Abstract
Prostate cancer (CaP) incidence and mortality rates in African and African American men are greatly elevated compared to other ethnicities. This disparity is likely explained by a combination of social, environmental, and genetic factors. A large number of susceptibility loci have been reported by genome-wide association studies (GWAS), but the contribution of these loci to CaP disparities is unclear. Here we investigated the population structure of 68 previously reported GWAS loci and calculated Genetic Disparity Contribution (GDC) statistics to identify SNPs that contribute the most to differences in CaP risk across populations. By integrating GWAS results with allele frequency data, we generated Genetic Risk Scores (GRS) for 45 African and 19 non-African populations. Tests of natural selection were used to assess why some SNPs have large allele frequency differences across populations. We report that genetic predictions of CaP risks are highest for West African men and lowest for East Asian men. These differences may be explained by the out-of-Africa bottleneck and natural selection. A small number of loci appear to drive elevated CaP risks in men of African descent including rs9632117, rs6983267, rs10896449, rs10993994, and rs817826. Although most CaP-associated loci are evolving neutrally, there are multiple instances where alleles have hitchhiked to high frequencies with linked adaptive alleles. For example, a protective allele at 2q37 appears to have risen to high frequency in Europe due to selection acting on pigmentation. Our results suggest that evolutionary history contributes to the high rates of CaP in African and African American men.
Keywords: Africa, health disparities, genomic medicine, population genetics, prostate cancer
Introduction
Men of African descent suffer disproportionately from prostate cancer (CaP) compared to men of other ethnicities (1,2). The International Agency for Research on Cancer GLOBOCAN program estimates that CaP has the highest incidence of any tumor site in African-American, Caribbean, and African men (3). CaP is also the leading cause of cancer-specific mortality in African and Caribbean men, and it is second only to lung cancer as the leading cause of cancer deaths in African-American men (3). In the United States, rates of CaP are substantially higher for African-American men than those from other groups, including a 69% higher incidence rate and a 134% higher death rate relative to European-American men (4). This health disparity is likely to be explained by the complex multifactorial effects of social, environmental, and genetic factors.
CaP has the highest familial risks of any major cancer (5,6). Heritability estimates of CaP also range from 42% to 58% (7,8), indicating that there is a strong genetic component to CaP etiology. To date, over 100 CaP susceptibility loci have been identified via genome-wide association studies (GWAS), the majority of which have been identified in studies of men with European or Asian ancestry (9). Admixture mapping has identified a large genomic region at 8q24 that plays an important role in the elevated CaP risk of African-American men (10), and novel CaP associations have been detected in studies of African and African-American men (11,12). There also is evidence that rare variants contribute to a substantial fraction of prostate cancer risk within each population (13). A previous study that included seven African populations from the Human Genome Diversity Project (HGDP) reported that men of African descent have elevated risks of CaP (14). However, HGDP populations capture only a small subset of the genetic diversity that exists in sub-Saharan Africa (15), and it is unknown how much the genetic risks of CaP vary across the continent. In addition, many additional CaP susceptibility loci have been identified since this report (9,14).
Allele frequency differences at disease susceptibility loci may contribute to differences in CaP risk across populations (16,17). Large allele frequency differences can quickly arise via founder effects and population bottlenecks (18). The great reshuffling of allele frequencies that occurred during the out-of-Africa bottleneck may have contributed to health disparities and differences in CaP risk across populations (19,20). Another possibility is that allele frequencies at CaP loci changed during the forced migration of African slaves to the new world (21). However, the middle passage did not involve a severe population bottleneck (22). Local adaptation is an alternative, but not mutually exclusive, mechanism that can result in large allele frequency differences between populations (23). Selective sweeps of adaptive alleles also lead to allele frequency changes at linked loci – a process known as genetic hitchhiking (24). The late age of onset of CaP implies that disease-causing alleles are likely to have negligible fitness effects, which is likely to limit the role of natural selection acting directly on CaP susceptibility loci. Therefore, we hypothesize that genetic hitchhiking of CaP risk alleles with closely linked adaptive alleles at other loci contributes to allele frequency differences and health disparities between populations.
In the present study, we combine GWAS results with allele frequencies from a broad panel of African and non-African populations to better understand the genetic architecture of CaP risk and disparities. We address three questions: 1) Which CaP susceptibility SNPs contribute the most to differences in CaP risk across continents? 2) Which populations have the highest genetic risk of CaP? 3) How much does natural selection contribute to CaP disparities and increased risk in African men?
Materials and Methods
Population genetic data
Allele frequencies were obtained for 64 global populations. This dataset includes 648 individuals from 38 African populations collected by the laboratory of Dr. Sarah Tishkoff and 2504 individuals from 26 populations from Phase 3 of the 1000 Genomes Project (25). Populations from the Tishkoff Lab dataset were genotyped using the Illumina1M-Duo array, and populations from Phase 3 of the 1000 Genomes Project were genotyped using low-coverage whole genome sequencing. These data were combined to produce a set of 1,034,074 genotyped SNPs across 64 global populations (Supplementary Table S1, Supplementary Table 2, and Supplementary Fig. S1). Genotypes at these SNPs were observed, rather than imputed.
GWAS-identified CaP susceptibility loci
We obtained information on all known CaP susceptibility loci based on genome-wide association studies archived in the NHGRI-EBI GWAS Catalog (9). We included one additional CaP study that was not in the GWAS Catalog (26). These 26 CaP GWAS yielded 169 unique SNPs that met a p-value cutoff of 1.0 × 10−5. This p-value cutoff was chosen because of prior evidence that GRS are more accurate if less stringent cutoffs are used (27,28). After excluding eight X-linked SNPs, 161 unique autosomal SNPs remained. X-linked SNPs were excluded from our analysis because genome-wide CMS scores (see below) are only available for autosomal loci. We then filtered out SNPs that lacked odds ratio (OR) information and SNPs that were not genotyped on the Illumina1M-Duo array. When CaP susceptibility loci were identified in multiple studies, we retained the OR from the CaP study reporting the strongest association (i.e. smallest p-value). When two or more candidate SNPs were found within 100kb of each other we retained the SNP with the strongest CaP association (i.e. smallest p-value). With this approach, we winnowed the candidate CaP-associated SNPs to 68 autosomal SNPs in linkage equilibrium. 63 of these SNPs had p-values below 5 × 10−8. Ancestral vs. derived states of risk alleles were inferred from dbSNP and 1000 Genomes Project data (25,29). After controlling for strand flipping issues, risk allele frequencies were found for each SNP and population (Supplementary Table S2).
