Abstract
Marine cyanobacteria are responsible for ~25% of the fixed carbon that enters the ocean biosphere. It is thought that abundant co-occurring viruses play an important role in regulating population dynamics of cyanobacteria and thus the cycling of carbon in the oceans. Despite this, little is known about how viral infections ‘play-out’ in the environment, particularly whether infections are resource or energy limited. Photoautotrophic organisms represent an ideal model to test this since available energy is modulated by the incoming light intensity through photophosphorylation. Therefore, we exploited phototrophy of the environmentally relevant marine cyanobacterium Synechococcus and monitored growth of a cyanobacterial virus (cyanophage). We found that light intensity has a marked effect on cyanophage infection dynamics, but that this is not manifest by a change in DNA synthesis. Instead, cyanophage development appears energy limited for the synthesis of proteins required during late infection. We posit that acquisition of auxiliary metabolic genes (AMGs) involved in light-dependent photosynthetic reactions acts to overcome this limitation. We show that cyanophages actively modulate expression of these AMGs in response to light intensity and provide evidence that such regulation may be facilitated by a novel mechanism involving light-dependent splicing of a group I intron in a photosynthetic AMG. Altogether, our data offers a mechanistic link between diurnal changes in irradiance and observed community level responses in metabolism, i.e., through an irradiance-dependent, viral-induced release of dissolved organic matter (DOM).
Introduction
Understanding the response of the biosphere to climate change requires a detailed knowledge of the biological transformation of carbon on Earth. It has become evident that the ocean represents an important sink for atmospheric carbon dioxide (CO2) [1]. Just two genera of picophytoplankton dominate open ocean regimes, Prochlorococcus and Synechococcus [2–4]. Altogether, these closely related genera are responsible for ~25% of oceanic CO2 fixation [2]. These organisms have become a model for studying the flow of carbon from CO2 to the microbial loop as well as the functioning of planktonic marine communities generally [5, 6]. However, biotic loss rates (antagonistic interactions, grazing, viral lysis) of Prochlorococcus and Synechococcus remain poorly understood. In the case of viruses, this view is nuanced by the discovery of viral-encoded genes that may act to maintain photosynthesis during infection [7, 8], such that despite the ultimate loss of fixed carbon to dissolved organic matter through lysis, CO2 fixation may be maintained transiently during relatively long viral latent periods. Recently, it has been shown that in fact cyanophage shut-down CO2 infection early during infection, yet maintain the photosynthetic light reactions [9]. The prevailing view is that cyanophage use excess ATP and reductant from photophosphorylation to fuel DNA replication while inhibiting a costly Calvin Benson cycle [10, 11]. Therefore, we speculated that DNA replication was the limiting factor to cyanophage morphogenesis and that increases in the rate of photophosphorylation resulting from increased irradiance would dramatically alter phage DNA replication kinetics and the resulting productivity of the phage. Such changes would have important consequences for our understanding of viral-induced lysis pressure of open ocean communities given the wide range in light flux in situ.
Results
Light intensity modules infection kinetics and photophysiology
We hypothesised that shifting infected Synechococcus cells to high light (HL) would provide increased energy for phage replication and as such, we would observe an increase in cyanophage DNA replication rate. To test this idea, we conducted light-shift experiments with Synechococcus sp. WH7803, infected with cyanophage S-PM2d [12], and monitored phage development. We conducted qPCR assays quantifying copies of the phage chromosome, from intracellular and extracellular fractions to measure the rate of DNA synthesis inside cells and to determine latent periods and burst sizes
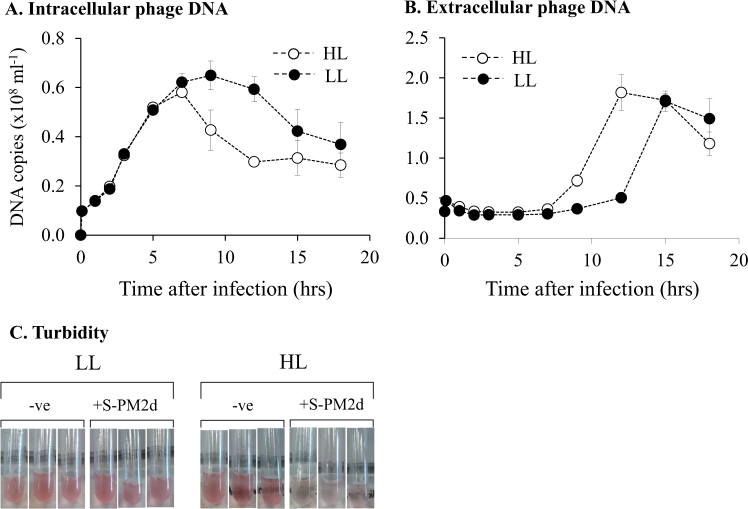
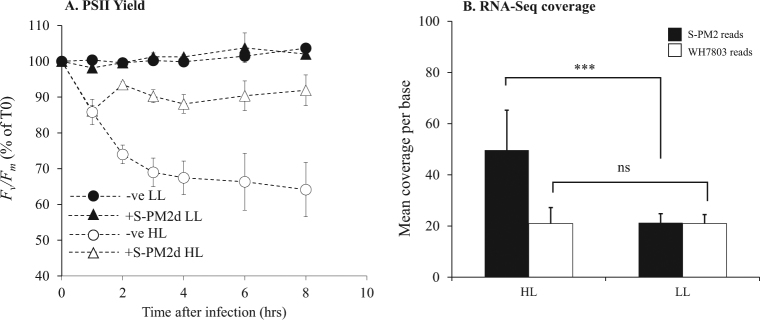
We observed a major difference in cyanophage development in HL compared with LL (Fig. 1). Under HL the latent period was reduced by 5 h (40% decrease in length), as evidenced both by an earlier reduction in copies of intracellular DNA (Fig. 1a), and an earlier increase in extracellular DNA copy number (Fig. 1b). This early burst under HL conditions was also evident from monitoring culture turbidity (Fig. 1c). In comparison, there was no significant difference in burst size between the different light treatments (Fig. 1b). Surprisingly, and contradicting our proposed hypothesis, there was no difference in the timing, length or rate of phage DNA synthesis between the two conditions (Fig. 1a). We had postulated that increased photochemically driven ATP synthesis under HL would supply more energy and resources to fuel phage DNA replication. Indeed, this idea has been implicated [13] and explicitly modelled [14, 15] as explaining cyanophage acquisition of photosynthesis genes, especially given that other stresses such as phosphate limitation show marked effects on cyanophage DNA synthesis [16]. Our hypothesis relies on the assumption that photochemistry is maintained such that excess absorbed photons can be used to generate a transmembrane potential for ATP synthesis. Therefore, we tested this by measuring photosystem II (PSII) maximum quantum yield (Fv/Fm) during the experiment. This assay measures the differences in the chlorophyll ‘fluorescent transient’ from PSII. As dark-adapted reaction centres become ‘closed’ by the reduction of the primary electron acceptor, there is a concomitant increase in chlorophyll fluorescence. Therefore, the Fv/Fm parameter is proportional to the number of open reaction centres at a given time [17]. Our measurements of PSII photochemical yield showed no change under LL conditions in infected or uninfected cells (Fig. 2a). In comparison, there was a rapid decline in Fv/Fm in uninfected cells at HL indicative of photodamage to the PSII reaction centre [18]. After 8 h at HL, Fv/Fm in uninfected cells stabilises at approximately 65% of the level of LL cultures. Similarly, under HL conditions there was an initial decline in Fv/Fm in infected cells, at a similar rate to uninfected cells (Fig. 2a). However, there was then an increase in Fv/Fm 2 h after infection to ~90% of the level at LL where it remained for the rest of the experiment. This observation suggests that not only are PSII reaction centres active in HL infected cells but also either their rate of damage is reduced or the turnover of PSII is much greater to support stable photochemistry at this irradiance.
