Abstract
P-starved plants scavenge inorganic phosphate (Pi) by developing elevated rates of Pi uptake, synthesizing extracellular phosphatases, and secreting organic acids. To elucidate mechanisms controlling these acclimation responses in photosynthetic organisms, we characterized the responses of the green alga Chlamydomonas reinhardtii to P starvation and developed screens for isolating mutants (designated psr [phosphorus-stress response]) abnormal in their responses to environmental levels of Pi. The psr1-1 mutant was identified in a selection for cells that survived exposure to high concentrations of radioactive Pi. psr1-2 and psr2 were isolated as strains with aberrant levels of extracellular phosphatase activity during P-deficient or nutrient-replete growth. The psr1-1 and psr1-2 mutants were phenotypically similar, and the lesions in these strains were recessive and allelic. They exhibited no increase in extracellular phosphatase activity or Pi uptake upon starvation. Furthermore, when placed in medium devoid of P, the psr1 strains lost photosynthetic O2 evolution and stopped growing more rapidly than wild-type cells; they may not be as efficient as wild-type cells at scavenging/accessing P stores. In contrast, psr2 showed elevated extracellular phosphatase activity during growth in nutrient-replete medium, and the mutation was dominant. The mutant phenotypes and the roles of Psr1 and Psr2 in P-limitation responses are discussed.
P is a nutrient that often limits plant growth in the natural environment. The primary source of P in soils is Pi, which is actively accumulated by both plants and microbes. However, most soil Pi is either covalently bonded to C molecules as Pi esters, or exists as Fe3+, Al3+, or Ca2+ salts. These Pi salts are relatively insoluble and, therefore, are not readily available for transport into microbial cells or plant roots (Halstead and McKercher, 1975).
When plants or microbes are starved for P, they exhibit increased Pi uptake (McPharlin and Bieleski, 1987; Furihata et al., 1992; Shimogawara and Usuda, 1995; Muchhal et al., 1996; Jeschke et al., 1997; Schachtman et al., 1998), secrete acid and alkaline phosphatases (Lefebvre et al., 1990; Duff et al., 1991) and RNases (Loffler et al., 1993; Bariola et al., 1994; Kock et al., 1995; Dodds et al., 1996), and exude low-Mr organic acids that help mobilize stores of Pi that are present in the soil as insoluble salts (Marschner, 1995). Studies with photosynthetic organisms have also demonstrated that glycolysis and photosynthetic activities are modified during P deprivation (Brooks, 1986; Duff et al., 1989; Dietz and Heilos, 1990; Jacob and Lawlor, 1993; Theodorou and Plaxton, 1993; Plesnicar et al., 1994; Wykoff et al., 1998). The mechanisms that control the acclimation of Escherichia coli and Saccharomyces cerevisiae to P limitation have been extensively studied (Wanner, 1993; Oshima et al., 1996; Oshima, 1997). In E. coli a two-component regulatory system governs the transcription of many genes that are responsive to the P levels of the environment (Wanner, 1993). Recently, similar regulatory systems have been identified in Bacillus subtilis and the cyanobacterium Synechococcus sp. strain PCC 7942 (Aiba et al., 1993; Hulett, 1996). In S. cerevisiae many mutants (pho series mutants) have been isolated that have lost their ability to regulate the synthesis of extracellular phosphatases in response to P starvation (Lenburg and O'Shea, 1996; Oshima, 1997). The lesions in these mutants define genes encoding both catalytic and regulatory functions that are important for the acclimation of S. cerevisiae to P limitation. Some of these gene products, including acid and alkaline phosphatases (Pho5 and Pho8) and a high-affinity phosphate transporter (Pho84), are involved in scavenging the limiting nutrient. Others function as transcriptional regulators (Pho4 and Pho2), a cyclin (Pho80), a cyclin-dependent kinase (Pho85), and a cyclin-dependent kinase inhibitor (Pho81); these regulators coordinate limited nutrient availability with the growth and metabolism of the cell. The existence of a similar regulatory pathway in Neurospora crassa has also been established (Kang and Metzenburg, 1990, 1993; Pelleg et al., 1996).
In an attempt to define mechanisms that control the acclimation of photosynthetic eukaryotes to low levels of P, we have identified mutants of Chlamydomonas reinhardtii with aberrant responses to P limitation. C. reinhardtii is a unicellular green alga that has been developed as a model organism for analyzing a number of different physiological processes in photosynthetic eukaryotes, and in particular for the dissection of photosynthesis (Harris, 1989). Many molecular techniques have been developed that allow for sophisticated molecular manipulation of this organism (Rochaix, 1995; Davies and Grossman, 1998; Shimogawara et al., 1998). To elucidate mechanisms that photosynthetic organisms use to sense and respond to P deprivation, we have characterized the responses of C. reinhardtii to P limitation (Quisel et al., 1996; Wykoff et al., 1998) and have isolated mutants of this alga that do not properly acclimate to P limitation. These mutants were selected in two different screens. The first screen involved the isolation of strains that survived high concentrations of radioactive Pi during starvation for P. The second screen identified mutants that were unable to accumulate extracellular phosphatases during P-limited growth or that were unable to completely repress the accumulation of extracellular phosphatase during nutrient-replete growth. Here we describe the physiological and genetic characteristics of these mutants and what they have revealed about the mechanisms that control the responses of C. reinhardtii to P limitation.
MATERIALS AND METHODS
Strains, Culture Medium, and Growth Condition
Chlamydomonas reinhardtii Dangeard strains CC125 (wild type mt+), CC124 (wild type mt−), and CC425 (cw15 arg7-8 mt+) were grown in TAP (Tris-acetate-Pi) medium (Harris, 1989) or TAP medium supplemented with 50 μmol mL−1 Arg. Cells grown in Erlenmeyer flasks were agitated on a gyratory shaker (120 rpm), maintained at 27°C, and illuminated at 80 μmol photons m−2 s−1 from cool-white fluorescent tubes. For P-starvation experiments Pi was eliminated from the culture medium and replaced with TA medium (1.5 mm KCl). To prepare TA solid medium, 0.5% (w/v) agarose (Agarose-I, electrophoretic grade, Dojindo, Kumamoto, Japan) was used instead of 1.2% agar, because there was Pi contamination in the latter.
