Abstract
Small cell lung cancer (SCLC) is the most deadly subtype of lung cancer due to its dismal prognosis. We have developed a lentiviral vector-mediated SCLC mouse model and have explored the role of both the NF-κB and CREB families of transcription factors in this model. Surprisingly, induction of NF-κB activity, which promotes tumor progression in many cancer types including non-small cell lung carcinoma (NSCLC), is dispensable in SCLC. Instead, suppression of NF-κB activity in SCLC tumors moderately accelerated tumor development. Examination of gene expression signatures of both mouse and human SCLC tumors revealed overall low NF-κB but high CREB activity. Blocking CREB activation by a dominant-negative form of PKA (dnPKA) completely abolished the development of SCLC. Similarly, expression of dnPKA or treatment with PKA inhibitor H89 greatly reduced the growth of SCLC tumors in syngeneic transplantation models. Altogether, our results strongly suggest that targeting CREB is a promising therapeutic strategy against SCLC.
Keywords: Small cell lung cancer, NF-κB, CREB, mouse model, lentiviral vector
Introduction
Lung cancer represents nearly 20% of all cancer-related deaths world-wide (1). Small cell lung cancer (SCLC) differs from other subtypes of lung cancer (altogether called non-small cell lung cancer, NSCLC) by histopathology and general treatment strategy in the clinic. SCLC is also unique among all lung cancers for its cell of origin, spectrum of genetic mutations and high potential to metastasize to distal organs. SCLC displays a neuroendocrine (NE) signature and is thus thought to be initiated from neuroendocrine cells scattered underneath Clara cell and ciliated cell layer of bronchioles (2). Two major tumor suppressors, p53 and Rb are found extensively mutated in >90% of SCLC patients (3). Though it accounts only for ~15% of all lung cancers in humans, SCLC is the most malignant subtype with five-year survival rate of <5%, due to its predisposition towards early metastasis (4). In contrast to NSCLC, against which several powerful targeted therapies have been developed, little innovation in treating SCLC has been achieved in the past decades (5).
SCLC has been modeled in mouse by conditional knockout of p53 and Rb genes in neuroendocrine cells with adenoviruses expressing Cre recombinase (6). These murine neuroendocrine tumors recapitulate the histopathology of the human disease; however, the latency for the developing tumor is extremely long (~400 days) which makes SCLC difficult to study via mouse models. We have in recent years expanded our lentiviral vector-mediated mouse cancer models to study SCLC (7, 8). Following transduction by lentiviral vectors with a combination of four common genetic mutations (Mycl1, p53, Rb and Pten) commonly found in human patients, mice develop advanced SCLC as early as 4 months.
The NF-κB pathway is an important player in almost all stages of tumorigenesis due to its essential roles in inflammation, cell proliferation and anti-apoptosis (9). Several studies including our own have shown that NF-κB activity is elevated in Kras-induced lung adenocarcinomas (ADC) and blocking NF-κB significantly reduces tumor growth in mice (7, 10, 11). However, little is known about NF-κB in SCLC and we wanted to study if the NF-κB pathway is also activated in this subtype of lung cancer and whether inhibition of NF-κB potentially benefit SCLC patients. We overexpressed IκBαM super-repressor to suppress NF-κB activity in these tumors and were surprised to find that they grew even faster than control tumors and led to an earlier clinical endpoint. A careful analysis of gene expression pattern of both mouse and human SCLC tumors revealed that the NF-κB pathway in SCLC is substantially down-regulated. In contrast CREB activity increases and likely helps to maintain the SCLC signature and cell proliferation in these cells. Our results thus suggest that SCLC may benefit from therapeutics targeting CREB transcriptional activity.
Materials and Methods
Mice
R26-M2rtTA knock-in mice was originally purchased from The Jackson Laboratory and backcrossed into pure FVB background. Wild-type FVB mice were used for syngeneic transplant model. All mice studies were carried out according to the protocols that were approved by the Institutional Animal Care and Use Committee (IACUC) of Salk Institute.
Clinical samples
Human lung adenocarcinoma and SCLC samples were collected from surgeries performed in the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University, with patient consent forms. All procedures were carried out under Institutional Review Board (IRB)-approved protocols. Written informed consent was obtained from all patients and the study was conducted in accordance with recognized ethical guidelines.
