New nanofluidic LC-TEM devices enable controlling and understanding electron irradiation history effects on liquid samples.
Abstract
In situ liquid cell transmission electron microscopy (LC-TEM) allows dynamic nanoscale characterization of systems in a hydrated state. Although powerful, this technique remains impaired by issues of repeatability that limit experimental fidelity and hinder the identification and control of some variables underlying observed dynamics. We detail new LC-TEM devices that improve experimental reproducibility by expanding available imaging area and providing a platform for investigating electron flux history on the sample. Irradiation history is an important factor influencing LC-TEM results that has, to this point, been largely qualitatively and not quantitatively described. We use these devices to highlight the role of cumulative electron flux history on samples from both nanoparticle growth and biological imaging experiments and demonstrate capture of time zero, low-dose images on beam-sensitive samples. In particular, the ability to capture pristine images of biological samples, where the acquired image is the first time that the cell experiences significant electron flux, allowed us to determine that nanoparticle movement compared to the cell membrane was a function of cell damage and therefore an artifact rather than visualizing cell dynamics in action. These results highlight just a subset of the new science that is accessible with LC-TEM through the new multiwindow devices with patterned focusing aides.
INTRODUCTION
Liquid cell transmission electron microscopy (LC-TEM) and liquid cell scanning transmission electron microscopy (LC-STEM) have proven to be powerful tools for investigating dynamics of nanoparticle growth (1), electrochemical processes (2), and biological structures (3, 4) at high spatial resolutions in a native hydrated environment. Although the concept is not new (5, 6), recently available commercial holders have been developed that considerably lower the barrier of entry to the field. These holders trap small volumes of liquid between thin (50 nm or less) membranes supported by a thicker substrate (7). Despite their utility, the field has struggled to control a large number of variables that can affect the outcome of LC-TEM experiments (8). Primary issues are related to understanding electron beam–liquid interactions, control of liquid thickness, flow variability, and reduced imaging area (9). Although a great deal has been achieved toward understanding the effects of the electron beam on a liquid sample, the devices and holders commonly used have remained largely unchanged. A number of purpose-built instruments have been detailed (10–12), but their function is either highly specific or not easily adoptable by the rest of the field. Here, we describe new multifunctional devices that solve some of the above-listed issues and enable new quantitative science. The devices are compatible with commercially available LC-TEM holders while maintaining a reasonable level of simplicity and flexibility so that others may immediately integrate or modify these designs to meet their own research needs.
RESULTS
Because electrons are ionizing radiation, LC-TEM experiments must contend with beam interactions on the sample under investigation. Known effects of electron irradiation include growth and etching of metal nanoparticles (1) and damage to soft materials (13) including biological samples (14). Understanding the role of electron-liquid interactions, however, has thus far been constrained to local irradiation events, although the question of how global irradiation affects electron-liquid interactions has not yet been described. To reliably quantify changes due to global irradiation history on a sample, increased reproducibility is required for sequential liquid cell experiments. Because of the inherent liquid variation that can occur when assembling multiple different liquid cells as a result of sample clumping or environmental contaminants, increasing the number of data sets that can be acquired within a single assembled device is preferential to repeating multiple experiments that provide one or few data points. The latter approach is the typical paradigm used for most LC-TEM experiments.
We have fabricated new devices that improve on currently available commercial devices by increasing the number of transparent membrane regions from one (Fig. 1, A and B), found on most devices currently, to five, as shown in Fig. 1C. These new five-membrane devices create a 5 × 5 array of windows (Fig. 1D) when assembled, with one rotated 90° with respect to the other, representing a 25-fold increase in imaging area over single-window devices. A detailed description of the fabrication process can be found in the Supplementary Materials and fig. S6. We additionally use liftoff deposition techniques to pattern features on the surface of the devices, such as spacer material or focusing aids (Fig. 1E). The device design and fabrication process allows a high degree of customization based on experimental needs. Although the results presented here use a Hummingbird liquid cell holder, we have also made multiwindow devices compatible with other holders such as the Protochips liquid cell holder (fig. S1, A to C), demonstrating the flexibility of our fabrication process for creating multiwindow liquid cell devices for different applications and holders.
Fig. 1. Overview of new multiwindow devices for improved LC-TEM.
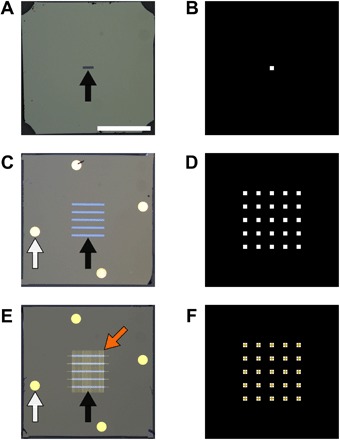
(A) Standard commercially available nanofluidic device with a single 200 × 50–μm window centered on the device (black arrow). (B) Schematic of available imaging area when devices from (A) are assembled with windows oriented perpendicular to each other. (C) Custom nanofluidic device with five windows of 50 μm width (black arrow). Gold spacer material can be seen on the devices (white arrow). (D) Schematic of available imaging area when devices from (B) are assembled with windows oriented perpendicular to each other, where a 5 × 5 grid of windows is created. (E) Nanofluidic devices from (C) that have been patterned with Au grid bars (orange arrow) crossing the windows for use as focusing aids. (F) Schematic of available imaging area when devices from (E) are assembled with windows oriented perpendicular to each other. Grid bars are shown bisecting the center of each window for focusing applications. Scale bar represents 1 mm for (A), (C), and (E), whereas (B), (D), and (F) are illustrations of window overlaps not shown to scale.
