less is known about these interactions in honey bees. Established axenic protocols can derive adult honey bees with microbial loads of less than 50,000 CFU via sterile handling techniques after larval emergence (38). However, the attainment of completely germfree adult honey bees is difficult due to the intricate developmental logistics (39–41), which makes mechanistic host-microbe associations challenging to investigate.
In this study, we used Drosophila melanogaster as a well-established insect model with established sensitivities to OP insecticides and a defined core microbiota dominated by Lactobacillus (which is a unique trait among both hymenopterans and dipterans) (35, 42–45). Importantly, this insect model can be derived germfree to demonstrate causal relationships between microbes and OP-induced insect toxicity. It was hypothesized that indigenous and probiotic lactobacilli affect CP metabolism and toxicity.
RESULTS
Indigenous gut bacteria of D. melanogaster increase CP toxicity.
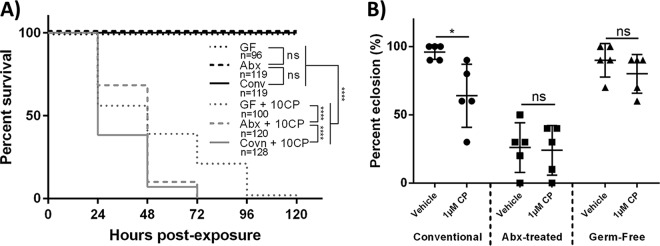
To determine how the D. melanogaster microbiota affects CP toxicity, survival with 10 μM CP (representing a minimal lethal dosage [17]) was compared between conventional, antibiotic (abx)-treated, and germfree wild-type (WT) Canton-S flies. abx-treated and germfree D. melanogaster flies had significantly increased overall survivals (log rank [Mantel-Cox], chi square = 34.2, df = 1, P < 0.0001 and chi square = 89.53, df = 1, P < 0.0001, respectively) and fewer early-time-point deaths (Gehan-Breslow-Wilcoxon test, chi square = 43, df = 1, P < 0.0001 and chi square = 66.19, df = 1, P < 0.0001, respectively) (Fig. 1A) than conventionally reared D. melanogaster flies. This suggested that the D. melanogaster microbiota could increase host toxicity to CP, likely due to altered CP metabolism.
FIG 1.
Indigenous gut bacteria of D. melanogaster increase CP toxicity. (A) Survival curves for newly eclosed conventionally reared, antibiotic-treated, and germfree flies that were exposed to a lethal concentration of CP (10 μM). Data are displayed from at least 3 independent experiments (consisting of 25 individuals each experiment). Statistical analyses shown are from log rank (Mantel-Cox) tests. (B) First-instar conventionally reared, antibiotic-treated, and germfree larvae were seeded on medium containing 1 μM CP (sublethal), and the percentages of larvae that subsequently eclosed were measured. Data are means ± standard deviations (two-way ANOVA) of results from 5 independent experiments (each dot represents 10 larvae). 10CP, 10 μM CP; *, P < 0.05; ****, P < 0.0001.
To evaluate the effects of the microbiota on CP toxicity during development, larval eclosion (emergence of adult flies from pupae) was compared between conventional, abx-treated, and germfree D. melanogaster larvae. Conventional larvae exposed to 1 μM CP exhibited a significantly reduced eclosion rate (two-way analysis of variance [ANOVA], P < 0.0463) compared to that of vehicle controls (Fig. 1B). Alternatively, significant differences in eclosion between vehicle- and 1 μM CP-exposed larvae that were germfree or treated with abx were not observed. However, abx-treated larvae had significantly reduced overall eclosion rates in both 1 μM CP-exposed and vehicle groups (two-way ANOVA, P < 0.0001 in both cases) compared to those of conventionally reared vehicle-treated and germfree larvae (Fig. 1B). The reduced eclosion rates in abx-treated larvae are believed to be attributed to antibiotic-mediated developmental toxicity (46). These results demonstrated that CP reduced eclosion rates in conventional but not abx-treated or germfree D. melanogaster larvae.
D. melanogaster-derived L. plantarum ISO is able to degrade CP but cannot utilize it as a carbon source for growth in the absence of glucose.
Since the D. melanogaster microbiota was shown to increase host toxicity to CP, we sought to evaluate whether this could be explained by microbe-mediated metabolic CP activation. Two cultured bacterial isolates from Canton-S fly stocks were sequenced to identify the predominant bacterial members in the D. melanogaster microbiota. One of the dominant bacterium-derived sequences best matched that of Acetobacter indonesiensis BCC15762 (accession number AB906398.1), with a maximum nucleotide alignment score of 2,464/2,464, an E value of 0.0, and 100% query coverage (referred to as A. indonesiensis ISO). The sequence of the other dominant bacterial member best matched that of Lactobacillus plantarum BC18 (accession number LC155900.1), with a maximum nucleotide alignment score of 2,436/2,436, an E value of 0.0, and 100% query coverage (referred to as L. plantarum ISO).
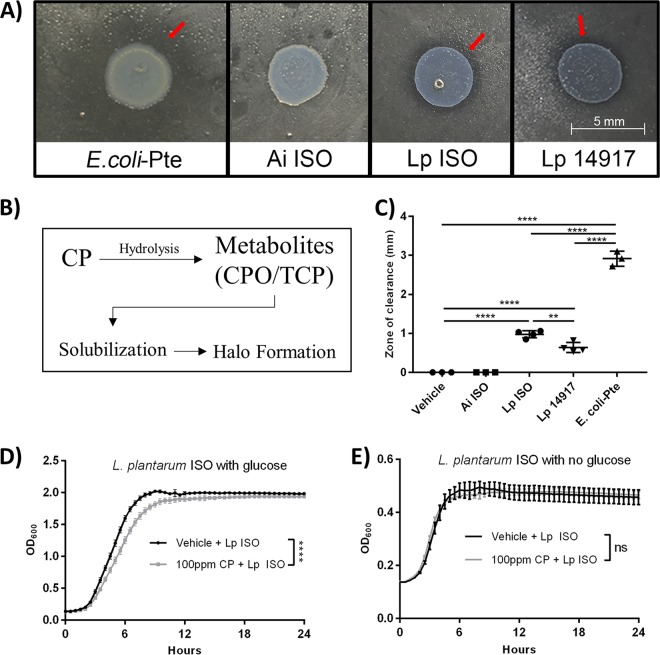
These findings are consistent with previous 16S rRNA-based microbiota study results demonstrating that Lactobacillus and Acetobacter were the dominant genera of the D. melanogaster microbiota (42). These isolates were subsequently tested for the ability to metabolize CP. It was observed that L. plantarum ISO was able to metabolize CP (Fig. 2A and C), as illustrated by a significantly larger zone of clearance in the CP hydrolysis assay compared to those of Acetobacter indonesiensis ISO and the vehicle (one-way ANOVA, P < 0.0001 in both cases). Additionally, Escherichia coli(pET20b-Pte) (positive control) and L. plantarum ATCC 14917 were both shown to have significantly larger zones of clearance (one-way ANOVA, P < 0.0001 in all cases) than A. indonesiensis and vehicle controls (Fig. 2C). These results indicated that L. plantarum ISO but not A. indonesiensis ISO could metabolize CP.
