Abstract
Indomethacin, a nonsteroidal anti-inflammatory drug, has been shown to induce white adipocyte differentiation; however, its roles in brown adipocyte differentiation and activation in brown adipose tissue (BAT) and obesity are unknown. To address this issue, we treated mouse brown preadipocytes with different doses of indomethacin, and delivered indomethacin to interscapular BAT (iBAT) of obese mice using implanted osmotic pumps. Indomethacin dose dependently increased brown preadipocyte differentiation and upregulated both mRNA and protein expression of uncoupling protein 1 (UCP1) and peroxisome proliferator-activated receptor (PPAR) γ coactivator 1-alpha. The mechanistic study showed that indomethacin significantly activated the reporter driven by the PPAR response element, indicating that indomethacin may work as a PPARγ agonist in this cell line. Consistently, indomethacin significantly decreased iBAT mass and fasting blood glucose levels in high-fat diet–induced obesity (DIO) mice. Histologic analysis showed that brown adipocytes of indomethacin-treated mice contained smaller lipid droplets compared with control mice, suggesting that indomethacin alleviated the whitening of BAT induced by the high-fat diet. Moreover, indomethacin significantly increased UCP1 mRNA expression in iBAT. Taken together, this study indicates that indomethacin can promote mouse brown adipocyte differentiation, and might increase brown fat and glucose oxidation capacity in DIO mice.
Introduction
In recent decades, obesity has become a major public health problem in developed, and even in developing countries (Ng et al., 2014). More than two-thirds of US adults are considered either overweight or obese (Flegal et al., 2012). Obesity is positively associated with a number of complications including type-2 diabetes, cardiovascular disease, gallbladder disease, osteoarthritis, and even some types of cancers (Jensen et al., 2014). Imbalance between energy intake and expenditure is thought to contribute to obesity (Spiegelman and Flier, 2001). Adipose tissue can be classified into white adipose tissue (WAT) and brown adipose tissue (BAT). WAT is the major site for energy storage, and BAT is the major site for thermogenic energy expenditure by metabolizing glucose and fatty acids to produce heat. Historically, it was believed that BAT only exists in human neonates. Newly developed technologies, especially positron emission tomography in combination with computed tomography, have demonstrated that adult humans do have BAT (Cypess et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009).
Despite extensive research, few drugs have been approved for treating obesity by the US Food and Drug Administration (Narayanaswami and Dwoskin, 2017). Currently approved drugs target energy intake either by suppression of appetite (phentermine) or by interfering with nutrient absorption (orlistat). However, these drugs have severe cardiovascular and/or psychiatric side effects, and obesity relapse may occur when stopping the drugs (Kakkar and Dahiya, 2015). Emerging evidence on the role of functional BAT in increasing energy expenditure in human adults opens a new direction for anti-obesity therapy.
Indomethacin was discovered in 1963 (Hart and Boardman, 1963) and approved as a nonsteroidal anti-inflammatory drug by the US Food and Drug Administration in 1965. Indomethacin’s anti-inflammatory effect is mainly mediated by inhibiting cyclooxygenases (COX), the key enzymes catalyzing the production of prostaglandins from arachidonic acid (Nalamachu and Wortmann, 2014). The adipogenic effect of indomethacin was first documented by Williams and Polakis (1977), who found that addition of insulin and indomethacin significantly induced ATP-citrate lyase activity and biosynthesis of triglycerides during 3T3-L1 adipocyte differentiation. Some researchers speculated that the adipogenic effects of indomethacin could be partly explained by the inhibited production of prostaglandins (Chen and London, 1981). Later, one study demonstrated that ob 17 cells treated with a combination of insulin and indomethacin differentiated into adipocytes more rapidly than the cells treated with insulin alone (Verrando et al., 1981). Other investigators (Négrel et al., 1989; Ye and Serrero, 1998) further confirmed the effect of indomethacin in promoting adipogenic differentiation. The mechanism for this effect of indomethacin was not revealed until Lehmann et al. (1997) reported that indomethacin induced C3H10T1/2 adipocyte differentiation by binding and activating peroxisome proliferator-activated receptor (PPAR) γ. It is well recognized that the differentiation of both white and brown adipocytes from adipose stem cells is regulated by transcriptional factors that include CCAAT/enhancer-binding proteins and PPARγ (Tontonoz et al., 1994; Wu et al., 1999). PPARγ is a critical and irreplaceable transcriptional factor in regulation of uncoupling protein 1 (UCP1) expression and brown adipocyte differentiation (Koppen and Kalkhoven, 2010). PPARγ agonists, such as rosiglitazone, are capable of inducing brown preadipocyte differentiation into mature brown adipocytes (Hwang et al., 2011). However, the effects and underlying mechanisms of indomethacin on brown adipocyte differentiation have not been investigated.
Although a number of studies have investigated adipogenic effects of indomethacin in vitro, only a few studies reported the effects of indomethacin on obesity in vivo. One study showed that indomethacin supplemented in food (16 mg/kg diet) prevented high-fat diet-induced obesity (DIO) in C57BL/6 mice, but failed to prevent hyperglycemia and impaired glucose tolerance (Fjære et al., 2014). Interestingly, the same group in an earlier study reported that in vitro treatment with indomethacin (1 µM) significantly inhibited UCP1 expression in primary inguinal adipocytes and dietary indomethacin supplementation (16 mg/kg diet) increased weight gain and prevented high-fat diet-induced UCP1 expression in inguinal WAT (iWAT) in male Sv129 mice (Madsen et al., 2010). While the discrepancy between the two in vivo studies might be related to different strains of mice used, it is also possible that indomethacin could enhance differentiation of both white and brown adipocytes in vivo. To further clarify this issue, we first investigated the effect of indomethacin on brown preadipocyte differentiation and its underlying mechanism in vitro. We further conducted a pilot animal study to deliver indomethacin to interscapular BAT (iBAT) of DIO mice to maximize brown differentiation while minimizing white differentiation, leading to desirable outcomes in treating obesity and its associated metabolic disorders.
Materials and Methods
Chemicals and Reagents.