The ability to acquire multiple data sets from a single device provides an ideal platform to study how increasing cumulative electron flux on a sample (both locally and globally) can affect subsequent behavior of particle growth experiments. The growth of metal nanoparticles from solution with the electron beam has been well characterized (15, 16), where radiolysis of water by the electron beam produces aqueous electrons and other radical species that reduce the precursor solution to precipitate particles from solution (17). Although this growth is expected to be constrained to the irradiated area, we have observed the precipitation/growth of particles outside of the illuminated region after the experiment, similar to other studies in the field (1). It currently remains unclear if these particles are the product of radiolysis products diffusing away from the illuminated area or if particle nuclei form within the illuminated region, diffuse outside of the irradiated area, and continue growth. Schneider et al. (18) have demonstrated through simulation that the most reactive radical species generated by electron beam–water interactions (such as aqueous electrons) react rapidly with the bulk solution immediately after leaving the irradiated area. Other more inert species, however, such as H2 and O2 may diffuse away from the irradiated area and can be present in significant concentrations several micrometers away from the area imaged (18). Because it is common practice to acquire multiple data sets from a single device, understanding how previous irradiation history can change growth behavior and kinetics (either via formation of reactive species or sample depletion) is critical for comparing observations between experiments. Although sample depletion effects have been observed qualitatively (9, 19) during localized beam exposure, a quantitative analysis of the effect of large cumulative electron flux on a sample has not been performed. This lack of quantification is surprising considering that the study of nanoparticle nucleation, growth, and interactions represents the largest fraction by far of published research in the LC-TEM field.
Effect of irradiation history on silver nanoparticle growth
Tracking the growth kinetics of metal nanoparticles precipitated from an aqueous precursor using the electron beam provides an ideal platform for studying irradiation history. There are a number of studies detailing kinetics for various precursor solutions and electron flux conditions for metal (1, 20, 21) and metal-organic (13, 22) nanoparticles. The precipitation of silver nanoparticles from a silver nitrate precursor using the electron beam has been especially well characterized (23, 24). For the current LC-STEM study tracking silver nanoparticle growth, we captured multiple data sets within a single multiwindow device by precipitating nanoparticles from a 0.1 mM AgNO3 solution, where each subsequent video was captured in adjacent window corners and approximately 50 μm away from the previous video. Each data set was captured with identical imaging conditions and equal number of STEM scans (200) to ensure that the incremental electron flux increase between replicate videos is consistent. The scan area was 1024 × 1024 pixels, the dwell time was 3 μs, the beam current was 5.85 pA, and the pixel size was 1.47 nm, yielding a final electron flux of 0.51 e−/Å2 per scan. At 200 scans per experiment, the total cumulative electron flux for each video capture was 101.4 e−/Å2. A total of 13 videos were taken for the data set (Fig. 2); the map of windows and the experiment locations relative to each other can be found in fig. S2 (A and B). Evidence of sample hydration is provided in fig. S2C, depicting the contrast gradient from corner to center typical of hydrated liquid cells in addition to an electron energy-loss spectroscopy (EELS) spectrum (fig. S2D), both of which were acquired after data collection to limit beam exposure. Figure 2A shows frames from three of the videos depicting the nucleation and growth of particles over time during irradiation. Particle tracking algorithms were used to track particle diameters for each video, where only particles that were in focus (located on the top membrane) and fully within the field of view were used for analysis (25, 26). Figure 2B shows the mean diameter for the particles of each video over time, where the growth of these particles follows approximately a t1/2 power law, indicating reaction-limited growth kinetics (23). The mean growth exponent for all videos was 0.5095 ± 0.0113 and was derived from a linear regression model following the form r = Ktβ, as has been described elsewhere (24, 27). Growth from AgNO3 has been shown to follow both a t1/3 (24) and a t1/2 (23) power law for identical precursors depending on the electron flux, where a lower electron flux results in a lower concentration of reducing species and reaction-limited growth behavior (23). The consistency of growth kinetics across multiple sequential videos suggests that for systems that are reaction-limited, the kinetics of growing particles are independent of increasing growth history within the lateral spatial distance between experiments investigated here (~50 μm). It is possible that there may be a minimum lateral distance between experiments such that, below a certain distance offset, subsequent experiments may be influenced by previous irradiation events. This minimum distance remains unknown, although it seems that data acquired in adjacent window corners (~50 um apart) are sufficient so that serial experiments do not affect growth kinetics of subsequent experiments.
Fig. 2. Cumulative electron flux effects on silver nanoparticle growth.
