Expression of AtBBX21 in potato causes morphological and physiological changes that improve photosynthetic rates in high-irradiance conditions without negatively affecting water use efficiency.
Abstract
B-box (BBX) proteins are zinc-finger transcription factors containing one or two B-box motifs. BBX proteins act as key factors in the networks regulating growth and development. The relevance of BBX21 to light and abscisic acid signaling in seedling development is well established; however, its importance in adult plant development and agronomic species is poorly understood. Therefore, we studied the effect of heterologous expression of Arabidopsis (Arabidopsis thaliana) BBX21 in potato (Solanum tuberosum) var Spunta. Three independent AtBBX21-expressing lines and the wild-type control were cultivated under sunlight and at controlled temperatures in a greenhouse. By anatomical, physiological, biochemical, and gene expression analysis, we demonstrated that AtBBX21-expressing plants were more robust and produced more tubers than wild-type plants. Interestingly, AtBBX21-expressing plants had higher rates of photosynthesis, with a significant increase in photosynthetic gene expression, and higher stomatal conductance, with increased size of the stomatal opening, without any associated decline in water use efficiency. Furthermore, AtBBX21-expressing potato plants had reduced photoinhibition associated with higher production of anthocyanins and phenolic compounds, and higher expression of genes in the phenylpropanoid biosynthesis pathway. To gain insights into the mechanism of BBX21, we evaluated the molecular, morphological, metabolic, and photosynthetic behavior in adult BBX21-overexpressing Arabidopsis. We conclude that BBX21 overexpression improved morphological and physiological attributes, and photosynthetic rates in nonoptimal, high-irradiance conditions, without associated impairment of water use efficiency. These characteristics of BBX21 may be useful for increasing production of potatoes, and potentially of other crops.
Sunlight is essential for autotrophic life. Radiation intensity (measured as photosynthetic photon flux density [PPFD]) is a determinant of plant photosynthesis, during which solar energy is converted into chemical energy and stored as organic carbon. The productivity of a crop depends not only on its photosynthetic rate, but also on other eco-physiological parameters related to crop architecture, such as intercepted radiation and radiation use efficiency (Sinclair and Muchow, 1999; Monteith and Moss, 1977).
Light also serves as a sensory cue for the adjustment of plant architecture (Casal, 2013). Molecular genetic studies of model plants in the past few decades have identified many key genes and pathways that control plant photomorphogenesis. Light absorption by different photoreceptors leads to modulation of core signaling networks, which further orchestrates specific hormone and metabolic signaling pathways to precisely affect growth and development (Quail, 2002; Lau and Deng, 2012). Photoreceptors activate many intermediary transcription factors that belong to diverse families that bind to light responsive elements to activate or repress transcription.
Several transcription factors that function downstream of one or multiple photoreceptors have been functionally characterized. For example, ELONGATED HYPOCOTYL5 (HY5) is a central positive regulator of photomorphogenic development in Arabidopsis (Arabidopsis thaliana; Gangappa and Botto, 2016). HY5 genetically and physically interacts with many signaling components to promote or suppress photomorphogenesis. One important partner of HY5 is CONSTITUTIVE PHOTOMOPHOGENIC1 (COP1), a key suppressor of photomorphogenesis (Hoecker, 2017; Lau and Deng, 2012). COP1 targets HY5 and many other transcription factors for degradation under dark conditions to promote skotomorphogenesis.
B-box (BBX) transcription factors also interact physically or genetically with HY5 and COP1. The BBX proteins are a functionally diverse family encoded by genes that are highly conserved across all multicellular plants, including blue-green algae and mosses (Khanna et al., 2009; Crocco and Botto, 2013; Gangappa and Botto, 2014). In Arabidopsis, 21 out of the 32 BBX proteins (BBX1-13 and BBX18-25) contain two B-boxes in tandem, whereas 11 (BBX14-BBX17 and BBX26-BBX32) contain one B-box. BBX21 regulates plant growth and development throughout the life cycle. BBX21 promotes seed germination in an abscisic acid-dependent manner by interfering with HY5 binding to the ABA-INSENSITIVE5 (ABI5) promoter to inhibit ABI5 expression (Xu et al., 2014). In addition, BBX21 binds to the T/G-box in the HY5 promoter and directly activates HY5 expression in the light-promoting seedling photomorphogenesis (Xu et al., 2016). The disruption of the second B-box in BBX21 impairs its ability to bind to the HY5 promoter (Xu et al., 2018). Interestingly, BBX32, a BBX that represses seedling photomorphogenesis, physically interacts with BBX21 leading to the inactivation of the BBX21-HY5 protein complex (Holtan et al., 2011). Furthermore, BBX21 reduces hypocotyl growth in seedlings exposed to shade light by down-regulating cell growth genes through the COP1 signaling pathway (Crocco et al., 2010).
The BBX proteins constitute a diverse group of transcription factors whose members have opposing functions; these functions contribute to the homeostatic integration of endogenous and environmental signals for the fine-tuning of several physiological processes (Gangappa and Botto, 2014; Crocco et al., 2015). It has also been suggested that BBX proteins mediate the adjustment of plant growth under stress (Nagaoka and Takano, 2003; Wang et al., 2013; Kiełbowicz-Matuk et al., 2014; Yang et al., 2014). The involvement of BBX21 in germination, seedling photomorphogenesis, and shade avoidance in Arabidopsis led us to investigate its role in determining desirable agronomic characteristics and its function in adult plants. The aim of this work was to study morphological, biochemical, physiological, and photosynthetic effects of the heterologous expression of AtBBX21 gene in potato (Solanum tuberosum) var Spunta and Arabidopsis.