Genetic Disparity Contribution (GDC) statistics
We developed the Genetic Disparity Contribution (GDC) statistic to quantify the contribution of each SNP to differences in CaP risk across populations. Here, we use an overbar notation to distinguish beta coefficients that refer to diploid genotypes, as opposed to alleles. Assuming additive allelic effects, beta coefficients are given by:
| (1) |
where pi,j,k is the frequency of the risk allele at the ith CaP susceptibility locus in individual j from population k, and ORi refers to the OR of the ith risk allele. ORs used to calculate beta coefficients were obtained from the NHGRI-EBI GWAS Catalog (9).
We generated GDC statistics by calculating the mean difference in beta coefficients for two different populations. The GDC of the ith SNP to differences in risk between African and non-African populations is given by:
| (2) |
where is the beta coefficient associated with the ith SNP in African populations, is the beta coefficient associated with the ith SNP in non-African populations. Beta coefficients in Equation 2 use the mean frequency of the risk allele at locus i in African and non-African populations. GDC statistics were also used to quantify which SNPs contribute the most to hereditary differences in disease risks between West and East African populations.
Genetic Risk Score (GRS) calculations
Genetic Risk Scores (GRS) were calculated to yield predictions of CaP risk across populations. These scores take into account allele frequency and OR information for each SNP. Assuming that all 68 CaP susceptibility loci are in linkage equilibrium, the GRS of individual j from population k is given by:
| (3) |
where is the beta coefficient associated with the ith CaP susceptibility locus in individual j from population k. An individual who is homozygous for the protective allele at all 68 loci would have a GRS of zero (although the likelihood of this occurring is exceedingly small). For each CaP-associated SNP, we assume that risk alleles have the same effect size in each population.
To compare CaP risks in different populations and obtain GRS distributions for each population, we simulated genotypes for one million individuals per population. These simulations used population-specific allele frequency information at each CaP susceptibility locus. Assuming linkage equilibrium and independent additive effects across loci, we combined genotype and OR information to generate GRS for each simulated individual. Bootstrap analyses were then used to determine whether population ranks of GRS statistics are robust to the inclusion of particular CaP SNPs. Bootstrapped sets of 68 CaP susceptibility loci were obtained by sampling SNPs with replacement, and a total of 200 bootstrap replicates were run. We also tested if population ranks of CaP risk are robust to weighting GRS calculations by OR. Unweighted GRS statistics were calculated using the same OR for each SNP (i.e. OR = 1.167, the mean OR of all 68 CaP risk alleles). Weighted GRS statistics were calculated using ORs from the GWAS Catalog (i.e. using the reported OR for each CaP SNP). Note that the median GRS of simulated individuals from each population is highly correlated (R2 = 0.965) with the median GRS of actual individuals from each population (Supplementary Fig. S2).
ADMIXTURE and genetic ancestry components
ADMIXTURE (30) was run for 3152 genotyped individuals in 64 study populations using 90,626 autosomal SNPs. The optimal number of genetic ancestry components was found by running ADMIXTURE for k = 1 to 15 and finding the value of k that minimizes cross-validation error (k = 12). For each ancestry component and individual, GRS was plotted against the proportion of an individual’s genome that has a particular ancestry. The lm and cor functions in R were used to fit data to a linear model and calculate correlations between GRS and ancestry proportions.
Ethnicity-specific estimates of CaP risk
Age-adjusted CaP mortality and incidence rates were obtained from the United States Centers for Disease Control and Prevention and the National Cancer Institute (CDC and NCI, 2012 data) (31). None of the 64 populations in our dataset came from American Indian or Alaska Native populations, so this category was removed from our analysis. We focused on a subset of populations from the 1000 Genomes Project that are representative of different ethnicities (African: ACB and ASW; Asian/Pacific Islander: CDX, CHB, CHS, KHV, and JPT; European: CEU, FIN, GBR, IBS, and TSI; Hispanic: CLM, MXL, PEL, and PUR). After converting CaP incidence and mortality rates to a natural log scale, ethnicity-specific estimates of CaP risk were plotted vs. median GRS for each population. Constraining the slope of the line to equal one, linear least squares regression was used to fit trend lines.
Tests of positive selection
Composite of Multiple Signals (CMS) scores (32,33) were used to detect selection in populations from Europe (CEU), Asia (CHB+JPT), and Africa (YRI). CMS scores integrate multiple signatures of natural selection, including extended haplotype homozygosity, high values of FST, and elevated frequencies of derived alleles. Genome-wide CMS scores were downloaded as a UCSC Genome Browser track (https://www.broadinstitute.org/cms/results). Approximately two-thirds (611,675) of the over 1 million genotyped SNPs in our data set had CMS scores. For the remaining SNPs, we selected the closest neighboring SNP with a CMS score as a representative value. The 95th percentile of this genome-wide set of CMS scores was used as a cutoff to identify outlier SNPs that are potential targets of positive selection. Because CaP SNPs need not be the direct targets of positive selection, we also obtained the maximum CMS score per 200kb window for each continent. Note that direct targets of selection tend to have CMS scores that are among the highest in each 200kb window (33), while alleles that are subject to genetic hitchhiking can have moderate CMS scores. CMS tests have an ability to detect positive selection when selection coefficients exceed 0.5% (33).
We used Berg and Coop’s framework to test whether CaP SNPs exhibit signatures of polygenic adaptation (34). This approach uses a weighted sum of allele frequencies to estimate genetic values for each population, and it assumes that allele frequency differences among populations should be uncorrelated with the sign and magnitude of CaP effect sizes if SNPs are evolving neutrally. Berg and Coop’s approach uses the test statistic QX to quantify the among population variance in estimated genetic values that is not explained by genetic drift, and QX statistics can be viewed as a type of QST/FST comparison. We converted QX statistics into p-values by comparing QX for our set of 68 independent CaP SNPs to an empirical null distribution of putatively neutral SNPs. To generate this null distribution, we matched SNPs by 2% minor allele frequency bin and distance to the nearest known gene.
Results
CaP allele frequencies
Risk allele frequencies at many CaP loci vary across the globe (Fig. 1). On a broad scale, allele frequencies differ for African and non-African populations. These patterns are due in part to population bottlenecks that followed the out-of-Africa migration. Allele frequency differences yield genetic risks of CaP that are elevated for West African populations, intermediate for East African populations, and low for non-African populations. However, risk allele frequencies also show extensive heterogeneity within each continent. Some CaP-associated disease SNPs have small risk allele frequency differences across populations while others have marked differences in risk allele frequencies across populations. Africa is not monomorphic with respect to the genetic risk of CaP, and there are some non-African populations with higher predicted risks of CaP than African populations.