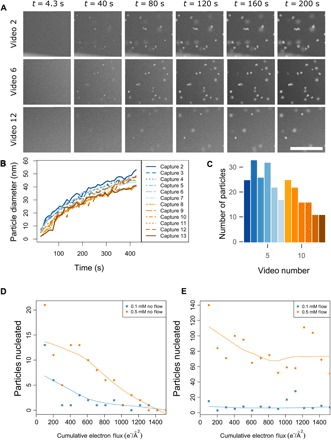
(A) Individual frames showing growth of silver nanoparticles by reducing a 0.1 mM AgNO3 aqueous precursor solution with the electron beam. LC-STEM frames taken from video captures 2, 6, and 12 are compared across time. The scan area was 1024 × 1024 pixels, the dwell time was 3 μs, the beam current was 5.85 pA, and the pixel size was 1.47 nm, resulting in an electron flux of 0.51 e−/Å2 per scan. (B) Diameter of particles from all videos plotted over the course of the 200 scan exposures. Only particles in focus, contained entirely within the viewing area, and nonoverlapping are used in the growth rate analysis. (C) Total number of particles nucleated in each capture video, where particles on both windows (in focus and out of focus) are counted. (D) Number of particles nucleated for a 0.1 mM AgNO3 precursor solution (blue squares) and a 0.5 mM AgNO3 precursor solution (orange circles) without flow of fresh precursor during imaging. Each experiment was performed in an adjacent window for 200 STEM scans. The electron flux was 0.51 e−/Å2 per scan and 101.4 e−/Å2 per video. (E) Number of particles nucleated for a 0.1 mM AgNO3 precursor solution (blue squares) and a 0.5 mM AgNO3 precursor solution (orange circles) while flowing fresh precursor solution at a rate of 0.5 μl/min. The electron flux is equivalent to that in (D). Scale bar, 1 μm (A).
Despite growth kinetics behaving consistently for increasing cumulative electron flux, it was noted that there was a gradual decrease in the number of particles formed with increasing cumulative electron flux (Fig. 2C). During the experiment described above, the liquid cell holder was used in a static regime, where there was no flow of fresh precursor solution during the experiment. We speculated that this reduction in number of nucleated particles could be the result of depletion of the precursor solution. As shown in Fig. 2 (D and E), we have repeated these experiments in the presence and absence of flow of fresh precursor solution while keeping variables such as electron flux and fluid thickness constant.
EELS measurements (28) were taken at the corner of each window to confirm flat device geometries. This was a priority because a thickness gradient resulting from two devices not being perfectly parallel to each other could cause a “wedge”-shaped liquid sample across the multiwindow area, which we know from previous studies can affect solution concentration and chemistry (23). In addition to changing solution chemistry, changing thickness can also influence growth by the physical confinement within the constrained space of the liquid cell that might differ from what may be observed in a bulk solution. Thus, for these experiments, only data sets where the assembled device geometry was equivalent were compared. We also tested a more concentrated precursor solution (0.5 mM) to determine whether depletion could be avoided by simply increasing the number of available silver ions in solution. Figure 2D shows particle counts for 0.1 mM (blue squares) and 0.5 mM (orange circles) solutions that were imaged without flow of precursor solution. The mean thickness of the 0.1 mM sample was 0.816 ± 0.266 inelastic mean free paths (IMFPs), whereas the mean thickness of the 0.5 mM sample was 1.650 ± 0.496 IMFPs. A map of window positions and IMFP measurements for the 0.1 mM experiment without flow is shown in fig. S3 (A and B). For both the 0.1 and 0.5 mM data sets, initial movies show high particle counts that fall off in subsequent movies. For the 0.1 mM condition without flow, after the third video, only one to three particles were nucleated for each subsequent video, whereas no particles were nucleated for videos 14 and 15. The 0.5 mM precursor resulted in growth of more particles and took longer to deplete for a comparable thickness. However, by the 10th video, only one to three particles were nucleated, and no particles were nucleated in videos 13 to 15.
Each experiment was then repeated under the same conditions but while flowing fresh precursor solution at a rate of 0.5 μl/min for the entire duration of the experiment (Fig. 2E). The mean thickness of the 0.1 mM flow sample was 0.906 ± 0.132 IMFPs, whereas that of the 0.5 mM flow sample was 1.610 ± 0.155 IMFPs. A map of window positions and IMFP measurements for the 0.1 mM experiment with flow is shown in fig. S3 (C and D). When fresh precursor solution was replenished to the imaging area, both sample concentrations continued to nucleate particles at the maximum cumulative electron flux (limited by the number of windows available in a single device). The 0.1 mM sample averaged 7 ± 5.7 particles nucleated per video, whereas the 0.5 mM sample nucleated an average of 71 ± 24.9 particles per video.
On the basis of nucleation theory, silver nanoparticle nucleation by LC-TEM or LC-STEM occurs as the concentration of free silver atoms increases due to precursor degradation or reduction by the electron beam (29). Once the concentration of available atoms reaches a critical threshold (the supersaturation point), nanoparticle nucleation and growth rapidly occurs, depleting the local concentration of available silver atoms (29). Nuclei that do not reach a critical size or free energy state are then dissolved back into solution, whereas those that reach above this barrier continue to grow through monomer addition (30). In our experiments, the observed decrease in particles formed as a function of increasing cumulative electron flux can be explained by the depletion of the sample where the precursor solution is globally depleted and fewer nuclei reach a stable size for growth. This can be explained by the concentration of silver atoms dropping below the supersaturation point faster with the progressively depleted precursor solution. This result would not have been possible without the ability to control for confounding variables such as thickness variation and equivalent irradiation history.
