Abstract
Lung cancer continues to be the leading topic concerning global mortality rate caused by can-cer; it needs to be further investigated to reduce these dramatic unfavorable statistic data. Non-coding RNAs (ncRNAs) have been shown to be important cellular regulatory factors and the alteration of their expression levels has become correlated to extensive number of pathologies. Specifically, their expres-sion profiles are correlated with development and progression of lung cancer, generating great interest for further investigation. This review focuses on the complex role of non-coding RNAs, namely miR-NAs, piwi-interacting RNAs, small nucleolar RNAs, long non-coding RNAs and circular RNAs in the process of developing novel biomarkers for diagnostic and prognostic factors that can then be utilized for personalized therapies toward this devastating disease. To support the concept of personalized medi-cine, we will focus on the roles of miRNAs in lung cancer tumorigenesis, their use as diagnostic and prognostic biomarkers and their application for patient therapy.
Keywords: ncRNAs, Lung cancer, Piwi-interacting RNA, Small nucleolar RNA, Long non-coding RNA, Circular RNA, RNA biomarkers
1. INTRODUCTION
Lung cancer has a leading mortality rate with regard to examining cancer on a global scale [1]. It is responsible for one out of every five deaths related to cancer and has an overall ratio of mortality to incidence of 0.87 [2]. This ratio not only encompasses lung cancer’s association to death but also its high reoccurrence [3] and metastatic potential [4]. The most representative and well-studied type of lung cancer is Non-Small Cell Lung Cancer (NSCLC), which accounts for 85% of cases, followed by Small Cell Lung Cancer (SCLC) for the remaining 15% of cases. For NSCLC, the three subtypes can be differentiated by: glandular location and production of significant amounts of mucus for adenocarcinoma (ADC); squamous cell location and keratin formation for Squamous Cell Carcinoma (SQC); and the presence of giant multinuclear cells for large cell carcinoma [5].
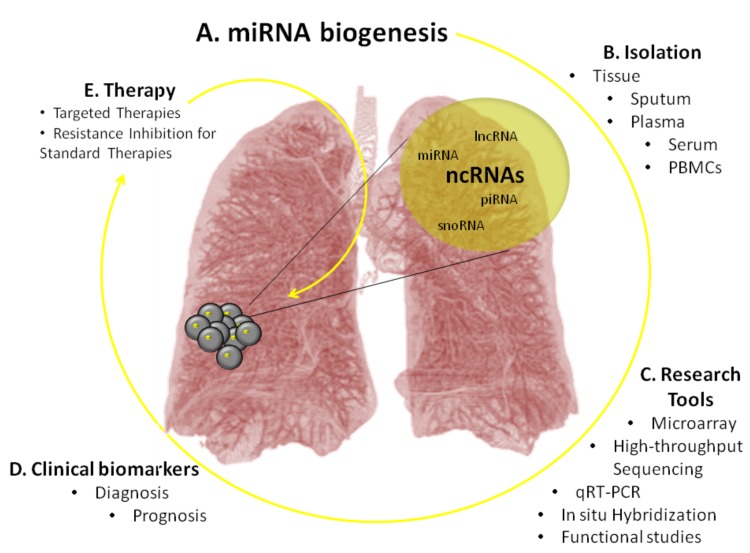
An estimated 98% of the genome is comprised of non-coding RNA, and what is called the “dark matter” of the genome is being slowly but surely understood [6]. Non-coding RNAs (ncRNAs) comprise a group of RNA molecules that are not translated into proteins. Non-coding RNAs are involved in diverse lung cancer cellular processes, some being of major functional importance and others having regulatory functions or pathological implications [7, 8]. Fig. (1) illustrates the methodology developed for the application of non-coding RNAs towards therapy. The process starts with determining what kind of patient sample can be used to isolate the particular non-coding RNA. Next using the current techniques, like qRT-PCR, high-thorough sequencing [9] or microarray [10, 11], the sample is analyzed to obtain quantifiable molecular profiles. These profiles are compared and patterns emerge that allow for the creation of biomarkers. These biomarkers can give insight into patient diagnosis or prognosis, as well as can be targeted for therapy.
Fig. (1).
The impact of ncRNAs on lung cancer patient management. Part (A) refers to the identification of specific ncRNAs and their expression levels, which are altered in lung cancer cells. Next in part (B) these molecules are isolated from patient samples of tissue, sputum, plasma, serum or PBMCs. Lastly, part (C) represents the methods used to generate molecular profiles (qRT-PCR, high-throughput sequencing and microarray) for each ncRNA. These profiles are utilized for the development of clinical biomarkers for diagnosis and prognosis (D). The combination of molecular profiles with diagnostic/prognostic biomarkers provides the basis for developing novel therapies (E) in patient-specific lung cancer.
Non-coding RNAs can be classified based on their function [11]. One class consists of housekeeping ncRNAs that are constantly expressed and perform genetic functions within the cell like telomerase RNA, transfer RNA, ribosomal RNA or small nuclear RNA (snoRNA) [12, 13]. However, the major group consists of regulatory ncRNAs that are only occasionally expressed like microRNA (miRNA), small interfering RNA (siRNA), small nucleolar RNA (snoRNA), piwi-interacting RNA (piRNA), long non-coding RNA (lncRNA), circular RNAs, extracellular RNAs (exRNA) and ultra-conserved regions [14-16].
Non-coding RNAs with regulatory functions are involved in physiological processes and have also been associated with a large number of diseases [13, 17]. These RNAs are of great clinical interest due to their expression profiles that correlate with the development and progression of different pathologies [10, 18-20]. This gives ncRNAs a great potential to be utilized as novel biomarkers for the diagnosis and prognosis of disease as well as therapeutic targets [21]. Moreover, they have been associated with numerous types of cancers, including lung cancer, where they can act either as oncogenes or tumor suppressors [15].
MiRNAs are small, 19-25 nucleotide long ncRNA, involved in gene regulation [11, 12]. They have also been used as targets for anticancer drug therapy [22] where oligonucleotide-based strategies were developed to either restore miRNA levels or block miRNA function [23]. These specific ncRNAs have proven to be suitable biomarkers for the diagnosis and prognosis of disease [24-26].
Another less known, large ncRNA subclass that plays a role in gene regulation is lncRNAs; called long non-coding RNAs because they are 200-base pairs long. Although their mode of action remains unclear, aberrant expression of the genes for these RNAs has been found to lead to the development of many diseases including lung cancer [27, 28]. In human lung cancer, researchers have identified one particular lncRNA, called metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which is conserved amongst all mammals. MALAT1 is highly expressed in the nucleus and has been shown to influence alternative splicing through its inverse relation with dephosphorylation of serine/arginine splicing factors in the cervical cancer cell line HeLa [29]. In lung cancer, MALAT1 is overexpressed and has a strong influence on metastasis. Furthermore, MALAT1 was found to be a strong regulator of NSCLC migration and invasion. This high MALAT1 expression in NSCLC indicates poor prognosis in patients suffering from this cancer, making it an important biomarker as well as a possible target for therapy [30].
Additional groups of ncRNAs called small nucleolar RNAs and piwi protein associated RNAs, have also been specifically identified in lung cancer. Firstly, small nucleolar RNAs or snoRNAs are a part of complexes responsible for sequence specific methylation and pseudouridylation of ribosomal RNAs [10, 31]. Secondly, “PIWI protein associated RNAs” or piRNAs have been associated with cancer related expression in somatic tissue, whereas normally these small ncRNAs are involved in transposon suppression in animal gonads [32]. Comparatively, in a recent study done on NSCLC investigating PIWI family expression during human lung embryogenesis, researchers had some interesting findings. Firstly, using 5-Aza-dC and bisulfate sequencing, the researchers observed that PIWIL1 expression was regulated by methylation. Secondly, the in silico study performed revealed a significant association between PIWIL1 expression and stem cell expression. Lastly, the multivariate analysis determined PIWIL1 expression as an independent prognostic factor indicating shorter relapse time and lower survival [33].
This review focuses on the non-coding RNAs found in lung cancer that have therapeutic applications and those that can serve as efficient biomarkers for the diagnosis and prognosis of this disease.
2. IMPORTANCE OF MICRO-RNA FOR LUNG CANCER
Table 1 summarizes all the findings on miRNAs mentioned in this chapter that serve as potential biomarkers for diagnosis and prognosis.
Table 1.
List of lung cancer miRNAs used as diagnostic and prognostic biomarkers based on their expression, chromosome location, function and their target genes controlling cancer mechanisms.
| MiRNA | Expression | Chromosome Location | Function | Cancer Associated Mechanism | Target Genes | Biomarker Role | References |
|---|---|---|---|---|---|---|---|
| miR-17-92 cluster | increased | 13q31.3 | Oncogene | proliferation | PTEN and RB2 | diagnostic | [64-66] |
| miR-21 | increased | 17q23.1 | Oncogene | proliferation | hMSH2 | diagnostic and prognostic | [108, 111, 117, 118-120] |
| miR-25 | increased (tissue) decreased (serum) |
7q22.1 | Oncogene/ tumor suppressor |
proliferation | CDC42/ (predicted) MAPK pathway genes | diagnostic | [67, 69, 96, 110] |
| miR-31 | increased | 9p21.3 | Oncogene | proliferation | BAP1 | diagnostic and prognostic | [77-79] |
| miR-155 | increased | 21q21.3 | Oncogene | proliferation | FoxO1 | diagnostic | [72-74, 76, 120] |
| miR-182 | increased | 7q32.2 | Oncogene | proliferation | PDCD4 | diagnostic | [72-74, 80, 114] |
| miR-197 | increased | 1p13.3 | Oncogene | apoptosis | FUS1 | diagnostic | [72, 75] |
| miR-210 | increased | 11p15.5 | Oncogene | angiogenesis | SDHD | diagnostic and prognostic | [98, 114, 117, 118, 120] |
| miR-223 | increased | Xq12 | Oncogene | acquired resistance | IGF1R/PI3K/Akt | diagnostic | [96, 185] |
| miR-328 | increased | 16q22.1 | Oncogene | metastasis | PRKCA | diagnostic and prognostic | [99] |
| miR-378 | increased | 5q32 | Oncogene | metastasis, angiogenesis, tumor growth | SuFu and Fus-1 | diagnostic and prognostic | [99, 100] |
| Let-7 family | decreased | 9 different locations | Tumor suppressor | apoptosis, proliferation, metastasis | CMYC, KRAS, HMGA2, ITGB3/MAP4K3, TGFBR3, CyclinD2, CDK6, CDC25A, IGF2BP1/IMP-1, IGF2BP2//IMP-2, Lin28, Lin41 | diagnostic | [66, 72, 91-95, 110] |
| miR-29 family | decreased | chr. 1 and 7 | Tumor suppressor | apoptosis | DNMT3A and DNMT3B | diagnostic | [90, 106] |
| miR-125a-5p | decreased | 19q13.41 | Tumor suppressor | metastasis | IL16 and CCL21 | diagnostic | [67, 68] |
| miR-126 | decreased | 9q34.3 | Tumor suppressor | angiogenesis, invasion, metastasis | Crk, VEGF | diagnostic and prognostic | [67, 70, 71, 80-87, 114] |
| miR-128 | decreased | 2q21.3 | Tumor suppressor | angiogenesis | VEGF-C | diagnostic | [89] |
| miR-133b | decreased | 6p12.2 | Tumor suppressor | proliferation, metastasis | EGFR | diagnostic and prognostic | [87, 88] |
2.1. miRNAs Definition and its Biogenesis
MicroRNAs are almost completely complementary with sequences of mRNA, giving them many, even thousands, of targets [34]. They act by binding to the target mRNAs, thereby, blocking translation at the ribosomal level. In addition, miRNAs can also regulate mRNA translation by promoting transcript degradation [35]. Their abilities can be explained from the fact that miRNAs precursors are commonly found in clusters throughout genome, specifically the intergenic regions and introns of protein-coding genes [36, 37]. These pre-miRNAs are then transcribed by RNA polymerase II or III. The intergenic miRNAs can be transcribed by either pol II or III while coding-intronic miRNAs are transcribed by pol II [37]. In addition, pol III was found to specifically synthesize small non-protein coding RNAs that are linked to regulating cell cycle and growth. While investigating a C19MC miRNAs cluster on human chromosome 19, pol III was found to specify transcription to microRNA that contained interspersed Alu repeats [38, 39]. Furthermore, a recent study by Gu et al. (2010) confirmed this specificity, identifying 68 miRNAs that were found to be transcribed by pol III in an Alu-dependent manner [40].
Subsequently, these primary miRNAs transcripts are further processed into the ~70 nucleotides long pre-miRNAs by microprocessor complex, comprised of RNase III endonuclease Drosha and the double-stranded RNA binding protein DiGeorge syndrome critical region gene 8 (DGCR8). This Drosha- DGCR8 microprocessor was recently described as self-regulating; the protein-protein interaction causes DGCR8 to stabilize Drosha as well as its protein levels while Drosha when a part of the microprocessor complex negatively regulates DGCR8 mRNA post-transcriptionally [41]. The nuclear cleavage of these pre-miRNAs by the microprocessor protein complex begins the maturation process. The most current model of microRNA processing suggests that DGCR8 recognizes the ssRNA-dsRNA junction followed by directing Drosha to the specific cleavage site where it cuts to make the pre-miRNAs (miRNAs hairpin precursor) [42-45].
It is important to note that for coding-intronic miRNAs, a different process occurs after transcription from pol II as a part of the pre-mRNA. It was initially reported that introns were excised out of the pre-mRNA followed by debranching by spiceosomal components. Recent studies propose a nuclear pathway hypothesis for those small excised debranched introns containing miRNAs. These introns, called mirtrons, were discovered in both invertebrates and mammals. In invertebrates, the mirtrons do not undergo microprocessor cleavage and enter maturation during the nuclear export to the cytoplasm for further processing [46-51]. Despite having been identified, it is unclear whether mammalian mirtrons undergo the same functional maturation pathway. An alternate miRNAs excision hypothesis states that there are pre-existing marked splice sites on the pre-mRNA that the microprocessor excises to liberate the pre-miRNAs while splicing continues normally to produce mature mRNA [46, 51, 52].
This ~70 nucleotide pre-miRNAs and possible mintron are then exported from the nucleus into the cytoplasm by the nucleocytoplasmic transport factor Exportin-5 and RanGTP. This transport protein has a dual function of preventing nuclear degradation as well as facilitating translocation to the cytoplasm [53-57].
In the cytoplasm, the pre-miRNAs are cut into a nucleotide duplex that is around 20 nucleotides long by the ribonucleoprotein complex that consists of Dicer and TRBP.
This cleavage pre-miRNAs process has been proposed to occur in either a single or double cleavage. The recent model of the single cleavage pre-miRNAs processing states that upon Ago2 binding to pre-miRNAs, the cleavage of the passenger strand occurs on the precursor hairpin 10 nucleotides upstream of the guide strand 5’end which facilitates unwinding. The remaining nucleotides undergo polyuridylation and nuclease-mediated trimming to generate mature miRNAs [58]. The double cleavage pre-miRNAs process proposed by Diedrichs and Haber (2007) states that Dicer, TRBP and Ago2 form a protein complex that recognizes and binds the pre-miRNAs through the PAZ domain of Dicer and Ago2 [59]. All these proteins have been shown to interact, however, as of yet there is no evidence demonstrating that all these proteins act as a complex.