RESULTS
Heterologous Expression of AtBBX21 Resulted in Short, Robust Potato Plants and an Increased Tuber Yield
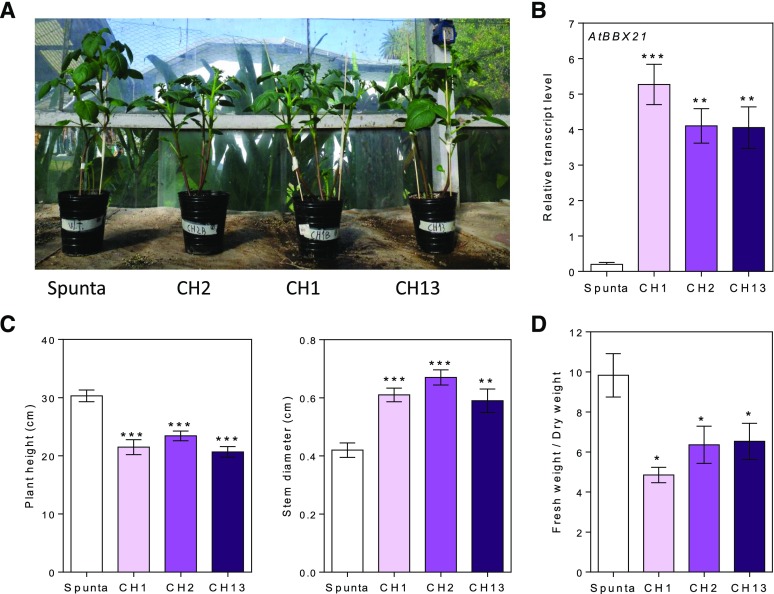
We expressed full-length BBX21 under control of a 35S promoter in S. tuberosum var Spunta. Transgenic potato plants were generated successfully by Agrobacterium tumefaciens-mediated transformation. We obtained 13 independent transgenic lines. To confirm the presence of BBX21 in transgenic potato plants, we conducted reverse transcription quantitative PCR (RT-qPCR) analysis on the BBX21 gene. Three independent transgenic lines (CH1, CH2, and CH13) displayed at least 4-fold higher levels of the BBX21 transgene than wild-type Spunta plants (Fig. 1, A and B). These three lines were used for further experiments according to the availability of tubers.
Figure 1.
Transgenic AtBBX21 potato lines are more robust and shorter than nontransformed plants. A, Representative photograph of 28-d-old nontransformed (Spunta) and transgenic potato plants (CH1, CH2, and CH13). B, AtBBX21 transcript levels of the three independent transgenic lines used in this study (n = 10). C, Plant height and stem diameter in 21-d-old plants (n = 10). D, Fresh weight relative to dry weight in 35-d-old plants (n = 10). n = number of biological replicates. Mean values are shown with error bars indicating se. Asterisks indicate significant difference between nontransformed and transgenic lines (*P < 0.05, **P < 0.01, and ***P < 0.001).
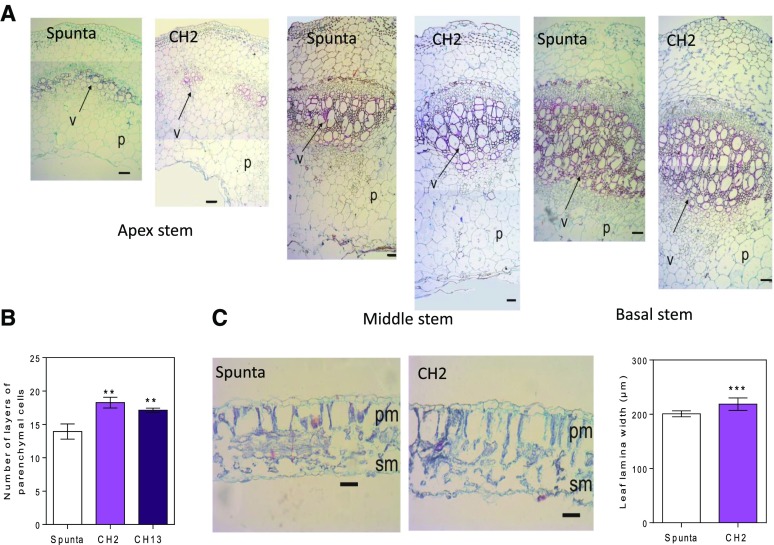
Transgenic and nontransformed plants were grown under natural radiation, a photoperiod of 13.5 h light, and a controlled temperature between 18 and 22°C in a greenhouse for 21 d. BBX21-overexpressing plants were on average significantly shorter and had significantly wider basal stems than control Spunta plants (P < 0.001; Fig. 1C). By the end of the experiment, BBX21-overexpressing plants showed a significant reduction in the fresh/dry weight ratio compared with the control and consequently had a higher dry weight (Fig. 1D; Supplemental Fig. 1). In addition, we did not detect differences in the leaf area between genotypes (Supplemental Fig. 2). Transgenic plants showed a wider vasculature (Fig. 2) and more layers of parenchyma cells than control plants (Fig. 2, A and B). These features were observed in the apical, medium, and basal sections of the stem (Fig. 2A) In addition, transgenic plants had slightly thicker leaf blades than control plants (Fig. 2C).
Figure 2.
Heterologous expression of AtBBX21 promotes stem vasculature and leaf thickening in potato plants. A, Cross section of apex, middle, and basal stems of nontransformed (Spunta) and transgenic plants (CH2). B, Layer number of parenchymal cells (n = 3). C, Leaf lamina width (n = 3). p, Parenchyma; v, vascular bundle; pm, palisade mesophyll cell; sm, spongy mesophyll cells. Bar = 50 µm. n = technical replicates for each genotype. Mean values are shown with error bars indicating se. Asterisks indicate significant difference between nontransformed and transgenic lines (**P < 0.01 and ***P < 0.001).
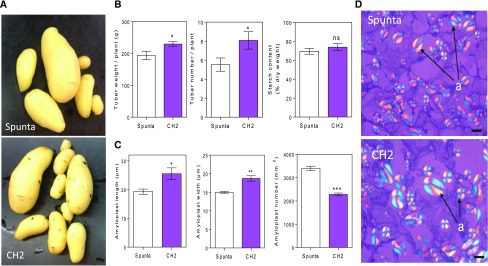
We hypothesized that the anatomical and morphological differences of BBX21-overexpressing plants could affect tuber yield at harvest. In an independent experiment, we evaluated the number and weight of tubers in transgenic and nontransformed plants at harvest. We found that transgenic plants had a 15% higher tuber yield and smaller tubers compared to control plants (Fig. 3, A and B). However, the tuber quality in terms of starch content was similar between genotypes (Fig. 3B). In addition, the number of amyloplasts contained in the tubers of transgenic lines was lower than in control Spunta plants, and the length and width of amyloplasts were higher than control plants (Fig. 3, C and D).
Figure 3.
Heterologous expression of AtBBX21 increases tuber yield in potato plants. A, Representative photographs of nontransformed (Spunta) and transgenic tubers (CH2) at the end of the experiment. B, Tuber weight, tuber number (n = 12), and starch content (n = 5). C, Amyloplast length, width, and number (n = 3). D, Cross section of tubers. a, Amyloplast. Bar = 10 µm. n = number of biological replicates. Mean values are shown with error bars indicating se. Data were analyzed by Student’s t tests, and asterisks indicate significant difference between nontransformed and transgenic lines (*P < 0.05, **P < 0.01, and ***P < 0.001; NS, not significant).
