SUMMARY
Beige and brown adipocytes generate heat in response to reductions in ambient temperature. When warmed, both beige and brown adipocytes exhibit morphological ‘whitening’, but it is unknown whether or to what extent this represents a true shift in cellular identity. Using cell type-specific profiling in vivo, we uncover a unique paradigm of temperature-dependent epigenomic plasticity of beige, but not brown, adipocytes, with conversion from a brown to a white chromatin state. Despite this profound shift in cellular identity, warm whitened beige adipocytes retain an epigenomic memory of prior cold exposure defined by an array of poised enhancers that prime thermogenic genes for rapid response during a second bout of cold exposure. We further show that a transcriptional cascade involving glucocorticoid receptor and Zfp423 can drive warm-induced whitening of beige adipocytes. These studies identify the epigenomic and transcriptional bases of an extraordinary example of cellular plasticity in response to environmental signals.
eTOC Blurb
Both beige and brown adipocytes “whiten” upon warming. XXX et al elegantly show that beige, but not brown, adipocytes undergo temperature-dependent reprogramming between brown- and white-like states, while retaining epigenomic memory of prior cold exposure. A transcriptional cascade underlies this plasticity response to environmental signals.
INTRODUCTION
All somatic cell types share the same genome, and yet each has a distinct identity manifested by a unique pattern of gene expression. These transcriptional profiles are themselves encoded by a highly cell type-specific pattern of genomic organization, often called chromatin state, comprised of a series of epigenomic modifications that determine which regions of the genome will be accessible for transcription. The key elements of this epigenomic profile are established during differentiation, and form a highly stable pattern that is unique to that cell type. An important corollary is that a given cell type can be identified based purely on knowledge of its chromatin state.
The chromatin state that gives a cell its unique identity is considered to be fairly immutable under normal conditions, although experimental genetic or epigenetic manipulations can induce mammalian cell dedifferentiation (e.g. fibroblasts to stem cells) (Takahashi and Yamanaka, 2006) and transdifferentiation (e.g. fibroblasts to neurons) (Vierbuchen et al., 2010), revealing the potential for epigenomic reprogramming of cell identity in differentiated mammalian cells. In contrast to the overall stability of their chromatin state, differentiated cells maintain a limited degree of epigenomic flexibility that allows them to fine tune their transcriptional output in response to environmental challenges. Such perturbations, including nutritional, chemical, hormonal, inflammatory, and thermal exposures, can have significant effects on gene expression, but they do not change the identity of cells (Feil and Fraga, 2012). Thus, for example, a fasted liver cell may increase gluconeogenic and reduce lipogenic gene expression, but it remains a hepatocyte.
Adipocytes provide an interesting test case for this general paradigm. Three distinct types of adipocyte are now recognized: white, brown, and beige. White adipocytes store energy in the form of triglyceride, while brown adipocytes exist in a discrete interscapular depot (in rodents) and primarily act to promote thermogenesis via the action of the mitochondrial uncoupling protein 1 (UCP1). Beige adipocytes are also UCP1-expressing thermogenic cells, but they arise within white fat pads in response to cold exposure; beige adipocytes can also support UCP1-independent thermogenesis (Harms and Seale, 2013; Kazak et al., 2015). Despite their phenotypic similarities, brown and beige cells arise from distinct lineages (Sanchez-Gurmaches et al., 2016; Wang and Seale, 2016). Thus, the three types of adipocytes are considered to be different cell types, with overlapping but distinct functions and gene expression profiles. Various studies have suggested that beige adipocytes exhibit significant plasticity following thermal stress. For example, initial cold exposure causes de novo differentiation of beige adipocytes from progenitor cells with the characteristics of vascular smooth muscle cells (Berry et al., 2016; Long et al., 2014; Wang et al., 2013). In response to warming, beige adipocytes undergo a ‘whitening’ process, adopting the unilocular morphology of white adipocytes. It has been proposed that such ‘whitened’ beige adipocytes can then return to the thermogenic state upon subsequent cold exposure (Rosenwald et al., 2013). Interestingly, interscapular brown adipocytes exhibit a similar morphological response to warming, developing a unilocular morphology and down-regulating thermogenic gene expression (Cui et al., 2016). It is, however, unclear for both brown and beige cells whether these morphological and transcriptional changes represent alterations in cellular state or cellular identity.
Recent studies have characterized epigenomic profiles of brown, beige and white adipocytes using in vitro culture models, particularly in the context of adipogenesis. Among other findings, epigenomic mechanisms have been elucidated that participate in brown adipocyte development in monolayer, such as a pathway involving the repressive histone mark H3K27me3 and the corresponding histone demethylase Jmjd3 (Inagaki et al., 2016; Pan et al., 2015). While such in vitro adipocyte models are useful in providing cell autonomous profiles, they often do not reflect in vivo chromatin states (Roh et al., 2017), and it is difficult to model physiologic perturbations such as response to temperature change. In vivo studies have been hampered by the fact that beige adipocytes are admixed with white fat cells and non-adipocytes within the fat depot, and isolating pure populations has not previously been possible.
In order to circumvent these issues, and to enable genomic and epigenomic studies in living animals generally, we have established a method for simultaneous isolation of transcripts and nuclei from specific cell types within complex tissues, involving a transgenic NuTRAP (Nuclear tagging and Translating Ribosome Affinity Purification) mouse which adopts the strategies of ribosome and nucleus tagging activated by cell type-specific Cre lines (Deal and Henikoff, 2010; Heiman et al., 2008; Roh et al., 2017). Here, we express the NuTRAP reporter in beige and brown adipocytes (Ucp1-Cre), or all adipocyte types (Adiponectin-Cre), to define transcriptomic and epigenomic profiles of beige, brown, and white adipocytes in vivo. This allows us to identify reliable markers for each cell type, and show that beige adipocytes undergo dramatic temperature-dependent chromatin remodeling that alters their cellular identity from brown to white. Brown adipocytes, in contrast, do not change cellular identity following warming to thermoneutrality, despite extensive morphological whitening. Further, we show that beige adipocytes retain an epigenomic memory of prior cold exposure that promotes rapid induction of thermogenic gene expression upon a repeat cold challenge. Finally, we identify a transcriptional cascade involving glucocorticoid receptor (GR) and Zfp423 as a key driver of temperature-dependent whitening of beige adipocytes.
RESULTS
Warming of brown and beige adipocytes causes morphological whitening
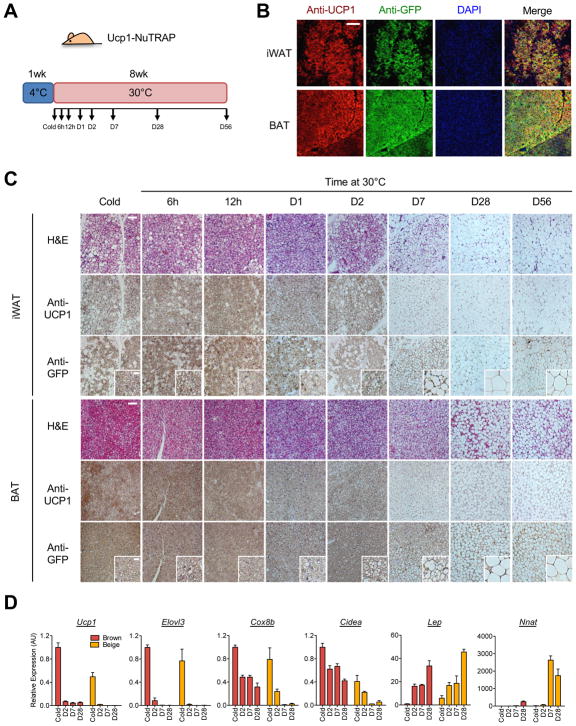
NuTRAP mice express GFP-tagged ribosomes and mCherry-tagged nuclei in a Cre recombinase-dependent manner (Roh et al., 2017). To investigate chromatin state dynamics in response to thermal stress, we crossed NuTRAP with Ucp1-Cre (to generate Ucp1-NuTRAP), which express the reporters in brown and beige adipocytes, allowing permanent labeling and subsequent tracing of the cells over the life of the cell (Kong et al., 2014; Roh et al., 2017). Ucp1-NuTRAP mice were housed at 4°C for 1 week to induce beige adipocyte formation and subsequently moved to thermoneutrality (30°C) for 8 weeks (Figure 1A). After cold exposure, the iWAT of Ucp1-NuTRAP mice exhibited a subset of GFP and mCherry-labelled (Figure S1A) multilocular UCP1-positive (Figure 1B) beige adipocytes within the PLIN1-positive total adipocyte population (Figure S1B). In BAT, nearly all PLIN1-positive adipocytes were labeled with GFP and mCherry (Figures S1A and S1B) and were positive for UCP1 (Figure 1B) in Ucp1-NuTRAP mice, indicating that Ucp1-NuTRAP mice faithfully label brown and beige adipocytes in cold conditions. Tracing the cells during the course of warming showed that both BAT and iWAT display an increasingly white-like morphology, with the appearance of unilocular adipocytes and reduced UCP1 expression. This process seemed to stabilize between 4 and 8 weeks after exposure to thermoneutral conditions (Figure 1C). Importantly, the new unilocular adipocytes seen in warming represent morphological conversion of preexisting mature beige or brown adipocytes, as these cells were GFP+ (Figure 1C). Quantitative analysis of the fraction of mCherry-labeled nuclei of beige adipocytes in iWAT showed no difference between cold and 4 weeks of warming (Figure S1C), confirming that beige adipocytes persist in warm iWAT, but take on a white adipocyte-like state, as visualized by fluorescence microscopy (Figure S1D).
Figure 1. Brown and beige adipocytes undergo morphologically whitening during warming.
(a) Ucp1-NuTRAP mice were exposed to 4°C for 1 week, subsequently incubated at 30°C for 8wk and studied at the indicated time points during warming.
(b) Whole mount immunofluorescence staining of iWAT and BAT of Ucp1-NuTRAP mice exposed to 4°C for 1wk. UCP1-positive brown and beige adipocytes are labeled by GFP. Scale bar: 100μm.
(c) Immunohistochemistry of BAT and iWAT from Ucp1-NuTRAP mice at the indicated time points during warming. H&E, anti-UCP1 and anti-GFP stained images are shown. Insets show higher magnification of GFP-labeled beige/brown adipocyte morphology. Scale bar: 50μm and 10μm (insets).
(d) Gene expression analysis by qRT-PCR with TRAP-isolated RNA from BAT and iWAT of Ucp1-NuTRAP mice at the indicated time points during warming. Bars indicate mean ± SEM (n=3 animals/group).
See also Figure S1.
At the molecular level, thermogenic marker gene expression profiles were assessed for both BAT and iWAT. Some cold-induced genes, such as Ucp1 and Elovl3, were rapidly and completely down-regulated during warming in both brown and beige adipocytes. In contrast, Cox8b and Cidea were more slowly down-regulated during warming, and they remained expressed in brown adipocytes, but not in beige adipocytes (Figure 1D). There were also discordances between cell types in genes considered to be white-selective, with Lep expression induced in both brown and beige adipocytes by warming, whereas Nnat was much more strongly induced in beige than brown adipocytes (Figure 1D). Taken together, these data suggest that brown and beige adipocytes undergo a morphologically similar whitening process in response to warming, with overlapping but distinct responses at the molecular level.