FIG 2.
D. melanogaster-derived L. plantarum (L. plantarum ISO) is able to degrade CP but cannot utilize it as a carbon source for growth in the absence of glucose. (A) Representative images showing results of the semiquantitative pesticide hydrolysis assay after 48-h aerobic (E. coli-Pte and Ai ISO) and anaerobic (L. plantarum Lp ISO and Lp 14917) incubations. Red arrows highlight halo formation. (B) Schematic diagram illustrating the semiquantitative pesticide hydrolysis assay. (C) Radii of halo formations were quantified following 48-h incubations. Data are presented as means ± standard deviations (one-way ANOVA) of results from at least 3 independent experiments performed in triplicates. Representative growth curves of L. plantarum ISO under glucose-rich (D) and glucose-limiting (E) conditions. Data are presented as means ± standard errors (unpaired, two-tailed t tests) from 3 independent experiments with triplicate technical replicates. Ai, A. indonesiensis; Lp, L. plantarum; **, P < 0.01; ****, P < 0.0001.
To determine the effects of CP on bacterial growth, L. plantarum ISO cultures were grown with 285 μM CP or vehicle, with or without dextrose as a carbon source. It was determined that 285 μM CP significantly reduced the L. plantarum ISO maximum growth rate in de Man, Rogosa, and Sharpe (MRS) medium during log phase compared to that of the vehicle controls (unpaired, two-tailed t test, t = 8.562, df = 6, P < 0.0001) (Fig. 2D). When grown in carbon-limited MRS medium, L. plantarum ISO showed no differences in maximum growth rate during log phase between 285 μM CP- and vehicle-treated groups (unpaired, two-tailed t test, t = 0.9852, df = 6, P < 0.3626) (Fig. 2E). These findings suggest L. plantarum ISO cannot utilize CP as a viable carbon source for growth in the absence of glucose.
L. plantarum ISO preferentially metabolizes CP into a more toxic metabolite, CPO.
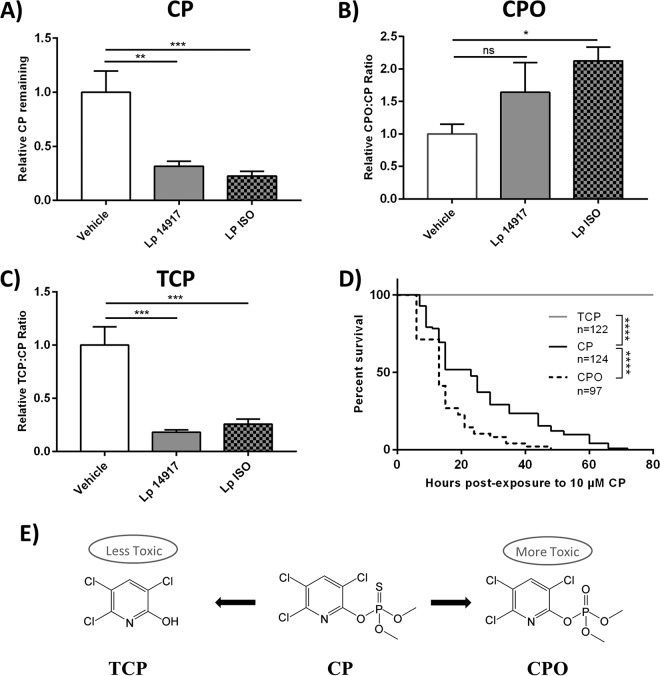
To further investigate CP metabolism by L. plantarum ISO, in vitro broth cultures containing 285 μM CP were analyzed for metabolite formation. The amounts of CP remaining after 24 h were significantly reduced in L. plantarum ISO and L. plantarum ATCC 14917 broth cultures relative to that in noninoculated vehicle controls spiked with CP (one-way ANOVA, P = 0.0005 and P = 0.0010, respectively) (Fig. 3A). The CP/CPO ratio after 24 h was significantly increased in L. plantarum ISO relative to that with the vehicle (Kruskal-Wallis test, P = 0.0338) (Fig. 3B). A trend toward an increased CP/CPO ratio was found in L. plantarum ATCC 14917 (Kruskal-Wallis test, P = 0.4081) (Fig. 3B). The TCP/CP ratios were significantly decreased in L. plantarum ISO and L. plantarum ATCC 14917 broth cultures relative to that with the vehicle (one-way ANOVA, P = 0.0003 and P = 0.0002, respectively) (Fig. 3C). This indicated that L. plantarum ISO preferentially produced CPO, rather than TCP, as its metabolic end product of CP metabolism.
FIG 3.
L. plantarum ISO preferentially metabolizes CP into a more toxic metabolite, CPO. (A to C) Relative CP (A), CPO (B), and TCP (C) remaining after exponential-phase growth of L. plantarum ISO and L. plantarum 14917 in MRS culture broth containing 285 μM CP. Data are presented as means ± standard deviations (one-way ANOVA [A and C] and Kruskal-Wallis [B]) from 3 independent experiments. (D) Survival curves of newly eclosed adult flies exposed to 10 μM CP, CPO, and TCP. Data displayed are from at least 3 independent experiments. Statistical analyses shown are from log rank (Mantel-Cox) tests. (E) Simplified schematic of CP and predominant metabolites. Lp, L. plantarum; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To demonstrate the differential toxicity profiles of CP, CPO, and TCP in vivo, conventional D. melanogaster flies were exposed to equimolar concentrations of each compound and monitored thereafter for survival. Newly eclosed D. melanogaster flies fed medium containing 10 μM CPO exhibited a significantly reduced overall survival (log rank [Mantel-Cox] test, chi square = 36.55, df = 1, P < 0.0001) and more early-time-point deaths (Gehan-Breslow-Wilcoxon test, chi square = 29.71, df = 1, P < 0.0001) in comparison to D. melanogaster flies fed medium containing 10 μM CP (Fig. 3D). However, D. melanogaster flies fed on medium containing 10 μM TCP showed no signs of toxicity, with a significantly increased overall survival (log rank [Mantel-Cox] test, chi square = 292.6, df = 1, P < 0.0001) and fewer early-time-point deaths (Gehan-Breslow-Wilcoxon test, chi square = 237, df = 1, P < 0.0001) than D. melanogaster flies fed medium with 10 μM CP (Fig. 3D). These results suggest CPO is more toxic to D. melanogaster than its parent compound CP, while TCP is less toxic.
Excess L. plantarum ISO increases toxicity of CP, while probiotic L. rhamnosus GG and E. coli(pET20b-Pte) decrease toxicity to CP.
