Summary
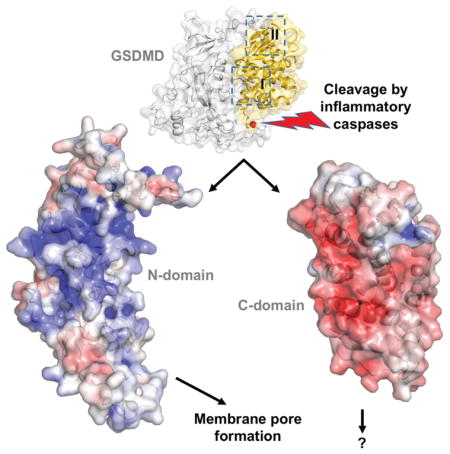
Pyroptosis is an inflammatory form of programmed cell death that plays important roles in immune protection against infections and in inflammatory disorders. Gasdermin D (GSDMD) is an executor of pyroptosis upon cleavage by caspases-1/4/5/11 following canonical and noncanonical inflammasome activation. GSDMD N-terminal domain assembles membrane pores to induce cytolysis, whereas its C-terminal domain inhibits cell death through intramolecular association with the N-domain. The molecular mechanisms of autoinhibition for GSDMD are poorly characterized. Here we report the crystal structures of the human and murine GSDMD C-terminal domains, which differ from those of the full-length murine GSDMA3 and the human GSDMB C-terminal domain. Mutations of GSDMD-C domain residues predicted to locate at its interface with the N-domain enhanced pyroptosis. Our results suggest that GSDMDs may employ distinct mode of intramolecular domain interaction and autoinhibition, which may be relevant to its unique role in pyroptosis downstream of inflammasome activation.
eTOC Blurb
The molecular mechanisms of autoinhibition for gasdermin D-mediated pyroptosis are poorly defined. Liu et al. report the crystal structures of the murine and human GSDMD C-terminal domains, which suggest that GSDMDs may employ distinct mode of intramolecular domain interaction to regulate pyroptosis.
Introduction
Pyroptosis is an inflammatory form of programmed cell death that plays important roles in immune defense against intracellular pathogens and in tissue damage from excessive inflammation (Cookson and Brennan, 2001; Latz et al., 2013; Miao et al., 2010). Gasdermin D (GSDMD) is an executor of pyroptosis upon cleavage by caspases-1/4/5/11 downstream of canonical and noncanonical inflammasome pathways (He et al., 2015; Kayagaki et al., 2015; J. Shi et al., 2015). The gasdermin family members contain N-terminal domains that are capable of forming membrane pores to induce cytolysis, whereas the C-terminal domains of gasdermins were reported to function as inhibitors of such cytolysis through intramolecular domain association (Aglietti et al., 2016; X. Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Rogers et al., 2017; Sborgi et al., 2016; Wang et al., 2017). It was proposed that both protease cleavage and release of autoinhibition are required for regulation of cytolysis: upon protease cleavage of the gasdermin N- and C- domain linker, the disruption of the intramolecular domain interaction in the presence of lipids releases the N-domain to assemble oligomeric membrane pores that trigger cell death. This is consistent with the observation that cell death induced by the N-domain from one gasdermin family member could be blocked by the C-domain from another, suggesting that the mode of autoinhibition among the gasdermin family members may be similar (P. Shi et al., 2015).
Structure of the full-length murine GSDMA3 (mGSDMA3) reveals an autoinhibited conformation with its N- and C-domains engaging in extensive intramolecular interactions through two distinct binding sites (Ding et al., 2016). Surprisingly, the crystal structure of the human GSDMB C-terminal domain (hGSDMB-C) demonstrates the lack of crucial structural elements at one binding site, and rearrangement of loops at the other (Chao et al., 2017). This called into question whether the mode of intramolecular domain interaction observed in mGSDMA3 is indeed conserved among other gasdermin family members or in other species. Furthermore, the full-length hGSDMB was shown to possess comparable lipid-binding capacity as its N-terminal domain (Chao et al., 2017), in stark contrast to the full-length mGSDMA3 and GSDMD that harbor minimal lipid binding activities (Aglietti et al., 2016; X. Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Sborgi et al., 2016). This implies that gasdermin family members may adopt distinct mechanisms of intramolecular domain interactions that modulate their lipid-binding and pore-forming activities. To shed light on the autoinhibition mechanisms of GSDMD as an effector of pyroptosis, we initiated structural studies of the human and murine GSDMD and determined the structures of their C-domains. Analyses of the GSDMD N- and C-domain interaction suggest that GSDMDs may employ distinct mode of intramolecular domain interaction and autoinhibition, which may be relevant to its unique role in pyroptosis downstream of inflammasome activation.
