Staphylococcus aureus is a major human pathogen worldwide in both community and health care settings. Surveillance for S. aureus strains is important to our understanding of their spread and to informing infection prevention and control. Confusion surrounding the strain nomenclature of one of the most prevalent lineages of S. aureus, clonal complex 8 (CC8), and the imprecision of current tools for typing S. aureus make surveillance and source tracing difficult and sometimes misleading. In this study, we clarify the CC8 strain designations and propose a new typing scheme for CC8 isolates that is rapid and easy to use. This typing scheme is based on relatively stable genomic markers, and we demonstrate its superiority over traditional typing techniques. This scheme has the potential to greatly improve epidemiological investigations of S. aureus.
KEYWORDS: CC8, MRSA, MSSA, phylogeny, Staphylococcus aureus, strain typing, assay development, whole genome
ABSTRACT
Strains of Staphylococcus aureus in clonal complex 8 (CC8), including USA300, USA500, and the Iberian clone, are prevalent pathogens in the United States, both inside and outside health care settings. Methods for typing CC8 strains are becoming obsolete as the strains evolve and diversify, and whole-genome sequencing has shown that some strain types fall into multiple sublineages within CC8. In this study, we attempt to clarify the strain nomenclature of CC8, classifying the major strain types based on whole-genome sequence phylogenetics using both methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) genomes. We show that isolates of the Archaic and Iberian clones from decades ago make up the most basal clade of the main CC8 lineages and that at least one successful lineage of CC8, made up mostly of MSSA, diverged before the other well-known strain types USA500 and USA300. We also show that the USA500 type includes two clades separated by the previously described “Canadian epidemic MRSA” strain CMRSA9, that one clade containing USA500 also contains the USA300 clade, and that the USA300-0114 strain type is not a monophyletic group. Additionally, we present a rapid, simple CC8 strain-typing scheme using real-time PCR assays that target single nucleotide polymorphisms (SNPs) derived from our CC8 phylogeny and show the significant benefit of using more stable genomic markers based on evolutionary lineages over traditional S. aureus typing techniques. This more accurate and accessible S. aureus typing system may improve surveillance and better inform the epidemiology of this very important pathogen.
IMPORTANCE Staphylococcus aureus is a major human pathogen worldwide in both community and health care settings. Surveillance for S. aureus strains is important to our understanding of their spread and to informing infection prevention and control. Confusion surrounding the strain nomenclature of one of the most prevalent lineages of S. aureus, clonal complex 8 (CC8), and the imprecision of current tools for typing S. aureus make surveillance and source tracing difficult and sometimes misleading. In this study, we clarify the CC8 strain designations and propose a new typing scheme for CC8 isolates that is rapid and easy to use. This typing scheme is based on relatively stable genomic markers, and we demonstrate its superiority over traditional typing techniques. This scheme has the potential to greatly improve epidemiological investigations of S. aureus.
INTRODUCTION
Staphylococcus aureus causes infection in both immunocompromised and healthy persons and in both health care and community settings. In the United States, most of the community-associated methicillin-resistant S. aureus (CA-MRSA) infections and a significant proportion of health care-associated (HA) infections are caused by strains in clonal complex 8 (CC8) (1–3). CC8 methicillin-susceptible S. aureus (MSSA) strains are also common agents of infection (4–6). Lineages within CC8 include the major so-called epidemic “clones” USA300, USA500, Archaic, Iberian, and the lineage identified by multilocus sequence typing as sequence type 239 (ST239) (7). ST239 is an HA lineage with distinct populations distributed throughout Asia, in Eastern Europe, South America, and Australia (1, 8, 9). ST239, a hybrid of strains ST8 and ST30 (10), is often classed in CC30, given its distant relationship to the rest of CC8 and its spa gene type similarity to ST30 isolates. The Archaic (ST250) and Iberian (ST247) strains are also HA; the Archaic clone was widespread in parts of Europe decades ago; however, it has largely disappeared with the appearance of other more antimicrobial-resistant CC8 lineages such as USA500 (11). Additional strains have also emerged and waned over time—often in geographically limited areas (12, 13) (e.g., the Hanover clone, ST254), adding to CC8’s epidemiological complexity. The CA-MRSA strain USA300 emerged clinically only around 2000 and is currently the most prevalent pathogenic strain circulating in the United States (2, 3). Despite its relatively recent emergence, USA300 has diverged into lineages distinct from its early branching ancestors (called the “Early Branching” clade here) (14). USA300 variants that lack the arginine catabolic mobile element (ACME) characteristic of USA300 strains have more recently been isolated in South American countries. These USA300-LV (for Latin American variant) isolates instead carry a copper and mercury resistance cassette, COMER, and were shown to belong to a monophyletic “sister” lineage, named USA300-SAE (for South American epidemic strain), to the USA300 North American epidemic (USA300-NAE) strain (14).
Distinguishing among the sublineages of CC8 is critical for purposes of epidemiology and surveillance, especially as the epidemiological separation between HA and CA strains disappears (1). Although strain-typing techniques have improved over time, they still have many limitations. Pulsed-field gel electrophoresis (PFGE), the method by which the “USA” strains were originally defined (15), is laborious, and determination of a strain type can be subjective. Heterogeneity in banding patterns and discordance with other typing methods is not uncommon (16). Sequencing and interpretation of the spa gene is relatively expensive, and spa types are not always consistent with evolutionary lineages (8, 16–19). Furthermore, PFGE and spa typing alone are often not able to distinguish among sublineages within CC8 or other clonal complexes (20). Currently, many laboratories use PCR typing that targets factors located on mobile genetic elements: e.g., Panton-Valentine leukocidin (PVL) genes, ACME genes, enterotoxin genes, and the SCCmec variants.
Confounding the issue is the multitude of names given to a strain type (21) and the use of one name for divergent strain types. The Iberian name identified a CC8 lineage that circulated in Europe in the 1990s, ST247 (22), but due to some shared genetic elements used for strain typing, “Iberian” has been used more recently to identify an ST8 strain closely related to USA500 (4). This confusion extends to the phylogenetic relatedness among the major strain types in CC8. Relatively imprecise methods of strain characterization and lack of consistency with regard to reference isolates have resulted in variation in the classification of the CC8 sublineages. Most strains were originally defined and deposited in repositories prior to the routine use of whole-genome sequencing (WGS) and WGS-based phylogenies, and relatedness to these type strains was inferred based on various criteria, resulting in inconsistent application of strain nomenclature. An influential study by Li et al. (7) on the evolution of virulence in CC8 suggested that USA300 is a lineage derived from USA500. In that study, the authors identified a now widely used set of genetic markers to distinguish between USA500 and Iberian strains, using a USA500 reference isolate called BD02-25. Two recent studies refuted the idea that a USA500 strain is the progenitor to USA300 using different USA500 isolate genomes as references: Jamrozy et al. (23) used 2395, originally described in a study on hypervirulence in a USA500 isolate (typing method unknown) (24), and Boyle-Vavra et al. (25) used NRS385 (aka 95938), the USA500 type strain described by McDougal in 2003 (15) (deposited at BEI Resources as USA500, catalog no. NR-46071). We postulate that not all of these isolates belong to the same phylogenetic clade, although they were previously described as the same strain, USA500.
In this study, our first goal was to closely examine the cladistics of CC8 with whole-genome sequence (WGS) data, illustrating the issues that have arisen from lack of consistency in type nomenclature, with the hopes of more clearly defining CC8 sublineages. Second, as significantly fewer studies address MSSA than MRSA, our goal was to gain a better understanding of MSSA’s role in CC8 epidemiology and its placement in the CC8 population structure. Our third goal was to develop a rapid and simple, yet robust strain-typing scheme based on more stable genomic markers: i.e., real-time PCR assays targeting canonical single nucleotide polymorphisms (canSNPs) or SNPs that define a lineage (20, 26). We hypothesized that as S. aureus shows clonal evolution with little evidence of recombination within lineages (8, 17–19), we could identify canSNPs from our CC8 phylogeny to target each of the major lineages, including the widely circulating USA300 subtype USA300-0114, an oft-cited etiologic cause for MRSA clusters (27). A canSNP-based approach eliminates the lineage confusion seen with PFGE, spa typing, and mobile genetic marker typing, as SNPs are relatively stable and quantify relatedness among strains.
RESULTS
Whole-genome phylogenetic analysis.