Identification of novel markers for white, brown, and beige adipocytes using cell type-specific transcriptomes
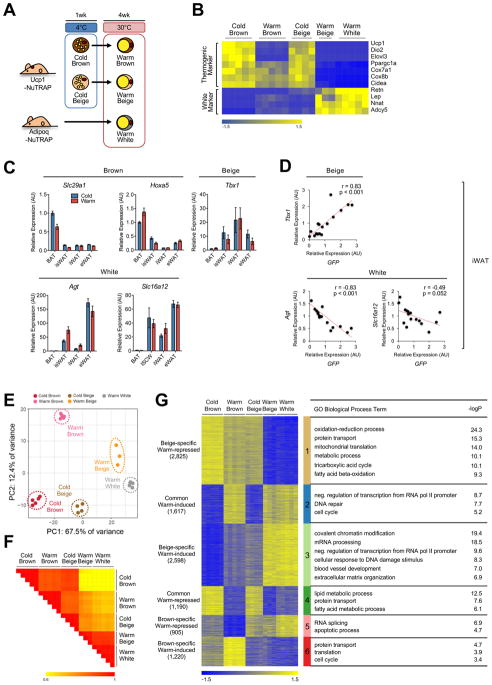
We defined transcriptional profiles of brown and beige adipocytes in both cold and warm conditions using TRAP followed by RNA-seq. BAT and iWAT were harvested from Ucp1-NuTRAP mice exposed to cold (4°C) for 1 week and then subsequently maintained at 30°C for 4 weeks. As a pure white adipocyte control, we used iWAT from Adiponectin-Cre;NuTRAP (Adipoq-NuTRAP) that were housed from birth at 30°C; while Adipoq-NuTRAP mice label all types of adipocytes, iWAT in this setting does not contain beige adipocytes, as they are not recruited at thermoneutrality (Figure 2A). As shown in Figure 2B, conventional brown/beige thermogenic adipocyte makers such as Ucp1, Dio2, Elovl3, Pgc1a, Cox7a1, Cox8b, and Cidea were highly expressed in cold brown and beige adipocytes and down-regulated in response to warming. White adipocyte markers such as Retn, Lep, Nnat, and Adcy5 exhibited the opposite pattern (Figure 2B).
Figure 2. Beige and brown adipocytes exhibit distinct transcriptomic changes upon warming.
(a) Experimental scheme showing how different types of adipocytes are collected. Brown and beige adipocytes (cold/warm) were collected from BAT and iWAT of Ucp1-NuTRAP mice, respectively, exposed to cold and after 4 weeks of warming at 30°C. White adipocytes were collected from iWAT of Adipo-NuTRAP mice housed at thermoneutrality from birth.
(b) Heatmap of marker gene expression for different adipocyte types from cold and warm conditions as described in (a). Columns represent biological replicates. Expression values (Counts Per Million; CPM) of each mRNA are represented by z-scores.
(c) Gene expression analysis by qRT-PCR of proposed brown, beige and white adipocyte markers in different fat depots (whole tissues) in response to cold (4°C, 1 week) and subsequent warming (30°C, 4 weeks). Bars indicate mean ± SEM (n=6 animals/group). BAT, interscapular brown adipose tissue; isWAT, interscapular white adipose tissue; iWAT, inguinal white adipose tissue; eWAT, epididymal white adipose tissue.
(d) Correlation analysis between expression of beige/white adipocyte markers and GFP within iWAT. Each dot indicates an individual sample.
(e) PCA of transcriptome of adipocyte types in cold and warm conditions. Each dot indicates an individual biological replicate (n=3–5 animals/replicate).
(f) Similarity matrix showing pairwise Pearson correlations for RNA-seq profiles. Pearson correlation coefficient r is represented in color as indicated.
(g) Heatmap of K-mean (K=6) clustering of transcriptomic profiles for differential genes. The number of genes in each cluster is in parentheses; each cluster is indicated by a different vertical color bar. Pathways (GO biological process) enriched in each cluster and their corresponding −log10 p-values are shown in the table at right.
Numerous marker genes have been proposed to differentiate among brown, beige, and white adipocytes, using either whole tissues or cells propagated in vitro (de Jong et al., 2015; Peirce et al., 2014). Our TRAP RNA-seq data has advantages over these prior approaches, because it allows for cell type-specific gene expression profiling in the in vivo setting, without confounding by culture artifacts or by non-adipocytes, which make up at least 50% of the cells in a fat pad (Roh et al., 2017). We first tested whether several commonly used brown and beige-specific markers show the expected cell type specificity in our RNA-seq data. Markers that have been proposed to be beige-specific, such as Tbx1, Shox2, Hoxc9 and Tmem26, were preferentially expressed in beige vs. brown adipocytes in cold conditions, while other proposed beige markers, such as Tnfrsf9 (CD137) were not specific. Under warm conditions, however, Tbx1 was the only beige marker that was not also expressed in inguinal white adipocytes (Figures S2A–C). This suggests that most of these genes are not, in fact, beige-specific markers, but rather reflect the anatomical location of the inguinal depot. White markers (Lep, Retn, Hoxc8, Adcy5, and Nnat) were similarly expressed in warm beige and white adipocytes (Figures S2B and S2C). Similarly, of several proposed brown adipocyte marker genes (Lhx8, Mpzl2, Fbxo31, and Ebf3), only Zic1 showed specificity for brown vs. beige adipocytes (Figure S2A). Zic1, however, was also expressed in interscapular white adipose tissue (isWAT) (Figure S2E), indicating that Zic1 is not a true brown adipocyte marker, but instead reflects anatomical position in the interscapular region, consistent with a previous report (de Jong et al., 2015).
These findings motivated us to identify new markers for adipocyte types using our RNA-seq data. We first screened for genes enriched ≥4-fold in one cell type vs. the other two types, and identified a set of 57 genes that could be clustered as either brown, beige or white (Figure S2D). Next, we tested the expression of several of these genes across multiple fat depots from different anatomical locations. Many brown adipocyte genes failed the latter test, including Ankrd33b and Foxp2, which were also identified in isWAT and epididymal white adipose tissue (eWAT), respectively (Figure S2E). Some genes, however, including Slc29a1 and Hoxa5, proved to be reasonably good markers of brown adipocytes (Figure 2C). We next focused on beige-selective genes; some, like Eln and Vwf, were poorly enriched in iWAT compared to BAT, and were also highly expressed in cold eWAT (Figure S2F). Tbx1, however, was highly expressed in the iWAT of Ucp1-NuTRAP mice in both cold and re-warmed conditions (Figure 2C). To support the notion that Tbx1 was being expressed specifically in beige adipocytes of the iWAT, we took advantage of the fact that beige adipocytes are unevenly distributed across the inguinal depot. Samples of iWAT from cold and then warm-exposed Ucp1-NuTRAP mice were harvested and then cut into small pieces representing different parts of the fat pad (Figure S2H); the high degree of correlation between expression of GFP and Tbx1 (R = 0.83) strongly suggests that Tbx1 expression is specific for beige adipocytes (Figure 2D). Finally, we focused on potential white adipocyte-selective markers. We found Agt and Slc16a12 were preferentially expressed in all tested white depots, de-enriched in brown and negatively correlated with beige adipocyte content (Figures 2C, 2D, S2F and S2G).
Beige and brown adipocytes display distinct transcriptional profile changes after warming
To compare temperature-dependent transcriptional changes of adipocyte cell types globally, we performed principal component analysis (PCA) of the RNA-seq data obtained from cold-exposed (1 week) and subsequently re-warmed (4 weeks) Ucp1-NuTRAP mice and from Adipoq-NuTRAP mice that had been maintained at thermoneutrality for their entire lives (Figure 2E). Of note, the transcriptomes of cold exposed beige and brown adipocytes are remarkably similar. Upon re-warming, beige and brown adipocytes diverge transcriptionally, with the former taking on a profile similar to that of warm white adipocytes. Re-warmed brown adipocytes, on the other hand, take on a profile that remains similar to cold brown adipocytes along PC1, which explains 67.5% of the variance (Figure 2E). Consistent with these results, global correlation analysis of adipocyte transcriptomes revealed that cold brown adipocytes, warm brown adipocytes, and cold beige adipocytes are very tightly correlated (Figure 2F). Warm beige adipocytes, on the other hand, are very strongly correlated with warm white adipocytes (Figure 2F). Taken together, these results indicate that beige adipocytes undergo a profound shift in their transcriptome from a brown to white pattern, while brown adipocytes show only modest changes in response to warming.
We next sought to understand which biological pathways might be affected by warming in brown and beige adipocytes. Using K-means clustering of RNA-seq profiles, we defined 6 patterns of gene expression; the largest two comprised genes that were induced (n= 2,598) or repressed (n= 2,825) by warming in a beige-specific manner. Additional clusters included brown-specific warm induced (n= 1,220), brown-specific warm repressed (n= 905), and commonly induced (1,617) and repressed (n= 1,190) genes (Figure 2G, left). Gene ontology (GO) analysis of warm-repressed clusters (Clusters 1, 4 and 5) revealed that lipid metabolism pathways were down-regulated by warming in both beige and brown adipocytes. Mitochondrial function-related pathways, including oxidation-reduction, tricarboxylic acid cycle and fatty acid oxidation, were specifically repressed in beige adipocytes, while the apoptosis-related pathway was specifically repressed in brown adipocytes (Figure 2G, right). Analysis of warm-induced clusters (Clusters 2, 3 and 6) also suggested distinct responses of beige and brown adipocytes upon warming. Whereas the DNA repair pathway was commonly induced in beige and brown adipocytes by warming, genes related to protein transport and translation were induced specifically in brown adipocytes. In contrast, beige adipocyte-specific induced pathways included genes of covalent histone modification. The complete transcriptomic data set across all cell types and temperatures is presented in Table S1.