Fig. 1.
: The effect of light intensity on the kinetics of cyanophage development. a qPCR quantification of cyanophage genome copies from the intracellular fraction over the infection period. b qPCR quantification of cyanophage genome copies from the extracellular fraction over the infection period. c Photographs of culture turbidity taken 10 h after infection
Fig. 2.
The effect of light intensity on Synechococcus photophysiology and cyanophage gene expression. a Maximum quantum yield of PSII photochemistry. Unfilled circles (uninfected) and triangles (infected) are samples subjected to the HL treatment and filled circles and triangles are LL. Samples are normalised to time 0. For raw data see Figure S4. b Mean per base coverage of the transcriptome across the Synechococcus host and cyanophage S-PM2d genome. Filled bars are S-PM2d reads and unfilled bars are Synechococcus sp. WH7803 reads. ***p < 0.001, NS not significant. Error bars represent SD from the mean
The maintenance of PSII photochemistry under increased irradiance suggests more energy was available in these cells through photophosphorylation [19]. Yet despite this, we observed no detectable change in the rate, timing or absolute amount of phage DNA synthesis (Fig. 1a, b). We did observe a reduction in the latent period however, and thus we speculate that there may be some temporal control of lysis in response to light. Indeed, bacteriophages have been shown to possess sophisticated mechanisms to delay lysis in response to potential stresses encountered during the infection process [20].
Cyanophage transcriptional response to high light conditions
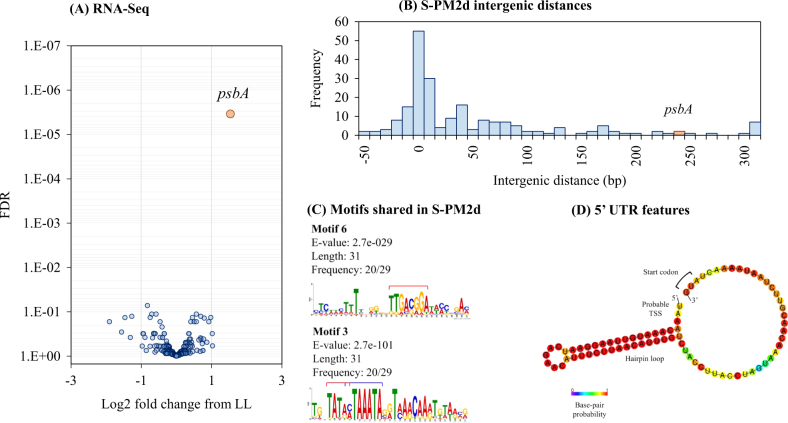
To decipher potential regulatory mechanisms occurring at the transcriptional level under HL compared to LL conditions, we conducted RNA-Seq analysis of transcriptomes from the same experiment. RNA was extracted from triplicate cultures at 1, 3, 6, and 9 h after infection in HL and LL infected cells and samples from each light treatment pooled in equimolar quantities to assess total changes in expression. Reads were mapped to both the S-PM2d and Synechococcus sp. WH7803 reference genomes and mean per base coverage estimated. There was a significant increase in coverage of the phage transcriptome under HL compared to LL conditions (2.34-fold increase, t3 = 3.98, p = 0.014), which was not apparent in the host (Fig. 2b). This was despite a fraction of the cells undergoing lysis at 9 h in HL compared with no lysis under LL conditions (Fig. 1a, b). This suggests a generalised increased rate of phage transcription at HL despite no change in DNA replication. We used differential expression tests to find light-responsive genes in the cyanophage genome. To our surprise, only one cyanophage gene was proportionately differentially expressed under HL, compared to LL, conditions. This was the photosynthetic AMG psbA encoding the D1 polypeptide located at the core of the PSII reaction centre (Fig. 3a).
Fig. 3.
The effect of light intensity on global gene expression. a A volcano plot showing the log2 fold change of gene expression relative to the LL treatment. The y-axis shows the false discovery rate corrected p-value calculated using edgeR. The orange circle shows statistically significant differential gene expression by edgeR. b Histogram of intergenic sequence lengths across the cyanophage S-PM2d genome. The bin containing psbA is shown in orange. c DNA sequence motifs found upstream of cyanomyoviral psbAs. Red bars indicate −35 and −10 elements of the σ70 binding sites and the blue bar shows the late σ factor Gp55-binding site. d Predicted hairpin loop folding structure of the S-PM2d psbA 5′-UTR
The cyanophage-encoded psbA is a ‘nearly core’ gene in cyanomyoviruses [21], the result of repeated acquisition events from their immediate hosts [22]. It is possible, therefore, that phage acquisition of host genes by recombination has also co-opted the same light-dependent regulatory features of psbA. Indeed, other cyanophages have been shown to regulate expression of phosphorus scavenging AMGs in response to phosphate limitation though exploitation of the host’s own PhoBR two-component system [16]. Cyanobacteria, including marine Synechococcus, display light-dependent transcription of psbAs [18, 23]. In model cyanobacteria, several factors influence the light-dependent accumulation of psbA transcripts, including alternative sigma factors [24–28] and DNA tertiary structure [29]. Moreover, D1 degradation products directly bind upstream of psbA sequences, such that light-induced damage could directly influence psbA transcription in a positive feedback manner [30]. We could not identify any conserved motifs in the upstream region of S-PM2d psbA that are shared with their Synechococcus hosts, perhaps suggesting divergent regulation strategies between the host and phage versions.
Furthermore, S-PM2d has an uncharacteristically long intergenic sequence upstream of psbA (232 bp, compared with a median of 6 bp across the genome; Fig. 3b). This difference is also true for other cyanomyoviruses where the psbA upstream regions vary between 125-453 bp. Therefore, we also sought to discover DNA sequence motifs shared amongst cyanomyoviruses that may be involved in regulation of psbA. We discovered six high confidence motifs (Fig. S1), of which two were present in S-PM2d (Fig. 3c). These two motifs contain the −35 (motif 6) and −10 (motif 3) elements of the σ70 transcription factor binding sites typical in T4-like early phage genes [31]. In addition, motif 3 contains the binding site for the T4-like late gene sigma factor Gp55 [32] (Fig. 3c). Thus, S-PM2d retains motifs required for coordinated expression of psbA in early and late infection.
However, the S-PM2d psbA region contains a novel 3′ antisense ncRNA, cfrI which is known to be expressed [33], as well as a previously unrecognised 36 bp inverted repeat producing a probable RNA hairpin loop in the 5′ untranslated region (5′-UTR; Fig. 3d). Structured 5′-UTRs are well known to affect mRNA stability in bacteria and bacteriophage [34] as are cis-encoded antisense RNAs [35], especially for cyanobacterial psbAs [36]. These features may therefore contribute to regulation of the cyanophage psbA in response to light.
In yet another twist, the S-PM2d psbA encodes a group I self-splicing intron within its psbA transcript, a feature found to be widespread in cyanophage psbA genes from marine metagenomic libraries [33, 37]. The intron interrupts the recognition sequence of a downstream homing endonuclease F-CphI therefore providing self-immunity while allowing the endonuclease to spread amongst intron-less alleles [38]. The intron contains several in-frame stop codons. Thus, it is unlikely that an unspliced transcript encodes a functional D1 polypeptide [33]. Nevertheless, the intron splices efficiently during infection [33].