UV Mutagenesis and 32Pi Suicide Selection of Mutants
Strain CC125 was grown to mid-logarithmic phase (2 × 106 cells mL−1), pelleted by centrifugation (4000g), and suspended in 10 mL of fresh TA medium. The cell suspension was placed in a Petri dish, exposed to UV irradiation from a germicidal UV tube (20 W, distance 50 cm, and 150 s), and then incubated in the dark for 1 d. High specific activity of 32Pi (10 μCi nmol−1) was added to the cultures of mutagenized cells to a final concentration of 10 μm to kill cells that developed an elevated capacity for Pi uptake during P limitation. The cell suspension was incubated in the light (50 μmol photons m−2 s−1) for 1 d and then placed at 4°C in the dark for 1 week. Cold treatment accelerated cell death by retarding processes involved in repairing damage caused by 32Pi accumulation. Surviving cells were spread onto solid medium containing 10 mm Pi and then screened for growth on solid medium containing high (10 mm) and low (10 μm) Pi. Strains that grew normally on high Pi but did not grow well on low Pi were further analyzed. Putative mutants were back-crossed four to five times with parental strains (CC124 and then CC125) before further characterizations.
Insertional Mutagenesis and Screening for Phosphatase Mutants
The plasmid pJD67, harboring the arginosuccinate lyase gene (ARG7) (Davies et al., 1994), was linearized by digestion with HindIII and transformed (Kindle, 1990) into the Arg auxotroph CC425. Mutagenized cells were screened for aberrant accumulation of extracellular phosphatases (Quisel et al., 1996) during P starvation. ARG transformants were replica plated at low density onto solid TAP medium with low (10 μm) or normal (1 mm) Pi and grown for 2 or 3 d in fluorescent light of 50 μmol photons m−2 s−1. Colonies were sprayed with an aqueous solution of 10 mm X-Pi as a visual assay for phosphatase activity (Davies et al., 1994).
Direct Measurement of Pi Uptake
Pi uptake was measured using a procedure similar to that described for the uptake of S (Yildiz et al., 1994). The cells were stirred as a dilute suspension in the light (200 μmol photons m−2 s−1) for 2 min before the addition of 33Pi. At varying times after the addition of the radiolabeled anion, the cells were vacuum filtered onto Supor-450 membranes (pore size 0.45 μm, Gelman Sciences, Ann Arbor, MI), and the membranes were washed with 10 mL of ice-cold TAP medium containing 20 mm Pi. The radioactivity on each filter was quantified in a liquid-scintillation counter (LKB Wallac, Turku, Finland).
Generation of Vegetative Diploids
Vegetative diploids were constructed according to the method of Harris (1989). NIT1 and NIT2 alleles were introduced into parental haploid strains, and diploid cells were selected for growth on solid medium containing nitrate as the sole N source (TAP-N +NO3; NH4Cl in the TAP medium was replaced by 3.5 mm KNO3).
O2 Evolution
Light-saturated (800 μmol photons m−2 s−1) photosynthesis was measured at 27°C as O2 evolution using a Clark-type O2 electrode (Hansatech, UK) as described elsewhere (Wykoff et al., 1998).
Secreted Phosphatase Activity and Periplasmic Protein Analysis
Cells were washed twice with TA medium and then resuspended in appropriate medium for growth. Phosphatase activity was measured at 27°C and pH 8.5 using p-nitrophenyl phosphate as the substrate, as previously described (Quisel et al., 1996). The hydrolysis of the substrate was limited by the amount of cells added to the assay mixture and was proportional to the incubation time for at least 1 h. The back-crossed (3–5 times) mutant strains were crossed to our cell wall-less wild-type strain (cw15), and periplasmic polypeptides were isolated from the mutant, cell wall-less progeny and resolved by SDS-PAGE according to the method of Davies et al. (1994). The polypeptides were visualized by silver staining (Porro et al., 1982).
RESULTS
Measurement of Pi Uptake
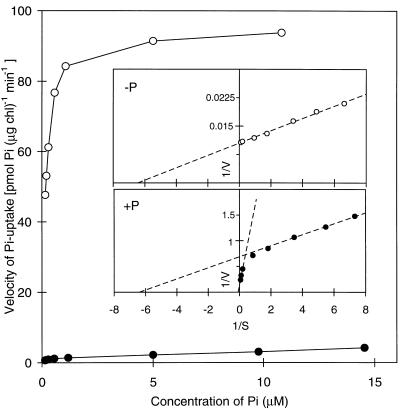
Filtration assays were used to determine the characteristics of Pi transport into C. reinhardtii cells grown under both nutrient-replete and P-starved conditions (Yildiz et al., 1994). The uptake of Pi by cells grown in nutrient-replete medium or starved for Pi for 24 h is shown as a function of the initial Pi concentration in Figure 1. The Vmax for Pi uptake increased by over 10-fold in cells starved for P (Fig. 1). The kinetic analysis of Pi uptake after nutrient-replete growth revealed two distinct kinetic components. The Km for one (low-affinity component) was approximately 10 μm, whereas that for the other (high-affinity component) was between 0.1 and 0.3 μm, as derived from the double-reciprocal plot (Fig. 1, inset). The low-affinity component comprised approximately 80% of total Pi uptake under nutrient-replete conditions. After 24 h of P starvation, all of the Pi uptake seemed to occur via the high-affinity system. Hence, P starvation of C. reinhardtii resulted in an enhanced capacity of the cells to take up Pi and an enhanced affinity for Pi. These results suggest that more than one Pi transport system is used by C. reinhardtii and that the high-affinity system is responsible for most of the transport observed in P-starved cells.
Figure 1.
The velocity of Pi uptake as a function of substrate concentration. 33Pi uptake of the wild-type strain CC125 was performed as described in Methods. Cultures in the early logarithmic phase of growth were either transferred to TAP (+P, •) or TA (−P, ○) medium and allowed to continue growth for 24 h before measuring Pi uptake. The insets are double-reciprocal plots of the data, and the Km values estimated from these plots are 0.16 μm for the high-affinity component and 10 μm for the low-affinity component. chl, Chlorophyll.
Isolation of Mutants Defective in Acclimation to P Limitation
The results presented in Figure 1 and previous data showing that P-starved cells synthesize high levels of extracellular phosphatases (Lien and Knudsen, 1972; Loppes, 1976a, 1976b; Matagne et al., 1976; Patni et al., 1977; Quisel et al., 1996) suggested possible screens for isolating mutants unable to properly acclimate to P limitation. One screen was based on a preferential killing of cells that attain the capacity for elevated 32Pi transport upon exposure to P limitation. The second screen was based on a colorimetric assay to identify mutants with abnormal levels of extracellular phosphatase activity (see Methods). The latter screen is conceptually similar to a screen previously used to isolate mutants in C. reinhardtii that were unable to acclimate to S deprivation (Davies et al., 1994).