Warming provokes a massive shift in the chromatin state of beige adipocytes from brown to white
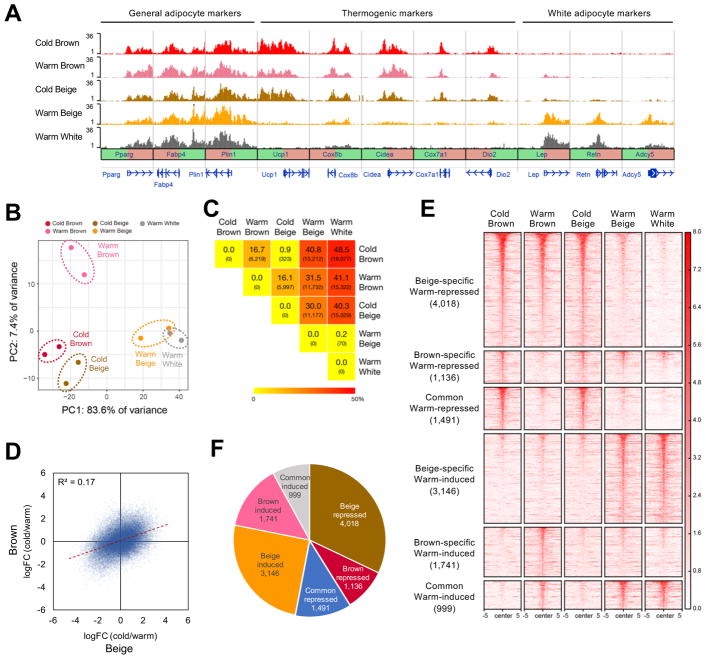
The temperature-dependent conversion of the beige adipocyte transcriptome from a brown to a white pattern, combined with the identification of chromatin modifying genes as a major up-regulated pathway in warmed beige adipocytes, suggested that warming may provoke a shift in cellular identity. In order to assess this directly, we exploited the cell type-specific nuclear labeling of NuTRAP mice to isolate nuclei from beige and brown adipocytes in cold and warm conditions (and white adipocytes in warm) using flow cytometry. H3K27ac ChIP-seq was then performed to define active promoter and enhancer regions. As expected, all adipocyte types had strong H3K27ac peak signals at general adipocyte genes, such as Pparg, Fabp4, and Plin1 (Figure 3A). Cold and warm brown adipocytes displayed strong H3K27ac signals at thermogenic genes, but no activity at white adipocyte marker loci. Also, as expected, warm white adipocytes have very little H3K27ac enrichment at thermogenic loci but extensive enrichment at white marker genes. Beige adipocytes, in contrast, show a pattern of H3K27ac enrichment that is remarkably similar to brown adipocytes in the cold, but become virtually indistinguishable from white adipocytes once warmed (Figure 3A).
Figure 3. Temperature-sensitive plasticity of beige adipocyte chromatin.
(a) H3K27ac ChIP-seq data centered on the TSS (±5kb) of general adipocyte, thermogenic, and white adipocyte marker genes.
(b) PCA of H3K27ac peaks from different adipocyte types in cold and warm conditions. Each dot indicates an individual biological replicate (a pool of 3–6 animals) analyzed in two independent experiments.
(c) Heatmap of pairwise comparisons for the number of differentially-regulated H3K27ac peaks. Values indicate the percentage and number (in parentheses) of differentially-regulated peaks in each comparison. Total number of H3K27ac peaks is 37,254.
(d) Scatter plot showing correlation of the changes of all individual H3K27ac peak signals upon warming between brown and beige adipocytes. The red dashed line is the trend line.
(e) Heatmap showing H3K27ac peaks (rows) across samples (columns). Peaks are classified into 6 clusters based on their patterns as labeled. Amplitude of each peak center (±5kb) is represented in color as indicated.
(f) Pie chart showing the number of H3K27ac peaks in each cluster. The number of H3K27ac peaks in each cluster is indicated.
See also Figures S3 and S4.
This finding was generalizable, as PCA of the H3K27ac profiles indicated that the chromatin states of cold beige and warm beige adipocytes resemble cold brown and warm white adipocytes, respectively, while warm brown adipocytes have a small number of unique H3K27ac peaks (Figure 3B). We also compared H3K27ac peaks between adipocytes in cold and warm conditions and found that cold beige adipocytes were highly similar to cold brown adipocytes, differing only at 323 out of 37,254 peaks (0.7% of total peaks). Incredibly, warm beige adipocytes are even more similar to white adipocytes, with only 70/37,254 peaks (0.2% of total) showing significant differences between these two cell types. Cold and warm beige adipocytes were very dissimilar, with 30% of H3K27ac peaks showing differential enrichment (Figure 3C). We observe a relatively weak correlation in the temperature-induced changes in H3K27ac enrichment between brown and beige adipocytes (R2=0.17) (Figure 3D). These results collectively suggest that the chromatin state of brown adipocytes is relatively static in the face of thermal stress, but beige adipocytes are highly plastic, with a near complete conversion of enhancer activity from brown to white during rewarming.
As with the transcriptional profiles, we classified differential H3K27ac peaks into 6 clusters (Figures 3E and 3F). Beige-specific warm-repressed and warm-induced peaks represented the largest fractions of differential peaks (Figures 3E and 3F). Analysis of the genomic distribution of H3K27ac peaks showed differential H3K27c peaks were generally located in promoter (~3–9%), intergenic (~35–45%), and intronic regions (~43–55%) (Figure S3). Intriguingly, we noted a somewhat distinct composition of the beige-specific warm-induced peak cluster; its promoter (8.8%), intronic (47.7%), and exonic (3.3%) fractions were the greatest among all the peak clusters, and its intergenic fraction (35.2%) was concomitantly smaller. The former regions are disproportionately likely to affect gene expression (Calo and Wysocka, 2013).
Brown adipocyte identity is maintained independently of sympathetic nerve activity
We were intrigued by the observation that warming causes a dramatic remodeling of chromatin state in beige, but not brown, adipocytes, despite similar morphological whitening in both cell types. This indicates that brown adipocytes maintain their identity in a temperature-independent manner, and suggests that there may be a mechanism that prevents brown adipocytes from undergoing the white conversion seen in their beige counterparts. Such a mechanism could be cell autonomous, or it could rely upon extrinsic factors within the brown adipose depot. One obvious candidate for the latter would be innervation, which is much denser in the interscapular region than in the inguinal depot (Vaughan et al., 2014). To test this hypothesis, we performed unilateral BAT denervation surgery and assessed chromatin state at different temperatures (Figure S4A). In mice housed at room temperature (RT) before and after surgery, the denervated pad was morphologically whitened (Figure S4B, top), and showed reduced expression of thermogenic genes and enhanced expression of white marker genes (Figure S4C, top), consistent with the idea that basal sympathetic tone prevents whitening at RT. In mice maintained at thermoneutrality for 4 weeks after surgery, the denervated BAT pad appeared morphologically undistinguishable from the sham-operated pad (Figure S4B, middle), although thermogenic gene expression was slightly lower on the denervated side (Figure S4C, middle). Mice housed at 30°C for both before and after surgery did not show any differences in BAT morphology or gene expression between the sham and denervated pads (Figures S4B and S4C, bottom), suggesting that sympathetic nerve signals were completely inactivated and brown adipocytes were maximally whitened by prolonged incubation in warm conditions.
We next assessed whether denervated brown adipocytes are “released” from their cellular identity under warm conditions, by looking at H3K27ac signals at thermogenic and white adipocyte marker loci. H3K27ac peak signals were indistinguishable between sham-operated and denervated brown adipocytes, and both were clearly distinct from warm white adipocytes (Figure S4D), despite their whitened morphology (Figure S4B). Hierarchical clustering analysis based on H3K27ac ChIP-seq profiles revealed that sham and denervated brown adipocytes form a cluster together with warm brown adipocytes which is distinct from cold brown and cold beige adipocytes. Most importantly, these clusters were formed completely away from warm white and warm beige adipocytes (Figure S4E). Taken together, these results indicate that the cellular identity of brown adipocytes is maintained independently of sympathetic nerve activity.
Warmed beige adipocytes retain an epigenomic memory of their thermogenic past
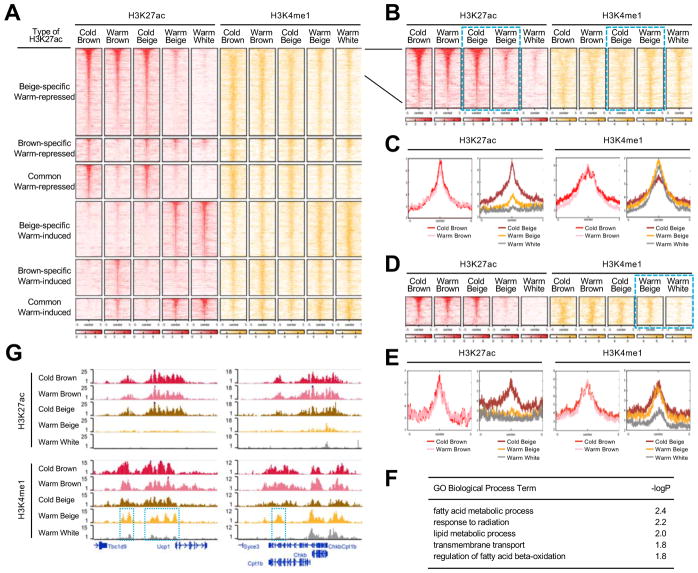
In addition to H3K27ac, which marks active promoters and enhancers, we also performed H3K4me1 ChIP-seq in brown, beige, and white adipocytes. H3K4me1 marks both transcriptionally active and inactive enhancers and promoters, and regulatory elements that are marked by H3K4me1 in the absence of H3K27ac are considered to be inactive, but “poised” (Creyghton et al., 2010). Such enhancers have been shown to serve a priming role during development and in response to environmental cues (Calo and Wysocka, 2013; Ghisletti et al., 2010). We found that 24,414 H3K27ac peaks (65.5% of total peaks) overlapped with 19,904 H3K4me1 peaks (35.2% of total peaks) (Figure S5A); changes in H3K27ac and H3K4me1 peak signals during warming were positively correlated in both beige and brown adipocytes (Figure S5B).
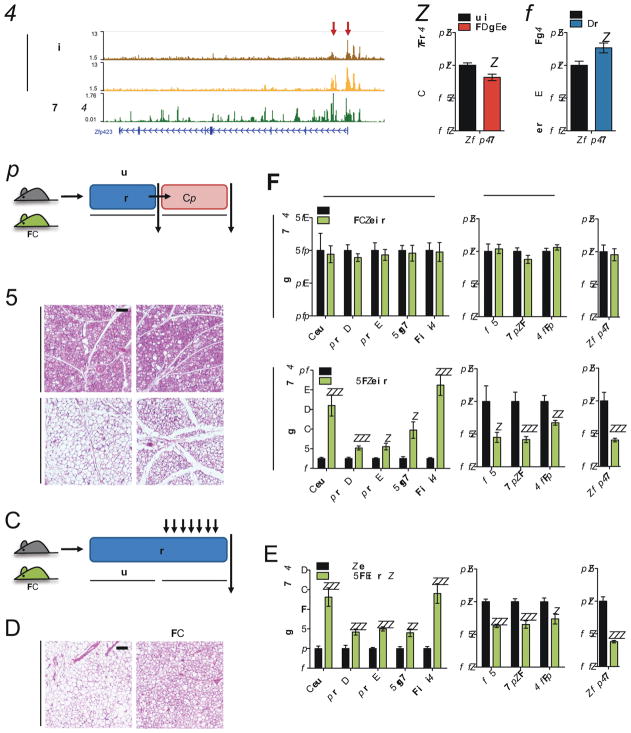
We organized H3K4me1 and H3K27ac peaks into 6 groups as described earlier for transcriptional and H3K27ac profiles (Figure 4A). Each cluster displays globally concordant changes between H3K27ac and H3K4me1 signals in both brown and beige adipocytes (Figures 4A and S5C). Next, we assessed whether beige adipocytes contain poised enhancers that correspond with warming. Within the beige-specific warm-repressed H3K27ac peak cluster, we discovered 692 poised elements (peaks in which H3K4me1 signals were unchanged or increased) (Figures 4B and 4C). Overall, this set of poised enhancers exhibited similar H3K4me1 enrichment in warm white adipocytes as well (Figures 4B and 4C), suggesting that these regions may define epigenomic features shared by all adipocytes. To determine poised enhancers that specifically reflect cold-experienced warm beige adipocytes, we identified a subset of 109 poised enhancers that exhibited stronger H3K4me1 signals in warm beige compared to warm white adipocytes (Figures 4D and 4E). GO analysis found that these poised enhancers were associated with genes involved in fatty acid metabolism and oxidation (Figure 4F), pathways critical to thermogenesis. For instance, the most well-known thermogenic gene, Ucp1, contains several poised enhancers in the upstream intergenic regions (Figure 4G, left). Cpt1b, which plays a critical role in fatty acid oxidation, contains an intronic poised enhancer (Figure 4G, right). Taken together, these results suggest that warm beige adipocytes retain an epigenomic memory from prior cold exposure at a small, but key subset of cis-elements.