Figure 1.
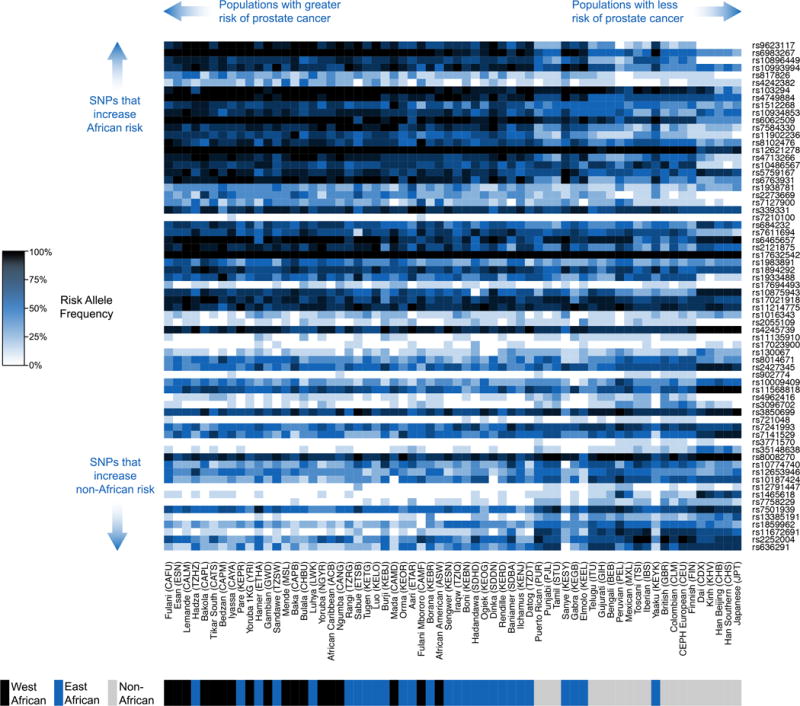
Allele frequencies at 68 independent CaP SNPs in 64 global populations. Higher frequencies of CaP risk alleles are indicated by darker shading. Autosomal CaP SNPs are ranked in terms of genetic disparity contribution (GDC) to health disparities, and populations are ranked by median genetic risk score (GRS).
Ranking SNPs by their contribution to CaP disparities
The relative contribution of different SNPs to CaP disparities depends on allele frequency differences between African and non-African populations and whether these SNPs have small or large ORs (Fig. 2). 38 out of 68 CaP SNPs have higher risk allele frequencies in African populations, and we are unable to reject the null hypothesis that this difference is due to chance (p-value = 0.3961, exact binomial test). However, nine of the 13 most divergent SNPs have higher risk allele frequencies in African populations than in non-African populations. In addition, the five SNPs with the largest ORs have higher risk frequencies in Africa. These patterns suggest that CaP health disparities between African and non-African populations may be driven by a relatively small number of SNPs. Each SNP has an ancestral allele that is shared with other primates, and a derived allele that is due to a recent mutation (35). Fig. 2 reveals that risk allele frequencies tend to be higher in Africa when risk alleles are ancestral, and risk allele frequencies tend to be higher in non-African populations when risk alleles are derived (p-value = 0.0005, Fisher’s exact test).
Figure 2.
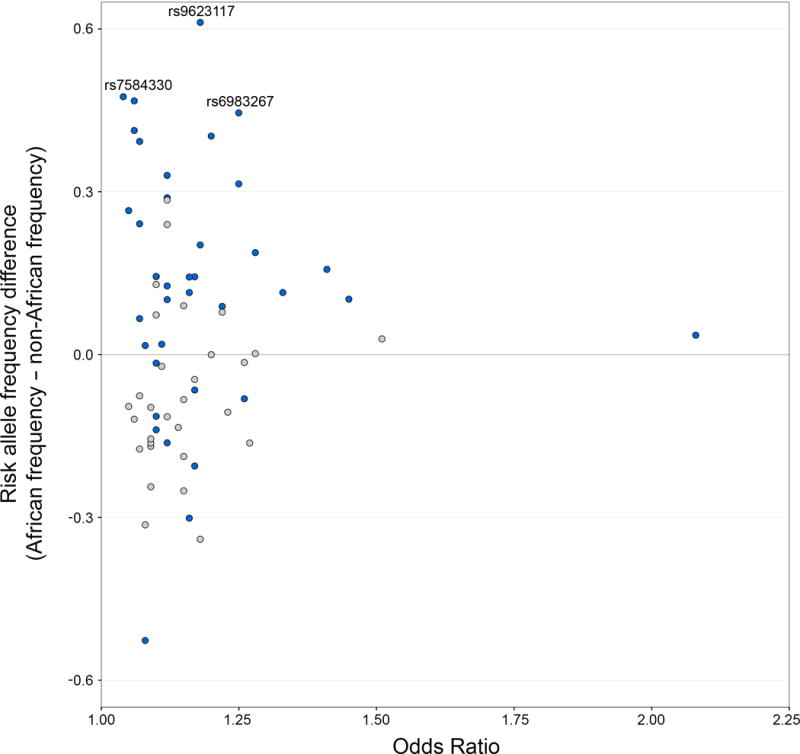
Allele frequency differences and ORs of CaP susceptibility loci. Differences in risk allele frequency between pooled African and non-African populations are plotted vs. ORs from published GWAS data (positive y-axis values indicate elevated risk in African populations). SNPs where the ancestral allele increases risk are labelled blue and SNPs where the derived allele increases risk are labelled gray.
A small number of loci appear to have a disproportionately large contribution to elevated CaP risks in African men (Fig. 3A). Some of these SNPs have risk allele frequencies that are over 40% higher in Africa than outside of Africa (e.g. rs7584330 near MLPH and rs9623117 near TNRC6B) while others have alleles of large effect (e.g. rs817826 near RAD23B and KLF4, which has an OR of 1.41). We developed a novel statistic, the Genetic Disparity Contribution (GDC), to rank order SNPs by how much they contribute to increased CaP risk in Africa. GDC scores are a function of ORs and the relative frequencies of risk alleles in African and non-African populations. SNPs with positive GDC scores have risk allele frequencies that are higher in African populations than non-African populations. SNPs that contribute the most to increased risk in African populations include rs9623117 at 22q13.1, rs6983267 at 8q24.1, rs10896449 at 11q13.3, rs10993994 at 10q11.23, and rs817826 at 9q31.2. Note that rs10993994 is near MSMB, which encodes PSP94, a major protein secreted by the prostate (36). Conversely, some SNPs increase the risk of CaP in non-African populations, including rs636291 at 1p36.22 and rs2252004 at 10q26.21. Most SNPs have minimal allele frequency differences between continents, and these SNPs are unlikely to make a major contribution to health disparities. Continental differences in CaP risk disappear if the six SNPs that contribute the most to elevated CaP risk in African men are ignored (i.e. the sum of 62 remaining GDC scores is close to zero: 0.0099). We also examined whether the SNPs that contribute most to CaP risk differences between African and non-African populations also contribute to differences in risk between West Africa and East Africa. Overall, GDC statistics between continents were positively correlated with GDC statistics within Africa (r2 = 0.217, Supplementary Fig. S3). Many of the same SNPs contribute to health disparities on multiple spatial scales.