Results
Overall structures of the GSDMD-C domains
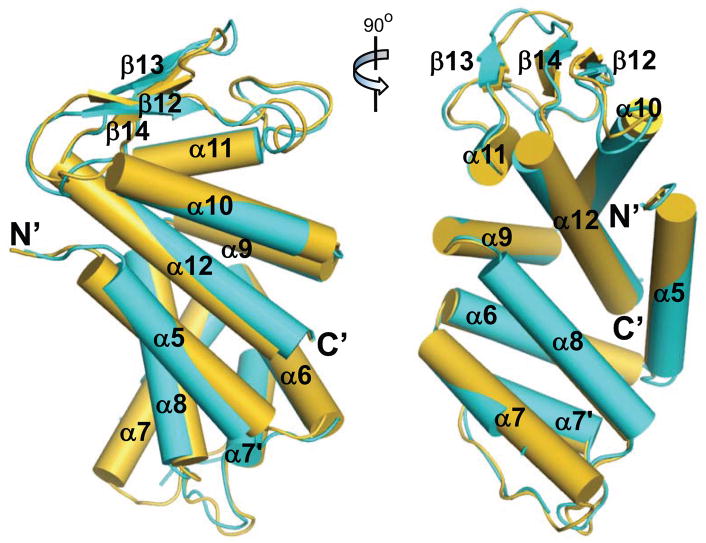
The C-terminal domains for both human and murine GSDMD were crystallized, and their structures determined using single wavelength anomalous diffraction (SAD) method (Table S1 and Figure S1A–B). The ~200 residue GSDMD-C domains are composed of nine helices capped by an anti-parallel β12–β14 sheet, and superimpose with an rmsd of 1.1 Å for 187 aligned residues (Figure 1). By contrast, these structures superimpose with that of mGSDMA3 (Ding et al., 2016) with rmsd of 1.7–2.0 Å for 170 aligned residues, and that of hGSDMB-C (Chao et al., 2017) with rmsd of 1.8–2.6 Å for 120–130 aligned residues, suggesting significant structural differences particularly between GSDMDs and hGSDMB. In agreement, both GSDMDs and mGSDMA3 contain the β12–β14 sheet and the α11 helix, which are absent in the hGSDMB-C domain (Figure S1C). Instead, hGSDMB-C contains a long α10–α12 loop, which along with its β11-α5 loop occupy the same space as the α1 helix and β1–β2 loop from mGSDMA3 (Figure S1D). The main structural difference between GSDMDs and mGSDMA3 is localized at the shorter α11 helices and an additional α helices between the α7 and α8 helices (hereafter referred to as α7′ helix) in GSDMDs, which was not anticipated in the previous GSDMD model (Ding et al., 2016) (Figure 1 and Figure S1E–S1F). Notably, structural superposition illustrates that the GSDMD α7′ helix is located distal to the N- and C-domain interface (Figure S1C). Whether this additional helix with poor sequence conservation (Data S1) contributes to GSDMD-specific function requires further investigation.
Figure 1. Crystal Structures of the Human and Murine GSDMD-C Domains.
Two orthogonal views of the crystal structures for the human (cyan) and murine (gold) GSDMD-C domains. Helices are represented as cylinders. The N- and C-termini are marked.
See also Figure S1.
Association of the GSDMD-C and GSDMD-N domains
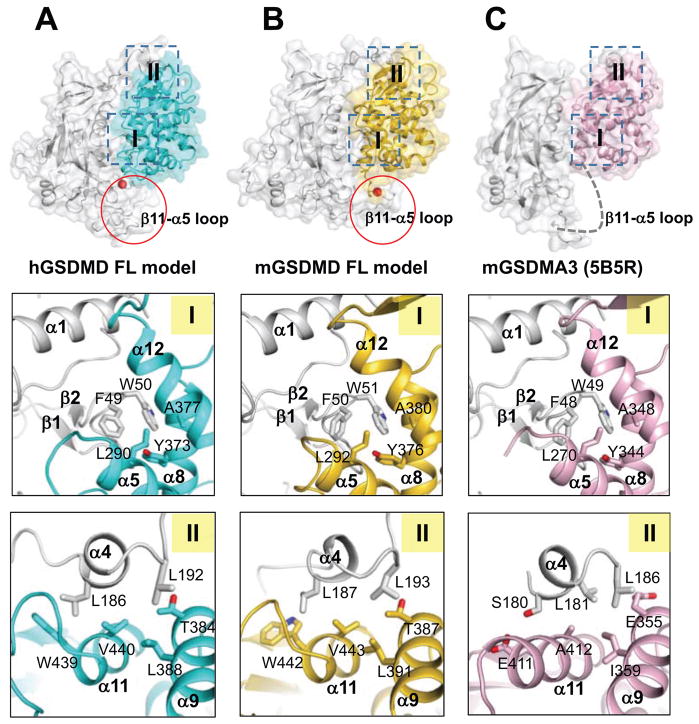
To guide analysis of the intramolecular domain interactions for GSDMDs, the full-length mGSDMA3 structure was used as a template to create models for the full-length hGSDMD and mGSDMD with the MMM server (http://manaslu.aecom.yu.edu/MMM/) (Rai et al., 2006). Inspection of the models revealed that the N- and C-terminal domains of both GSDMDs may associate with each other through two interaction sites, in a similar manner as the mGSDMA3 template (Figure 2). The first interaction site (“I”) involves the C-domain α5, α8, and α12 helices binding the N-domain α1 helix and the β1–β2 loop containing conserved residues 50-FW-51 (mGSDMD numbering), and possibly the β11-α5 loop connecting the two domains. This linker region harbors the caspase cleavage site but was not observed in the mGSDMA3 structure. The second interaction site (“II”) is composed of the α9 and α11 helices from the C-domain binding the α4 helix from the N-domain. This interface is located near the β 12–β14 sheet capping one side of the C-domain helical structure (Figure 2 and Data S1). Analysis of the charge surface of the GSDMD models and the mGSDMA3 structure shows that their C-domains are mostly negatively charged at their interface with the N-domains, which are largely positively charged (Figure S2A–C). This is consistent with the observation that the gasdermin N-domains participate in binding of negatively charged membrane lipid headgroups (Aglietti et al., 2016; X. Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Rogers et al., 2017; Sborgi et al., 2016; Wang et al., 2017). In contrast to GSDMDs and mGSDMA3, hGSDMB-C lacks either the β12–β14 sheet or the α11 helix (Chao et al., 2017) (Figure S1C and Data S1), crucial structural elements for the site II interface. Furthermore, superposition of the hGSDMB-C and mGSDMA3 structures reveals that the α10–α12 and β11-α5 loops from hGSDMB-C may clash with the site I α1 helix and β1–β2 loop from the N-domain (Figure S1D). Such major structural differences imply that the intramolecular domain interaction in hGSDMB may be drastically different from that in mGSDMA3 and possibly GSDMDs, suggesting distinct modes of autoinhibition for gasdermin family members.