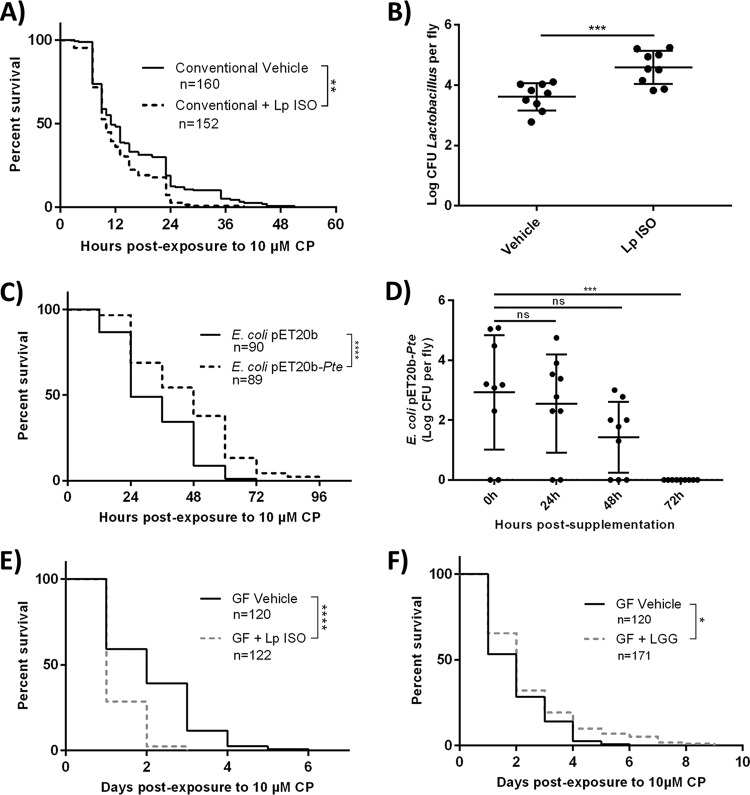
Since L. plantarum ISO was shown to metabolize CP into CPO, we sought to investigate whether increasing L. plantarum ISO abundances in D. melanogaster could increase host toxicity to CP exposure. Newly eclosed conventional D. melanogaster flies that were supplemented with L. plantarum ISO and fed medium containing 10 μM CP had a significantly decreased overall survival (log rank [Mantel-Cox] test, chi square = 10.65, df = 1, P = 0.0011) and more early-time-point deaths (Gehan-Breslow-Wilcoxon test, chi square = 4.424, df = 1, P = 0.0354) than vehicle controls fed medium containing 10 μM CP only (Fig. 4A). The log CFU for Lactobacillus spp. was found to be significantly increased (unpaired two-tailed t test, t = 4, df = 16, P = 0.0008) in L. plantarum ISO-supplemented conventional D. melanogaster flies compared to that in vehicle controls (Fig. 4B). These results suggest that supplementation with CP-metabolizing L. plantarum ISO increased the host toxicity to CP.
FIG 4.
Excess L. plantarum ISO increases toxicity of CP while probiotic L. rhamnosus GG and E. coli(pET20b-Pte) decrease toxicity to CP. (A) Survival curves of newly eclosed flies exposed to 10 μM CP with or without concurrent supplementation of L. plantarum ISO. (B) Surface-sterilized flies were homogenized and drop plated on MRS agar, and bacterial CFU per fly were enumerated after 48-h anaerobic incubations. Data are presented as means ± standard deviations (unpaired, two-tailed t test) from at least 3 independent experiments (each dot represents 10 individuals normalized to CFU per fly). (C and D) E. coli(pET20b-Pte) or vehicle E. coli(pET20b) was supplemented to D. melanogaster adults on normal food for 48 h, followed by the assessment of survival on 10 μM CP-spiked medium (C) or the enumeration of gut bacterial loads following transfer to fresh food (D). (E and F) Germfree (GF) flies were exposed to 10 μM CP with or without concurrent supplementation with L. plantarum ISO (E) or L. rhamnosus GG (F) and subsequent survival was recorded. Data represent at least 3 independent experiments. Statistical analyses shown for all survival curves are from log rank (Mantel-Cox) tests. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01, *, P < 0.05.
Since microbial bioactivation (CP to CPO conversion) by L. plantarum ISO increased the toxicity of CP (Fig. 4A), we sought to investigate whether microbial detoxification (CP to TCP conversion) by E. coli(pET20b-Pte) could reduce the toxicity of CP (Fig. 4C). Conventional D. melanogaster adults that were presupplemented with E. coli(pET 20b-Pte) for 48 h showed a significantly increased overall survival (log rank [Mantel-Cox], chi square = 21.63, P < 0.0001) and reduced early-time-point deaths (Gehan-Breslow-Wilcoxon test, chi square = 16.87, P < 0.0001) when challenged with 10 μM CP in comparison to CP-challenged vehicle controls that were presupplemented with background E. coli(pET20b) not expressing Pte. No deaths were observed in either group transferred to fresh food without 10 μM CP. Additionally, it was shown that the CFU value for E. coli(pET20b) per fly was unaltered at 24 h and 48 h (one-way ANOVA, P = 0.9377 and P = 0.1221, respectively) (Fig. 4D) but experienced a significant decrease at 72 h (one-way ANOVA, P = 0.0005) (Fig. 4D) after the flies were transferred to fresh food without CP postsupplementation. These findings suggest E. coli(pET20b) was unable to colonize D. melanogaster long term, but persisted as a transient colonizer for up to 48 h after supplementation.
To determine if L. plantarum ISO was sufficient to exacerbate CP toxicity, germfree D. melanogaster flies were supplemented with L. plantarum ISO and exposed to 10 μM CP. Newly eclosed germfree adults supplemented with L. plantarum ISO and fed medium containing 10 μM CP had a significantly reduced overall survival (log rank [Mantel-Cox] test, chi square = 126.9, df = 1, P < 0.0001) and more early-time-point deaths (Gehan-Breslow-Wilcoxon test, chi square = 102.8, df = 1, P < 0.0001) than germfree vehicle-treated flies that were fed medium containing 10 μM CP only (Fig. 4C). Germfree D. melanogaster flies were exposed to 10 μM CP and supplemented with L. rhamnosus GG, a probiotic lactobacillus strain shown previously to bind but not metabolize CP (17). Probiotic-supplemented flies exhibited a significantly increased overall survival (log rank [Mantel-Cox] test, chi square = 5.835, df = 1, P = 0.0157) and trended toward fewer early-time-point deaths (Gehan-Breslow-Wilcoxon test, chi square = 3.798, df = 1, P = 0.0513) than germfree vehicle-treated flies fed medium containing 10 μM CP only (Fig. 4D). Overall, these results suggest that Lactobacillus spp. differentially affect host CP toxicity in D. melanogaster, a phenomenon that appears to be due to differential CP metabolic capacity.