Figure 3.
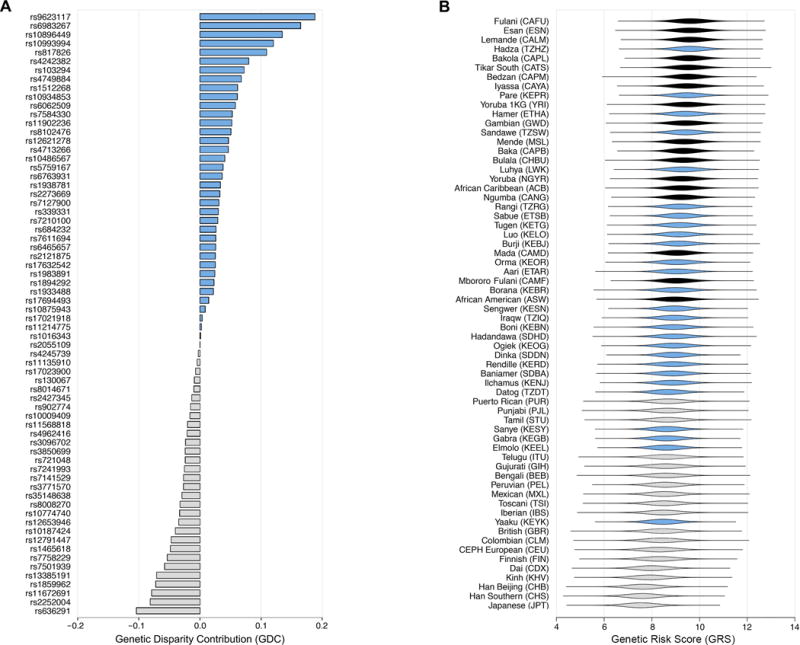
Relative contributions of different SNPs to African CaP disparities and predicted genetic risks of CaP in different populations. A, SNPs are ranked in terms of genetic disparity contribution (GDC) to health disparities, i.e. whether a SNP increases CaP risk in African populations relative to non-African populations. SNPs that have greater risk allele frequencies in African populations are labelled blue and SNPs that have greater risk allele frequencies in non-African populations are colored gray. B, Predicted risks of CaP are highest for West African populations and lowest for non-African populations. One million individuals per populations were simulated to generate distributions of predicted genetic risk, and populations are ranked by median genetic risk score (GRS). West African populations are labelled black, East African populations are labelled blue, and non-African populations are colored gray.
Genetic risk prediction in different populations
Using computer simulations, we calculated genetic risk scores (GRS) for one million simulated individuals from each population and ranked populations by median GRS (Fig. 3B). Here, we define an individual’s GRS as equal to the sum of single locus beta coefficients. Populations with higher median GRS scores have a higher predicted risk of CaP.
Overall, populations from West Africa have the highest predicted risk, followed by East African populations, and then non-African populations. Seven of the eight populations with the highest predicted risk are located in West Africa and the nine populations with the lowest predicted risk are non-African. Focusing on extreme populations, the Fulani from Cameroon have the highest median GRS and Japanese from Tokyo have the lowest median GRS. East African populations with the highest predicted risk include the Pare and Luhya of Kenya (which share Bantu ancestry with West-African populations (37)), as well as the Hadza of Tanzania. The Yaaku from Kenya have the lowest predicted risk of CaP of any East African population. Among non-African populations, predicted risk tends to be greater for South Asian populations, intermediate for European populations, and lower for East Asian populations. The GRS of different populations show substantial overlap. For example, although African-Americans (ASW) tend to have a higher predicted risk than Northern-Europeans (CEU), there is a 24.9% chance that an ASW genome will have a lower GRS than a CEU genome. Note that GRS only measure genetic contributions to disease risk, and it is likely that other factors, including environmental exposures, also contribute to CaP risk.
Populations with the highest predicted risk of CaP are enriched for specific genetic ancestry components. An ADMIXTURE plot of 3152 genotyped individuals reveals the effects of population structure (Fig. 4A). We find that orange (r = 0.4113, Fig. 4B), maroon (r = 0.3785, Fig. 4C), and green (r = 0.2096, Fig. 4D) ancestry components have the strongest positive correlation with GRS. These ancestry components are common in West African populations. Light purple (Fig. 4E), yellow (Fig. 4F), dark purple (Fig. 4G), dark blue (Fig. 4H), dark pink (Fig. 4I), and red (Fig. 4J) ancestry components have minimal effects. Conversely, light blue (r = 0.1791, Fig 4K), light pink (r = −0.3000, Fig. 4L) and light gray (r = −0.3963, Fig 4M) ancestry components exhibit the strongest negative correlation with GRS. Ancestry proportions vary within each population, and the extent of this heterogeneity reflects whether populations have experienced recent admixture (38). We note that the genetic risk of CaP for African Caribbean and African American individuals depends on the relative proportion of African (orange and maroon) and European (light blue) ancestry in each genome.
Figure 4.