These results demonstrate that the total electron flux on the sample before data collection is as important a factor as the total electron flux during the experiment. Because electron flux is reported in the literature only for the duration of the experiment, and not for previous exposures (such as for finding focus or setting up beam conditions), the variability observed for otherwise seemingly identical experiments in previously reported studies from our group and others may be the result of differing cumulative electron flux histories between the two samples. Future studies should minimize electron flux on the sample before data collection, in addition to documenting total electron flux on the sample not accounted for in the direct image acquisition. As an aside, although we find the above described behavior to be true for our reaction-limited system, other systems of different precursor solutions, concentrations, and electron fluxes or that are diffusion-limited may show increased or decreased sensitivity to a history of irradiation. Accounting for the sensitivity of a sample to global irradiation history should therefore be considered when attempting to understand the growth behavior and kinetics of other systems.
“Low-dose” imaging for biological samples
Investigating biological systems has additionally been a major interest of LC-TEM (31, 32), although the role of irradiation history on soft and organic samples is of even greater concern than for other systems. Thus, determining whether structural and morphological changes are actual physiological events or artifacts of the electron beam damaging the sample is a necessary consideration for any LC-TEM experiment on a biological sample. The cryogenic transmission electron microscopy (cryo-EM) field has shown a tolerable structural threshold of 5 to 20 e−/Å2 cumulatively for macromolecular complexes (33) and 50 to 200 e−/Å2 for whole cells depending on target resolution (34). However, although structural information may be intact at these electron fluxes, enzyme activity has been demonstrated to be altered with fluxes in the range of 0.05 e−/Å2 (35). To date, a rigorous investigation of the cumulative irradiation sensitivity to biological samples in the confined environment of thin liquid cells used for LC-TEM has not been performed, although several studies have attempted to demonstrate the viability of cells irradiated with the electron beam by using so-called live/dead cell fluorescent assays (3, 36). Concerns about the usage of these fluorescent assays have been raised (37), where it should be noted that an assay may report a cell as “alive,” even though significant damage has occurred to DNA, protein, or macromolecular complexes, resulting in the cell being altered from its natural physiological state. Problematically, the damage caused to a protein resulting in its inactivation can occur at significantly higher spatial resolutions than has been demonstrated by LC-TEM or LC-STEM on biological samples, meaning observations and conclusions about physiological events may be confounded by damage events. Because LC-TEM seeks to observe dynamics as they naturally occur, limiting electron exposure before imaging is critical. Unwanted irradiation most commonly occurs while finding focus for the sample before data collection. Although a window edge can be used to aid with focus, often the first few frames of an LC-TEM experiment are not quite in focus, and the sample is used to find the exact focus.
To improve reliability for low-dose focusing, we have patterned our multiwindow LC-TEM devices with thin grid bars of gold (Fig. 1, E and F) to serve as an in situ focusing aid. When assembled within a liquid cell holder, the grid bars bisect the center of each window (Fig. 3, A and B). Because imaging is most commonly performed at the corner of each window, focus can be found at the grid bars, and imaging can then be carried out at the adjacent corner on the sample of interest with minimal electron flux before imaging. This low-dose regime (similar to what is performed for cryo-EM) is depicted in Fig. 3C and movie S1, where 10-nm gold nanoparticles fixed to the top window were imaged near ideal focus in STEM up to 10 μm away from the area used for focusing and were captured from the very first frame at high magnification. This is highly useful because the highly convergent beam of a STEM probe has a very narrow depth of field, and a small defocus value can result in a significant loss in resolution. Time zero, in-focus imaging is a critical component for nanoparticle growth experiments and biological samples, in particular from minimizing electron flux on the sample before starting data collection.
Fig. 3. Low-dose image acquisition empowered by patterned in situ focusing aides.
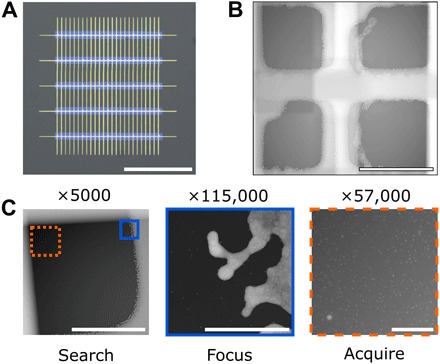
(A) Magnified view of five-window chip with grid bar patterning. (B) In situ image of grid bars over a window once assembled in a commercial LC-TEM holder. (C) Demonstration of low-dose capability allowed by using grid bars as focusing aids. Optimal focus is found on grid bars followed by imaging the sample area in a single shot with minimal previous electron exposure. Scale bars, 500 μm (A), 20 μm (B), 10 μm (C, Search), 500 nm (C, Focus), and 500 nm (C, Acquire).
Using these grid bars as focusing aids, we imaged a number of biological samples with both LC-TEM and LC-STEM. Cupriavidus metallidurans was imaged using TEM (fig. S4, A to C), whereas fig. S4D shows a bacterium labeled with 10-nm biotinylated gold nanoparticles imaged with STEM. The multiple windows in our devices were especially advantageous for imaging cells, which have a tendency to distribute nonuniformly over the surface of the LC-TEM devices, making them unlikely to rest over the single window present in conventional devices. This distribution problem is highlighted in fig. S5, which depicts the 25 windows of a device, where C. metallidurans was found in only 16 of the 25 windows. Having multiple imaging regions in this case allowed us to collect significantly more data from one assembled device than would have been possible with a conventional single window device.