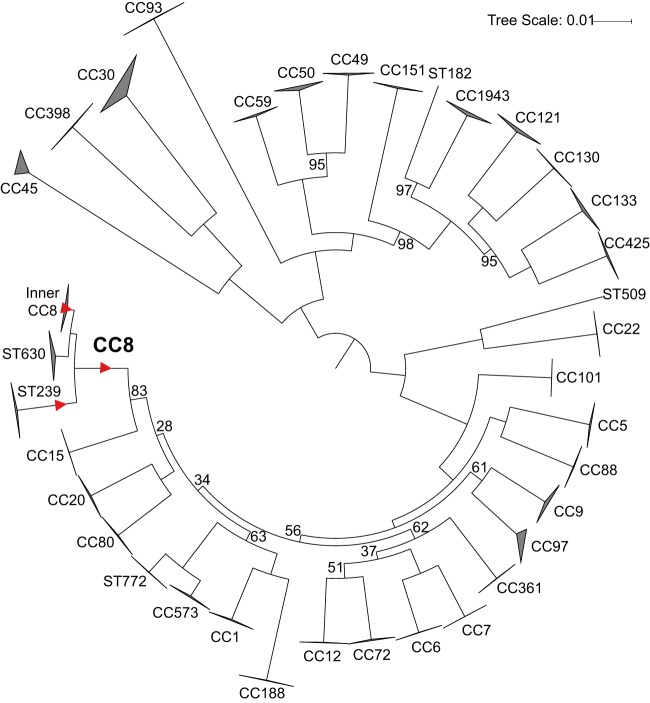
The overall S. aureus phylogeny (Fig. 1) shows the context of CC8 among other S. aureus lineages and shows that the CC8 strains in this tree all belong to one of three main lineages, ST239 (the HA SCCmec type III-carrying MRSA), ST630 (a lineage that branches off basal to the rest of CC8 and comprises five MSSA strains), and “Inner CC8” comprising the other known lineages. Table 1 shows common characteristics of these strain types. This phylogeny comprises 1.84 Mb shared by each genome and includes large regions exchanged among lineages that resulted in hybrid strains (e.g., ST34 and ST42 of CC30 and ST239 [10]). This tree, therefore, illustrates sum total relationships among lineages within S. aureus rather than within-lineage evolutionary history, as removal of these regions would imply a closer than actual relationship between a hybrid strain and one of its parent lineages.
FIG 1 .
WGS-based maximum likelihood phylogeny (using the best-fit model TVMe+ASC) of 497 S. aureus isolate genomes showing the CC8 group in the context of the whole of S. aureus. This analysis includes 1,000 bootstraps of 275,242 total SNPs in a core genome size (the length of the reference genome covered by all samples, excluding repeated regions) of 1.84 Mbp. Regions of chromosomal exchange among lineages resulting in hybrid strains (e.g., ST239) were not excluded. Bootstrap values are 100%, except where indicated. Branches of the phylogeny on which SNPs were selected for assay development are marked with a red triangle.
TABLE 1 .
Characteristics and reference isolates of lineages of CC8
| Traditional strain nomenclature |
Known isolate(s) (other names) |
Reference(s) | Main SCCmec type(s) |
Main spa type(s) |
Main sequence type |
WGS- based clade |
|---|---|---|---|---|---|---|
| ST239 | JKD6008, T0131, TW20 |
69–71 | III | t037, t431, t030 | ST239 | ST239 |
| ST630 | Unknown | Va | t377a, t4549a | ST630 | ST630 | |
| Archaic | Newman, COL, NCTC 10442 |
11, 15, 72, 73 | I | t051 | ST250 | CC8a |
| Iberian | HPV107, PER34, EMRSA5, E2125, NRS209 (28243, NR-46003) |
11, 15, 21, 54, 55, 74, 75 | I | t051 | ST247 | CC8a |
| NCTC 8325, BR-VRSA | 29, 75 | II, III, IVab | t334 | ST8, ST1181 | CC8b | |
| USA500/Iberianc | NRS385 (95938, NR-46071), BAA-1763 (GA229) |
21, https://www.atcc.org/Products/All/BAA-1763.aspx | IV | t064 | ST8 | CC8c |
| CMRSA9 | 01S-0965 | 31, 76 | VIII | t008 | ST8 | CC8d |
| USA500c | BD02-25, CA-224 (NRS645, NR-46174) |
7, 21 | IV | t008 | ST8 | CC8e |
| Early Branching USA300 |
V2200, HUV05 | 14 | IV | t008 | ST8 | CC8e |
| USA300-NAE | FPR3757, TCH1516 (USA300-HOU-MR) |
77–79 | IVa | t008 | ST8 | CC8f |
| USA300-SAE | M121, CA12 | 14 | IVc | t008 | ST8 | CC8e |
t377 is based on this study, and SCCmec type V and t4549 are based on one MRSA isolate (44).
Each SCCmec type is based on one MRSA isolate. Most isolates in this clade are MSSA.
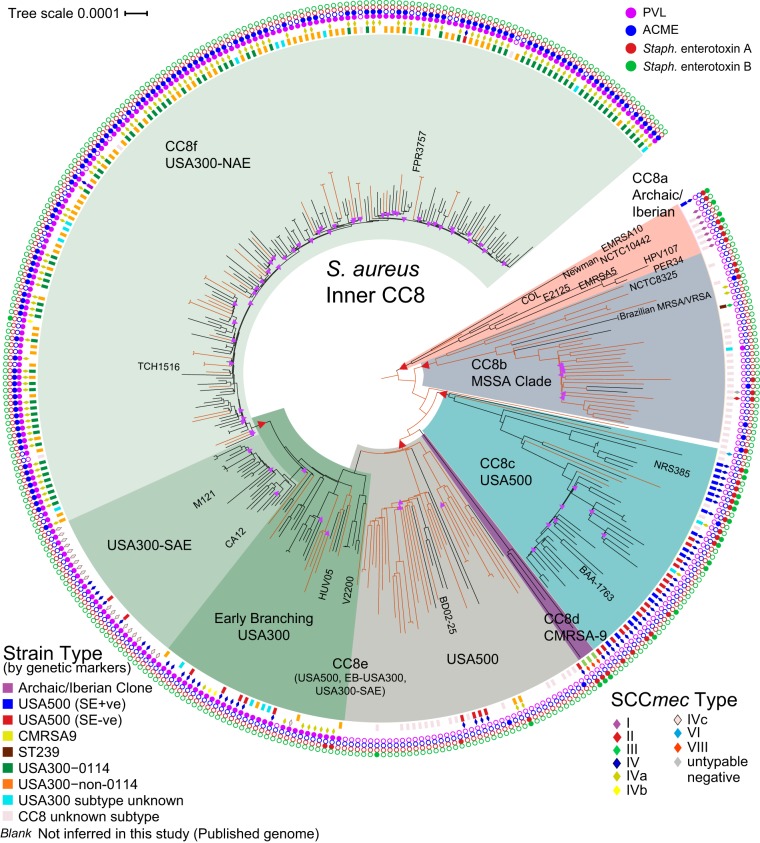
The topology of our Inner CC8 SNP-based phylogeny (excluding ST239 and ST630) comprising 348 genomes is similar to those reported recently (23, 28), showing multiple, distinct nested clades, with MSSA (orange branches) interspersed among the MRSA isolates (Fig. 2; Table 1). CC8a, which includes the Archaic and Iberian strains, is the most basal CC8 lineage, which supports the early circulation then disappearance of this lineage over time. All but one MRSA strain in CC8a carry SCCmec type I. To our knowledge, CC8b has not been characterized previously and contains the old strain NCTC 8325 and the Brazilian vancomycin-susceptible and resistant S. aureus isolates BR-VSSA and BR-VRSA, respectively, thought to be closely related to USA300 due to their carriage of SCCmec subtype IVa (29). The majority of the isolates in this clade are MSSA, a few of which carry ACME (suggesting previous SCCmec carriage [30]) or sea, and one of which has the PVL genes. Our phylogeny also shows that isolates known as USA500 fall into two distinct clades separated by CC8d, the Canadian HA-MRSA lineage, CMRSA9 (31): clade CC8c contains NRS385 (15) and BAA-1763 (ATCC), while group CC8e contains BD02-25 (7). This suggests that the CMRSA9 strains might be defined as USA500 by traditional typing methods. The CC8c clade includes an apparent rapidly expanded lineage (containing BAA-1763), illustrated as shallow branches with low bootstrap support, and several of these isolates were collected in Georgia in the United States. This clade is now known to be an epidemic lineage in Georgia (see the companion paper by Frisch et al. [32] and Fig. S1 in the supplemental material).
FIG 2 .
WGS-based maximum likelihood phylogeny (using the best-fit model TVMe+ASC) of 348 genomes of S. aureus (229 MRSA and 119 MSSA) belonging to the Inner CC8 clade (excluding ST239 and ST630 genomes), illustrating the relationship structure of clinically important CC8 groups and showing that genetic marker inference (GMI) strain typing is not always indicative of genetic relationship. MSSA genomes, on branches colored orange (Hex DF5000, RGB [223,80,0]), are interspersed among MRSA genomes. This analysis includes 1,000 bootstraps of 13,988 SNPs. Nodes with bootstrap values of <90% are marked with purple triangles. The core genome size is 2.26 Mbp (78.8% of reference genome FPR3757). Branches of the phylogeny on which SNPs were selected for assay development are marked with a red triangle.