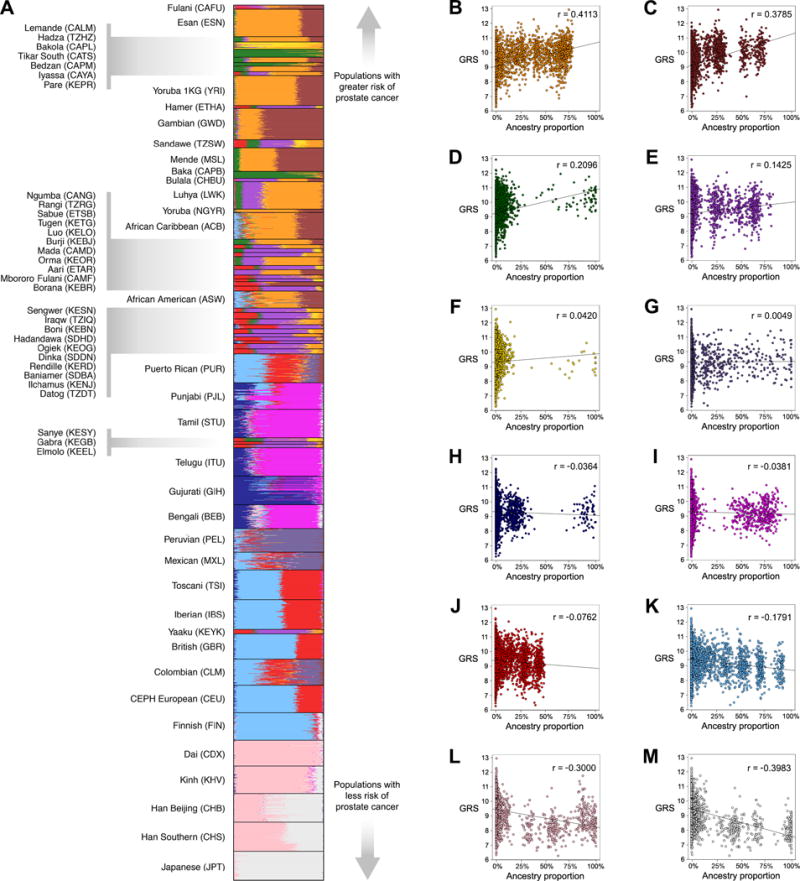
CaP risks vary for different genetic ancestries. A, ADMIXTURE plot (k = 12) of 3152 genotyped individuals. Each color corresponds to a different genetic ancestry component. Individuals are grouped by study population, and study populations are ranked by median GRS. B-M, GRS are plotted against the proportion of each individual’s genome that has a particular ancestry component. Data were fit to a linear model, and correlation coefficients indicate the extent to which each ancestry component is associated with GRS.
To test whether genetic information accurately predicts ethnicity-specific clinical disease risks we plotted median GRS vs. age-standardized CaP death and incidence rates from the United States (31). In general, we find that GRS predictions follow the same trend as public health data: African-American populations have the highest GRS and death rates, European and Hispanic populations have intermediate GRS and death rates, and Asian populations have the lowest GRS and death rates. Furthermore, genetic risk scores capture the magnitude of ethnic differences in clinical disease risks (Fig. 5). There is a strong positive relationship between GRS and CaP mortality (residual standard error = 0.1816, 15 df, Fig. 5A). Differences in GRS within each ethnicity appear to be due to admixture and different amounts of African ancestry components. There is also a strong positive relationship between GRS and age-standardized estimates of CaP incidence (residual standard error = 0.1862, 15 df, Fig. 5B). Taken together, our results indicate that GRS successfully quantify ethnic differences in CaP incidence and mortality rates.
Figure 5.
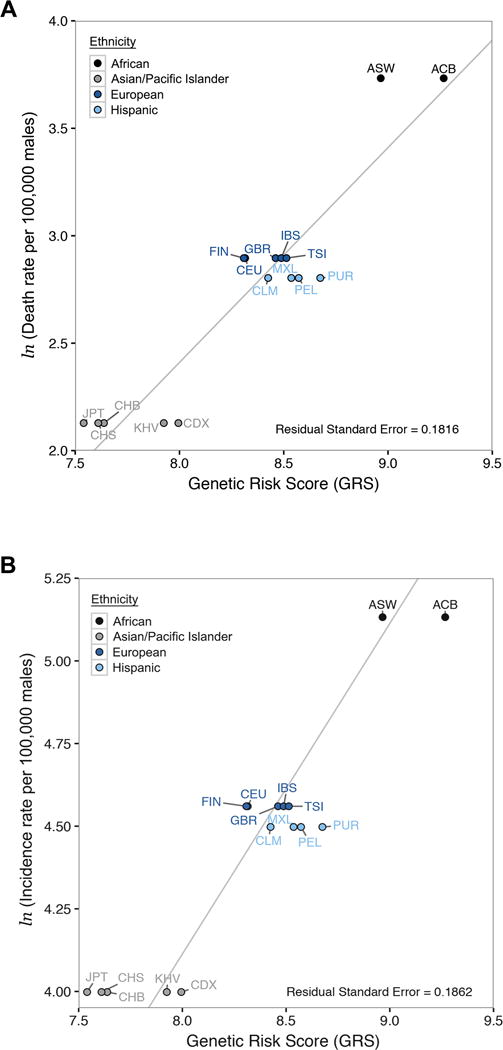
Strong concordance between ethnicity-specific estimates of CaP risk and GRS predictions. Age-adjusted CaP death rates and incidence rates from the United States were compared to the median GRS of each population. Ethnicity-specific data from CDC and NCI are grouped by ethnicity. The solid line in each panel is the best fit to data after constraining the slope to equal one. A, CaP death rates (natural log scale) vs. median GRS. B, CaP incidence rates (natural log scale) vs. median GRS
Tests of positive selection
Because local adaptation can result in large allele frequency differences between populations, we tested whether CaP hits were under selection in CEU, YRI, or JPT+CHB populations using the Composite of Multiple Signals (CMS) test (32,33). CMS scores combine several tests of positive selection into a single metric, with higher CMS scores indicating SNPs that are more likely to be adaptive. To distinguish between CaP loci that are direct targets of selection as opposed to linked hitchhiking loci, we found the CMS score nearest to each GWAS SNP and the largest CMS score within a 200kb window centered around each GWAS SNP (Fig. 6A). SNPs that are direct targets of selection are expected to be among the SNPs with the largest CMS scores found in any given window (33).
Figure 6.