Heterologous Expression of AtBBX21 Enhanced Photosynthesis and Stomata Opening in Potato Plants
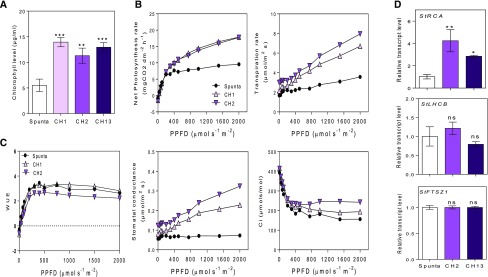
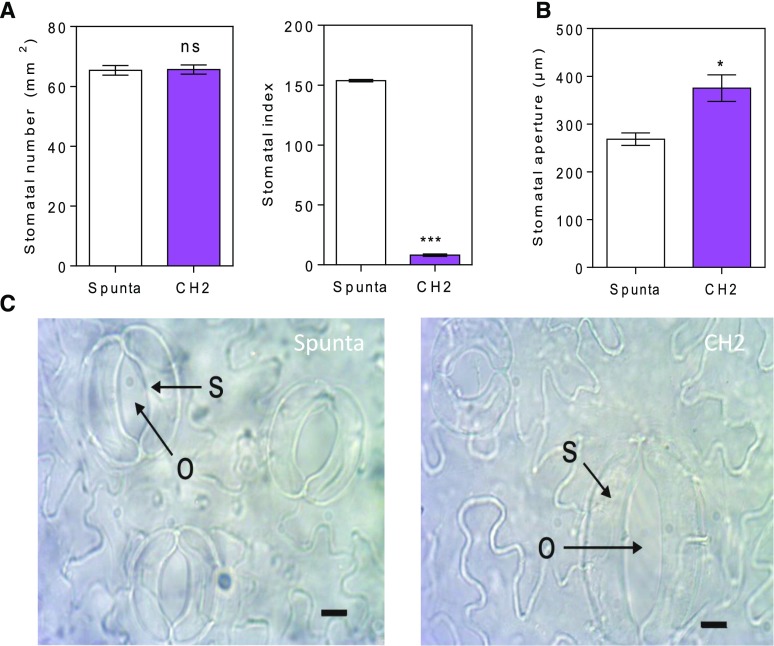
We speculated that the higher levels of chlorophyll detected in transgenic plants (Fig. 4A) may reflect improved photosynthesis. So, we measured the net photosynthetic rate and related photosynthetic traits in 28-d-old plants. We found that transgenic lines had higher photosynthetic rates than control Spunta plants between 400 µmol m−2 s−1 and 2,000 µmol m−2 s−1 PPFD, although no differences were found between genotypes under nonsaturating PPFD quantum yields (see slopes in Fig. 4B). In addition, these plants showed higher transpiration rates and stomatal conductance (Fig. 4B). Interestingly, the water use efficiency (WUE) was similar between genotypes, indicating that the higher photosynthetic rates of transgenic lines did not affect WUE (Fig. 4C). In addition, we found a positive relationship between higher rates of photosynthesis and an increase in the expression of the RCA (Rubisco ACTIVASE), but not LHCB (LIGHT-HARVESTING CHLOROPHYLL-B) or FTSZ1 (PLASTID-DIVIDING RING) photosynthetic genes (Fig. 4D). In addition, we reasoned that the increase of photosynthetic activity in transgenic lines compared with nontransformed plants could be associated with stomatal differences. Although the number of stomata was similar between genotypes, we found significant differences in the distribution of stomata in the leaf (Fig. 5A). The level of anphistomy (the increase of stomata density more intensively in the adaxial than in the abaxial side of the leaf) was higher in transgenic lines than control plants. This was indicated by a reduction in the stomatal index, calculated as the density of abaxial stomata/adaxial stomata (Fig. 5A; Supplemental Fig. 3A). More interestingly, the stomatal area was significantly higher in transgenic plants than in control plants (Fig. 5, B and C). This increase was associated with a higher stomata length but did not show a relationship with stomata width (Supplemental Fig. 3B). Thus, the higher stomatal area probably contributed to the increased photosynthetic activity observed in transgenic plants (Fig. 4B).
Figure 4.
Heterologous expression of AtBBX21 increases photosynthesis in potato plants. A, Chlorophyll levels of nontransformed (Spunta) and transgenic (CH1, CH2, and CH13) potato leaves (n = 10). B, Photosynthesis, transpiration rate, stomatal conductance, and internal concentration of CO2 as a function of PPFD (n = 4). C, WUE as function of PPFD (n = 4). D, StRCA, StLHCB, and StFTSZ1 transcript levels (n = 4). n = number of biological replicates. Mean values are shown with error bars indicating se. Data were analyzed by Student’s t tests, and asterisks indicate significant difference between nontransformed and transgenic lines (*P < 0.05, **P < 0.01, and ***P < 0.001; NS, not significant).
Figure 5.
Heterologous expression of AtBBX21 increases stomatal aperture in potato leaves. A, Stomatal density and stomatal index estimated as the number of abaxial stomata/adaxial stomata of nontransformed (Spunta) and transgenic (CH2) potato leaves (n = 3). B, Stomatal aperture (n = 3). C, Representative photographs of stomata in nontransformed and BBX21-overexpressing lines. S, Stomata; O, opercule. Bar = 10 µm. n = number of biological replicates. Mean values are shown with error bars indicating se. Data were analyzed by Student’s t tests, and asterisks indicate significant difference between nontransformed and transgenic lines (*P < 0.05 and ***P < 0.001; NS, not significant).
Heterologous Expression of AtBBX21 Increased the Abundance of Anthocyanin and Phenolic Compounds, and Reduced Photoinhibition of Potato Leaves
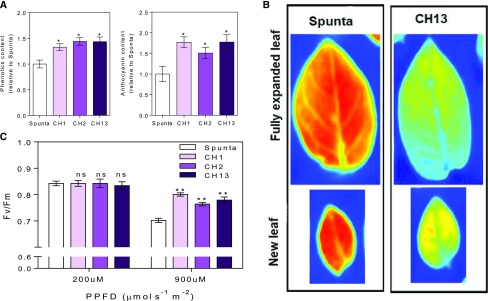
A classical photomorphogenic response is the synthesis of anthocyanins when plants are exposed to light (Mancinelli et al., 1991). We measured the abundance of anthocyanins and phenolic compounds in 21-d-old transgenic and nontransformed plants. The three transgenic lines showed a significant increase in anthocyanin and total phenolic compound levels (Fig. 6A), and this correlated with a lower fluorescence in the leaves (Fig. 6B).