These duplex stranded miRNAs then become a part of the RISC complex. In this complex, the RNA duplex is unwound and one of the strands becomes the mature miRNAs that guide the complex to the 3’ UTR of target mRNA, directing the silencing of the target mRNA [43, 60, 61]. The core components of the RISC complex are Dicer, Ago2, PACT and TRBP. It is important to note that of all the Argonaute proteins which can associate with miRNAs functioning to silence genes, only Ago2 has the “slicer” activity capable of cleaving mRNA [62]. Recent studies suggest that the unwinding associated with RISC activation in human miRNAs pathway is caused by RNA Helicase A [63]. This entire process is illustrated in detail in Fig. (2).
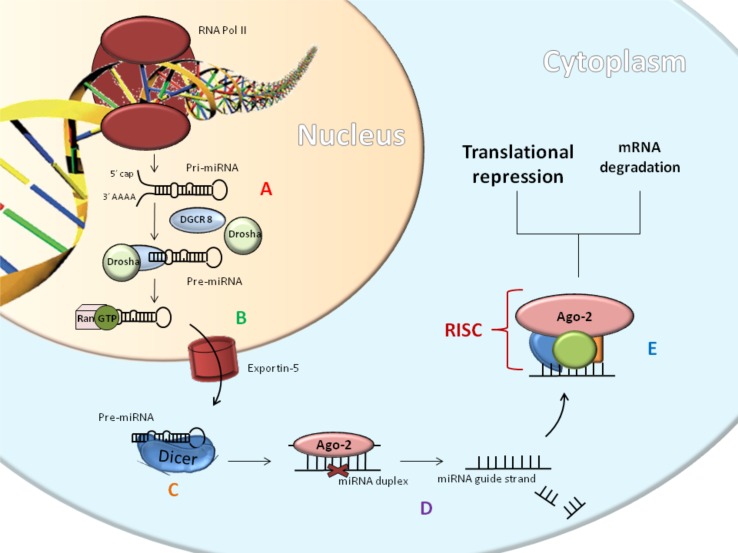
Fig. (2).
MiRNAs biogenesis: the RNA Polymerase transcribes the miRNAs gene and then Drosha and DGCR8 cleave the resulting primary transcript into the pre-miRNAs (A). Exportin 5 (B) carries out export to cytoplasm, where the RNase III endonuclease Dicer processes it. This forms duplex strands; the first strand is called the passenger strand and other is called the guide strand (C). The guide strand becomes incorporated into a complex called the miRNA-induced silencing complex, or RISC. The most important protein in this complex is Argonaute 2 (Ago-2) because it cleaves the miRNAs. The aforementioned Dicer is also a component of the RISC complex. RISC complex is involved in gene silencing through the processes of mRNA degradation or translational repression (D-E).
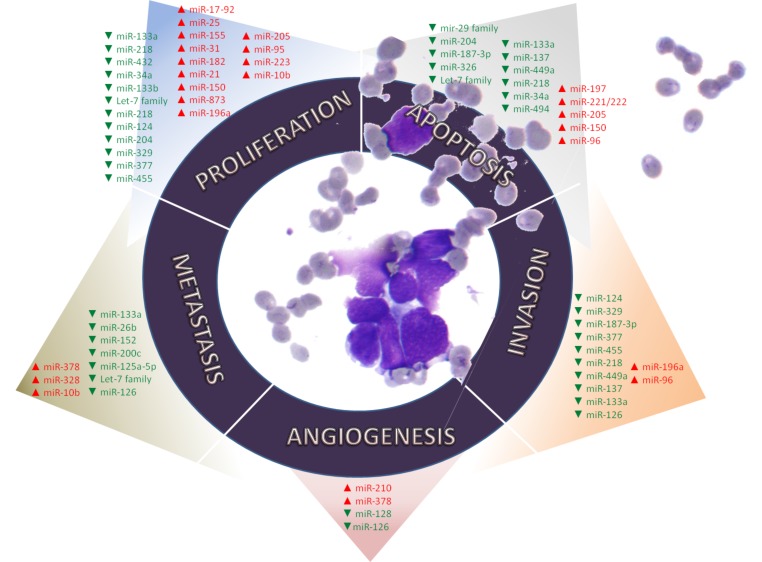
As shown in Fig. (3), these RNAs are of considerable interest because they have been identified as regulatory elements involved in the hallmarks of cancer, majority of which have been demonstrated in lung cancer.
Fig. (3).
MiRNAs associated to their corresponding lung cancer hallmarks. These hallmarks are cellular mechanisms acquired during tumor transformation and include proliferation, apoptosis, angiogenesis, invasion and metastasis.
2.2. miRNAs as Biomarkers for Prognosis and Diagnosis
At the center of the essential functions for the development of lung cancer is a miRNAs cluster known as miR-17-92; its location is at 13q31.3 in the C13orf25 gene within intron 3. The miR-17/92 cluster is overexpressed, particularly, in the SCLC subtype [64]. It was demonstrated that MYC oncogenes regulate this miRNAs cluster facilitating its oncogenic activity [65]. MYC and E2F1 positively regulate each other, thus, when MYC induces miR-17-92 expression, it negatively regulates E2F1; E2F1 expression levels regulate the entry into S-phase and its overexpression results in apoptosis induction. In addition, both MYC and E2F families transactivate miR-17-92 resulting in the suppression of E2F family expression by exerting a negative feedback loop [66].
In another study, three specific miRNAs from patient serum samples were useful in early detection of lung cancer; miR-125a-5p, miR-25, and miR-126 were found to be downregulated in serum from cancer patients versus controls [67].
This group of miRNAs had high sensitivity and specificity (0.936 AUC value with 87.5% sensitivity and 87.5% specificity) [67]. Firstly, miR-125a-5p was found to be involved in invasion and migration processes [68]. Secondly, miR-25 was reported to be involved in proliferation that correlated to high expression in NSCLC tissue samples [69]. Lastly, miR-126 regulates invasion and migration [70] as well as angiogenesis [71].
Zheng et al. identified three miRNAs, miR-155, miR-197, and miR-182, in patients’ plasma that showed high specificity in detecting lung cancer in its early stage [72]. Each of the miRNAs were found to regulate one hallmark of cancer: miR-155 and miR-182 were involved in proliferation [73, 74], and miR-197 in apoptosis [75]. A recent study has demonstrated how miR-155 induces drug resistance to multiple chemotherapeutic agents in lung cancer patients. Specifically, miR-155 and TP53 are linked in a negative feedback loop that leads to high expression of miR-155 and low TP53 expression. This association significantly correlated to shorter survival in lung cancer [76].
MiR-31 is an important miRNA receiving attention in the research of lung cancer pathology. MiR-31 can serve as a biomarker for the diagnosis of lung cancer from peripheral blood samples and its high expression is associated with poorer survival in lung cancer patients [77]. Additionally, miR-31 is notably overexpressed in adenocarcinoma lung patients who have lymph node metastasis, thereby, making this miRNA a good prognostic factor for poor survival in ADC lung patients [78]. A target for miR-31 is BAP1 which is a tumor suppressor gene involved in the proliferation of lung cancer [79].
Distinguishing primary lung tumors from metastatic ones is a challenging aspect of diagnosis. MiRNAs could prove to be reliable biomarkers for this purpose. Barshack et al. conducted a study on 79 FFPE primary and metastatic lung tumor tissues. The microarray data indicated that miR-182 was specifically over-expressed in primary lung tumor tissues while miR-126 was more over-expressed in metastatic tumors [80]. This overexpression of miR-126 in metastatic tumors comes as a surprise. From the research conducted at this point, it is clear that miR-126 plays a tumor suppressor role in proliferation, metastasis and tumor growth. A majority of the research indicate that because of its tumor suppressive effects miR-126 is often downregulated in breast, gastric, colon and lung cancers [81-84]. Conversely, miR-126 is also a key positive regulator of angiogenic growth factors like VEGF-A thereby promoting angiogenesis. This is done by repressing the negative regulators of signal transduction pathways [85, 86]. Therefore, these contradictory results indicate that miR-126 has several functions and is highly tissue specific. These discrepancies could also be due to the fact that the researchers did not distinguish between miR-126 and its complementary analog miR-126* [81]. In conclusion, miR-126 needs to be further investigated with a higher degree of specificity.
A study on NSCLC tissue samples identified miR-126 and miR-133b expression levels as potential biomarkers for this malignancy. Low levels of these RNAs have been correlated to tumor progression, metastasis and shorter overall survival in NSCLC patients [87, 88]. In addition, NSCLC tumorigenesis was linked to miR-128 because of its modulation of angiogenesis by directly targeting VEGF-C. It comes as no surprise that miR-128 is significantly downregulated in the NSCLC [89].
The miR-29 family (miR-29a, miR-29b and miR-29c) is downregulated in the NSCLC. The proposed mechanism of action for this family is the suppression of the WNT signaling pathway through the demethylation of WIF-1; miR-29s positively regulate WIF-1 expression by inhibiting the methylation of its promoter [90].
Many reports of deregulated miRNAs in lung cancer concern the let-7 family [91-93]. The Let-7 miRNAs family is downregulated in lung cancer causing its tumor suppressor function to be lost, which is important because of their regulation of RAS [94]. Therefore, concerning early stage detection, miRNAs expression levels dependent on let-7 can have significant positive clinical implications. For example, Wozniak et al. identified a group of 24 miRNAs, which included let-7, in samples from plasma for the efficient detection of stage I - III NSCLC. This group had a high AUC of 0.92 after screening a number of 742 miRNAs in 100 patients compared to 100 controls. In conclusion, the panel described in the study could serve as an efficient tool for the detection of both types of NSCLC [95].
The miRNAs profile in the serum of healthy versus cancer patients is distinguishably different, as demonstrated by Chen et al. in 2008 investigating the characterization of miRNAs as biomarkers. Interestingly, they found 28 miRNAs that were missing and discovered 63 new miRNAs expressed in lung cancer patients when compared to the control serum miRNAs expression profile. Two miRNAs in particular, miR-25 and miR223, were further studied for their use as biomarkers for NSCLC using Solexa sequencing and qRT-PCR techniques. The study also implemented an independent trial consisting of 152 lung cancer patients and 75 controls, which concluded miR-25 and miR-223, significantly overexpressed in the serum of lung cancer patients [96]. Incidentally, the study also found no difference in the serum expression levels of let-7a between lung cancer sera and normal sera [72]. With regard to serum biomarkers, a phase I/II biomarker study was implemented by Hennessey et al. (2010), identifying miR-15b and miR-27b as sensitive and effective biomarkers for early detection of NSCLC [97].
Additionally, another diagnostic and prognostic biomarker is miR-210, which was shown to be highly expressed in the late stages of lung cancer. One study conducted using ADC tissue samples and A549 cell line, examined the relationship between miR-210 and hypoxia. It was found that miR-210 was overexpressed in late stage NSCLC and it targeted SDHD, subunit D of succinate dehydrogenase complex (SDH). Furthermore, miR-210-mediated targeting of SDHD activated hypoxia-induced factor 1 (Hif-1). Previous studies have linked the loss-of-function of SDH to HIF-1 activation. Thus, the miR-210-mediated targeting of SDHD and subsequent HIF-1 activation can act as an intracellular 02 sensor. This means that this molecular mechanism may have a regulatory role in the phenomena causing increased glycolysis in tumors, known as the ‘Warburg effect’ [98].
Some miRNAs have been studied for their predictive role in brain metastasis in patients who underwent complete tumor resection for Stage III NSCLC. Specifically, the levels of miR-328 and miR-378 could be used to predict brain metastasis in patients with NSCLC. The mechanism of miR-328 involves the regulation of PRKCA [99], while the molecular mechanism of miR-378 is still unknown. However, investigations of miR-378 function on the primary glioblastoma cell line U87 revealed that it targets the tumor suppressors, SuFu and Fus-1 [100].
Table 2 describes the differential diagnosis of the NSCLC subtypes based on their specific biomarkers. From all the miRNAs mentioned, two miRNAs extracted from plasma samples that can potentially differentiate between the two NSCLC subtypes are miR-944 for operable SQC and miR-3662 for operable ADC [101].
Table 2.
List of miRNAs used for the differential diagnosis of histological subtypes of lung cancer, based on chromosome location and sample type.
| MiRNA |
Chromosome
Location |
Sample | Role | References |
|---|---|---|---|---|
| miR-944 | 3q28 | plasma | diagnostic operable SQC marker | [101] |
| miR-3662 | 6q23.3 | plasma | diagnostic operable ADC marker | [101] |
| miR-29b-3p, and MiR-19b-3p | 7q32.3 13q31 |
PBMCs | diagnostic/ higher sensitivity for SQC than ADC | [106] |
| miR-99b, miR-205 miR-102 miR-203, miR-202, and pre-miR-204 |
19q13.41 1q32.2 7q32.3 14q32 10q26.3 9q21.12 |
tissue | diagnostic group for differentiating SQC and ADC | [107] |
| miR-205 | 1q32.2 | FFPE tissue | SQC marker | [107-109] |
Generally speaking, it was found that the molecular marker genes that are altered in SCLC are associated with neuronal differentiation and growth such as human achaete-scute homolog 1 (ASCL1) or glycine receptor alpha1 subunit gene (GLRA1) [102, 103]. On the other hand, molecular marker genes in NSCLC were characterized by deregulated Epidermal Growth Factor Receptor (EGFR) or tumor-suppressor p53 [104, 105]. Similar specific miRNAs expressions were observed when comparing the various lung cancer subtypes. For example, differential miRNAs expression in peripheral mononucleated cells, involved in the first line of defense in cancer, could serve as a diagnostic tool for NSCLC, especially for its subtype, SQC. The panel of identified miRNAs is different from panels distinguished in other studies using tissue samples. It suggests that the miRNAs expression in PBMCs is not influenced by circulating tumor cells. The four miRNAs, miR-576-3p, miR-19b-3p, miR-29b-3p, and miR-29a-3p showed significantly different expression levels when compared to non-tumor samples. Furthermore, the expression levels of two of these miRNAs, miRs-19b-3p and 29b-3p, could serve as combined biomarkers for distinguishing between SQC and ADC [106]. In another study done by Yanaihara et al. in 2006, the researchers identified 43 miRNAs statistically different when comparing the phenotypic and histologic expression levels between NSCLC tumor and corresponding non-tumor tissue in 104 samples. Out of all of these miRNAs, six were found to be expressed differently between SQC and ADC: miR-205, miR-99b, miR-102, pre-miR-204, miR-203 and miR-202 [107].
By performing microarray and RT-PCR analyses, Solomides et al. (2012) studied miRNAs expression in archival formalin-fixed, paraffin-embedded tissues of NSCLC and normal lung tissues. Genome wide scans achieved by the microarray, revealed 34 differentially expressed miRNAs. Of these miRNAs, 5 distinctly identified miRNAs exhibited difference in normal tissue and NSCLC (either SQC or ADC), as well as SQC from ADC. In both tumor types when compared to normal tissue, miR-21 had significantly increased expression, while miR-451 and miR-486-5p expression levels were reduced [108]. SQC can be further differentiated from normal tissue and ADC by its increased miR-205 expression and decreased miR-26b expression [108]. A majority of the studies that have been done using microarray and RT-PCR have concluded that the overexpressed miR-205 is an accurate marker with high specificity for SQC [109]. Moreover, the altered expression of five miRNAs expression profiles (miR-25, miR-34c-5p, miR-191, let-7e and miR-34a) not only differentiated ADC from SQC but also strongly predicted survival for SQC [110]. Lastly, the overexpression of miR-21 has emerged to be a common characteristic of several types of cancers because of its involvement in the following mechanisms: proliferation, apoptosis, migration and invasion [111]. Therefore, it comes as no surprise that miR-21 is a prognostic factor in NSCLC linked to poor overall survival [60].