Figure 4. Beige adipocytes retain an epigenomic memory of cold exposure.
(a) Heatmap showing H3K27ac and H3K4me1 peaks, clustered as indicated. Each row represents an individual H3K27ac peak (red) and the co-localized H3K4me1 signal (orange) across different adipocyte types in cold and warm conditions.
(b) Heatmap of enhancers in beige adipocytes that display reduced H3K27ac signal but increased or constant H3K4me1 signal after warming (highlighted in dotted blue boxes).
(c) Distribution plots of poised enhancers in (b). H3K27ac and H3K4me1 are shown on the left and right, respectively. Brown adipocyte samples are shown separately from beige and white adipocytes.
(d) Heatmap of poised enhancers in beige adipocytes that are stronger in warm beige than warm white adipocytes.
(e) Distribution plots of poised peaks in (d).
(f) GO biological pathway analysis of genes associated with peaks in (d).
(g) H3K27ac and H3K4me1 signals at the Ucp1 (left) and Cpt1b (right) loci in different adipocyte types in cold and warm conditions. Warm beige adipocytes have H3K4me1 peaks that are higher than in white adipocytes (highlighted in dotted blue boxes).
See also Figure S5.
Poised enhancers prime thermogenic gene expression
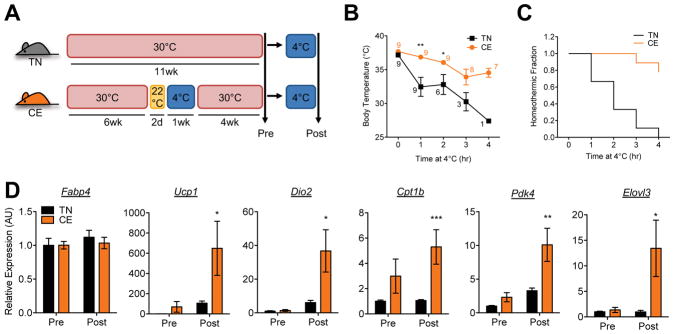
We postulated that poised enhancers in rewarmed beige adipocytes maintain a primed chromatin state at thermogenic gene loci, and might enable these cells to respond more rapidly to an acute cold challenge. To test this, we conducted a cold adaptation experiment; one group of mice was housed at thermoneutrality at all times (TN) while the other group was exposed to cold (CE) for 1 week and subsequently placed at thermoneutrality for 4 weeks (Figure 5A); the animals were then simultaneously exposed to cold conditions (first exposure for the TN group, and second exposure for the CE group). The CE group tolerated the cold challenge substantially better than the TN group, as shown by their higher body temperature and a higher fraction of animals able to maintain homeothermy (Figures 5B and 5C). Next, using an independent cohort of animals, we analyzed gene expression in iWAT during an acute (1hr) cold challenge. Pre-cold, the expression of a panel of thermogenic genes did not significantly differ between TN and CE in iWAT. Post-cold, however, thermogenic genes were more robustly induced in the iWAT of CE mice than in TN mice (Figures 5D), suggesting that poised enhancers in beige adipocytes may prime thermogenic gene expression and thus promote acute induction of gene expression upon cold challenge.
Figure 5. Poised enhancers prime thermogenic gene expression after repeat cold exposure.
(a) Experimental scheme. Wild-type mice were either housed at thermoneutrality for 11 weeks (TN) or were exposed to cold for 1 week after a 2-day transition period at RT (22°C), and then moved back to thermoneutrality for 4 weeks (CE). Both groups were then subjected to a short period of cold exposure as indicated in (b) and (c).
(b) Core body temperature during cold exposure. Animals with temperature below 30°C were removed to room temperature and excluded from further study. Numbers indicate the number of animals analyzed at the time points. Dots indicate mean ± SEM (n=9 animals/group) (*p<0.05; **p<0.01).
(c) Fraction of animals maintaining body temperature above 30°C during cold exposure at 4°C (n=9 animals/group).
(d) Gene expression analysis by qRT-PCR. iWAT from TN and CE groups were assessed pre- and post-cold exposure (1h) at 4°C. Bars indicate mean ± SEM (n=4–7 animals/group) (*p<0.05; **p<0.01; ***p<0.005). Fabp4 is included as a general adipocyte marker.
Glucocorticoid receptor mediates beige adipocyte whitening
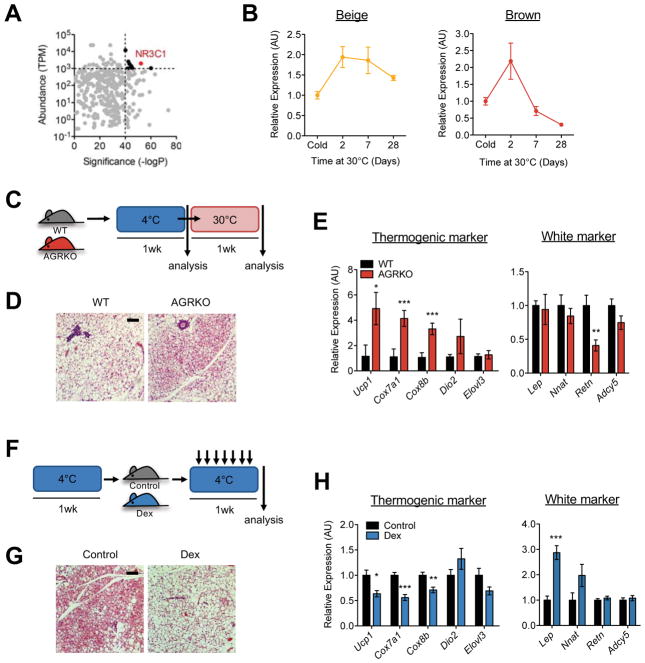
To identify transcriptional pathways that mediate beige adipocyte whitening, we conducted motif enrichment analysis using warm-induced H3K27ac peaks in brown and beige adipocytes. We observed a positive correlation in the significance of the warm-induced motifs between brown and beige adipocytes (Figures S6A and S6B), suggesting that many transcriptional pathways are shared in the whitening of both cell types. However, we also noted several motifs that were preferentially enriched in re-warmed beige adipocytes compared to brown adipocytes; these include NR3C1, NR3C2, AR and ZNF263 (Figures S6A–S6C). The first three of these are highly similar motifs known to bind the glucocorticoid receptor (GR), mineralocorticoid receptor (MR) and androgen receptor (AR), respectively. Integrating the motif results with expression data (Figure 6A) suggested the GR as a very strong candidate to promote beige adipocyte whitening.
Figure 6. The glucocorticoid receptor mediates beige adipocyte whitening.
(a) Motif enrichment analysis showing significance (−log10 p-value) of motifs plotted against the abundance (Transcripts Per Kilobase Million; TPM) of the corresponding transcription factor. Motifs that meet both significance and abundance cutoffs are shown in black. NR3C1 is highlighted in red.
(b) Hsd11b1 expression analyzed by qRT-PCR with TRAP-isolated RNA from BAT and iWAT of Ucp1-NuTRAP mice at the indicated different time points during warming after cold exposure.
(c) Experimental scheme of AGRKO experiment. WT (floxed) and AGRKO mice were exposed to cold (4°C) for 1 week and then moved to thermone utrality (30°C) for 1 week. Both groups are analyzed after cold exposure and after warming.
(d) H&E stained sections of iWAT from WT and AGRKO mice after warming. Scale bar: 20μm.
(e) Expression of thermogenic and white adipocyte genes in iWAT of WT and AGRKO mice after warming. Bars indicate mean ± SEM (n=6–7 animals/group) (*p<0.05; **p<0.01; ***p<0.005).
(f) Experimental scheme of Dex injection experiment. WT mice exposed to cold (4°C) for 1 week are divided into two groups; one injected daily with Dex (10mg/kg) for 1 week and the other injected with saline control; both groups were maintained at 4°C.
(g) H&E stained sections of iWAT from saline and Dex-injected mice. Scale bar: 20μm.
(h) Gene expression analysis of iWAT from saline and Dex-injected mice. Bars indicate mean ± SEM (n=5 animals/group) (*p<0.05; **p<0.01; ***p<0.005).
See also Figure S6.
GR activity is controlled by the availability of its glucocorticoid ligand, which in turn is regulated by tissue levels of 11β-hydroxysteroid dehydrogenase type 1 (encoded by Hsd11b1) (Chapman et al., 2013). Interestingly, Hsd11b1 was induced as early as 2 days after warming in both brown and beige adipocytes, but remained highly expressed over time only in the latter (Figure 6B). Consistently, corticosterone levels in plasma and iWAT trended up after 2 days of warming, but declined after 7 days (Figure S6D). Furthermore, a set of known GR target genes (Kang et al., 2015; Wang et al., 2004) were significantly induced at the onset of warming (Figure S6E). To definitively test whether GR mediates beige adipocyte whitening, we generated adipocyte-specific GR knockout (AGRKO) mice and tested them during a cold-warm temperature shift (Figure 6C). After incubation in the cold for 1 week, AGRKO mice exhibited no significant differences in morphology and only a few differences in gene expression in iWAT (Figures S6F and S6G), indicating that GR is not required for the response to cold in beige adipocytes. However, after re-warming, the iWAT of AGRKO mice display delayed whitening compared to control animals (Figure 6D). This was consistent with gene expression results, in which thermogenic genes such as Ucp1, Cox7a1, and Cox8b, remained higher while the white marker gene Retn was lower in iWAT of AGRKO compared to WT mice (Figure 6E).
To further confirm the role of GR in beige adipocyte whitening, we employed a gain-of-function approach using dexamethasone (Dex), a synthetic GR-specific ligand. We incubated wild-type mice at 4°C for 1 week to recruit beige a dipocytes and then injected Dex daily for 1 week while mice were kept in the cold (Figure 6F). While the iWAT of the control mice displayed numerous active beige adipocytes, Dex-injected mice show substantial whitening in this depot (Figure 6G). Consistently, thermogenic gene expression was significantly reduced and white marker gene expression was concomitantly elevated (Figure 6H). Taken together, these data indicate that GR plays an important role in beige adipocyte whitening.