Figure 2. The N- and C-domain Interface for hGSDMD, mGSDMD and mGSDMA3.
The full-length hGSDMD and mGSDMD structural models were created using the full-length mGSDMA3 as a template. (A) The N- and C-domains of an hGSDMD model are shown as gray and cyan surface, respectively. The two domain interaction sites I and II are outlined in the top panel, with the details shown on the lower two panels as ribbons and sticks. Similar representations are shown for mGSDMD in (B) with the C-domain colored gold and mGSDMA3 in (C) with the C-domain colored pink.
See also Figure S2.
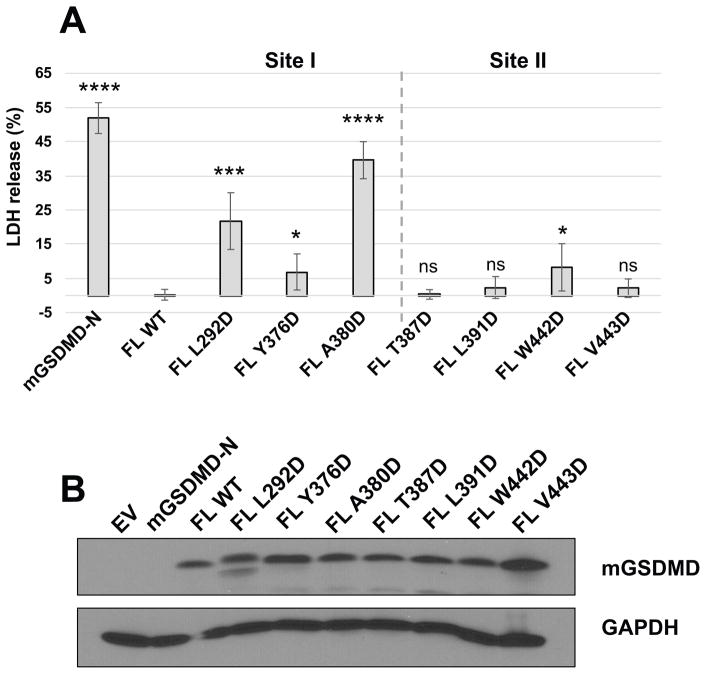
To investigate whether the GSDMD-C structural elements in the full-length GSDMD models indeed mediate domain interactions and modulate cell death, full-length wildtype or mutant mGSDMD were expressed in HEK293T cells. The mutation sites are located at the surface of the GSDMD-C domains (Figure 2), therefore they are unlikely to impact the folding of the GSDMD-C domain. Compared with the wildtype full-length GSDMDs, mGSDMD harboring mutations at site I induced significant LDH release (Figure 3), demonstrating spontaneous cytolysis due to compromised autoinhibition. In comparison, site II mutants induced low or minimal levels of LDH release. Notably, the levels of LDH release induced by the mutants were less than those by the GSDMD-N domains. This is in agreement with a previous report on mutant full-length GSDMDs (Ding et al., 2016), which suggests that GSDMD-C domains covalently attached to the GSDMD-N domains may impede optimal membrane pore assembly. Nonetheless, our data are consistent with the idea that the GSDMD-C domain serves important function in suppressing cell death by the GSDMD-N domain, and reduced autoinhibition may promote the assembly of membrane pores by GSDMD-N and spontaneous cell death.
Figure 3. Mutations at the N-/C- Domain Interface Impact Pyroptosis.
The WT or mutant mGSDMD was expressed in HEK293T cells. The LDH release induced by the expressed mGSDMD as percentage of total LDH content after Triton X-100 treatment is shown in (A). The gray dotted lines delineate mutations for site I (left) and site II (right). The average ± SD is shown for three independent experiments. Statistical analysis is calculated by two-tailed t-test, compared to the “FL WT” samples. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Expression levels for the WT and mutant mGSDMD as well as GAPDH are shown in (B).
See also Data S1.