Lentiviral production and mice infection
Lentiviruses were produced as described previously(12). The shRNA sequences used were: 5′-GTACATGTGTAATAGCTCC-3′ (p53); 5′-AGCATATCTCCGACTAAATA-3′ (Rb); 5′-AAGATCAGCATTCACAAATTA-3′ (Pten). Viral titer was estimated by p24 ELISA with serial diluted viruses, and verified by infecting 293-rtTA cells and examining Mycl1 expression by immunoblotting. For SCLC tumor model, 8-week old R26-M2rtTA mice were pretreated with naphthalene (200 mg/kg, i.p.) 44 h before viral infection, and then infected intra-tracheally with 2×105 lentiviral particles using a protocol described before(7). Mice were switched to doxycycline diet (200 mg/kg, Bio-Serv) one week after the viral infection.
Histology, immunofluorescence staining, and immunoblotting analysis
Mouse lung tumor samples collected at different time-points were fixed with 10% formalin, paraffin-embedded and sectioned for haematoxylin and eosin staining (H&E) and immunofluorescence staining. Elite ABC system (Vector labs) was applied where staining signal was weak. Immunoblotting analysis was performed according to standard protocol. Antibodies were purchased from Millipore (SPC, NCAM, both 1:2000), Sigma (CGRP, 1:2000), Abcam (NF-κB p65 E379, 1:500), Cell Signaling (phos-CREB Ser133, Sox2, both 1:1000), Vector Labs (Ki-67, 1:500; p53, 1:4000), Santa Cruz Biotechnology (L-Myc 1:1000).
Primary cell culture and syngeneic transplant model
Primary tumors isolated from SCLC lentiviral model were dissociated with trypsin and cultured in modified HITES medium DMEM/F12 (1:1) supplemented with Glutamax, 4 μg/ml Hydrocortisone (Sigma), 5 ng/ml murine EGF (Invitrogen), Insulin-Transferrin-Selenium mix/solution (Invitrogen) and 10% Fetal Bovine Serum. Culture dishes were pre-coated with Poly-HEMA (Poly 2-hydroxyethyl methacrylate, Sigma) to prevent cell adhesion and differentiation. Mycoplasma testing was performed using commercial kit (Lonza). SCLC cells (10^6) infected with different expression vectors were transplanted subcutaneously into flank regions of FVB hosts. Tumors were collected 4 weeks later, weighed and fixed for histopathological analysis. PKA inhibitor treatment (H89, 10 mg/kg in 75% DMSO/PBS, i.p.) was carried out 2 weeks after the transplantation for another 2 weeks.
Quantitative RT-PCR
Total RNA isolated from homogenized tumor samples or cell lines was reverse transcribed using qScript cDNA SuperMix (Quantabio). Quantitative PCR was performed in triplicate using 7900HT Fast Real-Time PCR system with SYBR green method (Applied Biosystems). Results were analyzed for the relative expression of mRNAs normalized against GAPDH. A complete list of primers used in the study is shown in Supplementary Table S2.
Electrophoretic mobility shift assay (EMSA)
EMSA was carried out using LightShift Chemiluminescent EMSA Kit from Thermo Fisher Scientific and their standard protocol. Consensus DNA binding sequences were listed as following. NF-κB 5′-AGT TGA GGG GAC TTT CCC AGG C-3′; CREB 5′-AGA GAT TGC CTG ACG TCA GAG AGC TAG-3′; OCT1 5′-TGT CGA ATG CAA ATC ACT AGA A-3′.
RNAseq and GSEA analysis
Total RNA isolated from homogenized primary tissue samples (8 normal lungs, 6 SCLC-luci tumors and 6 SCLC-IκBαM tumors) was used to make stranded mRNA library. Single-end RNAseq was carried out using standard protocol on Illumina HiSeq 2500 sequencer. Reads were then mapped to mouse (mm9) genome sequence using STAR. Differential gene expression was analyzed using Cuffdiff. Human SCLC RNAseq raw data of 19 normal lungs, 21 primary tumors and 21 cell lines shared from Dr. Seshagiri group(13) were reanalyzed using a similar method as described above. FPKM values of differentially expressed genes, NF-κB genes and CREB genes from multiple samples were clustered and heatmaps were drawn with Cluster/TreeView softwares.
Statistics
Statistical significance of the differences of gene expression or tumor size was evaluated using Student’s unpaired two-tailed t-test. The Kaplan-Meier curves were analyzed by Log-rank test.
Accession numbers
Raw data of microarray analysis have been deposited to NCBI Gene Expression Omnibus (GEO) with accession number GSE89857.