Finally, we used these devices to study the effect of electron flux on a biological sample. Using focus aids, we acquired images of C. metallidurans with minimal electron flux before initial exposure and captured a near-pristine state (Fig. 4A) at a cumulative electron flux of 1 e−/Å2. Cell membranes and internal structures are identifiable in this first image; in addition, metal nanoparticles are visible near, in, or on the cells. C. metallidurans is known for its high heavy metal resistance and role in gold biomineralization. The cells imaged here were grown in a 50 mM AuCl medium before imaging, which is likely the origin of the metal nanoparticles observed. After this first image, the beam is blanked for 20 min, and a second exposure is made at an electron flux of 1 e−/Å2 for a total cumulative flux of 2 e−/Å2. This is repeated two more times for a final cumulative flux of 4 e−/Å2 for the sample. Morphological changes in the cells from the first image to the last are immediately apparent (Fig. 4B). A mask of cell outlines calculated from the very first image is projected over the subsequent acquired images and illustrates the cells shrinking at a roughly equivalent rate centered on their initial position. Swelling of cells is a common imaging artifact in cryo-EM, indicative of beam damage due to a buildup of liberated hydrogen gas in the frozen sample (14). The shrinkage observed here is interesting due to its “opposite” apparent sample deformation. For these results, we are likely seeing mass loss as liberated hydrogen gas, and other species are able to freely diffuse away due to surrounding liquid medium rather than being trapped in a frozen state. More experiments are needed to calculate true dose thresholds for functional and morphological damage in a wide range of biological samples during LC-TEM to better understand the limits. The methods described here form a good basis for these future studies.
Fig. 4. Cumulative electron flux effects on biological cells.
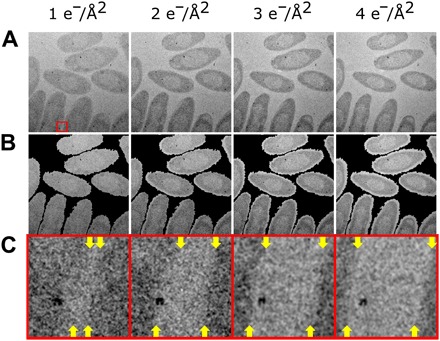
(A) Damage series of C. metallidurans, imaged using a low-dose imaging regime at an electron flux of 1 e−/Å2 per acquisition. Time between each subsequent acquisition was 20 min, where the beam was blanked during that time. Flux labeled is total cumulative flux for the frame. Total field of view, 3.5 μm × 3.5 μm. (B) Overlay of cell boundary from initial image (A) (1 e−/Å2) highlighting decreasing size and morphological changes of cells as a function of increasing cumulative electron flux. (C) Magnification (×10) of region depicted in (A), where the edge of the cells (yellow arrows at top and bottom of each frame) can be seen moving relative to the metal cluster with increasing electron flux.
Figure 4C further highlights a region from Fig. 4A, where a metal nanoparticle is observed near the edge of one of the cell membranes. Throughout the image series, the cell membrane (top and bottom positions labeled by yellow arrows in Fig. 4C) is seen to draw away from the nanoparticle as the cell shrinks. This observation highlights two important facts. The first is that, without careful consideration, our images could be interpreted as the cell dynamically “secreting” the nanoparticle, with the nanoparticle moving farther away from the cell over time. Other previous studies have attempted to draw physiological conclusions about cellular behavior based on tracking particle motion relative to the cell (32, 38). However, with our ability to acquire pristine time = 0 images due to the low-dose imaging regime, we can see that the nanoparticle actually remained static, whereas the cell periphery moves as a result of beam-induced shrinking at electron fluxes far below those typically used with most LC-TEM studies. Similar cell changes relative to nanoparticle location are seen in other areas of the images. Second, without careful control of irradiation before imaging, electron flux histories as small as 1 e−/Å2 are enough to cause beam-driven morphological changes in the cell. Future investigations of biological samples must be able to reliably start data acquisition on a cell (or purified protein complexes), which has experienced no or minimal irradiation before starting the experiment. Such an approach, in the future, will help simplify interpretation and ensure fidelity of any such dynamic experiments for soft materials, and is a defining capability empowered by the devices presented here.
DISCUSSION
In summary, we describe a fabrication process to create multiwindow and patterned devices for LC-TEM with a large increase in imaging area over conventional devices, and features that allow control over sample irradiation history. These devices empowered new quantitative studies on the impact of large cumulative electron fluxes to both inorganic and organic materials in liquid. We described experiments where all holder geometry and electron flux variables were controlled, allowing us to focus on understanding the impact of only flow versus no flow or precursor concentration differences. The ability to control all variables reproducibly for an LC-STEM experiment using the multiwindow devices allowed us to demonstrate clear evidence of depletion. Likewise, for biological samples, the ability to acquire images with minimal previous irradiation showed that a cumulative flux of 1 to 4 e−/Å2 is enough to significantly affect the ultrastructure of a whole intact organism. These results showcase new science enabled by the devices described here. They also reveal that considerable care needs to be taken to know and control total electron flux on the sample during the global experiment leading up to final image acquisition, rather than just the electron flux during single image collection, because this discrepancy can have an adverse impact on data analysis and interpretation.