Role of GSDMD intramolecular domain interaction in Salmonella infection
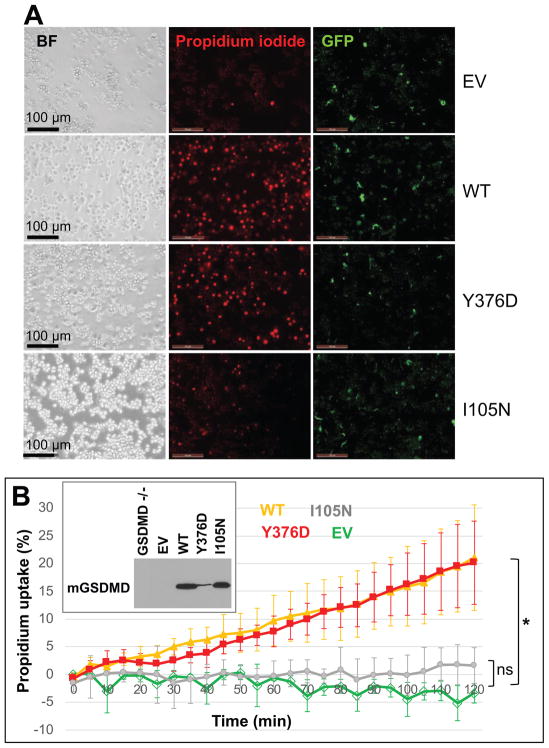
To probe the role of the GSDMD domain association in pyroptosis following inflammasome activation, we analyzed infection of macrophages by Salmonella enterica, which was known to induce macrophage pyroptosis through activation of canonical inflammasomes such as NAIP/NLRC4 and noncanonical inflammasomes (Baker et al., 2015; Broz et al., 2012; Casson et al., 2015; Fink and Cookson, 2006; Franchi et al., 2006; Mariathasan et al., 2004; Miao et al., 2006). Uptake of fluorescent propidium iodide (PI) by macrophages upon infection was tracked over time as an indicator of plasma membrane permeabilization or pyroptosis (Jorgensen et al., 2016; Russo et al., 2016). As previously described (Russo et al., 2016), Gsdmd was knocked out from an immortalized murine bone marrow derived macrophage (iBMDM) cell line (Stutz et al., 2013) using the CRISPR-Cas9 technology. The WT or mutant Gsdmd was then reconstituted in the Gsdmd-deficient iBMDM using a LentiCRISPRv2 plasmid (Chirieleison et al., 2016; Rathkey et al., 2017). During reconstitution of mGSDMD, despite our repeated efforts at screening for clones with high expression levels, we have failed to obtain a Y376D-expressing clone with comparable expression level as the WT and I105N mutants (Figure 4B insert). We suspect that high expression levels of Y376D may have induced spontaneous pyroptosis, similar to what we observed in HEK293T cells (Figure 3). As a result, clones with high expression of Y376D either died off or were growth retarded during the clone selection process. Nevertheless, upon infection with Salmonella, cells expressing the Y376D mutant induced comparable PI uptake as the WT macrophages, despite the much lower levels of expression, suggesting enhanced capacity of the Y376D mutant to mediate pyroptosis (Figure 4). Importantly, no PI uptake was induced by Salmonella infection of cells deficient for Gsdmd (“EV”), confirming that the expression of GSDMD was required for the observed PI uptake in our experimental setup. Cells expressing the I105N mutant showed minimal levels of PI uptake compared with the “EV” control, consistent with reports that this mutation led to attenuated membrane permeabilization and pyroptosis (Aglietti et al., 2016; Kayagaki et al., 2015). No PI uptake was observed in the absence of Salmonella infection (Figure S3), indicating minimal levels of spontaneous cell death within the infection period. Collectively, the above data establish that mutation at the GSDMD intramolecular domain interface significantly enhances the dissociation of the GSDMD-N and GSDMD-C domains, which may promote more efficient membrane pore formation by the former in response to bacterial infection.
Figure 4. Role of GSDMD Domain Interaction in Macrophage Pyroptosis upon Salmonella Infection.
Macrophages reconstituted with WT or mutant mGSDMD were infected with GFP-tagged Salmonella at an MOI of 10. PI uptake by the macrophages were tracked over time as an indicator of pyroptosis. Bright field and fluorescent images taken at 2-hour post-infection were shown in (A). The time course of PI uptake is plotted in (B). The average ± SD is shown for three independent experiments. Asterisk denotes P value <0.05 calculated by one-way ANOVA. Western blots for the reconstituted WT or mutant mGSDMD are shown in the insert.
See also Figure S3.
Discussion
The gasdermin family members were re-discovered recently as endogenous pore-forming proteins: their N-terminal domains assemble membrane pores to induce cytolysis, whereas their C-terminal domains inhibit cell death through intramolecular domain association. Previous structural studies of the full-length mGSDMA3 revealed extensive intramolecular domain interactions through two distinct binding sites (Ding et al., 2016). Surprisingly, the above mode of intramolecular domain interaction is not compatible with the hGSDMB-C domain structure, which showed a lack of crucial structural elements and rearrangement of loops at both of the binding sites (Chao et al., 2017). This called into question whether the mode of the autoinhibition observed in mGSDMA3 applies to other gasdermin family members or other species. Our work reports the crystal structures of the C-terminal domains from both human and murine gasdermin D. Our analysis of the association of the gasdermin D N- and C-domains offer mechanistic understanding of GSDMD autoinhibition through two binding sites.