DISCUSSION
This study demonstrated that abx-treated and germfree D. melanogaster flies were significantly more resistant to CP-induced toxicity than conventionally reared controls. These results suggested that certain D. melanogaster microbiota constituents promoted CP-induced host toxicity, which is a novel finding compared to those from previous reports of microbiota-mediated (36, 37) or probiotic-mediated (17) CP resistance. The contribution of the microbiota to variable host pharmacokinetic responses such as the absorption and biotransformation of xenobiotics is well documented (47). In this study, we demonstrated that one of the dominant D. melanogaster microbiota constituents, L. plantarum ISO, was responsible for converting CP to the more potent insecticidal metabolite CPO (Fig. 3A to C). In contrast, the other major microbiota constituent in our D. melanogaster stock microbiota, an A. indonesiensis isolate, could not metabolize CP. The metabolism of CP to CPO by L. plantarum ISO appears to be a common metabolic property in L. plantarum at the species level on the basis of the observation of similar CP-CPO production by L. plantarum ATCC 14917. We have demonstrated that L. plantarum ISO was necessary and sufficient to exacerbate CP-induced toxicity to D. melanogaster by utilizing germfree L. plantarum ISO monocolonization and conventional L. plantarum ISO supplementation experiments, respectively. Furthermore, supplementation with E. coli(pET20b-Pte) (which preferentially metabolizes CP to TCP) significantly improved D. melanogaster survival toward lethal CP exposure. These observations are comparable to those from other studies demonstrating microbiota-mediated alterations in melamine (48) and digoxin (49) toxicity in humans.
The development of CP resistance in pest insects is a common occurrence that has largely been attributed to host-level physiological adaptations (50–52). However, many pest organisms such as diamondback moths, alydid stinkbugs, and crucifer root maggots have shown increased insecticide resistance due to microbiota symbiont-mediated detoxification (16, 53, 54). Symbiotic-mediated pesticide resistance has yet to be reported in honey bees, but the aforementioned observations provide strong support for the potential to reduce off-target wildlife pesticide toxicity with microbiota-directed approaches. Alternatively, this study provides a basis to speculate on how potential “biopesticides,” or microorganisms that promote insecticide-induced toxicity, could be used to preferentially target pest organisms. More generally, the data support evaluating the effects of pesticides on off-target species prior to marketplace release and how the microbiota may contribute to pesticide tolerance (55). Future insecticide designs could benefit from understanding and targeting inherent differences in microbiota compositions between beneficial and pest insects, thereby minimizing off-target pesticide toxicity and reducing futile pest extermination attempts.
Furthermore, our findings suggest that an innovative approach to combating the causative factors of honey bee decline (e.g., pesticides and pathogens) may be to supplement honey bees with probiotic bacteria containing pesticide-detoxifying genes, similar to paratransgenesis (56, 57). Both honey bees and D. melanogaster flies have simple microbiotas that are not microbially diverse (1 to 30 species) and are typically dominated by Gram-positive Lactobacillus species (42). Lactobacillus symbionts can differ in their genome structures and biology depending on the insect species which they colonize (34) but generally confer their hosts with beneficial immune stimulation (58), growth (59), and pathogen exclusion (42), thereby combating the causal factors implicated in honey bee decline (25). Though some L. plantarum strains exert probiotic properties in insects, such as beneficial immune stimulation as seen with L. plantarum Lp39 (60), our results suggest the strains tested here would be poor probiotic candidates for the purposes of detoxification.
The present study has expanded on our previous work showing that L. rhamnosus GG could mitigate CP toxicity in conventionally raised D. melanogaster (17). Specifically, monocolonized germfree D. melanogaster experiments demonstrated that L. rhamnosus GG supplementation was sufficient for the mitigation of CP-induced toxicity in a microbiota-independent manner. These findings further exemplify species-level variation in Lactobacillus-mediated CP metabolism that has been previously reported (61). In particular, in vitro experiments have shown that Lactobacillus fermentum preferentially metabolizes CP into TCP, while L. lactis preferentially metabolizes CP to CPO (61). Our findings of L. plantarum ISO increasing and E. coli(pET20b-Pte) decreasing the toxicity of CP suggest that Lactobacillus strains able to metabolize CP to TCP may be even more effective than strains such as LGG that simply bind CP. Further research will be required to evaluate if these findings are translatable to honey bees, but the ability to fortify colonies with probiotic lactobacillus-containing pollen patties or honey (62) provides a convenient method for testing these promising findings.
In summary, this study has shown that (i) CP metabolism by an L. plantarum strain within the D. melanogaster microbiota exacerbates toxicity, and (ii) Lactobacillus spp. can have alternate effects on CP toxicity on the basis of differential CP metabolism. Future studies will benefit from determining the genetic or physiological basis for the differences in CP metabolism among species of Lactobacillus. It will be particularly interesting to determine if the functionality of an organophosphate-degrading gene(s) can fully account for the differences seen in CP metabolism or whether there are other unrecognized hydrolysis enzymes dispersed across the Lactobacillus genera. Moving forward, it will be imperative that our findings in D. melanogaster are validated in honey bees prior to implementation, to avoid potentially deleterious outcomes in commercial apiaries. However, the extension of these findings to honey bees is promising given that lactobacilli are affordable, convenient, and have already been shown to benefit honey bee colony growth (63), microbiota composition (57), and antimicrobial defenses (64). While organic farming is becoming more prevalent, it is difficult to avoid the use of pesticides for food production alongside a growing global population. A targeted approach to avoid collateral damage, as suggested here, may have appeal to farmers and help prevent the demise of a key pollinator species.
MATERIALS AND METHODS
Chemicals.
CP (catalog number 45395; Sigma-Aldrich), CPO (catalog number C425320; Toronto Research Chemicals), and TCP (catalog number 33972; Sigma-Aldrich) stock solutions were prepared at 100 mg/ml in dimethyl sulfoxide (DMSO) and stored frozen at −80°C until used.
Drosophila melanogaster husbandry.
Wild-type (WT) Canton-S stocks (stock number 1) were obtained from the Bloomington Drosophila Stock Center at Indiana University. All stocks were maintained on medium consisting of 7.6% corn syrup (vol/vol), 7.3% cornmeal (wt/vol), 1.73% yeast (wt/vol), 1.5% agar (wt/vol), and 0.58% propionic acid (vol/vol). All D. melanogaster stocks were maintained at 25°C under a constant 12 h light/dark cycle. The base food medium was autoclaved for all experimental groups (conventional, abx-treated, and germfree) to account for any nutrient losses. The antibiotic food medium contained an additional 500 μg/ml ampicillin, 50 μg/ml tetracycline, and 200 μg/ml rifamycin prior to solidification as previously described (46). For experimental procedures, the medium was supplemented with various concentrations of CP or vehicle (DMSO) prior to agar solidification. All experiments were performed in wide polystyrene Drosophila vials (GEN32-121 and GEN49-101; Diamed Lab Supplies Inc., Mississauga, ON, Canada) containing 10 ml of total medium.
Isolation of D. melanogaster gut bacteria.
Flies were surface sterilized with 70% ethanol and homogenized in sterile 0.01 M phosphate-buffered saline (PBS) using a motorized pestle. The homogenates were spread plated on de Man, Rogosa, and Sharpe (MRS; catalog number 288130; BD Difco), Acetobacter growth (ACE), and brain heart infusion (BHI; catalog number B11059; BD Difco) agar plates. The plates were incubated at 37°C and 25°C under aerobic and anaerobic conditions for 48 h. DNA was extracted from two seemingly unique colony types with different morphologies and growth patterns using the InstaGene Matrix protocol (catalog number 7326030; Bio-Rad). PCR was performed on extracted DNA using the established 16S rRNA gene protocol described, which is used for the phylogenetic characterization of most bacterial species (65). The primers were AGAGTTTGATCCTGGCTCAG (forward) and AAGGAGGTGATCCAGCCGCA (reverse). The PCR products were purified by 1% agarose gel electrophoresis and subsequently extracted with a QIAquick gel extraction kit (catalog number 28704; Qiagen). The PCR products were sequenced using the aforementioned primers with the Applied Biosystems 3730 Analyzer platform at the London Regional Genomics Centre (Robarts Research Institute, London, Canada).