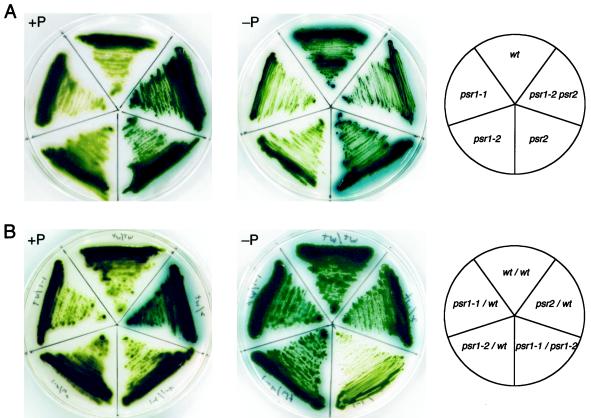
From the first screen we isolated the mutant psr1-1 (phosphorus-stress response), whereas the second screen yielded psr1-2 and psr2-1 (referred to as psr2 in the remainder of the text). Before physiological and biochemical analyses of the mutants, they were back-crossed four to five times to our wild-type strain (CC125) to ensure that the phenotypes analyzed were the result of a single lesion in a homogeneous genetic background. A qualitative analysis of extracellular phosphatase activity (based on the accumulation of a blue precipitate that forms around colonies capable of cleaving Pi from X-Pi) of the mutant and wild-type strains is presented in Figure 2A. In wild-type cells and the psr1-1 and psr1-2 mutants, little extracellular phosphatase activity was detected during growth on solid TAP medium (+P in Fig. 2A). In contrast, the psr2 and the psr1-2 psr2 double mutant accumulated relatively high levels of extracellular phosphatase activity during nutrient-replete growth. Upon P starvation (−P in Fig. 2A), wild-type cells (CC125) and psr2 accumulated high levels of extracellular phosphatase, as indicated by the intense blue halo that surrounds the cells. However, the psr1-1 and psr1-2 mutants showed almost no extracellular phosphatase activity. The psr1-2 psr2 double mutant appeared to accumulate similar levels of extracellular phosphatase activity under both nutrient-replete and P-starvation conditions.
Figure 2.
Qualitative analysis of phosphatase activity secreted by wild-type cells, mutant strains, and vegetative diploids. Wild-type cells (wt), psr1-1, psr1-2, psr2, and psr1-2 psr2 (A) and vegetative diploids of wild-type and the various mutant strains (B) were streaked onto TAP (+P) and TA (−P) solid media before spraying the plates with the colorimetric phosphatase substrate X-Pi. The plates were allowed to develop for 2 h before recording the results. The template shows the positions of the various mutants (A, right) and the different vegetative diploids (B, right) on the plates.
Genetic Characterization of the Mutants
Since the psr1-1 and psr1-2 strains were both unable to secrete active phosphatase during P limitation, we constructed vegetative diploids to determine if the lesions in the strains were allelic. Vegetative diploids of wild-type cells and the individual mutants were also constructed to determine if the mutations were dominant or recessive. The phenotype of a vegetative diploid of psr1-1 and psr1-2 was essentially identical, with respect to phosphatase activity, to that of the individual mutants; almost no phosphatase activity was observed when the diploid was starved for P (−P in Fig. 2B). In contrast, a vegetative diploid of the wild type and either psr1-1 or psr1-2 had a phenotype that was identical to that of wild-type cells; P starvation led to high-level accumulation of alkaline phosphatase activity. Furthermore, a cross of psr1-1 to psr1-2 yielded no wild-type cells in over 400 progeny that were tested. These results clearly demonstrate that the lesions in psr1-1 and psr1-2 are recessive and allelic.
A vegetative diploid of psr2 and wild-type cells exhibited a phenotype that was dominant with respect to psr2. The diploid cells accumulated phosphatase activity in TAP medium even in the presence of a wild-type copy of PSR2. Like the original psr2 mutant, the diploid appeared to have elevated phosphatase activity when starved for P. A similar phenotype was observed for the vegetative diploid of psr1-1 psr2 (data not shown). Furthermore, the psr1 and psr2 mutations segregated independently, indicating that they are nonallelic.
Quantitative Analysis of Phosphatase Activity in the Mutant Strains
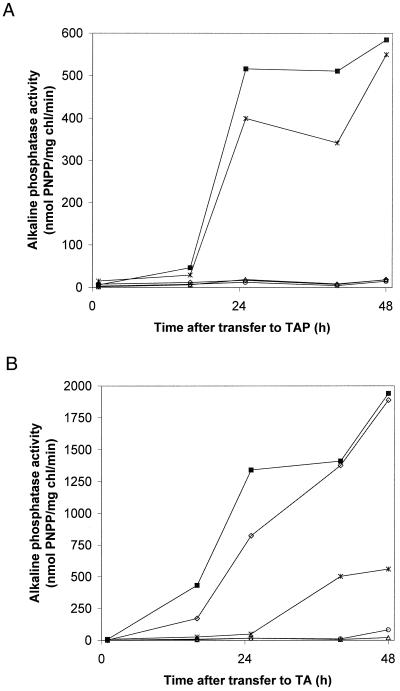
A quantitative analysis of the accumulation of phosphatase activity in the medium of wild-type cells and the mutant strains during nutrient-replete and P-limited growth is presented in Figure 3. Little phosphatase activity accumulated in cultures of wild-type cells grown on nutrient-replete medium (Fig. 3A). After the transfer of wild-type cells to medium devoid of P, a high level of phosphatase activity accumulated (Quisel et al., 1996); 24 to 48 h after the initiation of P deprivation, the alkaline phosphatase activity was at least 100-fold higher (Fig. 3B) than in cells that were not starved. The psr1-1 and psr1-2 mutants showed no extracellular phosphatase activity when grown on nutrient-replete medium (Fig. 3A) or after exposure to medium devoid of P (Fig. 3B). The psr2 strain exhibited constitutive phosphatase activity when grown on nutrient-replete medium that was approximately 25% of the level observed after starvation (compare activity for psr2 in Fig. 3). Most of the extracellular phosphatase activity associated with psr2 grown in nutrient-replete medium remained in the supernatant when the cells were washed by centrifugation (data not shown). This explains why the amount of extracellular phosphatase activity was initially low after the transfer of psr2 cells to fresh medium (Fig. 3, 0 time point). The psr1-2 psr2 double mutant accumulated approximately the same amount of phosphatase activity as the psr2 strain during growth on complete medium (Fig. 3A), however, this level did not change when the cells were transferred to medium lacking P (Fig. 3B). These results demonstrate that the psr1 lesion prevents the induction of phosphatase activity in the psr2 strain but does not prevent constitutive, extracellular phosphatase accumulation.