Results
A lentiviral mouse model of SCLC
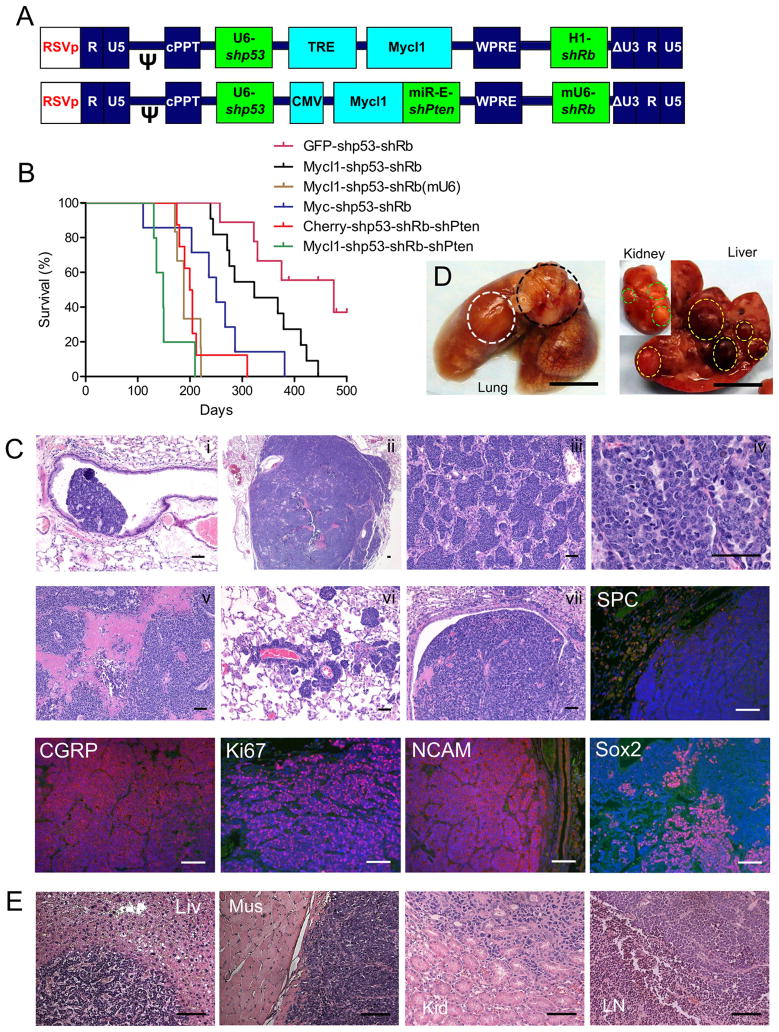
Currently available mouse models of SCLC are all based on conditional knockout in lung cells of different tumor suppressors including p53, Rb, p130 and Pten (6, 14–16). To faithfully recapitulate the human disease, we have successfully expanded our lentiviral vector-mediated cancer model to study SCLC in wild-type mice (Fig. 1A). Our result is consistent with earlier reports that loss of both p53 and Rb are indispensable to induce neuroendocrine tumors in lung while loss of Pten or overexpression of Mycl1 further accelerates tumor progression(6, 15). Interestingly, endogenous Mycl1 mRNA up-regulation and Pten down-regulation are readily detected in tumors induced with shp53-shRb vector (Supplementary Fig. S1A–B). Consistently, if Mycl1expression was lost upon the withdrawal of doxycycline, massive tumor shrinkage was observed (Supplementary Fig. S1C). All these observations indicated the requirement of enhanced Mycl1 expression and Pten suppression in SCLC development. Infected with a four-in-one vector (Mycl1-shp53-shRb-shPten), mice typically developed advanced SCLC within 4 months and had a medium survival time of 147 days (compared to 475 days with the shp53-shRb vector, Fig. 1B).
Figure 1. Lentiviral vector-mediated SCLC mouse model highly recapitulates human tumors.
(A) Schematic of transducing lentiviral vector. Mycl1 is controlled by a TRE promoter (top) and activated in Rosa26-M2rtTA mice upon doxycycline chow or directly by a CMV promoter (bottom). shPten is also driven by a CMV promoter in a miR-E backbone immediately after Mycl1, while other shRNAs are driven by U6 or H1 promoters. (B) Kaplan-Meier survival curves showing effects of lentiviral vector carrying different payloads. Median survival time of the groups was 475 days (GFP-shp53-shRb, n=9), 323 days (Mycl1-shp53-shRb, n=11), 188 days (Mycl1-shp53-shRb(mU6), n=6, shRb(mU6) has better Rb knockdown by mouse U6 promoter), 250 days (Myc-shp53-shRb, n=7), 201 days (Cherry-3shRNA, n=8), and 147 days (Mycl1-3shRNA, n=5), respectively. (C) Histopathological analysis of SCLC tumors induced by TRE-Mycl1-shp53-shRb vector, showing tumor initiating from bronchus (i), in low magnification view (ii), forming nested pattern (iii), in high magnification view (iv), forming massive necrosis (v), spreading along vasculature (vi), invading to big bronchus (vii). Tumors stained negative for SPC and positive for CGRP, Ki67, NCAM, Sox2. Scale bars, 100 μm. (D) View of whole lung, liver and kidney, showing primary lung tumor (white circle), metastases to mediastinal lymph node (black circle), liver (right, yellow circle) and kidney (right inset, green circle). Scale bars, 5 mm. (E) Histopathological analysis of metastases to liver, muscle, kidney and mediastinal lymph node. Scale bars, 100 μm.