Figure 6.
Heterologous expression of AtBBX21 increases anthocyanin and phenolic levels in potato leaves. A, Phenolics and anthocyanins in nontransformed (Spunta) and transgenic (CH1, CH2, and CH13) potato leaves (n = 5). B, Sunscreen pigment accumulation in young and totally expanded leaves. Sunscreen accumulation is revealed by a decrease in the intensity of UV-induced chlorophyll fluorescence (less fluorescence indicates higher accumulation of anthocyanin and phenolic compounds). C, Maximal photochemical efficiency (Fv/Fm) in 14-d-old plants acclimated at low irradiance (200 μmol m−2 s−1) and then exposed at high irradiance (900 μmol m−2 s−1) for 2 h (n = 6). n = number of biological replicates. Mean values are shown with error bars indicating se. Data were analyzed by Student’s t tests, and asterisks indicate significant difference between nontransformed and transgenic lines (*P < 0.05 and **P < 0.01; NS, not significant).
Previous studies reported that the increase in anthocyanin and phenols levels in the leaves improve the photoprotection at high irradiance (Gould et al., 1995; Manetas et al., 2002; Steyn et al., 2002; Harvaux and Kloppstech, 2001). To evaluate this possibility, we performed an experiment with 14-d-old plants that were acclimated at a low irradiance (200 μmol m−2 s−1) and then exposed to high irradiance (900 μmol m−2 s−1) for 2 h. The photoinhibition of photosystem II was estimated by measuring the maximal photochemical efficiency of photosystem II (Fv/Fm) at both levels of irradiance. We found that Fv/Fm was similar between genotypes at low irradiance, but the photoinhibition of the photosystem II was higher in nontransformed plants than in transgenic plants exposed to high irradiance (Fig. 6C). On average, the Fv/Fm was reduced by around 25% in control plants and 10% in the three transgenic lines. These results suggest a role for BBX21 in protecting against high irradiance, probably by inducing the accumulation of anthocyanin and phenolic compounds in their leaves.
Heterologous Expression of AtBBX21 Promoted the Phenylpropanoid Signaling Pathway in Potato Plants
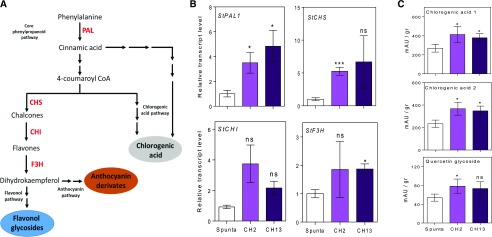
The phenylpropanoid signaling pathway induces the accumulation of flavonol glycosides, anthocyanin derivates, and chlorogenic acid by activating the expression of specific enzymes (Fig. 7A). We evaluated the expression of PAL1 (PHE AMMONIA LYASE1), CHS (CHALCONE SYNTHASE), CHI (CHALCONE ISOMERASE), and F3H (FLAVANONE 3-HYDROXYLASE) with RT-qPCR, as well as the accumulation of two chlorogenic acids and quercetin glycoside metabolites with HPLC. In general terms, all of these were higher in leaves of transgenic plants than control plants with the exception of CHI expression (Fig. 7, B and C). These results explain the higher levels of anthocyanins and phenolic compounds in transgenic potato plants compared to nontransformed plants.
Figure 7.
Heterologous expression of AtBBX21 in potato promotes gene expression and metabolites accumulation related with the phenylpropanoid pathway in leaves. A, Diagram of phenylpropanoid pathway and key enzymes. B, PAL, CHS, CHI, and F3H transcript levels (n = 4). C, Chlorogenic acid 1, chlorogenic acid 2, and quercetin glycoside metabolites measured by HPLC (n = 5). n = number of biological replicates. Mean values are shown with error bars indicating se. Data were analyzed by Student’s t tests, and asterisks indicate significant difference between nontransformed and transgenic lines (*P < 0.05; NS, not significant).
AtBBX21 Overexpression Promoted Rosette Expansion, Branching, and Accumulation of Anthocyanin and Phenolic Compounds in Arabidopsis Plants
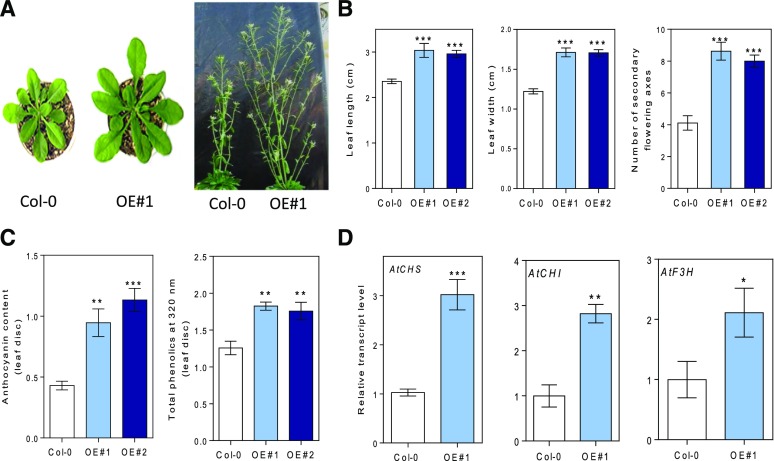
To gain insight into the role of BBX21 in adult plants, we cultivated two BBX21-overexpressing Arabidopsis lines (BBX21-OE#1 and OE#2) and the wild-type control (Col-0) in a greenhouse with natural radiation. We found that BBX21-OE lines produced bigger rosettes with longer and wider leaves than Col-0 (Fig. 8, A and B). In addition, BBX21-OE plants showed profuse flowering with a significantly higher number of secondary axes and seed yield than wild-type plants (Fig. 8A; data not shown). We did not identify any differences in leaf size and number of flowering axes between wild-type and bbx21-1 mutant plants (Supplemental Fig. 4). We evaluated the levels of anthocyanins and phenols in 35-d-old Arabidopsis plants and found that BBX21-OE plants produced higher levels of anthocyanins and phenols (Fig. 8C), corroborating the results observed in heterologous expression of AtBBX21 potato plants. Accordingly, the gene expression of CHS, F3H, and CHI was significantly higher in BBX21-OE than control plants (Fig. 8D). As expected, anthocyanin content and AtCHS gene expression in the null mutant of bbx21 were lower than in wild-type Arabidopsis plants (Supplemental Fig. 4).