Maximum likelihood SNP-based phylogeny (using the best-fit model TVMe+ASC) of 839 CC8 genomes: 348 genomes from this study (including the public genomes listed in Table S2) and 491 genomes from the companion article by Frisch et al. (32) (BioProject no. PRJNA342328). CC8 strain groups are labeled according to public literature isolate information and our canonical SNP state definitions. CC8a comprises all old isolates (1960s). CC8b is mostly MSSA and contains NCTC 8325 and the Brazilian BR-VSSA and BR-VRSA genomes (29). CC8c includes two clades, C1 and C2, described by Frisch et al. C2 primarily consists of the Georgia, USA, epidemic strain (32). One sample falls between CC8b and CC8c: SA-150 (as noted in Fig. 2). CC8c and CC8e, both considered USA500 groups, are separated phylogenetically by CC8d, the CMRSA9 clade, characterized by SCCmec type VIII carriage. Two related samples diverge between CC8d and CC8e: SRR3418706 and SRR3418948 (32). CC8e is a paraphyletic group with respect to USA300. Although USA300-SAE and USA300-NAE are considered monophyletic sister clades, it appears there may be other strains circulating that originate from their last common ancestor. (See the Early Branching sample located between USA300-SAE and USA300-NAE, also in Fig. 2.) Purple triangles mark nodes with bootstrap values of <90% of the 1,000 total. Download FIG S1, PDF file, 0.3 MB (277.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Accession numbers of previously sequenced S. aureus genomes used in this study, along with seven isolates sequenced in this study used solely for the overall S. aureus phylogeny (Fig. 1). Download TABLE S2, XLSX file, 0.1 MB (25.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Our data support the idea that USA500 in CC8e and USA300 share a direct common ancestor (Fig. 2). The WGS phylogeny indicates that the PVL genes were acquired by an early branching USA300 (14) ancestor (nested within CC8e) and passed down to the USA300 lineage, as most USA300 strains carry PVL, including USA300-SAE (14). As a predictor of USA300, the PVL genes have high sensitivity (97%) and specificity (99%) in our data; however, these genes are not confined to CC8 (see Table S1 in the supplemental material). The phylogeny also confirms that ACME was acquired by the USA300-NAE ancestor and passed vertically, as noted previously (14). ACME is present in six MSSA isolates in CC8f. As ACME is closely associated with SCCmec (30), Fig. 2 suggests at least four losses of SCCmec while retaining ACME. Spread across the CC8f USA300-NAE clade are 80 subtype USA300-0114 isolates interspersed with 41 non-0114 isolates, indicating that this important PFGE pattern subtype (27) is not a distinct lineage. Therefore, 0114 strains cannot be phylogenetically distinguished from other USA300 strains, and no canSNP marker can differentiate the 0114 strain type from non-0114 strains.
Typing information and screening results for all S. aureus isolates that were typed by genetic marker inference (GMI [Fig. 3]) and by whole-genome sequence (WGS) analysis in this study. Of the 295 isolates screened, 224 were typed by GMI and WGS phylogenetic analysis, and 89 of those were then used to validate the SNP strain typing assay panel. A total of 71 isolates were typed by GMI and SNP assay panel and then by WGS phylogenetic analysis for confirmation. Another 137 isolates (not in this table) were screened by GMI and SNP assay panel only (Table 3). Download TABLE S1, XLSX file, 0.1 MB (42.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The incorporation of a significant number of MSSA genomes in the CC8 phylogeny makes it apparent that MSSA was the founder of several of these CC8 strains. A majority of CC8b is MSSA, and the five MRSA isolates in this clade carry four different SCCmec types, suggesting independent acquisitions of the SCCmec cassettes and that much of CC8e remains or has reverted to MSSA. The mostly MRSA clades are each dominated by a single, different SCCmec type, indicating acquisition by the common ancestor to the clade, except in the Early Branching USA300 group, in which several different SCCmec types exist. All SCCmec types in the Early Branching USA300 group, however, are SCCmec IV subtypes. The MRSA strains in this clade could be a result of one acquisition event followed by recombination (33), or several separate SCCmec acquisitions. USA300-SAE comprises two SCCmec types, IV and IVc; however, it is not clear whether the typing schemes used always included an IVc subtype test. Although USA300-SAE is made up entirely of MRSA strains, this could be a sampling artifact. Besides their importance in CC8b and CC8e, MSSA genomes are interspersed with the MRSA genomes throughout CC8. The appearance of MSSA dispersed across the CC8 phylogeny supports the idea that the SCCmec cassette is highly mobile and upholds the notion that MSSA plays a principal role in S. aureus evolution and pathology.
GMI typing.
In general, the clades (Fig. 2) correlate with strain type inferred by the genetic marker inference (GMI) method (see Materials and Methods and Fig. 3), except in the case of CC8b and the USA500 groups. CC8b comprises mostly CC8-Unknown isolates, owing to our unfamiliarity with this clade and the clade’s makeup as mostly MSSA, for which GMI typing uses few markers. Using GMI, USA500 appears in two separate lineages, CC8c and CC8e, while GMI USA500/Iberian isolates fall within CC8c, with only one exception in CC8a. CC8c also contains two GMI USA300 isolates (positive for SCCmec subtype IVa) and three CC8-Unknown isolates. In group CC8e, the GMI typing was not consistent with WGS. For the 12 (out of 14 total) MRSA isolates characterized as USA500 by GMI, 5 were in the USA500 group that includes BD02-25, the reference USA500 isolate from Li et al. (7), 5 were in the Early Branching USA300 group, and 2 were in the USA300-SAE clade. Among the 37 isolates in CC8c, GMI inferred 16 as strain type USA500, while 15 were called USA500/Iberian. Evidence is inconclusive whether the CC8c lineage inherited the sea and seb genes from an Archaic or Iberian ancestor. The sea and seb genes are presumably frequently gained or lost. The mixture of enterotoxin-positive and -negative isolates in one lineage and the presence of sea or seb in other clades demonstrate that the sea and seb markers are not reliable indicators of a phylogenetically related group.
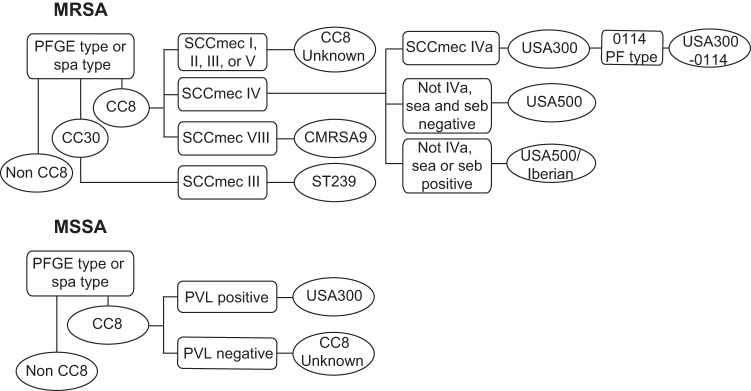
FIG 3 .
The genetic marker inference (GMI) methodology used for inferring S. aureus strain types by using genetic markers.
Within CC8f, almost all of the isolates typed by GMI in this study (157 of 158) typed as USA300 (Fig. 2). Only one isolate within CC8f was inferred as CC8-Unknown, an MSSA isolate lacking both PVL and ACME genes. Two other isolates in CC8f are negative for PVL, but both carry SCCmec subtype IVa and thus were typed as USA300. Four of the GMI-typed isolates outside CC8f carry SCCmec IVa: two in CC8e and two in CC8c. Interestingly, the addition of publicly available sequence data added eight genomes that carry SCCmec IVa that fall into CC8e in the early branching USA300 group and two that fall in the CC8b clade (BR-VSSA and BR-VRSA). Although SCCmec subtype IVa has been a hallmark of USA300-NAE, it is clear this trait is not unique to USA300-NAE. CC8e contains 10 isolates called by GMI as USA300. Besides the two that carry SCCmec subtype IVa, the remaining isolates are MSSA and PVL positive, which is the only criterion other than PFGE used to determine a USA300 strain type in MSSA (Fig. 3).
Assay screening.
The phylogenetically informative canSNPs identified using the genomic data presented above and used to design the assays are represented in Fig. 2. All assays (Table 2) can be used as stand-alone typing assays for any S. aureus, except for the CC8b assay, which must be used in combination with either the CC8 assay or the “Inner CC8” assay to confirm the phylogenetic placement of an isolate. Although the allelic state that the CC8b assay targets is unique within CC8, some isolates outside CC8 share this SNP state with the CC8b isolates—possibly due to recombination; therefore, an isolate positive for the CC8b SNP state should be screened across the CC8 or Inner CC8 assay to confirm (or refute) that it falls in CC8b.