Histopathology of tumors induced by these vectors had features of neuroendocrine tumors: small cell-size, minimal cytoplasm, salt and pepper chromatin, high mitotic index, massive necrosis, positive for NCAM and CGRP, and negative for SPC staining (Fig. 1C). These tumors were highly invasive within the lungs and metastasized to other organs (frequently to draining lymph nodes and liver, and rarely to muscle, kidney, Fig. 1D and 1E). Furthermore, cell lines derived from these SCLC tumors were easily transplantable in syngeneic hosts and grafts bore indistinguishable pathology as compared to the primary tumors (Supplementary Fig. S1D).
Inhibition of NF-κB pathway accelerates SCLC progression in mouse
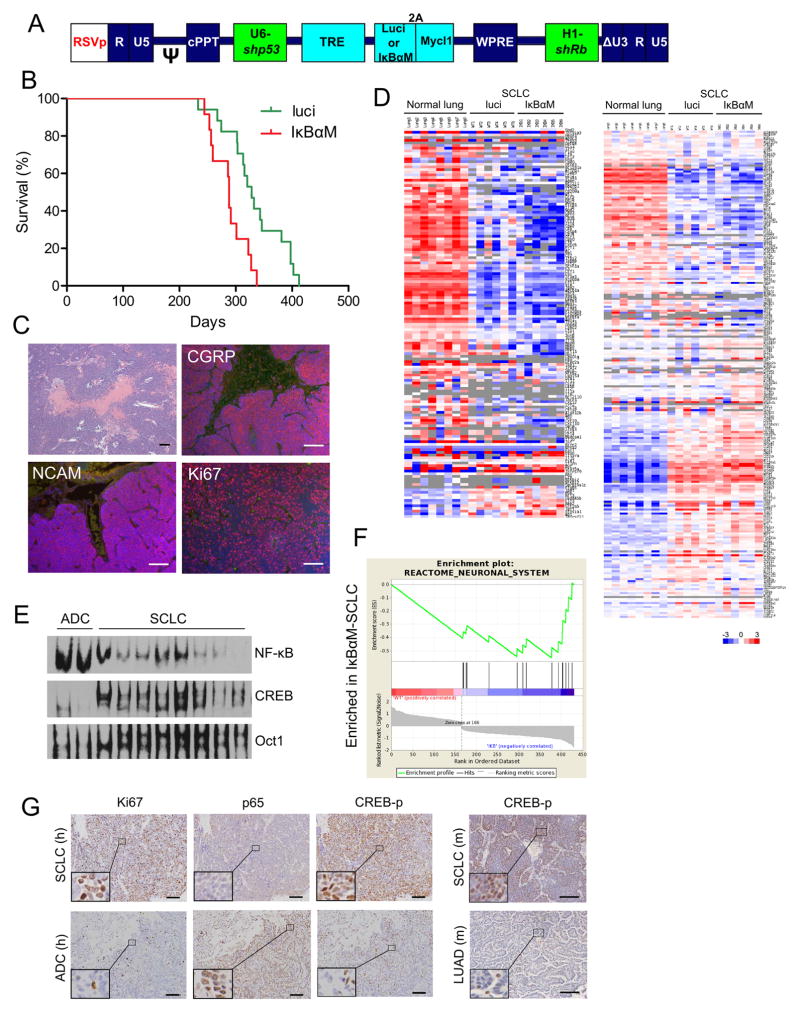
It has been shown that blocking the NF-κB pathway can limit tumor progression and prolong survival in many cancer types including lung adenocarcinoma harboring Kras mutation(9). We next asked whether this also happens in our SCLC model. We therefore generated a lentiviral vector containing the dominant negative form of IκBα (IκBαM) which cannot be phosphorylated or ubiquitinylated, and hence is resistant to proteosomal degradation thereby blocking NF-κB activation in cells (Fig. 2A). Surprisingly, inhibition of NF-κB moderately accelerated SCLC progression and resulted in a median survival time of 288 days as compared to 328 days in control group (Fig. 2B).
Figure 2. SCLC has low NF-κB and high CREB activity.