Bacterial strains and cultures.
Lactobacillus plantarum (obtained from the American Type Culture Collection [ATCC], number 14917) and D. melanogaster microbiota-derived Lactobacillus plantarum ISO and Lactobacillus rhamnosus GG were routinely cultured anaerobically at 37°C using MRS broth and agar, unless otherwise stated. Acetobacter indonesiensis derived from D. melanogaster was cultured aerobically at 25°C using mannitol-positive ACE medium containing 3 g/liter proteose peptone no. 3 (catalog number 211693; BD Difco), 5 g/liter yeast extract (catalog number 212750; BD Difco), and 25 g/liter d-mannitol (catalog number M9647; Sigma-Aldrich). A pET20b plasmid (EMD Millipore) containing a gene encoding an organophosphate-degrading phosphotriesterase (Pte) inserted between NdeI and EcoRI restriction sites (66) was obtained from Frank M. Raushel (Texas A&M University, USA) and cloned into chemically competent Escherichia coli BL21(DE3) as described previously (17). The subsequent culturing of E. coli(pET20b-Pte) was performed under aerobic conditions at 37°C using LB broth or agar containing 300 μg/ml ampicillin.
Generation of axenic D. melanogaster stocks.
Germfree WT Canton-S stocks were derived as previously described (67). Briefly, 1- to 2-h embryos were collected from grape agar plates dechorionated with 2.7% sodium hypochlorite for 2 min, washed twice with 70% ethanol, and washed twice with sterile double-distilled water (ddH2O). Sterilized embryos were seeded in sterile food vials under laminar flow conditions in a biological safety cabinet. The conventional Canton-S stocks used in this study were infected with Wolbachia (a bacterial endosymbiont commonly found in association with D. melanogaster and other arthropods); however, germfree stock lines were cured by treatment with 100 μg/ml tetracycline delivered in their food for four generations (68). Subsequent germfree stocks were fed sterile Drosophila medium (without the addition of antibiotics) under sterile conditions, and axenic conditions were routinely confirmed by performing PCR on whole fly homogenates using the previously described 16S rRNA primers (65). PCRs were screened for amplicons via 1% agarose gel electrophoresis, and axenic conditions were confirmed by the absence of any PCR product. Alternatively, adult homogenates were plated on MRS and ACE agar to verify germfree and monoassociation conditions with specified bacteria.
Adult D. melanogaster survival assays.
Twenty to twenty-five newly eclosed conventional, abx-treated, and germfree D. melanogaster flies were anesthetized by using CO2. Anesthetized flies were randomly assorted into the aforementioned standard vials containing experimental media. Flies were confirmed to be alive 1 h after transfer and subsequently monitored thereafter for daily survival (17). Experimental media contained the vehicle (DMSO) or various concentrations of CP, CPO, or TCP. For excess microbe experiments, overnight cultures of L. plantarum ISO, L. rhamnosus GG, and E. coli(pET20b-Pte) were centrifuged at 5,000 × g for 15 mins, washed twice with 0.01 M PBS, and resuspended in 0.01 M PBS to attain a 1010 CFU/ml bacterial suspension. The food medium was supplemented with 100 μl (109 CFU) of L. plantarum ISO, L. rhamnosus GG, E. coli pET20b-Pte, or vehicle [PBS or E. coli(pET20b) lacking Pte] and allowed to air dry before the flies were added. For E. coli(pET20b-Pte) experiments, the supplementation was stopped after 48 h to determine the subsequent ability to colonize the D. melanogaster intestinal tract. The surviving flies were transferred to fresh medium every 72 h for the duration of each experiment.
Larval D. melanogaster eclosion assays.
Eggs were collected on grape agar plates as outlined previously (69). First, instar larvae were transferred into standard vials (10 larvae/vial) containing experimental medium and incubated at 25°C. The larvae were monitored daily for up to 16 days for eclosion.
Pesticide hydrolysis assay.
CP-metabolizing bacteria were identified via semiquantitative culture plate assays by using a modified protocol as previously described (17, 37). Briefly, 5 μl of overnight broth cultures (106 CFU) of L. plantarum ISO, A. indonesiensis ISO, L. plantarum ATCC 14917, and E. coli(pET20b-Pte) (positive control) was spot plated on brain heart infusion (catalog number B11059; DB Difco) agar containing 2.85 mM emulsified CP (forms a precipitate). Following 48 h of incubation at 37°C under aerobic [A. indonesiensis ISO and E. coli(pET20b-Pte)] or anaerobic (L. plantarum ISO and L. plantarum ATCC 14917) conditions, the radii of halo formations (zones of clearance) were determined.
Pesticide tolerance assay.
Overnight broth cultures of D. melanogaster-derived Lactobacillus plantarum ISO (stationary phase) were subcultured (1:100 dilution) in 96-well plates (catalog number 351177; Falcon) containing MRS broth with the addition of CP (285 μM) or vehicle (DMSO). Alternatively, MRS broth containing minimal carbon sources (dextrose free) with the addition of CP (285 μM) or vehicle (DMSO) was used. The plates were incubated at 37°C in a Labsystems Multiskan Ascent microplate reader, and optical density (OD) measurements were taken every 30 min for 24 h at a wavelength of 600 nm.
Pesticide metabolism assay.
Stationary-phase Lactobacillus spp. were subcultured (1:100 dilution) in experimental media and supplemented with 285 μM CP or the vehicle (DMSO) and incubated for 24 h anaerobically at 37°C with gentle shaking (150 rpm) and protected from light. CP was then purified from culture suspensions via two-step ethyl acetate separation. A 2:1 ratio of ethyl acetate to bacterial culture was vortexed for 15 s, followed by organic layer extraction. Additional ethyl acetate was added in a 1:1 ratio to the remaining solution and vortexed for 15 s. The resulting organic layer was removed once again and added to the extracted material, followed by aspiration and evaporation under nitrogen. The samples were reconstituted in methanol (high-pressure liquid chromatography [HPLC] grade), filtered with a 0.22-μm-pore-size filter, and analyzed by liquid chromatography tandem-mass spectrometry (LC-MS/MS).
An Agilent 1290 Infinity HPLC system was coupled to a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, USA) with a heated electrospray ionization (HESI) source. Two microliters of each sample and standard was injected into a ZORBAX Eclipse plus C18 2.1 mm by 50 mm by 1.6 μm column. Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in acetonitrile. The initial composition of 100% mobile phase A was held constant for 1.5 min and decreased linearly to 0% over 4.5 min. Mobile phase A was held at 0% for 1.5 min then returned to 100% over 30 s. The system was reequilibrated at 100% mobile phase A for 1 min, for a total analysis time of 7.50 min.