TABLE 2 .
Assays designed and validated in this study
| Clade, group, or ST |
Assay | Primer/probe name |
Probe labelsa | Sequenceb | Product length (bp) |
|---|---|---|---|---|---|
| Clade CC8 (including ST239 and ST630) |
CC8_B+ | tCC8_F | CGAGTCAGCTAGTGGTCCGTT | 88 | |
| tCC8_R | ATGCATAGCTCTTGCTAAAGTGTA | ||||
| tCC8-A_FB+ | FAM, BHQ-1plus | ACCTATACCTGAACGTCAA | |||
| non-tCC8-G_TB+ | TET, BHQ-1plus | CTATACCTGAGCGTCAAA | |||
| Inner CC8 clade (excluding ST239 and ST630) |
inCC8_B+ | inCC8_F | TGCCCATAACACATTTGACACTTT | 79 | |
| inCC8_R1 | TTCGGCCACAGCTAAACTCG | ||||
| inCC8_R2 | GTTCGGCTACAGCTAAACTTGC | ||||
| inCC8_FB+ | FAM, BHQ-1plus | ATCGGACCCGGTAACC | |||
| non-inCC8_TB+ | TET, BHQ-1plus | TAATCGGACCTGGTAACC | |||
| Clade CC8a (Archaic and Iberian) |
CC8a_B+ | CC8a_F | CGCCAAATGACTCGCATTGT | 241 | |
| CC8a_R | GCATGTGCCTTTCCGAARTAAA | ||||
| CC8a-C_FB+ | FAM, BHQ-1plus | ATTACTGTAGCAGGGCTG | |||
| nonCC8a-T_TB+ | TET, BHQ-1plus | CTGTAGCAGGGTTGC | |||
| Clade CC8b | CC8b_B+ | CC8b_F | GATGACGTGATAACTGTACGTSGAT | 240 | |
| CC8b_R | CGCGATTGAGGGTGAATATTGC | ||||
| CC8b-C_FB+ | FAM, BHQ-1plus | AAGCTAACAAAATCACCTACTG | |||
| nonCC8b-T_TB+ | Tet, BHQ-1plus | CAAAGCTAACAAAATTACCTAC | |||
| Clade CC8c (USA500/Iberian) |
NewIber_B+ | NewIber_F | GCGCAACAGGGAAGCAA | 118 | |
| NewIber_R | TGCGGATGTCCTATGTCTGAAAG | ||||
| NewIber-T_FB+ | FAM, BHQ-1plus | TGCACTTACATATCATCCAT | |||
| nonNewIber-C_TB+ | Tet, BHQ-1plus | CACTTACATACCATCCATC | |||
| Group CC8ec (USA500, Early Branching USA300, and USA300-SAE) |
CC8e_B+ | CC8e_F | ACCTTATACRGAACATAGCAGACG | 106 | |
| CC8e_R | TCGATGCGCTTCTATCACTTC | ||||
| CC8e-C_FB+ | FAM, BHQ-1plus | TATTAGATGAAGGCCTCAATA | |||
| nonCC8e-T_TB+ | TET, BHQ-1plus | TTTATTAGATGAAGGCTTCAATA | |||
| Clade CC8fc (USA300-NAE) | CC8f_B+ | CC8f_F | CCTGAAGAAGAAGAGCGTTTAAGAA | 208 | |
| CC8f_R | RCATCCTACGATGGCCGAATC | ||||
| CC8f-T_FB+ | FAM, BHQ-1plus | TAAACGTCGTAAAGTAGAACAA | |||
| nonCC8f-A_TB+ | TET, BHQ-1plus | ACGTAAACGTCGTAAAGAAGAAC | |||
| ST239 | ST239_B+ | ST239_F | CATGACCGCCACTATAACCAGA | 99 | |
| ST239_R | ATGCAACATTAGCAGGAGGATG | ||||
| ST239-C_FB+ | FAM, BHQ-1plus | TACGACTGACCTGATGC | |||
| non239-T_TB+ | TET, BHQ-1plus | CGACTGACTTGATGCC |
FAM, 6-carboxyfluorescein; BHQ, black hole quencher; TET, tetrachlorofluorescein.
Nucleotides in boldface in each probe sequence are the phylogenetically informative canonical SNP state targeted by the assay.
USA300-NAE isolates will test positive on this assay.
Each assay was first validated across a set of isolates used to generate the original phylogeny (WGS followed by SNP assay in Table S1). In short, the SNP assays performed well and their results always agreed with the phylogeny (Table S1). A second set of 208 isolates that had not been sequenced was then screened, and results from here onward refer to this second set. Here, 144 MRSA and 64 MSSA isolates were compared between GMI and the SNP assay panel (Table 3). Out of the MRSA samples, both methods’ distinctions between CC8 and non-CC8 isolates were in full agreement; the PFGE/spa strain typing matched the CC8 SNP assay, where 114 isolates fell within CC8 while 30 were outside. Out of the MSSA samples, 61 were in agreement that all were CC8, but 3 isolates called CC8-Unknown by GMI were non-CC8 by the SNP assay (Table 3).
TABLE 3 .
Comparison of typing S. aureus isolates by the genetic marker inference assay and real-time PCR SNP assays on unknown (not sequenced) samples
| Genetic marker inference |
No. of isolates in category by real-time PCR SNP assaya |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC8 | Clade CC8a |
Clade CC8b |
Clade CC8c |
Group CC8e |
Clade CC8f |
ST239 | CC8 other |
Non- CC8 |
Total | |
| MRSA | ||||||||||
| CC8 | 114 | 7 | 0 | 34 | 11 | 57 | 4 | 1 | 0 | 114 |
| CC8-Unknown | 9 | 7 (5) | 0 | 2 (0) | 0 | 0 | 0 | 0 | 0 | 9 |
| USA500/Iberian | 21 | 0 | 0 | 21 (3) | 0 | 0 | 0 | 0 | 0 | 21 |
| CMRSA9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0) | 0 | 1 |
| USA500 | 22 | 0 | 0 | 11 (11) | 11 (3) | 0 | 0 | 0 | 0 | 22 |
| USA300 | 57 | 0 | 0 | 0 | 0 | 57 (1) | 0 | 0 | 0 | 57 |
| ST239 | 4 | 0 | 0 | 0 | 0 | 0 | 4 (0) | 0 | 0 | 4 |
| Non-CC8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 30 (0) | 30 |
| Total | 114 | 7 | 0 | 34 | 11 | 57 | 4 | 1 | 30 | 144 |
| MSSA | ||||||||||
| CC8 | 61 | 0 | 18 | 4 | 15 | 20 | 0 | 4 | 3 | 64 |
| CC8-Unknown | 45 | 0 | 18 (18) | 4 (4) | 15 (15) | 4 (4) | 0 | 4 (4) | 3 (3) | 48 |
| USA300 | 16 | 0 | 0 | 0 | 0 | 16 (0) | 0 | 0 | 0 | 16 |
| Total | 61 | 0 | 18 | 4 | 15 | 20 | 0 | 4 | 3 | 64 |
Numbers in parentheses are the number of isolates that were subsequently whole-genome sequenced to determine the true strain type.
Comparison of subtyping results within CC8 by GMI and the SNP assay panel gave fairly concordant results for MRSA isolates (Table 3). Out of the 114 CC8 isolates screened, 93 fell into their expected clade. Of the other 21, 11 were USA500 (SCCmec type IV, negative for sea and seb genes) and 2 were CC8-Unknown by GMI and typed as CC8c by the SNP panel. Eight isolates were typed as a strain for one method for which there was no assay by the other method: seven were CC8-Unknown by GMI and CC8a by the SNP panel, and one was CMRSA9 by GMI and CC8-Other by the SNP panel. Six of the seven CC8a MRSA isolates were collected in the 1960s and were SCCmec type I positive. This is the SCCmec type observed in the first Archaic and Iberian strains (11) (Table 1), but as these strains seem to have disappeared from circulation, the GMI approach does not account for them. For the 57 isolates typed as USA300 by GMI, all were typed in CC8f as expected (Table 2). All USA500/Iberian isolates by GMI were typed as CC8c by the SNP panel, and although testing was limited, all four ST239 isolates were concordant between the two typing methods. For MSSA, 45 of the total 64 isolates were typed as CC8-Unknown by GMI. These 45 by the SNP panel were typed as CC8f, CC8e, CC8c, non-CC8, or CC8-Other. No MSSA isolates were typed as non-CC8 by GMI, although three were by the SNP panel (Table 3).