(A) Diagram of lentiviral vector design. Luciferase or IκBαM gene is co-expressed with Mycl1 by 2A ribosomal skipping mechanism. (B) Kaplan-Meier curve showing mice survival. Median survival time of the two groups was 328 days (luci, n=17), and 288 days (IκBαM, n=12) respectively, p=0.0029. (C) Histopathological analysis of IκBαM tumors showed no difference compared to control tumors in Fig. 1. (D) Heatmap of the expression values of 146 NF-κB target genes (left) and 245 CREB target genes (right) in normal lung, luci tumor and IκBαM tumor. (E) EMSA showing SCLC tumors had lower NF-κB and higher CREB binding activity than lung adenocarcinomas (ADC). (F) GSEA analysis of gene sets enriched in IκBαM tumor vs. control luci tumors. (G) Human SCLC and lung adenocarcinoma samples were stained for Ki67, p65 and phos-CREB (left). Total 10 SCLC and 10 adenocarcinoma samples were analysed and the results were summarized in Supplementary Table S1. Mouse SCLC and lung adenocarcinoma (KrasG12D-shp53) samples were stained for phos-CREB (right). Insets were in higher magnification showing nuclear staining. Scale bars, 100 μm.
Histopathological analysis of these IκBαM tumors did not reveal any differences in neuroendocrine markers (CGRP, NCAM) and mitotic index, compared to control tumors (luciferase imaging) (Fig. 2C). Further, control tumors did not reveal significant differences in terms of differentially expressed genes (SCLC vs. normal lung) or the 36 SCLC signature genes defined by the Berns group (Supplementary Fig. S2A–B) (17). Interestingly, SCLC samples had substantially lower NF-κB inducible genes expression compared to normal lung tissues, even when IκBαM was not present in the vector (luci SCLC tumors, Fig. 2D; also see human SCLC tumors in Supplementary Fig. S3A). To examine the actual NF-κB activity in SCLC tumor cells, we carried out electrophoretic mobility shift assay (EMSA) using consensus probes for NF-κB as well as other common transcription factors. Surprisingly, we found that NF-κB binding activity was much lower in SCLC cells compared to lung adenocarcinoma cells induced by Kras-shp53 vector; however, SCLC cells had higher CREB binding activity (Fig. 2E; also see Fig. 2D and Supplementary Fig. S3B). This observation was consistent with many other reports that NF-κB and CREB pathways counteract each other at multiple levels(18). Besides, GSEA analysis showed that compared to control tumors, SCLC-IκBαM tumors were enriched for neuronal genes in line with the neuroendocrine nature of the tumors (Fig. 2F). We further examined NF-κB and CREB pathway activities in human lung cancer samples by immunohistochemistry. As shown in Fig. 2G and Supplementary Table S1, SCLC tissues indeed had low p65 nuclear staining but high phospho-CREB staining, which further supported the gene expression data described above. Thus the data presented here suggests that increase in tumor progression from down-regulation of NF-κB ultimately leads to high CREB activity which is required for maintaining neuronal signature and for promoting cell proliferation in SCLC tumors.
We have previously shown that knock out of IKK2 in tumor cells blocks NF-κB activation in a Kras-induced lung adenocarcinoma model leading to significantly impaired cell proliferation(7). This dichotomy in the role of the NF-κB family in SCLC and NSCLC supports the notion that mutations commonly found in SCLC and lung adenocarcinoma rely on different signal pathways for cell proliferation. To determine the generality of this hypothesis, we transformed mouse embryonic fibroblasts (MEFs) with similar vectors that induced in our NSCLC and SCLC mouse models. Consistent with in vivo data, IκBαM potently blocked transformation (or cell proliferation) induced by Kras-shp53 but not Mycl1-shp53-shRb (Supplementary Fig. S4A). More interestingly, Kras-shp53vector induced enhanced NF-κB DNA binding activity while Mycl1-shp53-shRb vector induced higher CREB activity even in these MEF cells (Supplementary Fig. S4B).