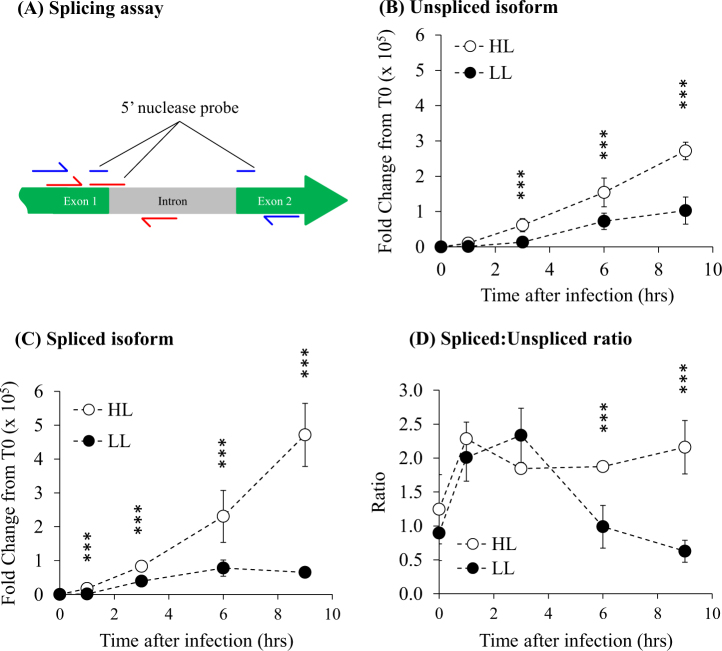
To understand whether intron splicing played a role in light-dependent psbA expression and indeed to validate our RNA-Seq data we conducted reverse transcriptase quantitative PCR (RT–qPCR) of the cyanophage psbA mRNA. We designed a splicing assay to detect relative expression of both isoforms of the transcript (Fig. 4a). We employed a 5′ nuclease qPCR assay which gives specificity to each transcript isoform using a 25–30 bp fluor-quencher linked probe that is either internal to the intron (unspliced isoform detection), or that bridges the exon 1/2 junction (spliced isoform detection). The reverse primer is also located in either the intron (unspliced isoform detection) or on exon 2 (spliced isoform detection) and is specific for cloned copies of each isoform.
Fig. 4.
Light-dependent splicing of the S-PM2d psbA intron. a Schematic of the splicing assay. The blue colour symbolises the primer/probe assay to detect the spliced transcript while red shows the primer/probe assay used to detect the unspliced transcript. b–c Relative expression of the unspliced isoform and spliced isoform, respectively, over the time-course of infection at HL and LL. d The ratio of spliced:unspliced isoforms over the time-course. Error bars represent the standard deviation of the mean. ***p < 0.001 as calculated using REST analysis
The RT–qPCR data validates that obtained from RNA-Seq, showing increased expression of both spliced and unspliced isoforms at HL (Fig. 4b, c). This increase is particularly pronounced in the spliced isoform during late infection, where we observed a 10.6-fold increase at HL compared with LL (Fig. 4c), compared with only a 3.37 fold increase of the unspliced isoform. Importantly, our assay allows determination of the spliced: unspliced isoform ratio (Fig. 4d), which shows that throughout early infection up to 4 h, the phage maintains a ~2-fold increase in the amount of the spliced isoform (Fig. 4d). At HL this is maintained throughout infection, whereas in LL we observed a rapid decline to approximately equimolar amounts by 6 h. Thus, the splicing of the intron appears to be responsive to light intensity. To our knowledge, this is the first report of a differentially spliced intron in a bacteriophage, though interestingly the eukaryotic alga Chlamydomonas reinhardtii, contains four group I introns in its psbA gene [39], which are known to be differentially spliced in response to light [40]. Since unspliced transcripts likely do not encode functional polypeptides, we suggest that differential splicing of the psbA mRNA represents a mechanism of gene regulation, potentially in coordination with the antisense ncRNA cfrI and/or the structured 5′-UTR affecting mRNA stability.
Differential expression of host genes reinforce translational bottlenecks
In addition to the differential expression of cyanophage genes, we also analysed light-responsive Synechococcus host genes. Of the 39 genes upregulated under high light conditions, 25 (64%) were involved in translation or posttranslational modification, protein turnover and chaperones (Table 1). This included 13/31 and 4/22 proteins forming the 50 S and 30 S subunits of the ribosome, respectively. Moreover, both subunits of the GroEL/GroES complex as well as the DnaJ and DnaK chaperones were upregulated (Table 1). Meanwhile, three of the four copies of the host’s psbA gene were also upregulated in high light (Table 1), perhaps complementing phage supplies of psbA mRNAs. Downregulated genes mostly included components of the light harvesting apparatus, potential cell motility genes and subunits involved in linear electron transport—most notably photosystem I reaction centre subunits (Table 1).
Table 1.
Differentially expressed Synechococcus host genes between high and low light conditions during infection with cyanophage S-PM2. Positive and negative log2 fold changes indicate up/down regulation respectively with respect to the low light treatment
| Locus Tag | Gene Name | Annotation | Functional group | Log2 Fold Change in expression | adj-p-value |
|---|---|---|---|---|---|
| Upregulated | |||||
| SYNWH7803_0424 | rplN | 50 S ribosomal protein L14 | Translation | 1.348 | 3.41E−02 |
| SYNWH7803_2372 | rplJ | 50 S ribosomal protein L10 | Translation | 1.246 | 2.41E−12 |
| SYNWH7803_0434 | rplC | 50 S ribosomal protein L3 | Translation | 1.240 | 1.69E−08 |
| SYNWH7803_0431 | rplB | 50 S ribosomal protein L2 | Translation | 1.209 | 4.96E−05 |
| SYNWH7803_1329 | rpsB | 30 S ribosomal protein S2 | Translation | 1.200 | 9.88E−06 |
| SYNWH7803_0429 | rplV | 50 S ribosomal protein L22 | Translation | 1.171 | 1.23E−02 |
| SYNWH7803_0422 | rplE | 50 S ribosomal protein L5 | Translation | 1.138 | 8.77E−05 |
| SYNWH7803_0427 | rplP | 50 S ribosomal protein L16 | Translation | 1.102 | 1.23E−02 |
| SYNWH7803_2374 | rplK | 50 S ribosomal protein L11 | Translation | 1.089 | 3.85E−04 |
| SYNWH7803_0591 | rpsD | 30 S ribosomal protein S4 | Translation | 1.072 | 3.53E−03 |
| SYNWH7803_0433 | rplD | 50 S ribosomal protein L4 | Translation | 1.033 | 5.82E−03 |
| SYNWH7803_0428 | rpsC | 30 S ribosomal protein S3 | Translation | 1.033 | 2.68E−03 |
| SYNWH7803_0418 | rpsE | 30 S ribosomal protein S5 | Translation | 1.003 | 5.55E−03 |
| SYNWH7803_0423 | rplX | 50 S ribosomal protein L24 | Translation | 1.001 | 1.76E−03 |
| SYNWH7803_0420 | rplF | 50 S ribosomal protein L6 | Translation | 0.972 | 1.66E−03 |
| SYNWH7803_2371 | rplL | 50 S ribosomal protein L7/L12 | Translation | 0.924 | 8.30E−06 |
| SYNWH7803_2373 | rplA | 50 S ribosomal protein L1 | Translation | 0.805 | 1.56E−02 |
| SYNWH7803_1863 | groEL | 60 kDa chaperonin 2 | Posttranslational modification, protein turnover, chaperones | 1.520 | 7.06E−15 |
| SYNWH7803_1118 | ahpC | Peroxiredoxin, AhpC/TSA family | Posttranslational modification, protein turnover, chaperones | 1.315 | 3.65E−05 |
| SYNWH7803_1998 | groEL | 60 kDa chaperonin 1 | Posttranslational modification, protein turnover, chaperones | 1.171 | 5.63E−12 |
| SYNWH7803_0023 | SynWH7803_0023 | DnaJ-class molecular chaperone | Posttranslational modification, protein turnover, chaperones | 1.043 | 3.61E−02 |
| SYNWH7803_2514 | dnaK | Molecular chaperone DnaK | Posttranslational modification, protein turnover, chaperones | 0.849 | 5.15E−07 |
| SYNWH7803_0016 | msrB | Peptide methionine sulfoxide reductase | Posttranslational modification, protein turnover, chaperones | 0.758 | 1.18E−03 |
| SYNWH7803_1999 | groES | 10 kDa chaperonin | Posttranslational modification, protein turnover, chaperones | 0.743 | 1.82E−02 |
| SYNWH7803_1555 | pepN | Aminopeptidase N | Posttranslational modification, protein turnover, chaperones | 0.603 | 2.50E−02 |