Furthermore, our study provides insights on the role of gasdermin D autoinhibition during macrophage pyroptosis in responses to Salmonella infection, using macrophages reconstituted with wildtype and mutant GSDMD. Our findings suggest that GSDMDs may employ distinct mode of intramolecular domain interaction and autoinhibition, which may be relevant to its unique role in pyroptosis downstream of inflammasome activation.
The site I and site II interface between the gasdermin N- and C-domains may play dual roles in both the autoinhibited and pore-forming states. The gasdermin N-domain was shown to associate with cardiolipin and phosphoinositides such as phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate (Aglietti et al., 2016; X. Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Rogers et al., 2017; Sborgi et al., 2016; Wang et al., 2017). A common feature of the phosphoinositide-binding motifs in various protein domains is the abundance of positively charged and large hydrophobic residues (Stahelin et al., 2014), which are well represented in the α1 helix and the β1–β2 loop region surrounding the FW residues at site I (Data S1). It is possible that this region may mediate the GSDMD N- and C-domain interaction similar to mGSDMA3, and the lipid-binding at or near the conserved FW residues may serve to release the N- and C-domain association. This is consistent with similar hypothesis that residues 1–56 from GSDME/DFNA5, encompassing the α1-β2 region, may function as the potential membrane targeting domain (Rogers et al., 2017). The site II interface involves the α4 helix region rich in Gly or small residues (Ala, Ser, or Cys) (Data S1). This is reminiscent of the GXXXG motif frequently observed in pore-forming cytolytic peptides and viral fusion peptides, which employ this particular motif at their transmembrane helix-helix interface (Del Angel et al., 2002; Kleiger et al., 2002; Russ and Engelman, 2000). Interestingly, mutation of L192 (hGSDMD) or C192 (mGSDMD) in the predicted α4 helices was reported to impair GSDMD-N oligomerization and pyroptosis (Ding et al., 2016; Liu et al., 2016), suggesting that the site II interface may participate in GSDMD-N self-association and pore formation.
The β11-α5 loops of GSDMDs connect their N- and C-domains, and harbor the cleavage site for caspases-1, 4, 5, and 11. Both cleavage at the β11-α5 loop and the presence of membrane lipids are required for the activation of GSDMDs as pore-forming proteins (Aglietti et al., 2016; X. Chen et al., 2016; Ding et al., 2016; Liu et al., 2016; Sborgi et al., 2016). Even though the β11-α5 loop is disordered in the mGSDMA3 structure, it is proximal to the N- and C-domain interface and likely shields the β1-β2 loop from the solvent in the autoinhibited state. While this manuscript was under review, Kuang et al reported the crystal structure of the human GSDMD-C domain, which reveals similar structural features and an important role for the F283 residue within the β11-α5 loop in maintaining autoinhibition (Kuang et al., 2017). Conceivably, cleavage of the β11-α5 loop by caspases may weaken its role in autoinhibition, and expose the β1-β2 loop to the solvent or membrane lipids, which then compete for association with the N-domain and promote the formation of GSDMD-N oligomers. It is not clear whether the β11-α5 loop per se is sufficient for GSDMD recognition by inflammatory caspases. Given the specific recognition of GSDMD by inflammatory caspases and GSDME/DFNA5 by caspase-3, it seems likely that structural elements outside of the β11-α5 loop may play a role in substrate recognition and binding by these caspases. An intriguing possibility is that besides autoinhibition, the GSDMD-C domain may participate in caspase recruitment, thus providing additional levels of pyroptosis regulation. Interestingly, the hGSDMB SNP sites (residues 299 and 306) associated with inflammatory diseases are located at its C-terminal domain distal from the predicted N- and C-domain interface. The SNP site mutations may alter the conformation of the C-domain surface loop and impact its association with an unknown partner protein, such as a protease (Chao et al., 2017). Alternatively, the gasdermin C-domains released from the N-domains may harbor other physiological functions through association with unknown partners.
In conclusion, our work reports the crystal structures of the C-terminal domains from both human and murine GSDMD and offers mechanistic insights on GSDMD autoinhibition that regulate pyroptosis. The gasdermin family members are dangerous molecules capable of forming membrane pores that induce cytolysis, and thus may have evolved autoinhibition mechanisms that regulate cell death through their C-terminal domains. It remains to be determined how most of the gasdermin family members are maintained in their autoinhibited states, and how they are activated through protease cleavage or other post-translational modifications, and ultimately how they function under physiological and pathological conditions. Understanding the molecular mechanisms of gasdermin transformation from soluble proteins to transmembrane pores will provide valuable insights into the roles of pyroptosis in immune protection against infections and in inflammatory disorders. These will pave the way for therapeutic targeting of various inflammatory disorders such as sepsis (Jorgensen and Miao, 2015), multiple sclerosis (Guo et al., 2015; Martin et al., 2016) and inflammatory bowel disease (Zaki et al., 2011).
STAR METHODS
Contact for reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tsan Sam Xiao, Ph.D. (tsx@case.edu).