Role of CREB in maintaining neuroendocrine signature and high proliferation in SCLC
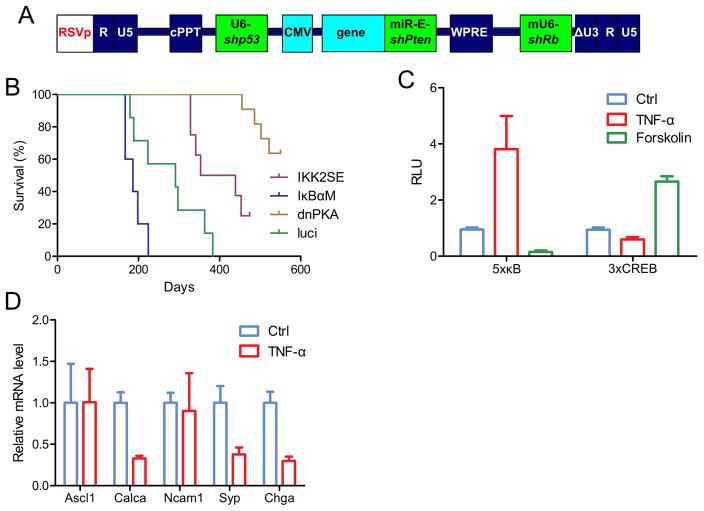
SCLC initiates from lung neuroendocrine cells (19) as tumors express neuronal markers (Fig. 1C). A number of cells of neuronal origin rely on CREB pathway for cell proliferation even though NF-κB and CREB pathways may counteract each other at multiple levels (20–22). To test the role of either signalling pathways in induction of SCLC, we used lentiviral vectors expressing either IκBαM (to inhibit NF-κB activity) or dnPKA (to inhibit CREB pathway) in addition to 3 shRNAs against tumor supressors (p53, Rb and Pten, Fig. 3A). Mice transduced with luciferase vector (as control) had median survival of 291 days. Mice expressing IκBαM had significantly accelerated tumor progression (median survival of 186 days), while expressing IKK2SE (constitutively active form of IKK2 leading to high basal NF-κB activity) delayed tumor progression (median survival of 396 days). Interestingly, expression of a dominant negative form of PKA (dnPKA) severely impaired tumor development and prolonged mouse survival (over 500 days, Fig. 3B).
Figure 3. SCLC relies on high CREB and low NF-κB to maintain neuroendocrine signature and high proliferation.
(A and B) Diagram of lentiviral vector designing. Kaplan-Meier curve showing mice survival. Median survival time of the four groups was 396 days (IKK2SE, n=8), 186 days (IκBαM, n=5), undefined (dnPKA, n=11), and 291 days (luci, n=7) respectively. p=0.0354 (IκBαM vs luci), p=0.0142 (IKK2SE vs luci), p<0.0001 (dnPKA vs luci). (C) primary SCLC cells derived from shp53-shRb-shPTEN model were infected with 5xκB reporter or 3xCREB reporter, and treated with TNF-α (10 ng/ml) or forskolin (5 μM) for 8 h, and luciferase activity was measured. (D) Same SCLC cells in C were treated with TNF-α (10 ng/ml) for 24 h, and the expression of five neuroendocrine signature genes were analysed with quantitative RT-PCR.
To demonstrate that NF-κB and CREB pathways in SCLC cells are antagonistic to each other, we infected them with 5xκB or 3xCREB luciferase reporter constructs. In these SCLC cells, TNF-α treatment activated the 5xκB reporter while inhibited the 3xCREB reporter. In contrast, forskoline treatment activated the 3xCREB reporter while inhibited the 5xκB reporter (Fig. 3C). Furthermore, TNF-α treatment remarkably down-regulated neuroendocrine signature genes, including Calca, Syp and Chga, in SCLC cells (Fig. 3D).
Targeting CREB pathway suppresses SCLC growth in vivo
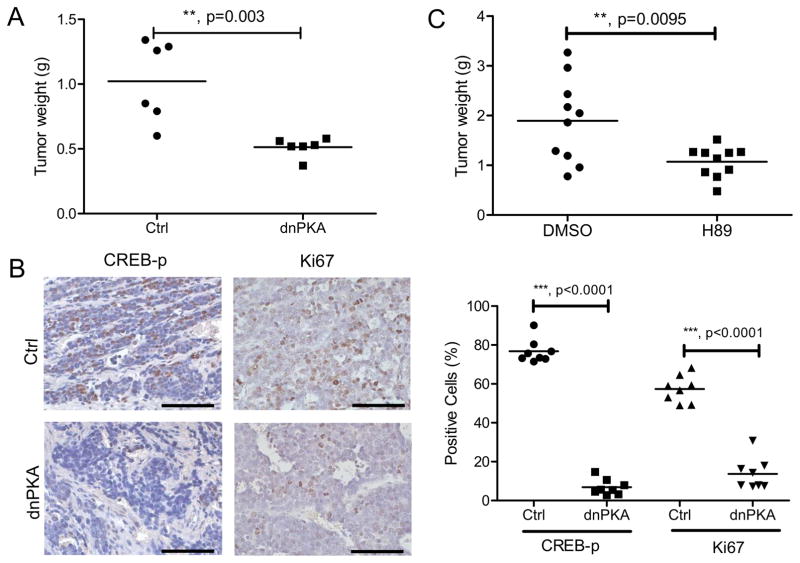
Based on these observations, we hypothesized that blocking the CREB pathway instead of the NF-κB pathway could benefit SCLC patients. Therefore, we tested this idea by examining the in vivo growth of a SCLC cell line derived from the 3-shRNA vector in a syngeneic transplant model. Upon induction of dnPKA, tumors were substantially reduced in size (Fig. 4A). Apparently, this tumor reduction was due to the impaired activity of CREB pathway (see reduced CREB phosphorylation as compared to control) and cell proliferation (Fig. 4B). As there are no specific PKA inhibitors available for in vivo study, we treated mice carrying SCLC grafts with the broad spectrum PKA inhibitor H89 for two weeks, and the compound reduced tumor size about 40% on average (Fig. 4C). All these results strongly suggest the possibility of targeting SCLC by inhibiting the CREB pathway.