Figure 3.
Quantitation of extracellular alkaline phosphatase activity in wild-type and mutant cultures. All strains were grown in TAP medium to early logarithmic phase, washed twice with TA medium, and then transferred to either TAP (A) or TA (B) liquid medium for 16, 25, 40, and 48 h before measuring the alkaline phosphatase activity. ◊, Wild type; ▪, psr2; ▵, psr1-1; ○, psr1-2; asterisks, psr1-2psr2. PNPP, p-Nitrophenylphosphate; chl, chlorophyll.
To determine if the aberrations in the psr1-1, psr1-2, and psr2 mutants were specific to P deprivation and not involved in global regulation of stress responses, we tested these strains for their ability to acclimate to S limitation. During S limitation C. reinhardtii synthesizes an extracellular arylsulfatase that can be assayed colorimetrically (Davies et al., 1994, 1996). None of the three mutant strains exhibited arylsulfatase activity before starving the cells for S, and they all accumulated normal levels of the extracellular arylsulfatase after S deprivation (D.D. Wykoff and A.R. Grossman, data not shown).
Pi Transport
Several tests were performed to determine if the lesions in the mutants resulted in aberrations in other responses observed in wild-type cells during P-limited growth. Initially, the mutant strains were tested for their ability to take up Pi after growth in TAP and TA media (Table I). Measurements of the Vmax for Pi uptake for both the wild-type and mutant strains grown in complete medium varied from 3.11 to 6.43 pmol Pi μg−1 chlorophyll min−1. After 24 h of P starvation, the wild-type and psr2 mutant cells exhibited a 14-fold increase in the Vmax for Pi uptake. In contrast, P starvation of psr1-1 or psr1-2 for 24 h resulted in little increase in the Vmax. The psr1-2 psr2 double mutant also exhibited little increase in the Vmax for Pi uptake after starvation. Finally, wild-type cells grown in nutrient-replete medium and the psr1 mutant strains maintained in either nutrient-replete or P-deficient medium exhibited both low- and high-affinity Pi transport (data not shown).
Table I.
Maximal rate of Pi uptake in wild-type and mutant strains
| Strain |
Vmax Pi Uptake
|
Increase in Vmax | |
|---|---|---|---|
| TAP medium 1d | TA medium 1d | ||
| pmol Pi μg−1 chlorophyll min−1 | -fold | ||
| Wild type | 6.43 (1.66) | 91.1 (3.9) | 14.2 |
| psr2 | 6.42 (0.24) | 90.8 (36) | 14.1 |
| psr1-1 | 3.11 (2.28) | 5.38 (3.1) | 1.7 |
| psr1-2 | 5.32 (1.64) | 7.71 (2.21) | 1.5 |
| psr1-2 psr2 | 4.12 (2.35) | 4.50 (2.6) | 1.1 |
The units below are the means of two independently grown cultures. The Vmax of Pi uptake was derived from at least three different Pi concentrations (i.e. 5, 10, and 15 μm). The values in parentheses are the difference from the mean in the same units.
Periplasmic Proteins
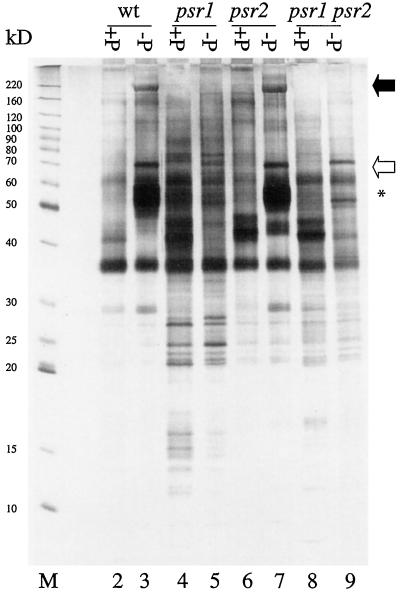
Profiles of periplasmic polypeptides from wild-type cells, psr1-1, psr2 and the double mutant psr1-1 psr2 grown in both complete medium and medium devoid of P are shown in Figure 4. For wild-type cells a periplasmic polypeptide of approximately 190 kD (marked by a filled arrow) accumulated as the cells grew in medium devoid of P (lanes 2 and 3). This polypeptide was previously shown to correspond to the major, derepressible extracellular phosphatase (Quisel et al., 1996). Cultures of the psr1-1 mutant did not accumulate the 190-kD polypeptide upon P starvation (compare lanes 4 and 5). Similar results were observed for the psr1-2 strain (data not shown). Hence, the 190-kD phosphatase is not synthesized, not exported, or rapidly degraded in the psr1 strains. In the psr2 mutant the 190-kD polypeptide accumulated only in the growth medium when the cells were starved for P (compare lanes 6 and 7). This suggests that the phosphatase activity that is constitutive in the psr2 strain is not a consequence of abnormal expression of the gene encoding the 190-kD species. Consistent with this interpretation is the finding that the constitutive phosphatase activity that accumulated in psr2 cultures in nutrient-replete medium was independent of Ca2+ (data not shown), whereas the 190-kD phosphatase requires Ca2+ for activity (Quisel et al., 1996). Furthermore, the psr1-1 psr2 double mutant did not accumulate the 190-kD polypeptide upon starvation for P (compare Fig. 4, lanes 8 and 9), which is consistent with the measurements of phosphatase activity in this strain (Fig. 3).
Figure 4.