A subset of isolates (n = 71) were sequenced and added to the CC8 or S. aureus overall phylogeny to determine their true strain type (Table 3; see Table S2 in the supplemental material). All samples in agreement between the two tests also agreed by WGS phylogenetic analysis (n = 7). For MRSA, the 11 samples called USA500 by GMI that were CC8c by the SNP panel were all typed as CC8c in the phylogeny. CC8-Unknown (GMI)/CC8a (SNP panel) isolates, of which five of the six typed in this study were sequenced, all fell into CC8a. Of the 45 MSSA samples that were labeled as CC8-Unknown by GMI, all the strain types called by the SNP panel were corroborated by phylogenetic analysis. The three non-CC8 isolates fell outside CC8 and were sequence typed as ST6. Of the four CC8-Unknown (GMI)/CC8-Other (SNP panel) isolates, two were sequence typed as ST630 (Fig. 1). The other two diverged after CC8b but before CC8c in the phylogeny (one of which is shown in Fig. 2), confirming that both GMI and SNP assay methods were correct but creating previously unseen lineages. It is likely that as we sequence more S. aureus strains, especially more MSSA strains, we will see additional CC8 lineages and a more complex CC8 tree topology develop.
Overall, the SNP assays were 100% specific and sensitive on the set of unknown isolates, according to the phylogeny generated through WGS; this result is expected due to the stability of SNPs. The genetic marker inference assay performed fairly well, except in the cases of the USA500 and USA500/Iberian types, as well as for MSSA isolates where the only genetic marker for CC8 subtyping was the PVL genes.
DISCUSSION
S. aureus remains an important pathogen in health care institutions as well as in healthy populations in the community. CC8 strains are among the most prevalent in both environments, especially USA300, and each sublineage has different clinical and pathological characteristics (1, 11, 25, 34, 35). Strain typing of S. aureus is important because of these phenotypic differences and their implications for virulence potential, and tracking strains and their prevalence in a health care system or network informs epidemiology and infection control practices to help focus resources effectively. Unfortunately, typing is not a routine practice in clinical microbiology laboratories, in part because of the cost, time, and expertise required, as well as the frequent inconclusiveness of results. PFGE, spa typing, and multilocus sequence typing (MLST) often do not provide the scale of resolution required to determine relationships among a given set of samples, and the presence of particular virulence factors—often located on mobile elements—can be misleading (16). The simple typing system we have developed here, based on presumably stable canSNPs, allows for wide use in clinical laboratories for robust tracking of both MRSA and MSSA infections. Additionally, this method can rapidly and inexpensively assess the possibility of an outbreak or transmission event. Isolates of the same strain type should be investigated further (by WGS), while isolates of different strain types would preclude an outbreak or transmission event, which is just as important (36).
The S. aureus CC8 strain nomenclature, including Iberian, Archaic, USA500, and USA300, was originally based on PFGE typing schemes that used an 80% banding pattern similarity threshold to classify isolates (15). Although adopted for tracking purposes, the continuous evolution and diversification of S. aureus over the years have rendered PFGE a misleading tool for this application. Strains that are within 80% banding pattern similarity may belong to multiple genetic lineages, as shown in this study. USA500 comprises at least two well-established lineages (see the companion paper by Frisch et al. [32]) and may encompass the Canadian CMRSA9 lineage. Strain BD02-25, called USA500 by Li et al. (7) and currently the CDC’s USA500 reference isolate (L. McDougal, unpublished observation), is not in the same lineage as strains NRS385 (the USA500 reference in the article by McDougal et al. [15]) and ATCC BAA-1763, although it is ≥80% similar, suggesting USA500 encompasses a wider genomic range than previously appreciated. Additionally, NRS385 and BAA-1763, which are sea and seb positive, share their clade with several isolates negative for these genes, which were used in the GMI typing scheme. It is necessary to exercise caution in interpretation of typing via mobile elements, as their sensitivity and specificity are not ideal. Likewise, the GMI typing system, although sensitive and specific for USA300-NAE, has limitations. The presence of SCCmec subtype IVa can be used for MRSA but not MSSA isolates, and we show that SCCmec subtype IVa is often found outside USA300-NAE. The presence of PVL, apparently vertically passed to USA300 from its progenitor (19), is a good predictor of USA300, as shown in other studies (16) as well as this one. However, the sequencing of the Early Branching USA300 and USA300-SAE genomes shows that PVL is inclusive of these newly understood strains and not specific to the highly clonal USA300-NAE (14). Also, we show that MSSA isolates are easily mistyped this way, and PVL is found in other CC8 strains as well as other clonal complexes (16, 37–39). The topologies of several whole-genome phylogenies recently generated for CC8 are in agreement (23, 25, 28), despite the differences in interpretations. Li et al. concluded that the USA500 strain is the progenitor of the widespread USA300 strain. Recent studies show that genomes labeled as USA500 fall into a more distant clade from USA300 (CC8c) but that there is an additional clade that shares an ancestor with USA300 (23, 25). We show here that both of these clades contain USA500 and surround the CMRSA9 clade, suggesting CMRSA9 might be considered a USA500 strain. By traditional typing methods, USA500 and other strains named for pulsed-field patterns do not represent monophyly. Future studies should note that different lineages contain “USA500” strains and use WGS phylogenetics or the assays presented here (or the SNPs they target) for strain typing within CC8.
The importance of MRSA is well known. MSSA, on the other hand, continues to have a critical impact on public health (5, 6, 40) and remains understudied. MRSA evolution evidences local selection and spread of particular strain types originating from successful MSSA lineages (12), and we demonstrate this within the CC8 lineage. Additionally, diverse MSSA strain types appear to be ubiquitous (6, 19, 41), and we show that MSSA strains are present in every major CC8 clade, advancing our understanding of the highly significant role that MSSA plays in S. aureus population structure. Importantly, MSSA may ultimately prove more of a challenge to clinically manage, as infection prevention measures targeting particular strain types of MRSA will be less effective against the more diverse MSSA strains (6). The MSSA strains in CC8 are interspersed with MRSA, further evidencing the significant mobility of SCCmec (12). Other species of Staphylococcus are likely active reservoirs of SCCmec, including the SCCmec subtype IVa strains characteristic of USA300 (42). The human carriage rate of SCCmec-positive, coagulase-negative staphylococcus (CoNS) can be relatively high, and cocolonization of MSSA and SCCmec-positive CoNS has been observed (42). Regardless of the directionality of SCCmec exchange among species and strains of Staphylococcus, the rate of SCCmec acquisition and/or excision may be higher than previously believed, and isolation of only MRSA in health care settings will not reveal the entire potential for MRSA carriage or infection.
Additionally, characterization of only MRSA isolates in CC8 (i.e., sampling bias) will give an incomplete evolutionary history of this important clonal complex. In our CC8 phylogeny, MSSA genomes add lineages not represented by MRSA alone, consistent with previous findings in CC8 (19). In our collection, ST630 comprises strictly MSSA isolates. ST630 may be an emerging strain of S. aureus, especially in China, where recently it reportedly caused a bloodstream infection (as MRSA) (43), endocarditis in a healthy person (as MRSA) (44), and several skin infections (as MSSA) (43, 45). CC8b comprises mostly MSSA, and the three MRSA strains appear to have emerged separately from different MSSA strains. This clade includes NCTC 8325, a strain isolated in 1943. The ancestor of CC8b diverged early in CC8 evolution, like the Archaic lineage. While the Archaic lineage expanded with SCCmec type I and has since apparently declined, CC8b does not appear to have acquired and maintained SCCmec, yet contains extant members that cause disease (included in this study). The study and WGS of more MSSA isolates will likely add complexity and clarity to the story of CC8 evolution.
Almost all of the USA300 isolates fall into a distinct clade with distinct features. PFGE profiling of USA300, which was not performed on many isolates in this study, in contrast with our genetic marker-inferred typing, may indeed be 100% concordant with our USA300 SNP-based assay currently. However, USA300 is a relatively young “clone,” and as more S. aureus lineages develop, a PFGE profiling system using similarity thresholds may soon prove obsolete as it has for other strains and species (46–48). Furthermore, we demonstrate that the PFGE type USA300-0114 is not a “clone” in the phylogenetic sense, as 0114 isolates do not form a monophyletic clade with a common ancestor as was previously believed (49). WGS is irreplaceable to determine if strains of the USA300-0114 PFGE type are part of a single outbreak.