An alternative strategy for the diagnosis of NSCLC using multiple biomarkers is combining miRNAs expression in a panel together with other types of noncoding RNAs, like lncRNAs. A recent study identified a panel of four ncRNAs that were differentially expressed in NSCLC compared to healthy serum samples. The three miRNAs are miR-485-5p, miR-574-5p and miR-1254, while lncRNA was MALAT1 [112]. MALAT1 is a particularly important lncRNA due to its function as a strong regulator of gene expression and splicing governing lung cancer metastasis [113]. This notion of combined biomarker signatures for better diagnosis was also emphasized in another study done by Zhu et al. Four new serum miRNAs were identified for early NSCLC diagnosis as well as further distinguishing current smokers from this cohort were miR-182, miR-183, miR-210 and miR-126 [114].
Functions of miR-30a in the development of lung cancer are also not clear, emphasizing the need for more research to understand its role. Conversely, miR-30a does have a diagnostic value and some studies have conducted examination on plasma samples from patients with NSCLC. It was shown to have a high level of expression in NSCLC compared to healthy or benign samples. However, miR-30a does not have significant differences between various NSCLC pathological features [115]. Up till now, MiR-30a has been found to be down-regulated in ADC cell lines and its overexpression was reported to inhibit cell migration and invasion by down-regulating EYA2 [116].
It has become apparent that there are a lot of variations in the expression of miRNAs for lung cancer samples extracted from tissue, plasma and serum; especially the expression levels detected between tissue and blood samples. However, two bioinformatics studies independently performed meta-analysis on miRNAs expression in lung cancer from all sample types did find some consensus; miR-21 and miR-210 are the most consistently expressed miRNAs in lung cancer. Therefore, these miRNAs represent a high potential for effective early diagnosis of lung cancer especially as circulating blood signatures [117, 118].
2.3. Minimally Invasive Sputum Biomarkers
Detection of accurate biomarkers in the most minimally invasive way is a highly sought goal. Sputum miRNAs expression profiles provide this minimally invasive approach, making them the most suitable representative liquid biopsy for diagnosis. In 2010, Yu et al. identified a group of 4 miRNAs from sputum samples from patients with high sensitivity (80.6) and specificity (91.7) for differentiating between ADC and normal samples. miR-21, miR-486, miR-375 and miR-200b expression levels could form a biomarker group that facilitates improved early detection of ADC [119]. Moreover, Roa and colleagues similarly used sputum samples and performed a cluster analysis that identified a panel of 5 miRNAs (miR-21, miR-372, MiR-155, miR-143, miR-210) with 83.3% sensitivity and 100% specificity to NSCLC. This sputum miRNAs panel could be used as another biomarker group for the early detection of NSCLC [120].
3. PiWI-interacting rna
Piwi-interacting RNAs or piRNAs are associated with Piwi proteins that are germline specific Argonaute proteins. Together they form piRNA-induced silencing complexes that function to repress transposons [121]. Moreover, they have been found to function beyond both transposon silencing and the germline. Increasing evidence shows that they are always expressed in somatic tissue and that they have new functions in diseases such as cancer. Although their role has yet to be fully understood, the aberrant expressions of piRNA and piRNA pathway components have been linked with cancer development [122]. They could be involved in tumorigenesis through transcriptional level gene silencing by epigenetic mechanisms and the post-transcriptional level gene silencing could be analogous to the miRNAs silencing mechanism [123]. The piRNA expression patterns are specific to malignancies and their clinical groups, such that the subgroup specific piRNA expression profiles from tumors of the same organ correlate to clinical features of each tumor type [122]. This does give piRNAs some prognostic or diagnostic value as a biomarker. However, some piRNAs are consistently expressed in all tissue types, suggesting that in somatic cells there could be a conserved general function [124].
The identification by Mei et al. of more than 550 piRNAs or piRNA-Ls in Human lung Bronchial Epithelium (HBE) and NSCLC cell lines provides evidence of the role these molecules play beyond the regulation of transposons. In this study, the expression patterns for piRNAs/piRNA-Ls were found to be different between HBE and lung cancer cells, as well as between ADCs and SQCs. This suggests piRNAs or piRNA-Ls could be used as biomarkers or therapeutic targets. Furthermore, piR-L-163, the top downregulated piRNA in lung cancer cells, was found to play an important regulatory role in the activation of ERM by phosphorylating ERM (p-ERM) [125].
Liang and fellow researchers conducted a study where the silencing of a Piwi human homologue, Hiwi, led to the inhibition of tumor growth xenografts murine model. The xenografts tumor model was created by subcutaneous inoculation with a rare population of lung cancer SSCloAldebr stem cells. This is relevant to the stem cell theory of cancer because Hiwi provides a link between cancer and stem cells. Hiwi is overexpressed in a number of human tumors and is involved in stem cell renewal [126].
The Piwi-piRNA pathway in tumorigenesis is poorly understood but more evidence for the function of Piwi proteins in cancer is being discovered. In a recent study, one out of four PIWI protein homologs (PIWIL1, PIWIL2, PIWIL3 and PIWIL4) was investigated to determine how the PIWI protein interaction with piRNA facilitates tumorigenesis. Firstly, PIWIL2 was found to be significantly expressed in malignant NSCLC tissue compared with non-tumor tissue. Furthermore, the high levels of PIWIL2 were associated with lower overall survival in NSCLC, suggestive of this molecule’s value as a prognostic biomarker. In the end, Qu and colleagues established the mechanism of action of PIWIL2 in the progression of NSCLC cells by its ability to regulate CDK2 and Cyclin A. [127]. A second major piRNA involved Piwi-piRNA pathway in tumorigenesis is piR-55490. Normally this piRNA is downregulated in lung cancer, but in a study where its expression levels were restored, there was inhibition of tumor growth through the suppression of Akt/mTOR pathway activation. These findings prove that piRNA can directly target an oncogene mRNA, thereby, extending the understanding of piRNA function [128]. This research shows that PIWI proteins and their associated RNAs represent a novel area of study for understanding cancer biology and for adding new tools in cancer diagnostics, prognostics and therapy.
4. Long non-coding rna
LncRNAs are the longest of the RNA molecules and play a predominant role in gene regulation. Although their mode of action remains largely unclear, aberrant expression of the genes for these RNAs has been found to lead to disease. The functions that can be attributed to lncRNAs are diverse, and include: transcription regulation, epigenetic regulation, chromatin remodeling, and post-transcriptional processing [27]. A number of lncRNAs have been identified whose dysregulation are involved in the development and progression of lung cancer and that can be seen in Table 3.
Table 3.
List of lncRNAs associated with lung cancer organized by chromosome location, regulation and the corresponding cancer-associated mechanism.
| LncRNA Name | Chromosome Location | Expression | Cancer-associated Mechanism | References |
|---|---|---|---|---|
| MALAT1 | 11q13 | increased | invasion/migration/metastasis | [28-30, 112, 113, 129-131, 145] |
| PVT1 | 8q24 | increased | metastasis | [142] |
| HOTAIR | 12q13.13 | increased | invasion/metastasis | [132-135, 145] |
| MVIH | 10q22 | increased | invasion | [136] |
| CARLo-5 | 8q24.21 | increased | metastasis/poor prognosis | [137] |
| H19 | 11p15.5 | increased | proliferation | [138] |
| SOX2ot | 3q26.33 | increased | proliferation/poor prognosis | [139, 140] |
| Linc00673 | 17q24.3 | increased | proliferation | [141] |
| RGMB-AS1 | 5q21.1 | increased | proliferation, migration, invasion | [144] |
| TATDN1 | 8q24.13 | increased | metastasis | [146] |
| CCAT2 | 8q24.21 | increased | metastasis/poor prognosis | [142, 143, 145] |
| PANDAR | 6p21.2 | decreased | proliferation, apoptosis | [147] |
| HMlincRNA717 | 18p11.22 | decreased | metastasis | [148, 149] |
Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1), also called noncoding Nuclear-Enriched Abundant transcript 2 (NEAT2), is a conserved lncRNA that is upregulated in many types of human cancers. Located in chromosome 11 at region q13, MALAT1 is involved in invasion and migration in NSCLC [30]. It has been associated with brain metastasis in NSCLC by inducing Epithelial-Mesenchymal Transition (EMT) [129]. MALAT1 has also demonstrated to be a great candidate for a blood-based biomarker used in the early detection of lung cancer. Firstly, Yao et al. found a four serum biomarker signature containing MALAT1 and three other proteins that showed a high accuracy for early stage detection for NSCLC [130]. Secondly, Weber and colleges reported that this long noncoding RNA is an effective biomarker in the early diagnosis of NSCLC that is also non-invasive [131]. Furthermore, MALAT1 also represents a promising therapeutic target because treatment with antisense oligonucleotides in murine xenograft models proved effective at inhibiting lung cancer metastasis [113]. Another lncRNA which is oncogenic and upregulated in lung cancer is HOX transcript antisense intergenic RNA (HOTAIR). This lncRNA functions as a gene expression repressor through the recruitment of chromatin modifiers and has proven to be a good biomarker correlating to poor prognosis and metastasis [132, 133]. In SCLC, it was linked with relapse and lymphatic invasion [134]. Whereas in NSCLC, it is correlated to lymph node metastasis and poor overall survival [135]. The best explanation to the way in which HOTAIR functions in NSCLC is by altering matrix metalloproteinases and HOXA5 expression; however, it does not deregulate EMT-induced marker expression [132].
A study on the role of lncRNAs in NSCLC development indicated that increased levels of lncRNA MVIH correlated to TMN stages, tumor size and lymph node metastasis. Moreover, MVIH overexpression in NSCLC patients had relatively poor prognosis and if we take all this information together, it could be a good prognostic marker for NSCLC. Ectopic overexpression of MVIH was found to promote cell proliferation and invasion by regulating MMP2 and MMP9 protein expression [136]. Another frequently upregulated lncRNA that arguably facilitates the development of NSCLC is CARLo-5. Principally, knockdown of this lncRNA led to lower the expression of EMT-related transcription factors in lung cancer cell lines [137]. With regard to the both development and progression of NSCLC, H19 was identified through its upregulated expression. Cui et al. researched the c-MYC regulation of the lncRNA H19 expression in this type of lung cancer. They found that H19 levels were higher in cells transfected with pcDNA3.1-c-Myc than in those transfected with empty vector plasmid pcDNA3.1. In addition, cells transfected with c-Myc siRNA had lower levels of H19. Luciferase reporter assays allowed Cui and colleges to conclude that c-Myc regulated the expression of lncRNA H19 by targeting its promoter. It was proposed that the regulatory function of lncRNA H19 in lung cancer was achieved by the down regulation of miR-107, which is involved in cell cycle regulation [138].
SOX2 overlapping transcript (SOXot) is a cancer-associated lncRNA that encodes a number of different transcripts spliced alternatively. Its roles have yet to be completely revealed in lung cancer but studies suggest that it is involved in cell, proliferation making it a useful prognosis indicator. Hou et al. investigated the expression levels of Sox2ot in 83 human lung cancer tissues and showed that in these samples, the level of Sox2ot was significantly increased versus non-tumor samples [139]. Additionally, the research also suggested that this lncRNA could serve as a useful marker for distinguishing between SQC and ADC because the levels of expression were higher in SQC. Furthermore, high levels of Sox2ot are predictive to be associated with a low overall survival [139]. In addition, an investigation of the preferential expression of different Sox2ot by qRT-PCR in twenty NSCLC patients revealed that two variants, Sox2ot4 and Sox2ot7, may have an oncogenic role in this type of cancer [140].
The notion of lncRNAs oncogenic activity is further supported by recent discovery of the long intergenic noncoding RNA 00673; it was found to be responsible for promotion tumor proliferation in NSCLC. In vitro and in vivo investigations on the cancer related effects of linc00673 revealed that knockdown of this RNA resulted in inhibition of cell proliferation. Moreover, overexpression of linc00673 also correlated to increased tumor cell growth. Further research on its mode of action demonstrated that it represses NCALD (neurocalcin delta) via LSD1 interaction, an epigenetic repressor. Taking these findings together, promotes linc00673 as a possible therapeutic target in NSCLC [141].
Significant overexpression in tumor tissues versus adjacent healthy tissue exposed an lncRNA specific to adenocarcinoma called CCAT2. Moreover, Qiu et al. also established the value of CCAT2 as a prognostic biomarker for NSCLC invasion and lymph node metastasis [142]. Conversely, a recent study done by Chen et al. found that it also serves as an independent unfavorable factor in the prognosis of SCLC patients [143]. With regard to regulatory functions in ADC, it was recently recognized that RGMB-AS1 has a potential use in targeted therapy. Results obtained by Li et al. suggest that this lncRNA facilitates the progression of adenocarcinoma by controlling cell proliferation, migration and invasion. Therefore, it comes as no surprise that RGMB-AS1 is upregulated in this pathology. Silencing of this lncRNA in both in vivo and in vitro studies led to a decrease of migration and invasion but there was also an evidence of G1/G0 phase cell cycle arrest. The proposed mechanism that could be responsible for these findings is the modulation of expression of RGMB; RGMB is also known as DRAGON and is a part of the RGM family [144]. Lastly, a recent study investigating the characteristic genetic polymorphisms of CCAT2, as well as the aforementioned MALAT1 and HOTAIR revealed that these lncRNAs were significantly associated with lung cancer susceptibility and platinum-based chemotherapy response; thereby, concluding that these lncRNAs could function as clinical biomarkers [145].
In vivo and In vitro studies investigating the activity of the lncRNA TATDN1 in murine models and cell lines demonstrated its influence over cell growth. These studies also propose that this overexpression of TATDN1 could also be correlated to lung cancer metastasis. The research suggests the potential pathways are the Wnt/β-catenin signal pathways and PI3K/AKT/MTOR pathway [146].
Low expression of some of the lncRNAs is important in the progression of this disease. For example, lncRNA PANDAR interacts with p53, NF-YA and BCL-2 responsible for disease progression in lung cancer. Its role as an intermediary could provide a target for developing therapies. Moreover, lncRNA PANDAR has been demonstrated to be a prognostic biomarker; its downregulation correlates to a poor prognosis of NSCLC [147]. Due to the fact that HMlincRNA717 is normally down-regulated in gastric cancer [148], this lncRNA was studied for its role in NSCLC. Coincidentally, the expression levels of HMlincRNA717 were also significantly decreased in NSCLC, when a comparison of tumor tissue versus adjacent healthy tissue was performed. Moreover, the expression levels of this lncRNAs can be associated with histological grade lymph node metastasis. The strong association between the poor overall survival for patients suffering from NSCLC and the decreased HMlincRNA717 expression means it could serve as a good prognostic biomarker [149]. In addition, the lower overall survival rates and lymph node metastasis in NSCLC have also been associated with the upregulation of PVT1 [142]. While the underlying mechanism is not clear there could be an overlap of activity between HMlincRNA717 and PVT1.
In conclusion, the present knowledge of long non-coding RNAs suggests that they are involved in the progression of cancer, starting at proliferation and ending with metastasis; an illustration of this can be seen in Fig. (4). This is particularly important for determining metastatic potential and survival rates which govern the prognosis of lung cancer patients.
Fig. (4).