Figure 4. Targeting CREB pathway suppresses SCLC growth in vivo.
(A) SCLC cells were infected with GFP (Ctrl) or dnPKA vector and transplanted into syngeneic hosts. Tumors were harvested after 4 weeks and weighed. (B) Immunohistochemistry staining of the tumors from (A). Scale bars, 100 μm. Quantification of positive staining cells were shown on the right. (C) SCLC cells were transplanted into syngeneic hosts. H89 treatment was started 2 weeks after the transplantation for another 2 weeks, and then tumors were harvested and weighed.
Discussion
Over quarter of a million people are diagnosed annually with small cell lung carcinoma (SCLC), one of the most deadly cancers with very poor prognosis. Targeted therapies have been rapidly developed in the last two decades against NSCLC with EGFR and ALK mutations; however, little progress has been made for SCLC. This, in part, is due to the fact that thoracic surgery is rarely performed in SCLC patients and as a result tumor sample accessibility is very limited. Previous genetic mouse models of SCLC by p53 and Rb knockout recapitulates human disease but the latency is extremely long. Although some improvement has been made by further knocking out p130 or Pten, time consuming, extensive mouse breeding is required (14–16). We leveraged our lentiviral delivery strategy to induce SCLC in wild-type mice with a single vector carrying all the necessary mutations (Fig. 1A). The vector can be further modified to include new combination of mutations for studying their roles in tumor initiation and development, or luciferase/GFP reporter genes for in vivo tracking tumor response upon treatment in preclinical study settings. Thus, this model system offers the flexibility for both basic and preclinical studies.
The role of NF-κB in cancer initiation, development and metastasis has been extensively studied, and in general promotes tumoriogenesis. Interestingly, NF-κB has been shown to also act as a tumor-suppressor in both a diethylnitrosamine (DEN)-induced hepatocellular carcinoma (HCC) model and a 7,12-Dimethylbenz[α]anthracene/12-O-tetradecanoylphorbol-13-acetate (DMBA/TPA)-induced skin squamous cell carcinoma (SCC) model(23, 24). These two seemingly dissimilar tumor models share one common feature: NF-κB and JNK are two major signalling pathways downstream of TNFR1 which usually counter-regulate each other. Upon chemical treatment and NF-κB inhibition in hepatocytes or keratinocytes, JNK signalling is unleashed, which leads to excessive oxidative stress, DNA damage and eventually induces tumorigenesis. In the scenario of SCLC, Mycl1 gene is usually amplified leading to overexpression which may lead to activation of PKA pathway by up-regulation of Prkacb expression(25). Indeed, we observed high Prkacb mRNA in mouse SCLC but not adenocarcinoma tumor samples (data not shown). Intriguingly, PKA and CREB activity has also been reported in NSCLC, which may contribute to cancer metastasis and drug resistance, however the mechanism of PKA activation in NSCLC could be very different from SCLC(26, 27).
Our current work indicates that, in contrast to many other types of cancer, SCLC does not benefit from high NF-κB activity with regard to tumor cell proliferation. Thus NF-κB inhibition may not directly benefit patients with SCLC in general, although we cannot exclude the possibility that NF-κB inhibitor could work well in certain scenarios, for example, to overcome chemo-resistance, which usually is linked to NF-κB activation. On the other hand, our data suggests that manipulating the PKA-CREB pathway should be further pursued for the sake of developing new therapeutic strategies against this deadly disease. The big hurdle for this therapy, however, is that most of the current available PKA inhibitors are ATP competitive antagonists and not very spe-cific. Furthermore, the PKA-CREB pathway is responsible for many other important biophysio-logical activities, especially for neuronal system. Interestingly, Jahchan et al. have recently reported that tricyclic antidepressants could effectively suppress SCLC tumor growth both in vitro and in animals through blocking autocrine survival signals involving neurotransmitters and their G protein-coupled receptors (GPCR). These GPCRs usually activate PKA pathway via Gαs, adenyl cyclase and cAMP and indeed they observed that tricyclic antidepressants could effectively reduce PKA-CREB activity in SCLC cells and thus attenuate tumor growth in animals(28). Altogether, our results support the notion that targeting the PKA-CREB pathway, is a novel strategy in fighting against SCLC—the most malignant form of human lung cancer.