Figure 8.
BBX21 overexpression promotes rosette expansion, branching, and accumulation of anthocyanins and phenolics in Arabidopsis. A, Representative photographs showing vegetative rosette (35-d-old plants, left) and secondary flowering ramifications (98-d-old plants, right) in wild-type (Col) and BBX21-overexpresed (BBX21-OE) Arabidopsis plants. B, Leaf length, width of the fourth expanded leaf, and number of secondary flowering axes (n = 8). C, Anthocyanin and phenolic content in leaves (n = 5). D, CHS, CHI, and F3H transcript levels (n = 3). n = number of biological replicates. Mean values are shown with error bars indicating se. Data were analyzed by Student’s t tests, and asterisks indicate significant difference between Col and BBX21-OE Arabidopsis plants (*P < 0.05, **P < 0.01, and ***P < 0.001).
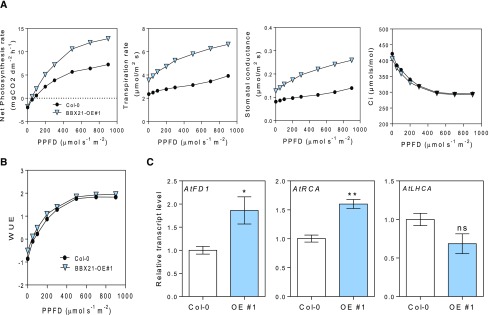
In other experiments, we examined the photosynthesis rates of BBX21-OE and wild-type plants cultivated in a greenhouse. We exposed 35-d-old Arabidopsis plants to different PPFDs and evaluated net photosynthesis rates and related photosynthetic traits. We found that BBX21-OE plants showed higher photosynthesis rates, transpiration rates and stomatal conductance than wild-type plants at high irradiances (200 µmol m−2 s−1; Fig. 9A). In accordance with our data in potato plants, the WUE was similar between genotypes (compare Figs. 9B and 4C). The expression of FD1 (FERRODOXIN-1) and RCA photosynthetic genes showed higher levels in the BBX21-OE than in wild-type plants, as expected for the higher activity of the photosynthetic apparatus (Fig. 9C).
Figure 9.
BBX21 overexpression promotes photosynthesis in Arabidopsis. A, Photosynthesis, transpiration rate, stomatal conductance, and internal concentration of CO2 as function of PPFD (n = 4). B, WUE as function of PPFD (n = 4). C, FD1, RCA, and LHCA transcripts (n = 3). n = number of biological replicates. Mean values are shown with error bars indicating se. Data were analyzed by Student’s t tests, and asterisks indicate significant difference between Col and BBX21-OE plants (*P < 0.05 and **P < 0.01; NS, not significant).
Heterologous Expression of AtBBX21 Resulted in Short Internodes in Potato Plants Cultivated under Simulated Shade
We demonstrated previously that BBX21 inhibits shade avoidance responses in Arabidopsis seedlings (Crocco et al., 2010). We hypothesized that the heterologous expression of AtBBX21 in potato plants might display a reduced shade avoidance response because of the reduced cell elongation in the stems. To test this idea, we cultivated 21-d-old potato plants under either white light or simulated shade in a growth chamber. As expected, we observed a significantly reduced shade avoidance response for internode stem elongation in transgenic plants compared to control plants (Supplemental Fig. 5). In addition, transgenic plants showed a constitutive phenotype with shorter and wider stems than nontransformed plants independently of light treatments (Supplemental Fig. 5). Interestingly, transgenic plants showed constitutively high levels of anthocyanins and phenols that was independent of light treatment. In opposition, the abundance of these compounds increased in control plants under shade (Supplemental Fig. 5).
DISCUSSION
Our knowledge of BBX proteins is still limited by the complexity and modularity of the molecular signaling pathways they act in and the scarce amount of available functional information (Gangappa and Botto, 2014). The BBX21 transcription factor belongs to structural group IV and contains two B-boxes domains. The biological roles for BBX21 had not been explored prior to this study with the exception of its roles in seed germination and seedling photomorphogenesis in the Arabidopsis model system. In Arabidopsis, BBX21 is a potent transcription factor that promotes photomorphogenesis (Datta et al., 2007) and inhibits shade avoidance elongation responses (Crocco et al., 2010). BBX21 is a pivotal component in the COP1-HY5 regulatory hub that regulates the seedling developmental program during the transition between dark and light (Xu et al., 2018). BBX21 is targeted for 26S proteasome-mediated degradation in darkness, and it is stabilized when seedlings grow in the light-promoting photomorphogenesis through HY5 activity. Interestingly, BBX21 has also been involved in the promotion of Arabidopsis seed germination by interfering with HY5 binding to the ABI5 promoter (Xu et al., 2014). Here, we clearly demonstrated that the heterologous expression of Arabidopsis BBX21 in potato resulted in short, robust plants with a higher tuber yield than nontransformed plants (Figs. 1–3). In addition, transgenic potato plants had a higher photosynthetic rate and stomatal conductance, which was associated with a larger stomatal opening (Figs. 4 and 5). These plants also exhibited reduced photoinhibition, which was related to a higher production of anthocyanins and phenolics, as well as an increase in expression of phenylpropanoid biosynthesis genes (Figs. 6 and 7). We also confirmed some of these features in mature BBX21-overexpressing Arabidopsis lines (Figs. 8 and 9), but not in the bbx21-1 mutant (Supplemental Fig. 4), probably because of redundancy within the BBX family.
Photosynthesis/PPFD relationships revealed that both heterologous expression of AtBBX21 in potato plants and BBX21-overexpresed Arabidopsis lines dramatically increased photosynthetic rates by up to 60% under saturating irradiances without altering quantum yield (Figs. 4B and 9A). In addition, RCA and FD1 photosynthesis related genes were up-regulated in both BBX21-overexpressing potato and Arabidopsis plants (Figs. 4D and 9C). These responses were closely related to stomatal conductance and the higher internal CO2 concentration in BBX21-overexpressing transgenic Arabidopsis and potato plants, suggesting that stomatal aperture is the major causal effect of the higher photosynthesis rate (Flexas et al., 2006). Although the stomatal distribution in the leaves differed between transgenic and nontransformed plants, no differences in the number of stomata were observed. Therefore, the effect of stomatal abundance is not associated with the higher photosynthesis rate of BBX21-overexpressing plants (Figs. 4 and 9). These results suggest that radiation use efficiency may be higher in a BBX21-overexpressing potato crop depending on plant architecture and light extinction across the canopy (Lambers et al., 1998). Further research is needed to test these working hypotheses in the field.