| SYNWH7803_0790 | psbA | Photosystem II protein D1 | Photosynthesis | 1.795 | 8.48E−04 |
| SYNWH7803_2084 | psbA | Photosystem II protein D1 | Photosynthesis | 1.695 | 5.50E−04 |
| SYNWH7803_0366 | psbA | Photosystem II protein D1 | Photosynthesis | 1.578 | 5.53E−04 |
| SYNWH7803_1861 | fabG | 3-oxoacyl-[acyl-carrier-protein] reductase | Fatty acid biosynthesis | 1.023 | 4.52E−02 |
| SYNWH7803_1870 | SynWH7803_1870 | Uncharacterised protein required for cytochrome oxidase assembly | Energy production and conversion | 1.599 | 3.16E−03 |
| SYNWH7803_2017 | atpA | ATP synthase alpha chain | Energy production and conversion | 0.718 | 1.21E−02 |
| SYNWH7803_2495 | cynS | Cyanate lyase | Cyanate metabolism | 0.616 | 3.14E−03 |
| SYNWH7803_0169 | ahcY | Adenosylhomocysteinase | Coenzyme transport and metabolism | 1.219 | 1.18E−03 |
| SYNWH7803_2069 | SynWH7803_2069 | Carbohydrate-binding protein; modular; contains a central CBM2 module | Carbohydrate transport and metabolism | 1.998 | 4.00E−11 |
| SYNWH7803_1930 | glgX | Glycogen debranching enzyme | Carbohydrate transport and metabolism | 1.814 | 1.08E−03 |
| SYNWH7803_1423 | SynWH7803_1423 | Conserved hypothetical protein | 1.490 | 4.99E−02 | |
| SYNWH7803_2070 | SynWH7803_2070 | Two-component system response regulator | 1.119 | 5.22E−03 | |
| SYNWH7803_0046 | SynWH7803_0046 | DHSS soluble hydrogenase, small subunit | 0.809 | 4.27E−02 | |
| SYNWH7803_1728 | SynWH7803_1728 | Integral membrane protein | 0.586 | 1.82E−02 | |
| Downregulated | |||||
| SYNWH7803_0611 | rpoD | RNA polymerase sigma factor RpoD, sigma-70 family | Transcription | −0.746 | 1.22E−04 |
| SYNWH7803_2501 | rpoD | Alternative RNA polymerase sigma factor, sigma-70 family | Transcription | −1.037 | 4.76E−04 |
| SYNWH7803_1592 | trxA | Thioredoxin | Redox homoeostasis | −0.882 | 7.75E−04 |
| SYNWH7803_2137 | SynWH7803_2137 | Predicted ATPase with chaperone activity | Posttranslational modification, protein turnover, chaperones | −1.957 | 9.76E−03 |
| SYNWH7803_0353 | psbO | Photosystem II manganese-stabilising protein PsbO | Photosynthesis | −0.500 | 4.78E−02 |
| SYNWH7803_0984 | petJ | Cytochrome C6 | Photosynthesis | −0.739 | 4.36E−02 |
| SYNWH7803_0252 | ndhC | NAD(P)H-quinone oxidoreductase chain 3 | Photosynthesis | −0.768 | 3.63E−02 |
| SYNWH7803_0391 | psaA | Photosystem I P700 chlorophyll A apoprotein A1 | Photosynthesis | −0.829 | 1.46E−06 |
| SYNWH7803_0534 | petD | Cytochrome b6-f complex subunit 4 | Photosynthesis | −0.869 | 1.18E−03 |
| SYNWH7803_1224 | psaK | Photosystem I reaction centre subunit X, PsaK | Photosynthesis | −0.983 | 1.45E−03 |
| SYNWH7803_0396 | psaL | Photosystem I reaction centre subunit XI | Photosynthesis | −1.001 | 3.18E−04 |
| SYNWH7803_0392 | psaB | Photosystem I P700 chlorophyll A apoprotein A2 | Photosynthesis | −1.078 | 2.41E−12 |
| SYNWH7803_0502 | mpeD | Phycobilisome linker polypeptide, C-phycoerythrin-associated | Light Harvesting | −0.543 | 2.88E−02 |
| SYNWH7803_0500 | SynWH7803_0500 | Conserved hypothetical protein in phycobilisome rod gene region | Light Harvesting | −0.884 | 6.12E−04 |
| SYNWH7803_0493 | mpeB | C-phycoerythrin class II beta chain | Light Harvesting | −1.034 | 7.27E−10 |
| SYNWH7803_0501 | cpeE | Phycobilisome linker polypeptide, C-phycoerythrin class I-associated | Light Harvesting | −1.042 | 4.63E−05 |
| SYNWH7803_0492 | mpeA | C-phycoerythrin class II alpha chain | Light Harvesting | −1.312 | 1.05E−09 |
| SYNWH7803_0670 | chlL | Protoporphyrin IX Mg-chelatase subunit ChlD | Chlorophyll biosynthesis | −1.493 | 5.53E−15 |
| SYNWH7803_0029 | gap1 | Glyceraldehyde-3-phosphate dehydrogenase | Central carbon metabolism | −0.944 | 5.50E−04 |
| SYNWH7803_0543 | SynWH7803_0543 | LysM-repeat protein | Cell wall/membrane biogenesis | −0.533 | 9.89E−03 |
| SYNWH7803_1746 | murA | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | Cell wall/membrane biogenesis | −1.010 | 1.37E−02 |
| SYNWH7803_1841 | pilT | Twitching motility protein | Cell motility | −1.435 | 3.85E−14 |
| SYNWH7803_1798 | SynWH7803_1798 | Uncharacterized conserved secreted protein, pili subunit superfamily | Cell motility | −1.524 | 4.63E−08 |
| SYNWH7803_1796 | SynWH7803_1796 | Uncharacterized conserved secreted protein, pili subunit superfamily | Cell motility | −1.540 | 1.21E−12 |
| SYNWH7803_1795 | SynWH7803_1795 | Uncharacterized conserved secreted protein, pili subunit superfamily | Cell motility | −1.658 | 9.13E−29 |
| SYNWH7803_0756 | SynWH7803_0756 | Alpha-glycosidase of family GH13; possible 1, 4-alpha-glucan branching enzyme | Carbohydrate transport and metabolism | −1.070 | 3.15E−03 |
| SYNWH7803_1937 | SynWH7803_1937 | Conserved hypothetical protein | −0.547 | 1.83E−02 | |
| SYNWH7803_0291 | SynWH7803_0291 | Conserved hypothetical protein | −0.657 | 5.56E−03 | |
| SYNWH7803_1897 | SynWH7803_1897 | Uncharacterized conserved secreted protein | −0.659 | 4.11E−02 | |
| SYNWH7803_0551 | SynWH7803_0551 | Hypothetical protein | −0.659 | 1.18E−02 | |
| SYNWH7803_2356 | SynWH7803_2356 | Uncharacterized conserved secreted protein | −0.750 | 1.18E−03 | |
| SYNWH7803_0849 | SynWH7803_0849 | Conserved hypothetical protein | −0.864 | 1.59E−02 | |
| SYNWH7803_0308 | SynWH7803_0308 | Secreted protein with pentapeptide repeats | −0.912 | 2.42E−03 | |
| SYNWH7803_2182 | SynWH7803_2182 | Conserved hypothetical protein | −0.922 | 1.77E−05 | |
| SYNWH7803_0069 | SynWH7803_0069 | Conserved hypothetical protein | −1.122 | 7.64E−04 | |
| SYNWH7803_2423 | SynWH7803_2423 | Conserved hypothetical protein | −1.160 | 2.20E−04 | |
| SYNWH7803_1348 | SynWH7803_1348 | Hypothetical protein | −1.325 | 9.08E−04 | |
| SYNWH7803_0705 | SynWH7803_0705 | Hypothetical protein | −1.393 | 3.38E−06 | |
| SYNWH7803_0839 | SynWH7803_0839 | Hypothetical protein | −1.394 | 1.56E−02 | |
| SYNWH7803_1797 | SynWH7803_1797 | Uncharacterized conserved secreted protein | −1.509 | 2.30E−03 | |
| SYNWH7803_2085 | SynWH7803_2085 | Uncharacterized conserved secreted protein | −1.612 | 2.27E−08 | |
| SYNWH7803_1502 | SynWH7803_1502 | Conserved hypothetical membrane protein | −1.629 | 9.89E−03 | |
| SYNWH7803_1800 | SynWH7803_1800 | Uncharacterized conserved membrane protein | −1.655 | 4.20E−17 | |
| SYNWH7803_0463 | SynWH7803_0463 | Uncharacterized conserved membrane protein | −1.757 | 3.86E−04 | |
| SYNWH7803_0464 | pex | Possible Pex protein (Period-extender gene product) | −2.178 | 9.21E−12 | |
| SYNWH7803_0940 | SynWH7803_0940 | Conserved hypothetical protein | −2.185 | 3.01E−14 | |
| SYNWH7803_2186 | SynWH7803_2186 | Conserved hypothetical protein | −2.274 | 7.21E−20 | |
| SYNWH7803_0934 | SynWH7803_0934 | Hypothetical protein | −3.056 | 2.56E−88 | |
Discussion