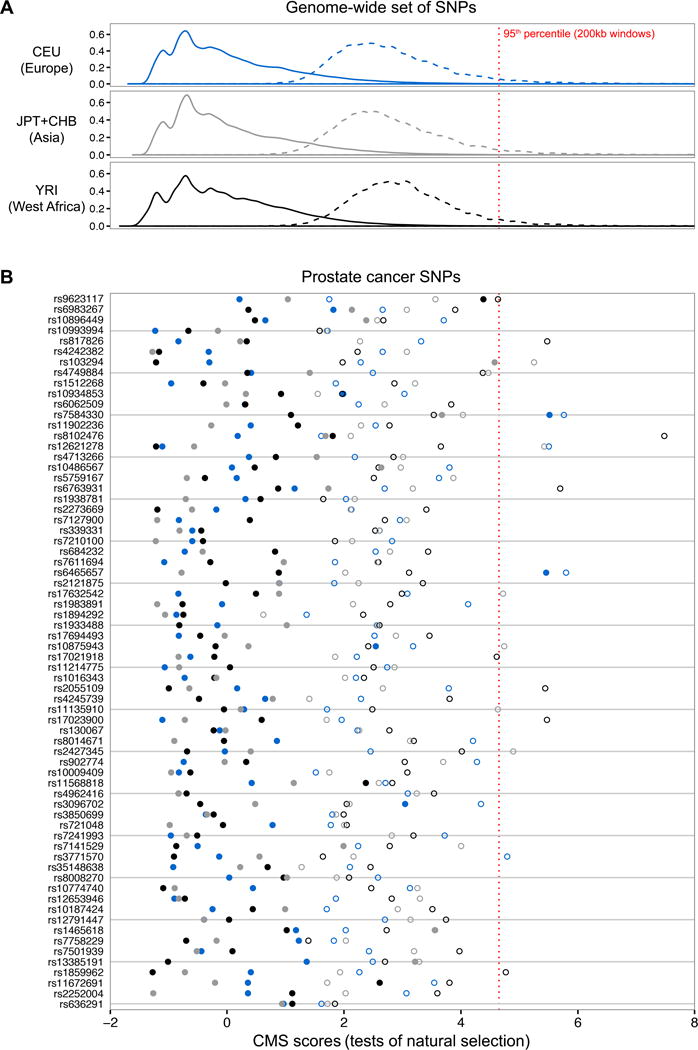
Scans of selection reveal that most CaP hits are in neutrally evolving regions of the human genome. Solid lines and filled circles indicate CMS (Composite of Multiple Signals (33)) scores of individual SNPs. Dashed lines and open circles indicate the maximum CMS score within a 200kb window centered around each SNP. SNPs that are direct targets of selection tend to have the highest CMS score in a 200kb window, and hitchhiking alleles have intermediate scores. West African (YRI) CMS scores are labelled black, European (CEU) CMS scores are labelled blue, and Asian (CHB+JPT) CMS scores are labelled gray. A, CMS score distributions for a genome-wide set of one million SNPs on the Illumina1M-Duo array. The 95th percentile of maximum CMS scores for each 200kb window are indicated by a dashed red line. B, CMS scores for CaP susceptibility loci (ordered by GDC statistics). A subset of CaP SNPs exhibit signatures of genetic hitchhiking.
We find that the majority of CaP-associated loci lie in neutrally evolving regions of the human genome (Fig. 6B). However, there are multiple instances where natural selection appears to have contributed to CaP disparities. CaP SNPs with the highest Northern European CMS scores include rs6465657 at 7q21.3 and rs7584330 at 2q37.3, CaP SNPs with the highest East Asian CMS scores include rs103294 at 19q13.42 and rs1465618 at 2p21, and CaP SNPs with the highest West African CMS scores include rs9623117 at 22q13.1 and rs11672691 at 19q13.2. We note that there are multiple CaP SNPs that have low CMS scores, but are found in genomic regions with high CMS scores. This is consistent with genetic hitchhiking of disease SNPs. Further support for the genetic hitchhiking hypothesis comes from the fact that there are no instances of a CaP SNP having the highest CMS score in its 200kb window.
We also tested whether CaP SNPs exhibit signatures of polygenic adaptation using the approach of Berg and Coop (34). For our collective set of 68 unlinked CaP SNPs, we find that the cumulative variance in CaP risk among African populations is not significantly higher than expected by genetic drift alone (p-value = 0.429, QX = 51.3). Qx statistics measure the extent to which trait-associated SNPs vary more across population than one would expect from genetic drift. Although some CaP SNPs are outliers with large allele frequency differences between populations, the overall set of CaP SNPs resembles the rest of the genome. Berg and Coop’s approach can also be used to determine if groups of populations have genetic values that deviate from neutral expectations (34). Focusing on different geographic regions of Africa, we find that East African populations have lower CaP risks than expected by genetic drift alone (p-value = 0.019, Z-score = −2.339) and that West African populations have slightly higher CaP risks than expected by drift alone (p-value = 0.087, Z-score = 1.711). This lends additional support to our inference that the genetic risk of CaP in East Africans differs from that of West Africans.
Evaluation of underlying assumptions
There are some caveats to our approach, including the use of published ORs and the fact that most reported CaP susceptibility loci have been identified in non-African populations (39). The GWAS Catalog provides reported ORs for study populations in which associations were detected, but these alleles may confer different ORs in other (yet unstudied) populations. To address the potential for OR heterogeneity across populations, we calculated GRS two different ways. Weighted GRS calculations used ORs reported in the GWAS Catalog, and unweighted GRS calculations assumed that each disease SNP confers the same OR magnitude of association on CaP risk in all populations. Comparing these two approaches, we find that the rank order of populations from different geographic regions is broadly conserved regardless of the OR used, and that GRS are highly correlated for both approaches (R2 = 0.967, Supplementary Fig. S4). We also tested whether GRS predictions are robust to inclusion of specific CaP SNPs via bootstrapping. West African populations consistently had higher predicted risk than East African populations (97.5% bootstrap support), and East African populations consistently had higher predicted risk than non-African (99.5% bootstrap support). The rank order of CaP risk for individual populations depends upon which SNPs are included in GRS calculations. Although Fulani (CAFU) had the highest median GRS when all 68 CaP SNPs were included, only 3% of the bootstrap runs resulted in Fulani having the highest GRS. By contrast, 25% of the bootstrap runs resulted in Lemande (CALM) having the highest median GRS. Finally, we note that the set of known disease loci is incomplete. In particular, European and Asian GWAS are likely to miss CaP susceptibility loci that have intermediate frequency alleles in African populations and rare alleles in non-African populations. To the extent that additional CaP SNPs will be found, disease risks may be underestimated for men of African descent in our current analysis. Nevertheless, we note that there is a striking concordance between our GRS predictions and clinical estimates of disease risk (Fig. 5).
Discussion
Like most of the genome, the majority of CaP susceptibility loci evolve neutrally with moderate allele frequency differences between populations. However, a small number of SNPs make a disproportionally large contribution to population-level differences in the genetic risks of CaP. Characteristics of these loci include larger effect sizes as estimated in GWAS studies and large allele frequency differences between African and non-African populations (Fig. 3). A relatively small number of SNPs have large GDC statistics, which in part explains how genetic health disparities can arise for a polygenic disease like CaP. We note that SNPs with the highest GDC statistics need not be same SNPs that contribute the most to heritability within a single population. Both neutral and selective evolutionary mechanisms appear to have contributed to disparities in the genetic risk of CaP. These mechanisms include founder effects due to the out-of-Africa migration and genetic hitchhiking of disease susceptibility alleles with locally adaptive alleles.