For the cell culture work, indomethacin, insulin, triiodothyronine, rosiglitazone, and dimethylsulfoxide were purchased from Sigma (St. Louis, MO). Primary antibody for UCP1 was purchased from Sigma Aldrich and primary antibody for PGC-1α was purchased from Millipore (Temecula, CA). ERK1/2 and horseradish peroxidase–conjugated goat anti-rabbit antibodies were purchased from Cell Signaling Technology (Danvers, MA). For the animal study, Kolliphor HS15 was a gift from BASF (Florham Park, NJ), and indomethacin was purchased from Cayman Chemical Company (Ann Arbor, MI).
Cell Culture and Treatment.
The murine brown fat cell line was a gift from Dr. Johannes Klein (University of Lubeck, Lubeck, Germany), who generated the cell line from iBAT of newborn C57BL/6 mice. Brown preadipocytes were maintained in Dulbecco’s modified Eagle’s medium supplemented with 20% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) at 37°C humidified atmosphere of 5% CO2 in air, until they reached confluence (designated as day 0). The cells were differentiated by incubation in the differentiation media containing Dulbecco’s modified Eagle’s medium supplemented with 20% fetal bovine serum, 1 nM triiodothyronine, and 20 nM insulin, and media were changed every other day until day 5. To study the effects of indomethacin on brown preadipocytes, indomethacin (2, 5, 10, 20, and 50 μM) or the vehicle control (dimethylsulfoxide) was added at day 0 and replaced with each change of the media during the differentiation process.
Western Blot Analysis.
Total cell lysates were prepared using 1X lysis buffer (Cell Signaling Technology), and protein concentrations were determined by the BCA Assay Kit (Thermo Scientific, Waltham, MA). Total cell lysates were subjected to 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). The membrane was blocked in 20 mM Tris·HCl, 137 mM NaCl, and 0.1% Tween 20 (pH 7.4) containing 5% nonfat milk. The membrane was immunoblotted with primary antibodies at 4°C overnight followed by secondary antibody conjugated with horseradish peroxidase for 1 hour. The membrane was exposed on X-ray film after being developed by enhanced chemilumescent western blot detection reagents (Pierce, Rockford, IL). The signal was quantified by densitometry using a ChemiDocXRS+ imaging system with ImageLab software (Bio-Rad).
Quantitative Real-Time Polymerase Chain Reaction (PCR) Analysis.
Total RNA was prepared using TRI Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s instructions. Total RNA abundance was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Reverse transcription was carried out using the High Capacity cDNA Reverse Transcription Kit (Thermo Scientific, Pittsburgh, PA) according to the manufacturer’s instructions. The mRNA expression of target genes and the housekeeping gene 36B4 was measured quantitatively using PowerUp SYBR Master Mix (Applied Biosystems, Austin, TX). PCR reactions were run in a 96-well format using an ABI 7300HT instrument. Cycle conditions were 50°C for 2 minutes and 95°C for 10 minutes, and then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Relative gene expression was calculated using the 2−ΔΔCt method, which normalizes against 36B4. All primer sequences are listed in Table 1.
TABLE 1.
Primer sequences for both mouse brown preadipocytes and C57BL6 mice
| Gene Name | Primer Sequence |
|---|---|
| 36B4 | Forward: GCGACATACTCAAGCAGGAGCA |
| Reverse: AGTGGTAACCGCTCAGGTGTTG | |
| UCP1 | Forward: GCTTTGCCTCACTCAGGATTGG |
| Reverse: CCAATGAACACTGCCACACCTC | |
| PGC-1α | Forward: GAATCAAGCCACTACAGACACCG |
| Reverse: CATCCCTCTTGAGCCTTTCGTG | |
| PPARγ | Forward: GTACTGTCGGTTTCAGAAGTGCC |
| Reverse: ATCTCCGCCAACAGCTTCTCCT | |
| Prdm16 | Forward: ATCCACAGCACGGTGAAGCCAT |
| Reverse: ACATCTGCCCACAGTCCTTGCA | |
| Tfam | Forward: GAGGCAAAGGATGATTCGGCTC |
| Reverse: CGAATCCTATCATCTTTAGCAAGC | |
| Nrf-1 | Forward: GGCAACAGTAGCCACATTGGCT |
| Reverse: GTCTGGATGGTCATTTCACCGC | |
| ATGL | Forward: TTGTCCTGCCCCACTAAGAG |
| Reverse: ACTCAGAGAACCCCGTGAAG | |
| HSL | Forward: AGACACTTCGCTGTTCCTCA |
| Reverse: CTTCAGCCTCTTCCTGGGAA | |
| CPT1 | Forward: GGACTCCGCTCGCTCATT |
| Reverse: GAGATCGATGCCATCAGGGG | |
| Acadm | Forward: TCAAGATCGCAATGGGTGCT |
| Reverse: GCTCCACTAGCAGCTTTCCA | |
| Plin1 | Forward: TATGCCCAGAGAGAAGCCTG |
| Reverse: AATCTGCCCACGAGAAAGGA | |
| Plin2 | Forward: AGCTCCAGACTACATGCCAG |
| Reverse: AGCTTTGACCTCAGACTGCT | |
| Cox4b | Forward: GGGCAGCTCTGGATAGTTCC |
| Reverse: TCAACGTAGGGGGTCATCCT | |
| CoxII | Forward: GCTCTCCCCTCTCTACGCAT |
| Reverse: AGCAGTCGTAGTTCACCAGG | |
| 18S | Forward: GATTCTCCGTTACCCGTCAA |
| Reverse: CGTGCTGACGACAGATCACT |
Total DNA was prepared using TRI Reagent (Molecular Research Center) according to the manufacturer’s instructions, and DNA concentrations were quantified using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Mitochondrial cytochrome C oxidase subunit 2 (COXII) and 18S rRNA control genes were measured quantitatively using PowerUp SYBR Master Mix (Applied Biosystems). PCR reactions were run in a 96-well format using the ABI 7300HT instrument (Applied Biosystems, Foster city, CA). Cycle conditions were 50°C for 2 minutes and 95°C for 10 minutes, and then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Relative gene expression was calculated using the 2−ΔΔCt method, which normalizes against 18s rRNA.