BBX21-overexpressing potato and Arabidopsis plants had enhanced rates of CO2 uptake and transpiration as well as an increase in stomatal conductance without any associated negative effect on WUE (Figs. 4C and 9B). An enhanced photosynthetic rate was previously observed in PHYTOCHROME B (PHYB)-overexpressing potato and Arabidopsis plants (Thiele et al., 1999; Boccalandro et al., 2003, 2009). However, in contrast to BBX21 overexpression, PHYB overexpression reduced the WUE in Arabidopsis plants (Supplemental Fig. 6; Boccalandro et al., 2009). Considering that BBX21 overexpression in potato and Arabidopsis did not affect the WUE (Figs. 4C and 9B), our results suggest a partial cross talk between BBX21 and PHYB signaling pathways. Several genes have been found to be involved in traits associated with WUE. The expression of some genes can increase or decrease the expression of others, forming a complex network of interactions. Such complex networks involve genotype-by-environment interactions as well as epistatic interactions. Future work is required to explore whether BBX21 overexpression has a negative effect on the WUE in different environmental contexts, beyond that of the controlled greenhouse conditions employed in this work. In addition, it would be interesting to explore how BBX21 affects WUE to further our understanding of the molecular mechanisms that define this trait.
Although anthocyanin and phenolic contents were higher in BBX21-overexpressing transgenic potato and Arabidopsis plants, a similar potential quantum yield was observed when transgenic or nontransformed plants were subjected to low acclimation radiation (i.e. Fv/Fm around 0.85; Fig. 6). Interestingly, a protective effect of these pigments was observed in BBX21-overexpressing plants after exposure to high PPFD, since only nontransformed plants were photoinhibited (Fv/Fm dropped below 0.7; Fig. 6C). These results suggest that the reduced photoinhibition of photosystem II in transgenic potato plants may be associated with the accumulation of photoprotective metabolites, leading to a higher photosynthetic rate. We demonstrated that high transcript levels of several genes involved in the phenylpropanoid pathway (PAL1, CHS, and F3H) led to an increase in anthocyanin and phenolic compound levels in potato and Arabidopsis plants (Figs. 6 and 8). Interestingly, the induction of a photoprotective acclimation mechanism to avoid the photosystem damage against excess visible radiation that is mediated by UV/blue-light-absorbing flavonols has been previously demonstrated using nonphotochemical quenching mutants of Arabidopsis (Harvaux and Kloppstech, 2001). In accordance with our results, PHYB overexpression increased the resistance of the potato photosynthetic apparatus to UV-B, which was associated with a higher accumulation of chlorophylls, carotenoids, and flavonoids in the leaves (Kreslavski et al., 2015).
Although the mechanism by which BBX21 regulates the expression of phenylpropanoid biosynthesis genes was not explored here, previous work has allowed us to discuss some of the molecular mechanisms of BBX21 action in the context of our results. BBX21 binds to the T/G-box of the HY5 promoter controlling its expression in Arabidopsis seedlings (Xu et al., 2016, 2018). Furthermore, HY5 directly binds to the promoter regions of CHS, CHI, and F3H to regulate their expression (Lee et al., 2007; Zhang et al., 2011). Furthermore, functional assays in protoplasts showed that BBX21 can activate CHI transcription through the G-box promoter element (Datta et al., 2007). These results suggest that BBX21 might positively regulate the phenylpropanoid gene expression pathway in HY5-dependent and -independent manners.
Despite many works having studied the function of CONSTANS/BBX1, few studies have focused on the functions of other BBX members in crop species (Gangappa and Botto, 2014). Our work reveals a role for BBX21 in crop species and its involvement in the regulation of photosynthetic rate and photoprotection responses in potato and Arabidopsis adult plants. Previous work showed that heterologous expression of Arabidopsis BBX32 in soybean (Glycine max) plants increased grain yield by altering the timing of reproductive development and thus extending the period between pod and seed development (Preuss et al., 2012). More recently, it has been demonstrated that BBX24 overexpression in Chrysanthemum morifolium produced plants with early flowering and increased tolerance to freezing and drought (Yang et al., 2014). Regarding the results of our and other’s works, we conclude that the overexpression of some members of BBXs (BBX21, BBX24, and BBX32) can contribute to improve crop plant phenotypes, apparently by affecting different physiological features (i.e. phenology, tolerance to stresses, and photosynthesis rates).
In the post-green revolution era, optimizing plant energy capture and use is a key target for improvement to increase crop yields (Murchie et al., 2009). Our work reveals the importance of BBX21 as a target transcription factor to increase photosynthetic efficiency and reduce photoinhibition by altering anatomical and biochemical features on potato plants with significant positive effects on tuber yield. The use of biotechnological tools to modify the expression levels of the BBX21 transcription factor can be a more useful strategy than photoreceptor transgenesis, to avoid nondesirable pleiotropic effects such as the decreased WUE previously documented in PHYB-overexpressing lines. Regarding the results that we obtained in potato plants cultivated under natural radiation in a greenhouse, new experiments should be designed to corroborate the BBX21-overexpressing phenotypes in the field, cultivating the plants in more variable and dynamic environmental conditions and using standard potato agronomic practices. Finally, the results presented here suggest that enhanced BBX21 levels could be used in breeding programs to increase tuber yield of other commercial potato genotypes.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Experimental Design
Potato (Solanum tuberosum) var Spunta and three AtBBX21 transgenic overexpressing lines (CH1, CH2, and CH13) were used in the experiments. To generate transgenic lines, the coding sequence of BBX21 was amplified by PCR from genomic Arabidopsis (Arabidopsis thaliana) DNA (Col-0 background) using the primers 35s172sen and 35s172ant (Supplemental Table 1). The full-length DNA was cloned into the pDONOR221 plasmid (Invitrogen) and inserted into the pB7WG2 binary plasmid under the control of the CaMV 35S promoter using Gateway technology (Invitrogen), and then introduced into Agrobacterium tumefaciens (GV3101). This construct was used to transform Arabidopsis plants of the Col-0 accession (Supplemental Fig. 7) by the floral dip method as previously described by Crocco et al. (2010), and S. tuberosum var Spunta as described by Beaujean et al. (1998).