Here we highlight three genomic regions that contribute to CaP health disparities: 2q37, 22q13, and 8q24. rs7584330 is a CaP SNP that is located at 2q37. For the populations studied in this paper, the mean frequency of the risk allele at rs7584330 is 47% higher in African populations compared to non-African populations. European CMS scores indicate that there is evidence of positive selection at 2q37 (Supplemental Fig. S5). This region contains the melanophilin (MLPH) and prolactin releasing hormone (PRLH) genes. MLPH mutations have been associated with diluted skin and hair pigmentation (40), and signatures of positive selection at 2q37 are consistent with previous studies that suggest variation at pigmentation genes is adaptive in non-African populations (41,42). The PRLH gene encodes prolactin-releasing peptide, and levels of prolactin are known to affect male fertility (43). These data suggest that a haplotype that protects against CaP has hitchhiked to high frequencies in Europe as an incidental byproduct of linkage and selection for lighter pigmentation.
We also observed risk allele frequencies differences and signatures of selection at CaP susceptibility loci located in the 22q13 region, which contains the CaP susceptibility locus rs9623117. For the populations studied in this paper, the mean frequency of the risk allele at rs9623117 is 61% higher in African populations compared to non-African populations. Elevated CMS scores at 22q13 suggest that there may have been recent positive selection in West Africa and East Asia (Supplementary Fig. S6). rs9623117 is close to the gene TNRC6B, the products of which play a role in RNA-mediated gene silencing (44). However, it is unclear what trait is responsible for the signatures of positive selection near rs9623117.
The 8q24 locus has been repeatedly implicated in CaP and other cancers. In this region alone, there are at least three independent CaP susceptibility loci and 21 additional CaP SNPs that have been excluded from our analysis of 68 SNPs due to linkage disequilibrium. Two of the independent 8q24 CaP loci have much higher risk allele frequencies in African populations (rs6983267and rs4242382) while a third locus has only a small contribution to health disparities, as indicated by a GDC statistic that is close to zero (rs1016343). Some of the excluded CaP SNPs in 8q24 have large effect sizes (e.g. rs116041037, rs188140481, and rs4242384 have ORs that exceed 1.8). The 8q24 region contains MYC, an oncogene that encodes a transcription factor that is associated with many human cancers (45). However, despite the importance of 8q24 to CaP risk and disparities, CMS scores do not reveal evidence of recent positive selection in this genomic region (Supplementary Fig. S7).
We report that the predicted genetic risks of CaP vary geographically, with African populations showing the highest risk. This is consistent with existing evidence that African ancestry is enriched in African-American CaP cases relative to African-American controls (10). Within Africa, GRS are highest for men of West African descent. However, there is substantial heterogeneity in GRS within and between populations. This heterogeneity is particularly relevant to the health of African-Americans, as individuals can trace their ancestry back to many different African and non-African source populations (15,46). We also note that ASW individuals represent only a subset of the diversity found in African-American populations. Finally, we note that Fulani who have settled in West African towns (CAFU) have higher GRS statistics than Mbororo Fulani (CAMF) who are nomadic pastoralists. Differences in risk among Fulani individuals may be due to differential admixture with West African populations that have Bantu ancestry (15).
Although GRS are lower for East African populations than West African populations, GLOBOCAN estimates of CaP incidence and mortality rates are similar for East African and West African countries (3). This pattern may be explained by environmental risk factors that have a larger contribution to CaP in East African populations, or because GLOBOCAL estimates of CaP incidence and mortality are based on limited data. Furthermore, we note that having a high genetic risk score does not guarantee that an individual will develop CaP. Genotype-by-environment interactions and socioeconomic factors also contribute to CaP risks and health disparities (47).
Going forward, there is a need to undertake GWAS and linkage studies in African populations to identify loci that may not be detectable in non-Africans, translate raw genetic risk scores into estimates of absolute risk (48), and identify the causal alleles in genomic regions that are associated with CaP (49,50). Future studies will increase our understanding of health disparities and the genetics of CaP as additional susceptibility loci are found in Africa.
Supplementary Material
Acknowledgments
We thank C. Hanson, M. Harlemon, M.S. Kim, and U. Martinez-Marigorta for helpful discussions during the writing of this paper. J. Lachance and T. R. Rebbeck are supported in part by NIH/NCI grant U01CA184374. M. E. B. Hanson was funded by a CEET training grant (T32ES019851). This work was also supported by NIH grant DP1ES022577 to S. A. Tishkoff. J. Lachance and T. R. Rebbeck are members of the MADCaP network (Men of African Descent, Carcinoma of the Prostate, https://www.madcapnetwork.org/).
Financial support: J. Lachance and T. R. Rebbeck are supported in part by NIH/NCI grant U01CA184374. M. E. B. Hanson was funded by a CEET training grant (T32ES019851). This work was also supported by NIH grant DP1ES022577 to S. A. Tishkoff. J. Lachance and T. R. Rebbeck are members of the MADCaP network (Men of African Descent, Carcinoma of the Prostate, https://www.madcapnetwork.org/).
Footnotes
Conflict of interest: The authors have no conflicts of interest to report.