Reporter Gene Assays.
Brown preadipocytes seeded on 48-well plates were transiently transfected with PPAR response element (PPRE)-luciferase reporter (Addgene plasmid #1015) and β-galactosidase expression plasmid with Lipofectamine 2000 Transfection Reagent and Plus Reagent (Thermo Fisher Scientific, Carlsbad, CA). After 24 hours of transfection, the cells were treated with indomethacin or the vehicle control for 18 hours. The cell lysate was prepared and reporter luciferase and β-galactosidase activities were measured using a GloMax Luminometer (Promega, Madison, WI). Relative luciferase activities were normalized by β-galactosidase activities.
Animal Study.
Six-week-old male C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) were housed in an animal facility with a 12-hour light/dark cycle. After 1 week of acclimation, mice were fed a high-fat diet (45% kcal from fat, D12451) (Research Diets, New Brunswick, NJ) for 9 weeks, and then the mice were weighed and randomly assigned to either the control or the indomethacin group (three mice/group). Indomethacin (10 mg/ml) dissolved in saline containing 18.75% Kolliphor HS15 (percentage of weight) was put into three ALZET mini-osmotic pumps (model 2006) (DURECT Corporation, Cupertino, CA), which were reported to have a delivery rate of 0.15 µl/h. In addition, three pumps delivered only saline and the same concentration of Kolliphor HS15. We connected a piece of silicon tubing at the end of each pump, and sutured the tubing on the mouse back muscle to ensure that the opening of the tubing was on the iBAT of the mouse during pump implantation. Mice had free access to water and food, and their body weight and food intake were recorded weekly. After 4 weeks, mice were euthanized by CO2 asphyxiation. Blood samples were collected in EDTA-coated tubes. Plasma was obtained by blood centrifugation at 2000g at 4°C for 20 minutes. Organs and tissues were immediately dissected, weighed, and flash-frozen in liquid nitrogen. A portion of WAT and iBAT was fixed with 10% phosphate-buffered saline formalin for histologic analysis. The animal protocol was approved by the Institutional Animal Care and Use Committee of Texas Tech University (protocol number: 16028).
Plasma Lipid Profile.
Plasma concentrations of triglycerides, total cholesterol, high-density lipoprotein cholesterol, and very low-density lipoprotein cholesterol were measured using the AU400 clinical chemistry analyzer with enzymatic reagents (Beckman Coulter, Inc., Brea, CA) by following the AU400 procedural manual. Low-density lipoprotein cholesterol concentrations were calculated using the Friedewald formula. Plasma concentrations of nonesterified fatty acids were measured using a nonesterified fatty acids quantification kit (ab65341) from Abcam (Cambridge, MA).
H&E Staining.
Tissues were fixed with 10% phosphate-buffered saline formalin for 48 hours. H&E staining was performed by the Department of Pathology at the Texas Tech University Health Sciences Center. Briefly, the paraffin sections of adipose tissues (5 µm) were dewaxed with xylene and rehydrated with alcohol. The samples were washed with water to remove any reagent residue, incubated with Gill’s Hematoxylin II for 4 minutes, and then washed several times using water. The samples were counterstained with eosin for 1 minute. Then the samples were dehydrated through absolute alcohol three times. Finally, samples were cleaned with xylene and mounted with xylene-based mounting medium.
Body Composition.
Body composition was measured at weeks 0, 2, and 4 using an EchoMRI Body Composition Analyzer (EchoMRI LLC, Houston, TX) according to the manufacturer’s instructions at the Texas Tech University animal facility.
Insulin Tolerance Test.
After fasting mice for 4 hours on week 4, blood glucose concentrations were measured by tail snip using the OneTouch Ultra2 Glucometer (LifeScan Europe, Zug, Switzerland) and blood glucose test strips. Insulin (0.75 U/kg body weight) was given to mice via intraperitoneal injection, and blood glucose concentrations were measured using the aforementioned method at 15, 30, 45, 60, and 120 minutes post injection.
Statistical Analysis.
All data presented in the text, Tables 1 and 2, and Figs. 1–5 are mean ± S.E.M. Measurements were performed in at least three independent experiments. Statistical analysis was performed using Prism 5 (GraphPad Software, Inc., San Diego, CA). One-way analysis of variance with repeated measures followed by multiple comparisons test (Student-Newman-Keuls method) was performed to determine the differences of group means between the treatment groups in the cell culture study. Student’s t test was performed to compare two group means in the animal study. The level of significance was set at P < 0.05.
TABLE 2.
Metabolic parameters of mice in both control and indomethacin-treated groups
| Metabolic Parameter | Control | Indomethacin | P value |
|---|---|---|---|
| Body weight (g) | 30.73 ± 1.57 | 31.30 ± 2.04 | 0.41 |
| Body composition | |||
| Fat mass (%) | 19.62 ± 3.05 | 13.32 ± 3.07 | 0.23 |
| Lean mass (%) | 79.51 ± 3.19 | 83.47 ± 2.88 | 0.20 |
| BAT of body weight (%) | 0.32 ± 0.01 | 0.28 ± 0.16 | 0.03 |
| rWAT of body weight (%) | 0.95 ± 0.17 | 0.81 ± 0.25 | 0.34 |
| eWAT of body weight (%) | 3.41 ± 0.64 | 2.25 ± 0.66 | 0.14 |
| iWAT of body weight (%) | 1.24 ± 0.38 | 1.29 ± 0.09 | 0.44 |
| Food intake (g/day) | 2.91 ± 0.16 | 3.02 ± 0.18 | 0.34 |
| Fasting glucose (mg/dl) | 229.30 ± 6.84 | 185.00 ± 13.58 | 0.02 |
| Blood lipid profile | |||
| TG (mg/dl) | 95.00 ± 5.00 | 85.00 ± 2.89 | 0.07 |
| TC (mg/dl) | 160.00 ± 32.79 | 133.30 ± 11.67 | 0.24 |
| LDL-C (mg/dl) | 63.33 ± 28.48 | 41.67 ± 7.27 | 0.25 |
| VLDL-C (mg/dl) | 18.33 ± 1.67 | 16.67 ± 1.67 | 0.26 |
| HDL-C (mg/dl) | 77.50 ± 5.35 | 74.67 ± 4.51 | 0.35 |
| NEFA (nm/ml) | 13.97 ± 0.49 | 13.88 ± 1.18 | 0.47 |
All data are expressed as mean ± SEM (n=3) and analyzed by student t tests. Abbreviations: rWAT, retroperitoneal white adipose tissue; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; NEFA, nonesterified fatty acids; TC, total cholesterol; TG, triglyceride; VLDL-C, very low-density lipoprotein cholesterol.