The experiments with potato and Arabidopsis plants were done in a greenhouse, located at the Faculty of Agronomy, University of Buenos Aires (34° 35’S, 58° 29’W), with natural radiation (PAR = 500 μmol m−2 s−1) and temperatures ranged between 18°C and 22°C. The experiments were carried out between March and November with a photoperiod around 13.5 h. For the potato experiments, sprouted tubers of similar size were sown in pots of 12 cm diameter × 12 cm height (500 cm3). The soil mix consisted of vermiculite (1/6), peat (1/6), perlite (1/6), and soil (3/6). The pots were watered daily at field capacity and fertilized twice during the experiment with TRIPLE 15 (15% of nitrogen, 15% of phosphorus, and 15% of potassium). For the Arabidopsis experiments, the seeds were sown on small pots with soil and stratified at 7°C for 3 d before moving them to the greenhouse.
For the shade avoidance experiment, potato tubers were sown in pots of 10 cm diameter × 10 cm height (250 cm3) using a mix soil of peat (1/3), perlite (1/3), and vermiculite (1/3). The plants were watered daily at field capacity and were fertilized with TRIPLE 15 twice during the experiment. The plants were cultivated in a white light chamber (PAR = 200 µmol m−2 s−1) with a photoperiod of 16 h light/8 h dark at 24°C. Simulated shade treatment consisted of white light plus far-red light disposed along both sides of plants during the photoperiod. Far-red light was provided with four incandescent lamps, a red acetate sheet, and two blue acrylic sheets (Paolini 2031) using the same protocol described previously (Botto, 2015). The plants received the same irradiance but different red/far-red ratios (white light = 2 and simulated shade = 0.5). The light quantity and quality were measured with a Li-Cor radiometer (Li-l88B).
A completely randomized ANOVA design was used for the experiments. Mean comparisons between control and transgenic lines were done with Student’s t test. Analyses were carried out using Prism (GraphPad Software).
Anatomical and Morphological Determinations
Samples from the central part of leaf were diaphanized by using Dizeo de Strittmater’s technique (Dizeo de Strittmatter, 1973) and stained with safranin. In addition, transverse sections of leafs and stems (apical, medium, and basal) were embedded in paraffin and serially cut at 10 to 12 µm with Minot-type rotary microtome, and sections were stained with safranin-fast green (Johansen, 1940). Sections were photographed with a Zeiss Axioplan optical microscope and analyzed with the Zeiss AxioCam ERc 5s software (Jena). The measurements were done in three biological samples with 10 technical replicates for each genotype. Photos were taken with polarized light.
Stem diameter, plant height, and dry and fresh weight of plants were assessed. The diameter was measured with a digital caliber at 2 cm of the soil surface, and the height of the principal stem was estimated using a ruler in 28-d-old plants. The aerial part of 35-d-old plants was harvested and weighed with a digital balance. The material was enveloped into a paper and incubated at 60°C during 72 h, and then weighted again to obtain the dry weight.
Chlorophyll, Anthocyanin, and Phenolic Determinations
Chlorophyll determinations were done in the last expanded leaves of 35-d-old plants using a SPAD 502 (Minolta) and calculating chlorophyll levels (Supplemental Fig. 8) according to the method described by Moran (1982). Leaf discs (6 mm diameter) of the third completely expanded leaf of 28-d-old plants were collected for anthocyanin and phenolic quantifications by spectrophotometry. Each biological sample was composed by two independent discs. Anthocyanin quantification was performed as described by Deikman and Hammer (1995). Total phenolic content was determined according to Waterman (1994). Sunscreen pigment accumulation in leaves by UV-induced chlorophyll fluorescence was measure as described by Mazza et al. (2000). Sunscreen accumulation is revealed by a decrease in the intensity of UV-induced chlorophyll fluorescence. For HPLC determinations, individual leaf phenolics were estimated as described by Keinänen et al. (2001) and Demkura et al. (2010). Phenolics were separated by HPLC (Knauer Euroline) on a Restek Pinnacle II C18 (5.0 μm, 4.6 × 150 mm) column. The retention times and UV-visible spectra of chlorogenic acid and quercetin were compared with those of true standards from Sigma-Aldrich.
Photosynthesis, Transpiration, and Stomata Conductance Measurements
Net photosynthesis rate, stomatal conductance, transpiration rate, and CO2 concentration of stomatal cavity were measured in 28-d-old plants using an open infrared gas analysis system (Li-Cor 6400). Light functions were measured at 0, 30, 50, 100, 200, 300, 400, 500, 750, 1,000, 1,500, and 2,000 μmol m−2 s−1. PPFD was done in fully expanded leaves using the 6400-02B LED light source chamber. Air flow and CO2 concentration in the reference cell (CO2R) were automatically controlled by the equipment at 300 µmol s−1 and 400 µmol s−1 (ppm), respectively. WUE (also called instantaneous water use efficiency) was estimated as the ratio of carbon assimilation and transpiration (net photosynthesis rate/transpiration rate).
Gene Expression Analysis
Leaf samples of 21-d-old plants were collected and ground in liquid nitrogen. Total RNA was extracted from the frozen samples using an RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1.5 μg of DNA-free RNA template using an oligo(dT) primer and SuperScript II reverse transcriptase (Invitrogen). RT-qPCR analysis was performed on an optical 96-well plate using SYBR Green PCR master mix (Applied Biosystems) and an ABI PRISM 7500 real-time PCR system (Applied Biosystems). Gene-specific primer pairs were designed using Beacon Designer 7.0 (Premier Biosoft). The primers used in this work are listed in Supplemental Table. 1. The expression of each gene was normalized to TUB or UBQ and each treatment was standardized to wild-type expression.
Accession Numbers
Accession numbers for genes used in this study are listed in Supplemental Table. 2.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used for quantification of genes in Arabidopsis and potato plants.
Supplemental Table S2. Gene codes for genes used in this study.
Supplemental Figure S1. Fresh and dry weight of 35-d-old nontransformed and transgenic potato plants (n = 10).
Supplemental Figure S2. Area of the totally expanded #5 leaf in 28-d-old nontransformed and transgenic potato plants (n = 10).
Supplemental Figure S3. Stomatal number in the abaxial and adaxial position and stomatal length and stomatal width of nontransformed and transgenic potato lines
Supplemental Figure S4. Representative photograph of 35-d-old wild-type and bbx21-1 plants and secondary flowering ramifications of the wild type and bbx21-1.
Supplemental Figure S5. Phenotype of nontransformed and transgenic potato plants under shade.