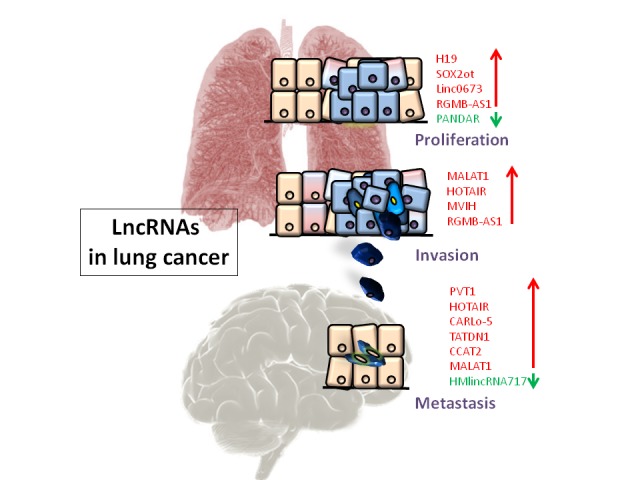
Respective lncRNAs involved in proliferation, invasion and metastasis in lung cancer, color-coded such that red represents upregulation and green represents downregulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
5. Small Nucleolar rna
Small nucleolar RNA activity has been correlated to the chemical modification of other RNAs. There are two main subdivisions for these snoRNA: firstly, the C/D box snoRNAs involved in methylation; and secondly, the H/ACA box snoRNAs associated with pseudouridylation.
Characterization of snoRNA expression in lung cancer by Next-Generation Deep Sequencing has generated certain characteristics that can facilitate the development of biomarkers for detection and prognostics in NSCLC. Using this Next-Generation Deep Sequencing technique, Gao et al. identified 458 snoRNAs. However, only snoRA47, snoRA68, snoRA78, snoRA21, snoRD28 and snoRD66 show the greatest potential for prognosis of NSCLC overall survival [150]. Another study identified snoRD33, snoRD66 and snoRD76 in plasma samples that could distinguish between NSCLC, healthy and chronic obstructive pulmonary disease samples. Higher expressions of these three snoRNAs produced 81.1% sensitivity and 95.8% specificity towards NSCLC when distinguishing between the samples [151].
One snoRNA was upregulated in lung cancer, snoRD78, was found to have oncogenic activity because it promoted cell invasion by the process of epithelial-mesenchymal transition. Therefore, high expression of this snoRNA could indicate poor prognosis in NSCLC patients and represent a potential therapeutic target [152]. Another similar example of a snoRNA with high expression levels in lung cancer is snoRD42. It was found to promote cell growth and proliferation. Coincidently, it can be found on region q22 on chromosome 1 which is frequently amplified in lung cancer. snoRD42 also represents a potential biomarker for prognosis and target for therapy because of it being associated with poor survival [153].
As previously mentioned, sputum biomarkers are very valuable tools in the clinical setting because they are noninvasive. Sputum snoRNA biomarkers were found to be useful in the diagnosis of NSCLC. snoRD66 and snoRD78 were identified as diagnostic biomarkers after qRT-PCR analysis produced a diagnostic specificity of 83.61% and sensitivity of 74.58%, from a sample set of 59 lung cancer patients and 61 cancer-free smokers [154].
6. Circular rna
Circular RNAs can simply be defined as RNAs with covalently closed continuous loops, whose unique structure makes them insensitive to ribonucleases. This promotes stability, long half-life period and resistance to enzymatic degradation which is clinically important because they can be found in tissues, serum, saliva and urine. Unlike linear RNAs and proteins, circRNAs are easier to extract and detect with a higher specificity. Changes to expression of circRNA have been detected using RNA sequences techniques for esophageal cancer, glioma, hepatocellular carcinoma and colorectal cancer. Moreover, expression levels of circRNAs were found to vary according to size, metastasis and TNM stage. In addition, testing circRNAs by rt-PCR and in situ hybridization was more specific and sensitive compared to detecting proteins via antigen-antibody reaction [155-162].
They have major functional roles: firstly, act as miRNAs sponges [163]; secondly, encode/translating peptides [164]; lastly, regulate RNA binding protein by modulating the expression of protein-encoding genes [165]. Of these, the main mechanism of circRNAs in tumor cells is its role as an miRNAs sponge. The several miRNAs binding domains give circRNA the “sponge” ability to regulate miRNAs as well as their downstream gene expressions through the ceRNA mechanism; critical to tumor progression [166].
ciRS-7 is the first and most reported circRNAs found to regulate miRNAs expression in tumor cells. It was found to be spliced from antisense transcript of CDR1 gene and contain more than 70 miRNAs binding sites, including for miR-7 and miR-671. CiRS-7 can downregulate miR-7 expression by binding as an miRNAs sponge. While the combination of ciRS-7 and miR-671 binding mediates the release of the absorbed miR-7 by the Ago2-mediated cleavage of ciRS-7. This suggests that ciRS-7 has a critical role for miR-671 diminishing the miR-7 expression. Not only does this explain increased levels of miR-7 in tumor cells but it proposes ciRS-7 promotion of vascularization, metastasis and reproduction of tumor cells by the increase in downstream target oncogenes EGFR and XIAP [167-171].
CircHIPK3 is an example of circRNA that was found to be expressed higher in prostate tumor samples than the normal ones. It is produced by the HIPK3 gene’s second exon, which was shown to affect the proliferation rate of tumor cells. Further investigations lead to the discovery that it binds to eighteen binding domains of nine miRNAs. More specifically, it influences proliferation of tumor cells by acting like an miRNAs sponge for miR-124 thereby inhibiting it [172].
Genomes of tumor cells are known to be unstable. Gene fusions in the case of circRNAs can lead to incorrectly spliced mRNAs. The PML/RARa and MLL genes for hematological malignancies exemplify this best. These gene fusions create f-circM9 and f-circPR which are associated to decreased apoptosis and drug sensitivity [166, 173]. However, the circRNAs created from gene fusion could be a potential target for drug treatments. The first example is for leukemia where fusion-circRNA was found to be chemoresistant; knocking out f-circM9 expression leads to apoptosis of the leukemia cells as well as decrease its drug resistance to Ara-C [173].
7. research focused on therapeutic applications
Non-coding RNAs could distinguish themselves as important players in cancer therapy [123]. Treatment of lung cancer is complex and dependent on the type of lung cancer. Standard treatments used include chemotherapy, radiotherapy or targeted therapy. Several miRNAs were found to be associated with resistance towards these standard therapies in lung cancer, as described in Table 4.
Table 4.
List of miRNAs with therapeutic implications associated with lung cancer based on expression level, chromosome location, function, cancer mechanism and the corresponding target gene for therapy.
| MiRNA | Expression | Location | Function | Cancer-Associated Mechanism | Target | References |
|---|---|---|---|---|---|---|
| miR-10b | increased | 2q31.1 | Oncogene | proliferation, metastasis | KLF4 | [209-213] |
| miR-95 | increased | 4p16.1 | Oncogene | radiosensitivity/ proliferation | SNX1 | [198, 199] |
| miR-96 | increased | 7q32.2 | Oncogene | cisplatin resistance/ apoptosis, invasion | SAMD9, FOXO3 | [195-197] |
| miR-150 | increased | 19q13.33 | Oncogene | proliferation, apoptosis | P53 | [189, 190] |
| miR-196a | increased | 17q21.32 | Oncogene | cisplatin resistance/ proliferation/ invasion | HOXA5 | [193, 194] |
| miR-205 | increased | 1q32.2 | Oncogene | proliferation, apoptosis, carboplatin resistance | PTEN | [107-109, 183] |
| miR-221/222 | increased | Xp11.3 | Oncogene | migration, apoptosis, TRAIL resistance | PTEN, TIMP3 | [176, 177] |
| miR-494 | increased | 14q32.31 | Oncogene | apoptosis, TRAIL resistance | Bim | [178] |
| miR-873 | increased | 9p21.1 | Oncogene | proliferation/ migration | SRCIN1 | [191] |
| miR-26b | decreased | 2q35 | Tumor Suppressor | metastasis | MIEN1 | [108, 206] |
| miR-34a | decreased | 1p36.22 | Tumor Suppressor | proliferation, apoptosis | TGFβR2, OCT4 | [110, 186-188] |
| miR-124 | decreased | 8p23.1 | Tumor Suppressor | proliferation, migration, invasion | SOX9 | [172, 192] |
| miR-133a | decreased | 18q11.2 | Tumor Suppressor | growth, invasion, metastasis, proliferation, TRAIL resistance | IGF-1R, TGFBR1 and EGFR | [182] |
| miR-137 | decreased | 1p21.3 | Tumor Suppressor | growth, invasion, TRAIL resistance |
BMP7 | [180] |
| miR-152 | decreased | 17q21.32 | Tumor Suppressor | metastasis | NRP1 | [208] |
| miR-187-3p | decreased | 18q12.2 | Tumor Suppressor | migration, invasion, apoptosis | BCL6 | [201] |
| miR-200c | decreased | 12p13.31 | Tumor Suppressor | metastasis | ZEB2 | [207] |
| miR-204 | decreased | 9q21.12 | Tumor Suppressor | proliferation, migration, apoptosis | ATF2 | [200] |
| miR-218 | decreased | 4p15.31 | Tumor Suppressor | proliferation, apoptosis/ carboplatin resistance | PARP, Caspase 3, Bcl-2 | [183] |
| miR-223 | decreased | Xq12 | Tumor Suppressor | chemoresistance, proliferation | IGF1R | [96, 185] |
| miR-326 | decreased | 11q13.4 | Tumor Suppressor | apoptosis | CCND1 | [202] |
| miR-329 | decreased | 14q32.31 | Tumor Suppressor | proliferation, migration, invasion | MET | [205] |
| miR-377 | decreased | 14q32.31 | Tumor Suppressor | proliferation, migration, invasion | AEG-1 | [203] |
| miR-432 | decreased | 14q32.2 | Tumor Suppressor | proliferation, cisplatin sensitivity | E2F3 and AXL | [184] |
| miR-449a | decreased | 5q11.2 | Tumor Suppressor | growth, invasion TRAIL resistance |
MAP2K1 | [181] |
| miR-455 | decreased | 9q32 | Tumor Suppressor | proliferation, migration, invasion | ZEB1 | [204] |
Resistance occurs when certain genetic or epigenetic changes make the treatment ineffective against the tumor cells [174, 175]. MiR-221 and miR-222 were first discovered to be involved in the development of some epithelial cancers [176]. Subsequently, these two miRNAs were found to be upregulated in NSCLC and further investigation revealed that they regulate the tumor suppressors PTEN and TIMP3 [177]. PTEN regulates the major cell survival pathway PI3K/AKT and plays a key role in the development of multiple drug resistance. TIMP3 induces activation of both apoptosis initiators caspase-8 and caspase-9. MiR-221 and miR-222 directly inhibit the expression of PTEN and TIMP3, thereby, facilitating the tumor cell migration, invasion and growth. However, the fascinating consequence of miR-221 and miR-222 overexpression is the induction of resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), thereby, rendering any treatment targeting the induction of apoptosis unsuccessful. TRAIL acts as a cytokine and is produced by almost all normal cells. The pro apoptotic gene TRAIL in tumor cells has been targeted in therapy by several drugs in order to initiate the apoptosis. The main conclusion being that either miR-221 /222 overexpression or the PTEN/TIMP3 downregulation, facilitates resistance to TRAIL-inducing apoptosis in NSCLC cells. Moreover, the finding that the MET oncogene regulates the miR-221 and mir-222 levels through the potent transcription factor c-Jun and JNK activation further supported this notion [177]. Resistance to TRAIL was also demonstrated in another upregulated miRNA that facilitates tumorigenesis by regulating apoptosis and cell proliferation. Overexpression of miR-494 by downregulation of BIM, (BCL2-like protein 11) an apoptosis regulator, increased the resistance to TRAIL-induced apoptosis in the lung cancer cell line H460 [178]. In addition, it was demonstrated that the transcription factors c-Jun and c-Fos, both directly bind to the miR-494 promoter. One commonality exposed in both of these TRAIL resistance studies was that the miRNAs mentioned exhibit the transcriptional activation by the activator protein 1 family, which include c-Jun and c-Fos. Despite being a quite promising biological agent for cancer treatment, the clinical application of TRAIL does pose problems because TRAIL has potent pro-apoptotic effect in all cells meaning that it can cause toxicity in healthy tissue. This means that for this type targeted therapy, TRAIL expression needs to be induced specifically in lung cancer cells. To address this issue, Wu and his colleagues generated a lung cancer specific TRAIL-expressing adenoviral vector that was regulated by miRNAs response elements. MiR-133a, miR-137 and miR-449a have consistently been reported to be downregulated in lung cancer [179-182] and, thus, their respective miRNAs response elements were inserted into the adenoviral vector to regulate the TRAIL expression. No TRAIL expression was detected in the normal cell lines, while this MRE-regulated vector was able to exert cytotoxicity in lung cancer cells. A tumor xenograft model was then used to further verify the selective TRAIL-mediated growth inhibiting effect [182].
Resistance does appear to be inherently related to miRNAs regulation, especially with regard to chemotherapy. In a study on the lung cancer cell lines A549 and H1975, two miRNAs were identified for carboplatin sensitivity and chemoresistance. The carboplatin sensitivity and chemoresistance were found to be associated with the overexpression of MiR-205 and the concurrent downregulation of the tumor suppressor miR-218. In particular, the overexpression of miR-205 resulted in the enhanced resistance to carboplatin in ADC cells by inhibiting apoptosis and increasing cell survival; while the ectopic miR-218 overexpression resulted in decreased cell proliferation, invasion and migration. In addition, there was increased inhibition efficiency of cancer cell migration and invasion when combining miR-218 and carboplatin. However, miR-205 can negate these suppressive effects of miR-218 by altering the expression of pro-apoptotic proteins PARP, Caspase 3 and Bax as well as upregulating the anti-apoptotic markers Survivin and Mcl-1 [183]. Another chemotherapy agent frequently used for the treatment of lung cancer is cisplatin. MiR-432 was initially studied for its role in cell proliferation and was later demonstrated to regulate sensitivity to cisplatin. It can function as a tumor suppressor because it targets E2F3 and AXL. MiR-432 is downregulated in ADC, thereby, representing a potential therapeutic target for upregulation leading to an increase in cisplatin sensitivity [184]. Lastly, resistance to EGFR-TK inhibitors is a critical issue concerning the treatment of EGFR mutant-positive NSCLC. MiR-223 downregulation was proposed to have a role in this resistance through regulation of the IGF1R/PI3K/Akt signaling pathway, thus, restoring its levels could restore chemotherapeutic sensitivity [185].
The most promising approaches to therapy is to use tumor suppressive miRNAs for cancer treatment. Reintroducing tumor suppressive miRNAs as inhibitors can lead to the normalizing cellular pathways (like apoptosis), stimulating a desirable therapeutic response. Mir-34a is one such tumor suppressor microRNA that is downregulated in lung cancer [186], Its expression in NSCLC cell lines H1299, A549, SPCA-1 and HCC827 was significantly lower than the non-tumorigenic bronchial epithelium cell line BEAS-2B. It targets TGFβR2 for downregulation which inhibits proliferation and promotes apoptosis [187]. The encoded transmembrane protein transforms growth factor beta receptor 2, which is responsible for phosphorylating proteins entering the nucleus that regulates the transcription of genes involved in cell proliferation. Interestingly, miR-34a also has demonstrated to have a role in oncogenic development in association with OCT4, a protein involved in self-renewal of undifferentiated stem cells. One study investigated the factors that promote or inhibit induced pluripotent stem cells in radiation-transformed human epithelial cells derived from different tissues. The major conclusion was that this functional positive feedback loop involves miR-34a and OCT4, which notably contribute to cell transformation. In essence, the inhibition of miR-34a upregulates OCT4; this promotes p63 while suppresses p53 which in turn further increases OCT4 upregulation by downregulating miR-34a [188]. Considering all this, Wiggins et al. decided to investigate the anti-oncogenic activity and therapeutic potential of miR-34a by developing a lipid-based delivery vehicle for chemically synthesized miR-34a. They demonstrated intravenous delivery causing accumulation of miR-34a in the tumor tissue leading to the downregulation of miR-34a targets, thereby, blocking tumor growth in the NSCLC mouse models [186]. This was done without inducing an immune response or elevating blood chemistries, determined by monitoring serum levels. The initially promising results lead to phase I clinical study in patients with advanced solid tumors. Unfortunately, the clinical trials had to be stopped due to severe immune-related adverse responses in five patients.