References
- 1.Rebbeck TR, Devesa SS, Chang BL, Bunker CH, Cheng I, Cooney K, et al. Global patterns of prostate cancer incidence, aggressiveness, and mortality in men of african descent. Prostate Cancer. 2013;2013:560857. doi: 10.1155/2013/560857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Odedina FT, Akinremi TO, Chinegwundoh F, Roberts R, Yu D, Reams RR, et al. Prostate cancer disparities in Black men of African descent: a comparative literature review of prostate cancer burden among Black men in the United States, Caribbean, United Kingdom, and West Africa. Infect Agent Cancer. 2009;4(Suppl 1):S2. doi: 10.1186/1750-9378-4-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, et al. Annual Report to the Nation on the Status of Cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122:1312–37. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemminki K, Sundquist J, Bermejo JL. How common is familial cancer? Annals of Oncology. 2008;19:163–7. doi: 10.1093/annonc/mdm414. [DOI] [PubMed] [Google Scholar]
- 6.Bruner DW, Moore D, Parlanti A, Dorgan J, Engstrom P. Relative risk of prostate cancer for men with affected relatives: systematic review and meta-analysis. Int J Cancer. 2003;107:797–803. doi: 10.1002/ijc.11466. [DOI] [PubMed] [Google Scholar]
- 7.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 8.Hjelmborg JB, Scheike T, Holst K, Skytthe A, Penney KL, Graff RE, et al. The heritability of prostate cancer in the Nordic Twin Study of Cancer. Cancer Epidemiol Biomarkers Prev. 2014;23:2303–10. doi: 10.1158/1055-9965.EPI-13-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdett T, Hall PN, Hasting E, Hindorff LA, Junkins HA, Klemm AK, et al. The NHGRI-EBI Catalog of published genome-wide association studies. Aug 3; doi: 10.1093/nar/gkw1133. < http://www.ebi.ac.uk/gwas>. Accessed 2016 August 3. [DOI] [PMC free article] [PubMed]
- 10.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook MB, Wang Z, Yeboah ED, Tettey Y, Biritwum RB, Adjei AA, et al. A genome-wide association study of prostate cancer in West African men. Hum Genet. 2014;133:509–21. doi: 10.1007/s00439-013-1387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–3. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mancuso N, Rohland N, Rand KA, Tandon A, Allen A, Quinque D, et al. The contribution of rare variation to prostate cancer heritability. Nat Genet. 2016;48:30–5. doi: 10.1038/ng.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corona E, Chen R, Sikora M, Morgan AA, Patel CJ, Ramesh A, et al. Analysis of the genetic basis of disease in the context of worldwide human relationships and migration. PLoS Genet. 2013;9:e1003447. doi: 10.1371/journal.pgen.1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–44. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–94. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Maitland ML, DiRienzo A, Ratain MJ. Interpreting disparate responses to cancer therapy: the role of human population genetics. J Clin Oncol. 2006;24:2151–7. doi: 10.1200/JCO.2005.05.2282. [DOI] [PubMed] [Google Scholar]
- 18.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 19.Blair LM, Feldman MW. The role of climate and out-of-Africa migration in the frequencies of risk alleles for 21 human diseases. BMC Genet. 2015;16:81. doi: 10.1186/s12863-015-0239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofer T, Ray N, Wegmann D, Excoffier L. Large allele frequency differences between human continental groups are more likely to have occurred by drift during range expansions than by selection. Ann Hum Genet. 2009;73:95–108. doi: 10.1111/j.1469-1809.2008.00489.x. [DOI] [PubMed] [Google Scholar]
- 21.Jin W, Xu S, Wang H, Yu Y, Shen Y, Wu B, et al. Genome-wide detection of natural selection in African Americans pre- and post-admixture. Genome Res. 2012;22:519–27. doi: 10.1101/gr.124784.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eltis D, Richardson D. Atlas of the transatlantic slave trade. New Haven: Yale University Press; 2010. [Google Scholar]
- 23.Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat Rev Genet. 2009;10:745–55. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 25.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akamatsu S, Takata R, Haiman CA, Takahashi A, Inoue T, Kubo M, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44:426–9. S1. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- 27.International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagenaars SP, Hill WD, Harris SE, Ritchie SJ, Davies G, Liewald DC, et al. Genetic prediction of male pattern baldness. PLoS Genet. 2017;13:e1006594. doi: 10.1371/journal.pgen.1006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.US Cancer Statistics Working Group. United States Cancer Statistics: 1999–2012 Incidence and Mortality Web-based Report. Atlanta: US Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2015. [Google Scholar]
- 32.Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–13. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grossman SR, Shlyakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–6. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 34.Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014;10:e1004412. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lachance J. Disease-associated alleles in genome-wide association studies are enriched for derived low frequency alleles relative to HapMap and neutral expectations. BMC Med Genomics. 2010;3:57. doi: 10.1186/1755-8794-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 37.Li S, Schlebusch C, Jakobsson M. Genetic variation reveals large-scale population expansion and migration during the expansion of Bantu-speaking peoples. Proc Biol Sci. 2014:281. doi: 10.1098/rspb.2014.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verdu P, Rosenberg NA. A general mechanistic model for admixture histories of hybrid populations. Genetics. 2011;189:1413–26. doi: 10.1534/genetics.111.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virlogeux V, Graff RE, Hoffmann TJ, Witte JS. Replication and heritability of prostate cancer risk variants: impact of population-specific factors. Cancer Epidemiol Biomarkers Prev. 2015;24:938–43. doi: 10.1158/1055-9965.EPI-14-1372. [DOI] [PubMed] [Google Scholar]
- 40.Menasche G, Ho CH, Sanal O, Feldmann J, Tezcan I, Ersoy F, et al. Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1) J Clin Invest. 2003;112:450–6. doi: 10.1172/JCI18264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hider JL, Gittelman RM, Shah T, Edwards M, Rosenbloom A, Akey JM, et al. Exploring signatures of positive selection in pigmentation candidate genes in populations of East Asian ancestry. BMC Evol Biol. 2013;13:150. doi: 10.1186/1471-2148-13-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myles S, Somel M, Tang K, Kelso J, Stoneking M. Identifying genes underlying skin pigmentation differences among human populations. Hum Genet. 2007;120:613–21. doi: 10.1007/s00439-006-0256-4. [DOI] [PubMed] [Google Scholar]
- 43.Gill-Sharma MK. Prolactin and male fertility: the long and short feedback regulation. Int J Endocrinol. 2009;2009:687259. doi: 10.1155/2009/687259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol Cell Biol. 2009;29:4144–55. doi: 10.1128/MCB.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baharian S, Barakatt M, Gignoux CR, Shringarpure S, Errington J, Blot WJ, et al. The Great Migration and African-American Genomic Diversity. PLoS Genet. 2016;12:e1006059. doi: 10.1371/journal.pgen.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taksler GB, Keating NL, Cutler DM. Explaining racial differences in prostate cancer mortality. Cancer. 2012;118:4280–9. doi: 10.1002/cncr.27379. [DOI] [PubMed] [Google Scholar]
- 48.Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han Y, Hazelett DJ, Wiklund F, Schumacher FR, Stram DO, Berndt SI, et al. Integration of multiethnic fine-mapping and genomic annotation to prioritize candidate functional SNPs at prostate cancer susceptibility regions. Hum Mol Genet. 2015;24:5603–18. doi: 10.1093/hmg/ddv269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gusev A, Shi H, Kichaev G, Pomerantz M, Li F, Long HW, et al. Atlas of prostate cancer heritability in European and African-American men pinpoints tissue-specific regulation. Nat Commun. 2016;7:10979. doi: 10.1038/ncomms10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