MATERIALS AND METHODS
Microfabrication
All microfabrication processes were performed in a class 1000 clean room to minimize surface contaminants of nanofluidic devices. Overall workflow (fig. S6) and a detailed step-by-step procedure are given in the Supplementary Materials. Silicon wafers of 200 μm thickness, 4-inch (100 mm) diameter, [100] orientation, double side–polished, and double side–coated with 10 nm of low-stress silicon nitride were purchased from Norcada. Masks were designed with AutoCAD (Autodesk) and printed using a direct-write lithography mask printer (SF-100, Intelligent Micro Patterning). Lithography for etching windows was rectangular, with dimensions of 700 μm × 50 μm spaced 150 μm (center-to-center distance) that resulted in five windows of ~40 μm × 690 μm final dimension centered on each device. This window pattern maximized the number and stability of windows while minimizing over etching by KOH. In addition, 40-μm-width channels were patterned around each 2.6 × 2.6–mm device, and individual devices were sectioned by etching concurrent with window etching by “perforating” the silicon wafer with the device outline. Individual devices could then be removed from the bulk wafer, where an entire 4-inch (100 mm) wafer is patterned with over 400 repeating devices. Gold grid bars were patterned to be 10 μm in width and to bisect each resulting window, while spacer posts were 200 μm in diameter. Detailed mask designs are available upon request to the corresponding author.
Liquid STEM imaging
All STEM imaging was performed on a 300-kV FEI Titan with a monochromator, probe corrector, Gatan imaging filter, and high-angle annular dark-field and bright-field detectors. The microscope was aligned using the standard gold waffle thin film, and the beam current was adjusted to 5.85 pA, an electron flux of 0.51 e−/Å2 per scan at ×57,000 magnification, 1024 × 1024 frame size, and 3-μs dwell time. Electron flux and imaging conditions were kept constant for each experiment, with gain autothresholded for both bright- and dark-field STEM. Electron energy-loss spectra were acquired using the same beam conditions, with a total collection ~0.1 s optimized for signal detection.
Fresh solutions of 0.1 or 0.5 mM AgNO3 (Sigma-Aldrich) were prepared and kept away from room lights with aluminum foil. Nanofluidic devices were plasma-cleaned in Ar for 60 s before loading to decrease the hydrophobicity of the device surface. A total of 0.3 μl of the 0.1 or 0.5 mM AgNO3 solution was placed on opposite sides of the nanofluidic holder tip (Hummingbird Scientific) to ensure a hydrated environment after loading nanofluidic devices. A single nanofluidic device was placed in the nanofluidic holder, and 0.3 μl of the AgNO3 precursor solution was pipetted on the surface. A second nanofluidic device was immediately placed on top of the first device, with the windows rotated 90° with respect to each other to form a 5 × 5 array of windows. The devices were sealed in the nanofluidic holder and checked for sealing in a Hummingbird pumping station. Upon observation that high vacuum was pulled on the device and no visible defects were seen in the windows, the nanofluidic holder was transferred to the microscope (during this time, the amount of light exposed to the loaded AgNO3 solution was minimized as much as possible).
After loading into the STEM, the nanofluidic holder was left to rest for 30 to 40 min to minimize stage drift. Imaging locations were then found by imaging at the lowest magnification (×5000) and a scan size of 512 × 512 with a 0.2-μs dwell time to minimize electron flux before imaging. Once a corner was located, imaging was performed at ×57,000 magnification and a scan size of 1024 × 1024 with a 3-μs dwell time, and videos were recorded for 200 scans. These imaging conditions were then repeated at each subsequent window corner, as depicted in fig. S2A. Individual and population kinetics for each data set were calculated using established particle tracking algorithms described below.
Liquid TEM imaging
All TEM imaging was performed on a 300-kV FEI Titan with an image corrector and a 2048 × 2048–pixel charge-coupled device (CCD) camera (Gatan). The microscope was aligned using the standard gold waffle thin film and various magnifications used in conjunction with the low-dose imaging function. C. metallidurans was cultured in standard nutrient broth (NB) with 50 mM AuCl medium until late log phase, harvested at 3000g, and resuspended in ~500 μl of medium to concentrate cells. Cells (0.3 μl) were loaded onto the five-window nanofluidic devices and sealed in the Hummingbird nanofluidic holder. After ensuring hermeticity of the devices with pumping station, the sample was loaded into the microscope and images were acquired with low-dose exposures of 1 s. The electron flux for images was either 0.5 or 1 e−/Å2.
Electron flux calculations
Scanning transmission electron microscopy
Electron flux for LC-TEM experiments was determined by measuring the beam current on the small fluorescent screen given in the TEM user interface. This value was calibrated using a Faraday cup holder to determine the actual beam current at the sample plane for the value given in the interface. All future experiments were then adjusted to this calibrated value for consistency between experiments. The calibrated beam current in picoamperes was then converted to electrons per second, and the known pixel dwell time and pixel size were used to find a flux of 0.51 e−/Å2 per scan.
Transmission electron microscopy
Electron flux for LC-TEM was determined by calibrating the CCD camera counts using a Faraday cup holder. Camera counts were calibrated for a corresponding beam current and illumination condition, and subsequent experiments were performed at the calibrated conditions. The calibrated beam current in picoamperes was then converted to electrons per second, and the known pixel size and exposure time were used to find a flux of 0.5 or 1.0 e−/Å2 depending on the experiment.
Video analysis and particle tracking
Particle formation was located, and growth was tracked in a video sequence of in situ liquid STEM images. An image segmentation method was first applied to locate individual particles in each image frame of the video (25). The detected particles were associated over different image frames to track growth trajectories of individual particles while identifying newly formed particles. The data association was formulated as a global optimization problem to minimize the association error (26). As illustrated in Fig. 2 (B and C), the main outcomes of the analysis are the diameters of particles and the number of newly formed particles versus time for the 13 different videos analyzed.