Profiles of periplasmic polypeptides from wild-type (wt) and the mutant strains after transfer to TAP (lanes 2, 4, 6, and 8) or TA (lanes 3, 5, 7, and 9) medium for 48 h. The samples (3 μg per lane) loaded in the different lanes are from wild type (lanes 2 and 3), psr1-1 (lanes 4 and 5), psr2 (lanes 6 and 7), and psr1-1 psr2 (lanes 8 and 9). Lane M, Benchmark Mr markers (GIBCO-BRL). The positions of the two alkaline phosphatases are marked with arrows (black arrow for the 190-kD species and white arrow for the 70-kD species), and a cluster of other prominent polypeptides that accumulate in the medium in response to P limitation is marked with an asterisk. The polypeptides between 10 and 30 kD in the psr1 strains were observed consistently throughout three independent protein isolations.
Quisel et al. (1996) reported the accumulation of a second extracellular alkaline phosphatase that accumulated during P limitation and migrates with an apparent molecular mass of 70 kD; the polypeptide marked with a white arrow may represent that phosphatase (Fig. 4). This polypeptide appeared to be absent in the psr1-1 strain (lanes 4 and 5) but, like the 190-kD species, accumulated in psr2 upon P deprivation (lanes 6 and 7). In the psr1-1 psr2 double mutant a polypeptide that migrated at a position that was slightly higher than the 70-kD phosphatase accumulated in the culture medium. It is unlikely that this species is the 70-kD phosphatase, although we cannot rule out that possibility.
Wild-type cells also synthesized a cluster of extracellular polypeptides with molecular masses ranging from 50 to 60 kD during P-limited growth (marked by an asterisk in Fig. 4). The functions of these polypeptides, which accumulated normally in psr2 but not in psr1-1 or the psr1-1 psr2, are not known. Finally, low-molecular-mass polypeptides observed (in three different periplasmic protein preparations) in the medium from psr1-1 and psr1-1 psr2 cultures were not apparent in extracellular protein preparations from wild-type cells. These polypeptides may arise from increased proteolysis, aberrant processing of extracellular proteins, or increased leakage of cytoplasmic proteins in the mutant strains.
Growth and Photosynthetic O2 Evolution
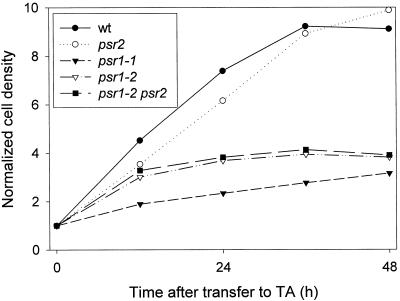
The psr1-1 and psr1-2 strains did not grow to the same extent as wild-type cells or the psr2 mutant when exposed to conditions of P deprivation (Fig. 5). Wild-type cells and the psr2 mutant doubled three to four times after they were placed in medium devoid of P. The psr1-1 mutant doubled only once, whereas the psr1-2 mutant doubled between one and two times after being placed in medium devoid of P. Growth characteristics of the psr1-2 psr2 double mutant were similar to those of psr1-2.
Figure 5.
Increase in cell density after the transfer of CC125 (wt), psr1-1, psr1-2, psr2, and psr1-2 psr2 to medium devoid of P. All of the cultures were grown to a density of 2 to 4 × 106 cells mL−1, washed twice with TA medium, and adjusted to a final density of 5 × 105 cells mL−1 in TA medium. The increase in cell density was determined at 12, 24, 36, and 48 h after the transfer. The data presented here are from one experiment, but identical trends were observed in two additional experiments.
Recently, it was shown that starvation for either P or S leads to a decline in photosynthetic electron transport activity (Wykoff et al., 1998). Within 4 d of P starvation and 1 d of S starvation, O2 evolution declined by approximately 75%. This decrease reflects damage to PSII and the generation of PSII QB-nonreducing centers. Furthermore, a mutant abnormal for many responses to S deprivation dies much more rapidly than wild-type cells during S stress. This death is light dependent and appears to reflect an inability of the mutant to down-regulate photosynthetic electron transport (Davies et al., 1996).
Wild-type cells and the psr2 mutant showed a similar decrease in photosynthetic O2 evolution during P deprivation. O2 evolution declined more rapidly in the psr1-1, psr1-2 (data not shown), and psr1-2 psr2 mutants than in wild-type cells (Table II), even though the viability of all of the mutant strains during P deprivation was similar to that of wild-type cells. The rapid decline in both photosynthesis and growth in the mutants suggests that they may more rapidly experience starvation when P is removed from the medium.
Table II.
Photosynthetic rate of wild-type and mutant strains
| Strain | Rate of O2 Evolution Relative to Wild-Type Unstarved Cells
|
||
|---|---|---|---|
| TAP medium | TA medium 1d | TA medium 2d | |
| Wild type | 100% (5.9%) | 89.7% (5.5%) | 59.8% (9.0%) |
| psr2 | 97.5% (5.0%) | 77.9% (3.8%) | 59.3% (5.0%) |
| psr1-1 | 115% (10.3%) | 36.8% (6.7%) | 26.5% (11.3%) |
| psr1-2 psr2 | 112% (10.0%) | 43.6% (7.9%) | 37.3% (3.9%) |
The means of three independently grown cultures are indicated below with the se in parentheses. The wild-type rate of O2 evolution (100%) was 204 μmol O2 mg−1 chlorophyll h−1.
DISCUSSION
Little is known about the ways in which photosynthetic eukaryotes perceive and respond to P limitation. Generally, when organisms are starved for P, they synthesize both phosphatases and RNases that help them scavenge Pi from external and internal pools. Vascular plants may also increase their root-to-shoot ratio, allowing for more effective mining of Pi from the soil (Lynch, 1995), associate with mycorrhizae, which would facilitate Pi uptake (Smith and Read, 1997), and secrete organic acids, which helps mobilize stores of bound Pi in the soil (Marschner, 1995). In C. reinhardtii there are two major extracellular phosphatases that accumulate in response to Pi limitation (Quisel et al., 1996). The most abundant of these phosphatases has an apparent molecular mass of approximately 190 kD and its activity is Ca2+ dependent. At pH 9.5 this phosphatase is responsible for between 90% and 95% of the extracellular phosphatase activity in wild-type cells that are starved for P; the pH optimum for this phosphatase is 9.5 with very low activity below pH 7.0. A second extracellular phosphatase, which accounts for most of the remaining activity, has a molecular mass of approximately 70 kD and its activity is independent of Ca2+. Some mutants of C. reinhardtii with impaired phosphatase activity have been isolated (Loppes, 1978; Bachir et al., 1996), but they have not been extensively characterized.