Zfp423 is a downstream transcriptional effector of GR during beige adipocyte whitening
To further identify downstream actors in the transcriptional cascade mediating beige adipocyte whitening, we looked into GR targets using a series of filtering criteria. First, we identified co-localization of beige-specific warm-induced H3K27ac peaks from our data with published GR binding data from adipocytes (Soccio et al., 2015); this yielded 1,186 likely target genes. We then focused on genes encoding transcription factors, and filtered further by transcript abundance, significance of the motifs for these transcription factors in warm-induced H3K27ac peaks in beige adipocytes, and preferential identification of the motif in beige vs. brown adipocytes. Two transcription factors met all criteria: Zfp423 and Rreb1 (Figure S7). Zfp423 was particularly interesting because it has been previously shown to control white adipocyte identity (Shao et al., 2016). The Zfp423 gene has H3K27ac peaks at the promoter and in intronic regions which were induced by warming in beige adipocytes, several of which co-localize with GR binding sites (Figure 7A). In addition, Zfp423 expression was lower in iWAT of AGRKO mice as compared to WT during rewarming (Figure 7B), and it was induced by Dex injection during cold exposure (Figure 7C), together indicating that Zfp423 is a direct GR target.
Figure 7. Zfp423, a downstream target of GR, mediates beige adipocyte whitening.
(a) H3K27ac and GR ChIP-seq peaks at the Zfp423 locus. Warm-induced H3K27ac peaks in beige adipocytes that overlap with GR binding sites are indicated by red arrows. GR ChIP-seq data is from Soccio et al. (2015).
(b) Zfp423 expression analyzed by qRT-PCR in iWAT of WT and AGRKO mice after warming. Bars indicate mean ± SEM (n=6–7 animals/group) (*p<0.05).
(c) Zfp423 expression analyzed by qRT-PCR in iWAT of cold-exposed control and Dex-injected mice. Bars indicate mean ± SEM (n=5 animals/group) (*p<0.05).
(d) Experimental scheme. WT (Adioponectin-rtTA; Zfp423 floxed) and Zfp423-iAKO (Adioponectin-rtTA; TRE-Cre; Zfp423 floxed) mice are exposed to cold at 6°C for 1 week and then maintained at thermoneutrality (30°C) for 1 week with Dox treatment (to induce loss of Zfp423). Both groups were studied before and after warming/Dox.
(e) H&E stained sections of iWAT from WT and Zfp423-iAKO mice before and after warming/Dox. Scale bar: 20μm.
(f) Expression of thermogenic and white adipocyte markers in iWAT of WT and Zfp423-iAKO mice before and after warming/Dox. Bars indicate mean ± SEM (n=5–6 animals/group) (*p<0.05; **p<0.01; ***p<0.005).
(g) Experimental scheme. WT and Zfp423-iAKO mice were exposed to cold at 6°C for 1 week and then injected daily with Dex (10mg/kg) for 1 week with Dox treatment. Both groups were maintained at 4°C.
(h) H&E stained sections of iWAT from WT and Zfp423-iAKO mice after Dex/Dox treatment. Scale bar: 20μm.
(i) Expression of thermogenic and white adipocyte markers in iWAT of WT and Zfp423-iAKO mice after Dex/Dox treatment. Bars indicate mean ± SEM (n=5 animals/group) (*p<0.05; **p<0.01; ***p<0.005).
See also Figure S7.
To determine whether Zfp423 mediates beige adipocyte whitening, we used reverse tetracycline-controlled transactivator (rtTA)-based inducible adipocyte-specific Zfp423 knockout (Zfp423 iAKO) mice (Shao et al., 2016) to specifically reduce Zfp423 in adipocytes during warming. We incubated the mice at 6°C for 1 week to activate beige adipocytes and subsequently shifted them to 30°C while treating with doxycycline (Dox) to ablate Zfp423 during warming (Figure 7D). Histological and gene expression analysis revealed no differences between Zfp423 iAKO and control mice in the cold (Figures 7E and 7F). In contrast, after warming, the iWAT of Zfp423 iAKO with ~60% reduced Zfp423 expression displayed delayed whitening (Figures 7E and 7F), consistent with gene expression profiles showing elevated expression of thermogenic genes (e.g. Ucp1, Cox7a1, Cox8b1, Dio2 and Elovl3) and reduced expression of white markers (e.g. Lep, Nnat and Retn) in iWAT of Zfp423 iAKO compared WT (Figure 7F). To verify that Zfp423 is a key downstream mediator of GR-induced whitening, we examined the effects of Dex in Zfp423 iAKO incubated in cold conditions (Figure 7G) and observed significantly delayed Dex-induced whitening in the iWAT of Zfp423 iAKO (Figures 7H and 7I). Taken together, these results suggest that Zfp423 functions in beige adipocyte whitening as a direct downstream target of GR.
DISCUSSION
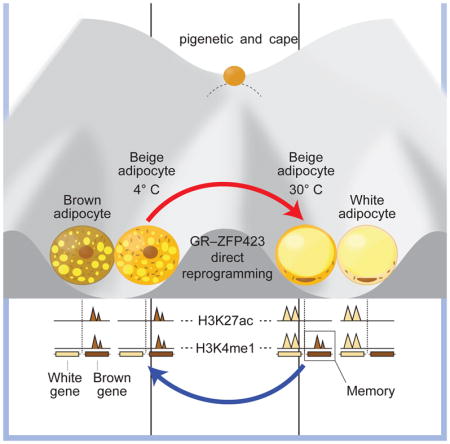
Few organs show the remarkable degree of morphological and functional plasticity demonstrated by adipose tissue. Adipose tissue is highly responsive to changes in environmental conditions, growing significantly larger in conditions of overnutrition and shrinking during food scarcity. Adipose tissue is also very sensitive to changes in temperature, increasing its thermogenic potential in response to cold by activating pre-formed interscapular brown adipocytes and by recruiting inducible beige adipocytes within white depots. The therapeutic potential of thermogenic brown and beige adipocytes in metabolic disease has led to intense investigation into how these cells are formed, and whether there might be significant functional differences between them. Here, we have utilized cell-type specific transcriptional and epigenomic profiling in vivo to define the unique gene expression and chromatin state patterns in brown, beige, and white adipocytes under different thermal conditions. Our results indicate that the response of beige adipocytes to changes in temperature represent an extraordinary example of natural reprogramming of cellular identity. Furthermore, we define a transcriptional cascade, involving the glucocorticoid receptor and Zfp423 as drivers of this process.
Trans-differentiation, also called ‘direct lineage reprogramming’, is a form of cellular reprogramming in which a mature cell type converts to another mature cell type without transitioning through an undifferentiated progenitor stage (Jopling et al., 2011). In point of fact, no true natural examples of mammalian trans-differentiation have been described, although the process can be induced experimentally. For example, introduction of key lineage driving transcription factors has been shown to reprogram mature cells and force them into a different developmental fate. This was first shown by converting cultured fibroblasts to myoblasts via ectopic expression of the transcription factor MyoD (Davis et al., 1987); analogous studies have been performed with hepatocytes, neurons, cardiomyocytes, and a variety of immature progenitor cells (Jopling et al., 2011). In vivo, expression of lineage-specific transcription factors has been used to generate pancreatic α-, β- and δ-cells, as well neurons and cardiomyocytes (Guo and Morris, 2017). More recently, direct lineage reprogramming has been accomplished via chemical induction, or by expression of chromatin-modifying proteins (Onder et al., 2012; Xie et al., 2017). An interesting experimental paradigm is the induced loss of virtually all β-cells by transgenic expression of diphtheria toxin receptor; the new β-cells that ultimately appear in this model are derived from mature α-cells (Thorel et al., 2010).
There have been several studies suggesting that the phenotypic plasticity of adipose tissue may result from trans-differentiation. Most of these studies have relied upon morphological data, including the existence of ‘transition-state’ adipocytes with intermediate morphology between brown and white adipocytes (Cinti, 2002). In some cases, these arguments have been bolstered by gene expression studies showing that beige adipocytes display brown- or white-like gene expression profiles at different temperatures (Rosenwald et al., 2013). However, none of these studies have examined chromatin state within different adipocyte types during thermal stress, and thus it has been difficult to disentangle temperature-dependent changes in cell state from changes in cell identity. Furthermore, we and others have noted that interscapular brown adipocytes display similar morphological and molecular whitening when challenged with thermoneutral temperatures (Cui et al., 2016), making it unclear whether beige adipocytes possess a unique form of cellular plasticity distinct from brown adipocytes. Here we have exploited newly developed technology to perform in vivo transcriptomic and epigenomic profiling in brown, beige, and white adipocytes under different environmental conditions. Our data indicate that beige adipocytes exhibit chromatin state profiles that are almost identical to those of brown adipocytes under cold conditions, and that this chromatin state converts almost completely to that of white adipocytes at thermoneutrality. Thus, these cells undergo a dramatic switch in cellular identity in response to warming, an effect not seen in interscapular brown adipocytes, which exhibit much more limited changes. This is consistent with our finding that chromatin modification was the top significantly upregulated pathway in whitened beige, but not brown, adipocytes. Some may argue that white, beige, and brown adipocytes are all forms of the same fundamental cell type, and thus interconversion between them should not be thought of as changed cellular identity. However, we note that a large part of the literature on trans-differentiation involves switching between α- and β-cells, which are both specialized forms of pancreatic neuroendocrine cells derived through a common lineage (Thorel et al., 2010). Brown and beige cells have distinct developmental origins, as do brown and white adipocytes, and these cells have overlapping but distinct adipokine profiles and alternative thermogenic mechanisms (Harms and Seale, 2013).
As noted above, the formal definition of trans-differentiation requires that the interconversion of cell types not pass through an immature progenitor stage. Our studies do not directly address this point, although the morphological changes shown in Figure 1 are not consistent with a ‘dedifferentiation-redifferentiation’ pathway. Wolfrum’s group noted that labeled beige adipocytes remain mature, and do not take on the appearance of immature progenitor cells during whitening (Rosenwald et al., 2013). Furthermore, the Kajimura group used a single cell monitoring system to observe individual beige adipocytes ex vivo following withdrawal of a β-3 agonist used to induce the formation of the cells (Altshuler-Keylin et al., 2016). Their conclusions are also in line with direct reprogramming, as no fibroblastic or pre-adipocyte-like cells were observed as individual adipocytes transitioned from a multilocular beige appearance to a unilocular white phenotype.
Our epigenomic analysis revealed that a subset of cis-regulatory elements that are active in cold beige adipocytes retain a mark suggestive of open chromatin after warming. These ‘poised’ enhancers are enriched near genes of thermogenesis and fatty acid oxidation, and are characterized by persistent H3K4me1 activity at thermoneutrality, even in the absence of H3K27ac. Such poised enhancers that persist in the absence of the original stimulation are known to function in priming gene expression in response to recurring stimulation (Calo and Wysocka, 2013). We therefore speculated that these enhancers remain poised in order to prime them for rapid activation following a second cold exposure. This notion is consistent with the observation by Wolfrum’s group that whitened beige adipocytes participate in the browning response upon second cold exposure (Rosenwald et al., 2013). Indeed, we found that cold-experienced animals exhibit improved cold tolerance, with greater induction of thermogenic genes in beige adipocytes. This result suggests that the epigenomic memory of prior cold exposure in whitened beige adipocytes is a major determinant of survival in subsequent exposures, although different epigenomic mechanisms other than H3K4me1 and other ‘learned’ responses to cold are certainly possible (e.g. enhanced shivering or piloerection). It is worth pointing out that retention of epigenomic marks from the parent cell has been noted in experimental models of trans-differentiation. For example, induced hepatocytes derived by ectopic expression of Hnf4a and Foxa factors were found to possess remnant signatures from their fibroblast precursors (Morris et al., 2014). While this may complicate the use of experimentally reprogrammed cells in therapeutic applications, our data argue that, in the context of temperature-dependent reprogramming of beige adipocytes, this incomplete conversion at the epigenomic level may be adaptive, and should perhaps be considered a feature of the system rather than a bug.