Supplementary Material
Acknowledgments
Funding: This work was supported by the Department of Energy’s Office of Biological and Environmental Research Molecules to Mesoscale Bioimaging project no. 66382 and was performed using Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. Effort for T.S. was supported by the NSF CAREER award (grant no. DMR-1564950). Effort for C.P. was partially supported by the NSF (grant no. 1334012). Author contributions: J.E.E. conceived the work. J.E.E., T.S., and T.H.M. planned the experiments. T.H.M. designed and fabricated all devices with assistance from H.M. and R.T.K. T.H.M. acquired all imaging data. T.H.M. and C.P. performed particle tracking and data analysis. All authors contributed to the writing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/4/eaaq1202/DC1
fig. S1. Multiwindow chips compatible with commercial LC-TEM holders.
fig. S2. Additional experimental data from Fig. 2.
fig. S3. Imaging path and thickness of multiwindow devices for no flow versus flow.
fig. S4. Low-dose imaging of biological samples.
fig. S5. Improved sampling of nonuniform samples with multiwindow areas.
fig. S6. Illustration of microfabrication workflow.
Supplementary protocol for fabrication of multiwindow devices
movie S1. Demonstration of in situ liquid cell “low-dose” imaging.
REFERENCES AND NOTES
- 1.Evans J. E., Jungjohann K. L., Browning N. D., Arslan I., Controlled growth of nanoparticles from solution with in situ liquid transmission electron microscopy. Nano Lett. 11, 2809–2813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu M., Parent L. R., Mehdi B. L., Unocic R. R, McDowell M. T., Sacci R. L, Xu W, Connell J. G, Xu P., Abellan P., Chen X., Zhang Y., Perea D. E., Evans J. E., Lauhon L. J., Zhang J.-G., Liu J., Browning N. D., Cui Y., Arslan I., Wang C.-M., Demonstration of an electrochemical liquid cell for operando transmission electron microscopy observation of the lithiation/delithiation behavior of Si nanowire battery anodes. Nano Lett. 13, 6106–6112 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Woehl T. J., Kashyap S., Firlar E., Perez-Gonzalez T., Faivre D., Trubitsyn D., Bazylinski D. A., Prozorov T., Correlative electron and fluorescence microscopy of magnetotactic bacteria in liquid: Toward in vivo imaging. Sci. Rep. 4, 6854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans J. E., Jungjohann K. L., Wong P. C. K., Chiu P.-L., Dutrow G. H., Arslan I., Browning N. D., Visualizing macromolecular complexes with in situ liquid scanning transmission electron microscopy. Micron 43, 1085–1090 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruska E., Beitrag zur übermikroskopischen Abbildung bei höheren Drucken. Kolloid-Z. 100, 212–219 (1942). [Google Scholar]
- 6.Abrams I. M., McBain J. W., A closed cell for electron microscopy. J. Appl. Phys. 15, 607–609 (1944). [DOI] [PubMed] [Google Scholar]
- 7.Williamson M. J., Tromp R. M., Vereecken P. M., Hull R., Ross F. M., Dynamic microscopy of nanoscale cluster growth at the solid–liquid interface. Nat. Mater. 2, 532–536 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Woehl T. J., Jungjohann K. L., Evans J. E., Arslan I., Ristenpart W. D., Browning N. D., Experimental procedures to mitigate electron beam induced artifacts during in situ fluid imaging of nanomaterials. Ultramicroscopy 127, 53–63 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Abellan P., Woehl T. J., Parent L. R., Browning N. D., Evans J. E., Arslan I., Factors influencing quantitative liquid (scanning) transmission electron microscopy. Chem. Commun. 50, 4873–4880 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Grogan J. M., Bau H. H., The nanoaquarium: A platform for in situ transmission electron microscopy in liquid media. J. Microelectromech. Syst. 19, 885–894 (2010). [Google Scholar]
- 11.Tanase M., Winterstein J., Sharma R., Aksyuk V., Holland G., Liddle J. A., High-resolution imaging and spectroscopy at high pressure: A novel liquid cell for the transmission electron microscope. Microsc. Microanal. 21, 1629–1638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leenheer A. J., Sullivan J. P., Shaw M. J., Harris C. T., A sealed liquid cell for in situ transmission electron microscopy of controlled electrochemical processes. J. Microelectromech. Syst. 24, 1061–1068 (2015). [Google Scholar]
- 13.Patterson J. P., Abellan P., Denny M. S. Jr, Park C., Browning N. D., Cohen S. M., Evans J. E. Gianneschi N. C., Observing the growth of metal–organic frameworks by in situ liquid cell transmission electron microscopy. J. Am. Chem. Soc. 137, 7322–7328 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Glaeser R. M., Specimen behavior in the electron beam. Methods Enzymol. 579, 19–50 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Abellan P., Parent L. R., Al Hasan N., Park C., Arslan I., Karim A. M., Evans J. E., Browning N. D., Gaining control over radiolytic synthesis of uniform sub-3-nanometer palladium nanoparticles: Use of aromatic liquids in the electron microscope. Langmuir 32, 1468–1477 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Abellan P., Moser T. H., Lucas I. T., Grate J. W., Evans J. E., Browning N. D., The formation of cerium(iii) hydroxide nanoparticles by a radiation mediated increase in local pH. RSC Adv. 7, 3831–3837 (2017). [Google Scholar]