AntagomiRs or antimiRs (AMO) were designed for targeted therapeutic strategy using antisense oligonucleotides to silence oncogenic miRNAs. This was demonstrated by Li and colleagues on A594 lung cancer cell lines and mouse models where tumor proliferation was suppressed after delivery of anti-miR-150 causing increased p53 expression [189]. Mir-150 has been found to be significantly upregulated in ADC cell lines A549 and H1975 and, more importantly, has been shown to have an inverse correlation with SRCIN1 protein levels [190]. SRC kinase signaling inhibitor 1 (SRCIN1 or p140CAP) is a newly identified tumor suppressor gene that is involved in the inactivation of the Src pathway. Src regulates multiple signaling pathways associated with tumor development and progression like cell proliferation, migration or invasion [190]. By inactivating Src, SRCIN1 then becomes the major regulator of multiple downstream pathways required for tumorigenesis which includes: Focal Adhesion Kinase (FAK), Epidermal Growth Factor Receptor (EGFR) and Ras/extracellular signal-regulated kinase (ERK) [139]. Coincidentally, SRCIN1 also has a similar inverse relationship with miR-873, another upregulated miRNAs found in ADC cell lines. Cell proliferation and migration are either promoted by overexpression of miR-873 or inhibited by upregulation of SRCIN1 [191]. In conclusion, either of these miRNAs, miR-150 or miR-873, could be silenced using antagomiRs to upregulate SRCIN1, thereby, inactivating Src pathway and promoting its tumor suppressive effects. Furthermore, it would be interesting to comparatively quantify the SRCIN1 upregulation and its downstream effects between the two miRNAs. Another potential candidate for targeted antagomiR-silencing therapy is miR-124, which is downregulated in ADC tissue compared to corresponding normal tissue. It was found to target SOX9 and its upregulation inhibited the processes of proliferation, migration and invasion in an experiment conducted on the lung cancer A549 cell line [192]. Lastly, some targeted therapies have silenced miRNAs involved in drug resistance to increase the sensitivity to standard therapies. One such therapy used anti-miR-196a to enhance sensitivity to cisplatin in cell lines and murine models of NSCLC [193]. Additionally, this anti-miR-196a targeted therapy would also inhibit proliferative and invasive effects caused by this miRNAs [194].
A different therapeutic approach could be using the knockdown of miRNAs that are involved in cancer pathways to increase the cancer cell’s sensitivity to chemotherapy or radiotherapy. MiR-96 is upregulated in lung cancer and it has been associated with mechanisms of apoptosis inhibition and invasion [195]. Furthermore, it has been linked to cisplatin chemoresistance in NSCLC by targeting the tumor suppressor SAMD9. Sterile α motif domain-containing 9 (SAMD9) is believed to be an inhibitor of tumor progression. Firstly, its deletion is commonly observed in the cells of patients with myeloid leukemia and myeloidysplastic syndrome. Secondly, its overexpression causes apoptosis and reduced proliferation of malignant cells. Lastly, downregulation of SAMD9 is associated with increased proliferation and tumor growth [196]. Thus, it was found that by knocking down miR-96 in NSCLC cell lines with antagomir-96, there was an increase in cisplatin induced apoptosis and SAMD9 expression [197]. In a similar manner, MiR-95 functions as an oncogene and miRNAs profiling of tissue samples from NSCLC patients following radiotherapy revealed its overexpression [198, 199]. Thus, when Ma et al. (2016) knocked down this miRNA, there was increased radiosensitivity and cell apoptosis leading to reduced tumor growth.
Current findings on the underlying mechanisms and, more specifically, the downstream effectors of miRNAs, have exposed the therapeutic potential of downregulated tumor suppressor miRNAs in NSCLC. A recent example was demonstrated in NSCLC cell lines by targeting miR-204 for overexpression and, through its transcription factor ATF2, researchers were able to suppress cell proliferation, induce apoptosis, cause G1-cell cycle arrest and inhibit cell migration [200]. Some more examples of downregulated tumor suppressor miRNAs in NSCLC found to exert antitumor effects are: miR-187-3p downregulation of the oncogene BCL6 [201], miR-326 downregulation of the oncogene CCND1 [202], miR-377 downregulation of the oncogene AEG-1 [203] and lastly, miR-455 upregulation of ZEB1 [204]. Finally, more investigation needs to be done on miR-329, a tumor suppressor in only some cancers. It was shown to have inhibitory role in proliferation, migration and invasion in NSCLC as well as an apoptosis promoting function through the downregulation of MET [205]. In conclusion, targeting downregulated tumor suppressor miRNAs in NSCLC patients could prove to be a successful treatment.
With regard to therapeutic applications of miRNAs, another thriving domain is the investigation of their mechanism of action in metastasis. Three such examples of downregulated miRNAs in NSCLC are: miR-26b suppression of metastasis by targeting MIEN1 through NF-κB/MMP-9/VEGF pathways [206], miR-200c inhibition of metastasis by targeting the epithelial mesenchymal transition regulator ZEB2 [207] and miR-152 which targets neuropilin-1, a mediator of cell invasiveness. Interestingly, miR-152 has an inverse relationship to its target in NSCLC cells, overexpression of miR-152 inhibited neuropilin-1 mediated cell invasiveness, while down-regulated expression of miR-152 increased neuropilin-1 mediated cell invasiveness [208].
One last suggestion for investigation of a promising therapeutic target is miR-10b. Initially, miR-10b was described as a regulator of metastasis in human esophageal cancer cell lines KYSE140 and KYSE450 by directly targeting KLF4. KLF4 is a known tumor suppressor gene that directly suppresses cell migration and invasion in esophageal cancer. miR-10b overexpression was proven to be correlated to human esophageal cancer samples; however, this overexpression was not significantly correlated to clinical metastasis statues [209]. This overexpression of miR-10b causing direct suppression of KLF4 and increased metastatic potential was also found to be the case in gastric carcinoma [210] as well as bladder cancer [211]. In a study on bladder cancer, the mechanism of action was expanded to include E-cadherin as a downstream factor of KLF4 [211]. Coincidently, all these findings are supported by the research conducted on NSCLC [212, 213]. Initially, Liu et al. performed qRT-PCR on A549 cells transfected with miR-10b leading to increased metastatic capabilities like proliferation, migration and invasion. Subsequently, Western blotting revealed KLF4 as a target of miR-10b but only being indirect [212]. The following year, Zhang et al. using the same techniques found that E-cadherin mRNA and its protein were overexpressed in miR-10b-suppressed NSCLC cell line A549 when compared to the control. Furthermore, the authors conclude that miR-10b overexpression is an independent prognostic factor for NSCLC patients [213]. In conclusion, miR-10b/KLF4/E-cadherin pathway does seem to remain consistent in various cancers and further illumination could prove to be very useful toward therapy.
Conclusion
Lung cancer is a complex disease that is often detected only in the late stages. Non-coding RNAs play various roles in the development and progression of lung cancer pathology, thereby, making their clinical implications quite valuable. The non-coding RNAs evaluated in this article, miRNAs, Piwi-interacting RNAs, small nucleolar RNAs and lncRNAs are involved in a number of pathologies including all lung cancer subtypes. The major function of ncRNAs concerns gene regulation. They have received much attention due to their utilization in diagnosis, their use as minimally invasive biomarkers isolated from bodily fluid or for their expression that could be inhibited or restored for patient therapy. Many studies relying on high-throughput techniques have identified a great number of ncRNAs that are expressed differently in normal and cancer samples as well as between the different types of cancer. Because ncRNAs are involved in molecular pathways that are oncogenic or tumor suppressive, they have been demonstrated to have great potential to be used as novel targeted therapeutic strategies. To this end, studies have shown that miRNAs could potentially be used to restore sensitivity to chemotherapeutics such as TRAIL by using a lung cancer specific TRAIL-expressing vector regulated by three miRNAs response elements [182].
Reintroducing tumor suppressive miRNAs into the cells leads to the reactivation of pathways that lead to a desired therapeutic response. One such study used a lipid based delivery system and mouse models of NSCLC for miR-34a [186], while other strategies involved the use of antisense oligonucleotides that silence oncogenic miRNAs. Many miRNAs have been identified as having an oncogenic role in lung cancer, distinguishing them as promising therapeutic targets. Piwi-interacting RNAs are a class of non-coding RNAs whose functions have not yet been fully explained or understood but whose aberrant expression has been associated with cancer progression. Research has revealed that piRNAs are up or downregulated in lung cancer and could be used in a similar manner to miRNAs for diagnosis or therapy. Small nucleolar RNAs are also emerging as valuable tools for the diagnosis, prognosis and therapy of lung cancer because they can be measured from sputum samples, distinguishing them as noninvasive biomarkers. LncRNAs have received much attention due to their involvement in the development and progression of lung cancer. Specifically, deregulated lncRNAs such as MALAT1, CARLo-5 or linc0067 could be considered desirable therapeutic targets because knockdown of these RNAs in lung cancer cell lines and murine models attained promising results [113, 137, 141]. Finally, circular RNAs have already proven to be great biomarkers due to their unique structure preventing degradation; however, in theory their function as miRNAs sponges could prove to be very successful for drug delivery.
Thus, revealing the molecular mechanisms of action has brought value to non-coding RNAs because of the potential for designing therapies. Above all, the development of novel non-coding RNA biomarkers for the detection and progression of lung cancer has promising implications for the personalized treatment of lung cancer.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
This study was financed by a POC-P_37_796/2016 grant, entitled “Clinical and economic impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance”-CANTEMIR.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Martini N., Bains M.S., Burt M.E., Zakowski M.F., McCormack P., Rusch V.W., Ginsberg R.J. Incidence of local recurrence and second primary tumors in resected stage I lung cancer. J. Thorac. Cardiovasc. Surg. 1995;109(1):120–129. doi: 10.1016/S0022-5223(95)70427-2. [DOI] [PubMed] [Google Scholar]
- 4.Nakazawa K., Kurishima K., Tamura T., Kagohashi K., Ishikawa H., Satoh H., Hizawa N. Specific organ metastases and survival in small cell lung cancer. Oncol. Lett. 2012;4(4):617–620. doi: 10.3892/ol.2012.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travis W.D., Brambilla E., Riely G.J. New pathologic classification of lung cancer: Relevance for clinical practice and clinical trials. J. Clin. Oncol. 2013;31(8):992–1001. doi: 10.1200/JCO.2012.46.9270. [DOI] [PubMed] [Google Scholar]
- 6.Diederichs S., Bartsch L., Berkmann J.C., Frose K., Heitmann J., Hoppe C., Iggena D., Jazmati D., Karschnia P., Linsenmeier M., Maulhardt T., Mohrmann L., Morstein J., Paffenholz S.V., Ropenack P., Ruckert T., Sandig L., Schell M., Steinmann A., Voss G., Wasmuth J., Weinberger M.E., Wullenkord R. The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. Med. 2016;8(5):442–457. doi: 10.15252/emmm.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling H., Pickard K., Ivan C., Isella C., Ikuo M., Mitter R., Spizzo R., Bullock M.D., Braicu C., Pileczki V., Vincent K., Pichler M., Stiegelbauer V., Hoefler G., Almeida M.I., Hsiao A., Zhang X., Primrose J.N., Packham G.K., Liu K., Bojja K., Gafa R., Xiao L., Rossi S., Song J.H., Vannini I., Fanini F., Kopetz S., Zweidler-McKay P., Wang X., Ionescu C., Irimie A., Fabbri M., Lanza G., Hamilton S.R., Berindan-Neagoe I., Medico E., Mirnezami A.H., Calin G.A., Nicoloso M.S. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut. 2016;65(6):977–989. doi: 10.1136/gutjnl-2015-309372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dechamethakun S., Muramatsu M. Long noncoding RNA variations in cardiometabolic diseases. J. Hum. Genet. 2017;62(1):97–104. doi: 10.1038/jhg.2016.70. [DOI] [PubMed] [Google Scholar]
- 9.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No. 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 11.Calin G.A., Liu C.G., Ferracin M., Hyslop T., Spizzo R., Sevignani C., Fabbri M., Cimmino A., Lee E.J., Wojcik S.E., Shimizu M., Tili E., Rossi S., Taccioli C., Pichiorri F., Liu X., Zupo S., Herlea V., Gramantieri L., Lanza G., Alder H., Rassenti L., Volinia S., Schmittgen T.D., Kipps T.J., Negrini M., Croce C.M. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Irimie A.I., Braicu C., Cojocneanu-Petric R., Berindan-Neagoe I., Campian R.S. Novel technologies for oral squamous carcinoma biomarkers in diagnostics and prognostics. Acta Odontol. Scand. 2015;73(3):161–168. doi: 10.3109/00016357.2014.986754. [DOI] [PubMed] [Google Scholar]
- 13.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Zaharie F., Muresan M.S., Petrushev B., Berce C., Gafencu G.A., Selicean S., Jurj A., Cojocneanu-Petric R., Lisencu C.I., Pop L.A., Pileczki V., Eniu D., Muresan M.A., Zaharie R., Berindan-Neagoe I., Tomuleasa C., Irimie A. Exosome-carried microRNA-375 inhibits cell progression and dissemination via Bcl-2 blocking in colon cancer. J. Gastrointestin. Liver Dis. 2015;24(4):435–443. doi: 10.15403/jgld.2014.1121.244.375. [DOI] [PubMed] [Google Scholar]
- 15.Pileczki V., Cojocneanu-Petric R., Maralani M., Neagoe I.B., Sandulescu R. MicroRNAs as regulators of apoptosis mechanisms in cancer. Clujul Med. 2016;89(1):50–55. doi: 10.15386/cjmed-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braicu C., Calin G.A., Berindan-Neagoe I. MicroRNAs and cancer therapy - from bystanders to major players. Curr. Med. Chem. 2013;20(29):3561–3573. doi: 10.2174/0929867311320290002. [DOI] [PubMed] [Google Scholar]