Altogether, our data show that light intensity plays a key role in modulating cyanophage development in their Synechococcus host. In contrast to our prediction, there was no change in the rate or amount of DNA replication. Instead, increased irradiance induced an early burst phenotype. Other than psbA, which was overexpressed under HL conditions, there was no change detectable in cyanophage gene expression. The encoded D1 protein binds all the proteins and cofactors required for charge separation leading to the oxidation of water [41]. The proximity to the charge separation event means D1 is particularly susceptible to damage and as such is rapidly turned over at HL [42]. Thus, upregulation of cyanophage psbA under HL conditions may increase the rate of delivery of D1 to PSII, restoring activity of the reaction centre, which in turn is manifest as the observed increase in PSII photochemistry (Fig. 2a). The resulting effect is an increase in the rate of photophosphorylation and more energy for cyanophage development. Taken together, and with no obvious change in expression of other cyanophage genes, we suggest that it is this increase in energy that supports an earlier burst under HL conditions, and therefore that translation is the rate limiting step for cyanophage development during late infection. In this respect, we calculated the energy requirement of cyanophage DNA replication assuming a burst size of 10. This calculation is based on known pathways of pyrimidine and purine deoxyribonucleotide biosynthesis in Synechococcus (Fig. S2). In total, we estimate a requirement of 2.2 × 10−17 mol ATP per infection (Supplementary Information). This assumes that all dNTPs are synthesised de novo, which is therefore an overestimate due to the several pathways predicted for nucleotide salvage in the S-PM2d genome [43]. In comparison, we computed the energy cost of production of just the detected structural proteins of cyanophage S-PM2 [44]. This includes both the biosynthesis (7.4 × 10−19 mol ATP per infection) but also crucially the polymerisation of amino acids (4.4 × 10−17 mol ATP per infection) (Supplementary Information). Thus, there is a 2-fold increase in energy required for translation of just the known structural proteins that form the phage particle compared with that of DNA synthesis. This must underestimate the overall increase in energy required as it does not include structural proteins that have yet to be identified, or the non-structural proteins critical for particle assembly, genome replication or subversion of host metabolism. Clearly then, translation represents the major energetic cost for cyanophage development. This conclusion is supported both by recent modelling studies into the energetics of development of coliphage T4 [45] and our own analysis of the host transcriptional response during infection under higher irradiance. The latter specifically showed upregulation of components of the bacterial ribosome as well as several chaperone proteins presumably involved in aiding correct folding of nascent polypeptides. We predict that upregulation of these genes will cause increases in the local concentration of ribosomes which would support an increased translation rate at higher light [46]. Unfortunately, this has largely been overlooked in previous cyanophage work since acquisition of AMGs has been proposed to overcome metabolic bottlenecks to increase cyanophage genome replication [7, 14, 15]. Our data suggests that the energy required for translation during late infection imposes such a metabolic block, and that acquisition and coordinated expression of the cyanophage S-PM2 psbA AMG overcomes this energy bottleneck. We predict that energy limitation scales with light intensity such that reduced latent periods would be expected during the day and at shallower depths (Fig. 5). Indeed, cyanophage development has been shown to be inhibited by darkness [7, 47] and infections with cyanosiphoviruses over a simulated diel cycle show that latent period length is correlated with light intensity [48]. Moreover, isolation studies show increased abundance of cyanophages at mid-day, but only at shallower depths with increased irradiance [49]. In addition, recent lagrangian metagenomics sampling of viral communities in waters of the north Pacific subtropical gyre suggest cyanophages exhibit diel rhythmicity, peaking in intracellular abundance between mid-day to early evening [50]. Given that Synechococcus and the closely related Prochlorococcus are dominant members of the photoautotrophic community in the global ocean, and that natural gradients of light intensity occur with depth, time of day and season, it is likely that energy limitation of cyanophage development would have important implications for the control of the Synechococcus community. For instance, it has recently been shown than Synechococcus exhibits a regular spring bloom in coastal waters off Woods Hole, USA [51]. This bloom is driven by increases in Synechococcus cell division rates in response to warmer water temperature (but importantly not light) and decoupling from loss rates in early spring. It may be that extended viral latent periods caused by light limitation are prominent in early spring. These would cause reduced Synechococcus loss rates, contributing to bloom formation. Conversely, the Synechococcus loss rate is maximal during mid-summer [51], when increased irradiance may shorten viral latent periods (Fig. 5a). This is particularly relevant in a warming ocean where Synechococcus blooms have already been shown to be occurring earlier in the year [51]. The effect of light on latent periods of Synechococcus likely also affect the daily and depth-dependent cycling of carbon in the oceans, with diurnal, depth and seasonal variability in the release of dissolved organic carbon (DOC) via the viral shunt (Fig. 5b) [52, 53]. Recently, whole-community metatranscriptomics has revealed reproducible daily trends in the expression of genes involved in oxidative phosphorylation and translation amongst the heterotrophic component of surface ocean communities [54, 55]. Peak expression coincides with daily maxima in irradiance and are subsequent to expression of photosynthetic genes by the phototrophic community members. Therefore, high light-induced lysis by viruses of phototrophs could link these two processes. Future studies should seek to determine the generality of this phenomenon amongst cyanophage and understand the relationship between rates of photophosphorylation and the energy requirement for lysis over a range of irradiances and varying host nutrient status so that such information can be integrated into models of oceanic C cycling.