- 17.Woehl T. J., Abellan P., Defining the radiation chemistry during liquid cell electron microscopy to enable visualization of nanomaterial growth and degradation dynamics. J. Microsc. 265, 135–147 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Schneider N. M., Norton M. M., Mendel B. J., Grogan J. M., Ross F. M., Bau H. H., Electron–water interactions and implications for liquid cell electron microscopy. J. Phys. Chem. C 118, 22373–22382 (2014). [Google Scholar]
- 19.Ahmad N., Le Bouar Y., Ricolleau C., Alloyeau D., Growth of dendritic nanostructures by liquid-cell transmission electron microscopy: A reflection of the electron-irradiation history. Adv. Struct. Chem. Imaging 2, 9 (2017). [Google Scholar]
- 20.Liao H.-G., Zherebetskyy D., Xin H., Czarnik C., Ercius P., Elmlund H., Pan M., Wang L.-W., Zheng H., Facet development during platinum nanocube growth. Science 345, 916–919 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Sutter E., Jungjohann K., Bliznakov S., Courty A., Maisonhaute E., Tenney S., Sutter P., In situ liquid-cell electron microscopy of silver-palladium galvanic replacement reactions on silver nanoparticles. Nat. Commun. 5, 4946 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Parent L. R., Robinson D. B., Cappillino P. J., Hartnett R. J., Abellan P., Evans J. E., Browning N. D., Arslan Ilke, In situ observation of directed nanoparticle aggregation during the synthesis of ordered nanoporous metal in soft templates. Chem. Mater. 26, 1426–1433 (2014). [Google Scholar]
- 23.Woehl T. J., Evans J. E., Arslan I., Ristenpart W. D., Browning N. D., Direct in situ determination of the mechanisms controlling nanoparticle nucleation and growth. ACS Nano 6, 8599–8610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woehl T. J., Park C., Evans J. E., Arslan I., Ristenpart W. D., Browning N. D., Direct observation of aggregative nanoparticle growth: Kinetic modeling of the size distribution and growth rate. Nano Lett. 14, 373–378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park C., Huang J. Z., Ji J. X., Ding Y., Segmentation, inference and classification of partially overlapping nanoparticles. IEEE Trans. Pattern Anal. Mach. Intell. 35, 669–681 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Park C., Woehl T. J., Evans J. E., Browning N. D., Minimum cost multi-way data association for optimizing multitarget tracking of interacting objects. IEEE Trans. Pattern Anal. Mach. Intell. 37, 611–624 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Liu Y., Tai K., Dillon S. J., Growth kinetics and morphological evolution of ZnO precipitated from solution. Chem. Mater. 25, 2927–2933 (2013). [Google Scholar]
- 28.Jungjohann K. L., Evans J. E., Aguiar J. A., Arslan I., Browning N. D., Atomic-scale imaging and spectroscopy for in situ liquid scanning transmission electron microscopy. Microsc. Microanal. 18, 621–627 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Xia Y., Xiong Y., Lim B., Skrabalak S. E., Shape-controlled synthesis of metal nanocrystals: Simple chemistry meets complex physics? Angew. Chem. Int. Ed. Engl. 48, 60–103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thanh N. T. K., Maclean N., Mahiddine S., Mechanisms of nucleation and growth of nanoparticles in solution. Chem. Rev. 114, 7610–7630 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Kennedy E., Nelson E. M., Tanaka T., Damiano J., Timp G., Live bacterial physiology visualized with 5 nm resolution using scanning transmission electron microscopy. ACS Nano 10, 2669–2677 (2016). [DOI] [PubMed] [Google Scholar]
- 32.de Jonge N., Peckys D. B., Kremers G. J., Piston D. W., Electron microscopy of whole cells in liquid with nanometer resolution. Proc. Natl. Acad. Sci. U.S.A. 106, 2159–2164 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bammes B. E., Jakana J., Schmid M. F., Chiu W., Radiation damage effects at four specimen temperatures from 4 to 100 K. J. Struct. Biol. 169, 331–341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson G. P., Gan L., Jensen G. J., 3-D ultrastructure of O. tauri: Electron cryotomography of an entire eukaryotic cell. PLOS ONE 2, e749 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hahn M., Seredynski J., Baumeister W., Inactivation of catalase monolayers by irradiation with 100 keV electrons. Proc. Natl. Acad. Sci. U.S.A. 73, 823–827 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peckys D. B., Mazur P., Gould K. L., de Jonge N., Fully hydrated yeast cells imaged with electron microscopy. Biophys. J. 100, 2522–2529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Jonge N., Peckys D. B., Live cell electron microscopy is probably impossible. ACS Nano 10, 9061–9063 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Dukes M. J., Peckys D. B., de Jonge N., Correlative fluorescence microscopy and scanning transmission electron microscopy of quantum-dot-labeled proteins in whole cells in liquid. ACS Nano 4, 4110–4116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/4/eaaq1202/DC1
fig. S1. Multiwindow chips compatible with commercial LC-TEM holders.
fig. S2. Additional experimental data from Fig. 2.
fig. S3. Imaging path and thickness of multiwindow devices for no flow versus flow.
fig. S4. Low-dose imaging of biological samples.
fig. S5. Improved sampling of nonuniform samples with multiwindow areas.
fig. S6. Illustration of microfabrication workflow.
Supplementary protocol for fabrication of multiwindow devices
movie S1. Demonstration of in situ liquid cell “low-dose” imaging.