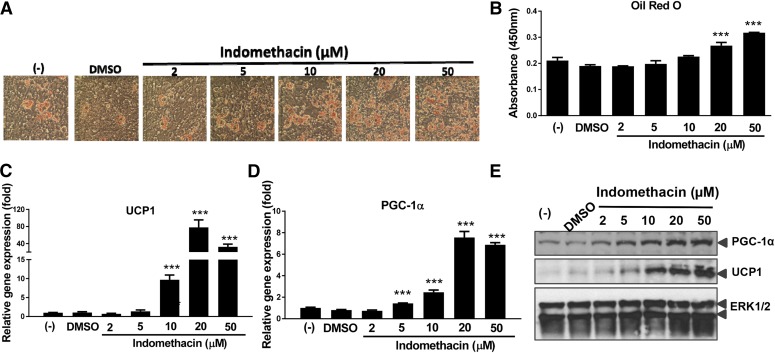
Fig. 1.
Indomethacin dose dependently induced brown preadipocyte differentiation. (A) Oil Red O stained cell morphology at day 5. (B) Lipid accumulation measured by Oil Red O absorbance. (C) UCP1 gene expression. (D) PGC-1α gene expression. (E) Western blotting of UCP1 and PGC-1α. Data = mean ± S.E.M. (n = 3). ***P < 0.001; **P < 0.01;*P < 0.05 compared with the dimethylsulfoxide (DMSO) group.
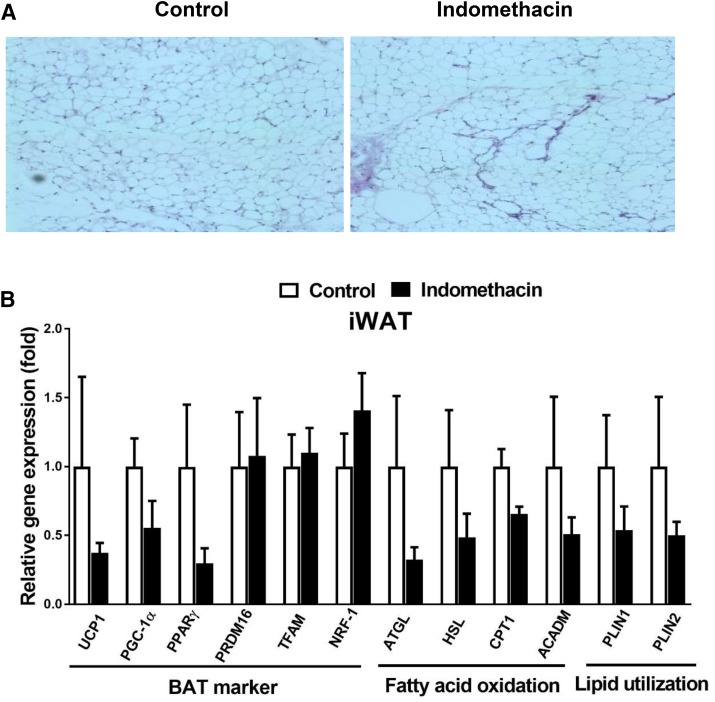
Fig. 5.
Indomethacin slightly inhibited browning markers in iWAT. (A) Representative H&E staining images of iWAT. (B) Gene expression analysis of BAT markers, fatty acid oxidation, and lipid utilization in iWAT. Data = mean ± S.E.M. (n = 3). *P < 0.05 compared with the control group.
Results
Indomethacin Stimulated Differentiation of Mouse Brown Preadipocytes.
We examined brown preadipocyte differentiation in the presence of increasing doses of indomethacin. Indomethacin dose dependently increased brown preadipocyte differentiation, as revealed by Oil Red O stained cell morphology (Fig. 1A). Quantitation of lipid accumulation by Oil Red O absorbance indicated that compared with the dimethylsulfoxide control, 20 and 50 µM of indomethacin significantly increased cellular lipid accumulation by 28% and 50%, respectively (Fig. 1B).
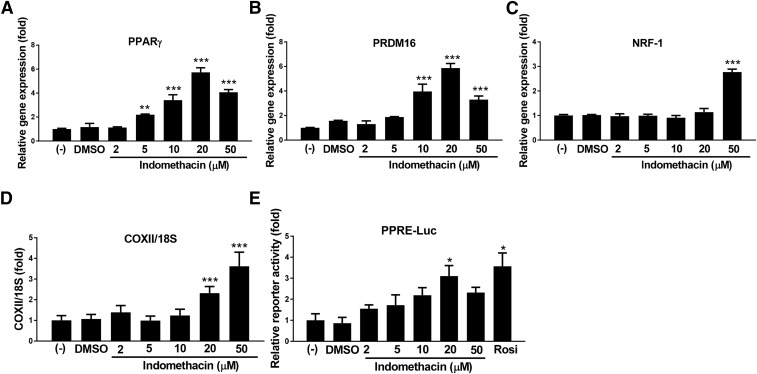
Real-time PCR was performed to measure mRNA expression of brown adipocyte related genes in response to indomethacin in brown adipocytes. Brown adipocyte selective markers, both mRNA (Fig. 1, C and D) and protein levels (Fig. 1E) of UCP1 and PPARγ coactivator 1-alpha (PGC-1α), were upregulated in a dose-dependent manner by indomethacin at 5, 10, and 20 µM (P < 0.001). Adipocyte differentiation markers, PPARγ and PR domain containing 16 (Prdm16) were also induced by indomethacin in a dose-dependent manner at 5–20 µM (Fig. 2, A and B), whereas nuclear respiratory factor 1 (Nrf-1) expression was significantly increased by indomethacin only at the high concentration (50 µM; P < 0.001) (Fig. 2C). Furthermore, the DNA content of the cytochrome c oxidase subunit 2 (COXII) gene, one of the mitochondrial biogenesis markers, was significantly increased when cells were treated with 20 or 50 µM of indomethacin (Fig. 2D).