Fig. 5.
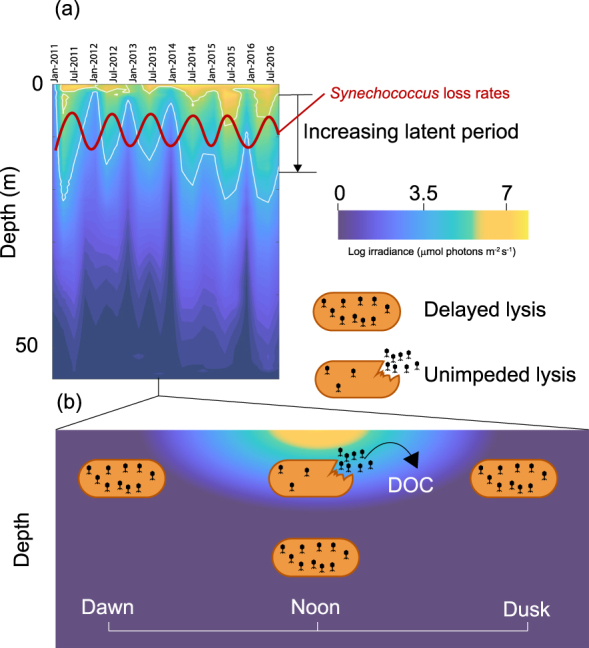
The ecological significance of light-dependent lysis delay. a Seasonal and inter-annual changes in irradiance with depth at a coastal observatory (the data shown here is from station L4 in the Western English Channel, the site of isolation of cyanophage S-PM2d). White lines correspond to the contours of the irradiances used in this study. The red line shows seasonal trends in Synechococcus loss rates [51]. b Diurnal changes in irradiance. Delayed lysis at low light results in a reduction in the rate of DOC release. Lysis at increased irradiances increases the rate of DOC release through the viral shunt
Methods
Infection conditions
Synechococcus sp. WH7803 was grown in ASW medium [56] to 1 × 108 cells ml−1 at 23˚C under continuous illumination of 15 µmol photons m−2 s−1. Cultures were then infected with cyanophage S-PM2d [12] at a VBR of 10 and samples incubated at LL (15 µmol photons m−2 s−1) for 1 h to allow adsorption. Three biological replicates were then shifted to 210 µmol photons m−2 s−1 (HL) while three control cultures were maintained at 15 µmol photons m−2 s−1 (LL). The HL treatment was selected based on the ability to induce transient photoinhibition in uninfected cells without being lethal. Photoinhibition was assessed by determining the PSII maximum photochemical yield (Fv/Fm) after treatment with lincomycin following [18]. The rate of photoinhibition for the HL treatment is shown in Fig. S3.
Assessment of infection dynamics
Infection dynamics were assayed using qPCR to enumerate both intracellular and extracellular cyanophage genome copy number (GCN) [11, 16, 57]. Samples were taken immediately after addition of cyanophage S-PM2d and then at 1, 2, 3, 5, 7, 9, 12, 15 and 18 h. For quantification of extracellular cyanophages 100 µl cell suspension was diluted to 500 µl by addition of ASW medium. This was syringe filtered through a 0.2 µm pore size disposable filter (Minisart, Sartorius, Goettingen, Germany). The filtrate was collected, snap frozen in liquid nitrogen, and then stored at −80 ˚C before quantification. Intracellular cyanophages were quantified by vacuum filtration of infected cells onto a 0.2 µm pore size polycarbonate filter (Isopore, Millipore, Billerica, USA). Filters were washed three times with 1 ml preservation solution (10 mM Tris-HCl, 100 mM EDTA, 500 mM NaCl, pH 8.0) and after their removal from the filtration tower added to a ribolyser tube (Lysing Matrix E, MP Bioproducts, CA, USA) and snap frozen in liquid nitrogen for DNA extraction.
DNA extraction
DNA extraction was extensively optimised and the following was found to yield optimal results. 650 µl Tris-HCl, pH 8.0 was added to a ribolyser tube (Lysing Matrix E, MP Bioproducts, CA, USA). The tube was then lysed three times for 30 s in a Tissue Lyser (Qiagen, Hilden, Germany) at 30 Hz. In between disruption the tubes were incubated on ice for 1 min. The lysate was subjected to centrifugation for 30 s at 10,000×g and 100 µl of the supernatant subsequently frozen at −80 ˚C. Samples were diluted 1/10 and 1 µl was used as the template for qPCR reactions.
qPCR
qPCR enumeration of cyanophage S-PM2d genome copies was achieved using a PrimeTime 5′ nuclease assay (IDT, Coralville, USA). Primer/probe sequences are shown in the Supplementary Information and were directed towards the psbA intron of S-PM2d. 20 µl reaction volumes were used containing 1 × Brilliant III Ultra-Fast qPCR Master Mix (Agilent, CA, USA), 500 nM primers and 250 nM probe, 1 µl template and nuclease free water. Each qPCR plate contained a triplicate dilution series from the initial phage stock from 2.5 × 1010 −2.5 × 102 pfu ml−1. Cycling conditions were 3 min at 95 ˚C, followed by 40 cycles of denaturing for 15 s at 95 ˚C and annealing for 15 s at 60 ˚C.
Photophysiology
The maximum quantum yield of PSII photochemistry (Fv/Fm) was measured according to [18]. All measurements were made using a pulse amplitude-modulated fluorometer (PhytoPAM, Walz, Effeltrich, Germany). Subsamples (2 ml) were incubated for ~5 min in the dark to completely oxidise the primary electron acceptor QA. 500 µl subsample was added to a PhytoPAM cuvette (Waltz, Effeltrich, Germany) and immediately diluted to 2 ml with ASW medium. A weak modulating light was then applied at 520 nm with intensity of 1 µmol photons m−2 s−1 and the basal fluorescence measured (F0). 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) was added to the cuvette to a final concentration of 100 µM, and an actinic light was supplied at ~1300 µmol photons m−2 s−1. Chlorophyll fluorescence rose rapidly followed by a slower period of increase until saturation. A saturating pulse of ~2600 µmol photons m−2 s−1 was then delivered for 200 ms to completely reduce QA. The fluorescence was simultaneously measured (Fm) and Fv/Fm calculated as (Fm−F0)/Fm.
RNA extraction and RNA-Seq library preparation
Total RNA was isolated following Logemann et al. [58]. Briefly, 50 ml of Synechococcus sp. WH7803 was centrifuged at 3220 × g at 4°C and flash frozen in liquid N2. Pellets were thawed in 1.5 ml Z buffer (8 M guanidinium hydrochloride; 50 mM β-mercaptoethanol; 20 mM EDTA) at room temperature for 30 min. 2 volumes of acidified phenol (pH 4.5) were added and heated to 65˚C for 30 min, followed by the addition of chloroform:isoamyl alcohol for 15 min. The aqueous phase was transferred to separate microcentrifuge tubes. RNA was precipitated by the addition of 1 vol. isopropanol followed by incubation overnight at 4˚C and subsequent centrifugation at 16,060×g (Biofuge Pico, Heraeus) at 4 ˚C for 30 min. The pellet was then washed with 70% (v/v) ethanol and further centrifuged for 15 min. Pellets were resuspended in 50 µl H2O. DNA was removed using the TURBO DNA-free kit (Ambion/Life Technologies, Carlsbad, USA) following the manufacturer’s instructions. gDNA contamination was tested by PCR using primers phoH_F/phoH_R (Table S1). Any samples that yielded a PCR product were further treated using the TURBO DNA-freeTM kit (Ambion, Forster City, USA).