Cellular identity is determined by a combination of cell autonomous intrinsic and non-autonomous extrinsic factors; this is no less true for adipocytes than any other cell type (Jeffery et al., 2016; Jiang et al., 2017; Macotela et al., 2012). We were struck by the observation that thermoneutrality induces reprogramming of cellular identity in beige, but not brown, adipocytes. Interscapular BAT has significantly more sympathetic innervation than inguinal WAT (Vaughan et al., 2014), and we speculated that at least part of the reason BAT maintains its cellular identity in the face of warming might be due to tonic effects of these nerves. To test this hypothesis, we performed BAT denervation operations, which had no effect on the chromatin state of warm brown adipocytes. This result suggests that there is little to no activity of these nerves at thermoneutrality, and strongly imply that intrinsic factors might play an important role in the maintenance of brown adipocyte identity. Alternatively, non-neural extrinsic factors, such as local immune cells or other components of the interscapular adipose niche, may be critical. It is also possible that longer incubation at thermoneutrality may further enhance whitening of brown adipocytes.
In the experimental models of trans-differentiation described above, effective reprogramming largely depends on the expression of lineage-specific transcription factors that are critical for the output cell type. In particular, pioneer factors play a particularly important role, as they can bind to compacted chromatin, increase local accessibility, and recruit additional transcription factors (Zaret and Carroll, 2011). Our bioinformatic analysis pointed to the GR as a regulator of beige adipocyte whitening; the GR is a well-known pioneer factor that binds to nucleosome-loaded DNA and provides access to additional factors (Voss et al., 2011). Glucocorticoids have been implicated in many aspects of adipocyte biology, including adipogenesis, lipolysis, thermogenesis, and insulin resistance (Kang et al., 2015; Peckett et al., 2011; Rosen and MacDougald, 2006). Recent studies, however, have challenged the notion that intra-adipocyte GR mediates many of these activities. For example, despite the inclusion of dexamethasone in virtually all canonical adipogenic cocktails, the GR is actually dispensable for both in vitro and in vivo adipocyte development (Desarzens and Faresse, 2016; Park and Ge, 2017). Furthermore, several groups have shown that, contrary to expectations, adipocyte GR does not play a role in obesity-associated insulin resistance (Bose et al., 2016; Shen et al., 2017). Finally, glucocorticoids exert opposing effects on adipocyte thermogenesis depending on the species and the duration of treatment (Ramage et al., 2016), and adipocyte-specific GR knockout mice display no obvious defects in cold tolerance (Shen et al., 2017). Our current data help to resolve this last point, as we find that the GR is most important during the shift from cold to warm, rather than the converse. Specifically, we noted that adipocyte-specific GR knockout mice exhibit defective beige adipocyte whitening during the return to thermoneutrality. This is supported by pharmacological data showing that systemic injection of dexamethasone is sufficient to induce whitening of beige adipocytes, even in cold-challenged animals. Our murine data are consistent with available human data demonstrating that glucocorticoid treatment inhibits brown adipose activity (Ramage et al., 2016). Of note, patients with glucocorticoid excess (i.e. Cushing’s syndrome) typically exhibit a ‘buffalo hump’ characterized by white fat accumulation in the dorsocervical and supraclavicular regions, where most human brown and beige adipocytes reside (Ferrau and Korbonits, 2015). We also note that the role of GR as a whitening factor could account for the belief, recently shown to be false (Park and Ge, 2017), that the GR is a critical adipogenesis factor, as it is easy to mistake lipid accumulation (‘morphological whitening’) for differentiation.
We further identified Zfp423 as a direct downstream effector of the GR during beige adipocyte whitening. Zfp423 was implicated in our study by a combination of temperature-dependent enhancer activity, transcript abundance, and the presence of GR binding sites documented by ChIP-seq. Zfp423 is a critical determinant of white adipocyte identity, and inducible ablation of this factor causes profound “beiging” of white adipose depots throughout the body (Shao et al., 2016). Of note, we did not detect the known Zfp423 motif as a top candidate in the motif analysis that identified the GR. There are several possible reasons for this. First, if Zfp423 binds to a small but critical subset of warm-induced enhancers, the significance of the motif may not supersede factors that bind a broader array of cis-regulatory elements. Additionally, Zfp423 can influence transcription in multiple ways beyond direct contact through its own DNA binding domain; these include interaction with Smad factors, EBFs, and likely other critical transcriptional drivers. We think it is likely that other transcription factors participate in temperature-dependent reprogramming of cellular identity in beige adipocytes. It should be pointed out, however, that simply because a transcription factor represses browning does not mean that it must participate in beige adipocyte whitening; for example, IRF3 did not mediate warm-induced beige adipocyte whitening (data not shown), despite its documented anti-browning effects (Kumari et al., 2016).
One of the strengths of our approach is the pure isolation of beige adipocytes from within a complex white fat pad in vivo. This affords us the opportunity to test the significance of several adipocyte type-specific markers that have been identified from either cultured systems or from unfractionated fat; additionally, it enables us to seek new markers that might have been missed in those systems. Our data confirm that commonly accepted thermogenic markers (e.g. Ucp1, Dio2, Elovl3, Pgc1a, Cox7a1, Cox8b, and Cidea) are shared between brown and beige cells, and also indicate the validity of several widely accepted white adipocyte markers (e.g. Retn, Lep, Nnat, and Adcy5). However, we also find that the majority of reported “brown-specific” and “beige-specific” markers do not adequately distinguish between these cell types. Rather, they are more indicative of anatomical position. Thus, Zic1, which has been reported to be highly brown-specific, is actually a dorsocervical marker also found in white adipocytes from that location (this study, and de Jong et al. 2015). Similarly, Tmem26, Tnfrsf9/CD137, Shox2 and Hoxc9 are markers for the inguinal region, and not for beige adipocytes per se. Somewhat disappointingly, our data suggests that there are no ‘absolute’ markers that are unique for any single adipocyte type. Instead, we find several relatively enriched markers, such as Slc29a1 and Hoxa5 for brown, Tbx1 for beige, and Agt and Slc16a12 for white adipocytes. Slc29a1 and Hoxa5 are novel brown markers with low expression in interscapular WAT. Tbx1 and Agt have been previously proposed as beige and white markers, respectively (Wang and Seale, 2016). Importantly, expression of these beige and white marker genes is tightly correlated with beige and white adipocyte content within heterogeneous WAT tissues, suggesting that these markers can be used to estimate the relative proportion of beige and white adipocytes in a bulk sample. None of these markers have been studied functionally in adipocytes, thus their characterization may provide new insights into the unique attributes of different adipocyte types. Of note, although these markers genes can distinguish among adipocyte types, most are also expressed in non-adipocyte cell types, complicating their use as drivers for adipocyte type-specific Cre lines.
Most studies on brown and beige adipocytes have focused on the induction of thermogenesis during the shift from room temperature (typically 22–23°C in most colonies) to cold conditions (4°C). In contrast, we have focused on the process of re-warming, as the animal moves from cold to thermoneutrality (30°C). Importantly, it is now recognized that standard rodent housing conditions represent a cold stress with substantial effects on metabolism, hemodynamics, immune response, and behavior (Ganeshan and Chawla, 2017). Because humans spend significant effort to maintain thermoneutrality, it has been proposed that animal studies might better model the human condition if they are performed at 30°C. In this light, it is reasonable to speculate that human beige adipocytes, once formed, may spend most of their lifetime in the whitened state. Accordingly, studies of beige adipocyte biology at thermoneutrality may be more relevant for human biology than more typical studies performed under standard housing conditions.
In summary, our study reveals that temperature-induced plasticity of beige adipocytes is a natural example of cellular trans-differentiation, defines components of epigenomic memory in whitened beige adipocytes that may persist in order to enhance adaptation to subsequent cold exposure, and identifies a GR-Zfp423 axis as a critical transcriptional driver of this unique environmentally driven example of reprogrammed cellular identity.
Limitations of Study
Our work identifies temperature-dependent changes in chromatin state in beige adipocytes, but there are several limitations to consider. First, there are several epigenomic marks that we did not assess. Important changes might be happening in any of dozens of other histone modifications, or in other forms of epigenomic status such as DNA methylation. Our study also does not fully define how whitened beige adipocytes and ‘normal’ or ‘unbrownable’ white adipocytes are fundamentally different from each other. The lack of white adipocyte-specific marker genes and genetic labeling systems has been the major technical limitation to addressing the question. However, recently developed single transcriptomic and epigenomic approaches, which are independent of genetic tools, may enable insights into deeper epigenomic mechanisms specific for each adipocyte type. Finally, all of our experiments were performed in mice. While our findings are expected to be conserved in human brown and beige adipocytes, this will need to be tested directly when the tools to do so become available.
STAR* Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Evan Rosen (erosen@bidmc.harvard.edu).
Experimental Model and Subject Details
Animals
All animal experiments were performed according to procedures approved by the BIDMC Institutional Animal Care and Use Committee. To label brown/beige adipocytes and pan-adipocytes, NuTRAP mice (Jackson Laboratory, 029899) (Roh et al., 2017) were crossed with Ucp1-Cre (Jackson Laboratory 024670) (Kong et al., 2014) and Adiponectin-Cre (Jackson Laboratory 010803) (Eguchi et al., 2011), respectively. To generate adipocyte-specific GR knockout mice, Adiponectin-Cre mice were crossed with GR floxed mice (Jackson Laboratory, 021021). To generate rtTA-based inducible adipocyte-specific Zfp423 knockout (Zfp423 iAKO) mice, Adiponectin-rtTA (Wang et al., 2013), TRE-Cre (Jackson Laboratory, 006224), and Zfp423 floxed mice (Shao et al., 2016) were crossed. In all cases, littermate controls were used.
Mice were maintained on a chow diet (8664 Harlan Teklad) under a 12h light/dark cycle at 22°C for standard room temperature housing or at 30°C for thermoneutrality. For cold-rewarming experiments, 6–8 week old male mice were individually housed at 4–6°C for 1 week and subsequently moved to 30°C for 4 weeks for the indicated length of time. For Dex treatment experiments, dexamethasone sodium phosphate (Santa Cruz Biotechnology, sc-204715) was injected intraperitoneally (10mg/kg daily) to wild-type C57Bl6/J or Zfp423 iAKO male mice at 6–8 week of age housed at 4–6°C for 1 week. Core body temperature was measured using a rectal probe (Yellow Spring Instruments).