- 17.Ling H., Fabbri M., Calin G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertoli G., Cava C., Castiglioni I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int. J. Mol. Sci. 2016;17(3):421. doi: 10.3390/ijms17030421. http://www.mdpi.com/ 1422-0067/17/3/421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren A., Dong Y., Tsoi H., Yu J. Detection of miRNA as non-invasive biomarkers of colorectal cancer. Int. J. Mol. Sci. 2015;16(2):2810–2823. doi: 10.3390/ijms16022810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enache L.S., Enache E.L., Ramiere C., Diaz O., Bancu L., Sin A., Andre P. Circulating RNA molecules as biomarkers in liver disease. Int. J. Mol. Sci. 2014;15(10):17644–17666. doi: 10.3390/ijms151017644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. https://biologicalproceduresonline.biomedcentral.com/articles/10.1186/1480-9222-16-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo F., Guo L., Li Y., Zhou Q., Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int. J. Clin. Exp. Pathol. 2015;8(12):15903–15910. [PMC free article] [PubMed] [Google Scholar]
- 23.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., Blencowe B.J., Prasanth S.G., Prasanth K.V. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt L.H., Spieker T., Koschmieder S., Schaffers S., Humberg J., Jungen D., Bulk E., Hascher A., Wittmer D., Marra A., Hillejan L., Wiebe K., Berdel W.E., Wiewrodt R., Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011;6(12):1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 25.Ni J., Tien A.L., Fournier M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89(4):565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 26.Mei Y., Clark D., Mao L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013;336(1):46–52. doi: 10.1016/j.canlet.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro A., Tejero R., Vinolas N., Cordeiro A., Marrades R.M., Fuster D., Caritg O., Moises J., Munoz C., Molins L., Ramirez J., Monzo M. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget. 2015;6(31):31544–31556. doi: 10.18632/oncotarget.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macfarlane L.A., Murphy P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 33.Hess J., Perez-Stable C., Wu G.J., Weir B., Tinoco I., Jr, Shen C.K. End-to-end transcription of an Alu family repeat. A new type of polymerase-III-dependent terminator and its evolutionary implication. J. Mol. Biol. 1985;184(1):7–21. doi: 10.1016/0022-2836(85)90039-7. [DOI] [PubMed] [Google Scholar]
- 34.Gu T.J., Yi X., Zhao X.W., Zhao Y., Yin J.Q. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics. 2009;10:563. doi: 10.1186/1471-2164-10-563. https://bmcgenomics.biomedcentral.com/ articles/10.1186/1471-2164-10-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136(1):75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. https://www.nature.com/articles/nature01957 [DOI] [PubMed] [Google Scholar]
- 38.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26(3):775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying S.Y., Lin S.L. Intron-derived microRNAs--fine tuning of gene functions. Gene. 2004;342(1):25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Lin S.L., Chang D., Wu D.Y., Ying S.Y. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochem. Biophys. Res. Commun. 2003;310(3):754–760. doi: 10.1016/j.bbrc.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 43.Berezikov E., Chung W.J., Willis J., Cuppen E., Lai E.C. Mammalian mirtron genes. Mol. Cell. 2007;28(2):328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rainer J., Ploner C., Jesacher S., Ploner A., Eduardoff M., Mansha M., Wasim M., Panzer-Grumayer R., Trajanoski Z., Niederegger H., Kofler R. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia. 2009;23(4):746–752. doi: 10.1038/leu.2008.370. [DOI] [PubMed] [Google Scholar]
- 45.Ruby J.G., Jan C.H., Bartel D.P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. https://www.nature.com/articles/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin S.L., Miller J.D., Ying S.Y. Intronic microRNA (miRNA). J. Biomed. Biotechnol. 2006;2006(4):26818. doi: 10.1155/JBB/2006/26818. https://www.hindawi.com/journals/bmri/2006/026818/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bohnsack M.T., Czaplinski K., Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. http://rnajournal.cshlp.org/content/10/2/185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lund E., Dahlberg J.E. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. http:// symposium.cshlp.org/content/71/59 [DOI] [PubMed] [Google Scholar]
- 49.Okada C., Yamashita E., Lee S.J., Shibata S., Katahira J., Nakagawa A., Yoneda Y., Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326(5957):1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 50.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng Y., Cullen B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32(16):4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D., Wolfe S.A., Giraldez A.J. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. http://science.sciencemag.org/content/328/5986/1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diederichs S., Haber D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131(6):1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 54.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 55.Ambros V. The evolution of our thinking about microRNAs. Nat. Med. 2008;14(10):1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 56.MacRae I.J., Ma E., Zhou M., Robinson C.V., Doudna J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA. 2008;105(2):512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robb G.B., Rana T.M. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26(4):523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 58.Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Choi P.S., Casey S.C., Dill D.L., Felsher D.W. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26(2):262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osada H., Takahashi T. let-7 and miR-17-92: Small-sized major players in lung cancer development. Cancer Sci. 2011;102(1):9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang P., Yang D., Zhang H., Wei X., Ma T., Cheng Z., Hong Q., Hu J., Zhuo H., Song Y., Jia C., Jing F., Jin Q., Bai C., Mao H., Zhao J. Early detection of lung cancer in serum by a panel of microRNA biomarkers. Clin. Lung Cancer. 2015;16(4):313–319. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Jiang L., Huang Q., Zhang S., Zhang Q., Chang J., Qiu X., Wang E. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318. doi: 10.1186/1471-2407-10-318. https://bmccancer.biomedcentral.com/articles/ 10.1186/1471-2407-10-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang T., Chen T., Li Y., Gao L., Zhang S., Wang T., Chen M. Downregulation of miR-25 modulates non-small cell lung cancer cells by targeting CDC42. Tumour Biol. 2015;36(3):1903–1911. doi: 10.1007/s13277-014-2793-0. [DOI] [PubMed] [Google Scholar]
- 64.Crawford M., Brawner E., Batte K., Yu L., Hunter M.G., Otterson G.A., Nuovo G., Marsh C.B., Nana-Sinkam S.P. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008;373(4):607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 65.Liu B., Peng X.C., Zheng X.L., Wang J., Qin Y.W. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169–175. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Zheng D., Haddadin S., Wang Y., Gu L.Q., Perry M.C., Freter C.E., Wang M.X. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int. J. Clin. Exp. Pathol. 2011;4(6):575–586. [PMC free article] [PubMed] [Google Scholar]
- 67.Hou L., Chen J., Zheng Y., Wu C. Critical role of miR-155/FoxO1/ROS axis in the regulation of non-small cell lung carcinomas. Tumour Biol. 2016;37(4):5185–5192. doi: 10.1007/s13277-015-4335-9. [DOI] [PubMed] [Google Scholar]
- 68.Wang M., Wang Y., Zang W., Wang H., Chu H., Li P., Li M., Zhang G., Zhao G. Downregulation of microRNA-182 inhibits cell growth and invasion by targeting programmed cell death 4 in human lung adenocarcinoma cells. Tumour Biol. 2014;35(1):39–46. doi: 10.1007/s13277-013-1004-8. [DOI] [PubMed] [Google Scholar]
- 69.Du L., Schageman J.J., Subauste M.C., Saber B., Hammond S.M., Prudkin L., Wistuba II, Ji L., Roth J.A., Minna J.D., Pertsemlidis A. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol. Cancer Res. 2009;7(8):1234–1243. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Roosbroeck K., Fanini F., Setoyama T., Ivan C., Rodriguez-Aguayo C., Fuentes-Mattei E., Xiao L., Vannini I., Redis R.S., D’Abundo L., Zhang X., Nicoloso M.S., Rossi S., Gonzalez-Villasana V., Rupaimoole R., Ferracin M., Morabito F., Neri A., Ruvolo P.P., Ruvolo V.R., Pecot C.V., Amadori D., Abruzzo L., Calin S., Wang X., You M.J., Ferrajoli A., Orlowski R., Plunkett W., Lichtenberg T.M., Davuluri R.V., Berindan-Neagoe I., Negrini M., Wistuba II, Kantarjian H.M., Sood A.K., Lopez-Berestein G., Keating M.J., Fabbri M., Calin G.A. Combining Anti-Mir-155 with chemotherapy for the treatment of lung cancers. Clin. Cancer Res. 2017;23(11):2891–2904. doi: 10.1158/1078-0432.CCR-16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan H.J., Ma J.Y., Wang L., Gu W. Expression and significance of circulating microRNA-31 in lung cancer patients. Med. Sci. Monit. 2015;21:722–726. doi: 10.12659/MSM.893213. https:// www.medscimonit.com/abstract/index/idArt/893213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meng W., Ye Z., Cui R., Perry J., Dedousi-Huebner V., Huebner A., Wang Y., Li B., Volinia S., Nakanishi H., Kim T., Suh S.S., Ayers L.W., Ross P., Croce C.M., Chakravarti A., Jin V.X., Lautenschlaeger T. MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clin. Cancer Res. 2013;19(19):5423–5433. doi: 10.1158/1078-0432.CCR-13-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu M., Liang H., Fu Z., Wang X., Liao Z., Zhou Y., Liu Y., Wang Y., Hong Y., Zhou X., Yan X., Yu M., Ma M., Zhang W., Guo B., Zhang J., Zen K., Zhang C.Y., Wang T., Zhang Q., Chen X. BAP1 suppresses lung cancer progression and is inhibited by miR-31. Oncotarget. 2016;7(12):13742–13753. doi: 10.18632/oncotarget.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barshack I., Lithwick-Yanai G., Afek A., Rosenblatt K., Tabibian-Keissar H., Zepeniuk M., Cohen L., Dan H., Zion O., Strenov Y., Polak-Charcon S., Perelman M. MicroRNA expression differentiates between primary lung tumors and metastases to the lung. Pathol. Res. Pract. 2010;206(8):578–584. doi: 10.1016/j.prp.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 75.Meister J., Schmidt M.H. miR-126 and miR-126*: New players in cancer. . Sci. World J. 2010;10:2090–2100. doi: 10.1100/tsw.2010.198. https://www.hindawi.com/journals/tswj/2010/257492/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miko E., Margitai Z., Czimmerer Z., Varkonyi I., Dezso B., Lanyi A., Bacso Z., Scholtz B. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585(8):1191–1196. doi: 10.1016/j.febslet.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 77.Sun Y., Bai Y., Zhang F., Wang Y., Guo Y., Guo L. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biochem. Biophys. Res. Commun. 2010;391(3):1483–1489. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- 78.Saito Y., Friedman J.M., Chihara Y., Egger G., Chuang J.C., Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem. Biophys. Res. Commun. 2009;379(3):726–731. doi: 10.1016/j.bbrc.2008.12.098. [DOI] [PubMed] [Google Scholar]
- 79.Fish J.E., Srivastava D. MicroRNAs: Opening a new vein in angiogenesis research. Sci. Signal. 2009;2(52):pe1. doi: 10.1126/scisignal.252pe1. http://stke.sciencemag.org/content/2/52/pe1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donnem T., Lonvik K., Eklo K., Berg T., Sorbye S.W., Al-Shibli K., Al-Saad S., Andersen S., Stenvold H., Bremnes R.M., Busund L.T. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer. 2011;117(14):3193–3200. doi: 10.1002/cncr.25907. [DOI] [PubMed] [Google Scholar]
- 81.Chen S.W., Wang T.B., Tian Y.H., Zheng Y.G. Down-regulation of microRNA-126 and microRNA-133b acts as novel predictor biomarkers in progression and metastasis of non small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015;8(11):14983–14988. [PMC free article] [PubMed] [Google Scholar]
- 82.Liu L., Shao X., Gao W., Zhang Z., Liu P., Wang R., Huang P., Yin Y., Shu Y. MicroRNA-133b inhibits the growth of non-small-cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 2012;279(20):3800–3812. doi: 10.1111/j.1742-4658.2012.08741.x. [DOI] [PubMed] [Google Scholar]
- 83.Hu J., Cheng Y., Li Y., Jin Z., Pan Y., Liu G., Fu S., Zhang Y., Feng K., Feng Y. microRNA-128 plays a critical role in human non-small cell lung cancer tumourigenesis, angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor-C. Eur. J. Cancer. 2014;50(13):2336–2350. doi: 10.1016/j.ejca.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 84.Tan M., Wu J., Cai Y. Suppression of Wnt signaling by the miR-29 family is mediated by demethylation of WIF-1 in non-small-cell lung cancer. Biochem. Biophys. Res. Commun. 2013;438(4):673–679. doi: 10.1016/j.bbrc.2013.07.123. [DOI] [PubMed] [Google Scholar]
- 85.Yang G., Zhang W., Yu C., Ren J., An Z. MicroRNA let-7: Regulation, single nucleotide polymorphism, and therapy in lung cancer. J. Cancer Res. Ther.Available from: http://www.cancerjournal.net/article.asp?issn=0973-1482. 2015;11(Suppl 1):C1–C6. doi: 10.4103/0973-1482.163830. [DOI] [PubMed] [Google Scholar]
- 86.Zhao B., Han H., Chen J., Zhang Z., Li S., Fang F., Zheng Q., Ma Y., Zhang J., Wu N., Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342(1):43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 87.Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J., Chin L., Brown D., Slack F.J. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 88.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 89.Wozniak M.B., Scelo G., Muller D.C., Mukeria A., Zaridze D., Brennan P. Circulating microRNAs as non-invasive biomarkers for early detection of non-small-cell lung cancer. PLoS One. 2015;10(5):e0125026. doi: 10.1371/journal.pone.0125026. http://journals. plos.org/plosone/article?id=10.1371/journal.pone.0125026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Zhang Y., Wang J., Jiang X., Xiang Y., Xu C., Zheng P., Zhang J., Li R., Zhang H., Shang X., Gong T., Ning G., Wang J., Zen K., Zhang J., Zhang C.Y. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 91.Hennessey P.T., Sanford T., Choudhary A., Mydlarz W.W., Brown D., Adai A.T., Ochs M.F., Ahrendt S.A., Mambo E., Califano J.A. Serum microRNA biomarkers for detection of non-small cell lung cancer. 2012 doi: 10.1371/journal.pone.0032307. http://journals.plos.org/plosone/article?id=10.1371/journal [DOI] [PMC free article] [PubMed]
- 92.Puissegur M.P., Mazure N.M., Bertero T., Pradelli L., Grosso S., Robbe-Sermesant K., Maurin T., Lebrigand K., Cardinaud B., Hofman V., Fourre S., Magnone V., Ricci J.E., Pouyssegur J., Gounon P., Hofman P., Barbry P., Mari B. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18(3):465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun D., Li X., Ma M., Liu J., Xu Y., Ye L., Hou H., Wang C., Li X., Jiang Y. The predictive value and potential mechanisms of miRNA-328 and miRNA-378 for brain metastases in operable and advanced non-small-cell lung cancer. J. Clin. Oncol. 2015;45(5):464–473. doi: 10.1093/jjco/hyv009. [DOI] [PubMed] [Google Scholar]