The samples were analyzed using a semitargeted, scheduled polarity-switching method. This method comprised a positive mode data-dependent acquisition (DDA) from 0 to 4.2 min, followed by a negative mode DDA from 4.2 to 4.8 min, and then returning to positive mode between 4.75 to 7.5 min. This was done to accommodate the known CP metabolite, TCP, which is detected in negative ionization mode. All DDA methods were top-3: scan range, m/z 100 to 1,000; resolution, 70,000; automatic gain control (AGC), 3 × 106; and maximum injection time (IT), 250 ms. Product ion spectra were acquired with a 2.0 m/z isolation window, a resolution of 17,500, AGC target of 1 × 105; max IT of 100 ms, normalized collision energy (NCE) of 30, threshold intensity of 1.0 × 105, a fixed first mass m/z 75, and with dynamic exclusion of 8 s. An inclusion list containing the m/z (± 8.56 μM) and retention times of CP, TCP, and CPO was used so that those m/z signals would be preferentially selected for MS/MS if detected above the threshold intensity. If the signals corresponding to those compounds were not detected, the three most intense ions found in the full MS scan were selected for MS/MS.
Enumeration of bacterial load in D. melanogaster guts.
Overnight cultures of D. melanogaster-derived L. plantarum ISO or E. coli(pET20b-Pte) were centrifuged at 5,000 × g for 15 min and washed twice with 0.01 M PBS, followed by resuspension in 0.01 M PBS to attain 1010 CFU/ml suspensions. Newly eclosed adult D. melanogaster flies were transferred to standard vials containing medium supplemented with 100 μl (109 CFU) of L. plantarum ISO, E. coli(pET20b-Pte), or the appropriate vehicle (PBS or E. coli pET20b lacking Pte) that had air dried. The flies were incubated at 25°C for 18 h and 48 h for L. plantarum ISO and E. coli(pET20b-Pte) experiments, respectively. The flies were subsequently surface sterilized with 70% ethanol and homogenized in sterile 0.01 M PBS using a motorized pestle. The homogenates were serially diluted and spot plated onto MRS or LB plus 300 μg/ml ampicillin agar plates in triplicates. CFU were enumerated following anaerobic (L. plantarum ISO) or aerobic [E. coli(pET20b-Pte)] incubation at 37°C for 48 h.
Statistical analyses.
All statistics were performed using GraphPad Prism 7.0 software. Data sets with unique values were tested for normality using the omnibus-based Shapiro-Wilk test, while data sets with ties (two or more identical values) were tested for normality using the D'Agostino-Pearson test. Normally distributed data were statistically compared with unpaired two-tailed t tests, one-way analysis of variance (ANOVA), or two-way ANOVA as indicated. ANOVA tests were complemented with Tukey's (data set with one categorical variable) or Sidak's (data set with two categorical variables) multiple-comparison tests when appropriate. Nonparametric data were statistically analyzed using Kruskal-Wallis tests with Dunn's multiple comparisons. Mantel-Cox tests were used to analyze overall D. melanogaster survival. Gehan-Breslow-Wilcoxon tests were used to analyze D. melanogaster survival with an emphasis on the early-time-point events. Multiple comparisons for Mantel-Cox and Gehan-Breslow-Wilcoxon tests were performed using Bonferroni's method.
Accession number(s).
The 16S rRNA gene sequences have been uploaded to the NCIB GenBank database (accession numbers MG774414.1, MG774413.1, MG774412.1, and MG774411.1).
ACKNOWLEDGMENTS
We thank Frank M. Raushel and Andrew N. Bigley for providing the pET20b-Pte plasmid. We also thank Justin Renaud for assisting with the LC-MS/MS methodology and interpretation.
This work was funded by the Government of Canada Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2014-05188 and CGSM-515651-2017).
REFERENCES
- 1.Davis RW, Kamble ST. 1992. Distribution of sub-slab injected Dursban TC (chlorpyrifos) in a loamy sand soil when used for subterranean termite control. Bull Environ Contam Toxicol 48:585–591. doi: 10.1007/BF00199078. [DOI] [PubMed] [Google Scholar]
- 2.Katz EJ, Cortes VI, Eldefrawi ME, Eldefrawi AT. 1997. Chlorpyrifos, parathion, and their oxons bind to and desensitize a nicotinic acetylcholine receptor: relevance to their toxicities. Toxicol Appl Pharmacol 146:227–236. doi: 10.1006/taap.1997.8201. [DOI] [PubMed] [Google Scholar]
- 3.United States Environmental Protection Agency. 2016. Revised human health risk assessment on chlorpyrifos. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 4.Porrini C, Mutinelli F, Bortolotti L, Granato A, Laurenson L, Roberts K, Gallina A, Silvester N, Medrzycki P, Renzi T, Sgolastra F, Lodesani M. 2016. The status of honey bee health in Italy: results from the Nationwide Bee Monitoring Network. PLoS One 11:e0155411. doi: 10.1371/journal.pone.0155411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiesa LM, Labella GF, Giorgi A, Panseri S, Pavlovic R, Bonacci S, Arioli F. 2016. The occurrence of pesticides and persistent organic pollutants in Italian organic honeys from different productive areas in relation to potential environmental pollution. Chemosphere 154:482–490. doi: 10.1016/j.chemosphere.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Calatayud-Vernich P, Calatayud F, Simó E, Suarez-Varela MM, Picó Y. 2016. Influence of pesticide use in fruit orchards during blooming on honeybee mortality in 4 experimental apiaries. Sci Total Environ 541:33–41. doi: 10.1016/j.scitotenv.2015.08.131. [DOI] [PubMed] [Google Scholar]
- 7.Cycoń M, Wójcik M, Piotrowska-Seget Z. 2009. Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere 76:494–501. doi: 10.1016/j.chemosphere.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Sparling DW, Fellers G. 2007. Comparative toxicity of chlorpyrifos, diazinon, malathion and their oxon derivatives to larval Rana boylii. Environ Pollut 147:535–539. doi: 10.1016/j.envpol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 9.Xu G, Zheng W, Li Y, Wang S, Zhang J, Yan Y. 2008. Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by a newly isolated Paracoccus sp. strain TRP. Int Biodeterior Biodegradation 62:51–56. doi: 10.1016/j.ibiod.2007.12.001. [DOI] [Google Scholar]
- 10.Racke KD, Laskowski DA, Scultz MR. 1990. Resistance of chlorpyrifos to enhanced biodegradation in soil. J Agric Food Chem 38:1430–1436. doi: 10.1021/jf00096a029. [DOI] [Google Scholar]
- 11.Croom EL, Wallace AD, Hodgson E. 2010. Human variation in CYP-specific chlorpyrifos metabolism. Toxicology 276:184–191. doi: 10.1016/j.tox.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Chishti Z, Hussain S, Arshad KR, Khalid A, Arshad M. 2013. Microbial degradation of chlorpyrifos in liquid media and soil. J Environ Manage 114:372–380. doi: 10.1016/j.jenvman.2012.10.032. [DOI] [PubMed] [Google Scholar]