Fig. 2.
Indomethacin dose dependently increased brown adipocyte related markers. (A–C) Gene expression of brown fat selected markers; negative control cells were set at fold 1. (D) Gene expression of mitochondrially encoded cytochrome c oxidase II. (E) Luciferase activity of PPAR reporter. Data = mean ± S.E.M. (n = 3). ***P < 0.001; **P < 0.01;*P < 0.05 compared with the dimethylsulfoxide (DMSO) group.
Induction of PPRE-Driven Luciferase Activity by Indomethacin.
The PPRE-driven luciferase activities were evaluated in mouse brown preadipocytes after transient transfection of PPRE-luciferase cells were treated with indomethacin at the concentrations indicated. The data showed that indomethacin dose dependently activated PPRE-luciferase reporter, reaching significance at 20 µM in mouse brown preadipocytes (Fig. 2E), indicating that indomethacin may work as a PPARγ agonist in this cell line.
Metabolic Effects of Local Delivery of Indomethacin to iBAT in Obese Mice.
We next investigated the metabolic effects of local delivery of indomethacin to iBAT using ALZET mini-osmotic pumps in DIO mice in vivo. The indomethacin treatment did not change food intake and body weight. However, compared with the control mice, indomethacin-treated mice had a 34% reduction in the percentage of epididymal WAT (eWAT) mass (P = 0.14), resulting in a 32% reduction in the percentage of body fat mass (P = 0.23) (Table 2). Additionally, indomethacin significantly decreased the percentage of iBAT mass by 13% compared with the control (P = 0.03). Fasting blood glucose concentrations were 19% lower in indomethacin-treated mice (185 ± 13.58 mg/dl) compared with the control mice (229 ± 6.84 mg/dl) (P = 0.02), although insulin tolerance tests showed no differences between the two groups (data not shown). Fasting blood concentrations of very low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, and nonesterified fatty acids were similar between the control and indomethacin groups. The indomethacin group had numerically lower fasting blood concentrations of triglyceride, total cholesterol, and low-density lipoprotein cholesterol compared with the control group, but the differences did not reach statistical significance, possibly due to the small sample size together with large variability in response.
Indomethacin Alleviated Brown Fat Whitening and Increased Expression of Genes Involved in Brown Fat Activation in Obese Mice.
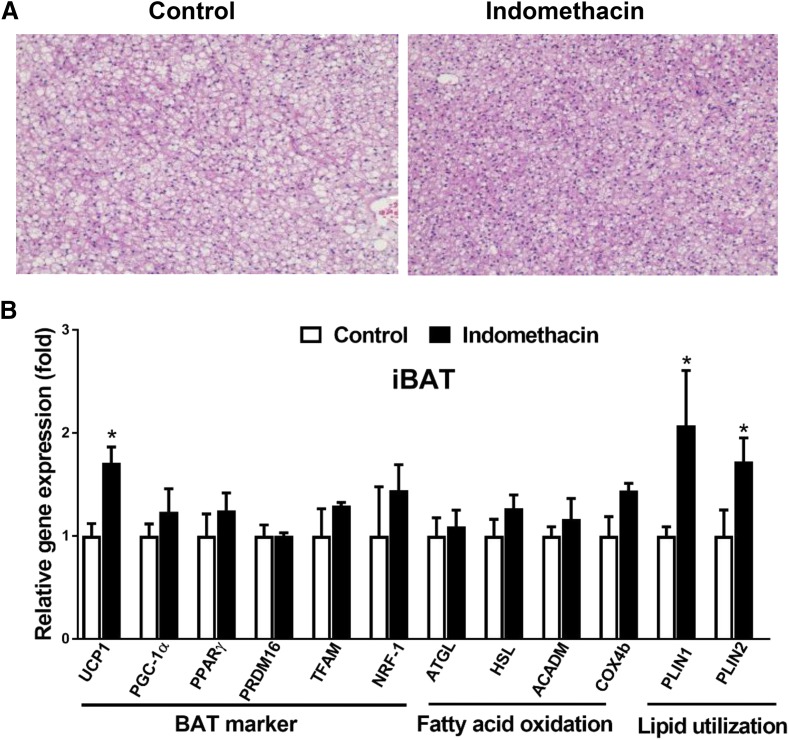
Since indomethacin was directly delivered to iBAT through ALZET osmotic mini-pumps, we specifically examined the effects of indomethacin on brown adipocyte morphology and expression of iBAT marker genes. H&E staining of iBAT cells in the control mice had enlarged lipid droplets and enhanced lipid deposition, giving the appearance of whitening of iBAT (Fig. 3A). In contrast, the whitening phenotype was not observed in iBAT cells in indomethacin-treated mice. Compared with the control, indomethacin significantly increased mRNA expression of UCP1, perilipin 1 (Plin1), and perilipin 2 (Plin2) (Fig. 3B). There were no significant differences in other brown markers including Prdm16, PGC-1α, PPARγ, mitochondrial transcription factor A (Tfam), and Nrf-1, in fatty acid β-oxidation genes including acyl-coenzyme A dehydrogenase (Acadm) and cytochrome c oxidase subunit IV isoform 2 (Cox4b), and in lipolysis genes including adipocyte triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) between the indomethacin-treated and control mice (Fig. 3B).
Fig. 3.
Indomethacin alleviated BAT whitening and increased thermogenesis related markers in DIO mice. (A) Representative H&E staining images of iBAT. (B) Relative mRNA levels of BAT markers, genes involved in fatty acid oxidation, and lipid utilization in iBAT. Data = mean ± S.E.M. (n = 3). *P < 0.05 compared with the control group.