The declining costs and increasingly common use of WGS and phylogenetic analysis allow for discovery of more phylogenetically informative and stable targets that can be used in rapid, relatively simple assays (28, 50, 51). Several advantages to the use of lineage-specific canSNPs as targets include the following: (i) they show stability over time, as they are passed vertically through generations, (ii) different SNPs provide different scales of resolution for identifying particular strains (e.g., a CC8-specific SNP versus a USA300-specific SNP) or even species in a given set of samples (51), or for use in global epidemiology (52), regional epidemiology (53), or local cluster analyses (36), and (iii) identification of canSNPs is a straightforward process using whole-genome sequence data and publicly available SNP matrix generators (e.g., NASP [50]), followed by parsing the SNPs by sample sets of interest. Here we use real-time PCR assays targeting canSNPs based on WGS to classify isolates into clear evolutionary lineages of CC8, and we illustrate their robustness (working with crude bacterial lysates) and high sensitivity and specificity. Inclusion of assays for SNPs on other branches in a hierarchical fashion, as we have done here, adds confidence to any typing scheme. The hierarchical scheme also provides opportunity to screen clinical or other complex specimens, which may harbor multiple strain types. Although WGS and phylogenetic analysis are irreplaceable in true outbreak situations, WGS is still relatively time-consuming and the analysis is complex. Robust real-time PCR assays can screen for isolates that may need further investigation with WGS. While WGS gains a foothold in both the public health laboratory and clinical laboratory, real-time PCR is a rapid, robust, easy, and therefore universal tool for clinical molecular biology and provides an excellent vehicle for the assays described here.
MATERIALS AND METHODS
Sample collection.
This study’s S. aureus isolates, mostly obtained from the CDC’s collection, were selected to represent the diversity of known CC8 strains, including USA300, USA500, Iberian, Archaic, Canadian MRSA9 (CMRSA9), and ST239 types, and to encompass both MRSA (313 isolates) and MSSA (119 isolates). Intentionally included were FPR3757 and TCH1516 (prototype USA300 isolates), BD02-25 (the USA500 reference isolate from Li et al. [7] and used in the CDC’s quality management system protocols), NRS385 (15) and ATCC BAA-1763 (two publicly available isolates typed as USA500), and the genomes of COL (an Archaic isolate from 1960 [11]), HPV107 and E2125 (ST247 Iberian strains from the 1960s [54, 55]), and NCTC 8325 (a laboratory strain originally isolated from a septic patient also around 1960). Also included were genomes belonging to the USA300 South American epidemic (USA300-SAE) strain type as well as samples considered Early Branching USA300 (14, 56, 57), and the Brazilian MRSA-turned-VRSA samples BR-VSSA and BR-VRSA (29). Table 1 lists several of the traditional CC8 strains and their characteristics. Table S1 in the supplemental material describes the isolates used in this study that were whole-genome sequenced.
Sequencing, SNP detection, and phylogenetic analysis.
Genome libraries for 288 S. aureus isolates were prepared with a 500-bp insert size using the Kapa library preparation kit with standard PCR library amplification (Kapa Biosystems) and sequenced on a 101-bp-read, paired-end Illumina GAIIx run or a 2× 250-bp Illumina MiSeq run (Tables S1 and S2). Additionally, 311 S. aureus genomes published in previous studies selected for sequence type diversity were used to generate the CC8 phylogeny and an overall S. aureus phylogeny encompassing several clonal complexes (Table S2) (18, 58).
The bioinformatics pipeline NASP (50) was used to detect SNPs among genomes. In brief, reads were aligned to the finished genome FPR3757 (GenBank accession no. CP000255) using Novoalign (Novocraft.com) and SNPs were called with GATK (59). Data filtered out included SNP loci with less than 5× coverage or less than 80% consensus in any one sample, SNP loci that were not present in all genomes in the data set, and any regions duplicated in the reference genome as identified by NUCmer (60). The results were formatted in an SNP matrix from a core genome common to all isolates in the analysis. Phylogenetic analysis model selection and generation of trees from the NASP SNP matrices were performed using IQ-TREE (61) and subsequently plotted with genetic marker data by means of ITOL v3 (62).
S. aureus typing.
The methods used for molecular typing of S. aureus were adopted from those previously described (4). These methods are based on a study conducted by the CDC (McDougal, unpublished) in which >350 CC8 isolates were tested for multiple genotypic and phenotypic markers, including the SCCmec type and the IVa subtype, Staphylococcus enterotoxin genes sea, seb, sek, and seq, PVL genes, ACME genes, and trimethoprim-sulfamethoxazole resistance. The markers with the greatest sensitivity and specificity for strain typing comprise the original typing algorithm (4).
For the purposes of this study, our modified genetic marker typing algorithm is shown in Fig. 3. In brief, traditional PFGE or spa type was used to infer clonal complex. Strain types of CC8 MRSA isolates were inferred based on SCCmec types and toxin gene profiles: SCCmec subtype IVa-positive isolates were called USA300, sea- and seb-negative isolates with SCCmec type IV (other than IVa) were called USA500, and isolates with SCCmec type VIII were called CMRSA9. We inferred that the presence of the sea and seb genes was indicative of a separate lineage, called Iberian by Li et al. in 2009 (7) and by the CDC in previous surveillance studies (4). However, as the SCCmec type I characteristic of the original Iberian strain has largely been replaced by SCCmec type IV, and because recent studies have referred to “Iberian” isolates (positive for sea or seb) as USA500 (NRS385 and BAA-1763), we called CC8 isolates positive for sea or seb that carry SCCmec type IV (other than subtype IVa) USA500/Iberian to distinguish them from the original Iberian clone. Isolates spa typed as CC30 with SCCmec type III were inferred to be ST239. CC8 MSSA isolates were called USA300 if they were PVL positive and called CC8-Unknown if they were PVL negative. Lastly, we noted whether the USA300 isolates were PF type 0114. This strain typing approach is herein termed the genetic marker inference (GMI) assay.
Multilocus sequence types (MLSTs) and spa types were determined by the traditional Sanger sequencing analysis or, when typing had not been performed and genomic sequence data were available, MLST was performed with SRST2 (63). SCCmec cassette typing using conventional methods was performed on a subset of isolates depending on the time of their collection (7, 64). To determine SCCmec types for isolates that did not have PCR results and to confirm previous conventional typing, WGS data were used: reads were assembled using SPAdes Genome Assembler (65), and an in silico PCR script using the BioPerl (66) toolkit was used to search for SCCmec typing PCR primer sequences (67) and analyze in silico amplicons. For 10 isolates for which conventional typing and WGS typing results were discordant, raw read data were aligned to sequences of several SCCmec cassette types using SeqMan NGen v.12.1.0 (DNAStar, Madison, WI). Types were confirmed by read coverage breadth and depth against the reference SCCmec type sequences.
SNP assays.
SNPs that differentiate specific clades of S. aureus (canSNPs), identified by NASP and phylogenetic analysis, were exploited for assay design. From the CC8 phylogenetic analysis, SNP loci at which the SNP state differed between a target lineage and the rest of the complex were selected. These loci were then checked in genomes from other clonal complexes to ensure the SNP state was unique to the targeted lineage. In this way, the potential for a shared SNP state across clonal complexes due to recombination (as has been observed [18]) was avoided. Eight sets of primers and probes targeting eight canSNPs were designed with Biosearch Technologies’ RealTimeDesign software (Biosearch Technologies, Petaluma, CA). Assay information is provided in Table 2.
Cell lysates of 311 isolates were prepared as previously described (68) and used to validate the assays. Reactions were run in 10 µl on the Applied Biosystems 7500 Fast real-time PCR instrument (Thermo Fisher Scientific) with 5 µl 2× TaqMan Universal PCR master mix (Thermo Fisher Scientific), 80 nM forward and reverse primers, 20 nM each probe, and 1 µl DNA template. Thermal conditions included denaturation at 95°C for 10 min and 40 cycles of 95°C for 15 s, 60°C for 1 min.
Accession number(s).
BioProject accession no. PRJNA374337 contains the whole-genome sequence read data generated in this study.
ACKNOWLEDGMENTS
We gratefully acknowledge the institutions that share their genome sequence data and the support staff that maintain the databases. We also thank the operations and administrative staff at TGen and the CDC for their support.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
This study was funded by NIH grant 5R01AI90782-5 and contract 200-2016-92313 with the Centers for Disease Control and Prevention under their Advanced Molecular Detection Initiative.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/mSphere.00571-17.