Experimental Model and Subject Details
HEK293T cell culture
HEK293T cells were cultured at 37 °C, 5% CO2 in DMEM plus 10% fetal bovine serum (FBS) supplemented with 100 U/ml Penicillin and 100 U/ml Streptomycin (Pen+Strep), and 2 mg/ml L-Glutamine, hereafter referred to as complete DMEM.
Immortalized mouse macrophage cell culture
Immortalized mouse bone marrow-derived macrophages (iBMDM) overexpressing NLRP3-FLAG and ASC-mCerulean were described previously (Stutz et al., 2013). They were cultured in complete DMEM.
Salmonella Typhimurium cell culture
Salmonella Typhimurium cells were grown in Luria broth at 37 °C.
Method Details
Protein expression and purification
The coding sequence for the C-terminal domains of human (residues 277–484) and murine (residues 277–487) GSDMD were cloned into a bacterial expression vector encoding a His6-SUMO tag (Mossessova and Lima, 2000). The transformed BL21 (DE3) Codon Plus RIPL cells (Agilent Technologies, Santa Clara, CA) were grown at 37 °C and protein expression was induced at 18 °C overnight with 0.2 mM IPTG. Cells were harvested and lysed by sonication in a lysis buffer containing 25 mM Tris-HCl (pH 8.0), 300 mM NaCl. The recombinant SUMO-fusion proteins in the cleared cell lysate were purified using Ni-NTA (Thermo Fisher Scientific, Waltham, MA) affinity chromatography. Elution fractions containing the purified SUMO-fusion proteins were pooled and incubated overnight with Ulp1 protease (Mossessova and Lima, 2000) at 4 °C, followed by a second Ni-NTA chromatography to remove the SUMO tag and uncleaved fusion protein. The GSDMD-C protein samples were further purified through size-exclusion chromatography in a buffer containing the lysis buffer plus 5 mM DTT. The purified protein was concentrated to 20–30 mg/ml before frozen in aliquots at −80 °C. The Se-Met derived mGSDMD-C was expressed using the BL21 (DE3) Codon Plus RIPL cells cultured in SelenoMet medium base plus nutrient mix (MD12-501, Molecular Dimensions, Maumee, OH) with the addition of L-selenomethionine (Thermo Fisher Scientific, Waltham, MA), followed by purification protocols as described for the wild-type protein.
Crystallization, X-ray diffraction data collection and structure determination
Screening of crystallization conditions for both hGSDMD-C and mGSDMD-C was performed using commercial screening kits from Hampton Research (Aliso Viejo, CA) and Molecular Dimensions (Maumee, OH) with sitting drop vapor diffusion method at 22 °C or 4 °C. The hGSDMD-C was crystallized with 2.1 M DL-malic acid, pH 7.0. The mGSDMD-C was crystallized with 25% (v/v) polyethylene glycol (PEG) 3350, 0.1 M Bis-Tris (pH 5.5), 0.2 M MgCl2. The Se-Met derived mGSDMD-C was crystallized in 27% (v/v) PEG3350 0.1 M Na-HEPES (pH 7.5), 0.2 M MgCl2. All crystals were frozen in their mother liquor plus 25% glycerol. X-ray diffraction data were collected at beamlines SER-CAT (22-ID) and GM/CA-CAT (23-ID) at the Advanced Photon Source, Argonne National Laboratory (Lemont, IL), and processed with program XDS (Kabsch, 2010).
The mGSDMD-C structure was determined using single wavelength anomalous diffraction (SAD) method with program Crank2 (Ness et al., 2004) within the CCP4 program suite (Potterton et al., 2003). Using a dataset collected at 0.97932 Å with a Se-Met derived mGSDMD-C crystal, eight Se-Met sites for the four mGSDMD-C molecules in the crystallographic asymmetric unit were located with program SHELXD (Schneider and Sheldrick, 2002). Automated model building and refinement were performed with program Buccaneer (Cowtan, 2006) and Refmac (Murshudov et al., 2011), respectively. In total, 805 out of 848 residues were built and refined by Crank2. The hGSDMD-C structure was determined using the mGSDMD-C structure as a search model. Manual model building and refinement were performed with program Coot (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010), respectively. The crystal structures were validated by the Molprobity server (V. B. Chen et al., 2010). Figures were prepared using PyMol (The PyMOL Molecular Graphics System, Version 1.8.2.3 Schrödinger, LLC.). The electrostatic charge surface was calculated with PDB2PQR (Dolinsky et al., 2004).
Cytotoxicity assay for mGSDMD mutants
HEK293T cells grown in DMEM plus 10% serum were transfected with vectors in the T-REx expression system (Thermo Fisher Scientific, Waltham, MA). Co-transfection of the pCDNA6/TR vector coding for the Tet repressor and the pCDNA4/TO vectors that code for mGSDMD N-domain, WT or mutant full-length proteins at 4:1 molar ratio was performed using 8 μg of plasmids mixed with calcium phosphate per 60 mm tissue culture plate. After overnight culture of the transfected cells, expression of mGSDMD was induced with 1 μg/ml tetracycline followed by collection of culture supernatants that were centrifuged to pellet detached cells. Lactate dehydrogenase (LDH) activities in the supernatants were measured using a Cytotoxicity Detection Kit (Roche Diagnostics Corporation, Indianapolis, IN) following the manufacturer’s instructions. The LDH release was expressed as a percentage of total LDH content upon 1% Triton X-100 treatment of the cells.