- 13.Anwar S, Liaquat F, Khan QM, Khalid ZM, Iqbal S. 2009. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J Hazard Mater 168:400–405. doi: 10.1016/j.jhazmat.2009.02.059. [DOI] [PubMed] [Google Scholar]
- 14.Sasikala C, Jiwal S, Rout P, Ramya M. 2012. Biodegradation of chlorpyrifos by bacterial consortium isolated from agriculture soil. World J Microbiol Biotechnol 28:1301–1308. doi: 10.1007/s11274-011-0879-z. [DOI] [PubMed] [Google Scholar]
- 15.Iyer R, Iken B, Damania A. 2013. A comparison of organophosphate degradation genes and bioremediation applications. Environ Microbiol Rep 5:787–798. doi: 10.1111/1758-2229.12095. [DOI] [PubMed] [Google Scholar]
- 16.Xia X, Zheng D, Zhong H, Qin B, Gurr GM, Vasseur L, Lin H, Bai J, He W, You M. 2013. DNA sequencing reveals the midgut microbiota of Diamondback Moth, Plutella xylostella (L) and a possible relationship with insecticide resistance. PLoS One 8:e68852. doi: 10.1371/journal.pone.0068852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trinder M, McDowell TW, Daisley BA, Ali SN, Leong HS, Sumarah MW, Reid G. 2016. Probiotic Lactobacillus rhamnosus reduces organophosphate pesticide absorption and toxicity to Drosophila melanogaster. Appl Environ Microbiol 82:6204–6213. doi: 10.1128/AEM.01510-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joly Condette C, Bach V, Mayeur C, Gay-Quéheillard J, Khorsi-Cauet H. 2015. Chlorpyrifos exposure during perinatal period affects intestinal microbiota associated with delay of maturation of digestive tract in rats. J Pediatr Gastroenterol Nutr 61:30–40. [DOI] [PubMed] [Google Scholar]
- 19.de Almeida LG, de Moraes LAB, Trigo JR, Omoto C, Cônsoli FL. 2017. The gut microbiota of insecticide-resistant insects houses insecticide-degrading bacteria: a potential source for biotechnological exploitation. PLoS One 12:e0174754. doi: 10.1371/journal.pone.0174754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collison E, Hird H, Cresswell J, Tyler C. 2016. Interactive effects of pesticide exposure and pathogen infection on bee health - a critical analysis. Biol Rev Camb Philos Soc 91:1006–1019. doi: 10.1111/brv.12206. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Zhang Y, Wang G, Han R, Xie X. 2016. Effects of chlorpyrifos on the gut microbiome and urine metabolome in mouse (Mus musculus). Chemosphere 153:287–293. doi: 10.1016/j.chemosphere.2016.03.055. [DOI] [PubMed] [Google Scholar]
- 22.Joly C, Gay-Quéheillard J, Léké A, Chardon K, Delanaud S, Bach V, Khorsi-Cauet H. 2013. Impact of chronic exposure to low doses of chlorpyrifos on the intestinal microbiota in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) and in the rat. Environ Sci Pollut Res Int 20:2726–2734. doi: 10.1007/s11356-012-1283-4. [DOI] [PubMed] [Google Scholar]
- 23.Blacquière T, Smagghe G, van Gestel CAM, Mommaerts V. 2012. Neonicotinoids in bees: a review on concentrations, side-effects and risk assessment. Ecotoxicology 21:973–992. doi: 10.1007/s10646-012-0863-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VanEngelsdorp D, Speybroeck N, Evans JD, Nguyen BK, Mullin C, Frazier M, Frazier J, Cox-Foster D, Chen Y, Tarpy DR, Haubruge E, Pettis JS, Saegerman C. 2010. Weighing risk factors associated with bee colony collapse disorder by classification and regression tree analysis. J Econ Entomol 103:1517–1523. doi: 10.1603/EC09429. [DOI] [PubMed] [Google Scholar]
- 25.Evans JD, Schwarz RS. 2011. Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol 19:614–620. doi: 10.1016/j.tim.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Henry M, Béguin M, Requier F, Rollin O, Odoux J-F, Aupinel P, Aptel J, Tchamitchian S, Decourtye A. 2012. A common pesticide decreases foraging success and survival in honey bees. Science 336:348–350. doi: 10.1126/science.1215039. [DOI] [PubMed] [Google Scholar]
- 27.Chaimanee V, Evans JD, Chen Y, Jackson C, Pettis JS. 2016. Sperm viability and gene expression in honey bee queens (Apis mellifera) following exposure to the neonicotinoid insecticide imidacloprid and the organophosphate acaricide coumaphos. J Insect Physiol 89:1–8. doi: 10.1016/j.jinsphys.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhu YC, Adamczyk J, Rinderer T, Yao J, Danka R, Luttrell R, Gore J. 2015. Spray toxicity and risk potential of 42 commonly used formulations of row crop pesticides to adult honey bees (Hymenoptera: Apidae). J Econ Entomol 108:2640–2647. doi: 10.1093/jee/tov269. [DOI] [PubMed] [Google Scholar]
- 29.Hester PG, Shaffer KR, Tietze NS, Zhong H, Griggs NL. 2001. Efficacy of ground-applied ultra-low-volume malathion on honey bee survival and productivity in open and forest areas. J Am Mosq Control Assoc 17:2–7. [PubMed] [Google Scholar]
- 30.Al Naggar Y, Codling G, Vogt A, Naiem E, Mona M, Seif A, Giesy JP. 2015. Organophosphorus insecticides in honey, pollen and bees (Apis mellifera L.) and their potential hazard to bee colonies in Egypt. Ecotoxicol Environ Saf 114:1–8. doi: 10.1016/j.ecoenv.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 31.Urlacher E, Monchanin C, Rivière C, Richard F-J, Lombardi C, Michelsen-Heath S, Hageman KJ, Mercer AR. 2016. Measurements of chlorpyrifos levels in forager bees and comparison with levels that disrupt honey bee odor-mediated learning under laboratory conditions. J Chem Ecol 42:127–138. doi: 10.1007/s10886-016-0672-4. [DOI] [PubMed] [Google Scholar]
- 32.Biddinger DJ, Robertson JL, Mullin C, Frazier J, Ashcraft SA, Rajotte EG, Joshi NK, Vaughn M. 2013. Comparative toxicities and synergism of apple orchard pesticides to Apis mellifera (L.) and Osmia cornifrons (Radoszkowski). PLoS One 8:e72587. doi: 10.1371/journal.pone.0072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu W, Schmehl DR, Mullin CA, Frazier JL. 2014. Four common pesticides, their mixtures and a formulation solvent in the hive environment have high oral toxicity to honey bee larvae. PLoS One 9:e77547. doi: 10.1371/journal.pone.0077547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yun J-H, Roh SW, Whon TW, Jung M-J, Kim M-S, Park D-S, Yoon C, Nam Y-D, Kim Y-J, Choi J-H, Kim J-Y, Shin N-R, Kim S-H, Lee W-J, Bae J-W. 2014. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol 80:5254–5264. doi: 10.1128/AEM.01226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamarit D, Ellegaard KM, Wikander J, Olofsson T, Vásquez A, Andersson SGE. 2015. Functionally structured genomes in Lactobacillus kunkeei colonizing the honey crop and food products of honeybees and stingless bees. Genome Biol Evol 7:1455–1473. doi: 10.1093/gbe/evv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng D, Guo Z, Riegler M, Xi Z, Liang G, Xu Y. 2017. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5:13. doi: 10.1186/s40168-017-0236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. 2012. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci U S A 109:8618–8622. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwong WK, Engel P, Koch H, Moran NA. 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A 111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. 2016. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc Natl Acad Sci U S A 113:13887–13892. doi: 10.1073/pnas.1610856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. 2017. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci U S A 114:4775–4780. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Näpflin K, Schmid-Hempel P. 2018. Host effects on microbiota community assembly. J Anim Ecol 87:331–340. doi: 10.1111/1365-2656.12768. [DOI] [PubMed] [Google Scholar]