REFERENCES
- 1.Bal AM, Coombs GW, Holden MTG, Lindsay JA, Nimmo GR, Tattevin P, Skov RL. 2016. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J Glob Antimicrob Resist 6:95–101. doi: 10.1016/j.jgar.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Carrel M, Perencevich EN, David MZ. 2015. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerg Infect Dis 21:1973–1980. doi: 10.3201/eid2111.150452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diekema DJ, Richter SS, Heilmann KP, Dohrn CL, Riahi F, Tendolkar S, McDanel JS, Doern GV. 2014. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol 35:285–292. doi: 10.1086/675283. [DOI] [PubMed] [Google Scholar]
- 4.Albrecht VS, Limbago BM, Moran GJ, Krishnadasan A, Gorwitz RJ, McDougal LK, Talan DA, EMERGEncy ID Net Study Group . 2015. Staphylococcus aureus colonization and strain type at various body sites among patients with a closed abscess and uninfected controls at U.S. emergency departments. J Clin Microbiol 53:3478–3484. doi: 10.1128/JCM.01371-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David MZ, Boyle-Vavra S, Zychowski DL, Daum RS. 2011. Methicillin-susceptible Staphylococcus aureus as a predominantly healthcare-associated pathogen: a possible reversal of roles? PLoS One 6:e18217. doi: 10.1371/journal.pone.0018217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miko BA, Hafer CA, Lee CJ, Sullivan SB, Hackel MA, Johnson BM, Whittier S, Della-Latta P, Uhlemann AC, Lowy FD. 2013. Molecular characterization of methicillin-susceptible Staphylococcus aureus clinical isolates in the United States, 2004 to 2010. J Clin Microbiol 51:874–879. doi: 10.1128/JCM.00923-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Zhou H, Wang H, Chen H, Leung KK, Tsui S, Ip M. 2014. Comparative genomics of methicillin-resistant Staphylococcus aureus ST239: distinct geographical variants in Beijing and Hong Kong. BMC Genomics 15:529. doi: 10.1186/1471-2164-15-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson DA, Enright MC. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J Bacteriol 186:1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs GW, Pearson JC, O’Brien FG, Murray RJ, Grubb WB, Christiansen KJ. 2006. Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg Infect Dis 12:241–247. doi: 10.3201/eid1202.050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planet PJ, Diaz L, Kolokotronis SO, Narechania A, Reyes J, Xing G, Rincon S, Smith H, Panesso D, Ryan C, Smith DP, Guzman M, Zurita J, Sebra R, Deikus G, Nolan RL, Tenover FC, Weinstock GM, Robinson DA, Arias CA. 2015. Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J Infect Dis 212:1874–1882. doi: 10.1093/infdis/jiv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2013. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. Medical Center. J Clin Microbiol 51:814–819. doi: 10.1128/JCM.02429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driebe EM, Sahl JW, Roe C, Bowers JR, Schupp JM, Gillece JD, Kelley E, Price LB, Pearson TR, Hepp CM, Brzoska PM, Cummings CA, Furtado MR, Andersen PS, Stegger M, Engelthaler DM, Keim PS. 2015. Using whole genome analysis to examine recombination across diverse sequence types of Staphylococcus aureus. PLoS One 10:e0130955. doi: 10.1371/journal.pone.0130955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strommenger B, Bartels MD, Kurt K, Layer F, Rohde SM, Boye K, Westh H, Witte W, De Lencastre H, Nübel U. 2014. Evolution of methicillin-resistant Staphylococcus aureus towards increasing resistance. J Antimicrob Chemother 69:616–622. doi: 10.1093/jac/dkt413. [DOI] [PubMed] [Google Scholar]
- 20.Engelthaler DM, Kelley E, Driebe EM, Bowers J, Eberhard CF, Trujillo J, Decruyenaere F, Schupp JM, Mossong J, Keim P, Even J. 2013. Rapid and robust phylotyping of spa t003, a dominant MRSA clone in Luxembourg and other European countries. BMC Infect Dis 13:339. doi: 10.1186/1471-2334-13-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O’Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campanile F, Bongiorno D, Borbone S, Stefani S. 2009. Hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) in Italy. Ann Clin Microbiol Antimicrob 8:22. doi: 10.1186/1476-0711-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamrozy DM, Harris SR, Mohamed N, Peacock SJ, Tan CY, Parkhill J, Anderson AS, Holden MT. 2016. Pan-genomic perspective on the evolution of the Staphylococcus aureus USA300 epidemic. Microb Genom 2:e000058. doi: 10.1099/mgen.0.000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson MA, Ohneck EA, Ryan C, Alonzo F III, Smith H, Narechania A, Kolokotronis SO, Satola SW, Uhlemann AC, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol 93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle-Vavra S, Li X, Alam MT, Read TD, Sieth J, Cywes-Bentley C, Dobbins G, David MZ, Kumar N, Eells SJ, Miller LG, Boxrud DJ, Chambers HF, Lynfield R, Lee JC, Daum RS. 2015. USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus. mBio 6:e02585-14. doi: 10.1128/mBio.02585-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keim P, Van Ert MN, Pearson T, Vogler AJ, Huynh LY, Wagner DM. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect Genet Evol 4:205–213. doi: 10.1016/j.meegid.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Nimmo GR. 2012. USA300 abroad: global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect 18:725–734. doi: 10.1111/j.1469-0691.2012.03822.x. [DOI] [PubMed] [Google Scholar]
- 28.Aanensen DM, Feil EJ, Holden MT, Dordel J, Yeats CA, Fedosejev A, Goater R, Castillo-Ramírez S, Corander J, Colijn C, Chlebowicz MA, Schouls L, Heck M, Pluister G, Ruimy R, Kahlmeter G, Åhman J, Matuschek E, Friedrich AW, Parkhill J, Bentley SD, Spratt BG, Grundmann H, European SRL Working Group . 2016. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. mBio 7:e00444-16. doi: 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi F, Diaz L, Wollam A, Panesso D, Zhou Y, Rincon S, Narechania A, Xing G, Di Gioia TS, Doi A, Tran TT, Reyes J, Munita JM, Carvajal LP, Hernandez-Roldan A, Brandão D, van der Heijden IM, Murray BE, Planet PJ, Weinstock GM, Arias CA. 2014. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med 370:1524–1531. doi: 10.1056/NEJMoa1303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goering RV, McDougal LK, Fosheim GE, Bonnstetter KK, Wolter DJ, Tenover FC. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J Clin Microbiol 45:1981–1984. doi: 10.1128/JCM.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christianson S, Golding GR, Campbell J, Canadian Nosocomial Infection Surveillance Program, Mulvey MR. 2007. Comparative genomics of Canadian epidemic lineages of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 45:1904–1911. doi: 10.1128/JCM.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frisch MB, Castillo-Ramirez S, Petit RA III, Farley MM, Ray SM, Albrecht VS, Limbago BM, Hernandez J, See I, Satola SW, Read TD. 2018. Invasive methicillin-resistant Staphylococcus aureus USA500 strains from the U.S. Emerging Infections Program constitute three geographically distinct lineages. mSphere 3:e00571-17. doi: 10.1128/mSphere.00571-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noto MJ, Kreiswirth BN, Monk AB, Archer GL. 2008. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J Bacteriol 190:1276–1283. doi: 10.1128/JB.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. 2006. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis 193:1495–1503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 35.Planet PJ, LaRussa SJ, Dana A, Smith H, Xu A, Ryan C, Uhlemann AC, Boundy S, Goldberg J, Narechania A, Kulkarni R, Ratner AJ, Geoghegan JA, Kolokotronis SO, Prince A. 2013. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4:e00889-13. doi: 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roe CC, Horn KS, Driebe EM, Bowers J, Terriquez JA, Keim P, Engelthaler DM. 2016. Whole genome SNP typing to investigate methicillin-resistant Staphylococcus aureus carriage in a health care provider as the source of multiple surgical site infections. Hereditas 153:11. doi: 10.1186/s41065-016-0017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlicková H, Coombs G, Kearns AM, Hill RL, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramírez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nübel U. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurt K, Rasigade JP, Laurent F, Goering RV, Žemličková H, Machova I, Struelens MJ, Zautner AE, Holtfreter S, Bröker B, Ritchie S, Reaksmey S, Limmathurotsakul D, Peacock SJ, Cuny C, Layer F, Witte W, Nübel U. 2013. Subpopulations of Staphylococcus aureus clonal complex 121 are associated with distinct clinical entities. PLoS One 8:e58155. doi: 10.1371/journal.pone.0058155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McAdam PR, Templeton KE, Edwards GF, Holden MT, Feil EJ, Aanensen DM, Bargawi HJ, Spratt BG, Bentley SD, Parkhill J, Enright MC, Holmes A, Girvan EK, Godfrey PA, Feldgarden M, Kearns AM, Rambaut A, Robinson DA, Fitzgerald JR. 2012. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 109:9107–9112. doi: 10.1073/pnas.1202869109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.See I, Gualandi N, Dumyati G, Koeck M, Lynfield R, Pasutti L, Schaffner W, Wright D, Magill SS. 2015. Public health importance of methicillin-sensitive Staphylococcus aureus (MSSA): results from pilot surveillance in five counties, 2014–2015. Open Forum Infect Dis 2:1121. doi: 10.1093/ofid/ofv133.833. [DOI] [Google Scholar]