Pi uptake in C. reinhardtii is also influenced by the P status of the medium. There appear to be two different kinetic components associated with the transport of Pi into cells grown under P-replete conditions. This suggests that at least two different Pi transport systems are present in C. reinhardtii. One of these systems has a much higher affinity for Pi than the other (0.1–0.3 μm compared with approximately 10 μm). When wild-type cells are starved for P, there is an over 10-fold increase in the rate of Pi transport and only the high-affinity system is detected.
Increased Pi uptake also occurs in vascular plants when Pi levels in the environment are low. The kinetics of Pi uptake by vascular plants is still controversial; most studies suggest the presence of multiple transport systems (Ullrich-Eberius et al., 1984; McPharlin and Bieleski, 1987; Nandi et al., 1987; Furihata et al., 1992), whereas others have argued for one transport system (Drew et al., 1984; Lefebvre et al., 1990; Shimogawara and Usuda, 1995). In the majority of studies there appears to be a constitutively expressed low-affinity Pi transport system and a second, high-affinity system that is derepressed during Pi-limited growth, although more than two systems may exist (Nandi et al., 1987). The high-affinity Pi transport system in plants has a Km of 3 to 7 μm, whereas the low-affinity system has a Km of 50 to 330 μm (Schachtman et al., 1998). Recently, a number of genes encoding Pi transport systems have been cloned from vascular plants (Muchhal et al., 1996; Kai et al., 1997; Leggewie et al., 1997; Mitsukawa et al., 1997; Smith et al., 1997; Daram et al., 1998; Liu et al., 1998; Okumura et al., 1998).
There appear to be both differences and similarities between Pi transport in vascular plants and C. reinhardtii. First, the Km values for the low- and high-affinity transporters in C. reinhardtii are 1 order of magnitude lower than the Km values for the corresponding transporters of vascular plants. Furthermore, whereas C. reinhardtii and at least some vascular plants (Drew et al., 1984; Schmidt et al., 1992; Shimogawara and Usuda, 1995) have high-affinity Pi transport when grown in P-replete medium, the level of induction of the high-affinity transporter during P starvation is higher for C. reinhardtii (more than 10-fold) than for vascular plants (2- to 5-fold). The detection of high-affinity Pi transport in nutrient-replete C. reinhardtii cultures suggests constitutive synthesis of this transport system. It is not known whether the increase in high-affinity Pi transport that accompanies P limitation is a consequence of increased synthesis of the constitutive system or induction of a second high-affinity Pi transporter. It is also possible that under the optimal growth conditions being used, the cells have the capacity for more rapid intracellular utilization of Pi than can be supplied by the low-affinity Pi transport system. Therefore, the cells would experience Pi limitation and high-affinity transport would be partially induced. Like wild-type cells, the psr1-1 and psr1-2 mutants have both low- and high-affinity Pi transport during nutrient-replete growth (data not shown). These results suggest that the high-affinity Pi transport activity observed in unstarved, wild-type cells is not regulated by the Psr1 polypeptide.
We have used two different approaches to isolate mutants that are unable to properly acclimate to P deprivation. One approach involved a suicide selection procedure using 32Pi. Cells that synthesize elevated levels of the high-affinity Pi transport system during P starvation would rapidly incorporate radiolabeled Pi into nucleic acids and phospholipids, which would result in lethality; mutants unable to synthesize the high-affinity transport system in response to P starvation would survive longer periods of exposure to the radioisotope. The second approach exploited the finding that P-starved cells secrete extracellular phosphatases that are readily detected by spraying the colonies with the chromogenic phosphatase substrate X-Pi. Colonies that cannot synthesize extracellular phosphatases during P starvation do not develop a blue ”halo” (e.g. psr1), whereas colonies that constitutively produce high levels of extracellular phosphatase develop a blue halo when grown in nutrient-replete medium (e.g. psr2).
Two mutants with a similar phenotype, psr1-1 and psr1-2, were isolated by the different screens described above. These mutants were defective in the synthesis of extracellular phosphatases and were unable to increase the rate of Pi transport upon exposure to P limitation. Whereas the psr1-1 allele appears to be null for both activities, the psr1-2 allele accumulates a small amount (less than 5% relative to wild-type cells) of extracellular phosphatase activity upon P starvation (Fig. 3B, 48 h). Furthermore, both strains failed to accumulate periplasmic polypeptides specifically associated with P-limited growth (Fig. 4). The finding that the phenotype of a psr1-1 psr1-2 vegetative diploid was essentially identical to that of each of the haploid strains demonstrated that psr1-1 and psr1-2 were alleles of the same gene.
Additional characterizations of the mutants demonstrated a decline in photosynthetic activity and growth, after transfer to medium devoid of P, that was more rapid than in wild-type cells; the extent of growth was slightly more for psr1-2 than for psr1-1 (again showing a slight difference in the phenotype of the two mutant alleles). The kinetics of the decline in growth and photosynthetic O2 evolution suggest that the psr1 mutants are more sensitive to P depletion than wild-type cells. This phenotype may result from the inability of these strains to access low levels of external Pi and/or to mobilize internal Pi stores. With a decreased ability to scavenge Pi, these mutants would more rapidly down-regulate metabolic processes such as photosynthetic O2 evolution, and growth would rapidly stop. Furthermore, whereas the decrease in photosynthetic activity in the psr1 mutants occurs more rapidly than in wild-type cells, the modes by which photosynthetic electron transport is down regulated appear to be similar to that of wild-type cells (D.D. Wykoff and A.R. Grossman, data not shown). There is a decrease in linear electron transport with the major site of inhibition being PSII. The inhibition results from decreased photochemical efficiency and the accumulation of reaction centers that can perform a charge separation but that have an extremely slow rate of electron transfer between QA and QB. These PSII, QB-nonreducing centers were previously shown to be major components in the down-regulation of photosynthetic activity in wild-type cells during both P and S deprivation (Wykoff et al., 1998). Finally, whereas the mutant cells stop growing more rapidly than wild-type cells upon elimination of P from the medium, they survive long periods of P limitation, just like wild-type cells. These results suggest that the “general responses” to nutrient deprivation (Davies et al., 1996), which lead to the cessation of cell division and decreased photosynthetic activity and allow for extended survival during nutrient limitation, can still occur in the psr1 strains.