Strand-specific library preparation
For each biological replicate, total RNA from time points 1, 3, 6, and 9 h after infection were pooled in equimolar quantities. RNA-Seq libraries were prepared using the ScriptSeq v2.0 kit (Illumina, San Diego, USA) by the Centre for Genomic Research, University of Liverpool and sequenced at the same facility using an Illumina GAIIx platform generating 100 bp paired-end reads. Raw reads have been submitted to ArrayExpress (EMBL-EBI) under accession E-MTAB-5840.
Read-mapping, read counts and differential expression tests
Paired-end reads were trimmed using Sickle [59] using default settings. Trimmed reads were mapped to the S-PM2d (E.B.I. Accession no. LN828717) and Synechococcus sp. WH7803 genome (Acc. No. NC_009481) using Bowtie 2 [60]. Mapping was done in –end-to-end mode using the following parameters: -D 20 -R 3 -N 0 -L 20 -i S,1,0.50. SAM files were converted to BAM files and sorted using SAMtools v0.1.18 [61] and reads counts of genomic features were calculated using BEDtools v2.26.0 [62]. Only reads which overlapped across 10% of the read length into the genomic feature were counted. Differential expression tests were conducted using EdgeR [63] in the Degust online form.
RT–qPCR
cDNA synthesis was carried out with SuperScript III reverse transcriptase (Life Technologies, Carlsbad, USA) in 20 µl volumes with 2 µg total RNA. Each reaction contained 0.5 mM dNTPs, 2 pM gene-specific primer, 1× SuperScript III buffer, 5 mM DTT and 200U SuperScript III. The RNA, dNTP and primers were mixed and heated to 65˚C for 5 mins before being cooled on ice for 5 mins. SuperScript III, DTT and buffer were then added, mixed, and incubated at 55˚C for 60 mins. The reaction was inactivated by heating to 70 ˚C for 15 mins. qPCR reactions were in 20 µl volumes containing 1× Brilliant III Ultra-Fast qPCR Master Mix (Agilent, Santa Clara, USA), 1× relevant primer/probe assay (500 nM primers and 250 nM probe), 1 µl cDNA and nuclease free water to 20 µl. An Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies, Carlsbad, USA) was used for quantification. Cycling parameters were 95 ˚C for 1 min followed by 40 cycles of 15 s at 95˚C and 15 s at 60˚C. Raw CT values were exported and analysed using the efficiency corrected ΔΔCT method [64] as computed using the REST software [65]. The calibrator gene was the host 16S rRNA gene when quantifying spliced and unspliced transcripts and the unspliced transcript when calculating the ratio shown in Fig. 4d. Primer sequences and assay validations are in the Supplementary Information. The specificity of the spliced and unspliced psbAs were validated using cloned copies of each isoform.
Motif discovery
Motifs upstream of psbAs were detected using MEME [66] using a minimum width of 6 bp and maximum of 50 bp and an eval cutoff of 10−5.
Electronic supplementary material
Acknowledgements
RJP was in receipt of a Natural Environment Research Council (NERC) PhD studentship and a Warwick University IAS Fellowship. This work was also supported in part by NERC grant NE/N003241/1 and Leverhulme Trust grant RPG-2014-354 to A.D.M., D.J.E., and D.J.S.
Author contributions
R.J.P. D.J.E., A.D.M., and D.J.S. designed the research; R.J.P. performed the research; R.J.P. analysed the data; R.J.P. D.J.E., A.D.M., and D.J.S. wrote the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Andrew D. Millard, Email: adm39@le.ac.uk
David J. Scanlan, Email: D.J.Scanlan@warwick.ac.uk
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0043-3) contains supplementary material, which is available to authorized users.
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary productivity of the biosphere: an integration of terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Flombaum P, Gallegos JL, Gordillo RA, Rincón, J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110: 9824–9829. [DOI] [PMC free article] [PubMed]
- 3.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 4.Bouman HA, Ulloa O, Scanlan DJ, Zwirglmaier K, Li WKW, Platt T, et al. Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science. 2006;312:918–921. doi: 10.1126/science.1122692. [DOI] [PubMed] [Google Scholar]
- 5.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol. 2015;13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 7.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 8.Mann NH, Cook A, Millard AD, Bailey S, Clokie M. Bacterial photosynthesis genes in a virus. Nature. 2003;424:741–742. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 9.Puxty RJ, Millard AD, Evans DJ, Scanlan DJ. Viruses inhibit CO2 fixation in the most abundant phototrophs on Earth. Curr Biol. 2016;26:1585–1589. doi: 10.1016/j.cub.2016.04.036. [DOI] [PubMed] [Google Scholar]
- 10.Puxty RJ, Millard AD, Evans DJ, Scanlan DJ. Shedding new light on viral photosynthesis. Photosynth Res. 2015;126:71–97. doi: 10.1007/s11120-014-0057-x. [DOI] [PubMed] [Google Scholar]
- 11.Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA. 2011;108:757–764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puxty RJ, Perez-Sepulveda B, Rihtman B, Evans DJ, Millard AD, Scanlan DJ. Spontaneous deletion of an “ORFanage” region facilitates host adaptation in a “photosynthetic” cyanophage. PLoS ONE. 2015;10:e0132642. doi: 10.1371/journal.pone.0132642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey S, Clokie MRJ, Millard A, Mann NH. Cyanophage infection and photoinhibition in marine cyanobacteria. Res Microbiol. 2004;155:720–725. doi: 10.1016/j.resmic.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Hellweger FL. Carrying photosynthesis genes increases ecological fitness of cyanophage in silico. Environ Microbiol. 2009;11:1386–1394. doi: 10.1111/j.1462-2920.2009.01866.x. [DOI] [PubMed] [Google Scholar]
- 15.Bragg JG, Chisholm SW. Modeling the fitness consequences of a cyanophage-encoded photosynthesis gene. PLoS ONE. 2008;3:e3550. doi: 10.1371/journal.pone.0003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Q, Chisholm SW. Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr Biol. 2012;22:124–128. doi: 10.1016/j.cub.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 17.Suggett DJ, Moore CM, Geider RJ. Estimating aquatic productivity from active fluorescence measurements. In Chlorophyll a fluorescence in aquatic sciences: Methods and applications. Dev Appl Phycol. 2010;4:103–127. [Google Scholar]
- 18.Garczarek L, Dufresne A, Blot N, Cockshutt AM, Peyrat A, Campbell DA, et al. Function and evolution of the psbA gene family in marine Synechococcus: Synechococcus sp. WH7803 as a case study. ISME J. 2008;2:937–953. doi: 10.1038/ismej.2008.46. [DOI] [PubMed] [Google Scholar]
- 19.Bornefeld T, Simonis W. Effects of light, temperature, pH, and inhibitors on the ATP level of the blue-green alga Anacystis nidulans. Planta. 1974;115:309–318. doi: 10.1007/BF00388613. [DOI] [PubMed] [Google Scholar]
- 20.Paddison P, Abedon ST, Dressman HK, Gailbreath K, Tracy J, Mosser E, et al. The roles of the bacteriophage T4 r genes in lysis inhibition and fine-structure genetics: a new perspective. Genetics. 1998;148:1539–1550. doi: 10.1093/genetics/148.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crummett LT, Puxty RJ, Weihe C, Marston MF, Martiny JBH. The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology. 2016;499:219–229. doi: 10.1016/j.virol.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulo P, Sakurai I, Aro EM. Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: From transcription to PSII repair. Biochim Biophys Acta Bioenerg. 2012;1817:247–257. doi: 10.1016/j.bbabio.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Imamura S, Yoshihara S, Nakano S, Shiozaki N, Yamada A, Tanaka K, et al. Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J Mol Biol. 2003;325:857–872. doi: 10.1016/S0022-2836(02)01242-1. [DOI] [PubMed] [Google Scholar]