- 41.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, European Staphylococcal Reference Laboratory Working Groups . 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med 7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbier F, Ruppé E, Hernandez D, Lebeaux D, Francois P, Felix B, Desprez A, Maiga A, Woerther PL, Gaillard K, Jeanrot C, Wolff M, Schrenzel J, Andremont A, Ruimy R. 2010. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis 202:270–281. doi: 10.1086/653483. [DOI] [PubMed] [Google Scholar]
- 43.Huang J, Ye M, Ding H, Guo Q, Ding B, Wang M. 2013. Prevalence of fusB in Staphylococcus aureus clinical isolates. J Med Microbiol 62:1199–1203. doi: 10.1099/jmm.0.058305-0. [DOI] [PubMed] [Google Scholar]
- 44.Zheng B, Jiang S, Xu Z, Xiao Y, Li L. 2015. Severe infective endocarditis with systemic embolism due to community associated methicillin-resistant Staphylococcus aureus ST630. Braz J Infect Dis 19:85–89. doi: 10.1016/j.bjid.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu FF, Hou Q, Yang HH, Zhu YQ, Guo XK, Ni YX, Han LZ. 2015. Characterization of Staphylococcus aureus isolated from non-native patients with skin and soft tissue infections in Shanghai. PLoS One 10:e0123557. doi: 10.1371/journal.pone.0123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. 2016. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 63:380–386. doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergholz TM, den Bakker HC, Katz LS, Silk BJ, Jackson KA, Kucerova Z, Joseph LA, Turnsek M, Gladney LM, Halpin JL, Xavier K, Gossack J, Ward TJ, Frace M, Tarr CL. 2016. Determination of evolutionary relationships of outbreak-associated Listeria monocytogenes strains of serotypes 1/2a and 1/2b by whole-genome sequencing. Appl Environ Microbiol 82:928–938. doi: 10.1128/AEM.02440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng X, Shariat N, Driebe EM, Roe CC, Tolar B, Trees E, Keim P, Zhang W, Dudley EG, Fields PI, Engelthaler DM. 2015. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J Clin Microbiol 53:212–218. doi: 10.1128/JCM.02332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, Driebe EM, Drees KP, Hicks ND, Williamson CHD, Hepp CM, Smith DE, Roe C, Engelthaler DM, Wagner DM, Keim P. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom 2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowers JR, Lemmer D, Sahl JW, Pearson T, Driebe EM, Wojack B, Saubolle MA, Engelthaler DM, Keim P. 2016. KlebSeq: a diagnostic tool for surveillance, detection, and monitoring of Klebsiella pneumoniae. J Clin Microbiol 54:2582–2596. doi: 10.1128/JCM.00927-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, Keim P, Limbago BM. 2015. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 10:e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engelthaler DM, Valentine M, Bowers J, Pistole J, Driebe EM, Terriquez J, Nienstadt L, Carroll M, Schumacher M, Ormsby ME, Brady S, Livar E, Yazzie D, Waddell V, Peoples M, Komatsu K, Keim P. 2016. Hypervirulent emm59 clone in invasive group A streptococcus outbreak, southwestern United States. Emerg Infect Dis 22:734–738. doi: 10.3201/eid2204.151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanches IS, Ramirez M, Troni H, Abecassis M, Padua M, Tomasz A, de Lencastre H. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J Clin Microbiol 33:1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Lencastre H, Chung M, Westh H. 2000. Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb Drug Resist 6:1–10. doi: 10.1089/mdr.2000.6.1. [DOI] [PubMed] [Google Scholar]
- 56.Glaser P, Martins-Simões P, Villain A, Barbier M, Tristan A, Bouchier C, Ma L, Bes M, Laurent F, Guillemot D, Wirth T, Vandenesch F. 2016. Demography and intercontinental spread of the USA300 community-acquired methicillin-resistant Staphylococcus aureus lineage. mBio 7:e02183-15. doi: 10.1128/mBio.02183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhlemann AC, Dordel J, Knox JR, Raven KE, Parkhill J, Holden MT, Peacock SJ, Lowy FD. 2014. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simões PM, Tristan A, Petersen A, Aziz M, Kiil K, Cirković I, Udo EE, del Campo R, Vuopio-Varkila J, Ahmad N, Tokajian S, Peters G, Schaumburg F, Olsson-Liljequist B, Givskov M, Driebe EE, Vigh HE, Shittu A, Ramdani-Bougessa N, Rasigade JP, Price LB, Vandenesch F, Larsen AR, Laurent F. 2014. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. mBio 5:e01044-14. doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delcher AL, Salzberg SL, Phillippy AM. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics Chapter 10:Unit 10.3. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I, Bork P. 2011. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L, Mediavilla JR, Oliveira DC, Willey BM, de Lencastre H, Kreiswirth BN. 2009. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec typing. J Clin Microbiol 47:3692–3706. doi: 10.1128/JCM.00766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehväslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howden BP, Seemann T, Harrison PF, McEvoy CR, Stanton JA, Rand CJ, Mason CW, Jensen SO, Firth N, Davies JK, Johnson PD, Stinear TP. 2010. Complete genome sequence of Staphylococcus aureus strain JKD6008, an ST239 clone of methicillin-resistant Staphylococcus aureus with intermediate-level vancomycin resistance. J Bacteriol 192:5848–5849. doi: 10.1128/JB.00951-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, Cao B, Zhang Y, Zhou J, Yang B, Wang L. 2011. Complete genome sequence of Staphylococcus aureus T0131, an ST239-MRSA-SCCmec type III clone isolated in China. J Bacteriol 193:3411–3412. doi: 10.1128/JB.05135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holden MT, Lindsay JA, Corton C, Quail MA, Cockfield JD, Pathak S, Batra R, Parkhill J, Bentley SD, Edgeworth JD. 2010. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J Bacteriol 192:888–892. doi: 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki E, Kuwahara-Arai K, Richardson JF, Hiramatsu K. 1993. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother 37:1219–1226. doi: 10.1128/AAC.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung M, de Lencastre H, Matthews P, Tomasz A, Adamsson I, Aires de Sousa M, Camou T, Cocuzza C, Corso A, Couto I, Dominguez A, Gniadkowski M, Goering R, Gomes A, Kikuchi K, Marchese A, Mato R, Melter O, Oliveira D, Palacio R, Sá-Leão R, Santos Sanches I, Song JH, Tassios PT, Villari P, Multilaboratory Project Collaborators . 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist 6:189–198. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 75.Crisóstomo MI, Westh H, Tomasz A, Chung M, Oliveira DC, de Lencastre H. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc Natl Acad Sci U S A 98:9865–9870. doi: 10.1073/pnas.161272898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:531–540. doi: 10.1128/AAC.01118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 78.Gonzalez BE, Martinez-Aguilar G, Hulten KG, Hammerman WA, Coss-Bu J, Avalos-Mishaan A, Mason EO Jr, Kaplan SL. 2005. Severe staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics 115:642–648. doi: 10.1542/peds.2004-2300. [DOI] [PubMed] [Google Scholar]
- 79.Highlander SK, Hultén KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood SNP-based phylogeny (using the best-fit model TVMe+ASC) of 839 CC8 genomes: 348 genomes from this study (including the public genomes listed in Table S2) and 491 genomes from the companion article by Frisch et al. (32) (BioProject no. PRJNA342328). CC8 strain groups are labeled according to public literature isolate information and our canonical SNP state definitions. CC8a comprises all old isolates (1960s). CC8b is mostly MSSA and contains NCTC 8325 and the Brazilian BR-VSSA and BR-VRSA genomes (29). CC8c includes two clades, C1 and C2, described by Frisch et al. C2 primarily consists of the Georgia, USA, epidemic strain (32). One sample falls between CC8b and CC8c: SA-150 (as noted in Fig. 2). CC8c and CC8e, both considered USA500 groups, are separated phylogenetically by CC8d, the CMRSA9 clade, characterized by SCCmec type VIII carriage. Two related samples diverge between CC8d and CC8e: SRR3418706 and SRR3418948 (32). CC8e is a paraphyletic group with respect to USA300. Although USA300-SAE and USA300-NAE are considered monophyletic sister clades, it appears there may be other strains circulating that originate from their last common ancestor. (See the Early Branching sample located between USA300-SAE and USA300-NAE, also in Fig. 2.) Purple triangles mark nodes with bootstrap values of <90% of the 1,000 total. Download FIG S1, PDF file, 0.3 MB (277.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Accession numbers of previously sequenced S. aureus genomes used in this study, along with seven isolates sequenced in this study used solely for the overall S. aureus phylogeny (Fig. 1). Download TABLE S2, XLSX file, 0.1 MB (25.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Typing information and screening results for all S. aureus isolates that were typed by genetic marker inference (GMI [Fig. 3]) and by whole-genome sequence (WGS) analysis in this study. Of the 295 isolates screened, 224 were typed by GMI and WGS phylogenetic analysis, and 89 of those were then used to validate the SNP strain typing assay panel. A total of 71 isolates were typed by GMI and SNP assay panel and then by WGS phylogenetic analysis for confirmation. Another 137 isolates (not in this table) were screened by GMI and SNP assay panel only (Table 3). Download TABLE S1, XLSX file, 0.1 MB (42.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.