Based on the phenotype of the psr1 mutants, the lesions in these strains are likely to be in a regulatory gene that is needed to activate the specific but not the general responses to P deprivation. The PSR1 gene product may be directly involved in sensing the P status of the environment, or a component of the signal transduction chain that transmits the P deprivation to the transcriptional machinery of the cell. The cloning and characterization of the PSR1 gene (data not shown) suggests that Psr1 may be a transcription factor. Recently, we have also identified several mutants of C. reinhardtii that are abnormal in their responses to S limitation (Davies et al., 1996, 1999). One such mutant has been designated sac1. This mutant, unlike the psr1 strain, is unable to control both the specific and general responses; this strain can neither regulate the synthesis of arylsulfatase (specific response) nor down-regulate photosynthetic electron transport (general response) during S starvation and, as a consequence, dies rapidly upon the imposition of S deprivation.
In contrast to the psr1 mutants, the psr2 mutant shows constitutive extracellular phosphatase activity and has a dominant phenotype. The extracellular phosphatase that accumulates during nutrient-replete growth does not appear to be the 190- or the 70-kD species, based on the metal dependence of the phosphatase activity and the analysis of periplasmic proteins in the mutant strains. The results are consistent with either relatively high-level constitutive expression of an extracellular phosphatase that is not normally abundant or the export of a phosphatase that is normally intracellular. Additional characterizations are required to elucidate the nature of the lesion that leads to constitutive extracellular phosphatase accumulation and the polypeptide that is responsible for this activity.
Abbreviation:
- X-Pi
5-bromo-4-chloro-3-indolyl-phosphate
Footnotes
This work was supported by the Japan-U.S. Cooperative Science Program from Japan Society of Plant Physiologists (JSPS) and the National Science Foundation (no. INT 9513 133 to H.U. and A.R.G.), the Research for the Future Program (no. JSPS-RFTF97R16001) from JSPS (to H.U.), the Asahi Glass Foundation (to K.S.), the Ministry of Education, Science, Sports and Culture, Japan (to K.S.), and the U.S. Department of Agriculture (grant no. 9302076 to A.R.G.). This is Carnegie Institution of Washington publication no. 1412.
LITERATURE CITED
- Aiba H, Nagaya M, Mizuno T. Sensor and regulator proteins from the cyanobacterium Synechococcus species PCC 7942 that belong to the bacterial signal-transduction protein families: implication in the adaptive response to phosphate limitation. Mol Microbiol. 1993;8:81–91. doi: 10.1111/j.1365-2958.1993.tb01205.x. [DOI] [PubMed] [Google Scholar]
- Bachir F, Baise E, Loppes R. Mutants impaired in derepressible alkaline phosphatase activity in C. reinhardtii. Plant Sci. 1996;119:93–101. [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Brooks A. Effects of phosphorus nutrition on ribulose-1,5-bisphosphate carboxylase activation, photosynthetic quantum yield and amounts of some Calvin-cycle metabolites in spinach leaves. Aust J Plant Physiol. 1986;13:221–237. [Google Scholar]
- Daram P, Brunner S, Persson BL, Amrhein N, Bucher M. Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta. 1998;206:225–233. doi: 10.1007/s004250050394. [DOI] [PubMed] [Google Scholar]
- Davies JP, Grossman AR. Responses to deficiencies in macronutrients. In: Rochaix J-D, Goldschmidt-Clermont M, Merchant S, editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 613–635. [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell. 1994;6:53–63. doi: 10.1105/tpc.6.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 1996;15:2150–2159. [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR (1999) Sac3, an Snf1-like serine-threonine kinase that positively and negatively regulates the responses of Chlamydomonas reinhardtii to sulfur limitation. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Dietz K-J, Heilos L. Carbon metabolism in spinach leaves as affected by leaf age and phosphorus and sulfur deprivation. Plant Physiol. 1990;93:1219–1225. doi: 10.1104/pp.93.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Clarke AE, Newbigin E. Molecular characterization of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Mol Biol. 1996;31:227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Drew MC, Saker LR, Barber SA, Jenkins W. Changes in the kinetics of phosphate and potassium absorption in nutrient-deficient barley roots measured by a solution-depletion technique. Planta. 1984;160:490–499. doi: 10.1007/BF00411136. [DOI] [PubMed] [Google Scholar]
- Duff SMG, Moorhead GBB, Lefebvre DD, Plaxton WC. Phosphate starvation inducible ‘bypasses’ of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol. 1989;90:1275–1278. doi: 10.1104/pp.90.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Plaxton WC, Lefebvre DD. Phosphate starvation response in plant cells: de novo synthesis and degradation of acid phosphatase. Proc Natl Acad Sci USA. 1991;88:9538–9542. doi: 10.1073/pnas.88.21.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Suzuki M, Sakurai H. Kinetic characterization of two phosphate uptake systems with different affinities in suspension-cultured Catharanthus roseus protoplasts. Plant Cell Physiol. 1992;33:1151–1157. [Google Scholar]
- Halstead RL, McKercher RB (1975) Biochemistry and cycling of phosphorus. In EA Paul, AD McLaren, eds, Soil Biochemistry, Vol 4. Marcel Dekker, New York, pp 50–54
- Harris EH. The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Hulett FM. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol Microbiol. 1996;19:933–939. doi: 10.1046/j.1365-2958.1996.421953.x. [DOI] [PubMed] [Google Scholar]
- Jacob J, Lawlor DW. In vivo photosynthetic electron transport does not limit photosynthetic capacity in phosphate-deficient sunflower and maize leaves. Plant Cell Environ. 1993;16:785–795. [Google Scholar]
- Jeschke W, Peuke A, Kirby E, Pate J, Hartung W. Effects of P defficiency on assimilation and transport of nitrate and phosphate in intact plants of castor bean (Ricinus communis L.) J Exp Bot. 1997;48:75–91. [Google Scholar]
- Kai M, Masuda Y, Kikuchi Y, Osaki M, Tadano T. Isolation and characterization of a cDNA from Catharanthus roseus which is highly homologous with phosphate transporter. Soil Sci Plant Nutr. 1997;43:227–235. [Google Scholar]