- 94.Lee D.Y., Deng Z., Wang C.H., Yang B.B. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl. Acad. Sci. USA. 2007;104(51):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Powrozek T., Krawczyk P., Kowalski D.M., Winiarczyk K., Olszyna-Serementa M., Milanowski J. Plasma circulating microRNA-944 and microRNA-3662 as potential histologic type-specific early lung cancer biomarkers. Transl. Res. 2015;166(4):315–323. doi: 10.1016/j.trsl.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 96.Taniwaki M., Daigo Y., Ishikawa N., Takano A., Tsunoda T., Yasui W., Inai K., Kohno N., Nakamura Y. Gene expression profiles of small-cell lung cancers: molecular signatures of lung cancer. Int. J. Oncol. 2006;29(3):567–575. [PubMed] [Google Scholar]
- 97.Taneja T.K., Sharma S.K. Markers of small cell lung cancer. World J. Surg. Oncol. 2004;2:10. doi: 10.1186/1477-7819-2-10. https://wjso.biomedcentral.com/articles/10.1186/1477-7819-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bangur C.S., Switzer A., Fan L., Marton M.J., Meyer M.R., Wang T. Identification of genes over-expressed in small cell lung carcinoma using suppression subtractive hybridization and cDNA microarray expression analysis. Oncogene. 2002;21(23):3814–3825. doi: 10.1038/sj.onc.1205480. [DOI] [PubMed] [Google Scholar]
- 99.Coate L.E., John T., Tsao M.S., Shepherd F.A. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10(10):1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- 100.Sonoki H., Sato T., Endo S., Matsunaga T., Yamaguchi M., Yamazaki Y., Sugatani J., Ikari A. Quercetin decreases Claudin-2 expression mediated by up-regulation of microRNA miR-16 in lung adenocarcinoma A549 cells. Nutrients. 2015;7(6):4578–4592. doi: 10.3390/nu7064578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T., Calin G.A., Liu C.G., Croce C.M., Harris C.C. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 102.Solomides C.C., Evans B.J., Navenot J.M., Vadigepalli R., Peiper S.C., Wang Z.X. MicroRNA profiling in lung cancer reveals new molecular markers for diagnosis. Acta Cytol. 2012;56(6):645–654. doi: 10.1159/000343473. [DOI] [PubMed] [Google Scholar]
- 103.Bishop J.A., Benjamin H., Cholakh H., Chajut A., Clark D.P., Westra W.H. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin. Cancer Res. 2010;16(2):610–619. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 104.Landi M.T., Zhao Y., Rotunno M., Koshiol J., Liu H., Bergen A.W., Rubagotti M., Goldstein A.M., Linnoila I., Marincola F.M., Tucker M.A., Bertazzi P.A., Pesatori A.C., Caporaso N.E., McShane L.M., Wang E. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 2010;16(2):430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhong Z., Dong Z., Yang L., Gong Z. miR-21 induces cell cycle at S phase and modulates cell proliferation by down-regulating hMSH2 in lung cancer. J. Cancer Res. Clin. Oncol. 2012;138(10):1781–1788. doi: 10.1007/s00432-012-1287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peng H., Wang J., Li J., Zhao M., Huang S.K., Gu Y.Y., Li Y., Sun X.J., Yang L., Luo Q., Huang C.Z. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–242. doi: 10.1016/j.lfs.2016.03.002. https://www.sciencedirect.com/science/article/pii/S0024320516301503 [DOI] [PubMed] [Google Scholar]
- 107.Gutschner T., Hammerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M., Zornig M., MacLeod A.R., Spector D.L., Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhu W., Zhou K., Zha Y., Chen D., He J., Ma H., Liu X., Le H., Zhang Y. Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS One. 2016;11(4):e0153046. doi: 10.1371/journal.pone.0153046. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0153046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun L., Chen Y., Su Q., Tang X., Liang Y., Che G., Luo F. Increased plasma miRNA-30a as a biomarker for non-small cell lung cancer. Med. Sci. Monit. 2016;22:647–655. doi: 10.12659/MSM.897330. https://www.medscimonit.com/abstract/index/idArt/897330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yuan Y., Zheng S., Li Q., Xiang X., Gao T., Ran P., Sun L., Huang Q., Xie F., Du J., Xiao C. Overexpression of miR-30a in lung adenocarcinoma A549 cell line inhibits migration and invasion via targeting EYA2. Acta Biochim. Biophys. Sin. (Shanghai) 2016;48(3):220–228. doi: 10.1093/abbs/gmv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guan P., Yin Z., Li X., Wu W., Zhou B. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J. Exp. Clin. Cancer Res. 2012;31:54. doi: 10.1186/1756-9966-31-54. https://jeccr.biomedcentral.com/articles/ 10.1186/1756-9966-31-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vosa U., Vooder T., Kolde R., Vilo J., Metspalu A., Annilo T. Meta-analysis of microRNA expression in lung cancer. Int. J. Cancer. 2013;132(12):2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 113.Yu L., Todd N.W., Xing L., Xie Y., Zhang H., Liu Z., Fang H., Zhang J., Katz R.L., Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int. J. Cancer. 2010;127(12):2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roa W.H., Kim J.O., Razzak R., Du H., Guo L., Singh R., Gazala S., Ghosh S., Wong E., Joy A.A., Xing J.Z., Bedard E.L. Sputum microRNA profiling: A novel approach for the early detection of non-small cell lung cancer. Clin. Invest. Med. 2012;35(5):E271. doi: 10.25011/cim.v35i5.18700. http://www.cimonline.ca/index.php/ cim/article/viewFile/18700/15645 [DOI] [PubMed] [Google Scholar]
- 115.Simonelig M. piRNAs, master regulators of gene expression. Cell Res. 2014;24(7):779–780. doi: 10.1038/cr.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martinez V.D., Vucic E.A., Thu K.L., Hubaux R., Enfield K.S., Pikor L.A., Becker-Santos D.D., Brown C.J., Lam S., Lam W.L. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. 2015 doi: 10.1038/srep10423. https://www.nature.com/articles/ [DOI] [PMC free article] [PubMed]
- 117.Braicu C., Catana C., Calin G.A., Berindan-Neagoe I. NCRNA combined therapy as future treatment option for cancer. Curr. Pharm. Des. 2014;20(42):6565–6574. doi: 10.2174/1381612820666140826153529. [DOI] [PubMed] [Google Scholar]
- 118.Ng K.W., Anderson C., Marshall E.A., Minatel B.C., Enfield K.S., Saprunoff H.L., Lam W.L., Martinez V.D. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol. Cancer. 2016;15:5. doi: 10.1186/s12943-016-0491-9. https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-016-0491-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mei Y., Wang Y., Kumari P., Shetty A.C., Clark D., Gable T., MacKerell A.D., Ma M.Z., Weber D.J., Yang A.J., Edelman M.J., Mao L. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. 2015 doi: 10.1038/ncomms8316. https://www.nature.com/articles/ [DOI] [PMC free article] [PubMed]
- 120.Liang D., Dong M., Hu L.J., Fang Z.H., Xu X., Shi E.H., Yang Y.J. Hiwi knockdown inhibits the growth of lung cancer in nude mice. Asian Pac. J. Cancer Prev. 2013;14(2):1067–1072. doi: 10.7314/apjcp.2013.14.2.1067. [DOI] [PubMed] [Google Scholar]
- 121.Qu X., Liu J., Zhong X., Li X., Zhang Q. PIWIL2 promotes progression of non-small cell lung cancer by inducing CDK2 and Cyclin A expression. J. Transl. Med. 2015;13:301. doi: 10.1186/s12967-015-0666-y. https://translational-medicine.biomedcentral.com/articles/ 10.1186/s12967-015-0666-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peng L., Song L., Liu C., Lv X., Li X., Jie J., Zhao D., Li D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2016;37(2):2749–2756. doi: 10.1007/s13277-015-4056-0. [DOI] [PubMed] [Google Scholar]
- 123.Shen L., Chen L., Wang Y., Jiang X., Xia H., Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neurooncol. 2015;121(1):101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 124.Yao Y., Fan Y., Wu J., Wan H., Wang J., Lam S., Lam W.L., Girard L., Gazdar A.F., Wu Z., Zhou Q. Potential application of non-small cell lung cancer-associated autoantibodies to early cancer diagnosis. Biochem. Biophys. Res. Commun. 2012;423(3):613–619. doi: 10.1016/j.bbrc.2012.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weber D.G., Johnen G., Casjens S., Bryk O., Pesch B., Jockel K.H., Kollmeier J., Bruning T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res. Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. https://bmcresnotes.biomedcentral.com/articles/10.1186/1756-0500-6-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu X.H., Liu Z.L., Sun M., Liu J., Wang Z.X., De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. https://bmccancer.biomedcentral.com/articles/ 10.1186/1471-2407-13-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Loewen G., Jayawickramarajah J., Zhuo Y., Shan B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. https://jhoonline.biomedcentral.com/articles/ 10.1186/s13045-014-0090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ono H., Motoi N., Nagano H., Miyauchi E., Ushijima M., Matsuura M., Okumura S., Nishio M., Hirose T., Inase N., Ishikawa Y. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Med. 2014;3(3):632–642. doi: 10.1002/cam4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao W., An Y., Liang Y., Xie X.W. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2014;18(13):1930–1936. [PubMed] [Google Scholar]
- 130.Nie F.Q., Zhu Q., Xu T.P., Zou Y.F., Xie M., Sun M., Xia R., Lu K.H. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 2014;35(8):7587–7594. doi: 10.1007/s13277-014-2009-7. [DOI] [PubMed] [Google Scholar]
- 131.Luo J., Tang L., Zhang J., Ni J., Zhang H.P., Zhang L., Xu J.F., Zheng D. Long non-coding RNA CARLo-5 is a negative prognostic factor and exhibits tumor pro-oncogenic activity in non-small cell lung cancer. Tumour Biol. 2014;35(11):11541–11549. doi: 10.1007/s13277-014-2442-7. [DOI] [PubMed] [Google Scholar]
- 132.Cui J., Mo J., Luo M., Yu Q., Zhou S., Li T., Zhang Y., Luo W. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015;8(10):12400–12409. [PMC free article] [PubMed] [Google Scholar]
- 133.Hou Z., Zhao W., Zhou J., Shen L., Zhan P., Xu C., Chang C., Bi H., Zou J., Yao X., Huang R., Yu L., Yan J. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int. J. Biochem. Cell Biol. 2014;53:380–388. doi: 10.1016/j.biocel.2014.06.004. https://www.sciencedirect.com/ science/article/pii/S1357272514002027?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 134.Jazi M.S., Samaei N.M., Ghanei M., Shadmehr M.B., Mowla S.J. Overexpression of the non-coding SOX2OT variants 4 and 7 in lung tumors suggests an oncogenic role in lung cancer. Tumour Biol. 2016;37(8):10329–10338. doi: 10.1007/s13277-016-4901-9. [DOI] [PubMed] [Google Scholar]
- 135.Shi X., Ma C., Zhu Q., Yuan D., Sun M., Gu X., Wu G., Lv T., Song Y. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7(18):25558–25575. doi: 10.18632/oncotarget.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qiu M., Xu Y., Yang X., Wang J., Hu J., Xu L., Yin R. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35(6):5375–5380. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 137.Chen S., Wu H., Lv N., Wang H., Wang Y., Tang Q., Shao H., Sun C. LncRNA CCAT2 predicts poor prognosis and regulates growth and metastasis in small cell lung cancer. 2016 doi: 10.1016/j.biopha.2016.05.017. direct.com/science/article/abs/pii/S0753332216304814 [DOI] [PubMed]
- 138.Li P., Zhang G., Li J., Yang R., Chen S., Wu S., Zhang F., Bai Y., Zhao H., Wang Y., Dun S., Chen X., Sun Q., Zhao G. Long noncoding RNA RGMB-AS1 indicates a poor prognosis and modulates cell proliferation, migration and invasion in lung adenocarcinoma. PLoS One. 2016;11(3):e0150790. doi: 10.1371/journal.pone.0150790. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0150790 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 139.Gong W.J., Yin J.Y., Li X.P., Fang C., Xiao D., Zhang W., Zhou H.H., Li X., Liu Z.Q. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37(6):8349–8358. doi: 10.1007/s13277-015-4497-5. [DOI] [PubMed] [Google Scholar]
- 140.Zequn N., Xuemei Z., Wei L., Zongjuan M., Yujie Z., Yanli H., Yuping Z., Xia M., Wei W., Wenjing D., Na F., Shuanying Y. The role and potential mechanisms of LncRNA-TATDN1 on metastasis and invasion of non-small cell lung cancer. Oncotarget. 2016;7(14):18219–18228. doi: 10.18632/oncotarget.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Han L., Zhang E.B., Yin D.D., Kong R., Xu T.P., Chen W.M., Xia R., Shu Y.Q., De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. https://www.nature.com/articles/cddis201530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shao Y., Chen H., Jiang X., Chen S., Li P., Ye M., Li Q., Sun W., Guo J. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 2014;35(10):9591–9595. doi: 10.1007/s13277-014-2243-z. [DOI] [PubMed] [Google Scholar]
- 143.Xie X., Liu H.T., Mei J., Ding F.B., Xiao H.B., Hu F.Q., Hu R., Wang M.S. LncRNA HMlincRNA717 is down-regulated in non-small cell lung cancer and associated with poor prognosis. Int. J. Clin. Exp. Pathol. 2014;7(12):8881–8886. [PMC free article] [PubMed] [Google Scholar]
- 144.Gao L., Ma J., Mannoor K., Guarnera M.A., Shetty A., Zhan M., Xing L., Stass S.A., Jiang F. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int. J. Cancer. 2015;136(6):E623–E629. doi: 10.1002/ijc.29169. [DOI] [PubMed] [Google Scholar]
- 145.Liao J., Yu L., Mei Y., Guarnera M., Shen J., Li R., Liu Z., Jiang F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer. 2010;9:198. doi: 10.1186/1476-4598-9-198. https://molecular-cancer.biomedcentral.com/articles/10.1186/1476-4598-9-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zheng D., Zhang J., Ni J., Luo J., Wang J., Tang L., Zhang L., Wang L., Xu J., Su B., Chen G. Small nucleolar RNA 78 promotes the tumorigenesis in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2015;34:49. doi: 10.1186/s13046-015-0170-5. https:// jeccr.biomedcentral.com/articles/10.1186/s13046-015-0170-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mei Y.P., Liao J.P., Shen J., Yu L., Liu B.L., Liu L., Li R.Y., Ji L., Dorsey S.G., Jiang Z.R., Katz R.L., Wang J.Y., Jiang F. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31(22):2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Su J., Liao J., Gao L., Shen J., Guarnera M.A., Zhan M., Fang H., Stass S.A., Jiang F. Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget. 2016;7(5):5131–5142. doi: 10.18632/oncotarget.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gao Y., Wang J., Zhao F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. https://genomebiology.biomedcentral.com/ articles/10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Su H., Lin F., Deng X., Shen L., Fang Y., Fei Z., Zhao L., Zhang X., Pan H., Xie D., Jin X., Xie C. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. 2016 doi: 10.1186/s12967-016-0977-7. tral.com/articles/10.1186/s129 [DOI] [PMC free article] [PubMed]