- 42.Blum JE, Fischer CN, Miles J, Handelsman J. 2013. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 4:e00860-. doi: 10.1128/mBio.00860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rangberg A, Diep DB, Rudi K, Amdam GV. 2012. Paratransgenesis: an approach to improve colony health and molecular insight in honey bees (Apis mellifera)? Integr Comp Biol 52:89–99. doi: 10.1093/icb/ics089. [DOI] [PubMed] [Google Scholar]
- 44.Rangberg A, Mathiesen G, Amdam GV, Diep DB. 2015. The paratransgenic potential of Lactobacillus kunkeei in the honey bee Apis mellifera. Benef Microbes 6:513–523. doi: 10.3920/BM2014.0115. [DOI] [PubMed] [Google Scholar]
- 45.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brummel T, Ching A, Seroude L, Simon AF, Benzer S. 2004. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A 101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee W-J, Brey PT. 2013. How microbiomes influence metazoan development: insights from history and Drosophila modeling of gut-microbe interactions. Annu Rev Cell Dev Bio 29:571–592. doi: 10.1146/annurev-cellbio-101512-122333. [DOI] [PubMed] [Google Scholar]
- 48.Zheng X, Zhao A, Xie G, Chi Y, Zhao L, Li H, Wang C, Bao Y, Jia W, Luther M, Su M, Nicholson JK, Jia W. 2013. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci Transl Med 5:172ra22. doi: 10.1126/scitranslmed.3005114. [DOI] [PubMed] [Google Scholar]
- 49.Lindenbaum J, Rund DG, Butler VP, Tse-Eng Saha DJR. 1981. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med 305:789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 50.Kasai S, Weerashinghe IS, Shono T, Yamakawa M. 2000. Molecular cloning, nucleotide sequence and gene expression of a cytochrome P450 (CYP6F1) from the pyrethroid-resistant mosquito, Culex quinquefasciatus Say. Insect Biochem Mol Biol 30:163–171. doi: 10.1016/S0965-1748(99)00114-9. [DOI] [PubMed] [Google Scholar]
- 51.Tabashnik BE, Carrière Y. 2010. Field-evolved resistance to Bt cotton: bollworm in the U.S. and pink bollworm in India. Southwest Entomol 35:417–424. doi: 10.3958/059.035.0326. [DOI] [Google Scholar]
- 52.Liu H, Xu Q, Zhang L, Liu N. 2005. Chlorpyrifos resistance in mosquito Culex quinquefasciatus. J Med Entomol 42:815–820. doi: 10.1093/jmedent/42.5.815. [DOI] [PubMed] [Google Scholar]
- 53.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 54.Lukwinski AT, Hill JE, Khachatourians GG, Hemmingsen SM, Hegedus DD. 2006. Biochemical and taxonomic characterization of bacteria associated with the crucifer root maggot (Delia radicum). Can J Microbiol 52:197–208. doi: 10.1139/w05-123. [DOI] [PubMed] [Google Scholar]
- 55.Nakasu EYT, Williamson SM, Edwards MG, Fitches EC, Gatehouse JA, Wright GA, Gatehouse AMR. 2014. Novel biopesticide based on a spider venom peptide shows no adverse effects on honeybees. Proc Biol Sci 281:20140619. doi: 10.1098/rspb.2014.0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trinder M, Bisanz JE, Burton JP, Reid G. 2015. Probiotic lactobacilli: a potential prophylactic treatment for reducing pesticide absorption in humans and wildlife. Benef Microbes 6:841–847. doi: 10.3920/BM2015.0022. [DOI] [PubMed] [Google Scholar]
- 57.Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC. 2012. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 7:e33188. doi: 10.1371/journal.pone.0033188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryu J-H, Kim S-H, Lee H-Y, Bai JY, Nam Y-D, Bae J-W, Lee DG, Shin SC, Ha E-M, Lee W-J. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 59.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 60.Daisley BA, Trinder M, McDowell TW, Welle H, Dube JS, Ali SN, Leong HS, Sumarah MW, Reid G. 2017. Neonicotinoid-induced pathogen susceptibility is mitigated by Lactobacillus plantarum immune stimulation in a Drosophila melanogaster model. Sci Rep 7:2703. doi: 10.1038/s41598-017-02806-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harishankar MK, Sasikala C, Ramya M. 2013. Efficiency of the intestinal bacteria in the degradation of the toxic pesticide, chlorpyrifos. 3 Biotech 3:137–142. doi: 10.1007/s13205-012-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alberoni D, Gaggìa F, Baffoni L, Gioia DD. 2016. Beneficial microorganisms for honey bees: problems and progresses. Appl Microbiol Biotechnol 100:9469–9482. doi: 10.1007/s00253-016-7870-4. [DOI] [PubMed] [Google Scholar]
- 63.Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol 78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans JD, Lopez DL. 2004. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J Econ Entomol 97:752–756. doi: 10.1093/jee/97.3.752. [DOI] [PubMed] [Google Scholar]
- 65.Falsen E, Pascual C, Sjödén B, Ohlén M, Collins MD. 1999. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int J Syst Bacteriol 49:217–221. doi: 10.1099/00207713-49-1-217. [DOI] [PubMed] [Google Scholar]
- 66.Tsai P-C, Fox N, Bigley AN, Harvey SP, Barondeau DP, Raushel FM. 2012. Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents. Biochemistry 51:6463–6475. doi: 10.1021/bi300811t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bakula M. 1969. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J Invertebr Pathol 14:365–374. doi: 10.1016/0022-2011(69)90163-3. [DOI] [PubMed] [Google Scholar]
- 68.Arbuthnott D, Levin TC, Promislow DEL. 2016. The impacts of Wolbachia and the microbiome on mate choice in Drosophila melanogaster. J Evol Biol 29:461–468. doi: 10.1111/jeb.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaffer CD, Wuller JM, Elgin SC. 1994. Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol 44:99–108. doi: 10.1016/S0091-679X(08)60908-5. [DOI] [PubMed] [Google Scholar]