Genome editing and reconstitution of Gsdmd
Using the CRISPR-Cas9 genome editing tool, Gsdmd was knocked out from an immortalized bone marrow derived macrophage (iBMDM) cell line overexpressing mCerulean-ASC (Stutz et al., 2013), as previously described (Russo et al., 2016). The iBMDM cell line was grown in DMEM plus 10% serum. Reconstitution of wildtype and mutant Gsdmd was achieved using the LentiCRISPRv2 plasmid (Chirieleison et al., 2016; Rathkey et al., 2017). Gsdmd-coding lentivirus were produced in HEK293T cells through calcium phosphate transfection of the Gsdmd reconstitution plasmid. The Gsdmd-coding lentivirus were harvested, filtered through a 0.45 μm filter, and transduced into the Gsdmd−/− iBMDM cells. Gsdmd expressing cells were selected with 1 mg/ml geneticin and maintained with 0.2 mg/ml geneticin. Western blot using a monoclonal mGSDMD-N-specific antibody (A-7, sc-393656, Santa Cruz Biotechnologies, Dallas, TX) was performed to analyze expression levels of the reconstituted mGSDMD.
Salmonella infection and propidium iodide uptake assay
The iBMDM cell lines reconstituted with wildtype or mutant Gsdmd were seeded in 24-well plates with 4 × 105 cells per well. After overnight culture, cells were washed with PBS before adding balanced salt solutions (25 mM Na-HEPES, pH 7.4, 130 mM NaCl, 4 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2) supplemented with 5 mM D-glucose and 0.1% BSA. Cells were then infected with a Salmonella enterica serotype Typhimurium expressing GFP (ATCC 14028GFP). The infection was carried out at 37°C with an MOI of 10:1 in the presence of 1 μg/ml propidium iodide (Thermo Fisher Scientific, Waltham, MA). The propidium iodide fluorescence (excitation/emission: 533/617 nm) in each well was measured every five to ten minutes for two hours with a SpectraMax i3x Multi Mode microplate reader (Molecular Devices, Sunnyvale, CA). The assays were terminated by adding 1% Triton X-100 to the wells and the recorded fluorescence was treated as the maximum for each well. PI uptake was plotted as a percentage of the maximum fluorescence after subtraction of basal fluorescence in control wells treated with propidium iodide without Salmonella.
Quantification and Statistical Analysis
Statistical analysis for LDH release assay and PI uptake assay reported in Figures 3, 4, and S3 are analyzed with program Excel (Microsoft Corporation, Redmond, WA) or program Prism (GraphPad software, Inc, La Jolla, CA).
Data and Software Availability
Coordinates and structural factors have been deposited with the Protein Data Bank with accession codes 6AO3 and 6AO4 for murine and human GSDMD-C domains, respectively.
Supplementary Material
Highly conserved residues are shown in red shade, and conserved residues in yellow shade. Residues are shown in black squares if their mutations are known to compromise activities. The caspase cleavage sites are shown in green shade. Secondary structural elements for known gasdermin structures are marked and underlined. Residues involved in site I interface are marked with magenta dots, those in site II with black dots. The black arrow indicates the minimal N-domain (residues 1–231 of hGSDMD and 1–234 of hGSDME/DFNA5) for pyroptosis induction (Rogers et al., 2017).
Highlights.
Crystal structures of the murine and human GSDMD C-domains are presented
Two interaction sites I and II mediate the association of the C- and N-domains
This domain-domain interaction regulates cell death upon Salmonella infection
The mode of GSDMD autoinhibition may be distinct from other gasdermins
Acknowledgments
Z.L. is funded by a Careers in Immunology postdoctoral fellowship from the American Association of Immunologists. T.S.X. is supported by NIH grant AR069908 and the Case Research Institute. J.K.R. is supported by NIH grant T32 GM007250. D.W.A. is supported by NIH grants DK091222 and GM086550. We are grateful for help from the Ramakrishnan lab on the use of the T-REx expression system. We would like to thank the Xiao, Abbott, Dubyak, and Ramakrishnan laboratories for insightful discussions. X-ray diffraction data were collected at the Advanced Photon Source (APS) SER-CAT beamline (Supporting institutions may be found at www.ser-cat.org/members.html) and GM/CA-CAT beamline, funded in whole or in part by funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the APS, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
Author Contributions
Conceptualization, Z.L. and T.S.X; Methodology, Z.L., C.W., G.R.D., D.W.A., and T.S.X.; Investigation, Z.L., C.W., J.R., and J.Y.; Writing – Original Draft, Z.L. and T.S.X.; Writing – Review & Editing, Z.L., and T.S.X.; Funding Acquisition, D.W.A. and T.S.X.; Resources, G.R.D., D.W.A., and T.S.X.; Supervision, D.W.A. and T.S.X.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Dueber EC. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci USA. 2016;113:7858–7863. doi: 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PJ, Boucher D, Bierschenk D, Tebartz C, Whitney PG, D’Silva DB, Tanzer MC, Monteleone M, Robertson AAB, Cooper MA, Alvarez-Diaz S, Herold MJ, Bedoui S, Schroder K, Masters SL. NLRP3 inflammasome activation downstream of cytoplasmic LPS recognition by both caspase-4 and caspase-5. Eur J Immunol. 2015;45:2918–2926. doi: 10.1002/eji.201545655. [DOI] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson CN, Yu J, Reyes VM, Taschuk FO, Yadav A, Copenhaver AM, Nguyen HT, Collman RG, Shin S. Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proc Natl Acad Sci USA. 2015;112:6688–6693. doi: 10.1073/pnas.1421699112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao KL, Kulakova L, Herzberg O. Gene polymorphism linked to increased asthma and IBD risk alters gasdermin-B structure, a sulfatide and phosphoinositide binding protein. Proc Natl Acad Sci USA. 2017;114:E1128–E1137. doi: 10.1073/pnas.1616783114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, He WT, Hu L, Li J, Fang Y, Wang X, Xu X, Wang Z, Huang K, Han J. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 2016;26:1007–1020. doi: 10.1038/cr.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirieleison SM, Kertesy SB, Abbott DW. Synthetic Biology Reveals the Uniqueness of the RIP Kinase Domain. J Immunol. 2016;196:4291–4297. doi: 10.4049/jimmunol.1502631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson BT, Brennan MA. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9:113–114. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1 Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Del Angel VD, Dupuis F, Mornon JP, Callebaut I. Viral fusion peptides and identification of membrane-interacting segments. Biochem Biophys Res Commun. 2002;293:1153–1160. doi: 10.1016/S0006-291X(02)00353-4. [DOI] [PubMed] [Google Scholar]
- Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006;8:1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Özören N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- Guo H, Callaway JB, Ting JPY. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev. 2015;265:130–142. doi: 10.1111/imr.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Zhang Y, Krantz BA, Miao EA. Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J Exp Med. 2016;213:2113–2128. doi: 10.1084/jem.20151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kleiger G, Grothe R, Mallick P, Eisenberg D. GXXXG and AXXXA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry. 2002;41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- Kuang S, Zheng J, Yang H, Li S, Duan S, Shen Y, Ji C, Gan J, Xu XW, Li J. Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis. Proc Natl Acad Sci USA. 2017;114:10642–10647. doi: 10.1073/pnas.1708194114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Martin BN, Wang C, Zhang CJ, Kang Z, Gulen MF, Zepp JA, Zhao J, Bian G, Do JS, Min B, et al. T cell-intrinsic ASC critically promotes TH17-mediated experimental autoimmune encephalomyelitis. Nat Immunol. 2016;17:583–592. doi: 10.1038/ni.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness SR, de Graaff RAG, Abrahams JP, Pannu NS. CRANK: new methods for automated macromolecular crystal structure solution. Structure. 2004;12:1753–1761. doi: 10.1016/j.str.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Rai BK, Madrid-Aliste CJ, Fajardo JE, Fiser A. MMM: a sequence-to-structure alignment protocol. Bioinformatics. 2006;22:2691–2692. doi: 10.1093/bioinformatics/btl449. [DOI] [PubMed] [Google Scholar]
- Rathkey JK, Benson BL, Chirieleison SM, Yang J, Xiao TS, Dubyak GR, Huang AY, Abbott DW. Live cell visualization of gasdermin D-driven pyroptotic cell death. J Biol Chem. 2017;292:14649–14658. doi: 10.1074/jbc.M117.797217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Biol Chem. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Russo HM, Rathkey J, Boyd-Tressler A, Katsnelson MA, Abbott DW, Dubyak GR. Active Caspase-1 Induces Plasma Membrane Pores That Precede Pyroptotic Lysis and Are Blocked by Lanthanides. J Immunol. 2016;197:1353–1367. doi: 10.4049/jimmunol.1600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborgi L, Rühl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Muller DJ, Broz P, Hiller S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016;35:1766–1778. doi: 10.15252/embj.201694696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Shi P, Tang A, Xian L, Hou S, Zou D, Lv Y, Huang Z, Wang Q, Song A, Lin Z, et al. Loss of conserved Gsdma3 self-regulation causes autophagy and cell death. Biochem J. 2015;468:325–336. doi: 10.1042/BJ20150204. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Scott JL, Frick CT. Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chem Phys Lipids. 2014;182:3–18. doi: 10.1016/j.chemphyslip.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz A, Horvath GL, Monks BG, Latz E. ASC speck formation as a readout for inflammasome activation. In: De Nardo CM, Latz E, editors. The Inflammasome Methods Mol Biol. Vol. 1040. Totowa, NJ: Humana Press; 2013. pp. 91–101. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, Shao F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a Gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- Zaki MH, Lamkanfi M, Kanneganti TD. The Nlrp3 inflammasome: contributions to intestinal homeostasis. Trends Immunol. 2011;32:171–179. doi: 10.1016/j.it.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Highly conserved residues are shown in red shade, and conserved residues in yellow shade. Residues are shown in black squares if their mutations are known to compromise activities. The caspase cleavage sites are shown in green shade. Secondary structural elements for known gasdermin structures are marked and underlined. Residues involved in site I interface are marked with magenta dots, those in site II with black dots. The black arrow indicates the minimal N-domain (residues 1–231 of hGSDMD and 1–234 of hGSDME/DFNA5) for pyroptosis induction (Rogers et al., 2017).