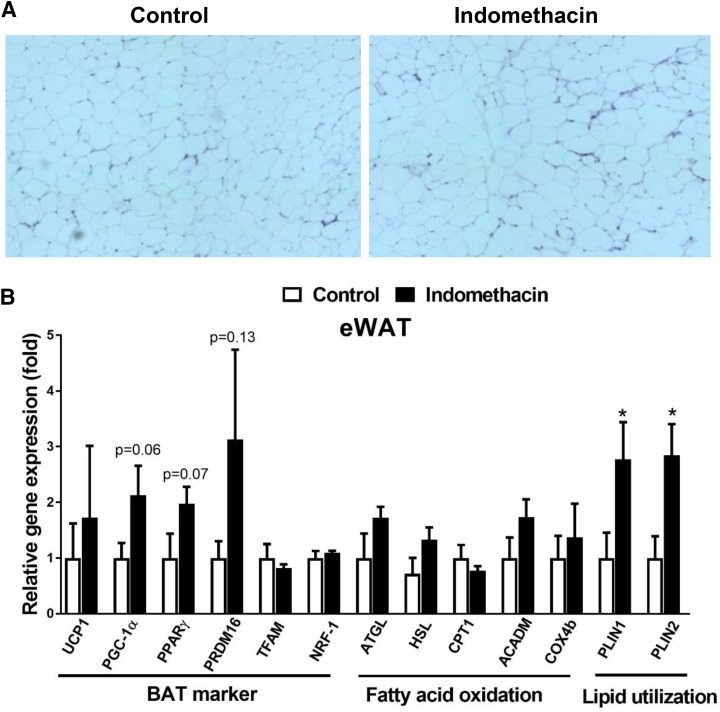
To evaluate browning of WAT, H&E staining and real-time PCR were performed in both eWAT and iWAT (Figs. 4 and 5). The H&E staining revealed similar morphology in both eWAT and iWAT in the indomethacin-treated mice (Figs. 4A and 5A) compared with the control mice. The expression of UCP1, PGC-1α, PPARγ, and Prdm16 was 80% higher in the eWAT isolated from the indomethacin-treated mice compared with the control mice, but this difference did not reach statistical significance (Fig. 4B). However, we observed that lipid utilization genes, Plin1 and Plin2, were significantly higher in eWAT isolated from the indomethacin-treated mice compared with the control mice (P < 0.05) (Fig. 4B). Unexpectedly, expression of browning related markers in iWAT tended to be lower in the indomethacin-treated mice compared with the control mice (Fig. 5B). We also measured the expression of genes related to fatty acid oxidation, and lipid utilization in iWAT, but found no significant difference between the treatment and control groups (Fig. 5B).
Fig. 4.
Indomethacin slightly increased browning markers in eWAT. (A) Representative H&E staining images of eWAT. (B) Real-time PCR analysis of the expression of BAT markers, genes involved in fatty acid oxidation, and lipid utilization in eWAT. Data = mean ± S.E.M. (n = 3). *P < 0.05 compared with the control group.
Discussion
The effects of indomethacin on brown adipocyte differentiation and activities have not been investigated. Our current study demonstrated that indomethacin stimulated mouse brown adipocyte differentiation by enhancing lipid accumulation and gene expression of brown adipocyte specific markers. We further showed that indomethacin activated the PPAR responsive reporter in mouse brown preadipocytes in vitro. In our pilot animal study, we validated that local delivery of indomethacin to iBAT increased expression of genes involved in thermogenesis in iBAT and improved hyperglycemia found in DIO mice.
Indomethacin stimulated mouse brown adipocyte differentiation via activating PPARγ, which is consistent with a previous study showing that indomethacin can bind to and activate PPARγ (Lehmann et al., 1997). In addition, the increased ratio of COXII to 18S rRNA DNA content (COXII/18S) in these cells suggests that mitochondrial biogenesis was enhanced by indomethacin. It has been proposed that browning effects of PPARγ agonist may be due to its ability to induce expression of PGC-1α (Wilson-Fritch et al., 2003; Hondares et al., 2006). In agreement with this, our in vitro study demonstrated that indomethacin increased both mRNA and protein expression of PGC-1α, a master regulator of mitochondrial biogenesis (Jornayvaz and Shulman, 2010).
To extend our in vitro findings into mice in vivo, we further performed a pilot study to deliver indomethacin directly to the iBAT of DIO mice. We found that indomethacin significantly decreased iBAT mass and induced UCP1 mRNA expression in iBAT. The expression of genes regulating lipid utilization, such as Plin 1 and 2, in iBAT was also increased by indomethacin. These data indicated that iBAT fat burning activity might be increased after local delivery of indomethacin. The results of the animal study demonstrated that delivery of indomethacin to the iBAT alleviated its whitening induced by the high-fat diet. One possible explanation is that indomethacin alleviated iBAT whitening via enhancing UCP1-mediated thermogenesis and fat utilization since our data showed that both UCP1 and lipid droplet proteins, such as Plin 1 and Plin 2, were significantly increased in the iBAT of indomethacin-treated mice. It is believed that lipid droplet proteins, including the Plin family and several others, play an important role in fat utilization in brown adipocytes (Townsend and Tseng, 2014). Local delivery of indomethacin appears to differentially affect white fat depending on the depot since the expression of browning markers was increased in eWAT but decreased in iWAT by the indomethacin treatment in this study. At this point, this depot-related difference in white fat browning in response to indomethacin is not well understood. Studies have shown that more beige adipocytes were formed in eWAT than in iWAT in response to some drugs/nutrients, such as β3-agonist CL316,243 (Warner et al., 2016) and retinaldehyde (Kiefer et al., 2012). We speculate that the concentrations of indomethacin reached in eWAT and iWAT may be different, resulting in different responses in indomethacin-targeted signaling events. For example, Lehmann et al. (1997) suggested that indomethacin at low concentrations may block COX activity but without activating PPARγ; however, at high concentrations (micromolar range) it may function as both a COX inhibitor and PPARγ agonist. Therefore, it is possible that indomethacin, depending on its concentrations in particular tissues, would differentially affect the signaling molecules generated from both COX and PPARγ pathways in the affected locations. Since COX-2 is required for recruitment of beige cells in WAT (Vegiopoulos et al., 2010), inhibition of COX-2 by indomethacin may lead to suppression of UCP1, which may well explain the lower expression of those browning markers in iWAT measured in our indomethacin-treated mice. Of note, a recent study showed that subcutaneous injection of indomethacin (2.5 mg/kg of body weight) suppressed cold-induced UCP1 gene expression in iWAT of C57BL/6 mice (Madsen et al., 2010).