Supplementary Material
Significance.
Activity of the transcription factor CREB is elevated in SCLC tumors, which helps to maintain its neuroendocrine signature and cell proliferation. Our results highlight the importance of targeting the CREB pathway to develop new therapeutics to combat SCLC.
Acknowledgments
We appreciate the generosity of Dr. S. Seshagiri for sharing human SCLC RNAseq data (accession: EGAS00001000334). We also thank M. Schmitt, G. Estepa, and B. Coyne for technical and administrative support.
Financial Support
I.M. Verma is an American Cancer Society Professor of Molecular Biology, and holds the Irwin and Joan Jacobs Chair in Exemplary Life Science. This work was supported in part by grants from the NIH (5R01CA195613-20 to I.M. Verma), the Cancer Center Core Grant (P30 CA014195-38), the H.N. and Frances C. Berger Foundation, and the Leona M. and Harry B. Helmsley Charitable Trust grant #2017-PG-MED001. This work was also supported by The National Natural Science Foundation of China (No. 81672268 to Y. Xia).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have nothing to declare.
References
- 1.Stewart BW, Wild C International Agency for Research on Cancer, World Health Organization. World cancer report. 2014. [Google Scholar]
- 2.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 3.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–96. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 5.William WN, Jr, Glisson BS. Novel strategies for the treatment of small-cell lung carcinoma. Nat Rev Clin Oncol. 2011;8:611–9. doi: 10.1038/nrclinonc.2011.90. [DOI] [PubMed] [Google Scholar]
- 6.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–9. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 7.Xia Y, Yeddula N, Leblanc M, Ke E, Zhang Y, Oldfield E, et al. Reduced cell proliferation by IKK2 depletion in a mouse lung-cancer model. Nat Cell Biol. 2012;14:257–65. doi: 10.1038/ncb2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, et al. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15:110–6. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia Y, Shen S, Verma IM. NF-kappaB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–30. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–7. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basseres DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;70:3537–46. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–5. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 13.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–6. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010;70:3877–83. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res. 2014;12:654–9. doi: 10.1158/1541-7786.MCR-13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19:244–56. doi: 10.1016/j.ccr.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 18.Gerlo S, Kooijman R, Beck IM, Kolmus K, Spooren A, Haegeman G. Cyclic AMP: a selective modulator of NF-kappaB action. Cell Mol Life Sci. 2011;68:3823–41. doi: 10.1007/s00018-011-0757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–64. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto K, Karelina K, Obrietan K. CREB: a multifaceted regulator of neuronal plasticity and protection. J Neurochem. 2011;116:1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ollivier V, Parry GC, Cobb RR, de Prost D, Mackman N. Elevated cyclic AMP inhibits NF-kappaB-mediated transcription in human monocytic cells and endothelial cells. J Biol Chem. 1996;271:20828–35. doi: 10.1074/jbc.271.34.20828. [DOI] [PubMed] [Google Scholar]
- 22.Dworkin S, Heath JK, deJong-Curtain TA, Hogan BM, Lieschke GJ, Malaterre J, et al. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Dev Biol. 2007;307:127–41. doi: 10.1016/j.ydbio.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–90. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–43. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 25.Wu KJ, Mattioli M, Morse HC, 3rd, Dalla-Favera R. c-MYC activates protein kinase A (PKA) by direct transcriptional activation of the PKA catalytic subunit beta (PKA-Cbeta) gene. Oncogene. 2002;21:7872–82. doi: 10.1038/sj.onc.1205986. [DOI] [PubMed] [Google Scholar]
- 26.Shaikh D, Zhou Q, Chen T, Ibe JC, Raj JU, Zhou G. cAMP-dependent protein kinase is essential for hypoxia-mediated epithelial-mesenchymal transition, migration, and invasion in lung cancer cells. Cell Signal. 2012;24:2396–406. doi: 10.1016/j.cellsig.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Troiani T, Vecchione L, Martinelli E, Capasso A, Costantino S, Ciuffreda LP, et al. Intrinsic resistance to selumetinib, a selective inhibitor of MEK1/2, by cAMP-dependent protein kinase A activation in human lung and colorectal cancer cells. Br J Cancer. 2012;106:1648–59. doi: 10.1038/bjc.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahchan NS, Dudley JT, Mazur PK, Flores N, Yang D, Palmerton A, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364–77. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.