Denervation
Male mice (8–10 week old) were anesthetized by intraperitoneal injection of ketamine/xylazine (80mg/ml and 12mg/ml, respectively) and kept on a heating pad during surgery. The skin in the upper dorsal surface was shaved, washed with Betadine, and opened with sterile scissors. The ventral surface of one BAT pad was gently exposed to visualize nerves beneath pad. Using a dissecting stereomicroscope, 5 branches of nerves bundles were identified and cut without disrupting the arterial supply or venous drainage. For sham-operation, the contralateral BAT pad was subjected to all of the same manipulations except for nerve cutting. The skin was closed by using tissue adhesive glue (Vetbond). Once awake, the mice were housed at 22°C or 30°C as indicated for 4 weeks until analysis.
Method Details
TRAP/RNA-seq
TRAP was conducted as previously described (Roh et al., 2017). Pieces of frozen adipose tissue (50–100mg) were homogenized by dounce (7ml) in homogenization buffer (50mM Tris [pH7.5], 12mM MgCl2, 100mM KCl, 1% NP-40; 100μg/ml cycloheximide, 1mg/ml sodium heparin, 2mM DTT, 0.2units/μl RNasin, 1x Roche Complete EDTA-free protease inhibitor); 1ml and 4ml for BAT and iWAT, respectively) and incubated on ice for 15 min with vortexing every 5 min. Lysates were centrifuged at 13,000rpm for 10min and the supernatant was collected after removing the top lipid layer. Anti-GFP antibody (5μg/ml, Abcam ab290) was added to the collected supernatant and rotated for 1h at 4°C. Protein G dynabeads washed twice with low-salt wash buffer (50mM Tris [pH7.5], 12mM MgCl2, 100mM KCl, 1% NP-40, 100μg/ml cycloheximide, 2mM DTT) were added to the lysates with antibody and further rotated for 30min at 4°C. The immunoprecipitates were washed three times in in high-salt wash buffer (50mM Tris [pH 7.5], 12mM MgCl2, 300mM KCl, 1% NP-40, 100μg/ml cycloheximide, 2mM DTT) on a magnet. After the final wash, immunoprecipitates were subjected to RNA extraction using the Qiagen Micro RNeasy kit according to the manufacturer’s instructions. For RNA-seq library construction, TRAP-isolated RNA (≤100ng) was treated with the Ribo-Zero rRNA removal kit (Epicentre) to deplete ribosomal RNA and converted into double stranded cDNA using NEBNext mRNA Second Strand Synthesis Module (E6111L). cDNA was subsequently tagmented and amplified for 12 cycles by using Nextera XT DNA Library Preparation Kit (Illumina FC-131). Sequencing libraries were analyzed Qubit and Agilent Bioanalyzer, pooled at a final concentration of 12pM, and sequenced on a HiSeq2500.
RNA isolation/Quantitative RT-PCR
We used TRIzol (Invitrogen) and Qiagen Micro RNeasy kit for total tissue RNA and TRAP-isolated RNA extraction, respectively, according to the manufacturers’ instructions. Extracted RNA (500ng) was converted into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR (qRT-PCR) was performed using an Applied Biosystems 7900 thermocycler and SYBR Green PCR Master Mix (Applied Biosystems). Fold change was determined by comparing target gene expression with the reference gene Tbp, which was chosen because its levels do not vary significantly across adipocyte types and temperatures, unlike other commonly used references genes, such as 36B4 (Rplp0), Rps18, Gapdh, cyclophilin A (Ppia) and β-actin (Actb). The primers used for qRT-PCR are shown in Table S2.
Nuclear Isolation/sorting
Nuclear isolation was performed as previously described (Roh et al., 2017). BAT or iWAT samples were collected, frozen in liquid nitrogen and stored at −80°C until used. Frozen tissues were homogenized using a dounce (7ml) in nucleus preparation buffer (NPB; 10mM HEPES [pH7.5], 1.5mM MgCl2, 10mM KCl, 250mM sucrose, 0.1% NP-40, 0.2mM DTT). Homogenates filtered through 100μm cell strainers were cross-linked with 1% paraformaldehyde (PFA) at room temperature for 4min while rotating and quenched by 125mM glycine for 10min. The homogenates were centrifuged at 1000g for 10min, and the nuclei-containing pellets were washed once by resuspending in NPB and spinning down at 1000g for 10min. The pellets were thoroughly resuspended by pipetting in nucleus sorting buffer (NSB; 10mM Tris [pH 7.5], 40mM NaCl, 90mM KCl, 2mM EDTA, 0.5mM EGTA, 0.1% NP-40, 0.2mM DTT) and filtered through 40μm cell strainers. Resuspended nuclei were sorted using a BD FACS Aria II, gating on FSC, SSC and mCherry fluorescence.
ChIP-seq
ChIP-seq was performed as previously described (Roh et al., 2017). Sorted nuclei were resuspended in nuclear lysis buffer (NLB, 10mM Tris [pH8], 1mM EDTA, 0.1% SDS) and sheared using a Covaris E220 using microTUBEs (130ul) with the following parameters: peak incident power 105W, duty factor 2%, cycles per burst 200 for 12 min. The sheared chromatin was centrifuged at 13,000rpm for 10min at 4°C to re move debris, and an aliquot (3%) was used for quantification. Sheared chromatin (1–2μg) was diluted in ChIP dilution buffer (CDB, 16.7mM Tris [pH8], 1.2mM EDTA, 167mM NaCl, 1.1% Triton X-100, 0.01% SDS), and 1μg/ml of an H3K27ac antibody (Active Motif, 39133) or 1μg/ml of an H3K4me1 antibody (Cell Signaling Technology, 5326) was added, followed by overnight rotation at 4°C. Protein A/G dynabeads were washed in PBS/1% BSA and added to the ChIP samples. After rotating for 1h at 4°C, the samples were washed in low salt wash buffer (20mM Tris [pH8], 1mM EDTA, 140mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS), high salt wash buffer (20mM Tris [pH8], 1mM EDTA, 500mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS), LiCl wash buffer (10mM Tris [pH8], 1mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate, 250mM LiCl) and TE buffer (10mM Tris [pH8], 1mM EDTA) sequentially twice in each step, and eluted and reverse cross-linked in elution buffer (10mM Tris [pH8], 50mM EDTA, 0.1% SDS, 300mM NaCl, 0.8mg/ml proteinase K, 10μg/ml RNase A) by incubating at 65°C for 8hr. Next, DNA was extracted using AMPure XP beads following the manufacturer’s manual; briefly, 2.2X volume of AMPure XP beads were added to the reverse cross-linked ChIP samples, resuspended and washed with 70% ethanol three times using the magnet. DNA was subsequently eluted in 10mM Tris [pH8.5] and quantified by Qubit (Thermo Fisher Scientific).
Sequencing libraries were constructed using the “on-bead” preparation method as previously described (Roh et al., 2017). ChIP-isolated DNA was subjected to end repair/phosphorylation using the End-It DNA End-Repair Kit (Epicentre), A-tailing using the Klenow Fragment (NEB M0212) and index adaptor ligation using Quick Ligase (NEB M2200). AMPure XP beads were left in all the reactions to allow isolation of DNA using PEG (Polyethylene Glycol 8000)/NaCl solution, thus minimizing loss between reactions. After ligation, DNA was eluted and amplified by PCR for 14–16 cycles using the PfuUltra II Hotstart PCR Master Mix (Agilent 600850). DNA sized between 250 and 600bp was selected by gel electrophoresis and extraction using E-Gel EX Agarose Gels (Invitrogen) and MinElute Gel Extraction kits (Qiagen). Libraries were analyzed using the Qubit and Agilent Bioanalyzer, pooled at a final concentration of 12pM and sequenced on a HiSeq2500 or NextSeq500 system.
Immunofluorescence and Immunohistochemistry
Dissected adipose tissues were fixed in 10% formalin for 1 day, washed in PBS and processed by the BIDMC histology core for paraffin-sectioning and immunohistochemistry in the following manner: sections were deparaffinized, treated with sodium citrate for antigen retrieval and then with hydrogen peroxide to inactivate endogenous peroxidase. Next, the sections were blocked with donkey serum and incubated with anti-GFP antibody (1:500) (Novus NB-100-1678) or anti-UCP1 antibody (1:1000) (Abcam ab10983) overnight at 4°C. After washing, the slides were incubated with anti-goat secondary antibody, enhanced, developed in DAB (diaminobenzidine) and counter-stained with hematoxylin.
For immunofluorescence, tissues were fixed in 10% formalin for 1 day, incubated in PBS/20% sucrose for 1 day and cryosectioned at 50 μm thickness. Slides were blocked and permeabilized in PBS/1% Triton X-100 supplemented with 10% FBS and incubated with anti-GFP (Novus NB-100-1678), anti-UCP1 (Abcam ab10983), anti-mCherry (Abcam ab167453), or anti-PLIN1 (Abcam ab61682) at 1/1000 dilution overnight. After washing in PBS/1% Triton X-100, slides were incubated with Alexa Fluor conjugated antibodies and Hoechst 33342 for nuclear staining. For whole mount staining, dissected fresh tissues were cut into small pieces (3mm × 3mm) and subjected to the same procedures as frozen section slides except that tissue pieces were held in 48-well plates for a longer incubation time: antibody incubation for 24h and each wash step for 1h. After the final wash, stained tissue pieces were placed in mounting solution on slides and covered with coverslips. Slides were visualized using a Zeiss LSM 510 Meta confocal microscope.
Corticosterone Measurement
Blood samples were collected in EDTA-coated blood collection tubes and plasma corticosterone levels were measured using an ELISA kit according to the manufacturer’s instructions (Abcam). For adipose tissue corticosterone analysis, frozen tissues were homogenized in PBS using Qiagen TissueLyzer and multiples cycles of freeze/thaw, and corticosterone was measured in tissue homogenates using an ELISA kit according to the manufacturer’s instructions (Cayman).
Data Analysis
For RNA-seq analysis, sequencing reads were demultiplexed using bcl2fastq and aligned to the mm10 mouse genome using HISAT2 (Kim et al., 2015). PCR duplicates and low-quality reads were removed by Picard (https://broadinstitute.github.io/picard). Filtered reads were assigned to the annotated transcriptome and quantified using feature Counts (Liao et al., 2014). Normalization and differential expression analysis were performed using EdgeR (Robinson et al., 2010). For differential gene expression analysis, we only tested genes that were detected in at least two samples with log2CPM≥2. Genes were considered significant if they passed a fold change (FC) cutoff of log2FC≥1 and a false discovery rate (FDR) cutoff of FDR≤0.05. Morpheus (https://software.broadinstitute.org/morpheus/) was used for K-means clustering, hierarchical clustering, Pearson correlation similarity analysis, and heatmap visualization. Principal Components Analysis (PCA) and volcano plots were generated in R. The volcano plots, specifically, were generated by an easy-to-use, interactive R Shiny app, we have developed for public use (https://bdawes.shinyapps.io/volcano_plotter/), which also includes functionality for depicting MA and abundance plots. Gene set enrichment analysis was performed using DAVID Bioinformatics Resources 6.8 (Huang da et al., 2009) and summarized by REVIGO (Supek et al., 2011).