We also found that indomethacin significantly decreased fasting blood glucose concentrations. In contrast to our results, Fjære et al. (2014) reported that indomethacin treatment (16 mg/kg of diet) prevented high-fat/high-sucrose induced obesity but deteriorated glucose intolerance in C57BL/6J mice; the different results could be explained by differences in the study design, such as the timing of indomethacin administration, diets, indomethacin doses, and delivery routes between two studies. It is worth pointing out that the increased concentrations of plasma glucose by oral delivery of indomethacin found in the Fjære et al. (2014) study was thought to be due to increased liver gluconeogenesis and impaired high-fat/high-sucrose induced glucose-stimulated insulin secretion, which is the possible consequence of systemic side effects of indomethacin. The decreased fasting blood glucose concentrations can be explained by the fact that local delivery of indomethacin might only induce the differentiation of BAT, not WAT. Activated BAT can take up and burn more circulating glucose (Townsend and Tseng, 2014). Emerging evidence suggests that BAT might protect against obesity and type-2 diabetes through BAT-derived endocrine factors (i.e., batokines) that act in a paracrine or autocrine manner (Townsend and Tseng, 2012). Therefore, indomethacin may stimulate the release of batokines, such as Fibroblast growth factor 21, interleukin-6, and neuregulin 4 (Villarroya et al., 2013, 2017), which might in turn contribute to activation of iBAT and lowered blood glucose concentrations. Future studies are needed to examine the effects of indomethacin on the release of batokines.
We recognize that the relatively small sample size is a limitation in this study. However, our animal pilot study does support the findings from our in vitro studies showing that indomethacin promoted brown adipocyte differentiation and enhanced gene expression of brown adipocyte specific markers in the iBAT. More animal studies are required to confirm our findings and to investigate additional underlying mechanisms of indomethacin in regulating brown adipocyte differentiation and activity. Another limitation is the DIO mouse model. Mice and humans differ significantly in terms of morphometry, physiology, and life history (Lai et al., 2014). For example, since mice are significantly smaller than humans, their basal metabolic rate is seven times greater than that of humans (Demetrius, 2005). Nevertheless, these DIO mice are believed to be the best choice for investigating novel anti-obesity drugs because they better mimic human obesity than most of the genetically modified models (Lutz and Woods, 2012).
In conclusion, the present studies demonstrated that indomethacin, a commonly used nonsteroidal anti-inflammatory drug, promoted mouse brown adipocyte differentiation in vitro. Local delivery of indomethacin to iBAT improved hyperglycemia, and increased brown fat thermogenesis capacity in DIO mice. Our studies highlighted the advantages of local or targeted BAT delivery of indomethacin to maximize its beneficial effects on promoting brown adipogenesis and to minimize its toxic side effects often associated with systemic delivery to WAT (increased white adipogenesis) and the liver (increased gluconeogenesis).
Acknowledgments
We thank Yujiao Zu, Jie Liu, and Md Shahjalal Khan for help in the animal study.
Abbreviations
- BAT
brown adipose tissue
- COX
cyclooxygenase
- COXII
cytochrome c oxidase subunit 2
- DIO
diet-induced obesity
- eWAT
epididymal white adipose tissue
- iBAT
interscapular brown adipose tissue
- iWAT
inguinal white adipose tissue
- PCR
polymerase chain reaction
- PPAR
peroxisome proliferator-activated receptor
- PPRE
peroxisome proliferator-activated receptor response element
- rWAT
retroperitoneal white adipose tissue
- WAT
white adipose tissue
Authorship Contributions
Participated in research design: Hao, Kearns, Zhao, Wang.
Conducted experiments: Hao, Kearns, Zhao, Wang, Scott, Wu.
Performed data analysis: Hao, Kearns, Sun, Zhao, Wang, Scott, Wu.
Wrote or contributed to the writing of the manuscript: Hao, Kearns, Zhao, Wang, Scott, Wu, Kodani, Morisseau, Hammock.
Footnotes
This work was supported by the National Center for Complementary and Integrative Health [Grants R15AT007013 and R15AT008733]; the National Institutes of Health National Institute of Environmental Health Sciences Superfund Research Program [Grants P42 ES0044699 and R01 ES002710]; the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01 DK103616]; the College of Human Sciences, Texas Tech University, Lubbock, TX; the U.S. Department of Agriculture (USDA)-Agriculture Research Service (ARS) under agreement #58-1950-0-014; and the University of Tennessee, Knoxville, TN.