- Kang S, Metzenburg RL. Molecular analysis of nuc-1 positive, a gene controlling phosphorus acquisition in Neurospora crassa. Mol Cell Biol. 1990;10:5839–5848. doi: 10.1128/mcb.10.11.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Metzenburg RL. Insertional mutagenesis in Neurospora crassa: cloning and molecular analysis of the preg+ gene controlling the activity of the transcriptional activator NUC-1. Genetics. 1993;133:193–202. doi: 10.1093/genetics/133.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K. High frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock M, Loffler A, Abel S, Glund K. cDNA structure and regulatory properties of a family of starvation induced ribonucleases from tomato. Plant Mol Biol. 1995;27:477–485. doi: 10.1007/BF00019315. [DOI] [PubMed] [Google Scholar]
- Lefebvre DD, Duff SMG, Fife CA, Julien-Inalsingh C, Plaxton WC. Response to phosphate deprivation in Brassica nigra suspension cells. Enhancement of intracellular, cell surface and secreted phosphatase activities compared to increases in Pi absorption rate. Plant Physiol. 1990;93:504–511. doi: 10.1104/pp.93.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggewie G, Wilmitzer L, Riesmeier JW. Two cDNAs from potato are able to complement a phosphate uptake deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell. 1997;9:381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, O'Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- Lien T, Knudsen G. Synchronous cultures of Chlamydomonas reinhardtii: synthesis of repressed and derepressed phosphatase during the life cycle. Biochim Biophys Acta. 1972;287:154–163. [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ. Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant-Microbe Interact. 1998;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- Loffler A, Glund K, Irie M. Amino acid sequence of an intracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur J Biochem. 1993;214:627–633. doi: 10.1111/j.1432-1033.1993.tb17962.x. [DOI] [PubMed] [Google Scholar]
- Loppes R. Release of enzymes by normal and wall free cells of Chlamydomonas. J Bacteriol. 1976a;128:114–116. doi: 10.1128/jb.128.1.114-116.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppes R. Genes involved in the regulation of the neutral phosphatase in Chlamydomonas reinhardtii. Mol Gen Genet. 1976b;15:1147–1157. doi: 10.1007/BF00332906. [DOI] [PubMed] [Google Scholar]
- Loppes R. A mutation altering some properties of the neutral phosphatase in Chlamydomonas reinhardtii: possible post-translational modification of phosphatase structure. J Bacteriol. 1978;135:551–558. doi: 10.1128/jb.135.2.551-558.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. Root architecture and plant productivity. Plant Physiol. 1995;109:7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. San Diego, CA: Academic Press; 1995. [Google Scholar]
- Matagne RF, Loppes R, Deltour R. Phosphatase of Chlamydomonas reinhardtii: biochemical and cytochemical approach with specific mutants. J Bacteriol. 1976;126:937–950. doi: 10.1128/jb.126.2.937-950.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPharlin IR, Bieleski RL. Phosphate uptake by Spirodela and Lemna during early phosphorus deficiency. Aust J Plant Physiol. 1987;14:561–572. [Google Scholar]
- Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc Natl Acad Sci USA. 1997;94:7098–7102. doi: 10.1073/pnas.94.13.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi SK, Pant RC, Nissen P. Multiphasic uptake of phosphate by corn roots. Plant Cell Environ. 1987;10:463–474. [Google Scholar]
- Okumura S, Mitsukawa N, Shirano Y, Shibata D. Phosphate transporter gene family of Arabidopsis thaliana. DNA Res. 1998;5:261–269. doi: 10.1093/dnares/5.5.261. [DOI] [PubMed] [Google Scholar]
- Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- Oshima Y, Ogawa N, Harashima S. Regulation of phosphatase synthesis in Saccharomyces cerevisiae. Gene. 1996;179:171–177. doi: 10.1016/s0378-1119(96)00425-8. [DOI] [PubMed] [Google Scholar]
- Patni NJ, Dhawale SW, Aaronson S. Extracellular phosphatases of Chlamydomonas reinhardtii and their regulation. J Bacteriol. 1977;130:205–211. doi: 10.1128/jb.130.1.205-211.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelleg Y, Aramayo R, Kang S, Hall JG, Metzenburg RL. NUC-2, a component of the phosphate-regulated signal transduction pathway in Neurospora crassa, is an ankyrin repeat protein. Mol Gen Genet. 1996;252:709–716. doi: 10.1007/BF02173977. [DOI] [PubMed] [Google Scholar]
- Plesnicar M, Kastori R, Petrovic N, Pankovic D. Photosynthesis and chlorophyll fluorescence in sunflower (Helianthus annuus L.) leaves as affected by phosphorus nutrition. J Exp Bot. 1994;45:919–924. [Google Scholar]
- Porro M, Viti S, Antoni G, Saletti M. Ultrasensitive silver-stain method for the detection of proteins in polyacrylamide gels and immunoprecipitates on agarose gels. Anal Biochem. 1982;127:316–321. doi: 10.1016/0003-2697(82)90179-8. [DOI] [PubMed] [Google Scholar]
- Quisel JD, Wykoff DD, Grossman AR. Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 1996;111:839–848. doi: 10.1104/pp.111.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J-D. Chlamydomonas reinhardtii as the photosynthetic yeast. Annu Rev Genet. 1995;29:209–230. doi: 10.1146/annurev.ge.29.120195.001233. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. Phosphorus uptake by plants. From soil to cell. Plant Physiol. 1998;116:447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M-E, Heim S, Wylegalla C, Helmbrecht C, Wagner KG. Characterization of phosphate uptake by suspension cultured Catharanthus roseus cells. J Plant Physiol. 1992;140:179–194. [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics. 1998;148:1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K, Usuda H. Uptake of inorganic phosphate by suspension cultured tobacco cells: kinetics and regulation by Pi starvation. Plant Cell Physiol. 1995;36:341–351. [Google Scholar]
- Smith FW, Ealing PM, Dong B, Delhaize E. The cloning of 2 Arabidopsis genes belonging to a phosphate transporter family. Plant J. 1997;11:83–92. doi: 10.1046/j.1365-313x.1997.11010083.x. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Theodorou ME, Plaxton WC. Metabolic adaptations of plant respiration to nutritional phosphate deprivation. Plant Physiol. 1993;101:339–344. doi: 10.1104/pp.101.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich-Eberius C, Novacky A, van Bel A. Phosphate uptake in Lemna gibba G1: energetics and kinetics. Planta. 1984;161:46–52. doi: 10.1007/BF00951459. [DOI] [PubMed] [Google Scholar]
- Wanner BL. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 1998;117:129–139. doi: 10.1104/pp.117.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Davies JP, Grossman AR. Characterization of a high-affinity sulfate transport system that accumulates during sulfur-limited growth of C. reinhardtii. Plant Physiol. 1994;104:981–987. doi: 10.1104/pp.104.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]