- 151.Song X., Zhang N., Han P., Moon B.S., Lai R.K., Wang K., Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):e87. doi: 10.1093/nar/gkw075. https://academic.oup.com/nar/article/44/9/e87/2462265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xuan L., Qu L., Zhou H., Wang P., Yu H., Wu T., Wang X., Li Q., Tian L., Liu M., Sun Y. Circular RNA: A novel biomarker for progressive laryngeal cancer. Am. J. Transl. Res. 2016;8(2):932–939. [PMC free article] [PubMed] [Google Scholar]
- 154.Qin M., Liu G., Huo X., Tao X., Sun X., Ge Z., Yang J., Fan J., Liu L., Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 155.Shang X., Li G., Liu H., Li T., Liu J., Zhao Q., Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore) 2016;95(22):e3811. doi: 10.1097/MD.0000000000003811. https://www.nature.com/articles/s41598-017-05432-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xie H., Ren X., Xin S., Lan X., Lu G., Lin Y., Yang S., Zeng Z., Liao W., Ding Y.Q., Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. https://www.nature.com/articles/nature11993 [DOI] [PubMed] [Google Scholar]
- 158.Granados-Riveron J.T., Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim. Biophys. Acta. 2016;1859(10):1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 159.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., Zhu P., Chang Z., Wu Q., Zhao Y., Jia Y., Xu P., Liu H., Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 160.Wang Y., Mo Y., Gong Z., Yang X., Yang M., Zhang S., Xiong F., Xiang B., Zhou M., Liao Q., Zhang W., Li X., Li X., Li Y., Li G., Zeng Z., Xiong W. Circular RNAs in human cancer. 2017 doi: 10.1186/s12943-017-0598-7. https://molecular-cancer.biomedcentral.com/articles/10.1186/s12 [DOI] [PMC free article] [PubMed]
- 161.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. https://www.nature.com/articles/nature11928 [DOI] [PubMed] [Google Scholar]
- 162.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. https:// www.nature.com/articles/srep12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chou Y.T., Lin H.H., Lien Y.C., Wang Y.H., Hong C.F., Kao Y.R., Lin S.C., Chang Y.C., Lin S.Y., Chen S.J., Chen H.C., Yeh S.D., Wu C.W. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70(21):8822–8831. doi: 10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- 164.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 166.Chen J., Xiao H., Huang Z., Hu Z., Qi T., Zhang B., Tao X., Liu S.H. MicroRNA124 regulate cell growth of prostate cancer cells by targeting iASPP. Int. J. Clin. Exp. Pathol. 2014;7(5):2283–2290. [PMC free article] [PubMed] [Google Scholar]
- 167.Guarnerio J., Bezzi M., Jeong J.C., Paffenholz S.V., Berry K., Naldini M.M., Lo-Coco F., Tay Y., Beck A.H., Pandolfi P.P. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 168.Tsvetkova E., Goss G.D. Drug resistance and its significance for treatment decisions in non-small-cell lung cancer. Curr. Oncol. 2012;19(Suppl. 1):S45–S51. doi: 10.3747/co.19.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Carter C.A., Giaccone G. Treatment of nonsmall cell lung cancer: overcoming the resistance to epidermal growth factor receptor inhibitors. Curr. Opin. Oncol. 2012;24(2):123–129. doi: 10.1097/CCO.0b013e32834ec6a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Garofalo M., Quintavalle C., Romano G., Croce C.M., Condorelli G. miR221/222 in cancer: Their role in tumor progression and response to therapy. Curr. Mol. Med. 2012;12(1):27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Garofalo M., Di Leva G., Romano G., Nuovo G., Suh S.S., Ngankeu A., Taccioli C., Pichiorri F., Alder H., Secchiero P., Gasparini P., Gonelli A., Costinean S., Acunzo M., Condorelli G., Croce C.M. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 172.Romano G., Acunzo M., Garofalo M., Di Leva G., Cascione L., Zanca C., Bolon B., Condorelli G., Croce C.M. MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc. Natl. Acad. Sci. USA. 2012;109(41):16570–16575. doi: 10.1073/pnas.1207917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lu F., Zhang J., Ji M., Li P., Du Y., Wang H., Zang S., Ma D., Sun X., Ji C. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int. J. Oncol. 2014;45(1):383–392. doi: 10.3892/ijo.2014.2390. [DOI] [PubMed] [Google Scholar]
- 174.Yang Y.R., Li Y.X., Gao X.Y., Zhao S.S., Zang S.Z., Zhang Z.Q. MicroRNA-137 inhibits cell migration and invasion by targeting bone morphogenetic protein-7 (BMP7) in non-small cell lung cancer cells. Int. J. Clin. Exp. Pathol. 2015;8(9):10847–10853. [PMC free article] [PubMed] [Google Scholar]
- 175.You J., Zhang Y., Li Y., Fang N., Liu B., Zu L., Zhou Q. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am. J. Cancer Res. 2015;5(9):2730–2744. [PMC free article] [PubMed] [Google Scholar]
- 176.Wu G., Ji Z., Li H., Lei Y., Jin X., Yu Y., Sun M. Selective TRAIL-induced cytotoxicity to lung cancer cells mediated by miRNA response elements. Cell Biochem. Funct. 2014;32(7):547–556. doi: 10.1002/cbf.3042. [DOI] [PubMed] [Google Scholar]
- 177.Zarogoulidis P., Petanidis S., Kioseoglou E., Domvri K., Anestakis D., Zarogoulidis K. MiR-205 and miR-218 expression is associated with carboplatin chemoresistance and regulation of apoptosis via Mcl-1 and Survivin in lung cancer cells. Cell. Signal. 2015;27(8):1576–1588. doi: 10.1016/j.cellsig.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 178.Chen L., Kong G., Zhang C., Dong H., Yang C., Song G., Guo C., Wang L., Yu H. MicroRNA-432 functions as a tumor suppressor gene through targeting E2F3 and AXL in lung adenocarcinoma. Oncotarget. 2016;7(15):20041–20053. doi: 10.18632/oncotarget.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Han J., Zhao F., Zhang J., Zhu H., Ma H., Li X., Peng L., Sun J., Chen Z. miR-223 reverses the resistance of EGFR-TKIs through IGF1R/PI3K/Akt signaling pathway. Int. J. Oncol. 2016;48(5):1855–1867. doi: 10.3892/ijo.2016.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Patrawala L., Brown D., Bader A.G. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ma Z.L., Hou P.P., Li Y.L., Wang D.T., Yuan T.W., Wei J.L., Zhao B.T., Lou J.T., Zhao X.T., Jin Y., Jin Y.X. MicroRNA-34a inhibits the proliferation and promotes the apoptosis of non-small cell lung cancer H1299 cell line by targeting TGFbetaR2. Tumour Biol. 2015;36(4):2481–2490. doi: 10.1007/s13277-014-2861-5. [DOI] [PubMed] [Google Scholar]
- 182.Ng W.L., Chen G., Wang M., Wang H., Story M., Shay J.W., Zhang X., Wang J., Amin A.R., Hu B., Cucinotta F.A., Wang Y. OCT4 as a target of miR-34a stimulates p63 but inhibits p53 to promote human cell transformation. 2014 doi: 10.1038/cddis.2013.563. https://www.nature.com/articles/cddis [DOI] [PMC free article] [PubMed] [Retracted]
- 183.Li Y.J., Zhang Y.X., Wang P.Y., Chi Y.L., Zhang C., Ma Y., Lv C.J., Xie S.Y. Regression of A549 lung cancer tumors by anti-miR-150 vector. Oncol. Rep. 2012;27(1):129–134. doi: 10.3892/or.2011.1466. [DOI] [PubMed] [Google Scholar]
- 184.Cao M., Hou D., Liang H., Gong F., Wang Y., Yan X., Jiang X., Wang C., Zhang J., Zen K., Zhang C.Y., Chen X. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur. J. Cancer. 2014;50(5):1013–1024. doi: 10.1016/j.ejca.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 185.Gao Y., Xue Q., Wang D., Du M., Zhang Y., Gao S. miR-873 induces lung adenocarcinoma cell proliferation and migration by targeting SRCIN1. Am. J. Transl. Res. 2015;7(11):2519–2526. [PMC free article] [PubMed] [Google Scholar]
- 186.Wang X., Liu Y., Liu X., Yang J., Teng G., Zhang L., Zhou C. MiR-124 inhibits cell proliferation, migration and invasion by directly targeting SOX9 in lung adenocarcinoma. Oncol. Rep. 2016;35(5):3115–3121. doi: 10.3892/or.2016.4648. [DOI] [PubMed] [Google Scholar]
- 187.Li Q., Yang Z., Chen M., Liu Y. Downregulation of microRNA-196a enhances the sensitivity of non-small cell lung cancer cells to cisplatin treatment. Int. J. Mol. Med. 2016;37(4):1067–1074. doi: 10.3892/ijmm.2016.2513. [DOI] [PubMed] [Google Scholar]
- 188.Liu X.H., Lu K.H., Wang K.M., Sun M., Zhang E.B., Yang J.S., Yin D.D., Liu Z.L., Zhou J., Liu Z.J., De W., Wang Z.X. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. https://bmccancer.biomedcentral.com/articles/10.11 86/1471-2407-12-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li J., Li P., Chen T., Gao G., Chen X., Du Y., Zhang R., Yang R., Zhao W., Dun S., Gao F., Zhang G. Expression of microRNA-96 and its potential functions by targeting FOXO3 in non-small cell lung cancer. Tumour Biol. 2015;36(2):685–692. doi: 10.1007/s13277-014-2698-y. [DOI] [PubMed] [Google Scholar]
- 190.Li C.F., MacDonald J.R., Wei R.Y., Ray J., Lau K., Kandel C., Koffman R., Bell S., Scherer S.W., Alman B.A. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics. 2007;8:92. doi: 10.1186/1471-2164-8-92. https://bmcgenomics.biomedcentral.com/ articles/10.1186/1471-2164-8-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu L., Pu X., Wang Q., Cao J., Xu F., Xu L.I., Li K. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9. Oncol. Lett. 2016;11(2):945–952. doi: 10.3892/ol.2015.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ma W., Ma C.N., Li X.D., Zhang Y.J. Examining the effect of gene reduction in miR-95 and enhanced radiosensitivity in non-small cell lung cancer. Cancer Gene Ther. 2016;23(2-3):66–71. doi: 10.1038/cgt.2016.2. [DOI] [PubMed] [Google Scholar]
- 193.Chen X., Chen S., Hang W., Huang H., Ma H. MiR-95 induces proliferation and chemo- or radioresistance through directly targeting sorting nexin1 (SNX1) in non-small cell lung cancer. Biomed. Pharmacother. 2014;68(5):589–595. doi: 10.1016/j.biopha.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 194.Zhang S., Gao L., Thakur A., Shi P., Liu F., Feng J., Wang T., Liang Y., Liu J.J., Chen M., Ren H. miRNA-204 suppresses human non-small cell lung cancer by targeting ATF2. Tumour Biol. 2016;37(8):11177–11186. doi: 10.1007/s13277-016-4906-4. [DOI] [PubMed] [Google Scholar]
- 195.Sun C., Li S., Yang C., Xi Y., Wang L., Zhang F., Li D. MicroRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6. Biochem. Biophys. Res. Commun. 2016;471(1):82–88. doi: 10.1016/j.bbrc.2016.01.175. [DOI] [PubMed] [Google Scholar]
- 196.Sun C., Huang C., Li S., Yang C., Xi Y., Wang L., Zhang F., Fu Y., Li D. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7(7):8341–8359. doi: 10.18632/oncotarget.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Meng F., Zhang L., Shao Y., Ma Q., Lv H. MicroRNA-377 inhibits non-small-cell lung cancer through targeting AEG-1. Int. J. Clin. Exp. Pathol. 2015;8(11):13853–13863. [PMC free article] [PubMed] [Google Scholar]
- 198.Li Y.J., Ping C., Tang J., Zhang W. MicroRNA-455 suppresses non-small cell lung cancer through targeting ZEB1. Cell Biol. Int. 2016;40(6):621–628. doi: 10.1002/cbin.10584. [DOI] [PubMed] [Google Scholar]
- 199.Sun C.C., Li S.J., Zhang F., Pan J.Y., Wang L., Yang C.L., Xi Y.Y. Li de, J. Hsa-miR-329 exerts tumor suppressor function through down-regulation of MET in non-small cell lung cancer. Oncotarget. 2016;7(16):21510–21526. doi: 10.18632/oncotarget.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li D., Wei Y., Wang D., Gao H., Liu K. MicroRNA-26b suppresses the metastasis of non-small cell lung cancer by targeting MIEN1 via NF-kappaB/MMP-9/VEGF pathways. Biochem. Biophys. Res. Commun. 2016;472(3):465–470. doi: 10.1016/j.bbrc.2016.01.163. [DOI] [PubMed] [Google Scholar]
- 201.Jiao A., Sui M., Zhang L., Sun P., Geng D., Zhang W., Wang X., Li J. MicroRNA-200c inhibits the metastasis of non-small cell lung cancer cells by targeting ZEB2, an epithelial-mesenchymal transition regulator. Mol. Med. Rep. 2016;13(4):3349–3355. doi: 10.3892/mmr.2016.4901. [DOI] [PubMed] [Google Scholar]
- 202.Zhang Y.J., Liu X.C., Du J., Zhang Y.J. MiR-152 regulates metastases of non-small cell lung cancer cells by targeting neuropilin-1. Int. J. Clin. Exp. Pathol. 2015;8(11):14235–14240. [PMC free article] [PubMed] [Google Scholar]
- 203.Tian Y., Luo A., Cai Y., Su Q., Ding F., Chen H., Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J. Biol. Chem. 2010;285(11):7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ma Z., Chen Y., Min L., Li L., Huang H., Li J., Yan Q., Song P., Dai L., Yao X. Augmented miR-10b expression associated with depressed expression of its target gene KLF4 involved in gastric carcinoma. Int. J. Clin. Exp. Pathol. 2015;8(5):5071–5079. [PMC free article] [PubMed] [Google Scholar]
- 205.Xiao H., Li H., Yu G., Xiao W., Hu J., Tang K., Zeng J., He W., Zeng G., Ye Z., Xu H. MicroRNA-10b promotes migration and invasion through KLF4 and HOXD10 in human bladder cancer. Oncol. Rep. 2014;31(4):1832–1838. doi: 10.3892/or.2014.3048. [DOI] [PubMed] [Google Scholar]
- 206.Liu Y., Li M., Zhang G., Pang Z. MicroRNA-10b overexpression promotes non-small cell lung cancer cell proliferation and invasion. Eur. J. Med. Res. 2013;18:41. doi: 10.1186/2047-783X-18-41. https:// eurjmedres.biomedcentral.com/articles/10.1186/2047-783X-18-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang J., Xu L., Yang Z., Lu H., Hu D., Li W., Zhang Z., Liu B., Ma S. MicroRNA-10b indicates a poor prognosis of non-small cell lung cancer and targets E-cadherin. Clin. Transl. Oncol. 2015;17(3):209–214. doi: 10.1007/s12094-014-1213-7. [DOI] [PubMed] [Google Scholar]
- 208.Zhang Y.J., Liu X.C., Du J., Zhang Y.J. MiR-152 regulates metastases of non-small cell lung cancer cells by targeting neuropilin-1. Int. J. Clin. Exp. Pathol. 2015;8(11):14235–14240. [PMC free article] [PubMed] [Google Scholar]
- 209.Tian Y., Luo A., Cai Y., Su Q., Ding F., Chen H., Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J. Biol. Chem. 2010;285(11):7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ma Z., Chen Y., Min L., Li L., Huang H., Li J., Yan Q., Song P., Dai L., Yao X. Augmented miR-10b expression associated with depressed expression of its target gene KLF4 involved in gastric carcinoma. Int. J. Clin. Exp. Pathol. 2015;8(5):5071–5079. [PMC free article] [PubMed] [Google Scholar]
- 211.Xiao H., Li H., Yu G., Xiao W., Hu J., Tang K., Zeng J., He W., Zeng G., Ye Z., Xu H. MicroRNA-10b promotes migration and invasion through KLF4 and HOXD10 in human bladder cancer. Oncol. Rep. 2014;31(4):1832–1838. doi: 10.3892/or.2014.3048. [DOI] [PubMed] [Google Scholar]
- 212.Liu Y., Li M., Zhang G., Pang Z. MicroRNA-10b overexpression promotes non-small cell lung cancer cell proliferation and invasion. Eur. J. Med. Res. 2013;18:41. doi: 10.1186/2047-783X-18-41. https:// eurjmedres.biomedcentral.com/articles/10.1186/2047-783X-18-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhang J., Xu L., Yang Z., Lu H., Hu D., Li W., Zhang Z., Liu B., Ma S. MicroRNA-10b indicates a poor prognosis of non-small cell lung cancer and targets E-cadherin. Clin. Transl. Oncol. 2015;17(3):209–214. doi: 10.1007/s12094-014-1213-7. [DOI] [PubMed] [Google Scholar]