For ChIP-seq analysis, demultiplexed ChIP-seq reads were aligned to the mm10 mouse genome using Bowtie2 (Langmead and Salzberg, 2012). PCR duplicates and low-quality reads were removed by Picard. Reads were processed using Samtools (Li et al., 2009) and subjected to peak-calling with MACS2 (Zhang et al., 2008); called peaks had ≥4-fold higher coverage than input genomic DNA. Reads assigned to peaks were quantified using featureCounts and normalization and differential analysis were performed using EdgeR. The same cutoff criteria used for RNA-seq analysis were applied for peak differential analysis: log2CPM≥2, log2FC≥1, and FDR≤0.05. To visualize ChIP-seq signals, reads were converted to the BigWig file format using bedtools (Quinlan and Hall, 2010) and bedGraphToBigWig (Kent et al., 2010). BigWig files were normalized based on duplication rate, library size, peak assignment efficiency and viewed in the WashU Epigenome Browser (Zhou and Wang, 2012). ChIP-seq peak heatmaps and distribution plots were generated with the normalized BigWig files within 10kb genomic regions surrounding peaks by using deepTools2 (Ramirez et al., 2016). Overlaps between H3K27ac and H3K4me1 were determined using bedtools. Peaks were assigned to genomic annotations and genes with HOMER (Heinz et al., 2010), and GO pathway analysis was performed by DAVID Bioinformatics Resources 6.8. Motif enrichment analysis was conducted using FIMO from the MEME package (Grant et al., 2011) with the JASPAR database (Mathelier et al., 2014); FIMO was used to locate and score the strongest instance of each JASPAR motif per peak and then, for each motif, we compared motif score distribution between differentially-regulated peaks and neutral peaks using a Wilcoxon test. Bonferroni correction was applied for multiple hypothesis testing. Neutral peaks were identified as those with |−log2FC|≤0.5 and CPM within the same interval as differentially-regulated peaks within the comparisons.
Quantification and Statistical Analysis
Two-tailed unpaired Student’s t-test was used for pair-wise comparisons, and p <0.05 was considered statistically significant, unless otherwise specified.
Data and Software Availability
The accession number for RNA-seq and ChIP-seq data reported in this paper is GEO: GSE108077. Bioinformatics software used in the study are publically available; the volcano plotter we developed is publicly available (https://bdawes.shinyapps.io/volcano_plotter/),
Supplementary Material
Table S1. (Related to Figure 2) mRNA abundance in adipocyte types at 4°C and 30°C
HIGHLIGHTS.
Cell type-specific RNA-seq identify markers for white, brown, and beige adipocytes
Beige adipocytes exhibit reprogramming between white and brown chromatin states
Warmed beige adipocytes retain an epigenomic memory of prior cold exposure
A GR-Zfp423 pathway mediates beige adipocyte whitening during warming
Acknowledgments
We thank James Granneman for instruction on denervation surgery, Qiong Wang and Philipp Scherer for help with rewarming experiments, and all members of the Rosen lab for their helpful advice and discussion. We are grateful to the Flow Cytometry Core, the Histology Core, and the Molecular Medicine Core at Beth Israel Deaconess Medical Center, as well as the Functional Genomics and Bioinformatics Core of the Boston Nutrition and Obesity Research Center (BNORC). This work was supported by an American Diabetes Association Postdoctoral Fellowship 1-17-PDF-133 to H.C.R., a Department of Defense grant W81XWH-14-PRMRP-DA to L.T-Y.T., an American Heart Association postdoctoral fellowship 16POST26420136 to M.S., NIH R01 DK104789 to R.K.G., and NIH R01 DK102173, DK102170, DK113669, and DK085171 to E.D.R.
Footnotes
AUTHOR CONTRIBUTIONS
Experiments were designed by H.C.R., L.T-Y.T., R.K.G. and E.D.R. and executed by H.C.R., M.S., D.T., Y.S. and M.K. Computational analysis was performed by H.C.R., A.L., C.J., and B.D. The manuscript was written by H.C.R. and E.D.R. with input from all other authors.
DECLARATION OF INTERESTS
These authors have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, Kajimura S. Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance. Cell Metab. 2016;24:402–419. doi: 10.1016/j.cmet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Jiang Y, Graff JM. Mouse strains to study cold-inducible beige progenitors and beige adipocyte formation and function. Nat Commun. 2016;7:10184. doi: 10.1038/ncomms10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose SK, Hutson I, Harris CA. Hepatic Glucocorticoid Receptor Plays a Greater Role Than Adipose GR in Metabolic Syndrome Despite Renal Compensation. Endocrinology. 2016;157:4943–4960. doi: 10.1210/en.2016-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K, Holmes M, Seckl J. 11beta-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol Rev. 2013;93:1139–1206. doi: 10.1152/physrev.00020.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti S. Adipocyte differentiation and transdifferentiation: plasticity of the adipose organ. J Endocrinol Invest. 2002;25:823–835. doi: 10.1007/BF03344046. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Nguyen NL, Zarebidaki E, Cao Q, Li F, Zha L, Bartness T, Shi H, Xue B. Thermoneutrality decreases thermogenic program and promotes adiposity in high-fat diet-fed mice. Physiol Rep. 2016;4 doi: 10.14814/phy2.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- de Jong JM, Larsson O, Cannon B, Nedergaard J. A stringent validation of mouse adipose tissue identity markers. Am J Physiol Endocrinol Metab. 2015;308:E1085–1105. doi: 10.1152/ajpendo.00023.2015. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desarzens S, Faresse N. Adipocyte glucocorticoid receptor has a minor contribution in adipose tissue growth. J Endocrinol. 2016;230:1–11. doi: 10.1530/JOE-16-0121. [DOI] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Ferrau F, Korbonits M. Metabolic comorbidities in Cushing’s syndrome. Eur J Endocrinol. 2015;173:M133–157. doi: 10.1530/EJE-15-0354. [DOI] [PubMed] [Google Scholar]
- Ganeshan K, Chawla A. Warming the mouse to model human diseases. Nat Rev Endocrinol. 2017;13:458–465. doi: 10.1038/nrendo.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Morris SA. Engineering cell identity: establishing new gene regulatory and chromatin landscapes. Curr Opin Genet Dev. 2017;46:50–57. doi: 10.1016/j.gde.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Wing A, Holtrup B, Sebo Z, Kaplan JL, Saavedra-Pena R, Church CD, Colman L, Berry R, Rodeheffer MS. The Adipose Tissue Microenvironment Regulates Depot-Specific Adipogenesis in Obesity. Cell Metab. 2016;24:142–150. doi: 10.1016/j.cmet.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Berry DC, Jo A, Tang W, Arpke RW, Kyba M, Graff JM. A PPARgamma transcriptional cascade directs adipose progenitor cell-niche interaction and niche expansion. Nat Commun. 2017;8:15926. doi: 10.1038/ncomms15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Kang S, Tsai LT, Zhou Y, Evertts A, Xu S, Griffin MJ, Issner R, Whitton HJ, Garcia BA, Epstein CB, et al. Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis. Nat Cell Biol. 2015;17:44–56. doi: 10.1038/ncb3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, Vetrivelan R, Lu GZ, Laznik-Bogoslavski D, Hasenfuss SC, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell. 2015;163:643–655. doi: 10.1016/j.cell.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell. 2014;158:69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M, Wang X, Lantier L, Lyubetskaya A, Eguchi J, Kang S, Tenen D, Roh HC, Kong X, Kazak L, et al. IRF3 promotes adipose inflammation and insulin resistance and represses browning. J Clin Invest. 2016;126:2839–2854. doi: 10.1172/JCI86080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014;19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macotela Y, Emanuelli B, Mori MA, Gesta S, Schulz TJ, Tseng YH, Kahn CR. Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes. 2012;61:1691–1699. doi: 10.2337/db11-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Huang L, Zhu LJ, Zou T, Ou J, Zhou W, Wang YX. Jmjd3-Mediated H3K27me3 Dynamics Orchestrate Brown Fat Development and Regulate White Fat Plasticity. Dev Cell. 2015;35:568–583. doi: 10.1016/j.devcel.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YK, Ge K. Glucocorticoid Receptor Accelerates, but Is Dispensable for, Adipogenesis. Mol Cell Biol. 2017;37 doi: 10.1128/MCB.00260-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckett AJ, Wright DC, Riddell MC. The effects of glucocorticoids on adipose tissue lipid metabolism. Metabolism. 2011;60:1500–1510. doi: 10.1016/j.metabol.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Peirce V, Carobbio S, Vidal-Puig A. The different shades of fat. Nature. 2014;510:76–83. doi: 10.1038/nature13477. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage LE, Akyol M, Fletcher AM, Forsythe J, Nixon M, Carter RN, van Beek EJ, Morton NM, Walker BR, Stimson RH. Glucocorticoids Acutely Increase Brown Adipose Tissue Activity in Humans, Revealing Species-Specific Differences in UCP-1 Regulation. Cell Metab. 2016;24:130–141. doi: 10.1016/j.cmet.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh HC, Tsai LT, Lyubetskaya A, Tenen D, Kumari M, Rosen ED. Simultaneous Transcriptional and Epigenomic Profiling from Specific Cell Types within Heterogeneous Tissues In Vivo. Cell Rep. 2017;18:1048–1061. doi: 10.1016/j.celrep.2016.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Rosenwald M, Perdikari A, Rulicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. 2013;15:659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Hung CM, Guertin DA. Emerging Complexities in Adipocyte Origins and Identity. Trends Cell Biol. 2016;26:313–326. doi: 10.1016/j.tcb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, MacPherson KA, Spurgin SB, Sun K, Holland WL, et al. Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab. 2016;23:1167–1184. doi: 10.1016/j.cmet.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Roh HC, Kumari M, Rosen ED. Adipocyte glucocorticoid receptor is important in lipolysis and insulin resistance due to exogenous steroids, but not insulin resistance caused by high fat feeding. Molecular Metabolism. 2017 doi: 10.1016/j.molmet.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccio RE, Chen ER, Rajapurkar SR, Safabakhsh P, Marinis JM, Dispirito JR, Emmett MJ, Briggs ER, Fang B, Everett LJ, et al. Genetic Variation Determines PPARgamma Function and Anti-diabetic Drug Response In Vivo. Cell. 2015;162:33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CH, Zarebidaki E, Ehlen JC, Bartness TJ. Analysis and measurement of the sympathetic and sensory innervation of white and brown adipose tissue. Methods Enzymol. 2014;537:199–225. doi: 10.1016/B978-0-12-411619-1.00011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2004;101:15603–15608. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Seale P. Control of brown and beige fat development. Nat Rev Mol Cell Biol. 2016;17:691–702. doi: 10.1038/nrm.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Fu Y, Liu J. Chemical reprogramming and transdifferentiation. Curr Opin Genet Dev. 2017;46:104–113. doi: 10.1016/j.gde.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang T. Using the Wash U Epigenome Browser to examine genome-wide sequencing data. Curr Protoc Bioinformatics. 2012;Chapter 10(Unit10):10. doi: 10.1002/0471250953.bi1010s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. (Related to Figure 2) mRNA abundance in adipocyte types at 4°C and 30°C