- 25.Imamura S, Asayama M, Shirai M. In vitro transcription analysis by reconstituted cyanobacterial RNA polymerase: roles of group 1 and 2 sigma factors and a core subunit, RpoC2. Genes Cells. 2004;9:1175–1187. doi: 10.1111/j.1365-2443.2004.00808.x. [DOI] [PubMed] [Google Scholar]
- 26.Ohkawa H, Pakrasi HB, Ogawa T. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J Biol Chem. 2000;275:31630–31634. doi: 10.1074/jbc.M003706200. [DOI] [PubMed] [Google Scholar]
- 27.Pollari M, Ruotsalainen V, Rantamäki S, Tyystjärvi E, Tyystjärvi T. Simultaneous inactivation of sigma factors B and D interferes with light acclimation of the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2009;191:3992–4001. doi: 10.1128/JB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimura T, Imamura S, Tanaka K, Shirai M, Asayama M. Cooperation of group 2 sigma factors, SigD and SigE for light-induced transcription in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 2007;581:1495–1500. doi: 10.1016/j.febslet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Asayama M, Kato H, Shibato J, Shirai M, Ohyama T. The curved DNA structure in the 5’-upstream region of the light-responsive genes: its universality, binding factor and function for cyanobacterial psbA transcription. Nucleic Acids Res. 2002;30:4658–4666. doi: 10.1093/nar/gkf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stelljes C, Koenig F. Specific binding of D1 protein degradation products to the psbAI promoter in Synechococcus sp. strain PCC 7942. J Bacteriol. 2007;189:1722–1726. doi: 10.1128/JB.01428-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller ES, Kutter E, Mosiq G, Arisaka F, Kunisawa T, Rüger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nechaev S, Geiduschek EP. Dissection of the bacteriophage T4 late promoter complex. J Mol Biol. 2008;379:402–413. doi: 10.1016/j.jmb.2008.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millard AD, Gierga G, Clokie MRJ, Evans DJ, Hess WR, Scanlan DJ. An antisense RNA in a lytic cyanophage links psbA to a gene encoding a homing endonuclease. ISME J. 2010;4:1121–1135. doi: 10.1038/ismej.2010.43. [DOI] [PubMed] [Google Scholar]
- 34.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 35.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakurai I, Damir S, Eisenhut M, Vuorio E, Steglich C, Hess WR, et al. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 2012;160:1000–1110. doi: 10.1104/pp.112.202127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Q, Bonocora RP, Shub DA. A free-standing homing endonuclease targets an intron insertion site in the psbA gene of cyanophages. Curr Biol. 2009;19:218–222. doi: 10.1016/j.cub.2008.11.069. [DOI] [PubMed] [Google Scholar]
- 39.Erickson JM, Rahire M, Rochaix J. Chlamydomonas reinhardii gene for the 32 000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 1984;3:2753–2762. doi: 10.1002/j.1460-2075.1984.tb02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshpande NN, Bao Y, Herrin DL. Evidence for light/redox-regulated splicing of psbA pre-RNAs in Chlamydomonas chloroplasts. RNA. 1997;3:37–48. [PMC free article] [PubMed] [Google Scholar]
- 41.Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 42.Tyystjärvi E, Aro EM. The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc Natl Acad Sci USA. 1996;93:2213–2218. doi: 10.1073/pnas.93.5.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann NH, Clokie MRJ, Millard A, Cook A, Wilson WH, Wheatley PJ, et al. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J Bacteriol. 2005;187:3188–3200. doi: 10.1128/JB.187.9.3188-3200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clokie MRJ, Thalassinos K, Boulanger P, Slade SE, Stoilova-McPhie S, Cane M, et al. A proteomic approach to the identification of the major virion structural proteins of the marine cyanomyovirus S-PM2. Microbiology. 2008;154:1775–1782. doi: 10.1099/mic.0.2007/016261-0. [DOI] [PubMed] [Google Scholar]
- 45.Mahmoudabadi G, Milo R, Phillips R. The energetic cost of building a virus. Proc Natl Acad Sci USA. 2017;114:E4324–E4333. doi: 10.1073/pnas.1701670114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vind J, Sorensen MA, Rasmussen MD, Pedersen S. Synthesis of proteins in Escherichia coli is limted by the concentration of free ribosomes. J Mol Biol. 1993;231:678–688. doi: 10.1006/jmbi.1993.1319. [DOI] [PubMed] [Google Scholar]
- 47.Thompson LR, Zeng Q, Chisholm SW. Gene expression patterns during light and dark infection of Prochlorococcus by cyanophage. PLoS ONE. 2016;11:1–20. doi: 10.1371/journal.pone.0165375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frois-Moniz K. Host/virus interactions in the marine cyanobacteriumProchlorococcus. PhD Thesis (Massachusetts Institute of Technology, Boston, USA, 2014).
- 49.Clokie MRJ, Millard AD, Mehta JY, Mann NH. Virus isolation studies suggest short-term variations in abundance in natural cyanophage populations of the Indian Ocean. J Mar Biol Assoc UK. 2006;86:499–505. doi: 10.1017/S0025315406013403. [DOI] [Google Scholar]
- 50.Aylward FO, Boeuf D, Mende DR, Wood-Charlson EM, Vislova A, Eppley JM, et al. Diel cycling and long-term persistence of viruses in the ocean’s euphotic zone. Proc Natl Acad Sci USA. 2017;114:11446–11451. doi: 10.1073/pnas.1714821114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunter-Cevera KR, Neubert MG, Olsen RJ, Solow AR, Shalapyonok A, Sosik HM. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science. 2016;354:326–329. doi: 10.1126/science.aaf8536. [DOI] [PubMed] [Google Scholar]
- 52.Weitz JS, Wilhelm SW. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol Rep. 2012;4:1–8. doi: 10.3410/B4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea. Bioscience. 1999;49:781–788. doi: 10.2307/1313569. [DOI] [Google Scholar]
- 54.Aylward FO, Eppley JM, Smith JM, Chavez FP, Scholin CA, DeLong EF. Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc Natl Acad Sci USA. 2015;112:5443–5448. doi: 10.1073/pnas.1502883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R, III, Schuster SC, et al. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345:207–212. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- 56.Wyman M, Gregory RPF, Carr NG. Novel role for phycoerythrin in a marine cyanobacterium, Synechococcus strain DC2. Science. 1985;230:818–820. doi: 10.1126/science.230.4727.818. [DOI] [PubMed] [Google Scholar]
- 57.Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 58.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 59.Joshi NA, Fass JN (2011). Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version1.33). Available at: https://github.com/najoshi/sickle.
- 60.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfaffl MW. Relative quantification. In: Dorak T, editor. Real-Time PCR. 1st ed. Abingdon, UK: Taylor and Francis; 2004. pp. 63–82. [Google Scholar]
- 65.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.