References
- Chen JJ, London IM. (1981) Hemin enhances the differentiation of mouse 3T3 cells to adipocytes. Cell 26:117–122. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. (2009) Identification and importance of brown adipose tissue in adult humans. N Engl J Med 360:1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetrius L. (2005) Of mice and men. When it comes to studying ageing and the means to slow it down, mice are not just small humans. EMBO Rep 6:S39–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjære E, Aune UL, Røen K, Keenan AH, Ma T, Borkowski K, Kristensen DM, Novotny GW, Mandrup-Poulsen T, Hudson BD, et al. (2014) Indomethacin treatment prevents high fat diet-induced obesity and insulin resistance but not glucose intolerance in C57BL/6J mice. J Biol Chem 289:16032–16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Kit BK, Ogden CL. (2012) Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA 307:491–497. [DOI] [PubMed] [Google Scholar]
- Hart FD, Boardman PL. (1963) Indomethacin: a new non-steroid anti-inflammatory agent. BMJ 2:965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondares E, Mora O, Yubero P, Rodriguez de la Concepción M, Iglesias R, Giralt M, Villarroya F. (2006) Thiazolidinediones and rexinoids induce peroxisome proliferator-activated receptor-coactivator (PGC)-1α gene transcription: an autoregulatory loop controls PGC-1α expression in adipocytes via peroxisome proliferator-activated receptor-γ coactivation. Endocrinology 147:2829–2838. [DOI] [PubMed] [Google Scholar]
- Hwang HH, Moon PG, Lee JE, Kim JG, Lee W, Ryu SH, Baek MC. (2011) Identification of the target proteins of rosiglitazone in 3T3-L1 adipocytes through proteomic analysis of cytosolic and secreted proteins. Mol Cells 31:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MD, Ryan DH, Apovian CM, Ard JD, Comuzzie AG, Donato KA, Hu FB, Hubbard VS, Jakicic JM, Kushner RF, et al. American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Obesity Society (2014) 2013 AHA/ACC/TOS guideline for the management of overweight and obesity in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and The Obesity Society. Circulation 129 (25 Suppl 2):S102–S138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jornayvaz FR, Shulman GI. (2010) Regulation of mitochondrial biogenesis. Essays Biochem 47:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkar AK, Dahiya N. (2015) Drug treatment of obesity: current status and future prospects. Eur J Intern Med 26:89–94. [DOI] [PubMed] [Google Scholar]
- Kiefer FW, Vernochet C, O’Brien P, Spoerl S, Brown JD, Nallamshetty S, Zeyda M, Stulnig TM, Cohen DE, Kahn CR, et al. (2012) Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med 18:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen A, Kalkhoven E. (2010) Brown vs white adipocytes: the PPARγ coregulator story. FEBS Lett 584:3250–3259. [DOI] [PubMed] [Google Scholar]
- Lai M, Chandrasekera PC, Barnard ND. (2014) You are what you eat, or are you? The challenges of translating high-fat-fed rodents to human obesity and diabetes. Nutr Diabetes 4:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, Lenhard JM, Oliver BB, Ringold GM, Kliewer SA. (1997) Peroxisome proliferator-activated receptors α and γ are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J Biol Chem 272:3406–3410. [DOI] [PubMed] [Google Scholar]
- Lutz TA, Woods SC. (2012) Overview of animal models of obesity. Curr Protoc Pharmacol 5:ph0561s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen L, Pedersen LM, Lillefosse HH, Fjaere E, Bronstad I, Hao Q, Petersen RK, Hallenborg P, Ma T, De Matteis R, et al. (2010) UCP1 induction during recruitment of brown adipocytes in white adipose tissue is dependent on cyclooxygenase activity. PLoS One 5:e11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalamachu S, Wortmann R. (2014) Role of indomethacin in acute pain and inflammation management: a review of the literature. Postgrad Med 126:92–97. [DOI] [PubMed] [Google Scholar]
- Narayanaswami V, Dwoskin LP. (2017) Obesity: current and potential pharmacotherapeutics and targets. Pharmacol Ther 170:116–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Négrel R, Gaillard D, Ailhaud G. (1989) Prostacyclin as a potent effector of adipose-cell differentiation. Biochem J 257:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Abera SF, et al. (2014) Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 384:766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS. (2001) Obesity and the regulation of energy balance. Cell 104:531–543. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. (1994) Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79:1147–1156. [DOI] [PubMed] [Google Scholar]
- Townsend K, Tseng YH. (2012) Brown adipose tissue: recent insights into development, metabolic function and therapeutic potential. Adipocyte 1:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend KL, Tseng YH. (2014) Brown fat fuel utilization and thermogenesis. Trends Endocrinol Metab 25:168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. (2009) Cold-activated brown adipose tissue in healthy men. N Engl J Med 360:1500–1508. [DOI] [PubMed] [Google Scholar]
- Vegiopoulos A, Müller-Decker K, Strzoda D, Schmitt I, Chichelnitskiy E, Ostertag A, Berriel Diaz M, Rozman J, Hrabe de Angelis M, Nüsing RM, et al. (2010) Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science 328:1158–1161. [DOI] [PubMed] [Google Scholar]
- Verrando P, Négrel R, Grimaldi P, Murphy M, Ailhaud G. (1981) Differentiation of ob 17 preadipocytes to adipocytes. Triggering effects of clofenapate and indomethacin. Biochim Biophys Acta 663:255–265. [DOI] [PubMed] [Google Scholar]
- Villarroya F, Cereijo R, Villarroya J, Giralt M. (2017) Brown adipose tissue as a secretory organ. Nat Rev Endocrinol 13:26–35. [DOI] [PubMed] [Google Scholar]
- Villarroya J, Cereijo R, Villarroya F. (2013) An endocrine role for brown adipose tissue? Am J Physiol Endocrinol Metab 305:E567–E572. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerbäck S, et al. (2009) Functional brown adipose tissue in healthy adults. N Engl J Med 360:1518–1525. [DOI] [PubMed] [Google Scholar]
- Warner A, Kjellstedt A, Carreras A, Böttcher G, Peng XR, Seale P, Oakes N, Lindén D. (2016) Activation of β3-adrenoceptors increases in vivo free fatty acid uptake and utilization in brown but not white fat depots in high-fat-fed rats. Am J Physiol Endocrinol Metab 311:E901–E910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams IH, Polakis SE. (1977) Differentiation of 3T3-L1 fibroblasts to adipocytes. The effect of indomethacin, prostaglandin E1 and cyclic AMP on the process of differentiation. Biochem Biophys Res Commun 77:175–186. [DOI] [PubMed] [Google Scholar]
- Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, Czech M, Corvera S. (2003) Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol 23:1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Spiegelman BM. (1999) Transcriptional activation of adipogenesis. Curr Opin Cell Biol 11:689–694. [DOI] [PubMed] [Google Scholar]
- Ye H, Serrero G. (1998) Stimulation of adipose differentiation related protein (ADRP) expression by ibuprofen and indomethacin in adipocyte precursors and in adipocytes. Biochem J 330:803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]