Supplemental Figure S6. Water use efficiency in upper leaves of wild-type and PHYB-overexpressing potatoes (Dara-5 and Dara-12).
Supplemental Figure S7. BBX21 transcript levels in two overexpressing lines of Arabidopsis.
Supplemental Figure S8. Calibration curve between SPAD levels and chlorophyll content (a+b) of potato leaves.
Supplemental Table S1. Primers used for quantification of genes in Arabidopsis and potato plants.
Supplemental Table S2. Gene codes for genes used in this study.
Acknowledgments
We thank Andrea Dengis for her technical collaboration in potato transformation and Patricia Demkura for the HPLC technical support.
Footnotes
Articles can be viewed without a subscription.
References
- Beaujean A, Sangwan RS, Lecardonnel A, Sangwan-Norreel BS (1998) Agrobacterium-mediated transformation of three economically important potato cultivars using sliced internodal explants: an efficient protocol of transformation. J Exp Bot 49: 1589–1595 [Google Scholar]
- Boccalandro HE, Ploschuk EL, Yanovsky MJ, Sánchez RA, Gatz C, Casal JJ (2003) Increased phytochrome B alleviates density effects on tuber yield of field potato crops. Plant Physiol 133: 1539–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccalandro HE, Rugnone ML, Moreno JE, Ploschuk EL, Serna L, Yanovsky MJ, Casal JJ (2009) Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiol 150: 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF. (2015) Plasticity to simulated shade is associated with altitude in structured populations of Arabidopsis thaliana. Plant Cell Environ 38: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Crocco CD, Holm M, Yanovsky MJ, Botto JF (2010) AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J 64: 551–562 [DOI] [PubMed] [Google Scholar]
- Crocco CD, Botto JF (2013) BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531: 44–52 [DOI] [PubMed] [Google Scholar]
- Crocco CD, Locascio A, Escudero CM, Alabadí D, Blázquez MA, Botto JF (2015) The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nat Commun 6: 6202. [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J, Hammer PE (1995) Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol 108: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkura PV, Abdala G, Baldwin IT, Ballaré CL (2010) Jasmonate-dependent and -independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant Physiol 152: 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizeo de Strittmatter CG. (1973) Nueva técnica de diafanización. Bol Soc Argent Bot 15: 126–129 [Google Scholar]
- Flexas J, Ribas-Carbó M, Bota J, Galmés J, Henkle M, Martínez-Cañellas S, Medrano H (2006) Decreased Rubisco activity during water stress is not induced by decreased relative water content but related to conditions of low stomatal conductance and chloroplast CO2 concentration. New Phytol 172: 73–82 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gould KS, Kuhn DN, Lee DW, Oberbauer SF (1995) Why leaves are sometimes red. Nature 378: 241–242 [Google Scholar]
- Harvaux M, Kloppstech K (2001) The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 213: 953–966 [DOI] [PubMed] [Google Scholar]
- Hoecker U. (2017) The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr Opin Plant Biol 37: 63–69 [DOI] [PubMed] [Google Scholar]
- Holtan HE, Bandong S, Marion CM, Adam L, Tiwari S, Shen Y, Maloof JN, Maszle DR, Ohto MA, Preuss S, et al. (2011) BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol 156: 2109–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen DA. (1940) Plant Microtechnique. McGraw-Hill, New York [Google Scholar]
- Keinänen M, Oldham NJ, Baldwin IT (2001) Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J Agric Food Chem 49: 3553–3558 [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbowicz-Matuk A, Rey P, Rorat T (2014) Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum double B-box gene. Ann Bot 113: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavski VD, Kosobryukhov AA, Shmarev AN, Aksenova NP, Konstantinova TN, Golyanovskaya SA, Romanov GA (2015) Introduction of the Arabidopsis PHYB gene increases resistance of photosynthetic apparatus in transgenic Solanum tuberosum plants to UV-B radiation. Russ J Plant Physiol 62: 204–209 [Google Scholar]
- Lambers H, Chapin FS III, Pons TL (1998) Plant Physiological Ecology. Springer, New York [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli AL, Rossi F, Moroni A (1991) Cryptochrome, phytochrome, and anthocyanin production. Plant Physiol 96: 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetas Y, Drinia A, Petropoulou Y (2002) High contents of anthocyanins in young leaves are correlated with low pools of xanthophyll cycle components and low risk of photoinhibition. Photosynthetica 40: 349–354 [Google Scholar]
- Mazza CA, Boccalandro HE, Giordano CV, Battista D, Scopel AL, Ballaré CL (2000) Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol 122: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteith JL, Moss CJ (1977) Climate and the efficiency of crop production in Britain. Philos Trans R Soc Lond B Biol Sci 281: 277–294 [Google Scholar]
- Moran R. (1982) Formulae for determination of chlorophyllous pigments extracted with n,n-dimethylformamide. Plant Physiol 69: 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Pinto M, Horton P (2009) Agriculture and the new challenges for photosynthesis research. New Phytol 181: 532–552 [DOI] [PubMed] [Google Scholar]
- Nagaoka S, Takano T (2003) Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot 54: 2231–2237 [DOI] [PubMed] [Google Scholar]
- Preuss SB, Meister R, Xu Q, Urwin CP, Tripodi FA, Screen SE, Anil VS, Zhu S, Morrell JA, Liu G, et al. (2012) Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS One 7: e30717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. (2002) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 14: 180–188 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, Muchow RC (1999) Radiation use efficiency. Adv Agron 65: 215–265 [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Thiele A, Herold M, Lenk I, Quail PH, Gatz C (1999) Heterologous expression of Arabidopsis phytochrome B in transgenic potato influences photosynthetic performance and tuber development. Plant Physiol 120: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Tu X, Zhang J, Chen X, Rao L (2013) Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol Biol Rep 40: 2679–2688 [DOI] [PubMed] [Google Scholar]
- Waterman PG, Mole S (1994) Analysis of Phenolic Plant Metabolites. Blackwell Scientific Publications, Oxford, UK [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M (2014) Convergence of Light and ABA signaling on the ABI5 promoter. PLoS Genet 10: e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA 113: 7655–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Holm M, Deng XW (2018) The B-box domain protein BBX21 promotes photomorphogenesis. Plant Physiol 176: 2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ma C, Xu Y, Wei Q, Imtiaz M, Lan H, Gao S, Cheng L, Wang M, Fei Z, Hong B, Gao J (2014) A zinc finger protein regulates flowering time and abiotic stress tolerance in Chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26: 2038–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]